Abstract
The fidelity of the poliovirus RNA-dependent RNA polymerase (3Dpol) plays a direct role in the genomic evolution and pathogenesis of the virus. A single site mutation (Gly64Ser) that is remote from the catalytic center results in a higher fidelity polymerase. NMR studies with [methyl-13C]methionine-labeled protein were used to compare the solution structure and dynamics of wild-type and Gly64Ser 3Dpol. The chemical shifts for the Met6 resonance were significantly different between wild-type and Gly64Ser 3Dpol when bound in ternary complexes with RNA and incorrect, but not with correct, nucleotide, suggesting that the Gly64Ser mutation induces structural changes in the N-terminal β-strand when the enzyme is bound to incorrect, but not correct nucleotide. We also observe changes in the transverse relaxation times for methionines near regions important for nucleotide and RNA binding, and catalysis. Our strategy to assign the [methyl-13C]methionine resonances involved separately mutating each of the seventeen methionines. Several substitutions produced additional resonances for both Met6 and Met187, a reporter for RNA binding and conformational changes in the highly conserved motif B loop, even though these methionines are greater than 20 Å apart. The results for Gly64Ser and the other mutants are intriguing considering that they can result in structural and/or dynamic changes to methionines distant from the site of mutation. We propose that there is a long-distance network operating throughout 3Dpol that coordinates ligand binding, conformational changes and catalysis. Mutation of Gly64 results in structural and/or dynamic changes to the network that may affect polymerase fidelity.
Positive-strand RNA viruses cause a variety of acute and chronic diseases in humans including the common cold, myocarditis, hepatitis, severe acute respiratory syndrome (SARS) and liver cancer (1–4). The enzyme responsible for replicating the genomes of RNA-based viruses is the virally encoded RNA-dependent RNA polymerase (3Dpol). The polymerase from poliovirus (PV) has emerged as an important model system for understanding the function of 3Dpol enzymes (5). Intriguingly, the error frequency or ‘fidelity’ of PV 3Dpol appears to be very important to the biology and pathogenesis of poliovirus. For example, restriction of genetic variability within a population of poliovirus through expression of a mutant high fidelity 3Dpol leads to lower tissue adaptability and viral pathogenicity (6–9). This suggests that understanding the function and fidelity mechanisms of 3Dpol has a direct bearing on disease intervention. Antiviral strategies may include targeting 3Dpol by small molecules to inhibit its activity and/or change its fidelity (10–13), or alternatively, virus encoding 3Dpol enzymes with altered fidelity may be potentially used as live, attenuated vaccine strains (7, 14).
PV 3Dpol has a ‘cupped right hand’ structure with fingers, palm and thumb subdomains that is common to many other RNA and DNA polymerases (15–17) (Figure 1). Most of the catalytic machinery is found in the palm region, including the conserved structural motifs A-E (5, 18). Asp233 in motif A and Asp328 in motif C help to coordinate magnesium and bind the phosphate groups of the incoming nucleotide (19, 20). The other highly conserved aspartate in motif A, Asp 238, along with Asn297 from motif B participate in hydrogen bond interactions with the ribose group of the incoming nucleotide; these interactions are critical for proper sugar selection (19–21).
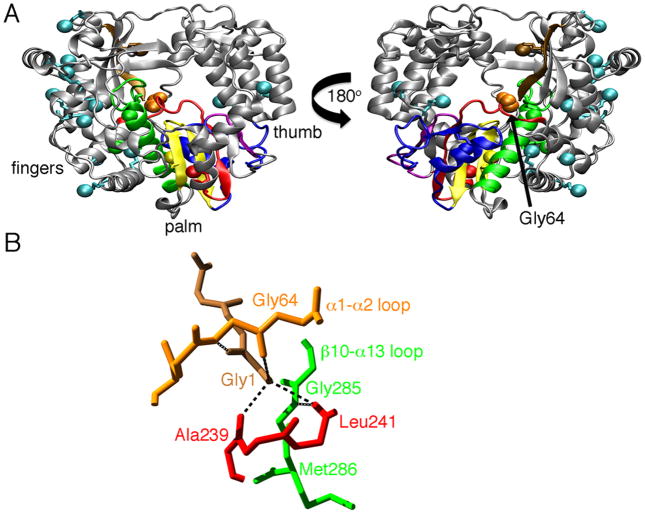
Figure 1.
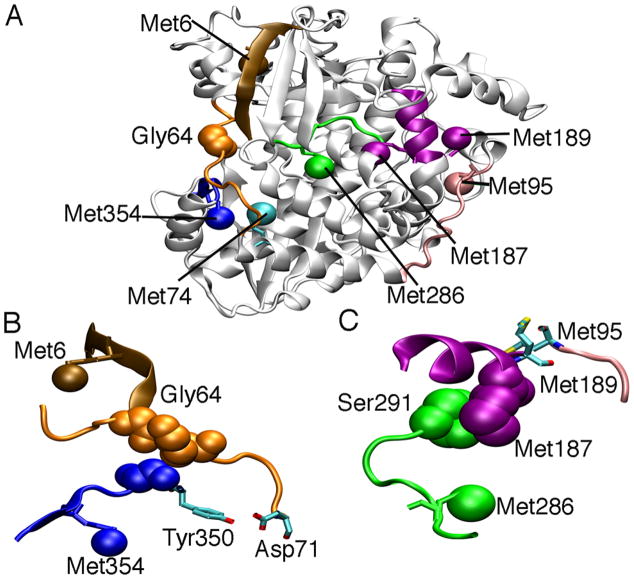
Structure of PV 3Dpol (PDB 1RA6). A. The locations of the terminal methyl groups of the methionines are indicated as colored spheres. The conserved structural motifs in the palm subdomain are colored as follows: motif A, red; motif B, green; motif C, yellow; motif D, blue; and motif E, purple. Gly64 is colored orange, and the N-terminal β-strand is colored brown. B. Hydrogen bond interactions involving Gly64, Gly1 on the N-terminal β-strand. Ala239 and Leu241 on motif A, and Gly285 on the β10-α13 loop. Motifs are colored as in A. Hydrogen bonds are indicated as black lines.
There are five proposed kinetic intermediates in the catalytic cycle of 3Dpol: enzyme binds RNA (E:Rn), followed by binding of nucleotide to form a ternary complex (E:RnNTP) that then undergoes a conformational change to form a complex competent for phosphoryl transfer (*E:RnNTP). Phosphodiester formation occurs to yield the ternary product complex (*E:Rn+1PPi) that then undergoes another conformational change (E:Rn+1PPi), followed by the release of pyrophosphate (E:Rn+1) (22, 23). In PV 3Dpol, the conformational change step prior to phosphoryl transfer and the phosphoryl transfer step itself are both partially rate limiting (22, 23). Thus, correct and incorrect nucleotide incorporation can be distinguished by kinetic differences in both of these steps.
The conformational change prior to chemistry is observed in most replicative nucleic acid polymerases (16, 24). However, there is some controversy concerning the nature of the conformational change (16, 25). It has been suggested that in some nucleic acid polymerases, the conformational change is a global ‘open-closed’ conformational change (16), but in 3Dpol, the conformational change step is likely to be more localized and traced to structural changes of motifs A and B (22, 23). For instance, the interaction between Asp238 and Asn297 observed in the unliganded enzyme occludes the ribose-binding pocket, and consequently, there must be a conformational change involving residues in motifs A and B in order for nucleotide to bind (19). Further structural changes involving Asp238, Asn297 and other residues in motif B such as Ser288 and Thr293 are also predicted to occur in order for the triphosphate group to align for catalysis (19, 22, 23). This suggestion is in agreement with crystal structures of foot-and-mouth disease virus (FMDV) 3Dpol that show localized structural changes to motifs A and B upon formation of the ternary complex with RNA and nucleotide triphosphate (NTP) (26). The highly conserved loop in motif B in FMDV 3Dpol (β10-α13 loop according to PV numbering and containing residues 285-293, β9-α11 loop according to FMDV) changes conformation to accommodate different nucleotides and RNA (26). The conformational change may also involve structural rearrangements in motif D to bring Lys359 into a position where it can act as a general acid to protonate the pyrophosphate leaving group (27, 28).
