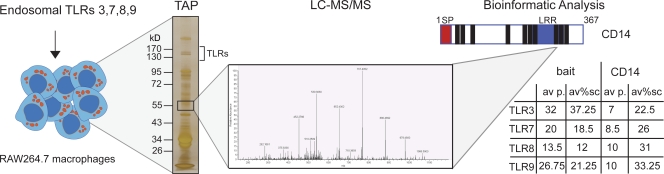
CD14 interacts with and is essential for the functions of endosomal TLR7 and TLR9 in mice.
Abstract
Recognition of pathogens by the innate immune system requires proteins that detect conserved molecular patterns. Nucleic acids are recognized by cytoplasmic sensors as well as by endosomal Toll-like receptors (TLRs). It has become evident that TLRs require additional proteins to be activated by their respective ligands. In this study, we show that CD14 (cluster of differentiation 14) constitutively interacts with the MyD88-dependent TLR7 and TLR9. CD14 was necessary for TLR7- and TLR9-dependent induction of proinflammatory cytokines in vitro and for TLR9-dependent innate immune responses in mice. CD14 associated with TLR9 stimulatory DNA in precipitation experiments and confocal imaging. The absence of CD14 led to reduced nucleic acid uptake in macrophages. Additionally, CD14 played a role in the stimulation of TLRs by viruses. Using various types of vesicular stomatitis virus, we showed that CD14 is dispensable for viral uptake but is required for the triggering of TLR-dependent cytokine responses. These data show that CD14 has a dual role in nucleic acid–mediated TLR activation: it promotes the selective uptake of nucleic acids, and it acts as a coreceptor for endosomal TLR activation.
Pattern recognition receptors are used by cells to scan for intruding pathogens. Whereas a series of pattern recognition receptors like RIG-I–like helicases and NOD-like receptors are present in the cytoplasm, Toll-like receptors (TLRs) are transmembrane proteins associated with either the plasma membrane or endosomes (Medzhitov, 2008; Takeuchi and Akira, 2010; Yoneyama and Fujita, 2010). Pathogen-associated molecular patterns like LPS are sensed by TLRs that are located at the plasma membrane. In contrast, nucleic acids from bacteria or viruses are sensed in acidified endosomes (O’Neill, 2008). Four TLR family members have been found in endosomal compartments of immune cells, sensing double-stranded RNA (TLR3), single-stranded RNA (ssRNA; TLR7/8), and nonmethylated DNA (TLR9; Takeda and Akira, 2005). Commonly used synthetic ligands to stimulate endosomal TLRs are Poly(I:C) for TLR3, the ssRNA nucleotide analogue imiquimod for TLR7, and nonmethylated CpG-DNA containing ssDNA for TLR9 (Takeda and Akira, 2005). Accordingly, endosomal TLRs recognize pathogens by their genomes. For example, ssRNA viruses such as influenza virus and vesicular stomatitis virus (VSV) are recognized by TLR7, whereas DNA viruses are sensed by TLR9 (Lund et al., 2003, 2004; Diebold et al., 2004).
It is thought that stimulation of TLRs induces a conformational change in receptor dimers associated with the formation of an intracellular platform able to recruit adaptor proteins important for intracellular signaling (Gay et al., 2006; Lin et al., 2010). TLR3 stimulation in macrophages and conventional DCs, for instance, recruits the adaptor protein TRIF (TIR domain–containing adaptor-inducing IFN-β), which leads to the TBK1 (Tank-binding kinase 1)–IRF3-dependent induction of type I IFNs as well as to the TRAF6-dependent induction of NF-κB. However, in the same cells, the activation of TLR7 and TLR9 induces the recruitment of the adaptor MyD88, which by recruiting IRAK (IL-1 receptor [IL-1R]–associated kinase) kinases mediates the induction of proinflammatory cytokines. An additional pathway exists in plasmacytoid DCs, where TLR7 and TLR9 use MyD88 to induce high amounts of type I IFNs directly via the transcription factor IRF7 (Blasius and Beutler, 2010).
Although the recently solved crystal structures of several TLR–ligand interfaces (Jin et al., 2007; Liu et al., 2008; Park et al., 2009) have aided our understanding for the molecular basis of pathogen recognition, the identity and role of proteins participating to the fully functional TLR molecular machines have only been understood satisfyingly for the LPS receptor TLR4. This receptor requires the concerted action of at least four proteins, each of which is essential for LPS recognition: LBP (LPS-binding protein), MD2, TLR4, and CD14 (cluster of differentiation 14; Moore et al., 2000; Fitzgerald et al., 2004). CD14 is a glycosylphosphatidylinositol-anchored, membrane-associated protein that functions to aid the delivery of various ligands to TLRs, including LPS, lipoteichoic acid, ceramide, or Poly(I:C)/double-stranded RNA (Schmitz and Orsó, 2002; Lee et al., 2006; Akashi-Takamura and Miyake, 2008). In addition, CD14 has been proposed to mediate the uptake of Poly(I:C) into TLR3-containing endosomes, thereby promoting TLR3 activation (Lee et al., 2006).
Two classes of cofactors have been described for the endosomal TLRs: proteins of the ER are important for proper TLR localization and folding, such as the chaperones gp96/Grp94, Prat4A, and Unc93B (Brinkmann et al., 2007; Akashi-Takamura and Miyake, 2008). Other proteins that directly bind to TLRs in the endosome are involved in DNA ligand delivery such as HMGB-1, a histone-like protein, and LL37, a secreted antimicrobial peptide. These proteins are believed to convey DNA from dying cells to TLR9-containing endosomes (Lande et al., 2007; Tian et al., 2007). Because of the necessity of few TLRs to recognize a variety of structurally different ligands, it is reasonable to expect that ancillary proteins concur in a modular and combinatorial fashion to ensure functional variety to the different TLR molecular machines (Gavin et al., 2006; Zak and Aderem, 2009).
RESULTS
An interaction proteomic approach identifies CD14 as a partner of TLR7/9
The characterization of cellular protein complexes by purification and mass spectrometry (MS) analysis has previously been successfully used to identify new components of signaling pathways in immunity both in a focused fashion and at a large scale (Kim et al., 2002; Andrejeva et al., 2004; Bouwmeester et al., 2004; Häcker et al., 2006; Soulat et al., 2008). In this study, we sought to analyze protein complexes formed by the endosomal TLRs. To this end, we subjected the endosomal TLRs to the tandem affinity purification (TAP)/MS procedure (Bürckstümmer et al., 2006; Köcher and Superti-Furga, 2007). TAP-tagged versions of endosomal murine TLR3, TLR7, TLR8, and TLR9 were expressed in murine RAW264.7 macrophages by retroviral transduction (Fig. 1). Protein complexes of TLRs were purified using a two-step purification protocol (Bürckstümmer et al., 2006). Eluates were analyzed by one-dimensional SDS-PAGE, and the proteins were visualized by silver staining (Fig. 1 and Fig. S1; Shevchenko et al., 1996). 20 slices were excised from each of the gel lanes, the proteins were digested in situ with trypsin (Shevchenko et al., 1996), and the peptides were extracted and analyzed by liquid chromatography (LC)–tandem MS (MSMS). All purifications were performed as biological duplicates and analyzed as technical duplicates (n = 4 for each TLR). Two strategies were used to obtain a confidence score for bona fide interactors. Semiquantitative information on identified proteins (peptide counts/sequence coverage) was used to compute enrichment of the protein over the initial cell lysate of RAW264.7 cells. In addition, specificity was calculated by statistical comparison with ∼1,000 unrelated protein complex purifications and analyses in our laboratory. After applying these criteria, one of the identified high-confidence interactors present in all TLR-precipitations was CD14 (UniProtKB/Swiss-Prot ID P10810, CD14_MOUSE). From four analyses, CD14 was identified with 7–10 peptides that correspond to a sequence coverage of 22.5–33.25% (Fig. 1 and Fig. S1 B).
Figure 1.
Schematic illustration of the TLR interaction proteomic approach. Murine endosomal TLR3, TLR7, TLR8, and TLR9 were stably expressed in the macrophage cell line RAW264.7 by retroviral transduction. After TAP of the receptor complexes, eluates were analyzed by one-dimensional SDS-PAGE and silver stained. 20 gel slices were excised from each lane, the proteins were digested, and the peptides were analyzed by LC-MSMS. All bait TLRs were recovered with the average amount of peptides and sequence coverage as indicated in the table. At ∼55 kD, several peptides were identified from CD14. A schematic illustration of CD14 is depicted with the domains shown (signal peptide in red and leucine-rich repeats in blue) and the location of the identified peptides (black bars). For each TLR, the average number of peptides (av p.) identified and the sequence coverage (av%sc) of CD14 are indicated in the table.
