In humans and mice, CD8α+ conventional dendritic cells are the primary source of interferon-λ released in response to the adjuvant and Toll-like receptor 3 agonist poly IC.
Abstract
Polyinosinic:polycytidylic acid (poly IC), a double-stranded RNA, is an effective adjuvant in vivo. IFN-λs (also termed IL-28/29) are potent immunomodulatory and antiviral cytokines. We demonstrate that poly IC injection in vivo induces large amounts of IFN-λ, which depended on hematopoietic cells and the presence of TLR3 (Toll-like receptor 3), IRF3 (IFN regulatory factor 3), IRF7, IFN-I receptor, Fms-related tyrosine kinase 3 ligand (FL), and IRF8 but not on MyD88 (myeloid differentiation factor 88), Rig-like helicases, or lymphocytes. Upon poly IC injection in vivo, the IFN-λ production by splenocytes segregated with cells phenotypically resembling CD8α+ conventional dendritic cells (DCs [cDCs]). In vitro experiments revealed that CD8α+ cDCs were the major producers of IFN-λ in response to poly IC, whereas both CD8α+ cDCs and plasmacytoid DCs produced large amounts of IFN-λ in response to HSV-1 or parapoxvirus. The nature of the stimulus and the cytokine milieu determined whether CD8α+ cDCs produced IFN-λ or IL-12p70. Human DCs expressing BDCA3 (CD141), which is considered to be the human counterpart of murine CD8α+ DCs, also produced large amounts of IFN-λ upon poly IC stimulation. Thus, IFN-λ production in response to poly IC is a novel function of mouse CD8α+ cDCs and their human equivalents.
The IFN-λ1, IFN-λ2, and IFN-λ3 cytokine family, also called IL-29, IL-28A, and IL-28B, respectively, has recently been identified (Kotenko et al., 2003; Sheppard et al., 2003) and shown to be related to type I IFNs (IFN-Is) and the IL-10 family of cytokines. IFN-λs signal via a heterodimeric receptor, consisting of one chain unique for IFN-λ (IFN-λR1 or IL-28Rα) and another chain (IL-10R2), which is shared with IL-10–related cytokines. Similar to IFN-Is, IFN-λs possess antiviral, antitumor, and various immune-modulating functions (Li et al., 2009). In contrast to the ubiquitous expression of the IFN-I receptor (IFN-IR), the expression of the IFN-λ receptor is restricted to certain cell types, including epithelial cells and plasmacytoid DCs (pDCs; Ank et al., 2008; Sommereyns et al., 2008). Exposure to viruses or analogues of nucleic acids such as CpG-oligonucleotide (ODN), conditions known to trigger the production of IFN-Is, also induces IFN-λs and largely depends on similar signaling components (Coccia et al., 2004; Onoguchi et al., 2007; Osterlund et al., 2007; Ank et al., 2008). The precise role of IFN-λs in viral infections is not yet known. However, it plays a role in Toll-like receptor (TLR)–induced protection against mucosal viral infections, and recent studies link the IL28B gene with an ability to clear and recover from Hepatitis C infection (Ank et al., 2008; Ge et al., 2009; Suppiah et al., 2009; Tanaka et al., 2009; Thomas et al., 2009). Thus, it is of utmost importance to understand the cellular origin of IFN-λs and the regulation of its production.
Several cell types, including PBMCs, monocyte-derived DCs, and pDCs, stimulated with various agents in vitro have been described to produce IFN-λ, mostly analyzed by gene transcripts (Kotenko et al., 2003; Sheppard et al., 2003; Coccia et al., 2004; Osterlund et al., 2005; Ank et al., 2008). Polyinosinic:polycytidylic acid (poly IC) is used as an important immune stimulant and is an excellent adjuvant for the induction of TH1 CD4 T cell responses in a DC-targeted vaccine model (Schulz et al., 2005; Longhi et al., 2009). It is a mimic of viral double-stranded RNA (dsRNA) and is recognized by TRIF-dependent TLR3 or Cardif (also known as IPS-1, MAVS, and VISA)-dependent Rig-like helicases (RLHs). Injection of poly IC in vivo induces large systemic levels of IFN-α (Field et al., 1967). Nonhematopoietic cells using the RLH MDA5 have been identified to be responsible for the majority of this systemic IFN-α (Gitlin et al., 2006; Longhi et al., 2009).
IFN-I production in response to RLH or TLR signaling depends on the presence and the activation of transcription factors of the IFN regulatory factor (IRF) family. IRF3 and IRF7 have been described to be important for the optimal production of IFN-I (Tamura et al., 2008). However, depending on the cellular source and the pattern recognition receptor (PRR) used, the role of individual IRFs varies (Tamura et al., 2008). For the production of IFN-λs, much less is known, but a role for IRF3 and IRF7 has been suggested for different stimuli and different cells, including DCs (Coccia et al., 2004; Onoguchi et al., 2007; Osterlund et al., 2007).
Based on phenotypic, functional, and developmental differences, the DCs of the spleen are divided into several subsets. A major distinction can be made between pDCs and conventional DCs (cDCs). pDCs are best known for their outstanding capacity for IFN-α production. cDCs are further grouped into CD8α+ and CD8− cDC subsets (Vremec et al., 1992). The CD8− cDCs can be further segregated by the expression of CD4 into CD4+ cDCs and double negative (DN) cDCs (Vremec et al., 2000), but other surface markers including CD11b and CD172a are similarly highly expressed by CD4+ and DN cDCs. Quantitative proteomic analysis of spleen cDC subsets clearly demonstrated the relatedness of the two CD8− cDC subsets distinguishing these DCs from CD8α+ cDCs (Luber et al., 2010).
CD8α+ cDCs are well known for the production of IL-12p70 in various organs, including spleen, lymph nodes, thymus, and liver (Reis e Sousa et al., 1997; Hochrein et al., 2001; Pillarisetty et al., 2004). Another important function of CD8α+ cDCs is their outstanding capacity for cross-presentation (Shortman et al., 2009). Cross-presentation is not confined to spleen cDCs bearing the CD8α molecule because a rare subset of CD8− cDCs with the cross-presenting ability of CD8α+ cDCs has been identified in spleen. They lack CD11b and CD172a expression and express CD205 and high levels of CD24 (Bedoui et al., 2009). Upon treatment with Fms-related tyrosine kinase 3 ligand (FL), the numbers of those equivalent of CD8α+ (eCD8α) cDCs can be greatly enhanced in vivo (Bedoui et al., 2009). Likewise, FL treatment of BM cells can drive eCD8α cDCs (as well as eCD8− cDCs and pDCs) in vitro. Recent work on mice deficient for, or carrying mutations in, transcription factors such as IRF8 or Batf3 demonstrated the absence of CD8α+ cDCs in lymphoid organs and lack of eCD8α cDCs in peripheral organs as well as the failure to drive eCD8α cDCs in FL cultures (Schiavoni et al., 2002; Aliberti et al., 2003; Hildner et al., 2008; Edelson et al., 2010). Recently, cross-presenting CD8− cDCs in other organs such as lymph nodes have also been characterized (Henri et al., 2010).
CD8 is not expressed on human DCs, whereas CD4 is expressed on all DC subsets. A set of antibodies designated BDCA1–4 has been developed to differentiate between pDCs and subsets of cDCs (Dzionek et al., 2000). The human BDCA3-positive cDCs have been proposed as the human eCD8α cDCs. Common to the mouse CD8α+ DCs, BDCA3-positive cDCs selectively express high levels of Clec9a and Necl2 but low amounts of CD11b (Shortman et al., 2009). Genome-wide transcriptional analysis substantiated a close relationship of murine CD8α+ cDCs with human BDCA3+ cDCs (Robbins et al., 2008). Like mouse CD8α+ and eCD8α cDCs, human BDCA3+ cDCs have been found in various organs, including blood, spleen, tonsils, lymph nodes, and liver (Dzionek et al., 2000; Lindstedt et al., 2005; Velásquez-Lopera et al., 2008; Bamboat et al., 2009; Poulin et al., 2010). Recently, the functional hallmarks of murine CD8α+ cDCs, IL-12p70 production, cross-presentation ability, and TLR3 expression, have also been shown to align with human BDCA3+ cDCs. This strongly suggests that BDCA3+ cDCs are the human equivalents of murine CD8α+ cDCs (Bachem et al., 2010; Crozat et al., 2010; Jongbloed et al., 2010; Poulin et al., 2010; Villadangos and Shortman, 2010).
Expression of the different nucleic acid–sensing systems TLR3, TLR7, or TLR9 and the RLHs varies among DC subsets (Hochrein and O’Keeffe, 2008). The downstream functions after engagement of these receptors also differ among the different DCs. pDCs predominantly use TLR7 and TLR9 for nucleic acid sensing, resulting in the high production of IFN-I and IFN-λs. Among cDCs, CD8α+ cDCs highly express TLR3 but lack expression of TLR7 (Edwards et al., 2003) and, in stark contrast to CD8− cDCs, hardly express the RLHs and as a consequence are unable to detect the single-stranded RNA viruses Sendai or influenza virus (Luber et al., 2010).
DNA viruses such as poxviruses or herpesviruses are recognized by pDCs predominantly via a TLR9- and MyD88 (myeloid differentiation factor 88)-dependent pathway, whereas other cells including cDCs use MyD88-independent, poorly defined recognition pathways (Lund et al., 2003; Hochrein et al., 2004; Krug et al., 2004; Samuelsson et al., 2008; Siegemund et al., 2009; Wilkins and Gale, 2010).
In this study, we found that poly IC induced systemic production of IFN-λ, and we investigated the molecular and cellular events required for this production. Mice that lacked CD8α+ cDCs were unable to produce IFN-λ to poly IC in vivo. This production depended on TLR3, IRF3, IRF7, and IFN-IR. We identified mouse CD8α+ and eCD8α cDCs as the major producers of IFN-λ in response to poly IC in vitro. Similar to mice, among human cDCs, human eCD8α cDCs isolated via expression of BDCA3 were found to produce large quantities of IFN-λs upon stimulation with poly IC. Thus, IFN-λ production to poly IC was identified as a hallmark function of eCD8α cDCs in mice and man.
RESULTS
Poly IC induces systemic IFN-λ production in vivo, which depends on TLR3 and IFN-IR
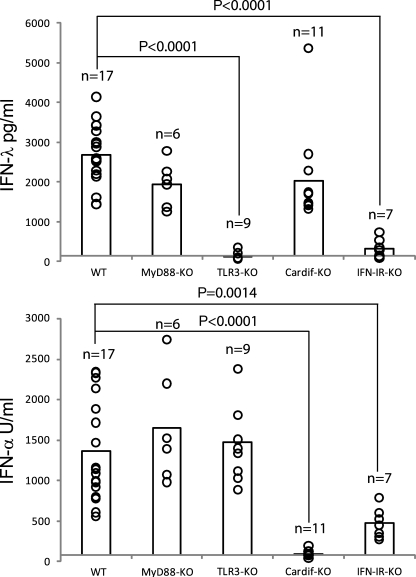
Poly IC is sensed by redundant pathways, and roles for RLHs as well as TLR3 have been described previously (Alexopoulou et al., 2001; Gitlin et al., 2006). Injection of poly IC is known to induce large systemic levels of IFN-α. In this study, we found that the injection of poly IC in WT mice also induced large quantities of IFN-λ (Fig. 1). To determine the PRRs involved in this IFN-λ production, we injected poly IC into mice deficient for various PRRs or their adaptor molecules, specifically TLR3, MyD88, or Cardif (also known as MAVS, IPS-1, or VISA), and measured IFN-λ as well as IFN-α in the corresponding sera (Fig. 1). Large amounts of IFN-λ and IFN-α were induced in WT and MyD88-KO mice (Fig. 1), demonstrating that MyD88-dependent TLRs were not involved. This also suggests that pDCs, which largely depend on MyD88 for IFN production, unlikely contributed to the production of both cytokines under those conditions. However, deficiency of TLR3 resulted in abrogated IFN-λ production with no effect on the production of IFN-α (Fig. 1). In contrast, Cardif deficiency revealed no effects on IFN-λ production but, consistent with previous studies, using MDA5-deficient mice, complete abrogation of serum IFN-α (Fig. 1; Gitlin et al., 2006; Longhi et al., 2009). Thus, whereas poly IC induced large systemic levels of both IFN-λ and IFN-α in WT mice, the dependence on TLR3 or Cardif for their production seems to be mutually exclusive.