Some insight into the function and fidelity of PV 3Dpol can be gained by the functional and structural characterization of mutant enzymes that either increase or decrease polymerase fidelity. For example, mutation of the highly conserved Gly64 present on the α1-α2 loop (containing residues 61-71) in the fingers domain to Ser results in a higher fidelity polymerase. The rate of correct nucleotide incorporation is ~6000 fold greater than incorrect nucleotide incorporation for wild-type 3Dpol, whereas for Gly64Ser 3Dpol, correct nucleotide incorporation is ~9000 fold greater than incorrect nucleotide incorporation (i.e. the fidelity of Gly64Ser 3Dpol is ~1.5 fold greater than wild-type 3Dpol) (6). Thus, for Gly64Ser 3Dpol, nucleotide incorporation is more stringent, and this leads to a lower error-rate and decreased diversity within the viral population (6, 7). The differences in fidelity are not due to differences in nucleotide binding or to the rates of phosphodiester formation, but rather appear to be due to alterations in the conformational change step that brings the enzyme into an active conformation (6).
Gly64 is involved in a hydrogen bonding network that includes residues in the N-terminal β-strand, motif A and the β10-α13 loop in motif B (Figure 1B). This suggests that mutation of Gly64 to Ser may result in structural changes to the N-terminal β-strand, motif A and/or the β10-α13 loop. However, comparison of the X-ray crystal structure of the unliganded forms of wild-type (WT) and Gly64Ser 3Dpol did not show any significant structural differences in these regions (29). The crystal structures of WT and Gly64Ser 3Dpol overlapped to a great extent (rmsd= 0.28 Å for the protein backbone) and there were no obvious structural differences that pointed towards changes in function and/or fidelity (29, 30). These results prompted the suggestion that the functional differences may arise not because of structural changes but due to differences in the internal protein motions between WT and Gly64Ser 3Dpol (29, 30). However, it should be noted that structural differences might only become apparent when the enzymes are bound to RNA and/or form ternary complexes competent for phosphodiester formation. Unfortunately, there are currently no available structures of WT or Gly64Ser PV 3Dpol bound with RNA that would shed more insight.
We have used solution NMR to gain insight into the structural changes that occur when PV 3Dpol binds RNA and nucleotide, and to identify structural and/or dynamic differences between WT and Gly64Ser 3Dpol that might help to explain their differences in function and fidelity. 3Dpol is challenging to study by solution NMR, considering that the protein is relatively large by NMR standards (52 kDa). However, there are now a number of established strategies to study larger proteins by NMR (31, 32), including TROSY (transverse relaxation optimized spectroscopy)-based methods to study both backbone amide groups and side-chain methyl groups (33). In our case, we have isotopically labeled the terminal methyl groups of the methionine residues in the protein. This allowed us to quickly adapt existing protein expression and purification procedures for isotope labeling (34). During our studies, we identified a complex, long distance interaction network operating throughout 3Dpol that might be responsible for coordinating nucleotide and nucleic acid binding and catalysis. Moreover, the Gly64Ser mutation results in structural and/or dynamic changes to residues on this network. Further delineation of this amino acid network will bring new insights into the fidelity and function of PV 3Dpol and related polymerases, and can be used as leverage in rational viral vaccine and/or inhibitor design.
EXPERIMENTAL PROCEDURES
Site directed mutagenesis
Site-directed mutagenesis was carried out using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). Forward and reverse primers were designed for each of the Met site mutations (Supporting Information Table S1). Mutations were confirmed by DNA sequencing (Nucleic Acid Facility, Pennsylvania State University).
Expression of 3Dpol
Expression and purification of WT and mutant 3Dpol enzymes followed previous procedures (34–36) with some minor modifications. For the production of [methyl-13C]methionine-labeled 3Dpol, plasmids expressing 3Dpol were transformed into Met-auxotroph E. coli B834(DE3) cells, and grown in auto-inducible minimal medium containing [ε-13CH3]methionine as described in ref (37). 3Dpol is expressed as a fusion protein to SUMO and an N-terminal polyhistidine tag that increases protein production, eases purification and allows for production of 3Dpol with the naturally occurring Gly1 (34, 36).
Protein Purification
Buffer B (100 mM potassium phosphate pH 8.0, 500 mM NaCl, 5 mM imidazole, 5 mM β-mercaptoethanol, 60 μM ZnCl2, and 20% w/v glycerol) and buffer C (100 mM potassium phosphate pH 8.0, 500 mM NaCl, 60 μM ZnCl2, 5 mM β-mercaptoethanol and 20% w/v glycerol) were prepared. Cell pellets were resuspended in 50 mL lysis buffer (50 mL buffer B, 1.4 μg/mL pepstatin A, 1 μg/mL leupeptin, 1 mM PMSF, and 0.1% N-P40) and subjected to sonication. Cell lysates were centrifuged at 30,000xg and 4°C for 30 min. Supernatant was applied to Ni-NTA (Invitrogen) columns pre-equilibrated with buffer C1 (buffer C, 5 mM imidazole and 0.1% N-P40). The resin was washed with three bed volumes each of buffer C1 and buffer C2 (buffer C, 5 mM imidazole), and protein was eluted using high imidazole buffers C3 (buffer C, 50 mM imidazole) and C4 (buffer C, 500 mM imidazole). The polyhistidine tag and SUMO protein domain were cleaved from 3Dpol using the protease Ulp1. Protein solutions were dialyzed against 80 mM Tris-HCl pH 8.0, 500 mM NaCl, 20% w/v glycerol, 10 mM β-mercaptoethanol and 60 μM ZnCl2 overnight (optimal buffer for protease cleavage), and then dialyzed against 100 mM potassium phosphate pH 8.0, 20% w/v glycerol, 10 mM β-mercaptoethanol and 60 μM ZnCl2 for 2–3 hours. A second Ni-NTA column was used to separate the purified 3Dpol from the cleaved polyhistidine tag/SUMO domain, using procedures identical to the first Ni-NTA column. For WT and Gly64Ser 3Dpol, additional phospocellulose and Q-sepahrose columns were used to ensure protein solutions were free of trace contaminants of nuclease and phosphatase activities (34–36). Proteins were > 95% homogeneous as estimated by coomassie-blue staining of SDS PAGE gels. Protein is stable at 4°C for 3–4 months under high salt conditions (80 mM Tris-HCl pH 8.0, 500 mM NaCl, 20% w/v glycerol, 10 mM β-mercaptoethanol, and 60 μM ZnCl2).
Preparation of RNA oligonucleotides
10-mer single-stranded RNA oligonucleotides (5′-GCAUGGGCCC-3′) were purchased from Dharmacon RNAi Technologies and deprotected before use. Deprotection was performed by resuspending the RNA into 100 mM acetic acid adjusted to pH 3.8 with tetramethylethylenediamine (TEMED) and incubating for 30 min at 60°C. The RNA solutions were dried by SpeedVac, and the deprotected RNA pellets were then resuspended in 2H2O. The final stock concentration of ~ 7 mM was determined via the absorbance of the RNA at 260 nm using a calculated extinction coefficient of 90000 M−1cm−1. Importantly, the RNA oligonucleotide was previously designed such that it forms a six base pair duplex flanked by two 4-nucleotide 5′ overhangs (38). This symmetrical primer/template substrate (sym/sub) RNA has been used extensively in the kinetic analysis of PV 3Dpol (22, 23).
Activity assay of 3Dpol-RNA complex formation
To ensure we could achieve complex formation with our purified protein ([methyl-13C]methionine-labeled WT and Gly64Ser 3Dpol) and the RNA substrate using NMR buffer (10 mM HEPES pH 8.0, 200 mM NaCl, 5 mM MgCl2, 20 μM ZnCl2, 0.02% NaN3−) in 2H2O, reactions were carried out with 500 μM ATP or 3′-dATP and 10 μM 32P-labeled RNA. For comparison, reactions were also performed in H2O containing 50 mM HEPES, pH 7.5, 10 mM 2-mercaptoethanol, 5 mM MgCl2, 20 μM ZnCl2, 500 μM ATP or 3′-dATP and 10 μM RNA and without 13C-labeled protein. Reactions were initiated by the addition of 3Dpol to a final concentration of 1 μM and incubated at 30°C for 10 min. 3Dpol was diluted immediately prior to use in 50 mM HEPES pH 7.5, 10 mM β-mercaptoethanol, 60 μM ZnCl2, and 20% glycerol. Reaction volumes were 50 μl. Reactions were quenched by the addition of ethylenediaminetetraacetic acid (EDTA) to a final concentration of 100 mM. Products were resolved from substrate by electrophoresis through a denaturing polyacrylamide gel, visualized by phosphorimaging, and quantified by using the ImageQuant software (Molecular dynamics) (see Supporting Information Figure 1).