Coimmunoprecipitation and colocalization of CD14 with endosomal TLRs
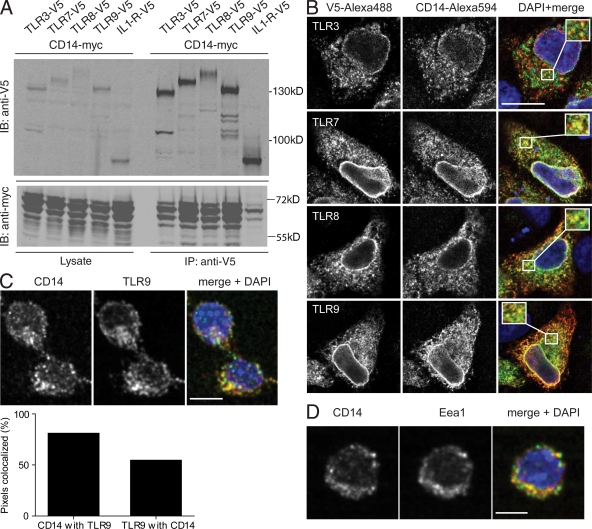
We addressed whether the TAP/MS–detected association could be confirmed by coimmunoprecipitation of V5-tagged TLRs with myc-tagged CD14 from double transfected Hek293T cells (Fig. 2 A). CD14 bound to all four endosomal TLRs, showing the strongest association with the MyD88-dependent TLR7, TLR8, and TLR9. In contrast, CD14 did not coprecipitate with IL-1R, which served as negative control. Differently migrating forms of CD14 (Fig. 2 A and see Fig. 5 A) may be attributed to different isoforms (soluble CD14/membrane-bound CD14) and have been observed previously (von Schlieffen et al., 2009).
Figure 2.
CD14 is an interactor of TLR3, TLR7, TLR8, and TLR9. (A) Myc-tagged CD14 was double transfected in Hek293T cells with either one V5-tagged endosomal TLR or V5–IL-1R as a negative control. 48 h after transfection, V5-tagged proteins were immunoprecipitated out of cell lysates using V5 agarose. Coprecipitation of CD14 was analyzed by Western blotting for myc and V5. IB, immunoblot. (B) Myc-CD14 was cotransfected with V5-TLR3, -TLR7, -TLR8, or -TLR9 into HeLaS3 cells. Colocalization was analyzed by confocal imaging. Representative areas of overlapping localization are shown magnified in the insets in the merge panel. (C) Images of a confocal section of endogenous CD14 and TLR9 in unstimulated RAW264.7 macrophages. (bottom) Quantification of three-dimensional colocalization analysis. Graph shows the percentage of pixels positive for CD14 colocalizing with pixels positive for TLR9 and the percentage of pixels positive for TLR9 colocalizing with pixels positive for CD14. (D) Colocalization analysis of the endosomal marker protein Eea1 and CD14 in unstimulated RAW264.7 macrophages. Data in A–D are representative of three independent experiments. Quantitation in C is a mean of two independent experiments with each n = 600. Bars: (B) 10 µm; (C and D) 5 µm.
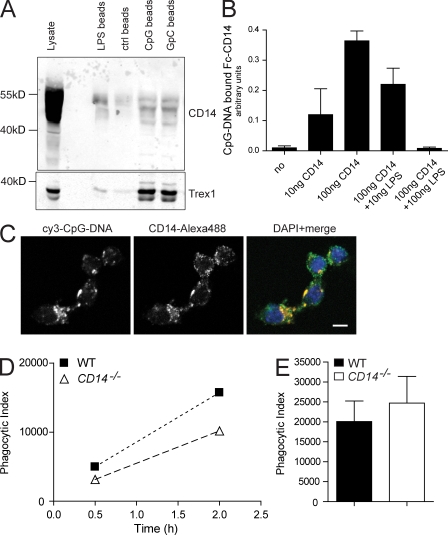
Figure 5.
Association of CD14 with DNA is required for DNA internalization and IL-6 induction. (A) Lysates from untreated RAW264.7 macrophages were incubated for 2 h with streptavidin beads that were coated with either biotinylated LPS or biotinylated CpG-DNA or with GpC-DNA. After four wash steps, bead-bound CD14 or a DNA-binding protein control (Trex1) was visualized by Western blotting. (B) For ELISA-based assay, CpG-DNA–coated ELISA plates were incubated with a soluble CD14-Fc chimera and LPS as competitor. Absorbance of anti–mouse horseradish peroxidase antibody indicates DNA-bound CD14. Data are mean ± SD. (C) 1 µM cy3-CpG-DNA was added to the culture medium of RAW264.7 macrophages. CD14 colocalization with endocytosed fluorescent DNA was visualized by immunostaining and analyzed by confocal imaging. Bar, 5 µm. (D) 1 µM cy3-CpG-DNA was added to the supernatant of WT- or CD14−/−-derived peritoneal macrophages for the indicated time periods (x axis). The uptake of fluorescent DNA by F4/80-positive macrophages was measured by FACS analysis and depicted as phagocytic index (y axis). (E) The uptake of heat-killed E. coli by thioglycollate-elicited peritoneal macrophages of WT and CD14−/− mice was tested and depicted as phagocytic index. Data in E are presented as mean ± SD. All experiments in A–E are representative of at least two independent experiments.
We further tested physical proximity of epitope-tagged CD14 and endosomal TLRs by confocal imaging of transfected HeLa cells (Fig. 2 B). CD14 clearly colocalized with the MyD88-dependent TLR7, TLR8, and TLR9 (Fig. 2 B) but less for TLR3 (Fig. 2 B). We next addressed whether colocalization could also be observed by analyzing endogenous CD14 and TLR9. Therefore, we chose to use mouse RAW264.7 macrophages that are known to be competent for TLR signaling. Indeed, the endogenous proteins colocalized in nonstimulated cells (Fig. 2 C, top). Quantitation of colocalized pixels of CD14 and TLR9 in >600 cells revealed that ∼82% of detected CD14 was present in the same areas as TLR9, and, vice versa, ∼54% of TLR9 colocalized to the same region as CD14 (Fig. 2 C, bottom). We did not observe changes in this localization pattern upon stimulation with different ligands (unpublished data). By costaining for the endosomal marker Eea1, we addressed whether CD14 would colocalize to endosomal compartments in unstimulated cells (Fig. 2 D) and indeed observed significant co-distribution. Thus, CD14 is a constitutive endosomal protein that associates with all endosomal TLRs in unstimulated cells.
CD14 promotes CpG-DNA and imiquimod recognition in Hek293-TLR9 and in isolated mouse immune cells
Next, we set out to explore the functional consequences of the association between CD14 and TLR7 and TLR9. First, we tested whether Hek293 cells that express TLR9 but not CD14 can be modulated in their proinflammatory cytokine response by the addition of soluble recombinant CD14 (sCD14) to the medium. sCD14 alone did not induce cytokine secretion (Fig. 3 A). As expected, CpG-DNA elicited low but detectable levels of IL-8. Intriguingly, this CpG-DNA response could be amplified up to fourfold by exogenous addition of sCD14 (Fig. 3 A). We conclude that sCD14 has the ability to sensitize cells for DNA recognition by TLR9.
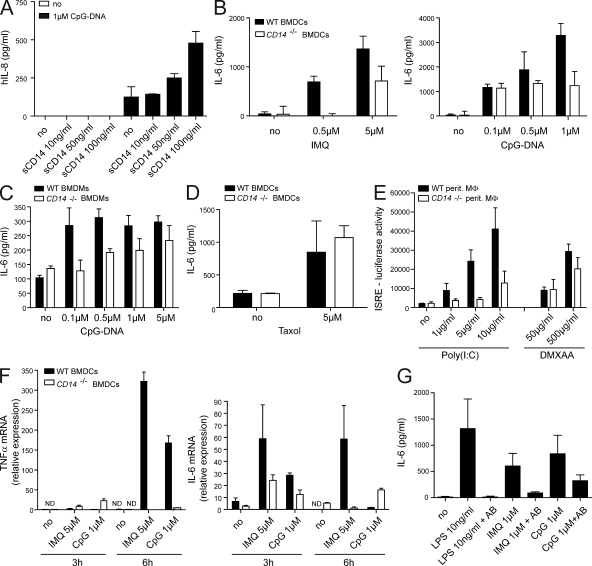
Figure 3.
CD14 is required for the proinflammatory response to imiquimod and CpG-DNA. (A) Hek293-TLR9 cells were preincubated with the indicated concentrations of sCD14 30 min before stimulation with 1 µM CpG or mock control (no). Culture supernatants were harvested 8 h later, and the levels of human IL-8 were determined. Data are presented as mean ± SD and are representative of two independent experiments, each performed in biological duplicates. (B–D) BMDCs (B and D) or BMDMs (C) from WT or CD14−/− mice were stimulated with CpG-DNA, imiquimod (IMQ), or Taxol for 6 h. IL-6 in supernatants was measured by ELISA. Data are presented as mean ± SD. (E) Peritoneal macrophages were stimulated with Poly(I:C) or DMXAA for 16 h, and type I IFN levels were determined using the IFN luciferase reporter cell line LL171. (F) BMDCs were stimulated for 3 and 6 h as indicated. TRIZOL-extracted RNA was reverse transcribed and used for quantitative real-time PCR to analyze the abundance of transcripts encoding TNF and IL-6. (G) BMDMs from WT mice were incubated with a CD14-specific inhibitory antibody (AB) where indicated 30 min before stimulation with LPS, imiquimod, or CpG-DNA. After 12 h of stimulation, culture supernatants were harvested, and IL-6 protein was measured by ELISA. Data are presented as mean ± SD and are representative of at least three independent experiments, each performed in biological triplicates.
