Abstract
Cerebral gene expression changes in response to traumatic brain injury will provide useful information in the search for future trauma treatment. In order to characterize the outcome of mild brain injury, we studied C57BL/6J mice in a weight-drop, closed head injury model. At various times post-injury, mRNA was isolated from neocortex and hippocampus and transcriptional alterations were studied using quantitative reverse transcriptase PCR and gene array analysis. At three days post-injury, the results showed unilateral injury responses, both in neocortex and hippocampus, with the main effect seen on the side of the skull hit by the dropping weight. Upregulated transcripts encoded products characterizing reactive astrocytes, phagocytes, microglia, and immune-reactive cells. Markers for oligodendrocytes and T-cells were not altered. Notably, strong differences in the responses among individual mice were seen (e.g., for the Gfap transcript expressed by reactive astrocytes and the chemokine Ccl3 transcript expressed by activated microglial cells). In conclusion, mild TBI chiefly activates transcripts leading to tissue signaling, inflammatory processes, and chemokine signaling, as in focal brain injury, suggesting putative targets for drug development.
Key words: closed head injury, GeneChip array, inflammation, mild trauma, quantitative RT-PCR, weight drop impact
Introduction
Traumatic brain injury (TBI) represents a major public health problem, and is the most common cause of mortality and disability in younger Western populations. At present, no effective pharmaceutical therapies for TBI are available and current treatment chiefly involves optimized intensive care management (Johnston and Gupta, 2002). When also considering milder forms of injury not demanding intensive care, an even higher fraction of the population is affected, indicating a multitude of differential functional responses to brain injury (Byrne, 2000; Comper et al., 2005). The current study presents data on gene transcripts regulated in response to brain injury in a mouse model of a mild TBI (mTBI), produced by a closed head weight drop injury (Zohar et al., 2003; Milman et al., 2005; Tweedie et al., 2007). This injury model in mice is based on an anteriolateral impact to the region between the right ear and the corner of the eye, and results in an injury without any damage to skull, scalp, or dura but with widespread and diffuse symptoms. In previous studies, no morphological changes have been seen. In contrast, when using parameters for cognitive disabilities and depression, clear effects were detected (Pan et al., 2003), which also can be seen in humans suffering from TBI without any obvious neuroanatomical changes. The cortical and cerebrovascular effects and the inflammatory components also mimic human TBI.
Patients with post-concussion syndrome display a variety of different responses (Kibby and Long, 1996; Milman et al., 2005). In order to gain insights into the post-injury pathophysiological processes in TBI and to suggest new treatments targeting mediators of cellular interactions, we turned to molecular characterization of a closed skull injury model. Hence, we performed an unbiased genome-wide search for transcripts regulated in the mildly injured brain, aiming to further characterize secondary injury processes. To achieve this, the GeneChip Mouse Genome 430 2.0 Array (Affymetrix, Santa Clara, CA) was utilized to define transcriptional alterations in neocortex three days after injury. In addition, we studied transcriptional changes in the injured neocortex and hippocampus in mice using quantitative reverse transcriptase PCR (qRT-PCR).
We recently showed that inflammatory reactions and robust activation of microglia and phagocytes are key events in the injured cortex after a severe focal traumatic brain injury (Israelsson et al., 2008). The present data provide additional information on injury-induced cell-type specific gene profiles in response to the mild injury model. In line with the present findings, inflammation plays an important role in the secondary injury cascade of TBI (McIntosh et al., 1998; Potts et al., 2006), with an emphasis on the upregulation of chemokines (Hausmann et al., 1998; Esche et al., 2005; Charo and Ransohoff, 2006). Hence, it may become possible to improve the outcome of mild TBI after pharmacotherapy targeting inflammatory mediators.
Materials and Methods
Mild traumatic brain injury
C57BL/6J male mice (body weight 25–30 g) were subjected to a closed head injury as previously described (Zohar et al., 2003; Milman et al., 2005; Tweedie et al., 2007). The weight drop apparatus includes a vertical 80 cm long metal tube (diameter 13 mm) with a 30 g weight (diameter 10 mm) dropped through the upper end of the tube. Mice were anesthetized with isofluran placed under the metal tube's lower end and the weight dropped anteriolaterally at the right side without damaging the sagittal sinus. Mice were sacrificed at different post-injury time points (uninjured, 1 h, 4 h, 22 h, 3 days, and 7 days; five animals per time point, following mTBI). The skull was opened and neocortex and hippocampus were dissected separately from the ipsilateral and contralateral hemisphere, and preserved in RNAlater (Qiagen, Valencia, CA). RNA was isolated using RNeasy Mini kit for isolation of total RNA from animal tissues (Qiagen). RNA concentration (ng/μL) was normalized from the absorbance at 260 nm, determined by a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) for each sample of total RNA. The experimental protocol was approved by the NIDA/ACUC.
