Abstract
Autophagy is a fundamental and phylogenetically conserved self-degradation process that is characterized by the formation of double-layered vesicles (autophagosomes) around intracellular cargo for delivery to lysosomes and proteolytic degradation. The increasing significance attached to autophagy in development and disease in higher eukaryotes has placed greater importance on the validation of reliable, meaningful and quantitative assays to monitor autophagy in live cells and in vivo in the animal. To date, the detection of processed LC3B-II by western blot or fluorescence studies, together with electron microscopy for autophagosome formation, have been the mainstays for autophagy detection. However, LC3 expression levels can vary markedly between different cell types and in response to different stresses, and there is also concern that over-expression of tagged versions of LC3 to facilitate imaging and detection of autophagy interferes with the process itself. In addition, the realization that it is not sufficient to monitor static levels of autophagy but to measure ‘autophagic flux’ has driven the development of new or modified approaches to detecting autophagy. Here, we present a critical overview of current methodologies to measure autophagy in cells and in animals.
Keywords: autophagy, method, technique, analysis, process, mechanism, LC3, autophagic flux
Detection of autophagy by electron microscopy
Autophagy was first described by transmission electron microscopy (TEM) approximately 50 years ago [1-3] and TEM remains one of the most widely used and sensitive techniques to detect the presence of autophagic vesicles [4,5]. TEM has recently been used to great effect in combination with tomographical approaches to identify regions of the endoplasmic reticulum as the likely origin of autophagosomes in mammalian cells [6,7]. TEM characterizes autophagy qualitatively, as early autophagic compartments (autophagosomes) containing morphologically intact cytosol or organelles (Figure 1A), or as late, degradative autophagic structures (autolysosomes) containing partially degraded cytoplasmic as well as organelle material. However, this sub-classification of the autophagic process requires expertise and experience to reproducibly define ultrastructural features that is not always readily available and has on occasion led to erroneous identification of autophagosomes [8].
Figure 1.
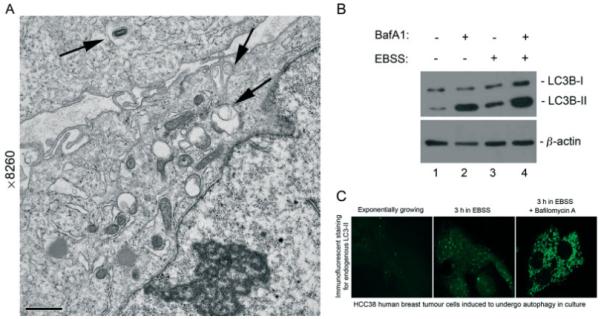
Technical approaches for measuring autophagy. (A) An example of TEM to identify autophagosomes. TEM remains an important tool for detecting autophagosomes and, while not readily quantifiable, can provide significant insight to the extent of on-going autophagy in cells. This image shows human HCC-38 breast tumour cells that have been exposed to hypoxia (1% oxygen) for 24 h, undergoing selective autophagy of mitochondria, a process referred to as mitophagy. Mitochondria are seen inside double-membrane vesicles (black arrows). (B) The use of LC3B processing to measure autophagy. Measuring processing of endogenous LC3B by western blot is one of the most commonly used approaches to detecting increased autophagy in cells. In the example shown, HCC38 breast tumour cells are starved of nutrients by growth for several hours in Earle’s balanced salt solution (EBSS) in the presence or absence of Bafilomycin A1 (lanes 3, 5). Starvation induces LC3B processing (lane 4) compared to growth in regular DMEM medium (lanes 1, 4) and treatment with Bafilomycin A1 increases the amount of processed LC3B-II detected (lanes 3, 5), indicating that these cells have a high basal rate of autophagy (lane 3) that is further increased by starvation (lane 5). (C) Formation of LC3-positive puncta as evidence of autophagy. Processed LC3 can also be detected by immunofluorescence for LC3B-II, using epitope-specific antibodies on methanol-fixed cells. In the example shown, HCC38 cells were starved by growth in EBSS in the presence or absence of Bafilomycin A1. Starvation induced increased punctate staining for LC3 that was further increased by the addition of Bafilomycin A to block turnover at the lysosome.
Perhaps the major criticism of TEM to analyse autophagy is that it is not objectively quantitative. Some efforts have been made to quantify autophagy by TEM, for example, by measuring the ratio of early to late autophagic compartments or autophagic volume as a percentage of cytoplasmic volume [9], but again this depends on the trained eye and is arguably subjective. Thus, while TEM remains an important qualitative approach to monitor steady-state levels of autophagy and to gain structural insight to the unique inter-relationship between phagophore membranes and other organelles, additional techniques are needed in conjunction with TEM to quantify steady-state levels of autophagy and autophagic flux.
Molecular assays permit a more quantitative approach to monitoring autophagy
Researchers have exploited unique molecular features of autophagosome formation and turnover to derive more quantitative and meaningful assays for measuring autophagy [5,10]. Central to these approaches has been the detection of processed LC3B-II as a cellular readout of autophagy levels [1,11-13]. LC3 is the commonly used name for microtubule-associated protein 1 light chain 3, a ubiquitin-like molecule that is the mammalian homologue of the autophagy-related Atg8 encoded product in yeast [11,14]. Following translation, the unprocessed form of LC3 (proLC3) is proteolytically cleaved by Atg4 protease, resulting in the LC3-I form with a carboxyterminal exposed glycine. Upon induction of autophagy, the exposed glycine of LC3-I is conjugated by Atg7 (an E1-like activity), Atg3 (an E2-like conjugating activity) and by Atg12-Atg5-Atg16L multimers (E3-like ligase activity) to the highly lipophilic phosphatidylethanolamine (PE) moiety to generate LC3-II [15,16]. The PE group promotes integration of LC3-II into lipid membranes at the phagophore and autophagosomes. At the autophagosome, LC3-II has been shown to play a role both in selecting cargo for degradation (eg interaction of LC3-II with p62/SQTM1 targets-associated protein aggregates for turnover) but has also been reported to promote membrane tethering and fusion in vitro [17], supporting a possible role in fusion of other membrane compartments, such as endosomes or even mitochondria with autophagosomes. To date, LC3-II is the only well-characterized protein that is specifically localized to autophagic structures throughout the process from phagophore to lysosomal degradation [16].