Having established that sCD14 positively impacted DNA recognition via TLR9, we tested whether loss of CD14 would affect cytokine responses after TLR stimulation. For this purpose, we analyzed the induction of proinflammatory cytokines in macrophages and DCs derived from WT or CD14-deficient (CD14−/−) mice that were treated with TLR ligands. The TLR7 and TLR9 ligands imiquimod and CpG-DNA, respectively, elicited less IL-6 in CD14-deficient BM-derived DCs (BMDCs) and BM-derived macrophages (BMDMs) as compared with WT cells (Fig. 3, B and C; and Fig. S3). As shown previously (Moore et al., 2000), LPS-induced IL-6 secretion was completely abrogated in CD14-deficient cells (Fig. S3). Importantly, stimulation with Taxol, a ligand for TLR4 which does not require CD14 (Kawasaki et al., 2000; Zanoni et al., 2009), resulted in equal amounts of IL-6 in WT and CD14−/− BMDCs (Fig. 3 D). Similarly, DMXAA, a compound which activates the TBK1–IRF3 pathway via a yet unclear mechanism (Roberts et al., 2007), induced almost equal amounts of type I IFN in peritoneal macrophages (Fig. 3 E) and IL-6 in BMDCs (see Fig. 6 G), indicating integrity of the cells and specificity of the CD14-dependent reduction of cytokines. In addition, quantitative real-time PCR analysis of CpG and imiquimod-stimulated DCs (BMDCs) showed decreased levels of TNF and IL-6 messenger RNA levels in CD14-deficient cells as compared with WT cells (Fig. 3 F).
To obtain further evidence for a role of CD14 in DNA recognition, we made use of a monoclonal antibody that selectively blocks LPS binding to CD14 by competitive binding to the LPS-binding site on the ectodomain of CD14 (see Materials and methods). Imiquimod- and CpG-DNA–induced IL-6 secretion was strongly reduced upon preincubation of both DCs and macrophages with the inhibitory antibody (Fig. 3 G and Fig. S4 A). This suggested competitive binding of the antibody and of imiquimod/CpG-DNA to the LPS-binding region of the CD14 ectodomain. Thus, CD14 is a critical cofactor for the recognition of imiquimod and DNA by the endosomal TLR7 and TLR9 ex vivo.
Because CD14 is important for LPS recognition, we rigorously tested our stimuli for endotoxin contamination: a Limulus amebocyte lysate test showed no detectable endotoxin levels (Fig. S2 A). Addition of eritoran, a lipid A analogue which functions as an LPS antagonist on TLR4 (Kim et al., 2007), was able to inhibit LPS-induced IL-6 secretion but did not affect imiquimod- or CpG-DNA–induced IL-6 cytokine production (Fig. S2 B). Similarly, using macrophages from TLR4-deficient mice, CpG-DNA–, imiquimod-, or Poly(I:C)-induced IL-6 releases were similar in the absence of TLR4, whereas LPS-induced responses were abolished in cells lacking TLR4 (Fig. S2 C). Moreover, challenge of TLR7-deficient DCs (BMDCs) with LPS and CpG-DNA but not with the TLR7 ligand imiquimod led to strong IL-6 production, arguing against endotoxin contamination of this ligand (Fig. S2 D). Collectively, these data strongly suggest that the blunted cytokine response in CD14-deficient cells stimulated with imiquimod or CpG-DNA was not caused by endotoxin contamination.
CD14 promotes CpG-DNA–induced inflammation in vivo
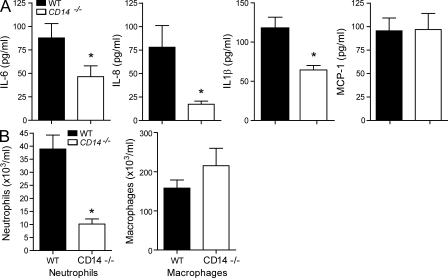
To test whether the induction of proinflammatory cytokines by CpG-DNA also required CD14 in vivo, we injected WT or CD14−/− mice i.p. with CpG-DNA. Peritoneal lavage fluid was harvested 4 h after injection, and the accumulation of proinflammatory cytokines was analyzed. Consistent with the aforementioned results, CD14−/− mice showed decreased IL-6 levels as compared with WT mice (Fig. 4 A). In addition, the levels of IL-1β and keratinocyte chemoattractant (KC; IL-8) were significantly reduced, whereas MCP-1 was unaltered. The cytokines KC and IL-1β are markers for neutrophil influx (Rogers et al., 1994; Wengner et al., 2008), whereas MCP-1 is involved in the regulation of macrophage recruitment (Takahashi et al., 2009). To test whether the reduction in cytokine levels was reflected in immune cell recruitment into the peritoneal cavity, we measured neutrophil and macrophage influx into the peritoneum of WT and CD14-deficient mice after CpG stimulation. CD14−/− mice exhibited lower amounts of neutrophils compared with WT mice upon CpG-DNA challenge, whereas the numbers of macrophages were not changed (Fig. 4 B). We concluded that CD14 is necessary for the proinflammatory response to CpG-DNA in vivo, which is consistent with its role as a cofactor of TLR9.
Figure 4.
CD14 is important for the proinflammatory response to CpG-DNA in vivo. WT or CD14−/− mice were i.p. injected with 8 nmol CpG-DNA. After 4 h, mice were sacrificed. (A) The levels of the proinflammatory cytokines IL-6, KC (IL-8), IL-1β, and MCP-1 in peritoneal lavage were analyzed by ELISA. (B) Peritoneal lavage was taken, and total cells, neutrophils, and macrophages were counted. Differences in cytokine values of WT and CD14−/− stimulation pairs were tested for significance using the one-way analysis of variance and Tukey’s post test. Significant alterations in pattern are marked with an asterisk (*, P < 0.05 vs. WT mice; Mann–Whitney T test). In A, P = 0.0434, P = 0.0005, and P = 0.0029 for IL-6, IL-8, and IL-1β, respectively. In B, P = 0.0002 for neutrophils. Data are presented as mean ± SD and are representative of two independent experiments, each performed with eight mice/group.
CD14 associates with CpG-DNA and is required for its phagocytic uptake
One possible explanation for the decreased cytokine response in CD14-deficient cells could be reduced delivery of the ligand into TLR-containing endosomal compartments. CD14 has previously been reported to bind to and promote the phagocytic uptake of Poly(I:C) (Lee et al., 2006), and thus, a similar role for CD14 in DNA binding and endosomal delivery could be anticipated. To test this, we first asked whether CD14 has affinity for CpG-DNA. We precipitated endogenous CD14 from RAW264.7 macrophages with streptavidin beads that were coated with biotinylated CpG-DNA, GpC-DNA, or LPS (Fig. 5 A). Trex1, a known DNA-binding protein (Stetson et al., 2008), served as control. Western blot analysis of precipitated material showed that CD14 was significantly enriched on beads that were coupled with either DNA species, indicating that CD14 associates with DNA. The LPS control precipitated CD14 as well, as expected. Similarly, peritoneal macrophages from CD14−/− mice that were shifted to 4°C to inhibit active phagocytosis showed ∼50% reduction in DNA association as compared with WT control, suggesting that CD14 promotes the binding of DNA to the plasma membrane (Fig. S5).
However, the association of CD14 with DNA could be mediated by other proteins and thus might be indirect. Therefore, we tested for a direct association of CD14 with DNA in an ELISA-based assay. Recombinant CD14-Fc chimera bound to CpG-coated plates, and this association could be blocked by using LPS as competitor (Fig. 5 B). This suggests that CD14 binds to CpG-DNA directly and indicates that LPS and CpG-DNA might bind to overlapping regions on CD14, which was already suggested by the antibody inhibition experiments (Fig. 3 G).
A CD14–DNA interface could be required for the active transport of free nucleic acids into the TLR-containing endosome. Fluorescently labeled CpG-DNA that was added to the culture medium of RAW264.7 macrophages colocalized with endogenous CD14 and the endosomal markers Eea1 and Lamp1 (Fig. 5 C and Fig. S4 B). These results suggest an association of CD14 with CpG-DNA in endosomal compartments. Moreover, when we compared the uptake of fluorescent DNA by WT and CD14−/− peritoneal macrophages after 30 min and 2 h (Fig. 5 D), an impaired internalization of CpG-DNA was observed in cells lacking CD14 as compared with control cells. This suggests that the process of DNA/nucleic acid internalization is partly dependent on CD14. Importantly, there was no difference between WT and CD14-deficient thioglycolate-elicited macrophages in the uptake of heat-killed Escherichia coli (Fig. 5 E). In summary, these results suggest that CD14 promotes the selective uptake of DNA into the endosome.