RNA Isolation and Affymetrix microarray profiling
Neocortical RNA samples of seven uninjured mice and five injured mice sacrificed three days after mTBI were analyzed individually using GeneChip Mouse Genome 430 2.0 Arrays (Affymetrix). Sample handling was carried out at the Uppsala Array Platform (Uppsala University Hospital, Uppsala, Sweden). Gene expression data were analyzed using Affymetrix Microarray Suite, version 5.1 (MAS 5.1), applying the percentile algorithm (parameters = percentile: 75). The experimental material, including transcripts with significant presence of the hybridization signal, significant upregulation at a three-fold or higher level, and annotated genes, was analyzed (after exclusion of all ESTs and control RNA). Thereafter, the data were sorted after detectable RNA levels (present) in each brain analyzed. Transcripts evaluated on the basis of a signal intensity of 38 or higher were saved and statistical significance levels were determined. Mouse Genome Informatics (MGI; www.informatics.jax.org) was used as a resource for assigning gene symbols/names, accession Ids, and gene function (Gene Ontology, GO).
Quantitative reverse transcriptase PCR
In order to study changes in transcriptional levels after the closed head injury model, the following primer pairs were used (GenBank accession numbers and upper as well as lower primer stated): Gfap (NM_010277, 5′-CGG GAG TCG GCC AGT TAC CAG-3′ and 5′-TTT CCT GTA GGT GGC GAT CTC-3′); Ccl3 (NM_011337, 5′-GCC TGC TGC TTC TCC TAC AG-3′ and 5′-TCT GCC GGT TTC TCT TAG TC-3′); Cxcl10 (NM_021274, 5′-ACC CAA GTG CTG CCG TCA TT-3′ and 5′-ATT CTC ACT GGC CCG TCA TC-3′); Itgax (NM_021334, 5′-ACA CAG TGT GCT CCA GTA TGA-3′ and 5′-GCC CAG GGA TAT GTT CAC AGC-3′); Lyz2 (NM_017372, 5′-ATG GGT GGC ATG GCG AGC AC-3′ and 5′-TGA GAA AGA GAC CGA ATG AG-3′); Selp (NM_011347, 5′-TGC CAG CCT GGA TAT AGA GC-3′ and 5′-GGA GGT TCA CAC GCA ATA GC-3′); Ccl12 (NM_011331, 5′-TGC CTC CTG CTC ATA GCT AC-3′ and 5′-TCA GCT TCC GGA CGT GAA TC-3′); Tcra (U07662, 5′-GAT GCC ACG TTG ACC GAG AA-3′ and 5′-GAC CAC AGC CTC AGC GTC AT-3′); Cnp1 (NM_009923, 5′-CAA GAT GGT GTC CGC TGA TG-3′ and 5′-TCA TGT CCC GGC GGC AGT AG-3′); and 28S rRNA (X00525, 5′-GGG AGA GGG TGT AAA TCT CGC-3′ and 5′-CTG TTC ACC TTG GAG ACC TGC-3′). All primer pairs, except Gfap, Tcra, and Cnp1, span exon-intron boundaries. In order to check for possible DNA contamination, three primer pairs for Gfap, spanning exon-intron boundaries, were tested (5′-CGC CAA GCC AAG CAC GAA G-3′ and 5′-GCC GCT CTA GGG ACT CGT TC-3′; 5′-GAC TAT CGC CGC CAA CTG-3′ and 5′-TCC TGG TAA CTG GCC GAC TC-3′; 5′-CGG CAC GAA CGA GTC CCT A-3′ and 5′-CCT CCT CCA GCC GAG CAA GT-3′). The Bio-Rad iScript One-Step RT-PCR Kit (Bio-Rad Laboratories, Hercules, CA) with SYBR Green (reverse transcription at 50°C for 10 min) was used yielding amplified fragments of about 100 base pairs. qRT-PCR was run for 36 cycles (95°C for 10 s, 60°C for 30 s) using a MyIQ thermal cycler (Bio-Rad). After completion of the cycles, melting curves obtained by increasing the temperature from 55.0 to 94.5°C in increments of 0.5°C were examined to ascertain specificity of the PCR products (a symmetrical single peak). In each microwell, 10 ng of total RNA was analyzed. Threshold cycle values (Ct) for the injured brain RNA samples were subtracted from average Ct values obtained from uninjured brain tissue analyzed in the same run. The resulting differences (ΔΔCt) were transformed to linear fold increase with the normal (uninjured) tissue as a reference and subjected to statistical analysis. In addition to absorbance at 260 nm, we determined any possible changes in “household” gene activity due to the mild brain trauma, by examining levels of 28S ribosomal RNA in samples using qRT-PCR.