NMR sample preparation
For NMR studies, proteins were concentrated (~200–400 μM 3Dpol) and then desalted using spin columns (Thermo Scientific) to exchange into NMR buffer. For our initial studies with WT and methionine mutant apoenzymes, and with our CoCl2 titrations and NMR relaxation experiments, protein was exchanged into 25 mM potassium phosphate pH 8.0, 150 mM NaCl, 1mM DTT, 0.02% w/v NaN3 in 2H2O. For the RNA and nucleotide binding studies, the buffer used was 10 mM HEPES pH 8.0, 200 mM NaCl, 5 mM MgCl2, 10 μM ZnCl2 and 0.02% NaN3 in 2H2O. Importantly, there were no significant chemical shift differences for WT or Gly64Ser 3Dpol between the two buffer systems. For complex formation, the typical final concentrations were 200–250 μM for 3Dpol, 200–500 μM for duplex RNA, 3.1 mM for 3′-dATP and 4 mM for UTP. In general, a 2:1 duplex RNA:protein ratio was required to saturate 3Dpol with sym/sub RNA (i.e. for resonances corresponding to the unliganded enzyme to disappear) (see Supporting Information Figure S2). Desalting of the ternary complexes bound with UTP through spin columns did not result in changes to the NMR spectrum, suggesting that this complex is quite stable under NMR conditions.
NMR spectroscopy
Most NMR experiments were performed on a Bruker Avance III 600MHz spectrometer equipped with a 5 mm “inverse detection” triple resonance (1H/13C/15N) single axis gradient TCI-cryoprobe. We also attempted to perform 13C-methyl R2 relaxation dispersion experiments using a similarly equipped 850 MHz spectrometer. 1H-13C HSQC spectra were generally acquired as 64 (t1) × 512 (t2) complex matrix, with 64–128 scans per increment and 1.0 sec recovery delay. A spectrum for WT 3Dpol was also acquired using a recovery delay of 7.0 sec, but was not qualitatively different (i.e. no new resonances were observed) than spectra acquired with a shorter delay. The spectra of the methionine mutants, along with WT 3Dpol were acquired at 298 K. All other experiments were conducted at 293 K. 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) was used as an external chemical shift reference.
For the metal titrations, CoCl2 was added to protein in a varying ratios (1:0.06 to 1:1.7 for protein:metal), and the 1H-13C HSQC recorded. T1 and T2 relaxation experiments were performed as described (39, 40). Data for T1 delays were acquired out to 6.4 s, and data for the T2 delays were acquired out to 157 ms. T1 and T2 experiments were repeated 3–4 times on 2 or 3 separately expressed and purified samples of WT and Gly64Ser 3Dpol. R2 relaxation dispersion experiments were performed according to the pulse schemes in refs (41, 42) using a total relaxation delay of 10 ms. The Rex values (i.e. R2 contribution from conformational exchange) were estimated by subtracting the R2 value estimated under ‘fast pulsing conditions’ (the time between consecutive 180° pulses, τ = 500 μs, 1/τ = 2000 s−1) from the R2 value estimated under ‘slow pulsing conditions’ (τ = 2500 μs, 1/τ = 400 s−1). All NMR spectra and relaxation data were processed with NMRPipe (43) and analyzed with NMRView (44).
RESULTS AND DISCUSSION
NMR assignments of [methyl-13C] methionine-labeled 3Dpol
We have produced [methyl-13C]methionine-labeled 3Dpol through overpression of PV 3Dpol in a methionine auxotrophic strain of E. coli by adapting procedures from previously established protocols (34, 36, 37). Importantly, we are able to obtain 0.2 – 0.4 mM protein samples suitable for NMR. Dynamic light scattering measurements indicated that the protein exists as a monomer under the NMR solution conditions (data not shown).
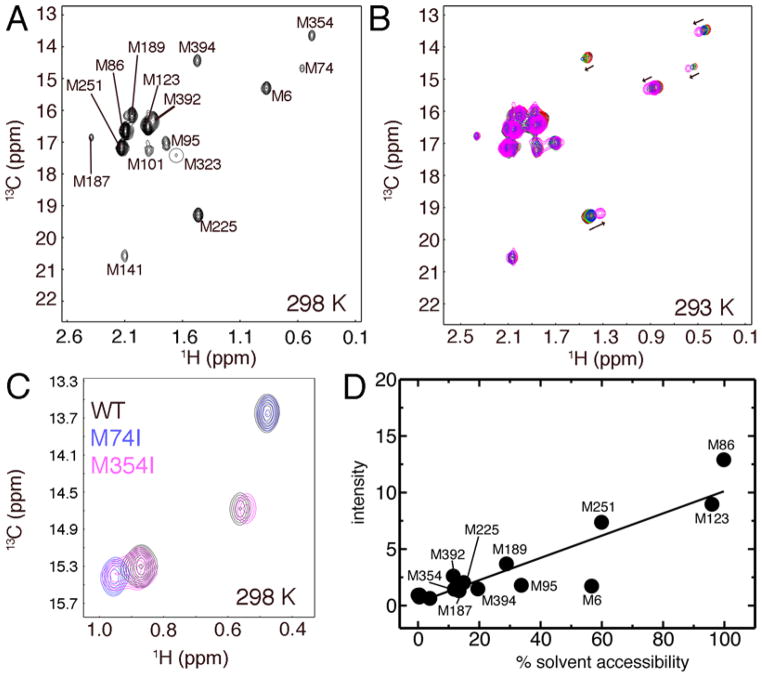
There are seventeen methionines scattered throughout the structure of 3Dpol, including methionines in the fingers, palm and thumb subdomains of 3Dpol (Figure 1A, see also Supporting Information Table S1). It should be noted that the overexpression and purification procedure yields 3Dpol with the correct amino acid (i.e. glycine) at position 1 rather than a methionine (36). It has been previously observed that mutations at the amino terminus results in enzymes with altered enzyme kinetics, due in part to structural pertubations to Asp238 that is responsible for properly positioning the incoming nucleotide (36, 45). To assign the resonances in the 1H-13C HSQC spectrum of [methyl-13C]methionine-labeled 3Dpol (Figure 2), we generated mutants in which each methionine residue was substituted with an isoleucine residue (or leucine in the case of Met123Leu). Most of the site mutants resulted in the elimination of a single resonance, allowing for straightforward assignment of all 15 resonances observed for the WT spectrum (Figure 2A). Met286Ile and Met299Ile mutations did not result in the elimination of any resonances, although they did result in some chemical shift changes (Supporting Information Table S2). Both methionine residues belong to motif B and are in close structural vicinity to each other. Mutation of the other methionines did not reveal any other peaks that might be obscured in the WT spectrum. This suggests that the peak intensities for Met286 and Met299 are quite low compared to the other methionines. It should also be noted that binding of RNA and/or nucleotide did not result in any new resonances that might be ascribed to Met286 or Met299.
Figure 2.
Assignment of the [methyl-13C]-methionine WT 3Dpol NMR spectrum. A. The 1H-13C HSQC spectra of WT 3Dpol obtained at 298 K (300 μM 3Dpol in 25 mM potassium phosphate pH 8.0, 150 mM NaCl, 1mM DTT, 0.02% w/v NaN3 in 2H2O). B. WT 3Dpol titrated with the paramagnetic metal Co2+ at 293 K. The protein concentration of 3Dpol was 180 μM. Co2+ concentrations were 0 (black), 10 (red), 50 (green), 100 (blue) and 300 (magenta) μM. C. Comparion of the 1H-13C HSQC spectra of WT (black), Met74Ile (blue) and Met354Ile (magneta) 3Dpol at 298 K. D. Correlation between solvent accessibility and peak intensity. The percent solvent accessibilities were determined using the GETAREA program with the default radius for the water probe (1.4 Å) (57).
Paramagnetic shifts resulting from addition of Co2+
We also recorded the 1H-13C HSQC of [methyl-13C]methionine-labeled 3Dpol upon addition of stoichiometric amounts of MgCl2 and CoCl2. It has been previously observed that 3Dpol can use Co2+ cofactor for catalysis (35). Addition of Mg2+ had little effect on the 3Dpol spectrum, other than a small chemical shift change to Met6 (data not shown), but addition of the paramagnetic ion Co2+ resulted in chemical shift changes for Met225, Met323, Met392 and Met394, with smaller chemical shift changes for Met74 and Met354 (Figure 2B). These findings are consistent with the resonance assignments of the methionines. Met225, Met323 and Met354 are all located in the palm region of 3Dpol (Supporting Information Table S1), and are relatively near the metal binding sites (12–16 Å) compared to most of the other methionines (> 25 Å). Met392 is also relatively close to the metal binding site, although the orientation of Met394 results in its β-methyl group being ~20–21 Å from the metal binding sites. Movement of Met394 or structural changes within the thumb domain may bring Met394 into closer proximity to the metal binding sites. Unfortunately, direct comparison between methionines is problematic considering that the pseudocontact shift resulting from the paramagnetic ion has an angular dependence (46, 47), which is different for each of the effected methionines.