CD14 is dispensable for virus uptake but required for virus recognition
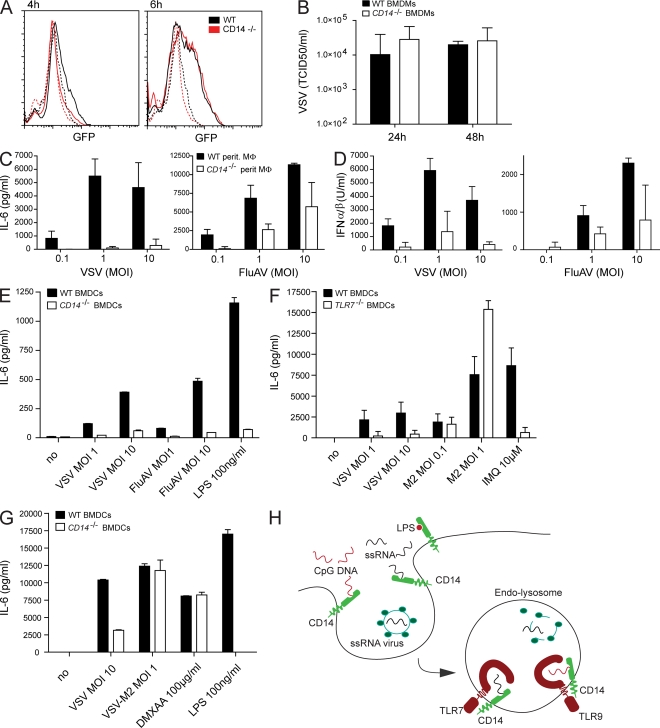
Unlike free-floating synthetic ligands, pathogen-associated DNA and RNA are released only in the late endosome/lysosome because of the acidification and activity of proteases (Blasius and Beutler, 2010). We asked whether CD14-dependent induction of proinflammatory cytokines would entirely be attributable to defective uptake of TLR agonists or whether CD14 has a more promiscuous role in endosomal recognition of nucleic acids by TLR7/9. To distinguish between these two hypotheses, we decided to use influenza A virus (FluAV) and VSV, which are both taken up by an active cellular process and are sensed by TLR7 in the endosome (Diebold et al., 2004; Lund et al., 2004). Uptake and replication of VSV bearing a green fluorescently tagged glycoprotein was indistinguishable in CD14-deficient and WT peritoneal macrophages (Fig. 6 A). Furthermore, supernatants from VSV-infected WT and CD14−/− macrophages (Fig. 6 B) and BMDCs (Fig. S6 A) contained similar amounts of infectious particles, suggesting that virus growth was not changed in CD14-deficient cells. However, FluAV- and VSV-infected CD14-deficient peritoneal macrophages generated less IL-6 and type I IFN as compared with WT control cells (Fig. 6, C and D). Equally, BMDCs from CD14−/− mice produced much less IL-6 than WT control cells. This blunted cytokine response in CD14-deficient cells was strongest for VSV and weaker but still significant after infection with FluAV (Fig. 6, C–E). Similarly, IL-6 responses induced by both viruses were strongly inhibited in the presence of the CD14 antibody but not when an isotype control was used (Fig. S6 B). These data suggest that CD14 is dispensable for the uptake of pathogens but important for their ability to stimulate proinflammatory cytokines.
Figure 6.
CD14 is dispensable for virus uptake but required for the induction of cytokines in ssRNA virus-infected macrophages. (A) Peritoneal macrophages from WT or CD14−/− mice were infected with VSV that expresses a GFP-fused glycoprotein (VSV-GFP; MOI: 1) and analyzed by FACS. The histogram shows GFP positivity of uninfected cells (dashed lines) or cells that were infected for either 4 or 6 h (solid lines). Cells were gated on forward and sideward scatter. (B) Virus accumulation in supernatant of BMDMs that were infected with VSV (MOI: 1) for the indicated time periods. The graph shows the mean virus titer from three independent experiments. Data are mean ± SD. (C–E) Accumulation of IL-6 or type I IFN 14 h after infection of WT and CD14-deficient peritoneal macrophages (C and D) or BMDCs (E) with the indicated MOI of VSV and FluAV or stimulated with LPS. (F and G) BMDCs from WT or TLR7-deficient (F) or WT and CD14-deficient (G) mice were infected with the indicated amounts of VSV or VSV-M2, a mutant which is mainly recognized in the cytoplasm, or stimulated with imiquimod, LPS, or DMXAA. IL-6 accumulation was tested 14 h after stimulation. Data in C–G are presented as mean ± SD and are representative of at least two independent experiments. (H) The model shows that CD14 associates with DNA/ssRNA at the plasma membrane and promotes their endocytosis. ssRNA viruses enter the endosome in a CD14-independent way. Acidification destroys the viral envelope, releasing the viral ssRNA genome. In the endosome, CD14 functions as a coreceptor for TLR7 and TLR9 in the recognition of ssRNA and DNA.
Unlike defined synthetic ligands, viruses can activate multiple pathways, including the TLR and the cytoplasmic RIG-like receptor pathway. Therefore, we tested whether IL-6 production in response to VSV and FluAV depends on TLR activation. We could partially inhibit VSV- and FluAV-induced IL-6 in BMDCs by applying z-FA-FMK (benzyloxycarbonyl–phenyl-alanyl–fluoromethylketone), a cysteine protease inhibitor which blocks endosomal cathepsin activity and thereby inhibits full activation of endosomal TLRs (Park et al., 2008). In contrast, LPS-induced IL-6 levels were unaltered by this drug, suggesting that VSV and FluAV recognition depended on the activation of endosomal TLRs (Fig. S6 D). As z-FA-FMK could additionally affect the degradation of viral particles in the endosome and therefore could reduce the amount of nucleic acid ligand, we proved that VSV is indeed recognized by TLR7 because TLR7-deficient BMDCs only generated minimal amounts of IL-6 after stimulation with VSV (Fig. 6 F).
We established that TLR7 requires CD14 for proper activation and hypothesized that a virus that is recognized by TLR7-independent mechanisms should therefore not require CD14 for cytokine induction. To test this hypothesis, we used a mutant version of VSV (VSV-M2), which bears a point mutation in the VSV matrix protein that serves as suppressor of the cytoplasmic recognition pathway in the WT virus (Stojdl et al., 2003). As expected, after VSV-M2 infection, TLR7-deficient and WT cells generated comparable amounts of IL-6 (Fig. 6 F), probably elicited through activation of the cytoplasmic sensor RIG-I (Kato et al., 2006). We next used this virus to stimulate CD14-deficient as well as WT BMDCs and peritoneal macrophages. As shown in Fig. 6 (C and E), VSV required CD14 for proper induction of IL-6 (Fig. 6 G and Fig. S6 C). In contrast, VSV-M2 induced similar levels of IL-6 in CD14−/− and WT BMDCs (Fig. 6 G and Fig. S6 C). Collectively, these data strongly suggest that CD14 is required for TLR-dependent recognition of viruses but is dispensable for the induction of proinflammatory cytokines in the cytoplasmic sensing of virus infection.
DISCUSSION
CD14 is a well established component of the protein complexes conferring ligand binding and signaling competence to the cell surface–expressed TLR4 (Ulevitch and Tobias, 1999). Itself a protein containing leucine-rich repeats, as the TLRs, it shares some of the structural and ligand-binding properties of TLRs and, in the case of LPS, may appear to act as a dimeric TLR coreceptor, effectively enhancing the ability of TLR4 to be triggered by LPS. The three-dimensional structure of CD14 is known, and together with mutational analysis, it has been possible to locate the LPS-binding region in the N terminus of a horseshoe-like structure (Kim et al., 2005). We have identified CD14 as interactor of all four endosomal TLRs. Our data extend the current list of receptors that require CD14 for proper function to add the MyD88-dependent TLR7 and TLR9 and thus establish a wider role for CD14 in innate immunity. We provide a series of experiments, including a mouse model, that show that CD14 is involved in the recognition and subsequent innate immune response to the ligands DNA and imiquimod/ssRNA.
It is interesting to note that CD14 has been shown to bind a whole variety of microbial products through overlapping but not identical portions of the molecule (Pugin et al., 1994; Kim et al., 2005). We now show that CD14 is not only associated with TLR7 and TLR9 but also engages their ligands. LPS and CpG-DNA appeared to compete for binding to recombinant CD14 in vitro, and nucleic acid–induced cytokine production was inhibited by antibodies that recognize the LPS-binding portion of CD14, suggesting that the N-terminal domain of CD14 may also be involved in nucleic acid recognition (Kim et al., 2005). This notwithstanding, the common region involved in binding could at the same time mediate interaction with different coreceptor proteins. How a full or partial recognition of nucleic acids by CD14 complements the nucleic acid binding of endosomal TLRs will be an interesting subject of further studies.