Statistical analysis
SigmaStat software, version 3.1 (SPSS, Inc., Richmond, CA), was used for calculations. For parametric data, two-tailed Student's t-test and ANOVA were used, and for non-parametric data, Mann-Whitney and Kruskal-Wallis tests were used. Additional post-hoc tests such as Holm-Sidak and Dunn's were performed when appropriate. Correlation among data sets was analyzed using the Pearson Product Moment Correlation procedure (SigmaStat).
Results
Genome-wide analysis of closed head injury-induced transcriptional changes in neocortex
Considering the transcriptional changes previously found by qRT-PCR three days after CCI (Israelsson et al., 2008), we chose this time point to extend our genome-wide GeneChip array (Affymetrix) search for changes in the mouse neocortex also subjected to mild traumatic brain injury. Applying unbiased, stringent inclusion criteria, we identified 37 genes significantly activated in the injured ipsilateral neocortex (Table 1). Assigning gene functions (gene ontology, GO; Table 1) to the 37 transcripts shows that more than half of the genes upregulated encode proteins involved in stress responses (Serpina3n, Loxl2, and Timp1), inflammation, immunity, and defense responses (C3ar1, Cd44, Cd52, Cd68, Cd84, Clec7a, Csf2rb1, Ctsc, Fcgr2b, Fcrls, H2-Aa, Ifi30, Itga6, Lgals1, Lilrb4, Ly86, Lyz1, Lyz2, Mpeg1, Ncf1, Ptx3, and Spp1). Fc receptor-like S, scavenger receptor (Fcrls, previously known as macrophage scavenger receptor 2, Msr2) is among the inflammatory transcripts upregulated. Moreover, the GeneChip data show that in addition to Lyz2, the related gene Lyz1 is also strongly upregulated in the mildly injured neocortex (Table 1). Furthermore, transcripts involved in cell migration, cytoskeletal processes, and cell adhesion or comprising extracellular matrix (Capg, Cd93, Col3a1, Gfap, Tgfbi, and Vim) are also upregulated. A single transcript (Pycard) linked to proapoptotic functions was found to be upregulated. In addition, a single gene associated with growth and signal transduction (Osmr) was increased as was a singleton associated with cell proliferation (Rrm2). No increases were found in transcripts encoding transcription factors, whereas three genes classified as having “other functions” were upregulated (Adfp, Pbk, and S100a4).
Table 1.
The 37 Genes Identified as Significantly Upregulated (3-fold or more) after mTBI in an Unbiased Analysis of GeneChip Data
| Gene symbol | Gene title | GO classification | Fold increase | GenBank | Probe set ID |
|---|---|---|---|---|---|
| Adfp | Adipose differentiation related protein | Long-chain fatty acid transport | 3.0 | NM_007408 | 1448318_at |
| C3ar1 | Complement component 3a receptor 1 | Chemotaxis | 4.1 | NM_009779 | 1419483_at |
| Capg | Capping protein (actin filament), gelsolin-like | Cell projection biogenesis | 3.8 | NM_001042534 | 1450355_a_at |
| Cd44 | CD44 antigen | Healing during inflammatory response | 3.5 | NM_009851 | 1423760_at |
| Cd52 | CD52 antigen | Anchored to membrane | 5.0 | NM_013706 | 1460218_at |
| Cd68 | CD68 antigen | Integral to membrane | 3.3 | NM_009853 | 1449164_at |
| Cd84 | CD84 antigen | Cell adhesion | 3.1 | NM_013489 | 1422875_at |
| Cd93 | CD93 antigen | Cell adhesion | 3.0 | NM_010740 | 1456046_at |
| Clec7a | C-type lectin domain family 7, member a | Inflammatory response | 7.0 | NM_020008 | 1420699_at |
| Col3a1 | Procollagen, type III, alpha 1 | Extracellular matrix structural constituent | 5.9 | NM_009930 | 1427883_a_at |
| Csf2rb1 | Colony-stimulating factor 2 receptor, beta 1, low-affinity (granulocyte-macrophage) | Cytokine and chemokine mediated signaling pathway | 3.2 | NM_007780 | 1455660_at |