Figure 1.
Poly IC–induced IFN-λ production in vivo depends on TLR3 and IFN-IR but not on MyD88 or Cardif. Mice with the indicated genotype were injected i.v. with 100 µg poly IC. After 3–4 h, sera were analyzed for IFN-λ and IFN-α. Circles indicate the results of individual mice, and their total number (n) is indicated in the graph. The bars represent the mean of all mice per genotype. At least three independent experiments have been performed.
It has been described that optimal IFN-α production to poly IC in vivo requires expression of a functional IFN-IR (Barchet et al., 2002). A role for IFN-IR has also been proposed for the production of IFN-λ in response to viruses (Ank et al., 2008). In this study, we found that systemic production of IFN-λ and IFN-α in response to poly IC was largely dependent on the presence of the IFN-IR (Fig. 1).
IFN-λ production to poly IC in vivo depends on IRF3 and IRF7
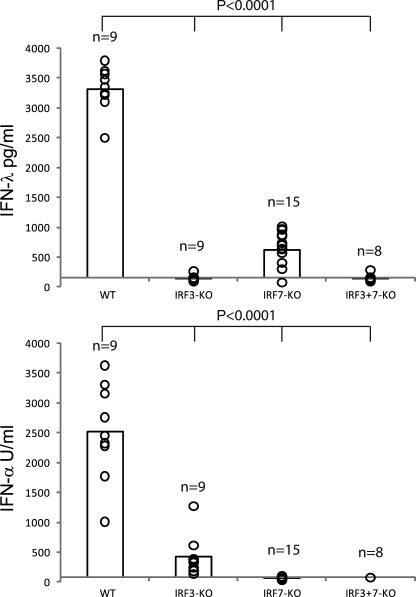
To get an insight into the transcription factors involved in the IFN-λ production to poly IC in vivo, we challenged mice deficient for either IRF3 or IRF7 or both IRF3+7 (Fig. 2). Mice that lacked IRF3 had a completely abrogated IFN-λ production and an 80% reduction in the production of IFN-α. Mice lacking IRF7 showed the inverse pattern with a complete lack of IFN-α production and an 80% reduction in the IFN-λ production. Mice deficient for both transcription factors (IRF3+7) produced neither IFN-λ nor IFN-α. Thus, the full production of poly IC–induced IFN-λ and IFN-α in vivo depended on the presence of IRF3 and IRF7 (Fig. 2).
Figure 2.
IFN-λ production to poly IC in vivo depends on IRF3 and IRF7. Mice with the indicated genotype were injected i.v. with 100 µg poly IC. After 3–4 h, sera were analyzed for IFN-λ and IFN-α. Circles indicate the result of individual mice, and their total number (n) is indicated in the graph. The bars represent the mean of all mice per genotype. Three independent experiments have been performed.
IFN-λ production to poly IC in vivo depends on hematopoietic cells, FL, and IRF8 but not on lymphocytes
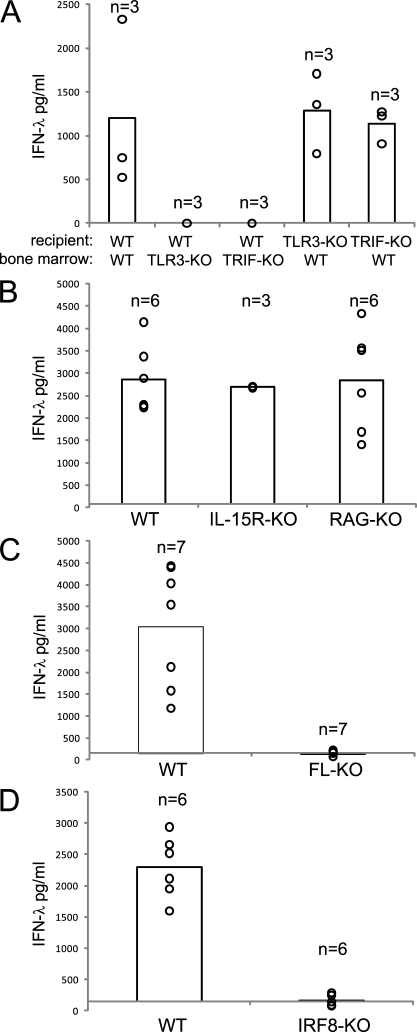
Systemic IFN-α in response to poly IC originates from nonhematopoietic cells via MDA5 recognition (Gitlin et al., 2006; Longhi et al., 2009). To test for the origin of systemic IFN-λ, we made BM chimeras of WT and TLR3 or TRIF mice. The data show that TLR3-KO and TRIF-KO mice, which received a WT BM, gained systemic IFN-λ–producing capacity upon poly IC injection (Fig. 3 A). In contrast, WT mice reconstituted with TLR3-KO or TRIF-KO BM were unable to produce systemic IFN-λ to poly IC (Fig. 3 A). Thus, hematopoietic cells are the origin of this IFN-λ.
Figure 3.
IFN-λ production to poly IC in vivo depends on hematopoietic cells, FL, and IRF8. Mice with the indicated genotype were injected i.v. with 100 µg poly IC, and after 3–4 h, sera were analyzed for IFN-λ. (A) BM reconstituted mice as indicated. (B) WT, IL-15R–KO, and RAG1-KO. (C) WT and FL-KO. (D) WT and IRF8-KO. Circles indicate the result of individual mice, and their total number (n) is indicated in the graph. The bars represent the mean of all mice per genotype. One (BM chimeras in A and IL-15R–KO in B), two (WT and RAG-KO in B and WT and IRF8-KO in D), and three (WT and FL-KO in C) independent experiments have been performed.
To rule out lymphocytes as a source of the systemic IFN-λ production in vivo, we tested IL-15R–KO and RAG-KO mice, which lack NK cells and B cells and T cells, respectively, but found that both strains were able to produce normal WT levels of IFN-λ (Fig. 3 B) and IFN-α (Fig. S4 A). To identify the cellular source of the systemic IFN-λ, we tested FL-KO mice, which are known to have greatly reduced numbers of DCs (McKenna et al., 2000). In contrast to WT mice, poly IC–injected FL-KO mice were unable to produce IFN-λ (Fig. 3 C), whereas IFN-α was produced (Fig. S4 B), suggesting a role for DCs in the production of IFN-λ to poly IC in vivo. Application of recombinant FL into FL-KO mice not only restored but even increased their IFN-λ–producing capacity above the WT level (unpublished data). Along those lines, FL-treated WT mice, which display elevated DC numbers, skewed toward increased CD8α+ cDCs and eCD8α cDCs (O’Keeffe et al., 2002; Bedoui et al., 2009), had a greatly increased systemic IFN-λ response to poly IC challenge (unpublished data). The FL dependence suggests that IFN-λ production to poly IC in vivo is largely mediated by DCs.
To clarify which DC subset would be the source of IFN-λ production to poly IC in vivo, we used mice lacking the transcription factor IRF8 and as a consequence CD8α+ and eCD8α cDCs (Schiavoni et al., 2002; Aliberti et al., 2003; Edelson et al., 2010). In contrast to WT mice, IRF8-KO mice were unable to produce IFN-λ to poly IC challenge in vivo, strongly suggesting that CD8α+ and eCD8α cDCs were the source of IFN-λ (Fig. 3 D).
Poly IC injection in vivo induces IFN-λ production in spleen cells with the phenotype of CD8α+ cDCs
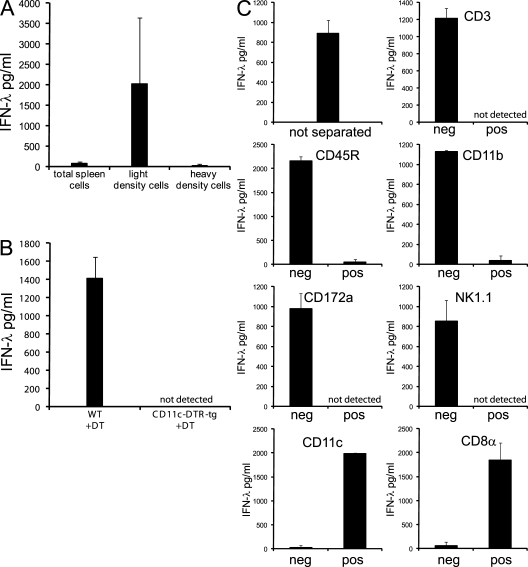
Because gene deletions of the aforementioned mice may affect several functions, we decided to confirm the conclusion obtained so far. For this, we injected poly IC i.v. into WT mice, dissected the spleen, and cultured the splenocytes in vitro. IFN-λ was detected in the supernatant of cultured total spleen cells (Fig. 4, A–C). Separation of the splenocytes with a density centrifugation, a step used to enrich DCs in the light density fraction, revealed that most IFN-λ was associated with these light density cells. (Fig. 4 A).
Figure 4.
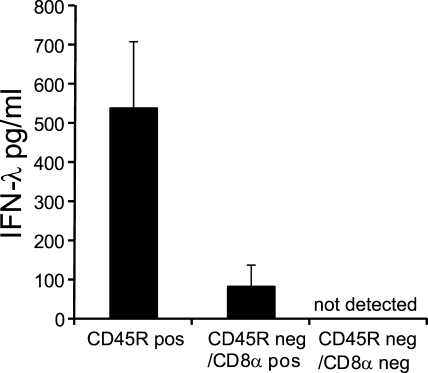
The IFN-λ production to poly IC injection in vivo separates with CD45R−/CD11c+/CD8α+ splenocytes. 1.5–2 h after i.v. injection of poly IC, spleens were harvested and processed. Cell-free supernatants were analyzed for IFN-λ after in vitro culture for 18 h. (A) 5 × 106 cells/ml of total spleen cells or cells separated by density centrifugation into light density cells or heavy density cells. (B) 25 × 106 cells/ml of total spleen cells of WT or CD11c-DTR-tg mice treated 2 d before with DT. (C) Total spleen cells before separation or after magnetic bead separation into the denoted populations. The initial cell number of splenocytes added onto the column was 20 × 106. Without further counting, each fraction was distributed into 2 wells with 200 µl of medium/well. Bars represent the mean ± SD of two independent experiments (A and C) or one experiment (B) using two mice per experiment.