A requirement for CD14 in Poly(I:C) ligand endocytosis for efficient TLR3-mediated cytokine induction has been reported previously (Lee et al., 2006). This, together with our uptake assays, suggests that CD14 has a role in enhancing the selective internalization of different nucleic acids, which in turn promotes their active delivery to TLR-containing endosomes. CD14-dependent shuttling processes have been the object of previous studies and allowed for the attribution of further roles to CD14, including clearance of endotoxin (Dunzendorfer et al., 2004), Golgi trafficking of the TLR4 receptor complex (Latz et al., 2002), and the internalization of the TLR2 ligand FSL-1 after signal induction (Shamsul et al., 2010). This suggests a dual function of CD14 in the uptake as well as trafficking of ligands and receptor complexes, so far for plasma membrane–associated TLRs.
A previous functional study on Poly(I:C) uptake and TLR3 colocalization showed a strict requirement for CD14 in reconstituted Chinese hamster ovary cells (Lee et al., 2006). In this study, we tested the uptake of nucleic acids/DNA in primary murine CD14-deficient phagocytes. Interestingly, our data suggest that, even in the total absence of CD14, phagocytes still internalize ∼60% of DNA compared with their WT counterparts (Fig. 5 D). This suggests that CD14 contributes but may not be strictly necessary for DNA uptake.
We show that the uptake of heat killed E. coli and ssRNA viruses, which are internalized similarly to nucleic acids (Sun et al., 2005), does not require CD14 (Figs. 5 E and 6 A). Also, virus growth was indistinguishable in CD14−/− and WT cells, suggesting that uptake of viral pathogens is independent of CD14 (Fig. 6 B and Fig. S6 B). Functional dependence on TLRs for the recognition of ssRNA allowed for the investigation of CD14-dependent TLR activation without the complication of reduced ligand uptake. TLR7 signal induction by ssRNA viruses unequivocally required CD14 in both macrophages and DCs. The requirement of CD14 for induction of proinflammatory cytokines after virus challenge could be rescued using the VSV-M2 mutant, which is sensed by the cytoplasmic recognition machinery. These data strongly suggest a selective role of CD14 in endosomal but not cytosolic recognition of ssRNA viruses. Given that CD14 appeared to be a constitutive interactor of endosomal TLRs even in unstimulated cells, we believe that CD14 functions as a coreceptor for endosomal TLRs.
CD14 was also involved in the induction of type I IFN in peritoneal macrophages (Fig. 6 D). As the activation of TLR7/9 in macrophages is believed to exclusively trigger the MyD88-dependent induction of proinflammatory cytokines (Blasius and Beutler, 2010), it is possible that CD14 has, in addition to its function as a TLR coreceptor, a role in the cytoplasmic induction of type I IFNs in macrophages. MAVS (mitochondrial antiviral signaling protein), a platform for cytoplasmic RNA receptors, exerts signaling from the membranes of different cellular organelles (Dixit et al., 2010), and thus, it is tempting to speculate that the membrane-associated, glycosylphosphatidylinositol-anchored CD14 could be involved in the organization of those membrane rafts. In summary, we suggest two roles for CD14 in endosomal TLR activation: (1) a function in the selective nucleic acid uptake and (2) a function as a coreceptor of TLR7 and TLR9 (Fig. 6 H).
In the future, it will be important to further dissect the activities of CD14 in the endosomal recognition of pathogens. Urgent questions to be addressed are how CD14 grants ligand sensitivity to endosomal TLRs and how the LPS-binding pocket contributes to this. CD14 could contribute to the engagement of nucleic acids by a holo-receptor in analogy to LPS recognition by the TLR4–CD14 complex (Fitzgerald et al., 2004). Alternatively, CD14 might act indirectly by providing a physical platform for the recruitment of factors required to assemble a fully functional receptor complex (Schmitz and Orsó, 2002).
The finding that CD14 is required for both ssRNA/imiquimod as well as DNA recognition by TLR7 and TLR9 offers the possibility that CD14 contributes to DNA-mediated immunization and autoimmune processes. Free host DNA that can cause the development of autoantibodies and contributes to diseases like systemic lupus erythematosus has been linked to TLR9 (Krieg and Vollmer, 2007). For instance, HMGB-1, although not membrane associated, binds and enhances nucleic acid uptake into the endosome, such that a role in autoimmunity has been suggested (Tian et al., 2007). We speculate that CD14 levels affect endosomal TLR-mediated adjuvant effects and thus may impact certain anticancer therapies or antiviral responses.
MATERIALS AND METHODS
Reagents.
ELISA kits were purchased from BD. The Limulus amebocyte lysate endotoxin test was purchased from Cambrex Bio Science Verviers. Murine GM-CSF and M-CSF were purchased from PeproTech. Taxol was obtained from Sigma-Aldrich. z-FA-FMK was purchased from Sanova. All synthetic TLR ligands, Poly(I:C), imiquimod, CpG-DNA-ODN1826, CpG-DNA-ODN1585, and LPS (E. coli K12) as well as biotinylated LPS and Hek293-TLR9 cells were obtained from InvivoGen. Biotinylated and cy3-labeled oligonucleotides were synthesized at Microsynth. Protein G–Sepharose was purchased from GE Healthcare, and streptavidin beads were purchased from Thermo Fisher Scientific. Rat anti-CD14 antibody was obtained from BD, and mouse anti-V5 antibody was obtained from Invitrogen. Rabbit anti-myc was purchased from Sigma-Aldrich, mouse anti-TLR9 was purchased from Imgenex (IMG-305A), mouse anti-Lamp1 was purchased from Stressgen, and mouse anti-Eea1 was purchased from BD. Inhibitory CD14 antibody Big53 and endotoxin-free recombinant, soluble murine and human CD14 were purchased from Biometec (http://www.biometec.de/index.php?id=137), recombinant Fc-CD14 chimera was obtained from R&D Systems. F4/80 antibody was obtained from Abcam, and CD86 and CD11c antibodies were obtained from BD. All secondary antibodies were purchased from Invitrogen (Alexa Fluor 488, 546, 594, 680, and 800 nm). RT-PCR reagents were bought from GeneXPress, and buffers were bought from Sigma-Aldrich.
MS sample preparation.
Samples were alkylated with iodoacetamide and separated by one-dimensional SDS-PAGE on a 4–12% bis-Tris gel (NuPAGE; Invitrogen). After visualization of the proteins by silver staining, entire lanes were excised into 20 pieces and digested in situ with modified porcine trypsin (Promega) essentially as described by Shevchenko et al. (1996). Before analysis by nano-LC–electrospray ionization–MSMS, peptides were purified and concentrated via customized reversed-phase columns adapted from Rappsilber et al. (2003).
MS and data analysis.
Tryptically digested samples were analyzed by data-dependent nanocapillary reversed-phase LC-MSMS using customized 50 µm i.d. × 16 cm analytical column packed with C18 3-µm diameter Reprosil beads (Maisch) on a nano-LC system (1200 series; Agilent Technologies) coupled to a hybrid mass spectrometer (LTQ-Orbitrap; Thermo Fisher Scientific). Data-dependent acquisition was performed for 100 min using one MS channel for every four MSMS channels and a dynamic exclusion for selected ions of 60 s.
The acquired data were processed and searched against the mouse IPI database version 3.41 with the search engine MASCOT (Matrix Science). Submission to the search engine was via a Perl script that performs an initial search with relatively broad mass tolerances on both the precursor and fragment ions (±10 ppm and ±0.6 D, respectively). High-confidence peptide identifications were used to recalibrate all precursor and fragment ion masses before a second search with narrower mass tolerances (±4 ppm and ±0.3 D). A false-positive detection rate of <1% was estimated by searching the dataset against a reversed database. Criterion for a positive protein identification was detection of peptides with ≥6 aa and a minimum of two peptides with a MASCOT peptide score of ≥20.
Plasmids.
All constructs were cloned using the gateway system. Murine and human TLR constructs (available from GenBank/EMBL/DDBJ under the following accession nos.: mTLR3, NM_126166; mTLR7, NM_133211; mTLR8, NM_133212; mTLR9, NM_031178; hTLR3, NM_003265; hTLR7, NM_016562; hTLR8, NM_138636; and hTLR9, NM_017442) and mCD14 (available from GenBank under accession no. NM_009841) were cloned from InvivoGen vectors (pUno-HA vectors). Human IL-1R (available from GenBank under accession no. NM_000877.2) and CD14 (available from GenBank under accession no. NM_000591) were amplified from a human testis cDNA library (Takara Bio Inc.) using the following primers: CD14 5′, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGGAGCGCGCGTCCTGCTTGTTGCTGCTG-3′; CD14 3′, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTAGGCAAAGCCCCGGGCCCCTTGGAGC-3′; IL-1R 5′, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGAAAGTGTTACTCAGACTTATTTG-3′; and IL-1R 3′, 5′-CCCGAGAGGCACGTGAGCCTCTCTTTGGGGGACCACTTTGTACAAGAAAGCTGGGTT-3′. All gateway entry clones for C-terminal tagging of full-length cDNAs were created using 20–25 bp of flanking regions and the following gateway primer sequences: sense attB1 primer, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTAGACTGCCATG(NNN)5–10-3′; and antisense attB1 primer, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTNOSTOP(NNN)10–15-3′. Gateway LR Clonase (Invitrogen) target vectors for protein expression were pTracer-V5 (Invitrogen), pcDNA6myc, and pCeMM CTAP(SG).