| Ctsc | Cathepsin C | Proteolysis | 4.1 | NM_009982 | 1416382_at |
| Fcgr2b | Fc receptor, IgG, low affinity IIb | Immune response | 3.0 | NM_001077189 | 1435477_s_at |
| Fcrls | Fc receptor-like S, scavenger receptor | Receptor activity | 4.1 | NM_030707 | 1448891_at |
| Gfap | Glial fibrillary acidic protein | Intermediate filament-based process | 5.3 | NM_010277 | 1426509_s_at |
| H2-Aa | Histocompatibility 2, class II antigen A, alpha | Positive regulation of T cell differentiation | 3.2 | NM_010378 | 1435290_x_at |
| Ifi30 | Interferon gamma inducible protein 30 | Antigen processing and presentation | 4.2 | NM_023065 | 1422476_at |
| Itga6 | Integrin alpha 6 | Leukocyte migration | 13.4 | NM_008397 | 1422444_at |
| Lgals1 | Lectin, galactose binding, soluble 1 | Heterophilic cell adhesion | 4.4 | NM_008495 | 1419573_a_at |
| Lilrb4 | Leukocyte immunoglobulin-like receptor, subfamily B, member 4 | Immune response | 15.8 | NM_013532 | 1420394_s_at |
| Loxl2 | Lysyl oxidase-like 2 | Oxidation reduction | 3.5 | NM_033325 | 1431004_at |
| Ly86 | Lymphocyte antigen 86 | Inflammatory response | 3.8 | NM_010745 | 1422903_at |
| Lyz1 | Lysozyme 1 | Cytolysis | 20.9 | NM_013590 | 1439426_x_at |
| Lyz2 | Lysozyme 2 | Cytolysis | 8.1 | NM_017372 | 1423547_at |
| Mpeg1 | Macrophage expressed gene 1 | Integral to membrane | 3.7 | NM_010821 | 1427076_at |
| Ncf1 | Neutrophil cytosolic factor 1 | Inflammatory response | 3.1 | NM_010876 | 1451767_at |
| Osmr | Oncostatin M receptor | Cytokine receptor activity | 3.5 | NM_011019 | 1418674_at |
| Pbk | PDZ binding kinase | Protein amino acid phosphorylation | 5.6 | NM_023209 | 1448627_s_at |
| Ptx3 | Pentraxin related gene | Positive regulation of phagocytosis | 3.1 | NM_008987 | 1418666_at |
| Pycard | PYD and CARD domain containing | Induction of apoptosis | 3.1 | NM_023258 | 1417346_at |
| Rrm2 | Ribonucleotide reductase M2 | DNA replication | 3.5 | NM_009104 | 1434437_x_at |
| S100a4 | S100 calcium binding protein A4 | Calcium ion binding | 4.1 | NM_011311 | 1424542_at |
| Serpina3n | Serine (or cysteine) peptidase inhibitor, clade A, member 3N | Acute-phase response | 9.9 | NM_009252 | 1419100_at |
| Spp1 | Secreted phosphoprotein 1 | Cell adhesion | 12.2 | NM_009263 | 1449254_at |
| Tgfbi | Transforming growth factor, beta induced | Cell adhesion | 3.7 | NM_009369 | 1448123_s_at |
| Timp1 | Tissue inhibitor of metalloproteinase 1 | Metalloendopeptidase inhibitor activity | 5.4 | NM_001044384 | 1460227_at |
| Vim | Vimentin | Intermediate filament-based process | 4.3 | NM_011701 | 1450641_at |
The 37 upregulated genes are also identified as upregulated in the established controlled cortical impact (CCI) injury model in mice. This is based on a comparison of the present gene list with a recent publication from our laboratory (Israelsson et al., 2008) demonstrating that 146 genes were upregulated three days after CCI. In addition, we have now updated this list by increasing the number of samples being analyzed and by using the most current assignments from GeneChip arrays in a fully independent study of CCI samples from the mouse to include over 250 genes (data not shown).
The only gene in Table 1 that in our previous CCI material did not increase at least three-fold is Loxl2. However, analysis of the CCI data shows that the Loxl2 transcript was upregulated 2.3-fold (p < 0.001) in this injury model. We therefore conclude that there are no major differences in the response between the closed head injury and the controlled cortical impact injury. In fact, a substantial fraction of upregulated genes encode products involved in inflammatory and tissue remodeling responses in both models.