Solvent accessibility of the methionines
We also compared the solvent accessibility of the ε-methyl groups of the methionines to the peak intensities (Figure 2D). The peak intensity generally correlated with the solvent accessibility of the methyl probe, as has been previously observed in the DNA damage recognition protein UvrB (48). This makes intuitive sense considering that more solvent exposed residues should be less motionally restricted, leading to longer transverse relaxation times (T2) and more intense peaks (48). Of course, one major caveat to this analysis is that exchange broadening can significantly reduce the peak intensities. The Met6 resonance has a much lower signal intensity than what would be expected, suggesting that Met6 might be experiencing exchange broadening, which is consistent with our R2 relaxation dispersion experiments (see below). The more solvent-exposed methionines, Met86, Met123, Met189, Met251 and Met392, also have chemical shifts closer to that of free methionine.
Mutational analysis of 3Dpol
Although the assignment of the [methyl-13C]methionine spectrum of PV 3Dpol was relatively straightforward using the methionine mutants, some of the methionine mutants led to additional spectral changes (Figure 2C, Supporting Information Tables S2–S4). Some of these spectral changes could be readily rationalized because these methionines make direct interactions or they are spatially close. For example, the backbone nitrogen of Met95 hydrogen bonds to the backbone carbonyl oxygen of Met189. The Met95Ile mutation leads to a small chemical shift change to the Met189 resonance (Supporting Information Table S3), and likewise the Met189Ile mutation leads to a small chemical shift change to the Met95 resonance (Supporting Information Table S4). Met225 and Met323 are in van der Waals contact distance. The Met225Ile mutation leads to a chemical shift change to the Met323 resonance, and likewise the Met323Ile mutation leads to a chemical shift change to the Met225 resonance (Supporting Information Table S4).
However, other spectral changes are less readily rationalized. For example, the Met299Ile mutation leads to chemical shift changes to the Met74 and Met354 resonances (Supporting Information Tables S3, S4), despite Met299 being ~12 and ~17 Å (distances between α-carbons) away from Met74 and Met354, respectively. An even more dramatic example is the Met74Ile mutation that results in a chemical shift change to the Met95 resonance, despite the α-carbons of these amino acids being ~30 Å apart.
Half of the methionine mutants (9 of 17) also show spectral changes to the Met6 and Met187 resonances. The Met74Ile, Met86Ile and Met95Ile mutants show similar chemical shift changes to Met6 and Met187 (Supporting Information Table S2). Other methionine mutants (i.e. Met141Ile, Met189Ile, Met251Ile, Met323Ile and Met354Ile) show two resonances for both Met6 and Met187 - one set of resonances overlap that of wild-type 3Dpol, whereas the other set of resonances overlap that of Met74Ile/Met86Ile/Met95Ile 3Dpol (e.g. Figure 2C, also see Supporting Information Table S2). Mutations leading to this behavior are found throughout 3Dpol, including amino acid positions in the fingers (i.e. Met74, Met86, Met95, Met141, Met189, Met251) and palm (i.e. Met323 and Met354) regions of the enzyme. Some of the mutations are near Met187 (e.g. Met95, Met189), but the mutations are all distant (> 25 Å) from Met6. Intriguingly, the methionine mutations result in similar behavior to both the Met6 and Met187 resonances, that is, whenever there is chemical shift change, or the appearance of two resonances for Met6, there is a corresponding change for Met187. Moreover, the peak ratios between the “major” and “minor” resonances of Met6 and Met187 are very similar for all the mutants that show two sets of resonances for Met6 and Met187 (Supporting Information Table S5). This suggests that Met6 and Met187 are somehow conformationally coupled despite being > 26 Å apart (α-carbon distance).
Analysis of RNA-binding and ternary complexes of wild-type 3Dpol
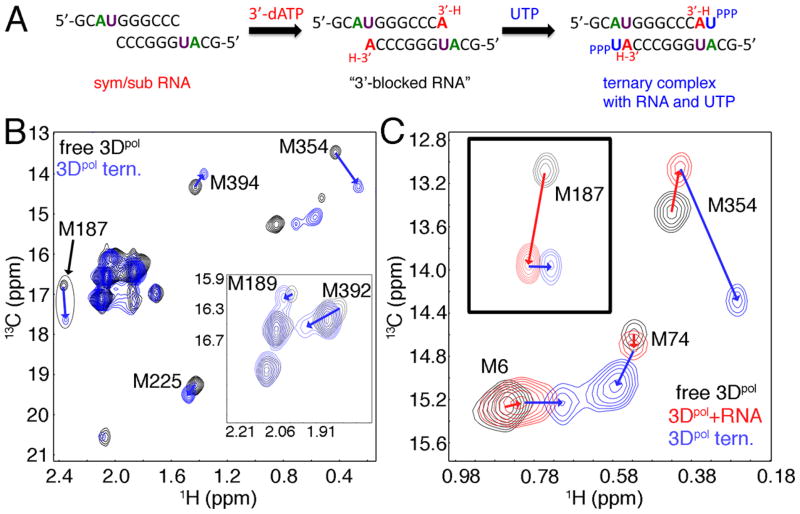
To gain more insight into the structural changes to 3Dpol that accompany RNA binding and formation of the ternary complex with both RNA and incoming nucleotide nucleotide bound, we titrated 3Dpol with the symmetrical primer/template substrate (sym/sub RNA) previously developed to study the kinetic mechanism of PV 3Dpol, and nucleotide (Figure 3). Our strategy to “capture” the ternary complex involved first adding 3′-dATP to the enzyme/RNA mixture that would become incorporated into the RNA, but then would prevent phosphodiester formation with subsequent nucleotide additions (“3-blocked RNA”) (Figure 3A). Importantly, there were no significant chemical shift differences between the enzyme bound to the original sym/sub RNA and after 3′-dATP was added for WT 3Dpol (Supporting Information Table S6, Figure S3), but there were significant chemical shift changes after the addition of the second nucleotide, UTP (Figure 3B,C, Supporting Information Table S6), suggesting that we were successful in capturing the ternary complex bound with 3′-blocked RNA and UTP.
Figure 3.
Conformational changes upon binding RNA and nucleotide to WT 3Dpol. A. Strategy to generate 3Dpol complexes bound with RNA and nucleotide. Addition of 3′-dATP to the 3Dpol/RNA reaction mixture will result in nucleotide incorporation into the sym/sub RNA, but will prevent additional phosphodiester bond formation with subsequent addition of UTP nucleotide. This will essentially “trap” the enzyme in a ternary complex bound with 3′-blocked RNA and UTP. B. 1H-13C HSQC comparison of WT 3Dpol in the unliganded state (black) and in the ternary complex with 3′-blocked RNA and UTP (blue). C. Closeup of the Met6, Met74 and Met354 resonances showing a comparison between unliganded enzyme (black), enzyme bound with sym/sub RNA (before 3′-dATP addition) (red) and enzyme bound with 3′-blocked RNA and UTP (blue). 3Dpol (245 μM) was in 2H2O-based 10 mM HEPES pH 8.0, 200 mM NaCl, 5 mM MgCl2, 10 μM ZnCl2 and 0.02% NaN3. Sym/sub RNA and UTP were at concentrations of 490 μM (duplex concentration) and 4 mM, respectively. Spectra were acquired at 293 K.
We observed chemical shift changes when the sym/sub RNA was added and additional chemical shift changes upon formation of the ternary complex, although some methionines showed chemical shift changes only for RNA addition or after ternary complex formation (Supporting Information Table S6). For example, there were chemical shift changes to the Met392 and Met394 resonances when RNA was bound, but there were not any additional chemical shift changes to these resonances once the ternary complex was formed (Supporting Information Table S6). Met392 and Met394 are both on the thumb of the enzyme and the chemical shift changes may be reporting on the closing of the active site cleft as the fingers and thumbs “wrap” around the RNA. The lack of additional chemical shift changes to these methionine resonances upon formation of the ternary complex with UTP suggests that the conformational change step prior to chemistry is not a global closing of the active-site (i.e. any closing of the active-site cleft occurs after RNA binding, but before ternary complex formation) consistent with previous suggestions for PV 3Dpol (5). In contrast, the resonance for Met225 only showed a chemical shift change once both RNA and UTP were bound (Supporting Information Table S6). This observation is consistent with rearrangements in motif A (and motif B) that are predicted to occur in order to bring the triphosphate group of the incoming nucleotide into proper alignment for phosphodiester formation.