Cell culture and viruses.
Cell lines used were RAW264.7 murine macrophages, Hek293, Vero, and HeLaS3 cells. Cells were cultured in DME (PAA) supplemented with 10% FCS (Invitrogen) and antibiotics (100 U/ml penicillin and 100 µg/ml streptomycin) at 37°C, 5% CO2. BM was retrieved from C57BL/6J (breeding facility of the Medical University of Vienna, Vienna, Austria) and B6.129S-Cd14tm1Frm/J CD14−/− (The Jackson Laboratory) by flushing the tibia and femur with RPMI (PAA). BMDMs were obtained by differentiating BM in RPMI supplemented with 10% FCS, 1% penicillin and streptomycin (Pen/Strep), and 20 ng/ml M-CSF for 7 d. BMDCs were obtained by differentiating BM with RPMI supplemented with 10% FCS, 1% Pen/Strep, 20 ng/ml GM-CSF, and 50 µM β-mercaptoethanol. Primary peritoneal macrophages were obtained using peritoneal lavage with 5 ml of sterile saline. Lavage fluid was collected in sterile tubes and placed on ice. Peritoneal macrophages were resuspended in RPMI supplemented with 10% FCS and 1% Pen/Strep at the appropriate cell density and allowed to adhere overnight. 1 h before stimulations, all primary cells were washed twice with serum-free medium and retaken in RPMI without FCS to avoid cell reconstitution with serum-derived soluble CD14.
For influenza virus production, Vero cells were cultured in VP-SFM serum-free medium (Invitrogen) in the presence of 3 µg/ml trypsin (Sigma-Aldrich). Influenza A/PR/8/34 was propagated on Vero cells adapted from serum-free medium and supplemented with 3 µg/ml trypsin. Semliki forest virus (a gift from I. Kerr, Cancer Research UK, London, England, UK), VSV (strain: Indiana; a gift from F. Weber, University Hospital Freiburg, Freiburg, Germany), and the VSV-M2 mutant strain (provided by J. Bell and U. Kalinke) were grown on Vero cells under standard conditions, and virus stocks were titrated by plaque assay on Vero cells.
C57BL/6J mice were obtained from the breeding facility of the Medical University of Vienna, and B6.129S-Cd14tm1Frm/J CD14−/− were ordered from The Jackson Laboratory. BM of mice originally generated in the lab of S. Akira (Osaka University, Suita, Osaka, Japan) was provided by C. Lassnig (University of Veterinary Medicine, Vienna, Austria; TLR4−/−) and by B. Drobits and M. Sibilia (Medical University of Vienna; TLR7−/−). The BM was retrieved by flushing the tibia and femur with RPMI (PAA). BMDMs were obtained by differentiating BM in RPMI supplemented with 10% FCS, 1% Pen/Strep, and 20 ng/ml M-CSF for 7 d.
Total IFN-α/β was measured by titration on LL171 cells and compared with recombinant IFN-β used as a cytokine standard (Habjan et al., 2009). To measure VSV growth on primary cells, BMDMs and BMDCs were infected with VSV (multiplicity of infection [MOI]: 1), and accumulation of infectious virus after 24 and 48 h was measured by serial dilution on Vero cells.
RT-PCR.
RNA from primary cells for quantitative real-time PCR analysis was extracted using TRIZOL. 100 ng RNA/sample was reversely transcribed using reagents/RT (Fermentas). cDNA was diluted 1:20, and transcript abundance of murine IL-6 and TNF was analyzed using SYBR green (GeneXPress) and the following primers: IL-6 5′, 5′-TTCCATCCAGTTGCCTTCTT-3′; IL-6 3′, 5′-ATTTCCACGATTTCCCAGAG-3′; TNF 5′, 5′-CAAAATTCGAGTGACAAGCCTG-3′; TNF 3′, 5′-GAGATCCATGCCGTTGGC-3′; cyclophilin B 5′, 5′-CAGCAAGTTCCATCGTGTCATCAAGG-3′; and cyclophilin B 3′, 5′-GGAAGCGCTCACCATAGATGCTC-3′. Quantitative real-time PCR analysis was performed on a Rotor Gene 6000 (QIAGEN). Measured TNF and IL-6 cycles were normalized to the ones of cyclophilin B.
In vivo experimental procedures.
Pathogen-free 9–11-wk-old female C57BL/6J (breeding facility of the Medical University of Vienna) and B6.129S-Cd14tm1Frm/J CD14−/− were used in all in vivo experiments, which were approved by the Animal Care and Use Committee of the Medical University of Vienna. Mice were injected i.p. with 8 nmol CpG-DNA and 20 mg DGALN (Sigma-Aldrich) in 200 µl of saline per mouse. After 4 h, mice were sacrificed, and peritoneal lavage was taken. Cell counts were determined on each peritoneal lavage sample stained with Turks solution, and differential cell counts were performed on cytospin samples stained with Giemsa.
Phagocytosis assay and FACS.
For analysis of phagocytosis of fluorescent DNA, either RAW264.7 or peritoneal macrophages were plated at a density of 106 on 6-well plates. 1 µM cy3-CpG-DNA was added to the cell culture medium for the indicated time points. 1 well of each cell type was shifted to 4°C before the addition of oligonucleotides and served as a phagocytosis-negative control. After three washes with ice cold PBS, cells were harvested and resuspended in PBS, and cy3-positive/F4-80–positive cells were analyzed on a FACSCalibur flow cytometer (BD). A set of 20,000 gated cells was analyzed for their GMI (geometric mean intensity of Cy3 fluorescence). The phagocytoid index was calculated as follows: GMI sample × amount of gated cells − GMI 4°C control × amount of gated cells.
Immunoprecipitation and Western blotting.
1.5 × 109 RAW264.7 macrophages per endosomal TLR construct have been used for TAP. All steps have been performed as described in Bürckstümmer et al. (2006), despite the use of lysis buffer (50 mM Tris-HCl, pH 6, 150 mM NaCl, 1 mM EDTA, 7.5% glycerol, 25 mM NaF, 0.2% NP-40, 1 mM DTT, and 1 mM Na3VO4, adjusted to pH 6.0 with HCl).
Immunoprecipitation of tagged TLRs and CD14 was performed as follows: 6 × 106 Hek293T cells were seeded on 10-cm dishes and double transfected with 7.5 µg of plasmid DNA encoding human CD14-myc and 7.5 µg of plasmid DNA encoding either the C-terminally V5-tagged human TLR3, TLR7, TLR8, or TLR9 or IL-1R protein. 48 h later, cells were lysed with lysis buffer (1% NP40, 20 mM Tris-Hcl, pH 7.4, 150 mM NaCl, 5 mM EDTA, and 5 mM EGTA), and the soluble lysate was used for immunoprecipitation using V5 agarose (Sigma-Aldrich). After a 2-h incubation of beads on a rotating wheel for 2 h, beads were washed four times with cold lysis buffer and taken in Laemmli buffer. Boiled eluates were run on SDS-PAGE and transferred on membranes by Western blotting. V5 and myc epitopes were visualized by Western blotting and detected using an Odyssey system (LI-COR Biosciences). For immunoprecipitation of endogenous CD14, biotinylated CpG-DNA or GpC-DNA or biotinylated LPS was coupled to streptavidin beads and washed twice before incubation with RAW264.7 macrophage lysates. After a 2-h incubation at 4°C on a rotating wheel, beads were washed four times with lysis buffer and taken and boiled in Laemmli buffer. Precipitation of endogenous CD14 and Trex1 was analyzed by Western blotting using anti-CD14 and anti-Trex1 antibodies. To investigate the specificity of recombinant CD14 to CpG-DNA, streptavidin-coated 96-well plates (Thermo Fisher Scientific) were coated with biotinylated CpG-DNA overnight. After three washes of the plates with PBS/0.1% Tween, the wells were incubated with the indicated amounts of Fc-CD14 chimera (R&D Systems) and LPS competitor for 2 h. After three washes, bound CD14 was incubated with anti–mouse horseradish peroxidase antibody and, after another five washes of the plates, detected with tetramethylbenzidine substrate (BD).
Immunofluorescence analysis.