Closed head injury induces modest effects on transcription levels in ipsilateral and contralateral cortex
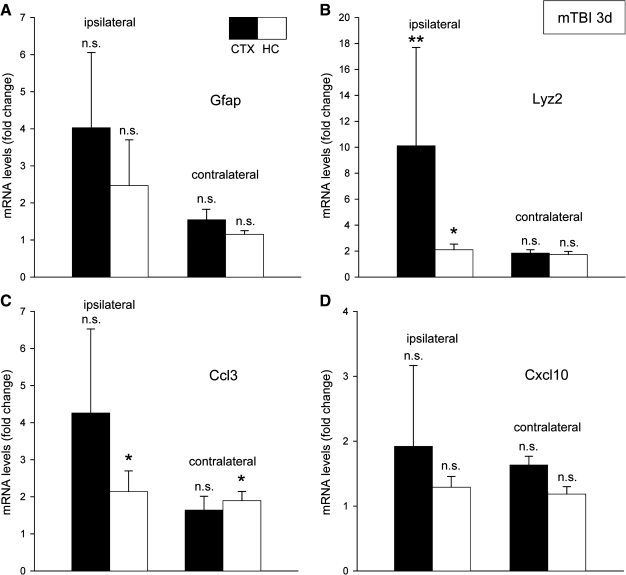
Quantitative RT-PCR (qRT-PCR) analysis was used to further study the injury-induced regulation of Gfap and Lyz2 transcripts three days post-injury (Fig. 1A and B). Increased expression of the Gfap transcript, characteristic of reactive astrocytes, was seen on the side of the brain injured by the dropping weight. The response was more marked in the neocortex compared to the hippocampus (Fig. 1A). Using three additional primer pairs encoding various exons of Gfap gave no evidence for DNA contamination of the isolated total RNA samples. However, due to the large individual variations in the response to the injury, the mean increase in the group of five mice studied three days after injury did not reach statistical significance (p > 0.05). It is noteworthy that the individual variability was strongly correlated in pairwise comparisons among the total of four primer pairs used (0.915 < r < 0.953; p < 0.001 in all cases). Furthermore, comparing Gfap transcript increases in individual mice by qRT-PCR and GeneChip array also showed a strong correlation (r = 0.905; p < 0.001).
FIG. 1.
Transcriptional changes three days after mTBI in neocortex (CTX) and hippocampus (HC) ipsilaterally and contralaterally analyzed by qRT-PCR. Individual samples from five mice were measured at least twice in duplicate reactions. (A) Gfap expression showed increases on the injured side that did not reach significance. (B) Lyz2 expression showed an increase on the ipsilateral side in both neocortex and hippocampus. (C) Ccl3 expression showed a significant response in hippocampus on both sides and a marked increase in the ipsilateral neocortex. (D) Cxcl10 expression increased but did not reach significant levels. Mean value ± SEM is indicated. Statistical differences to samples from uninjured mice are shown (*p < 0.05; **p < 0.01).
The largest expression differences were seen in the lysozyme 2 (Lyz2) transcript, characterizing phagocytic cells in the injured brain. Both neocortex and hippocampus showed significant increases ipsilaterally (Fig. 1B). Considering the profound upregulation of chemokines previously found in the CCI injury model (Israelsson et al., 2008), we chose to study the expression of Ccl3 and Cxcl10 in the current closed head injury model (Fig. 1C and D). The microglia-associated expression of chemokine Ccl3 transcript was significantly increased in the hippocampus on both sides. Again, the neocortex on the ipsilateral side showed varying degrees of increased transcript levels in response to the injury, with two of the five mice contributing to the high standard error of the mean value (Fig. 1C). A similar pattern was seen for the T-cell attractant chemokine Cxcl10 transcript, again not reaching levels of significant increase when analyzed as a group (Fig. 1D). No changes in ribosomal RNA levels (28S rRNA; selected to represent “household” gene activity) correlated with the trauma. Thus, we regard the changes detected by qRT-PCR as real injury-induced shifts in transcript levels. This is also supported by the Affymetrix GeneChip data from analysis of the same brains used for qRT-PCR.
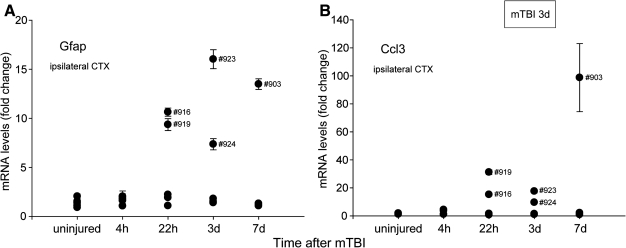
Temporal changes in reactive astrocytes and microglia following mTBI
We investigated the temporal pattern of injury-induced Gfap and Ccl3 transcriptional changes individually in five mice at each time point (4 h, 22 h, 3 days, and 7 days) post-injury and in uninjured controls (Fig. 2A and B). The differences in individual responses in the injured ipsilateral neocortex at 22 h, 3 days, and 7 days post-injury are shown, with five of the 15 mice analyzed at these time points showing more severe damage than the other animals. At the 3-day time point, it is of note that the same subjects (mouse #923 and mouse #924) showed robust increases in the levels of Gfap in reactive astrocytes (Fig. 2A) as well as in the levels of Ccl3 expressed by microglia (Fig. 2B), which was confirmed by the GeneChip data for individual mice (data not shown).