As an alternative approach to investigate the role of DCs in the IFN-λ production to poly IC in vivo, we used mice transgenic for the diphtheria toxin (DT) receptor (DTR) under the control of the CD11c promoter (CD11c-DTR-tg; Jung et al., 2002). CD11c is highly expressed in cDCs and to a minor extent in pDCs, some lymphocytes, and certain macrophages of the spleen (Jung et al., 2002; Probst et al., 2005). Our initial unpublished experiments confirmed that splenocytes of DT-untreated CD11c-DTR-tg mice produced IFN-λ upon poly IC injection in vivo. Next, we treated WT and CD11c-DTR-tg mice with DT to ablate DCs in vivo. After injection of poly IC in vivo, spleen cells of the DT-treated WT but not DT-treated CD11c-DTR-tg mice produced IFN-λ (Fig. 4 B), suggesting a role of CD11c+ cells, probably DCs, as the main source of IFN-λ.
The previous two separation experiments indicated that among splenocytes of poly IC–injected mice, light density DCs that highly expressed CD11c were the major source of IFN-λ. To rule out other cell types and get an insight into the subset of DCs involved, poly IC was injected in WT mice, and 1.5–2 h later, total spleen cells were separated with a set of antibodies and magnetic beads into positive and negative fractions. The IFN-λ production separated with cells negative for CD3 (T cells), CD45R (B cells and pDCs), CD11b (macrophages and CD8− cDCs), CD172a (macrophages, pDCs, and CD8− cDCs), and NK1.1 (NK cells) but positive for CD11c (DCs) and CD8α (CD8α cDCs and CTLs). Thus, upon poly IC injection in vivo, the IFN-λ production in splenocytes segregated with cells of light density and the phenotype CD3−, CD45R−, NK1.1−, CD11b−, CD172a−, CD11c+, and CD8α+. This phenotype exactly describes CD8α+ cDCs and excludes other cell types of the spleen such as lymphocytes or macrophages as major producers of IFN-λ to poly IC injection in vivo.
CD8α+ cDCs are the major producers of IFN-λ in response to poly IC in vitro
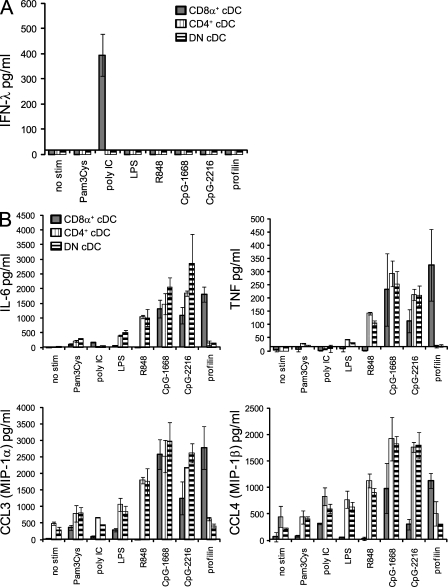
Stimulation of fractionated spleen cells with different TLR ligands in vitro revealed that the major lymphocyte fractions consisting of T and B lymphocytes were unable to produce IFN-λ, and its production was confined to enriched preparations of DCs (unpublished data). Therefore, we used a DC isolation protocol, which included density enrichment, depletion of T cells, B cells, NK cells, and granulocytes, and multiparameter fluorescence-activated cell sorting. Those highly purified DC subsets were stimulated with a panel of TLR ligands in vitro and IFN-λ, and other cytokines and chemokines were determined in the supernatants. pDCs were the major source of IFN-λ in response to the A-type ODN CpG-2216 (Fig. S1), as previously suggested by messenger RNA analysis of human pDCs (Coccia et al., 2004). However, in response to poly IC stimulation, the CD8α+ cDCs were the major producers of IFN-λ, with CD8− cDCs, either expressing CD4 (CD4+ cDCs) or not (DN cDCs), and pDCs being largely unable to participate in IFN-λ production (Fig. 5 A and Fig. S1 A). The IFN-λ production was selective for poly IC stimulation and for CD8α+ cDCs even though many different TLR ligands induced robust activation in CD8α+ and CD8α− cDCs, as seen by the induction of the other cytokines (Fig. 5 B). As described previously, some TLR ligands were selective for certain DC subsets, with CD8α+ cDCs being unresponsive to the TLR7 stimulation via R848, but in contrast being the sole responder to the TLR11 ligand profilin (Fig. 5 B and Fig. S1 B; Edwards et al., 2003; Yarovinsky et al., 2005).
Figure 5.
Splenic CD8α+ cDCs are the major producers of IFN-λ in response to poly IC in vitro. Highly purified 5 × 105 cells/ml of splenic cDC subsets were stimulated in the presence of IL-3 and GM-CSF with the stimuli as indicated. (A and B) After 18 h, supernatants were analyzed for IFN-λ (A) or IL-6, TNF, CCL3, and CCL4 (B). Bars represent the mean ± SD of three (A) or two (B) independent experiments using the pool of at least eight mice per experiment.
IFN-λ and IL-12p70 production by CD8α+ cDCs depends on the type of stimulus and the cytokine conditions
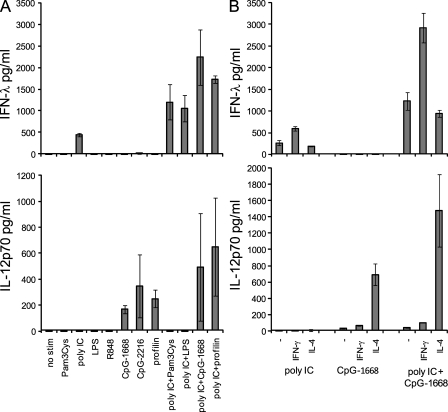
CD8α+ cDCs are well known for their exceptional capacity for IL-12p70 production. Because we found that the CD8α+ cDCs were also able to produce large amounts of IFN-λ, we next elucidated the conditions that would govern IFN-λ compared with IL-12p70 production. By using a panel of different TLR stimuli, we found that TLR ligands known for their high IL-12p70 induction such as CpG-ODN or profilin of toxoplasma (Hochrein et al., 2000; Yarovinsky et al., 2005) induced large amounts of IL-12p70 as expected, but surprisingly the CD8α+ cDCs did not produce any IFN-λ under these conditions (Fig. 6 A). In contrast, poly IC stimulation induced IFN-λ but not IL-12p70 production by CD8α+ cDCs (Fig. 6 A). Combinations of poly IC together with Pam3Cys, LPS, CpG-ODN, or profilin, ligands for TLR2, TLR4, TLR9, or TLR11, respectively, synergistically increased IFN-λ production (Fig. 6 A). The data demonstrate a synergistic increase of poly IC–induced IFN-λ with MyD88-dependent stimuli and confirm described synergistic effects on the production of IL-12p70 by CD8α+ cDCs (Fig. 6 A; Napolitani et al., 2005).
Figure 6.
The production of IFN-λ or IL-12p70 by CD8α + cDCs depends on the stimuli and the cytokine conditions. 5 × 105 cells/ml of sorted splenic CD8α+ cDCs were stimulated, and supernatants were analyzed after 18 h for IFN-λ and IL-12p70. (A) Stimulation in the presence of IL-3 and GM-CSF with the stimuli as indicated. (B) Stimuli and cytokines as indicated. Bars represent the mean ± SD of two independent experiments using a pool of at least eight mice per experiment.
Previously, we and others have shown that the cytokine milieu during stimulation is highly influential for IL-12p70 production in murine and human DCs, with IL-4 being a major enhancer for bioactive IL-12 production (Hochrein et al., 2000; Kalinski et al., 2000). Using poly IC or CpG-1668 or a combination of poly IC and CpG-1668 as stimuli, we analyzed the effect of the addition of either IFN-γ or IL-4 on the production of IFN-λ and IL-12p70. IFN-γ enhanced the production of IFN-λ to poly IC or the combination of poly IC and CpG-1668, whereas still no IFN-λ was induced to CpG-1668 only (Fig. 6 B). As described previously, IFN-γ had little effect on the IL-12p70 production, whereas IL-4 increased IL-12p70 but not IFN-λ production (Fig. 6 B; Hochrein et al., 2000).
In vivo and in vitro FL-induced equivalents of CD8α+ cDCs produce IFN-λ to poly IC
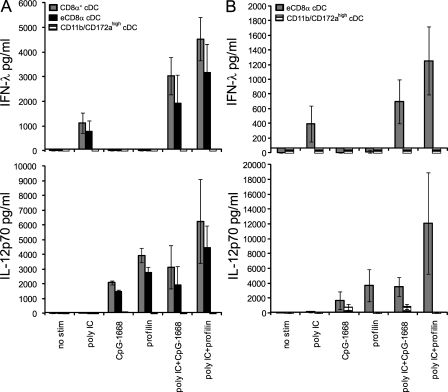
Spleen cDCs contain a very small fraction of DCs, which do not express CD8α but otherwise resemble the CD8α+ cDCs in phenotype and their cross-presentation ability and are thus regarded as eCD8α cDCs (Bedoui et al., 2009). These cells are expanded by repeated FL injections. To test whether ex vivo isolated eCD8α cDCs share other functional similarities with CD8α+ cDCs, we expanded DCs with FL in vivo and sorted CD8α+ cDCs, eCD8α cDCs, and the remaining cDCs, which highly express CD11b and CD172a. Both, the CD8α+ and eCD8α cDCs but not the CD11b/CD172a-expressing cDCs produced large amounts of IFN-λ to poly IC and high IL-12p70 to CpG-1668 or profilin, indicating that eCD8α cDCs share the high IFN-λ– and IL-12p70–producing capacity of CD8α+ cDCs (Fig. 7 A).
Figure 7.
In vivo and in vitro FL-generated CD8α+ cDCs and eCD8α cDCs are major producers of IFN-λ and IL-12p70. (A and B) 5 × 105 cells/ml of highly purified splenic DCs from FL-treated mice (A) or FLDCs (B) were stimulated in the presence of IL-3 + GM-CSF + IL-4 + IFN-γ with the stimuli as indicated. After 18 h, supernatants were analyzed for IFN-λ and IL-12p70. Bars represent the mean ± SD of two independent experiments each using a pool of at least two mice per experiment.
The culture of BM cells with FL in vitro induces the development of DC subsets, including eCD8α cDCs, with phenotypic, developmental, and functional similarities with CD8α+ cDCs (Hildner et al., 2008; Naik et al., 2010). Comparing eCD8α cDCs and CD11b/CD172a-expressing cDCs revealed that similar to ex vivo isolated cDC subsets, the mutually exclusive stimulus-dependent production of IFN-λ or IL-12p70 was mostly confined to the eCD8α subset (Fig. 7 B). Thus, eCD8α cDCs either ex vivo isolated or in vitro generated share with CD8α+ cDCs the ability for high IFN-λ as well as IL-12p70 production.
In vivo, we had found that poly IC–induced IFN-λ was independent of the presence of the adaptor molecules MyD88 and Cardif but depended on TLR3 and IFN-IR (Fig. 1). Because the in vitro generated eCD8α cDCs showed such a similar production of IFN-λ as their ex vivo isolated counterparts, we generated and sorted eCD8α cDCs from the FL cultures of WT, MyD88-KO, Cardif-KO, TLR3-KO, and IFN-IR–KO mice. The production of IFN-λ by FL-eCD8α cDCs upon stimulation with poly IC was dependent on TLR3 and IFN-IR but was independent of MyD88 and Cardif (Fig. S3, A–D). Thus, the systemic production of IFN-λ in vivo and the production in vitro by directly stimulated eCD8α cDCs were governed by the same set of receptors, especially TLR3, which is highly expressed and known to be used for poly IC detection by CD8α+ cDCs (Schulz et al., 2005). These data strongly suggest that CD8α+ and eCD8α cDCs are the source of poly IC–induced IFN-λ in vivo.