Other methionines showed chemical shift changes both after RNA and then UTP nucleotide were added (Figure 3). The resonance for Met187 showed a large chemical shift change when RNA was added, and then a smaller chemical shift change after forming the ternary complex. Met187 thus appears to be sensitive to RNA binding, which is consistent with crystal structures of FMDV 3Dpol that show the equivalent residue to Arg188 in PV 3Dpol interacting with RNA (26). Smaller changes to this region and/or RNA binding may occur once the ternary complex has formed. Met6 and Met74 also show chemical shifts changes upon RNA binding and formation of the ternary complex. Met6 is on the N-terminal β-strand that makes hydrogen bond interactions with motifs A and B (Figure 1B), and may be reporting indirectly on structural changes in these motifs. Met74 is in the fingers region, but makes hydrogen bond interactions with Asp71 and van der Waals contacts with other residues on the α1-α2 loop that also interacts with motifs A and B (Figure 1B).
The Met354 resonance has a small chemical shift change when RNA is bound, but a much larger chemical shift change upon formation of the ternary complex. Met354 is in motif D that has been suggested to be the most dynamic structural element of 3Dpol enzymes (28). Motif D has been predicted to undergo a conformational change in order to properly orient the active-site acid Lys359 for catalysis (28). The large chemical shift change for Met354 upon formation of the ternary complex is consistent with structural rearrangements in motif D.
Solution structure comparison between WT and Gly64Ser 3Dpol
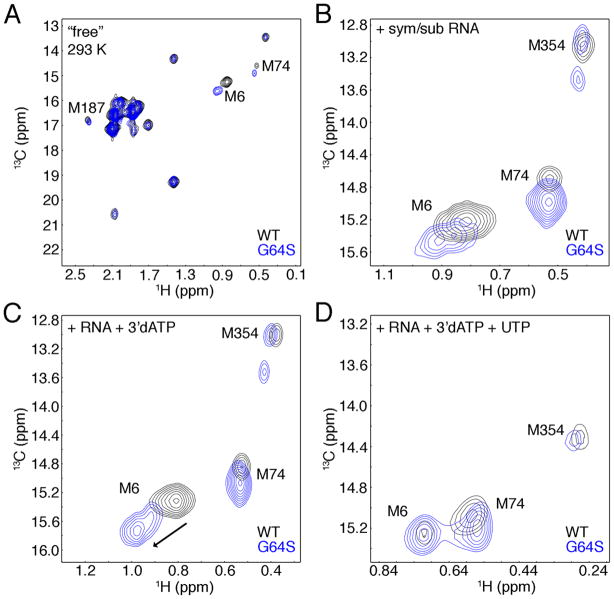
We also performed similar RNA and nucleotide binding experiments with Gly64Ser 3Dpol (Figure 4). The 1H-13C HSQCs of Gly64Ser 3Dpol were nearly identical to that of WT 3Dpol (Figure 2, Supporting Information Table S6), suggesting that the solution structures of WT and Gly64Ser 3Dpol are very similar, consistent with the finding that the crystal structures of the unliganded enzyme are nearly identical (29). The only significant chemical shift changes for the unliganded enzymes were for the Met6 and Met74 resonances (Figure 4A). Gly64 makes hydrogen bond interactions with Gly1 on the N-terminal β-strand that includes Met6 (Figure 1B). Upon mutation of Gly64 to Ser, there are additional hydrogen bond interactions with the side-chain hydroxyl of Ser64 and the carboxylate group of Glu2 (29). This may result in a small conformational change that can be transmited to Met6 through the β-strand resulting in the observed chemical shift change. However, it should be noted that the N-terminal β-strands of WT and Gly64Ser 3Dpol essentially overlap in the crystal structures (rmsd = 0.45 Å for Cα), and the orientation of the side-chain for Met6 is identical between WT and Gly64Ser 3Dpol. This suggests that the structural and/or dynamic changes for Met6 may only be apparent in solution. Likewise, as previously stated, Met74 is in contact with the α1-α2 loop containing Gly64. Mutation to Ser64 may result in structural and/or dynamic changes to the α1-α2 loop that results in chemical shift changes to Met74.
Figure 4.
Comparison of WT (black) and Gly64Ser (blue) 3Dpol in A. the unliganded form, B. bound with sym/sub RNA, C. following reaction with 3′-dATP, D. bound with 3′-blocked RNA and UTP. WT (245 μM) and Gly64Ser (200 μM) 3Dpol were in 2H2O-based 10 mM HEPES pH 8.0, 200 mM NaCl, 5 mM MgCl2, 10 μM ZnCl2 and 0.02% NaN3. Sym/sub RNA concentrations for WT (Gly64Ser) 3Dpol were 490 (200) μM (B, C) or 490 (400) μM (D). 3′-dATP and UTP concentrations were 3.1 mM and 4 mM, respectively. Spectra were acquired at 293 K.
Similar chemical shift differences in Met6 and Met74 were observed when RNA was added (Figure 4B). However, there was an additional chemical shift change to Met6 for Gly64Ser 3Dpol once 3′-dATP was added that was not observed for the WT enzyme (Figure 4C). Upon addition of UTP, the Met6 resonance then shifted such that it was essentially identical to that of the WT peak position, and there were only minor chemical shift differences for the Met74 and Met354 resonances (Figure 4D, Supporting Information Table S6). This suggests that there are no substantial structural differences between WT and Gly64Ser 3Dpol when bound to RNA and UTP in the ternary complex, at least assessed by the conformations and chemical environments of the methionine probes.
It should be noted that when 3′-dATP is added to the reaction mixture, it is in far excess (~ 3 mM) compared to protein or RNA (~0.2 – 0.4 mM). Thus, even after the enzyme catalyzes the chemical reaction to form 3′-blocked RNA, 3′-dATP is still in excess and could potentially bind to the protein-RNA complex as an ‘incorrect’ nucleotide. The 1H-13C HSQC analysis suggests that WT and Gly64Ser 3Dpol form nearly identical ternary complexes when bound with correct nucleotide (i.e. in this case, UTP) (Figure 4D) consistent with their nearly identical rates of phosphodiester formation (6), but their ternary complexes with incorrect nucleotide (i.e. with 3′-dATP) are more dissimilar (Figure 4C), at least for the N-terminal β-strand.
Dynamic comparison between the apoenzymes of WT and Gly64Ser 3Dpol
As has been suggested before, the functional differences between WT and Gly64Ser 3Dpol may also be related to the differences in the internal motions of the polymerases (29, 30). NMR provides atomic detail of protein motion over a broad range of timescales (ps-ms) (49). For example, the longitudinal (R1) and transverse (R2) relaxation rates, along with the heteronuclear nuclear Overhauser effect (heteronuclear NOE), have been used in Lipari-Szabo model-free analysis (50, 51) to yield order parameters (S2) on the ps-ns timescale. Unfortunately, model-free analysis of methyl dynamics using this strategy, especially CH3 (i.e. no deuterium), is not nearly as robust as that for 15N backbone dynamics (52). Moreover, our heteronuclear NOE results were insufficient for quantitative analysis. Instead, we focused on a qualitative comparison between WT and Gly64Ser 3Dpol to guage any differences in their internal protein dynamics that may be related to their functional differences (Figure 5).
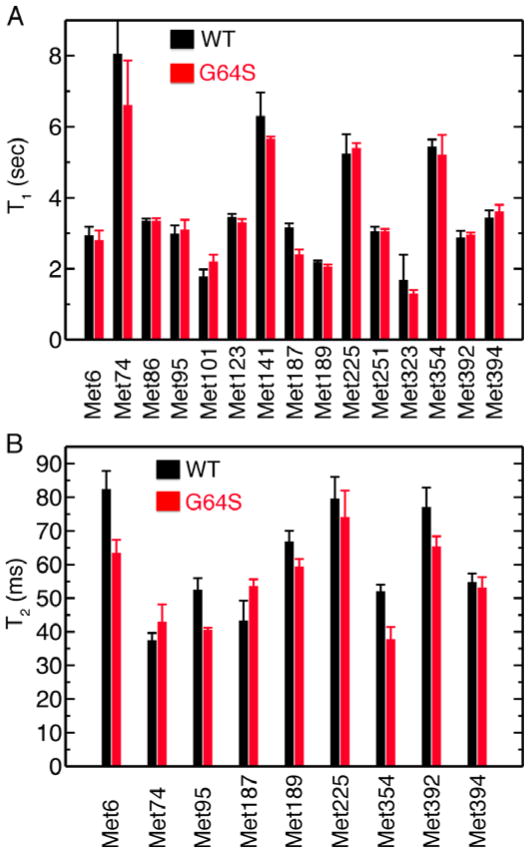
Figure 5.