FIG. 2.
Ipsilateral gene expression at different time points after mTBI shown by qRT-PCR (uninjured, 4 h, 22 h, 3 days, 7 days). The same mice show elevated levels with both markers. Individual samples from five mice were measured at least twice in duplicate reactions. (A) Gfap levels peak after three days. (B) Ccl3 levels show the largest individual result after seven days. SEM is indicated.
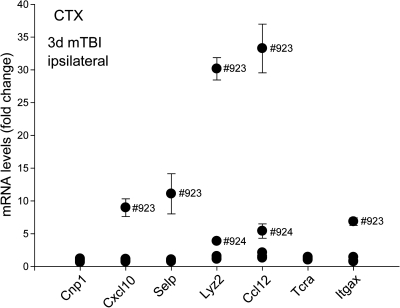
Ipsilateral changes in inflammation-related transcripts three days after mTBI
In addition to the increased Lyz2 expression (Table 1), other transcripts characteristic of immune responses were analyzed by qRT-PCR in the ipsilateral neocortex three days after mTBI in individual mice (Fig. 3). Apart from Lyz2, transcripts showing the highest expression levels at this time point are factors involved in inflammatory reactions in immune cells and endothelial cells. Cxcl10, Selp (P-selectin), Ccl12, and Itgax (integrin alpha X) produced a higher mean value because of a specific expression in some animals. In contrast, Cnp1 (marker for oligodendrocytes) and Tcra (T-cell receptor alpha) did not react to the trauma in any of the animals using this model (Fig. 3). From these data, it is suggested that mouse #923 was the most severely injured, followed in rank by mouse #924, in line with the GeneChip results.
FIG. 3.
Neocortical changes using qRT-PCR in several different markers indicating a large variety in response to mTBI. Mean and SEM of samples from five mice were measured individually at least twice in duplicate reactions.
Discussion
The present study makes use of a noninvasive closed head weight-drop mouse model to produce mild TBI. The injured mice suffer long-lasting learning and memory deficits that are injury magnitude- and time-dependent (Zohar et al., 2003). The cognitive deficits occur without overt neurological damage, brain edema, damage to the blood-brain barrier, or clear morphological changes to the brain as assessed by magnetic resonance imaging (Zohar et al., 2003) or triphenyltetrazolium chloride staining (Tweedie et al., 2007). This mTBI model thus produces a specific, significant, and irreversible long-term learning and memory impairment in mice. We used this weight-drop model of mTBI in the anesthetized mouse designed to deliver sheer force to the right temporal region while the dura, skull, and scalp remain intact. The model with mTBI to the right temporal lobe in the mouse has been shown to result in time-dependent mild behavioral changes without distinct changes in cerebral blood flow (Pan et al., 2003).
The current study examined transcriptional changes induced by mTBI, offering the possibility of comparing data from earlier studies of a severe focal brain injury (Israelsson et al., 2008). The temporal pattern of injury responses in the mouse neocortex and hippocampus was characterized, with RNA samples collected at various time points after mTBI inflicted by closed head weight-drop injury. The transcriptional responses to mTBI were examined and demonstrated similar changes in neocortex and hippocampus of the mouse, although with a different amplitude in hippocampus. Unbiased genome-wide high-stringency analysis of the injured neocortex three days after mTBI identified a distinct group of upregulated gene transcripts. The present list of genes affected by injury was compared with previously reported data from the mouse and rat cerebral cortex subjected to traumatic or ischemic injuries (Kobori et al., 2002; Natale et al., 2003; Raghavendra Rao et al., 2003; Hedtjärn et al., 2004a, 2004b; Küry et al., 2004; Poulsen et al., 2005; von Gertten et al., 2005).
Although the currently used mild head injury model is based on a closed skull procedure, the resulting injury is strikingly unilateral with the greater effect seen on the side of the brain closest to the weight drop. A marked unilateral response to the mTBI was previously noted (Pan et al., 2003), demonstrating increased uptake of radioactively labeled tumor necrosis factor alpha (TNFα) in the hippocampus ipsilateral to the injury one day post-injury. Previous studies have also defined biochemical changes accompanying the mTBI in this mouse model and demonstrated increased levels of the proapoptotic protein Bax (Tweedie et al., 2007). The mTBI model used here has previously been found to be associated with increased apoptosis developing over three days following injury, and most prominent in the anterior cingulate cortex and hippocampus (Tashlykov et al., 2007). These areas were not included in our current GeneChip analysis that was instead focusing on the neocortex. This fact may account for why we detected only a single upregulated transcript involved in apoptosis (Pycard). This gene was also found upregulated in the CCI model (Israelsson et al., 2008). Nevertheless, the possibility exists that post-injury apoptosis may not be strongly controlled at the transcriptional level.