Production of IFN-λ and IL-12p70 upon selective stimulation indicates the presence of CD8α+ or eCD8α cDCs in the liver
Based on the data accumulated thus far, we hypothesized that the production of IFN-λ and IL-12p70 exclusively induced by certain stimuli in CD8α+ and eCD8α cDCs might be used to identify the presence of these DCs in complex mixtures of cells. For the highest production of both factors in vitro, we used a combination of cytokines (IL-3 + GM-CSF + IL-4 + IFN-γ), which promotes both IFN-λ and IL-12p70 production. As stimuli, we used poly IC or profilin or the combination of poly IC + profilin, which are stimuli highly selective for CD8α+ and eCD8α cDCs (Fig. 5 and Fig. S1). First, we wanted to test the selectivity of these stimuli for CD8α+ cDCs within splenic cDC subsets. Indeed, we confirmed the stimulus-dependent production of IFN-λ to poly IC, IL-12p70 to profilin, or the combined production of IFN-λ and IL-12p70 to the combination of poly IC + profilin exclusively by CD8α+ cDCs (Fig. S2 A). Stimulations of nonparenchymal liver cells under these conditions revealed the same pattern of IFN-λ and IL-12p70 production (Fig. S2 B), indicating the presence of CD8α+ or eCD8α cDCs as previously shown (Sumpter et al., 2007). Liver cells from FL-KO mice, known to have drastically reduced numbers of DCs, were largely unable to produce IFN-λ or IL-12p70 under these conditions (Fig. S2 B).
CD8α+ cDCs, eCD8α cDCs, and pDCs produce IFN-λ to HSV-1 and parapoxvirus
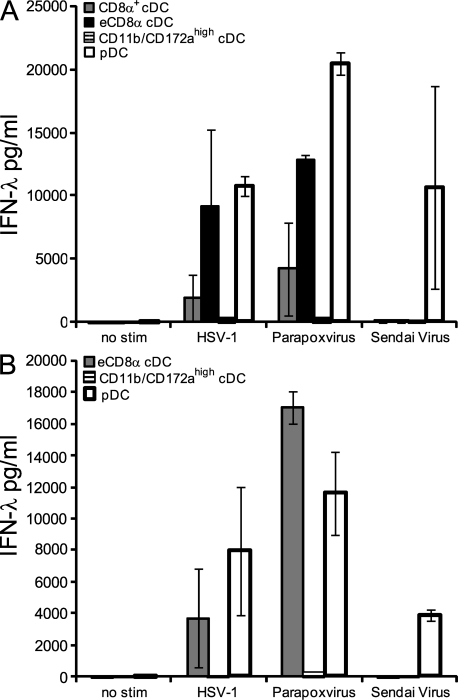
To extend our analysis of IFN-λ production in response to the dsRNA viral mimic poly IC to bona fide viruses, we stimulated ex vivo isolated splenic DC subsets from FL-expanded mice and DC subsets of in vitro generated FLDCs with HSV-1, parapoxvirus, and Sendai virus and analyzed the supernatants for IFN-λ. Among the spleen DC subsets, CD8α+ cDCs, eCD8α cDCs, and pDCs produced large amounts of IFN-λ in response to HSV-1 and parapoxvirus (Fig. 8 A). The CD8− cDCs sorted as CD11b/CD172ahigh cDCs produced only very limited but, in contrast to the stimulation with poly IC, detectable amounts (between 100 and 400 pg/ml) of IFN-λ upon stimulation with HSV-1 or parapoxvirus. Only pDCs and none of the cDC subsets produced IFN-λ in response to Sendai virus (Fig. 8 A). FLDC subsets generated in vitro displayed a very similar pattern with IFN-λ production to HSV-1 and parapoxvirus mostly associated with eCD8α cDCs and pDCs with a very limited contribution by CD11b/CD172ahigh cDCs (Fig. 8 B). Again, all IFN-λ to Sendai virus was selectively produced by pDCs but not from the cDC subsets (Fig. 8 B). Thus, like pDCs, CD8α+ and eCD8α cDCs are able to respond in vitro to some viruses with the production of large amounts of IFN-λ.
Figure 8.
In vivo and in vitro FL-generated CD8α+ cDCs, eCD8α cDCs, and pDCs are major producers of IFN-λ to HSV-1 and parapoxvirus. (A and B) 5 × 105 cells/ml of highly purified splenic DCs from FL-treated mice (A) or FLDCs (B) were stimulated in the presence of IL-3 + GM-CSF + IL-4 + IFN-γ with the stimuli as indicated. After 18 h, supernatants were analyzed for IFN-λ. Bars represent the mean ± SD of two independent experiments each using a pool of at least two mice per experiment.
HSV-1 injection in vivo indicates that among splenocytes, pDCs and CD8α+ cDCs produce IFN-λ
Similar to our approach to identify the IFN-λ producers among spleen cells in response to poly IC injection (Fig. 4 C), we performed experiments injecting HSV-1. In contrast to the results with poly IC, the majority of the HSV-1–induced IFN-λ was within the CD45R-positive fraction (Fig. 9). This suggested that probably pDCs, which beside B cells express CD45R, are the major producers. We separated the CD45R-negative fraction into a CD8α-positive and -negative fraction of cells. All remaining IFN-λ production within the CD45R-negative fraction segregated with the CD8α-positive cells (Fig. 9). These data suggest that the CD8α+ cDCs, which are CD45R−/CD8α+, participate in the IFN-λ production upon HSV-1 inoculation in vivo.
Figure 9.
The IFN-λ production to HSV-1 injection in vivo separates with CD45R+ and CD45R−/CD8α+ splenocytes. Spleen cells 1.5 h after in vivo injection with DISC–HSV-1 were separated with anti-CD45R and magnetic beads into positive and negative fractions. The CD45R-negative fraction was further separated into cells positive or negative for CD8α. Separated cells were cultured in vitro for the next 18 h, and cell-free supernatants were analyzed for IFN-λ. Bars represent the mean ± SD of two independent experiments using one mouse per experiment.
Human BDCA3+ DCs are the major producers of IFN-λs upon poly IC stimulation
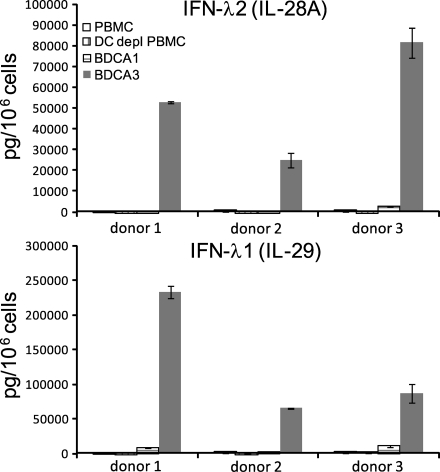
We found that the IFN-λ production in response to poly IC in mice is a CD8α+ cDC subset–specific feature, and therefore, we wanted to determine whether this function correlated with any human DC subset. Given that human BDCA3+ DCs are attributed as functional correlates of CD8α+ cDCs, we stimulated PBMCs, PBMCs depleted of cDCs, and highly enriched fractions of BDCA1+ and BDCA3+ cDCs and measured IFN-λ1 and IFN-λ2 in the supernatants. In PBMCs (Fig. 10) and fractions of DC-enriched PBMCs (not depicted), we found that poly IC induced IFN-λ1 (IL-29) and IFN-λ2 (IL-28A). Separation of cDC subsets using the markers BDCA1 or BDCA3 revealed that the BDCA3+ cells were the major producers of IFN-λ1 as well as IFN-λ2 (Fig. 10). Thus, IFN-λ production upon poly IC stimulation is another functional attribute that the human BDCA3+ cDCs share with murine CD8α+ and eCD8α cDCs.
Figure 10.
Human BDCA3+ cDCs are major producers of IFN-λ upon poly IC stimulation. PBMCs, PBMCs depleted of BDCA1 and BDCA3, or cells positively selected either for BDCA1 or BDCA3 were stimulated in the presence of IL-3, GM-CSF, and IFN-γ with 100 µg/ml poly IC + 10 µg/ml Pam3Cys + 10 µg/ml LPS (donor 1) or with 100 µg/ml poly IC for 18–24 h (donors 2 and 3). Supernatants were analyzed for IFN-λ1 and IFN-λ2. The experiments are shown for the individual donors, and data represent mean ± SD of duplicate samples.
DISCUSSION
In this study, we have shown that poly IC, an outstanding adjuvant, induces IFN-λ production by human BDCA3+ (CD141+) and mouse CD8α+ cDCs, subsets which are also specialized in IL-12p70 production and cross-presentation. In response to poly IC, IFN-λ was not coexpressed with IL-12p70 unless MyD88-dependent stimuli were also sensed by these cells. Because poly IC is shown in this study to directly target CD8α+ cDCs for IFN-λ induction, it is tempting to speculate that this cytokine constitutes an important component of the poly IC–induced adjuvant response. Understanding the individual adjuvant components of poly IC may permit the design of vaccines with enhanced antiviral responses but minimal side effects. Moreover, poly IC is a mimic of viral dsRNA, and the IFN-λ responses to this adjuvant are potentially mirrored in the response to numerous DNA and RNA viruses that produce dsRNA intermediates or potentially other pathogen-associated patterns. The recent studies linking IFN-λ to hepatitis C viral clearance highlight the potential importance of this cytokine in viral disease (Ge et al., 2009; Suppiah et al., 2009; Tanaka et al., 2009; Thomas et al., 2009). Moreover, the role of IFN-λ extends beyond viral disease. It is reported that TH1-inducing properties (Jordan et al., 2007) could have potential use as an adjuvant in airway diseases in which its production is possibly deficient (Contoli et al., 2006). It has also been shown that IFN-λ induces apoptosis in epithelial cell tumors (Brand et al., 2005), raising the possibility that it may function as a tumor suppressor.
Poly IC is a well-known agent that induces systemic levels of IFN-α in experimental animals. Until recently, the molecular and cellular bases for this IFN-α were unknown. We and others have described that CD8α+ cDCs but not the CD8− cDCs were producers of IFN-α in response to poly IC stimulation in vitro (Hochrein et al., 2001; Longhi et al., 2009). Recently, it became clear that DCs are not responsible for the systemic production of IFN-α. Poly IC–driven IFN-α induction depends on the RLH MDA5 and originates from cells of nonhematopoietic origin (Gitlin et al., 2006; Longhi et al., 2009). These data were further corroborated in TLR3-KO and Batf3-KO mice, which lack the dsRNA recognition receptor predominantly used by CD8α+ cDCs or CD8α+ cDCs per se, respectively. Induction of poly IC–induced systemic IFN-α was not impaired in these mice (Gitlin et al., 2006; Longhi et al., 2009; McCartney et al., 2009; Edelson et al., 2010).
In this study, we have provided evidence that poly IC induces early systemic IFN-λ production. To our knowledge, this is the first report of directly measured systemic IFN-λ in response to poly IC in vivo. Indirect evidence of systemic poly IC–induced IFN-λ was provided by Ank et al. (2008), who nicely demonstrated that antiviral protection induced by poly IC partially depended on the presence of the IFN-λ receptor (IL-28R).