For colocalization analysis, HeLaS3 cells were plated on coverslips at a density of 3 × 105/well on a 6-well plate. 24 h later, cells were transfected with 1 µg human CD14-myc and 1 µg human TLR-V5 construct using Lipofectamine (QIAGEN). 24 h later, cells were washed and fixed with 4% PFA/PBS for 10 min at room temperature. After three washes with PBS, slides were permeabilized with 0.1% Triton/PBS for 10 min. Then, coverslips were blocked with 3% BSA/PBS for 30 min, and incubations with primary and secondary antibody were each performed for 1 h in 3% BSA/PBS at room temperature. Primary antibodies were stained with the fluorescence-coupled antibodies Alexa Fluor 488–goat anti–rabbit (A11008) and Alexa Fluor 594–goat anti–mouse (A11005; both from Invitrogen). DNA was visualized by DAPI (Roth). After another three wash steps with PBS, coverslips were mounted on glass slides using ProLong Gold (Invitrogen). Stainings of endogenous proteins in RAW264.7 macrophages were performed equally, except that direct primary antibodies binding to the respective proteins were used. The localization of proteins was analyzed using a confocal microscope (LSM510; Carl Zeiss, Inc.), equipped with the 63× numerical aperture 1.4 Plan-Apochromat lens. DAPI was excited with the 405-nm laser line and the emitted fluorescent light was detected in the range of 420–470 nm. Green represents fluorescent signal collected in the range of 505–550 nm upon excitation with a 488-nm laser. Red corresponds to a signal in the range of 575–620 nm and resulted from 561-nm excitation. Confocal stacks with pinhole of 1 airy unit were recorded, with the section spacing of 150 nm. Threshold-based colocalization was performed on the three-dimensional datasets using the Definiens Software Suite (Definiens AG). Fixed thresholds were used to determine the green, red, and colocalizing population.
For colocalization analysis of phagocytosed cy3-CpG-DNA with CD14 in RAW264.7 macrophages, a confocal microscope (TCS SP5; Leica) with a 63× oil immersion objective (Leica) was used. Fluorochromes were excited using an argon laser at 488 nm and a HeNe laser at 568 nm for Texas red. Detector slits were configured to minimize any cross talk between the channels. Z stacks (optical sections) of the images were collected with an optical thickness of 0.2 mm. Images were processed using the LAS system software (Leica).
All other pictures were taken with a microscope (Eclipse 80i; Nikon) with a Plan-Fluor 40× objective (Nikon) with a numerical aperture of 0.75. All images were processed with Photoshop (Adobe) and assembled in Illustrator software (Adobe) with identical processing for all images in one experiment.
Online supplemental material.
Fig. S1 shows representative silver stain gels of the TLR TAP eluates and the peptides of CD14 that were detected by MS. Fig. S2 contains several lines of evidence that the TLR ligands imiquimod and CpG-DNA were endotoxin free. Fig. S3 shows the requirement of CD14 for IL-6 production upon CpG and imiquimod stimulation in peritoneal macrophages. Fig. S4 shows inhibition of IL-6 levels upon addition of an inhibitory CD14 antibody to WT BMDCs before stimulation with imiquimod and also shows that cy3-CpG is sufficiently taken up into early and late endosomes of RAW264.7 macrophages. Fig. S5 shows the relative adhesion of free cy3-CpG-DNA to WT or CD14−/− peritoneal macrophages. Fig. S6 shows that VSV titers are equally high after infection of BMDCs, that VSV and FluAV recognition in macrophages depends on CD14 and can be inhibited in WT cells with a CD14-specific antibody, and that the protease inhibitor z-FA-FMK decreases the proinflammatory response to VSV and FluAV but not LPS. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20101111/DC1.
Acknowledgments
We thank John Bell and Ulrich Kalinke for the VSV-M2 virus, Friedemann Weber for providing VSV-GFP, and Eisai Inc. for supplying eritoran. We thank Shizuo Akira for TLR4−/− and TLR7−/− mice and Caroline Lassnig, Barbara Drobits, and Maria Sibilia for providing BM. We are grateful to Birgit Niederreiter for help with Leica confocal imaging. We thank Tilmann Bürckstümmer for expert advice on TAP and reading of the manuscript and the members of the Superti-Furga and Knapp laboratories for helpful discussions.
C.L. Baumann was supported by a Marie Curie Fellowship (PIEF-GA-2008-220596). A. Pichlmair was supported by a European Molecular Biology Organization long-term fellowship (ATLF 2008-463).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- BMDC
- BM-derived DC
- BMDM
- BM-derived macrophage
- FluAV
- influenza A virus
- IL-1R
- IL-1 receptor
- KC
- keratinocyte chemoattractant
- LC
- liquid chromatography
- MOI
- multiplicity of infection
- MS
- mass spectrometry
- sCD14
- soluble recombinant CD14
- ssRNA
- single-stranded RNA
- TAP
- tandem affinity purification
- TLR
- Toll-like receptor
- VSV
- vesicular stomatitis virus
References
- Akashi-Takamura S., Miyake K. 2008. TLR accessory molecules. Curr. Opin. Immunol. 20:420–425 10.1016/j.coi.2008.07.001 [DOI] [PubMed] [Google Scholar]
- Andrejeva J., Childs K.S., Young D.F., Carlos T.S., Stock N., Goodbourn S., Randall R.E. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA. 101:17264–17269 10.1073/pnas.0407639101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius A.L., Beutler B. 2010. Intracellular toll-like receptors. Immunity. 32:305–315 10.1016/j.immuni.2010.03.012 [DOI] [PubMed] [Google Scholar]
- Bouwmeester T., Bauch A., Ruffner H., Angrand P.O., Bergamini G., Croughton K., Cruciat C., Eberhard D., Gagneur J., Ghidelli S., et al. 2004. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat. Cell Biol. 6:97–105 10.1038/ncb1086 [DOI] [PubMed] [Google Scholar]
- Brinkmann M.M., Spooner E., Hoebe K., Beutler B., Ploegh H.L., Kim Y.M. 2007. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J. Cell Biol. 177:265–275 10.1083/jcb.200612056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürckstümmer T., Bennett K.L., Preradovic A., Schütze G., Hantschel O., Superti-Furga G., Bauch A. 2006. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods. 3:1013–1019 10.1038/nmeth968 [DOI] [PubMed] [Google Scholar]
- Diebold S.S., Kaisho T., Hemmi H., Akira S., Reis e Sousa C. 2004. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 303:1529–1531 10.1126/science.1093616 [DOI] [PubMed] [Google Scholar]
- Dixit E., Boulant S., Zhang Y., Lee A.S., Odendall C., Shum B., Hacohen N., Chen Z.J., Whelan S.P., Fransen M., et al. 2010. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 141:668–681 10.1016/j.cell.2010.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunzendorfer S., Lee H.K., Soldau K., Tobias P.S. 2004. TLR4 is the signaling but not the lipopolysaccharide uptake receptor. J. Immunol. 173:1166–1170 [DOI] [PubMed] [Google Scholar]
- Fitzgerald K.A., Rowe D.C., Golenbock D.T. 2004. Endotoxin recognition and signal transduction by the TLR4/MD2-complex. Microbes Infect. 6:1361–1367 10.1016/j.micinf.2004.08.015 [DOI] [PubMed] [Google Scholar]
- Gavin A.C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L.J., Bastuck S., Dümpelfeld B., et al. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature. 440:631–636 10.1038/nature04532 [DOI] [PubMed] [Google Scholar]
- Gay N.J., Gangloff M., Weber A.N. 2006. Toll-like receptors as molecular switches. Nat. Rev. Immunol. 6:693–698 10.1038/nri1916 [DOI] [PubMed] [Google Scholar]
- Habjan M., Pichlmair A., Elliott R.M., Overby A.K., Glatter T., Gstaiger M., Superti-Furga G., Unger H., Weber F. 2009. NSs protein of rift valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J. Virol. 83:4365–4375 10.1128/JVI.02148-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häcker H., Redecke V., Blagoev B., Kratchmarova I., Hsu L.C., Wang G.G., Kamps M.P., Raz E., Wagner H., Häcker G., et al. 2006. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 439:204–207 10.1038/nature04369 [DOI] [PubMed] [Google Scholar]
- Jin M.S., Kim S.E., Heo J.Y., Lee M.E., Kim H.M., Paik S.G., Lee H., Lee J.O. 2007. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 130:1071–1082 10.1016/j.cell.2007.09.008 [DOI] [PubMed] [Google Scholar]
- Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J., et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 441:101–105 10.1038/nature04734 [DOI] [PubMed] [Google Scholar]
- Kawasaki K., Akashi S., Shimazu R., Yoshida T., Miyake K., Nishijima M. 2000. Mouse toll-like receptor 4.MD-2 complex mediates lipopolysaccharide-mimetic signal transduction by Taxol. J. Biol. Chem. 275:2251–2254 10.1074/jbc.275.4.2251 [DOI] [PubMed] [Google Scholar]