All of the 37 transcripts currently found upregulated in our mTBI model are also represented in focal brain injury (Israelsson et al., 2008, and unpublished data). As in the focal injury model, the main functions for upregulated transcripts are associated with inflammation, phagocytosis and immune reactions, defense and stress responses, and cell adhesion and cytoskeletal functions.
In the present study, upregulation of chemokines and their receptors were less prominent than that in the focal injury model (Israelsson et al., 2008). To a large extent, the closed head injury model results in individual variations obscuring the upregulation clearly seen in the more severely affected mice. Thus, GeneChip analysis of mice #923 and #924 revealed considerable overlap to previous CCI data (Israelsson et al., 2008). Of the 15 mildly injured mice currently analyzed transcriptionally at 22 h to 7 days post-injury, five were more severely affected by the trauma (33%). The wide range in outcome measures is also manifested among mTBI patients (Comper et al., 2005). In comparison with our earlier study of severe focal brain injury (Israelsson et al., 2008), the present mTBI model resulted in the upregulation of far fewer transcripts in the neocortex.
The similarity in response among different experimental models used suggests a fairly stereotyped response to a variety of brain insults. Thus, the majority of the currently identified transcripts are among those previously characterized in focal injury, strengthening the idea of a common mechanism activated by various insults to the brain. This will aid in identification of target candidates for pharmacological neuroprotection using these techniques.
Acknowledgments
This work was supported by the Swedish Research Council (grant no. 2006-3014 [T.E.]), the Swedish Brain Research Foundation/Sparbanksstiftelsen Upland (T.E. and C.I.), and intramural grants from NIH/NIDA (B.J.H.). Hanna Göransson and Maria Rydåker carried out the GeneChip procedures.
Author Disclosure Statement
No competing financial interests exist.
References
- Byrne E. The post concussional syndrome after mild head injury: some practical considerations. J. Clin. Neurosci. 2000;7:473–474. doi: 10.1054/jocn.2000.0779. [DOI] [PubMed] [Google Scholar]
- Charo I.F. Ransohoff R.M. The many roles of chemokines and chemokine receptors in inflammation. N. Engl. J. Med. 2006;354:610–621. doi: 10.1056/NEJMra052723. [DOI] [PubMed] [Google Scholar]
- Comper P. Bisschop S.M. Carnide N. Tricco A. A systematic review of treatments for mild traumatic brain injury. Brain Inj. 2005;19:863–880. doi: 10.1080/02699050400025042. [DOI] [PubMed] [Google Scholar]
- Esche C. Stellato C. Beck L.A. Chemokines: key players in innate and adaptive immunity. J. Invest. Dermatol. 2005;125:615–628. doi: 10.1111/j.0022-202X.2005.23841.x. [DOI] [PubMed] [Google Scholar]
- Hausmann E.H. Berman N.E. Wang Y.Y. Meara J.B. Wood G.W. Klein R.M. Selective chemokine mRNA expression following brain injury. Brain Res. 1998;788:49–59. doi: 10.1016/s0006-8993(97)01160-8. [DOI] [PubMed] [Google Scholar]
- Hedtjärn M. Mallard C. Eklind S. Gustafson-Brywe K. Hagberg H. Global gene expression in the immature brain after hypoxia-ischemia. J. Cereb. Blood Flow Metab. 2004a;24:1317–1332. doi: 10.1097/01.WCB.0000141558.40491.75. [DOI] [PubMed] [Google Scholar]
- Hedtjärn M. Mallard C. Hagberg H. Inflammatory gene profiling in the developing mouse brain after hypoxia-ischemia. J. Cereb. Blood Flow Metab. 2004b;24:1333–1351. doi: 10.1097/01.WCB.0000141559.17620.36. [DOI] [PubMed] [Google Scholar]
- Israelsson C. Bengtsson H. Kylberg A. Kullander K. Lewén A. Hillered L. Ebendal T. Distinct cellular patterns of upregulated chemokine expression supporting a prominent inflammatory role in traumatic brain injury. J. Neurotrauma. 2008;25:959–974. doi: 10.1089/neu.2008.0562. [DOI] [PubMed] [Google Scholar]