In vitro, the induction of IFN-λ transcripts was reported by the first papers that described the discovery of the IFN-λs (Kotenko et al., 2003; Sheppard et al., 2003). Many studies have since extended those findings in several cells types in vitro, including GM-CSF–derived murine and human monocyte-derived DCs (GM-DCs), macrophages, and pDCs (for review see Ank and Paludan, 2009). One study demonstrated induction of IFN-λ upon stimulation with Sendai virus, HSV-2, poly IC, and B-type ODN (CpG-1826) in GM-DCs (Ank et al., 2008) and another with LPS or CD40L (Wolk et al., 2008). In contrast, we could only find limited amounts of IFN-λ in ex vivo isolated or in vitro with FL-generated CD11b/CD172ahigh cDCs induced by the DNA viruses HSV-1 and parapoxvirus but none to Sendai virus (Fig. 8), to poly IC, or the other TLR ligands (Figs. 5 and 7), even though the viruses induced some IFN-α in those CD11bhigh cDCs (Hochrein et al., 2004; Siegemund et al., 2009; Luber et al., 2010). We have also analyzed GM-DCs, but again we could only find limited amounts of IFN-λ to HSV and parapoxvirus but none to Sendai virus or poly IC or a panel of TLR ligands including B-type ODN (CpG-1668; unpublished data). However, comparing the protocols for the generation of GM-DCs, we realized substantial differences in the duration of culture and thus the maturation state of these DCs. It will be interesting to elucidate the basis of the differences in the IFN-λ production capacity of non-CD8α+ cDCs. It is important to note that our in vivo analyses and ex vivo isolated DC subset analyses and organ stimulations were performed using healthy pathogen-free mice. Under these noninflammatory steady-state conditions, all of our in vivo and in vitro data point to CD8α+ and eCD8α cDCs as the sole and nonredundant producers of early IFN-λ upon poly IC exposure. It has been shown previously that these steady-state conditions have no substantial numbers of inflammatory DCs (e.g., monocyte-derived DCs or TIP DCs; for review see Shortman and Naik, 2007). This situation may change dramatically upon infection or inflammation, circumstances which we have not analyzed. It is known that inflammatory cytokines such as IFN-Is, IFN-γ, GM-CSF, or TNF dramatically alter the cytokine production capacity of many immune and nonimmune cells, and some studies indicate that this might also be the case for the production of IFN-λs (Sirén et al., 2005; Ank et al., 2006; Megjugorac et al., 2010).
We found that systemic IFN-λ originated from hematopoietic cells and among those exclusively from CD8α+ and eCD8α cDCs, and its production was fully dependent on TLR3 but independent of the adaptor molecules for other TLRs or RLHs, MyD88 or Cardif, respectively. In contrast, and confirming previous data (Gitlin et al., 2006; Longhi et al., 2009), we found that the production of systemic IFN-α was independent of TLR3 but fully dependent on Cardif. CD8α+ cDCs highly express and recognize poly IC via TLR3 (Edwards et al., 2003; Schulz et al., 2005), whereas they hardly express the RLHs (Luber et al., 2010). It is interesting to see that the same danger signal (dsRNA) induces early and high systemic levels of two different IFN families. This danger signal is not only sensed by two different families of PRRs (TLRs and RLHs) but also by completely different cellular sources (CD8α+ cDCs and nonhematopoietic cells).
Mice deficient in FL have drastically reduced but not fully absent numbers of DCs, including CD8α+ cDCs. Based on the reduced numbers of DCs, we expected to detect residual IFN-λ production upon poly IC stimulation. However, the production of IFN-λ by FL-KO mice was nearly completely abolished in vivo (Fig. 3 C) and in liver cells in vitro (Fig. S2 B). Previously, we found that pDCs isolated from FL-KO mice were inhibited in their production of IFN-α upon TLR9 stimulation (unpublished data). This suggested that DCs from FL-deficient mice might also be functionally impaired or alternatively that these non-FL–dependent DCs are a subset of the normal steady-state DC populations and do not share the same functional attributes with FL-dependent DCs. In line with our observations, it was found recently that mice with a mutation in the receptor for FL (flt3) demonstrated a severely reduced production of cytokines upon TLR stimulation of their DCs, including CD8α+ cDCs (Eidenschenk et al., 2010).
Mice deficient for IRF8 have defects in the development of pDCs and CD8α+ and eCD8α cDCs (Aliberti et al., 2003; Edelson et al., 2010). In line with the absence of CD8α+ cDCs, we found that IRF8-KO mice were unable to produce systemic IFN-λ to poly IC. Like FL, IRF8 seems to play an additional functional role, as demonstrated by the impaired IFN-α production by pDCs (Tailor et al., 2007), which was not further examined in our study. However, it is interesting to note that both DC subsets able to produce large amounts of IFN-λ, pDCs and CD8α+ cDCs, not only depend on IRF8 for their development but also express high levels of IRF8 when isolated as end-stage cells from spleen (Luber et al., 2010). Whether the highly expressed IRF8 of these two professional IFN-λ–producing DC subsets is somehow linked to the IFN-λ producing capacity grants further examination.
In this study, we describe that IRF3 and IRF7 play important roles in the poly IC–induced IFN-λ and IFN-α production in vivo (Fig. 2). A role for IRF3 has been previously described for the induction of IFN-I downstream of both PRRs involved, the RLHs as well as for TLR3 (Tamura et al., 2008). Interestingly, in the absence of IRF3, no IFN-λ but some remaining IFN-α was produced, whereas in the absence of IRF7, we found a remaining IFN-λ but completely abrogated IFN-α production. An essential role for IRF7 has been demonstrated previously for MyD88-dependent IFN-α production by pDCs, and a participation of IRF7 in TRIF-dependent IFN-I production by DCs has been proposed (Honda et al., 2005; Tamura et al., 2008). Our in vivo findings of a prominent role for both IRF3 and IRF7 for the production of IFN-λ in response to poly IC are in line with previous promoter-based data (Osterlund et al., 2007). We have performed some preliminary experiments with eCD8α cDCs sorted from FLDCs, which confirmed the complete dependence on IRF3 for the production of IFN-λ in response to poly IC (unpublished data). For stimulations of eCD8α cDCs from IRF7-KO with poly IC, the role of IRF7 seemed to be less prominent, which makes it possible that the role of IRF7 in vivo is indirect, for example via the complete absence of IFN-α (Fig. 2), but this has to be confirmed with more detailed experiments. We have shown that the IFN-λ production in IFN-IR–deficient mice is not only reduced upon poly IC in vivo (Fig. 1) but also by eCD8α cDCs stimulated with poly IC in vitro (Fig. S3 D), suggesting that the lack of IFN-IR on CD8α+ cDCs is directly responsible for this effect. The expression of IFN-IR by DCs and especially by CD8α+ cDCs was previously shown to be important for their function in antigen presentation (Longhi et al., 2009; Cucak et al., 2009), and a positive feedback mechanism by IFN-I on the production of IFN-λ has previously been described (Ank et al., 2008). Because the IFN-λ production to poly IC in vivo and in vitro was not fully abrogated in the absence of the IFN-IR, this leaves a window for the activity of poly IC–induced IFN-λ in the absence of the IFN-IR. Indeed, Ank et al. (2008) have shown that the protective effect of poly IC to HSV infection was partially dependent on the IFN-λR and functional in the absence of IFN-IR. However, IRF8 is highly expressed by CD8α+ DCs (Luber et al., 2010), and both IRF3 and IRF7 play a role in IFN-λ production by CD8α+ DC in response to poly IC. It is possible that the role of IFN-IR is not only limited to an IFN-I feedback signaling at the time of stimulation but that it might play a role in the expression and/or regulation of essential IRFs such as IRF8, IRF3, or IRF7 in the DCs during their development. This is currently under investigation.
We found that upon poly IC injection in vivo, only splenic CD11c+/CD8α+ cells but not other splenic cell populations such as lymphocytes or macrophages could produce IFN-λ. Highly purified splenic DCs stimulated in vitro confirmed that the CD8α+ cDCs selectively produced IFN-λ to poly IC but not to any other sole TLR ligand, whereas pDCs produced IFN-λ to CpG-2216. Beside ex vivo isolated CD8α+ cDCs, equivalents of those cells (eCD8α cDCs) expanded by FL in vivo or in vitro shared the high IFN-λ– and IL-12p70–producing capacity, a feature previously not described for ex vivo isolated FL-expanded eCD8α cDCs. eCD8α cDCs have been identified based on their dependence on the transcription factors IRF8 or Batf3 to be present in several lymphoid and nonlymphoid organs (Hildner et al., 2008; Edelson et al., 2010). Our selective stimulations of liver cells indicated that eCD8α or CD8α+ cDCs were present, and cells with this phenotype have been previously identified in the liver (Sumpter et al., 2007). Thus, IFN-λ production seems to be a function of lymphoid and nonlymphoid organ eCD8α or CD8α+ cDCs.
Research on human cDC subsets has, until recently, relied on phenotypical analysis, which indicated that certain surface markers such as Clec9a or Necl2 selectively expressed by mouse CD8α+ cDCs were associated with a human blood DC subset that could be identified by expression of CD141 (BDCA3). We had previously shown that the IL-12p70 production of thymic human cDC subsets was preferentially localized to the CD11b− subset (Vandenabeele et al., 2001). Recently, the evidence for human BDCA3+ cDCs as the mouse eCD8α equivalents was substantiated in parallel by four groups who demonstrated that among human cDCs, BDCA3+ cDCs, like their mouse counterparts, highly express TLR3, produce IL-12p70, and possess superior cross-presenting activity (Bachem et al., 2010; Crozat et al., 2010; Jongbloed et al., 2010; Poulin et al., 2010; Villadangos and Shortman, 2010). We have now shown that the newly discovered function of mouse CD8α+ and eCD8α as the major producers of IFN-λ in response to poly IC is conserved throughout evolution to the human eCD8α (BDCA3+) cDCs.
We do not yet know all of the consequences of the dedication of a single cell type to produce IFN-λ to dsRNA, nor do we know the full consequences of IFN-λ in the network of defense and immunoregulation. However, the strong functional conservation of similar cell types between mice and man is an indication of importance, and it strengthens the likelihood that certain aspects of IFN-λ biology, including its regulation and novel therapeutic strategies based on experimental work performed in mice, will translate into humans.
MATERIALS AND METHODS
Mice.
Mice were bred and maintained either in the animal facilities at Bavarian Nordic GmbH, the Max Planck Institute of Biochemistry, the Max Planck Institute of Immunobiology, the Amrep Animal facility, the Walter and Eliza Hall Institute of Medical Research, the University of Melbourne, or at the University of Zurich according to institutional guidelines. Breeding stocks were originally obtained from MyD88-KO and TRIF-KO mice from S. Akira (Osaka University, Suita, Osaka, Japan; Adachi et al., 1998), Cardif-KO mice from J. Tschopp (University of Lausanne, Epalinges, Switzerland; Meylan et al., 2005), FL-KO mice (McKenna et al., 2000), transgenic mice that express the primate DTR under the control of the CD11c promoter from S. Jung (The Weizmann Institute of Science, Rehovot, Israel; Jung et al., 2002), TLR3-KO (Alexopoulou et al., 2001), IL-15R–KO (Lodolce et al., 1998) and RAG1-KO (Mombaerts et al., 1992) mice from The Jackson Laboratory, IRF3-KO and IRF7-KO mice from T. Taniguchi (University of Tokyo, Bunkyo-ku, Tokyo, Japan; Honda et al., 2005), IRF8-KO from I. Horak (Leibniz Institut für Molekulare Pharmakologie, Berlin, Germany; Holtschke et al., 1996), and C57BL/6 WT mice purchased from Harlan Winkelmann GmbH, Elevage Janvier, or the Walter and Eliza Hall Institute of Medical Research bred C57BL/6. Protocols for animal experimentation were approved by the government of upper Bavaria (Regierung von Oberbayern) or by the Walter and Eliza Hall Institute of Medical Research or University of Melbourne Animal Ethics Committees.