Comparion of (A) T1 and (B) T2 methyl 13CH3 relaxation times for WT (black) and Gly64Ser (red) [methyl-13C]-methionine-labeled PV 3Dpol at 293 K. Protein (250 μM) was in 2H2O-based 25 mM potassium phosphate pH 8.0, 150 mM NaCl, 1mM DTT and 0.02% w/v NaN3. The error bars correspond to standard deviations taken from 3–4 separate experiments.
We observed significant differences between unliganded WT and Gly64Ser 3Dpol in the transverse (T2) relaxation times for Met6, Met95, Met354 and Met392 (Figure 4). As noted, the N-terminal β-strand containing Met6 interacts with Gly64, and thus, it is reasonable that mutation of Gly64 could result in changes to the structure and/or dynamics of Met6 and its β-strand. The remaining methionines are more distant from the site of mutation than Met6. The methionines that show differences in T2 values include residues in the fingers (Met6, Met95), palm (Met354) and thumb (Met392) domains (Supporting Information Table 1), suggesting that the effects of the Gly64Ser mutation are not localized to a single region of the protein.
We did not compare the T2 values for Met86, Met123 and Met251; these ε-methyl groups are fully solvent exposed, consistent with their T2 values being much larger (>190 ms) than the other methionines. We were also unable to compare the T2 values for Met101, Met141 and Met323; these resonances are too broadened for reliable intensity measurements.
We also attempted to measure the R2 contribution from conformational exchange (Rex) on the μs-ms timescale by employing 13C-methyl R2 relaxation dispersion experiments (41, 42). For the free enzymes, we were only able to observe Rex for Met6, Met187 and Met189. Unfortunately, due to the short T2 times for the methionines at higher field strength (1H 850 MHz), we were unable to acquire relaxation data at two field strengths that would enable us to quantitatively describe differences in terms of kinetics, thermodynamics and/or chemical shift differences between exchanging conformations. Nonetheless, we observed significant differences in Rex for Met187 and Met189 between WT (11 s−1 for Met187, 7 s−1 for Met189) and Gly64Ser 3Dpol (27 s−1 for Met187, 12 s−1 for Met189) by measurement at a single magnetic field strength (1H 600 MHz).
As described with some of the methioninine mutants, we observed either chemical shift changes to Met6 and Met187, or two sets of resonances for Met6 and Met187. Together, the results with the methionine mutants and the relaxation data suggest that there is an exchange process involving the conformations of Met6 and Met187, and it is likely that the methionine mutants disturb this conformational equilibrium. Some insight into the conformational equilibrium can be gained by comparing the spectra of methionine mutant 3Dpol (e.g. Met251Ile) with wild-type 3Dpol in the unliganded and RNA-bound states (Supporting Information Figure S4, Tables S2, S6). The second Met187 resonance for Met251Ile 3Dpol (and other methionine mutants that show this behavior) is near the Met187 resonance when WT 3Dpol is bound to RNA. This suggests that the second conformation for Met187, and the surrounding region, might be involved in the RNA binding mechanism of 3Dpol. If the structural differences are assumed to be similar between the exchanging conformations in WT and Gly64Ser 3Dpol (i.e. the chemical shift difference between the exchanging conformations are similar), the increased Rex values for Gly64Ser 3Dpol may be due to a change in the exchange kinetics or change in the relative populations of the exchanging conformers. Kinetic data comparing the rates of formation of the RNA binding complexes between WT and methionine mutant 3Dpol would likely shed more insight into this conformational exchange process and its role (if any) in RNA recognition and/or binding. These studies can also be accompanied with R2 relaxation dispersion studies with other probes (e.g. Val, Leu, Ile) that might be more amenable to this larger protein system (32), as has been performed with the bacteriophage φ6 RNA-dependent RNA polymerase (53).
Long-range interactions in the function and fidelity of PV 3Dpol
Taken together, our results with Gly64Ser 3Dpol and the methionine mutants suggest that there are long-range interaction networks operating throughout PV 3Dpol that might be important for its function. One motivation for this study was to gain more insight into the effect of the Gly64Ser mutation on the structure and/or conformational dynamics of 3Dpol. Our studies indicate that the Gly64Ser mutation not only affects the conformational environment of those methionines (Met6 and Met74) closest to the site of mutation, but it can have long range effects on the dynamics of distant amino acid residues. For example, the Gly64Ser mutation can lead to changes to the ps-ns timescale dynamics of Met95 and the μs-ms timescale dynamics of Met187 and Met189, at least in the unliganded enzyme. Close inspection of the 3Dpol structure suggests potential path(s) through which changes to Gly64 may affect Met95, Met187 and Met189. Met187 makes van der Waals contact with Ser291 on the highly conserved β10-α13 loop in motif B (Figure 6). The other end of the loop is involved in a hydrogen bond network involving the N-terminal β-strand, Gly64 and motif A (Figure 1B). The β10-α13 loop, including Ser288 and Thr293, is intimately involved in the conformational change that is predicted to reorient the triphosphates of the incoming nucleotide (19, 22, 23, 26). A change in the structure and/or dynamics of the loop would help to both explain the effect that Gly64Ser has on the kinetics of the conformational change step and the resulting effect on the dynamics of Met95, Met187 and Met189. A similar conclusion has been recently drawn from crystal structures of FMDV 3Dpol. The equivalent Gly64Ser mutation in FMDV 3Dpol (Gly62Ser) results in a structural change to the motif B loop resulting in significant changes to the RNA binding-mode (54). The authors also note extensive hydrogen bond interactions between the α1-α2 loop, the N-terminal β-strand, and motifs A and B that would be able to transmit changes from Gly64 to the active-site (54). Unfortunately, we were unable to observe a resonance for Met286 that is on the β10-α13 loop that might have revealed additional information about changes to the structure and/or dynamics of the loop upon mutation of Gly64.
Figure 6.
Transmission of long distance signals across the protein structure of PV 3Dpol. A. Mutation of Gly64 to Ser affects the structure and/or dynamics of distant amino acids, including Met95, Met187 and Met189 on the far side of the protein. B. The α1-α2 loop (orange) interacts with motif D (blue), including a hydrogen bond interaction between the sidechains of Asp71 and Tyr350. C. Met187 interacts with residues on the β10-α13 loop of motif B, including Ser291. This suggests that Met187 is an indirect probe for the structure/dynamics of the β10-α13 loop. The backbone amide of Met95 hydrogen bonds to the backbone carbonyl of Met189. These interactions, including the hydrogen bonding network outlined in Figure 1B, allows the transmission of signals from Gly64 all the way to Met95 over 30 Å away.
It is intriguing that other mutations in 3Dpol also caused changes to Met6 and Met187. As stated, although Met6 and Met187 are distant from one another, they appear to be conformationally coupled. Changes to Met6 (or Met187) may be transmitted through a similar network of interactions as suggested for Gly64Ser, considering that Met6 is on the N-terminal β-strand that is involved in the hydrogen bonding network between motifs A, B and Gly64. Mutations to methionines that are interacting with structural elements containing Met6 or Met187 may induce changes along this long-range network to affect the resonances of both Met6 and Met187. For example, the Met95Ile and Met189Ile mutations induce changes to both Met6 and Met187, likely due to their close interactions with Met187. Likewise, Met141 makes contact with residues (e.g. Leu181, Asn182, Val185) on the α-helix containing Met187. Met323 in motif C interacts with residues in the motif B helix (e.g. Asn 301, Asn302, Ile305, Arg306) that can in turn affect the motif B loop and the Met6/Met187 resonances. Met74 and Met251 make contact with residues in the α1-α2 loop (e.g. Val70), and other residues within the same α-helix as Met251 make contact with residues in the β10-α13 loop. The loop immediately preceding Met354 also makes contact with the α1-α2 loop, including a hydrogen bond interaction between the sidechains of residues Asp71 and Tyr350 (Figure 6). The chemical shift changes to Met74 and Met354 induced by the Met299Ile (Met299 being in motif B) mutation may also be explained in terms of this network of interaction.
It should be pointed out that although we can trace these networks of interaction through PV 3Dpol, there may be other interactions more important than the interactions we have identified and/or a more complex set of interactions may be responsible for these long-range responses. Moreover, some long-range interactions still remain difficult to rationalize. For example, the Gly64Ser mutation can lead to small changes to the T2 relaxation time for Met392 (Figure 5), and the Met392Ile mutation can lead to a small chemical shift change to the Met6 resonance (Supporting Information Table S3), even though Met392 is on the thumb domain of 3Dpol far from Met6 and Gly64 without an easily traceable, direct path of interaction.