- Kim D.H., Sarbassov D.D., Ali S.M., King J.E., Latek R.R., Erdjument-Bromage H., Tempst P., Sabatini D.M. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 110:163–175 10.1016/S0092-8674(02)00808-5 [DOI] [PubMed] [Google Scholar]
- Kim J.I., Lee C.J., Jin M.S., Lee C.H., Paik S.G., Lee H., Lee J.O. 2005. Crystal structure of CD14 and its implications for lipopolysaccharide signaling. J. Biol. Chem. 280:11347–11351 10.1074/jbc.M414607200 [DOI] [PubMed] [Google Scholar]
- Kim H.M., Park B.S., Kim J.I., Kim S.E., Lee J., Oh S.C., Enkhbayar P., Matsushima N., Lee H., Yoo O.J., Lee J.O. 2007. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 130:906–917 10.1016/j.cell.2007.08.002 [DOI] [PubMed] [Google Scholar]
- Köcher T., Superti-Furga G. 2007. Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nat. Methods. 4:807–815 10.1038/nmeth1093 [DOI] [PubMed] [Google Scholar]
- Krieg A.M., Vollmer J. 2007. Toll-like receptors 7, 8, and 9: linking innate immunity to autoimmunity. Immunol. Rev. 220:251–269 10.1111/j.1600-065X.2007.00572.x [DOI] [PubMed] [Google Scholar]
- Lande R., Gregorio J., Facchinetti V., Chatterjee B., Wang Y.H., Homey B., Cao W., Wang Y.H., Su B., Nestle F.O., et al. 2007. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 449:564–569 10.1038/nature06116 [DOI] [PubMed] [Google Scholar]
- Latz E., Visintin A., Lien E., Fitzgerald K.A., Monks B.G., Kurt-Jones E.A., Golenbock D.T., Espevik T. 2002. Lipopolysaccharide rapidly traffics to and from the Golgi apparatus with the toll-like receptor 4-MD-2-CD14 complex in a process that is distinct from the initiation of signal transduction. J. Biol. Chem. 277:47834–47843 10.1074/jbc.M207873200 [DOI] [PubMed] [Google Scholar]
- Lee H.K., Dunzendorfer S., Soldau K., Tobias P.S. 2006. Double-stranded RNA-mediated TLR3 activation is enhanced by CD14. Immunity. 24:153–163 10.1016/j.immuni.2005.12.012 [DOI] [PubMed] [Google Scholar]
- Lin S.C., Lo Y.C., Wu H. 2010. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 465:885–890 10.1038/nature09121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Botos I., Wang Y., Leonard J.N., Shiloach J., Segal D.M., Davies D.R. 2008. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 320:379–381 10.1126/science.1155406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund J., Sato A., Akira S., Medzhitov R., Iwasaki A. 2003. Toll-like receptor 9–mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J. Exp. Med. 198:513–520 10.1084/jem.20030162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund J.M., Alexopoulou L., Sato A., Karow M., Adams N.C., Gale N.W., Iwasaki A., Flavell R.A. 2004. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. USA. 101:5598–5603 10.1073/pnas.0400937101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. 2008. Origin and physiological roles of inflammation. Nature. 454:428–435 10.1038/nature07201 [DOI] [PubMed] [Google Scholar]
- Moore K.J., Andersson L.P., Ingalls R.R., Monks B.G., Li R., Arnaout M.A., Golenbock D.T., Freeman M.W. 2000. Divergent response to LPS and bacteria in CD14-deficient murine macrophages. J. Immunol. 165:4272–4280 [DOI] [PubMed] [Google Scholar]
- O’Neill L.A. 2008. The interleukin-1 receptor/Toll-like receptor superfamily: 10 years of progress. Immunol. Rev. 226:10–18 10.1111/j.1600-065X.2008.00701.x [DOI] [PubMed] [Google Scholar]
- Park B., Brinkmann M.M., Spooner E., Lee C.C., Kim Y.M., Ploegh H.L. 2008. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat. Immunol. 9:1407–1414 10.1038/ni.1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B.S., Song D.H., Kim H.M., Choi B.S., Lee H., Lee J.O. 2009. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 458:1191–1195 10.1038/nature07830 [DOI] [PubMed] [Google Scholar]
- Pugin J., Heumann I.D., Tomasz A., Kravchenko V.V., Akamatsu Y., Nishijima M., Glauser M.P., Tobias P.S., Ulevitch R.J. 1994. CD14 is a pattern recognition receptor. Immunity. 1:509–516 10.1016/1074-7613(94)90093-0 [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Ishihama Y., Mann M. 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75:663–670 10.1021/ac026117i [DOI] [PubMed] [Google Scholar]
- Roberts Z.J., Goutagny N., Perera P.Y., Kato H., Kumar H., Kawai T., Akira S., Savan R., van Echo D., Fitzgerald K.A., et al. 2007. The chemotherapeutic agent DMXAA potently and specifically activates the TBK1–IRF-3 signaling axis. J. Exp. Med. 204:1559–1569 10.1084/jem.20061845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers H.W., Tripp C.S., Schreiber R.D., Unanue E.R. 1994. Endogenous IL-1 is required for neutrophil recruitment and macrophage activation during murine listeriosis. J. Immunol. 153:2093–2101 [PubMed] [Google Scholar]
- Schmitz G., Orsó E. 2002. CD14 signalling in lipid rafts: new ligands and co-receptors. Curr. Opin. Lipidol. 13:513–521 10.1097/00041433-200210000-00007 [DOI] [PubMed] [Google Scholar]
- Shamsul H.M., Hasebe A., Iyori M., Ohtani M., Kiura K., Zhang D., Totsuka Y., Shibata K.I. 2010. The Toll-like receptor 2 (TLR2) ligand FSL-1 is internalized via the clathrin-dependent endocytic pathway triggered by CD14 and CD36 but not by TLR2. Immunology. 130:262–272 10.1111/j.1365-2567.2009.03232.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850–858 10.1021/ac950914h [DOI] [PubMed] [Google Scholar]
- Soulat D., Bürckstümmer T., Westermayer S., Goncalves A., Bauch A., Stefanovic A., Hantschel O., Bennett K.L., Decker T., Superti-Furga G. 2008. The DEAD-box helicase DDX3X is a critical component of the TANK-binding kinase 1-dependent innate immune response. EMBO J. 27:2135–2146 10.1038/emboj.2008.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson D.B., Ko J.S., Heidmann T., Medzhitov R. 2008. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 134:587–598 10.1016/j.cell.2008.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojdl D.F., Lichty B.D., tenOever B.R., Paterson J.M., Power A.T., Knowles S., Marius R., Reynard J., Poliquin L., Atkins H., et al. 2003. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell. 4:263–275 10.1016/S1535-6108(03)00241-1 [DOI] [PubMed] [Google Scholar]
- Sun X., Yau V.K., Briggs B.J., Whittaker G.R. 2005. Role of clathrin-mediated endocytosis during vesicular stomatitis virus entry into host cells. Virology. 338:53–60 10.1016/j.virol.2005.05.006 [DOI] [PubMed] [Google Scholar]
- Takahashi M., Galligan C., Tessarollo L., Yoshimura T. 2009. Monocyte chemoattractant protein-1 (MCP-1), not MCP-3, is the primary chemokine required for monocyte recruitment in mouse peritonitis induced with thioglycollate or zymosan A. J. Immunol. 183:3463–3471 10.4049/jimmunol.0802812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K., Akira S. 2005. Toll-like receptors in innate immunity. Int. Immunol. 17:1–14 10.1093/intimm/dxh186 [DOI] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. 2010. Pattern recognition receptors and inflammation. Cell. 140:805–820 10.1016/j.cell.2010.01.022 [DOI] [PubMed] [Google Scholar]
- Tian J., Avalos A.M., Mao S.Y., Chen B., Senthil K., Wu H., Parroche P., Drabic S., Golenbock D., Sirois C., et al. 2007. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat. Immunol. 8:487–496 10.1038/ni1457 [DOI] [PubMed] [Google Scholar]
- Ulevitch R.J., Tobias P.S. 1999. Recognition of gram-negative bacteria and endotoxin by the innate immune system. Curr. Opin. Immunol. 11:19–22 10.1016/S0952-7915(99)80004-1 [DOI] [PubMed] [Google Scholar]
- von Schlieffen E., Oskolkova O.V., Schabbauer G., Gruber F., Blüml S., Genest M., Kadl A., Marsik C., Knapp S., Chow J., et al. 2009. Multi-hit inhibition of circulating and cell-associated components of the toll-like receptor 4 pathway by oxidized phospholipids. Arterioscler. Thromb. Vasc. Biol. 29:356–362 10.1161/ATVBAHA.108.173799 [DOI] [PubMed] [Google Scholar]
- Wengner A.M., Pitchford S.C., Furze R.C., Rankin S.M. 2008. The coordinated action of G-CSF and ELR + CXC chemokines in neutrophil mobilization during acute inflammation. Blood. 111:42–49 10.1182/blood-2007-07-099648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M., Fujita T. 2010. Recognition of viral nucleic acids in innate immunity. Rev. Med. Virol. 20:4–22 10.1002/rmv.633 [DOI] [PubMed] [Google Scholar]
- Zak D.E., Aderem A. 2009. A systems view of host defense. Nat. Biotechnol. 27:999–1001 10.1038/nbt1109-999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanoni I., Ostuni R., Capuano G., Collini M., Caccia M., Ronchi A.E., Rocchetti M., Mingozzi F., Foti M., Chirico G., et al. 2009. CD14 regulates the dendritic cell life cycle after LPS exposure through NFAT activation. Nature. 460:264–268 10.1038/nature08118 [DOI] [PubMed] [Google Scholar]