Cells and flow cytometric sorting.
DC subsets were isolated from pooled mouse spleens as described previously (Vremec et al., 2007). In brief, spleens were chopped, digested with collagenase (Worthington Biochemical) and DNase (Roche) at room temperature, and treated with EDTA. Low-density cells were enriched by density centrifugation; non-DC lineage cells were coated with mAbs (anti-CD3 [KT3-1.1], anti–Thy-1 [T24/31.7], anti–Gr-1 [1A8], anti-CD19 [ID3], anti-erythrocytes [TER119], and anti–NK cells [DX5]) and depleted using anti–rat Ig magnetic beads (QIAGEN). Dead cells were excluded by propidium iodide staining. cDC populations were sorted based on the expression of CD11c, CD45RA, CD4, CD8α, and CD172a, and pDCs were purified based on CD11c, CD45RA, and CD172a (all from BD) expression. Cell sorting was performed on a FACS Aria instrument (BD).
To generate FL-induced DCs in vivo, mice were treated i.p. with nine doses of FL (10 µg/day), and spleen DCs were enriched on day 10 as described in the previous paragraph. pDCs were sorted as CD11cint, CD45RAhigh, CD11bneg, and CD172aint cells; CD8α+ cDCs were sorted as CD11cpos, CD45RAneg, CD11bneg/low, CD172aneg, CD24high, and CD8apos cells; eCD8α cDCs were sorted as CD11cpos, CD45RAneg, CD11bneg/low, CD172aneg, CD24high, and CD8αneg cells; CD11b/CD172ahigh cDCs were sorted as CD11cpos, CD45RAneg, CD11bhigh, CD172ahigh, CD24neg/low, and CD8αneg cells.
FL BM culture–derived DCs (FLDCs) were prepared as described previously (Hochrein et al., 2004). pDCs and eCD8α and CD11b/CD172high cDC subsets were sorted based on the expression of CD11c, CD45R, CD11b, CD24, and CD172a (all from BD).
In vitro stimulation and cytokine detection.
Cells were stimulated in vitro with single TLR agonists or combinations thereof containing 10 µg/ml Pam3Cys (InvivoGen), 100 µg/ml poly IC (Axxora), 10 µg/ml LPS (Escherichia coli; Sigma-Aldrich or Axxora), 10 µg/ml R848 (Axxora), 1 µM CpG-1668 or CpG-2216 (TIB-Molbiol), and 1 µg/ml profilin of toxoplasma (Axxora). The recombinant cytokines mouse IL-3, mouse IL-4, rat IFN-γ (PeproTech), and mouse GM-CSF (Tebu-Bio; 10 ng/ml each) were added as indicated. The addition of IL-3 and GM-CSF was based on our previous observations that GM-CSF promoted the production of IL-12p70 (Hochrein et al., 2000) and that the combination of IL-3 and GM-CSF increased virus-induced IFN-α production in pDCs and cDCs (Hochrein et al., 2004). DISC–HSV-1 (HSV-1 disabled infectious single cycle) is a replication-deficient form of HSV-1, which lacks the gene for glycoprotein H and has to be propagated on transgenic feeder cells. In noncomplementing cells, it can perform only a single cycle of infection (McLean et al., 1994). The virus has been shown to be equally potent as a WT HSV-1 to induce IFN-α in pDCs and cDCs in vitro (Hochrein et al., 2004). Inactivated parapoxvirus ovis used in veterinary science (Pfizer Animal Health) was described to induce IFN-α/β in pDCs and cDCs (Siegemund et al., 2009). Sendai virus strain Cantell (Charles River) was described to induce IFN-α in pDCs and CD11bhigh but not CD8α+ cDCs (Luber et al., 2010). IFN-λ in supernatants was analyzed by ELISA, and IL-12p70, IL-6, TNF, CCL3, and CCL4 were determined by FlowCytomix bead assay (Bender MedSystems) according to the manufacturer’s protocol. Note that IFN-λ was analyzed by an IFN-λ3 (IL-28B) ELISA (R&D Systems), but we found that this ELISA is largely cross-reactive to IFN-λ2 (IL-28A) and thus were not able to differentiate between these two mouse IFN-λs.
Liver cell preparation.
To obtain liver cell suspensions for stimulation cultures, livers were harvested from mice, chopped into small pieces, and incubated with 1 ml collagenase D (1 mg/ml; Roche) at 37°C for 30 min. Single-cell suspensions were then prepared by mechanically disrupting the organs through a 70-µm filter. Nonparenchymal liver cells were further isolated by centrifugation in 35% Percoll (GE Healthcare) and then subjected to RBC lysis.
In vivo challenge and magnetic bead separation.
Mice were injected i.v. into the lateral tail vein with 100 µg poly IC (Axxora), and serum was collected 3–4 h after challenge. Sera were prediluted 1:5, and IFN-λ (R&D Systems) and IFN-α (PBL) were analyzed by ELISA.
To separate splenocytes after i.v. poly IC or DISC–HSV-1 injection, spleens were dissected 1.5–2 h after challenge, erythrocytes were lysed (RBC lysing buffer; Sigma-Aldrich) and stained with the corresponding fluorochrome-labeled antibodies (against CD3, CD8α, CD11b, CD11c, CD45R, CD172a, and NK1.1 [BD or eBioscience]), and the cells were separated with antifluorochrome magnetic beads as described by the manufacturer (Miltenyi Biotec) into a negative and positive enriched fraction. The initial cell number of splenocytes added onto an individual column per separation was 20 × 106. Without further counting or analyzing the purity, each fraction was distributed into 2 wells with 200 µl of medium/well. For some experiments, the negative fraction was incubated with another round of beads using beads against a different fluorochrome as used for the first round of selection. All of the cells were incubated in vitro for the next 18 h, and cell-free supernatants were analyzed for IFN-λ as described in In vitro stimulation and cytokine detection. For some experiments, the mice, either C57BL/6 WT or CD11c-DTR-tg, were treated with 100 ng DT (Sigma-Aldrich) i.p. 2 d before challenge with poly IC to deplete CD11chigh transcribing cells.
Isolation and stimulation of human DC.
PBMCs were prepared from peripheral blood of nonatopic blood donors by density gradient centrifugation, and BDCA3+ DCs were purified from PBMCs using the BDCA3/CD141+ DC isolation kit (Miltenyi Biotech) on an AutoMACS separator (Miltenyi Biotec). Subsequently, BDCA1+ DCs were purified from the BDCA3-depleted PBMCs using the BDCA1/CD1c+ DC isolation kit (Miltenyi Biotech). Preliminary experiments with PBMCs and DC-enriched fractions of PBMCs have indicated that the addition of the recombinant human cytokines IL-3, GM-CSF, and IFN-γ (10 ng/ml each; all PeproTech) enhanced the IFN-λ1 and IFN-λ2 production, and accordingly, this combination of cytokines was added to all stimulations shown. After stimulation for 18–24 h, the supernatants were analyzed for IFN-λ1 and IFN-λ2 by ELISA according to the manufacturer’s recommendations (Tebu-Bio).
BM reconstitution.
Groups of three C57BL/6, TRIF-KO, or TLR3-KO mice were irradiated with 2 × 550 rads, 2 h apart. They were rested for 3 h and then reconstituted by i.v. injection with 5 × 106 red and dead cell–depleted BM cells of the required genotype. Mice were maintained on antibiotic-supplemented drinking water, and 4 wk after reconstitution, the mice were injected i.v. with 100 µg poly IC. 4 h later, blood was taken for IFN-λ assays.
Statistical analysis.
Statistical significance was calculated using a two-tailed Student’s t test. Results are expressed as means ± SD.
Online supplemental material.
Fig. S1 shows that splenic pDCs produce large amounts of IFN-λ to CpG-2216. Fig. S2 shows that the production of IFN-λ and IL-12p70 in response to specific stimulation suggests the presence of CD8α+ or eCD8α cDCs in liver of WT but not FL-KO mice. Fig. S3 shows that IFN-λ production to poly IC by FLDC-derived eCD8α cDCs depends on TLR3 and IFN-IR but not on MyD88 or Cardif. Fig. S4 shows IFN-α production to poly IC in vivo by WT, IL-15R–KO, RAG-KO, and FL-KO mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20092720/DC1.
Acknowledgments
We thank Shizuo Akira, Tadatsugu Taniguchi, Steffen Jung, and Jurg Tschopp for providing mice; Juliane Paetzold and Ronny Kassub for excellent technical assistance; and the animal services facilities at the University of Zurich, Max Planck Institute of Biochemistry, Max Planck Institute of Immunobiology, the Walter and Eliza Hall Institute of Medical Research, the University of Melbourne, and Bavarian Nordic GmbH for animal husbandry.