Many of the methionines involved in these long-range interaction networks also show chemical shift changes upon binding RNA or formation of the ternary complex with RNA and nucleotide, including Met6, Met74, Met187, Met189, Met354 and Met392, and/or are in regions known to be important for the function and fidelity of 3Dpol, including regions important for nucleotide (motif B), magnesium (motif C i.e. Asp328 on the same α-helix as Met323) and RNA (e.g. Arg188, Ser291) binding, and catalysis (e.g. motif D). Thus, we propose that these long-distance interactions are functionally important, and may help to coordinate ligand binding and enzyme catalysis. Similar interaction networks in 3Dpol have also been suggested to be important for the regulation of polymerase activity by the viral protein 3CD and the formation of higher order replication machinery within the cell (55). Long-range interaction networks may also explain the increased thermal stability of 3Dpol in the presence of nucleotide (56). Recent NMR studies of the bacteriophage φ6 RNA-dependent RNA polymerase (53) have revealed long-range interactions operating on the μs-ms timescale important for the function of this structurally similar enzyme (53).
Structural and/or dynamic changes to this network of interactions may thus affect the function and fidelity of PV 3Dpol. In the case of Gly64Ser 3Dpol, these differences include changes to regions important for RNA binding and recognition (e.g. Met95, Met187, Met189), nucleotide recognition (Met6 insofar as it indirectly reports on structural changes in motifs A and B) and phosphodiester bond formation (Met354 in motif D that also contains the general acid Lys359). It is also intriguing that there are more significant chemical shift differences between WT and Gly64Ser 3Dpol in the ternary complex when bound to the incorrect nucleotide (i.e. E:RNA:3′-dATP) than the correct nucleotide (E:RNA:UTP). Met6 on the N-terminal β-strand especially appears to be responsive to differences between correct versus incorrect ternary complex formation. It was previously observed that mutations to Gly1 result in enzymes with significantly reduced catalytic activity (36, 45), and moreover, these mutations (e.g. Gly1Ala) can significantly affect the interactions with motifs A and B, and in particular, change the orientation of Asp238 (45) that is responsible for making hydrogen bond interactions with the incoming nucleotide to properly position the nucleotide for phosphodiester bond formation (19–21, 45). Thus, Met6 may be an indirect reporter on the structural changes in motif A. Our findings suggest that there is a conformational change when incorrect nucleotide binds to Gly64Ser 3Dpol that does not occur when incorrect nucleotide binds to WT 3Dpol, and this difference can be traced to the N-terminal β-strand and likely, motif A. For Gly64Ser 3Dpol, this may result in a decrease to the conversion rate between ‘inactive’ conformation to catalytically competent conformation when incorrect nucleotide binds compared to what occurs with WT 3Dpol, resulting in a higher fidelity polymerase. NMR studies with other probes (e.g. Val, Leu, Ile) may reveal additional structural and/or dynamic differences between WT and Gly64Ser 3Dpol and further highlight the roles of the long-range interaction network in the function and fidelity of 3Dpol.
Supplementary Material
Acknowledgments
We thank Drs. Craig E. Cameron, Ibrahim Moustafa and Eric Smidansky for their insight into polymerase function and fidelity, and for the critical reading of this manuscript. We also thank Drs. Alan Benesi and Bernie O’Hare for their expertise in the collection of the NMR data, and members of the Boehr lab for their valuable input and support.
Abbreviations Footnote
- NMR
nuclear magnetic resonance
- HSQC
heteronuclear single quantum coherence
- NOE
nuclear Overhauser effect
- TROSY
transverse relaxation optimized spectroscopy
- RNA
ribonucleic acid
- DNA
deoxyribonucleic acid
- 3Dpol
RNA-dependent RNA polymerase
- PV
poliovirus
- NTP
nucleotide triphosphate
- FMDV
foot and mouth disease viru
- rmsd
root mean square deviation
- WT
wild-type
- PMSF
phenylmethylsulfonyl fluoride
- SUMO
small ubiquitin-like modifying protein
- UTP
uracil triphosphate
- 3′-dATP
3′deoxyadenine triphosphate
- TEMED
tetramethylethylenediamine
- EDTA
ethylenediaminetetraacetic acid
- DSS
4,4-dimethyl-4-silapentane-1-sulfonic acid
Footnotes
This work was supported by a Pennsylvania State start-up fund to D.D.B. and NIH grant AI45818 to J.J. Arnold and Craig E. Cameron.
SUPPORTING INFORMATION AVAILABLE. Table S1 contains a list of all methionines, their locations on the 3Dpol structure and primers designed to generate single-site methionine mutants. Tables S2–S4 contain chemical shift data for WT and methionine mutant 3Dpol. Table S5 contains a list of relative peak intensities for the Met6 and Met187 resonances for various methionine mutant 3Dpol, Table S6 contains chemical shift data for WT and Gly64Ser 3Dpol bound to RNA and nucleotide. Figure S1 compares 3Dpol activity under various buffer conditions. Figure S2 contains titration data to determine the optimal ratio of protein and RNA to get fully complexed 3Dpol. Figure S3 compares WT and Gly64Ser 3Dpol spectra with different RNA and nucleotide additions. Figure S4 compares the NMR spectra of unliganded Met251Ile 3Dpol to WT 3Dpol in the unliganded state and in the ternary complex. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hayden FG. Update on influenza and rhinovirus infections. Adv Exp Med Biol. 1999;458:55–67. doi: 10.1007/978-1-4615-4743-3_6. [DOI] [PubMed] [Google Scholar]
- 2.Kim WR. The burden of hepatitis C in the United States. Hepatology. 2002;36:S30–34. doi: 10.1053/jhep.2002.36791. [DOI] [PubMed] [Google Scholar]
- 3.Weiss SR, Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayden FG. Respiratory viral threats. Curr Opin Infect Dis. 2006;19:169–178. doi: 10.1097/01.qco.0000216628.51563.b1. [DOI] [PubMed] [Google Scholar]
- 5.Castro C, Arnold JJ, Cameron CE. Incorporation fidelity of the viral RNA-dependent RNA polymerase: a kinetic, thermodynamic and structural perspective. Virus Res. 2005;107:141–149. doi: 10.1016/j.virusres.2004.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnold JJ, Vignuzzi M, Stone JK, Andino R, Cameron CE. Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J Biol Chem. 2005;280:25706–25716. doi: 10.1074/jbc.M503444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfeiffer JK, Kirkegaard K. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc Natl Acad Sci U S A. 2003;100:7289–7294. doi: 10.1073/pnas.1232294100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfeiffer JK, Kirkegaard K. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog. 2005;1:e11. doi: 10.1371/journal.ppat.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harki DA, Graci JD, Galarraga JE, Chain WJ, Cameron CE, Peterson BR. Synthesis and antiviral activity of 5-substituted cytidine analogues: identification of a potent inhibitor of viral RNA-dependent RNA polymerases. J Med Chem. 2006;49:6166–6169. doi: 10.1021/JM060872x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harki DA, Graci JD, Korneeva VS, Ghosh SK, Hong Z, Cameron CE, Peterson BR. Synthesis and antiviral evaluation of a mutagenic and non-hydrogen bonding ribonucleoside analogue: 1-beta-D-Ribofuranosyl-3-nitropyrrole. Biochemistry. 2002;41:9026–9033. doi: 10.1021/bi026120w. [DOI] [PubMed] [Google Scholar]
- 12.Collett MS, Neyts J, Modlin JF. A case for developing antiviral drugs against polio. Antiviral Res. 2008;79:179–187. doi: 10.1016/j.antiviral.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Crotty S, Maag D, Arnold JJ, Zhong W, Lau JY, Hong Z, Andino R, Cameron CE. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 14.Vignuzzi M, Wendt E, Andino R. Engineering attenuated virus vaccines by controlling replication fidelity. Nat Med. 2008;14:154–161. doi: 10.1038/nm1726. [DOI] [PubMed] [Google Scholar]
- 15.Ng KK, Arnold JJ, Cameron CE. Structure-function relationships among RNA-dependent RNA polymerases. Curr Top Microbiol Immunol. 2008;320:137–156. doi: 10.1007/978-3-540-75157-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joyce CM, Benkovic SJ. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 17.Beard WA, Wilson SH. Structural insights into the origins of DNA polymerase fidelity. Structure. 2003;11:489–496. doi: 10.1016/s0969-2126(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 18.O’Reilly EK, Kao CC. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology. 1998;252:287–303. doi: 10.1006/viro.1998.9463. [DOI] [PubMed] [Google Scholar]