- Johnston A.J. Gupta A.K. Advanced monitoring in the neurology intensive care unit: microdialysis. Curr. Opin. Crit. Care. 2002;8:121–127. doi: 10.1097/00075198-200204000-00006. [DOI] [PubMed] [Google Scholar]
- Kibby M.Y. Long C.J. Minor head injury: attempts at clarifying the confusion. Brain Inj. 1996;10:159–186. doi: 10.1080/026990596124494. [DOI] [PubMed] [Google Scholar]
- Kobori N. Clifton G.L. Dash P. Altered expression of novel genes in the cerebral cortex following experimental brain injury. Brain Res. Mol. Brain Res. 2002;104:148–158. doi: 10.1016/s0169-328x(02)00331-5. [DOI] [PubMed] [Google Scholar]
- Küry P. Schroeter M. Jander S. Transcriptional response to circumscribed cortical brain ischemia: spatiotemporal patterns in ischemic vs. remote non-ischemic cortex. Eur. J. Neurosci. 2004;19:1708–1720. doi: 10.1111/j.1460-9568.2004.03226.x. [DOI] [PubMed] [Google Scholar]
- McIntosh T.K. Saatman K.E. Raghupathi R. Graham D.I. Smith D.H. Lee V.M. Trojanowski J.Q. The Dorothy Russell Memorial Lecture. The molecular and cellular sequelae of experimental traumatic brain injury: pathogenetic mechanisms. Neuropathol. Appl. Neurobiol. 1998;24:251–267. doi: 10.1046/j.1365-2990.1998.00121.x. [DOI] [PubMed] [Google Scholar]
- Milman A. Rosenberg A. Weizman R. Pick C.G. Mild traumatic brain injury induces persistent cognitive deficits and behavioral disturbances in mice. J. Neurotrauma. 2005;22:1003–1010. doi: 10.1089/neu.2005.22.1003. [DOI] [PubMed] [Google Scholar]
- Natale J.E. Ahmed F. Cernak I. Stoica B. Faden A.I. Gene expression profile changes are commonly modulated across models and species after traumatic brain injury. J. Neurotrauma. 2003;20:907–927. doi: 10.1089/089771503770195777. [DOI] [PubMed] [Google Scholar]
- Pan W. Kastin A.J. Rigai T. McLay R. Pick C.G. Increased hippocampal uptake of tumor necrosis factor alpha and behavioral changes in mice. Exp. Brain Res. 2003;149:195–199. doi: 10.1007/s00221-002-1355-7. [DOI] [PubMed] [Google Scholar]
- Potts M.B. Koh S.E. Whetstone W.D. Walker B.A. Yoneyama T. Claus C.P. Manvelyan H.M. Noble-Haeusslein L.J. Traumatic injury to the immature brain: inflammation, oxidative injury, and iron-mediated damage as potential therapeutic targets. NeuroRx. 2006;3:143–153. doi: 10.1016/j.nurx.2006.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen C.B. Penkowa M. Borup R. Nielsen F.C. Caceres M. Quintana A. Molinero A. Carrasco J. Giralt M. Hidalgo J. Brain response to traumatic brain injury in wild-type and interleukin-6 knockout mice: a microarray analysis. J. Neurochem. 2005;92:417–432. doi: 10.1111/j.1471-4159.2004.02877.x. [DOI] [PubMed] [Google Scholar]
- Raghavendra Rao V.L. Dhodda V.K. Song G. Bowen K.K. Dempsey R.J. Traumatic brain injury-induced acute gene expression changes in rat cerebral cortex identified by GeneChip analysis. J. Neurosci. Res. 2003;71:208–219. doi: 10.1002/jnr.10486. [DOI] [PubMed] [Google Scholar]
- Tashlykov V. Katz Y. Gazit V. Zohar O. Schreiber S. Pick C.G. Apoptotic changes in the cortex and hippocampus following minimal brain trauma in mice. Brain Res. 2007;1130:197–205. doi: 10.1016/j.brainres.2006.10.032. [DOI] [PubMed] [Google Scholar]
- Tweedie D. Milman A. Holloway H.W. Li Y. Harvey B.K. Shen H. Pistell P.J. Lahiri D.K. Hoffer B.J. Wang Y. Pick C.G. Greig N.H. Apoptotic and behavioral sequelae of mild brain trauma in mice. J. Neurosci. Res. 2007;85:805–815. doi: 10.1002/jnr.21160. [DOI] [PubMed] [Google Scholar]
- von Gertten C. Flores Morales A. Holmin S. Mathiesen T. Nordqvist A.C. Genomic responses in rat cerebral cortex after traumatic brain injury. BMC Neurosci. 2005;6:69. doi: 10.1186/1471-2202-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohar O. Schreiber S. Getslev V. Schwartz J.P. Mullins P.G. Pick C.G. Closed-head minimal traumatic brain injury produces long-term cognitive deficits in mice. Neuroscience. 2003;118:949–955. doi: 10.1016/s0306-4522(03)00048-4. [DOI] [PubMed] [Google Scholar]