H. Lauterbach, B. Bathke, P. Chaplin, M. Suter, M. O’Keeffe, and H. Hochrein are employees of Bavarian Nordic GmbH. All other authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- cDC
- conventional DC
- DN
- double negative
- dsRNA
- double-stranded RNA
- DT
- diphtheria toxin
- DTR
- DT receptor
- FL
- Fms-related tyrosine kinase 3 ligand
- IFN-IR
- IFN-I receptor
- IRF
- IFN regulatory factor
- ODN
- oligonucleotide
- pDC
- plasmacytoid DC
- poly IC
- polyinosinic:polycytidylic acid
- PRR
- pattern recognition receptor
- RLH
- Rig-like helicase
- TLR
- Toll-like receptor
References
- Adachi O., Kawai T., Takeda K., Matsumoto M., Tsutsui H., Sakagami M., Nakanishi K., Akira S. 1998. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 9:143–150 10.1016/S1074-7613(00)80596-8 [DOI] [PubMed] [Google Scholar]
- Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 413:732–738 10.1038/35099560 [DOI] [PubMed] [Google Scholar]
- Aliberti J., Schulz O., Pennington D.J., Tsujimura H., Reis e Sousa C., Ozato K., Sher A. 2003. Essential role for ICSBP in the in vivo development of murine CD8alpha + dendritic cells. Blood. 101:305–310 10.1182/blood-2002-04-1088 [DOI] [PubMed] [Google Scholar]
- Ank N., Paludan S.R. 2009. Type III IFNs: new layers of complexity in innate antiviral immunity. Biofactors. 35:82–87 10.1002/biof.19 [DOI] [PubMed] [Google Scholar]
- Ank N., West H., Bartholdy C., Eriksson K., Thomsen A.R., Paludan S.R. 2006. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J. Virol. 80:4501–4509 10.1128/JVI.80.9.4501-4509.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ank N., Iversen M.B., Bartholdy C., Staeheli P., Hartmann R., Jensen U.B., Dagnaes-Hansen F., Thomsen A.R., Chen Z., Haugen H., et al. 2008. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J. Immunol. 180:2474–2485 [DOI] [PubMed] [Google Scholar]
- Bachem A., Güttler S., Hartung E., Ebstein F., Schaefer M., Tannert A., Salama A., Movassaghi K., Opitz C., Mages H.W., et al. 2010. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J. Exp. Med. 207:1273–1281 10.1084/jem.20100348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamboat Z.M., Stableford J.A., Plitas G., Burt B.M., Nguyen H.M., Welles A.P., Gonen M., Young J.W., DeMatteo R.P. 2009. Human liver dendritic cells promote T cell hyporesponsiveness. J. Immunol. 182:1901–1911 10.4049/jimmunol.0803404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchet W., Cella M., Odermatt B., Asselin-Paturel C., Colonna M., Kalinke U. 2002. Virus-induced interferon α production by a dendritic cell subset in the absence of feedback signaling in vivo. J. Exp. Med. 195:507–516 10.1084/jem.20011666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedoui S., Prato S., Mintern J., Gebhardt T., Zhan Y., Lew A.M., Heath W.R., Villadangos J.A., Segura E. 2009. Characterization of an immediate splenic precursor of CD8+ dendritic cells capable of inducing antiviral T cell responses. J. Immunol. 182:4200–4207 10.4049/jimmunol.0802286 [DOI] [PubMed] [Google Scholar]
- Brand S., Beigel F., Olszak T., Zitzmann K., Eichhorst S.T., Otte J.M., Diebold J., Diepolder H., Adler B., Auernhammer C.J., et al. 2005. IL-28A and IL-29 mediate antiproliferative and antiviral signals in intestinal epithelial cells and murine CMV infection increases colonic IL-28A expression. Am. J. Physiol. Gastrointest. Liver Physiol. 289:G960–G968 10.1152/ajpgi.00126.2005 [DOI] [PubMed] [Google Scholar]
- Coccia E.M., Severa M., Giacomini E., Monneron D., Remoli M.E., Julkunen I., Cella M., Lande R., Uzé G. 2004. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur. J. Immunol. 34:796–805 10.1002/eji.200324610 [DOI] [PubMed] [Google Scholar]
- Contoli M., Message S.D., Laza-Stanca V., Edwards M.R., Wark P.A., Bartlett N.W., Kebadze T., Mallia P., Stanciu L.A., Parker H.L., et al. 2006. Role of deficient type III interferon-lambda production in asthma exacerbations. Nat. Med. 12:1023–1026 10.1038/nm1462 [DOI] [PubMed] [Google Scholar]
- Crozat K., Guiton R., Contreras V., Feuillet V., Dutertre C.A., Ventre E., Vu Manh T.P., Baranek T., Storset A.K., Marvel J., et al. 2010. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8α+ dendritic cells. J. Exp. Med. 207:1283–1292 10.1084/jem.20100223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucak H., Yrlid U., Reizis B., Kalinke U., Johansson-Lindbom B. 2009. Type I interferon signaling in dendritic cells stimulates the development of lymph-node-resident T follicular helper cells. Immunity. 31:491–501 10.1016/j.immuni.2009.07.005 [DOI] [PubMed] [Google Scholar]
- Dzionek A., Fuchs A., Schmidt P., Cremer S., Zysk M., Miltenyi S., Buck D.W., Schmitz J. 2000. BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J. Immunol. 165:6037–6046 [DOI] [PubMed] [Google Scholar]
- Edelson B.T., Kc W., Juang R., Kohyama M., Benoit L.A., Klekotka P.A., Moon C., Albring J.C., Ise W., Michael D.G., et al. 2010. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8α+ conventional dendritic cells. J. Exp. Med. 207:823–836 10.1084/jem.20091627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A.D., Diebold S.S., Slack E.M., Tomizawa H., Hemmi H., Kaisho T., Akira S., Reis e Sousa C. 2003. Toll-like receptor expression in murine DC subsets: lack of TLR7 expression by CD8 alpha+ DC correlates with unresponsiveness to imidazoquinolines. Eur. J. Immunol. 33:827–833 10.1002/eji.200323797 [DOI] [PubMed] [Google Scholar]
- Eidenschenk C., Crozat K., Krebs P., Arens R., Popkin D., Arnold C.N., Blasius A.L., Benedict C.A., Moresco E.M., Xia Y., Beutler B. 2010. Flt3 permits survival during infection by rendering dendritic cells competent to activate NK cells. Proc. Natl. Acad. Sci. USA. 107:9759–9764 10.1073/pnas.1005186107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field A.K., Tytell A.A., Lampson G.P., Hilleman M.R. 1967. Inducers of interferon and host resistance. II. Multistranded synthetic polynucleotide complexes. Proc. Natl. Acad. Sci. USA. 58:1004–1010 10.1073/pnas.58.3.1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge D., Fellay J., Thompson A.J., Simon J.S., Shianna K.V., Urban T.J., Heinzen E.L., Qiu P., Bertelsen A.H., Muir A.J., et al. 2009. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 461:399–401 10.1038/nature08309 [DOI] [PubMed] [Google Scholar]
- Gitlin L., Barchet W., Gilfillan S., Cella M., Beutler B., Flavell R.A., Diamond M.S., Colonna M. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA. 103:8459–8464 10.1073/pnas.0603082103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henri S., Poulin L.F., Tamoutounour S., Ardouin L., Guilliams M., de Bovis B., Devilard E., Viret C., Azukizawa H., Kissenpfennig A., Malissen B. 2010. CD207+ CD103+ dermal dendritic cells cross-present keratinocyte-derived antigens irrespective of the presence of Langerhans cells. J. Exp. Med. 207:189–206 10.1084/jem.20091964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildner K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S., et al. 2008. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 322:1097–1100 10.1126/science.1164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochrein H., O’Keeffe M. 2008. Dendritic cell subsets and toll-like receptors. Handb. Exp. Pharmacol. 183:153–179 10.1007/978-3-540-72167-3_8 [DOI] [PubMed] [Google Scholar]
- Hochrein H., O’Keeffe M., Luft T., Vandenabeele S., Grumont R.J., Maraskovsky E., Shortman K. 2000. Interleukin (IL)-4 is a major regulatory cytokine governing bioactive IL-12 production by mouse and human dendritic cells. J. Exp. Med. 192:823–833 10.1084/jem.192.6.823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochrein H., Shortman K., Vremec D., Scott B., Hertzog P., O’Keeffe M. 2001. Differential production of IL-12, IFN-alpha, and IFN-gamma by mouse dendritic cell subsets. J. Immunol. 166:5448–5455 [DOI] [PubMed] [Google Scholar]
- Hochrein H., Schlatter B., O’Keeffe M., Wagner C., Schmitz F., Schiemann M., Bauer S., Suter M., Wagner H. 2004. Herpes simplex virus type-1 induces IFN-alpha production via Toll-like receptor 9-dependent and -independent pathways. Proc. Natl. Acad. Sci. USA. 101:11416–11421 10.1073/pnas.0403555101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtschke T., Löhler J., Kanno Y., Fehr T., Giese N., Rosenbauer F., Lou J., Knobeloch K.P., Gabriele L., Waring J.F., et al. 1996. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 87:307–317 10.1016/S0092-8674(00)81348-3 [DOI] [PubMed] [Google Scholar]
- Honda K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., Shimada N., Ohba Y., Takaoka A., Yoshida N., Taniguchi T. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 434:772–777 10.1038/nature03464 [DOI] [PubMed] [Google Scholar]
- Jongbloed S.L., Kassianos A.J., McDonald K.J., Clark G.J., Ju X., Angel C.E., Chen C.J., Dunbar P.R., Wadley R.B., Jeet V., et al. 2010. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J. Exp. Med. 207:1247–1260 10.1084/jem.20092140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan W.J., Eskdale J., Srinivas S., Pekarek V., Kelner D., Rodia M., Gallagher G. 2007. Human interferon lambda-1 (IFN-lambda1/IL-29) modulates the Th1/Th2 response. Genes Immun. 8:254–261 10.1038/sj.gene.6364382 [DOI] [PubMed] [Google Scholar]
- Jung S., Unutmaz D., Wong P., Sano G., De los Santos K., Sparwasser T., Wu S., Vuthoori S., Ko K., Zavala F., et al. 2002. In vivo depletion of CD11c+ dendritic cells abrogates priming of CD8+ T cells by exogenous cell-associated antigens. Immunity. 17:211–220 10.1016/S1074-7613(02)00365-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinski P., Smits H.H., Schuitemaker J.H., Vieira P.L., van Eijk M., de Jong E.C., Wierenga E.A., Kapsenberg M.L. 2000. IL-4 is a mediator of IL-12p70 induction by human Th2 cells: reversal of polarized Th2 phenotype by dendritic cells. J. Immunol. 165:1877–1881 [DOI] [PubMed] [Google Scholar]
- Kotenko S.V., Gallagher G., Baurin V.V., Lewis-Antes A., Shen M., Shah N.K., Langer J.A., Sheikh F., Dickensheets H., Donnelly R.P. 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 4:69–77 10.1038/ni875 [DOI] [PubMed] [Google Scholar]
- Krug A., Luker G.D., Barchet W., Leib D.A., Akira S., Colonna M. 2004. Herpes simplex virus type 1 activates murine natural interferon-producing cells through toll-like receptor 9. Blood. 103:1433–1437 10.1182/blood-2003-08-2674 [DOI] [PubMed] [Google Scholar]
- Li M., Liu X., Zhou Y., Su S.B. 2009. Interferon-lambdas: the modulators of antivirus, antitumor, and immune responses. J. Leukoc. Biol. 86:23–32 10.1189/jlb.1208761 [DOI] [PubMed] [Google Scholar]
- Lindstedt M., Lundberg K., Borrebaeck C.A. 2005. Gene family clustering identifies functionally associated subsets of human in vivo blood and tonsillar dendritic cells. J. Immunol. 175:4839–4846 [DOI] [PubMed] [Google Scholar]
- Lodolce J.P., Boone D.L., Chai S., Swain R.E., Dassopoulos T., Trettin S., Ma A. 1998. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity. 9:669–676 10.1016/S1074-7613(00)80664-0 [DOI] [PubMed] [Google Scholar]