- 19.Gohara DW, Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol): kinetic, thermodynamic, and structural analysis of ribonucleotide selection. Biochemistry. 2004;43:5149–5158. doi: 10.1021/bi035429s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gohara DW, Crotty S, Arnold JJ, Yoder JD, Andino R, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol): structural, biochemical, and biological analysis of conserved structural motifs A and B. J Biol Chem. 2000;275:25523–25532. doi: 10.1074/jbc.M002671200. [DOI] [PubMed] [Google Scholar]
- 21.Korneeva VS, Cameron CE. Structure-function relationships of the viral RNA-dependent RNA polymerase: fidelity, replication speed, and initiation mechanism determined by a residue in the ribose-binding pocket. J Biol Chem. 2007;282:16135–16145. doi: 10.1074/jbc.M610090200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+ Biochemistry. 2004;43:5126–5137. doi: 10.1021/bi035212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arnold JJ, Gohara DW, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mn2+ Biochemistry. 2004;43:5138–5148. doi: 10.1021/bi035213q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berdis AJ. Mechanisms of DNA polymerases. Chem Rev. 2009;109:2862–2879. doi: 10.1021/cr800530b. [DOI] [PubMed] [Google Scholar]
- 25.Showalter AK, Tsai MD. A reexamination of the nucleotide incorporation fidelity of DNA polymerases. Biochemistry. 2002;41:10571–10576. doi: 10.1021/bi026021i. [DOI] [PubMed] [Google Scholar]
- 26.Ferrer-Orta C, Arias A, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N. Sequential structures provide insights into the fidelity of RNA replication. Proc Natl Acad Sci U S A. 2007;104:9463–9468. doi: 10.1073/pnas.0700518104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castro C, Smidansky E, Maksimchuk KR, Arnold JJ, Korneeva VS, Gotte M, Konigsberg W, Cameron CE. Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc Natl Acad Sci U S A. 2007;104:4267–4272. doi: 10.1073/pnas.0608952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castro C, Smidansky ED, Arnold JJ, Maksimchuk KR, Moustafa I, Uchida A, Gotte M, Konigsberg W, Cameron CE. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat Struct Mol Biol. 2009;16:212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marcotte LL, Wass AB, Gohara DW, Pathak HB, Arnold JJ, Filman DJ, Cameron CE, Hogle JM. Crystal structure of poliovirus 3CD protein: virally encoded protease and precursor to the RNA-dependent RNA polymerase. J Virol. 2007;81:3583–3596. doi: 10.1128/JVI.02306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cameron CE, Moustafa IM, Arnold JJ. Dynamics: the missing link between structure and function of the viral RNA-dependent RNA polymerase? Curr Opin Struct Biol. 2009;19:768–774. doi: 10.1016/j.sbi.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sprangers R, Velyvis A, Kay LE. Solution NMR of supramolecular complexes: providing new insights into function. Nat Methods. 2007;4:697–703. doi: 10.1038/nmeth1080. [DOI] [PubMed] [Google Scholar]
- 32.Korzhnev DM, Kloiber K, Kanelis V, Tugarinov V, Kay LE. Probing slow dynamics in high molecular weight proteins by methyl-TROSY NMR spectroscopy: application to a 723-residue enzyme. J Am Chem Soc. 2004;126:3964–3973. doi: 10.1021/ja039587i. [DOI] [PubMed] [Google Scholar]
- 33.Tugarinov V, Kay LE. An isotope labeling strategy for methyl TROSY spectroscopy. J Biomol NMR. 2004;28:165–172. doi: 10.1023/B:JNMR.0000013824.93994.1f. [DOI] [PubMed] [Google Scholar]
- 34.Arnold JJ, Bernal A, Uche U, Sterner DE, Butt TR, Cameron CE, Mattern MR. Small ubiquitin-like modifying protein isopeptidase assay based on poliovirus RNA polymerase activity. Anal Biochem. 2006;350:214–221. doi: 10.1016/j.ab.2005.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnold JJ, Ghosh SK, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3D(pol)). Divalent cation modulation of primer, template, and nucleotide selection. J Biol Chem. 1999;274:37060–37069. doi: 10.1074/jbc.274.52.37060. [DOI] [PubMed] [Google Scholar]
- 36.Gohara DW, Ha CS, Kumar S, Ghosh B, Arnold JJ, Wisniewski TJ, Cameron CE. Production of “authentic” poliovirus RNA-dependent RNA polymerase (3D(pol)) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr Purif. 1999;17:128–138. doi: 10.1006/prep.1999.1100. [DOI] [PubMed] [Google Scholar]
- 37.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 38.Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3D(pol)). Assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub) J Biol Chem. 2000;275:5329–5336. doi: 10.1074/jbc.275.8.5329. [DOI] [PubMed] [Google Scholar]
- 39.Yamazaki T, Muhandiram R, Kay LE. NMR Experiments for the measurement of carbon relaxation properties in highly enriched, uniformly 13C,15N-labeled proteins: Application to 13C.alpha. carbons. J Am Chem Soc. 1994;116:8266–8278. [Google Scholar]
- 40.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 41.Skrynnikov NR, Mulder FA, Hon B, Dahlquist FW, Kay LE. Probing slow time scale dynamics at methyl-containing side chains in proteins by relaxation dispersion NMR measurements: application to methionine residues in a cavity mutant of T4 lysozyme. J Am Chem Soc. 2001;123:4556–4566. doi: 10.1021/ja004179p. [DOI] [PubMed] [Google Scholar]
- 42.Mulder FA, Hon B, Mittermaier A, Dahlquist FW, Kay LE. Slow internal dynamics in proteins: application of NMR relaxation dispersion spectroscopy to methyl groups in a cavity mutant of T4 lysozyme. J Am Chem Soc. 2002;124:1443–1451. doi: 10.1021/ja0119806. [DOI] [PubMed] [Google Scholar]
- 43.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 44.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 45.Thompson AA, Peersen OB. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 2004;23:3462–3471. doi: 10.1038/sj.emboj.7600357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bose-Basu B, DeRose EF, Kirby TW, Mueller GA, Beard WA, Wilson SH, London RE. Dynamic characterization of a DNA repair enzyme: NMR studies of [methyl-13C]methionine-labeled DNA polymerase beta. Biochemistry. 2004;43:8911–8922. doi: 10.1021/bi049641n. [DOI] [PubMed] [Google Scholar]
- 47.Emerson SD, La Mar GN. NMR determination of the orientation of the magnetic susceptibility tensor in cyanometmyoglobin: a new probe of steric tilt of bound ligand. Biochemistry. 1990;29:1556–1566. doi: 10.1021/bi00458a029. [DOI] [PubMed] [Google Scholar]
- 48.DellaVecchia MJ, Merritt WK, Peng Y, Kirby TW, DeRose EF, Mueller GA, Van Houten B, London RE. NMR analysis of [methyl-13C]methionine UvrB from Bacillus caldotenax reveals UvrB-domain 4 heterodimer formation in solution. J Mol Biol. 2007;373:282–295. doi: 10.1016/j.jmb.2007.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boehr DD, Dyson HJ, Wright PE. An NMR perspective on enzyme dynamics. Chem Rev. 2006;106:3055–3079. doi: 10.1021/cr050312q. [DOI] [PubMed] [Google Scholar]
- 50.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J Am Chem Soc. 1982;104:4546–4559. [Google Scholar]
- 51.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J Am Chem Soc. 1982;104:4559–4570. [Google Scholar]
- 52.Choy WY, Kay LE. Model selection for the interpretation of protein side chain methyl dynamics. J Biomol NMR. 2003;25:325–333. doi: 10.1023/a:1023065310430. [DOI] [PubMed] [Google Scholar]
- 53.Ren Z, Wang H, Ghose R. Dynamics on multiple timescales in the RNA-directed RNA polymerase from the cystovirus {phi}6. Nucleic Acids Res. 2010;38:5105–5108. doi: 10.1093/nar/gkq210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferrer-Orta C, Sierra M, de la Higuera I, Agudo R, Arias A, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N. Structure of foot-and-mouth disease virus mutant polymerases with reduced sensitivity to ribavirin. J Virol. 2010;84:6188–6199. doi: 10.1128/JVI.02420-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boerner JE, Lyle JM, Daijogo S, Semler BL, Schultz SC, Kirkegaard K, Richards OC. Allosteric effects of ligands and mutations on poliovirus RNA-dependent RNA polymerase. J Virol. 2005;79:7803–7811. doi: 10.1128/JVI.79.12.7803-7811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson AA, Albertini RA, Peersen OB. Stabilization of poliovirus polymerase by NTP binding and fingers-thumb interactions. J Mol Biol. 2007;366:1459–1474. doi: 10.1016/j.jmb.2006.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fraczkiewicz R, Braun W. Exact and efficicient analytical calculation of the accessible surface areas and their gradients for macromolecules. J Comp Chem. 1998;19:319–333. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.