- Longhi M.P., Trumpfheller C., Idoyaga J., Caskey M., Matos I., Kluger C., Salazar A.M., Colonna M., Steinman R.M. 2009. Dendritic cells require a systemic type I interferon response to mature and induce CD4+ Th1 immunity with poly IC as adjuvant. J. Exp. Med. 206:1589–1602 10.1084/jem.20090247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luber C.A., Cox J., Lauterbach H., Fancke B., Selbach M., Tschopp J., Akira S., Wiegand M., Hochrein H., O’Keeffe M., Mann M. 2010. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity. 32:279–289 10.1016/j.immuni.2010.01.013 [DOI] [PubMed] [Google Scholar]
- Lund J., Sato A., Akira S., Medzhitov R., Iwasaki A. 2003. Toll-like receptor 9–mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J. Exp. Med. 198:513–520 10.1084/jem.20030162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney S., Vermi W., Gilfillan S., Cella M., Murphy T.L., Schreiber R.D., Murphy K.M., Colonna M. 2009. Distinct and complementary functions of MDA5 and TLR3 in poly(I:C)-mediated activation of mouse NK cells. J. Exp. Med. 206:2967–2976 10.1084/jem.20091181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna H.J., Stocking K.L., Miller R.E., Brasel K., De Smedt T., Maraskovsky E., Maliszewski C.R., Lynch D.H., Smith J., Pulendran B., et al. 2000. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 95:3489–3497 [PubMed] [Google Scholar]
- McLean C.S., Erturk M., Jennings R., Challanain D.N., Minson A.C., Duncan I., Boursnell M.E., Inglis S.C. 1994. Protective vaccination against primary and recurrent disease caused by herpes simplex virus (HSV) type 2 using a genetically disabled HSV-1. J. Infect. Dis. 170:1100–1109 [DOI] [PubMed] [Google Scholar]
- Megjugorac N.J., Gallagher G.E., Gallagher G. 2010. IL-4 enhances IFN-lambda1 (IL-29) production by plasmacytoid DCs via monocyte secretion of IL-1Ra. Blood. 115:4185–4190 10.1182/blood-2009-09-246157 [DOI] [PubMed] [Google Scholar]
- Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 437:1167–1172 10.1038/nature04193 [DOI] [PubMed] [Google Scholar]
- Mombaerts P., Iacomini J., Johnson R.S., Herrup K., Tonegawa S., Papaioannou V.E. 1992. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 68:869–877 10.1016/0092-8674(92)90030-G [DOI] [PubMed] [Google Scholar]
- Naik S.H., O’Keeffe M., Proietto A., Hochrein H., Shortman K., Wu L. 2010. CD8+, CD8-, and plasmacytoid dendritic cell generation in vitro using flt3 ligand. Methods Mol. Biol. 595:167–176 10.1007/978-1-60761-421-0_10 [DOI] [PubMed] [Google Scholar]
- Napolitani G., Rinaldi A., Bertoni F., Sallusto F., Lanzavecchia A. 2005. Selected Toll-like receptor agonist combinations synergistically trigger a T helper type 1-polarizing program in dendritic cells. Nat. Immunol. 6:769–776 10.1038/ni1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keeffe M., Hochrein H., Vremec D., Pooley J., Evans R., Woulfe S., Shortman K. 2002. Effects of administration of progenipoietin 1, Flt-3 ligand, granulocyte colony-stimulating factor, and pegylated granulocyte-macrophage colony-stimulating factor on dendritic cell subsets in mice. Blood. 99:2122–2130 10.1182/blood.V99.6.2122 [DOI] [PubMed] [Google Scholar]
- Onoguchi K., Yoneyama M., Takemura A., Akira S., Taniguchi T., Namiki H., Fujita T. 2007. Viral infections activate types I and III interferon genes through a common mechanism. J. Biol. Chem. 282:7576–7581 10.1074/jbc.M608618200 [DOI] [PubMed] [Google Scholar]
- Osterlund P., Veckman V., Sirén J., Klucher K.M., Hiscott J., Matikainen S., Julkunen I. 2005. Gene expression and antiviral activity of alpha/beta interferons and interleukin-29 in virus-infected human myeloid dendritic cells. J. Virol. 79:9608–9617 10.1128/JVI.79.15.9608-9617.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund P.I., Pietilä T.E., Veckman V., Kotenko S.V., Julkunen I. 2007. IFN regulatory factor family members differentially regulate the expression of type III IFN (IFN-lambda) genes. J. Immunol. 179:3434–3442 [DOI] [PubMed] [Google Scholar]
- Pillarisetty V.G., Shah A.B., Miller G., Bleier J.I., DeMatteo R.P. 2004. Liver dendritic cells are less immunogenic than spleen dendritic cells because of differences in subtype composition. J. Immunol. 172:1009–1017 [DOI] [PubMed] [Google Scholar]
- Poulin L.F., Salio M., Griessinger E., Anjos-Afonso F., Craciun L., Chen J.L., Keller A.M., Joffre O., Zelenay S., Nye E., et al. 2010. Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8α+ dendritic cells. J. Exp. Med. 207:1261–1271 10.1084/jem.20092618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst H.C., Tschannen K., Odermatt B., Schwendener R., Zinkernagel R.M., Van Den Broek M. 2005. Histological analysis of CD11c-DTR/GFP mice after in vivo depletion of dendritic cells. Clin. Exp. Immunol. 141:398–404 10.1111/j.1365-2249.2005.02868.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis e Sousa C., Hieny S., Scharton-Kersten T., Jankovic D., Charest H., Germain R.N., Sher A. 1997. In vivo microbial stimulation induces rapid CD40 ligand-independent production of interleukin 12 by dendritic cells and their redistribution to T cell areas. J. Exp. Med. 186:1819–1829 10.1084/jem.186.11.1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins S.H., Walzer T., Dembélé D., Thibault C., Defays A., Bessou G., Xu H., Vivier E., Sellars M., Pierre P., et al. 2008. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 9:R17 10.1186/gb-2008-9-1-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsson C., Hausmann J., Lauterbach H., Schmidt M., Akira S., Wagner H., Chaplin P., Suter M., O’Keeffe M., Hochrein H. 2008. Survival of lethal poxvirus infection in mice depends on TLR9, and therapeutic vaccination provides protection. J. Clin. Invest. 118:1776–1784 10.1172/JCI33940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavoni G., Mattei F., Sestili P., Borghi P., Venditti M., Morse H.C., III, Belardelli F., Gabriele L. 2002. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8α+ dendritic cells. J. Exp. Med. 196:1415–1425 10.1084/jem.20021263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz O., Diebold S.S., Chen M., Näslund T.I., Nolte M.A., Alexopoulou L., Azuma Y.T., Flavell R.A., Liljeström P., Reis e Sousa C. 2005. Toll-like receptor 3 promotes cross-priming to virus-infected cells. Nature. 433:887–892 10.1038/nature03326 [DOI] [PubMed] [Google Scholar]
- Sheppard P., Kindsvogel W., Xu W., Henderson K., Schlutsmeyer S., Whitmore T.E., Kuestner R., Garrigues U., Birks C., Roraback J., et al. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4:63–68 10.1038/ni873 [DOI] [PubMed] [Google Scholar]
- Shortman K., Naik S.H. 2007. Steady-state and inflammatory dendritic-cell development. Nat. Rev. Immunol. 7:19–30 10.1038/nri1996 [DOI] [PubMed] [Google Scholar]
- Shortman K., Lahoud M.H., Caminschi I. 2009. Improving vaccines by targeting antigens to dendritic cells. Exp. Mol. Med. 41:61–66 10.3858/emm.2009.41.2.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegemund S., Hartl A., von Buttlar H., Dautel F., Raue R., Freudenberg M.A., Fejer G., Büttner M., Köhler G., Kirschning C.J., et al. 2009. Conventional bone marrow-derived dendritic cells contribute to toll-like receptor-independent production of alpha/beta interferon in response to inactivated parapoxvirus ovis. J. Virol. 83:9411–9422 10.1128/JVI.02362-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirén J., Pirhonen J., Julkunen I., Matikainen S. 2005. IFN-alpha regulates TLR-dependent gene expression of IFN-alpha, IFN-beta, IL-28, and IL-29. J. Immunol. 174:1932–1937 [DOI] [PubMed] [Google Scholar]
- Sommereyns C., Paul S., Staeheli P., Michiels T. 2008. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 4:e1000017 10.1371/journal.ppat.1000017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpter T.L., Abe M., Tokita D., Thomson A.W. 2007. Dendritic cells, the liver, and transplantation. Hepatology. 46:2021–2031 10.1002/hep.21974 [DOI] [PubMed] [Google Scholar]
- Suppiah V., Moldovan M., Ahlenstiel G., Berg T., Weltman M., Abate M.L., Bassendine M., Spengler U., Dore G.J., Powell E., et al. 2009. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat. Genet. 41:1100–1104 10.1038/ng.447 [DOI] [PubMed] [Google Scholar]
- Tailor P., Tamura T., Kong H.J., Kubota T., Kubota M., Borghi P., Gabriele L., Ozato K. 2007. The feedback phase of type I interferon induction in dendritic cells requires interferon regulatory factor 8. Immunity. 27:228–239 10.1016/j.immuni.2007.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T., Yanai H., Savitsky D., Taniguchi T. 2008. The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 26:535–584 10.1146/annurev.immunol.26.021607.090400 [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Nishida N., Sugiyama M., Kurosaki M., Matsuura K., Sakamoto N., Nakagawa M., Korenaga M., Hino K., Hige S., et al. 2009. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat. Genet. 41:1105–1109 10.1038/ng.449 [DOI] [PubMed] [Google Scholar]
- Thomas D.L., Thio C.L., Martin M.P., Qi Y., Ge D., O’Huigin C., Kidd J., Kidd K., Khakoo S.I., Alexander G., et al. 2009. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 461:798–801 10.1038/nature08463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele S., Hochrein H., Mavaddat N., Winkel K., Shortman K. 2001. Human thymus contains 2 distinct dendritic cell populations. Blood. 97:1733–1741 10.1182/blood.V97.6.1733 [DOI] [PubMed] [Google Scholar]
- Velásquez-Lopera M.M., Correa L.A., García L.F. 2008. Human spleen contains different subsets of dendritic cells and regulatory T lymphocytes. Clin. Exp. Immunol. 154:107–114 10.1111/j.1365-2249.2008.03734.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villadangos J.A., Shortman K. 2010. Found in translation: the human equivalent of mouse CD8+ dendritic cells. J. Exp. Med. 207:1131–1134 10.1084/jem.20100985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vremec D., Zorbas M., Scollay R., Saunders D.J., Ardavin C.F., Wu L., Shortman K. 1992. The surface phenotype of dendritic cells purified from mouse thymus and spleen: investigation of the CD8 expression by a subpopulation of dendritic cells. J. Exp. Med. 176:47–58 10.1084/jem.176.1.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vremec D., Pooley J., Hochrein H., Wu L., Shortman K. 2000. CD4 and CD8 expression by dendritic cell subtypes in mouse thymus and spleen. J. Immunol. 164:2978–2986 [DOI] [PubMed] [Google Scholar]
- Vremec D., O’Keeffe M., Hochrein H., Fuchsberger M., Caminschi I., Lahoud M., Shortman K. 2007. Production of interferons by dendritic cells, plasmacytoid cells, natural killer cells, and interferon-producing killer dendritic cells. Blood. 109:1165–1173 10.1182/blood-2006-05-015354 [DOI] [PubMed] [Google Scholar]
- Wilkins C., Gale M., Jr 2010. Recognition of viruses by cytoplasmic sensors. Curr. Opin. Immunol. 22:41–47 10.1016/j.coi.2009.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk K., Witte K., Witte E., Proesch S., Schulze-Tanzil G., Nasilowska K., Thilo J., Asadullah K., Sterry W., Volk H.D., Sabat R. 2008. Maturing dendritic cells are an important source of IL-29 and IL-20 that may cooperatively increase the innate immunity of keratinocytes. J. Leukoc. Biol. 83:1181–1193 10.1189/jlb.0807525 [DOI] [PubMed] [Google Scholar]
- Yarovinsky F., Zhang D., Andersen J.F., Bannenberg G.L., Serhan C.N., Hayden M.S., Hieny S., Sutterwala F.S., Flavell R.A., Ghosh S., Sher A. 2005. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science. 308:1626–1629 10.1126/science.1109893 [DOI] [PubMed] [Google Scholar]