Based on the importance of LC3 processing for autophagosome formation and function, antibodies to LC3-I and LC3-II are widely used in western blotting techniques to monitor autophagy [12,13,18]. LC3-I and LC3-II can be readily distinguished based on their differential mobility in SDS–PAGE (Figure 1B). Despite increased molecular weight than LC3-I, LC3-II migrates more rapidly in SDS–PAGE compared to LC3-I, likely due to higher hydrophobicity associated with the PE group [13]. Technically, western blotting for LC3-I and LC3-II is straightforward, with reliable antibodies now available from various commercials sources, including Cell Signalling Technologies (http://www.cellsignal.com/products/2775.html) and Novus (http://www.novusbio.com/cart/products/NB100-2331). It should be noted that LC3 is expressed as three isoforms in mammalian cells, LC3A, LC3B and LC3C, but only LC3B-II correlates with increased levels of autophagic vesicles, and therefore it is recommended to use anti-LC3B antibodies for analysis. Additional aspects to successful blotting for LC3-I and -II include the use of PVDF membrane as opposed to nitrocellulose and extraction in a detergent based buffer (Barth and Macleod, unpublished).
Perhaps the greatest controversy has surrounded how LC3 western blot data is interpreted [18]. Previously, the western blot signal ratio between LC3-I and LC3-II was used to determine changes in the extent of autophagy. However, due to differential affinities of antibodies for LC3-I compared to LC3-II, as well as different expression levels of LC3-I and LC3-II, depending on cell line and tissue, this approach gave numerous false-positive or false-negative results. Additionally, LC3-II itself is subject to autophagic degradation at the lysosome. Thus, a consensus has emerged whereby overall levels of LC3-II are normalized to a loading control, such as β-actin or α-tubulin [19].
Increased LC3-II levels can be associated with either enhanced autophagosome synthesis or reduced autophagosome turnover, perhaps due to delayed trafficking to the lysosomes, reduced fusion between compartments or impaired lysosomal proteolytic activity. To better interpret changes in levels of processed LC3-II, it is now de rigeur in the field to perform western blotting on control extracts harvested from cells treated with inhibitors, such as Bafilomycin A1 (Figure 1B), hydroxychloroquine or pepstatin A/E64d, that inhibit degradation of autolysosome content by inhibiting the Na+H+ pump at the lysosome, increasing lysomal pH and inhibiting acidic lysosomal proteases, respectively [10]. In the presence of such inhibitors, accumulation of LC3-II-positive autophagosomes would be evidence of efficient autophagic flux, while failure of LC3-II protein to increase in the presence of such inhibitors, would indicate a defect or delay earlier in the process, prior to degradation at the autolysosome. However, it is important to use such inhibitors appropriately, since their activity becomes non-specific and influences protein turnover at the proteasome, as well as at the autolysosome, when used at too high a concentration or for extended periods of time [20].
In addition to the ubiquitin-like conjugation system (Atg7, Atg3, Atg16L) involved in processing LC3B in autophagic cells, a second ubiquitin-like conjugation system (Atg7, Atg12, Atg10) is involved in linking Atg12 to cleaved Atg5 to generate Atg5–Atg12, that complexes with Atg16 and associates with the growing phagophore [15,16]. However, the conjugation of Atg12 to Atg5 is not dependent on induction of autophagy, although it is essential for it, and Atg5–Atg12 dissociates from the autophagosome, making it an unsuitable molecular read-out of autophagy. Similarly, other proteins that are critical for autophagy are not commonly used to measure changes in autophagy, since their expression (eg induction of Beclin-1) or activity (eg the cysteine protease activity of Atg4) is not a unique readout of elevated autophagy.
Other adaptor molecules implicated in activating autophagy or in targeting cargo to autophagosomes are increasingly being used to measure selective autophagy [1]. For example, changes in p62 (SQSTM1/sequestosome 1) protein levels are used to indicate a defect in the turnover of poly-ubiquitinated protein aggregates [21]. p62/SQSTM1 interacts with poly-ubiquitinated protein aggregates through a ubiquitin-binding domain and with LC3 through its LC3-binding domain, thereby targeting these aggregates for degradation at the autolysosome [22]. P62-associated protein aggregates accumulated in Atg7/autophagy-deficient mouse liver and targeted deletion of p62 prevented the accumulation of these protein aggregates, suggesting that p62 accumulation is a good measure of defects in selective autophagy of ubiquitinated aggregates [23,24]. However, p62 is regulated at the transcriptional level by oxidative stress and by the Ras oncogene, and also feeds back to regulate NF-κB activity, so again, additional methods, such as measurement of LC3-II levels, will be required to validate changes in protein aggregate turnover by autophagy [25].
Selective clearance of mitochondria by autophagy is a process referred to as mitophagy [26]. Mitophagy is an integral part of reticulocyte maturation and defects in autophagy can lead to anaemia [27-29]. Mitophagy is also induced as an adaptive response to hypoxia, allowing the cell to reduce its mitochondrial mass to limit production of damaging reactive oxygen species from the mitochondria at a time when oxygen is not available to accept electrons from the respiratory chain [30,31]. Changes in levels of mitochondrial proteins (TOM-20, ANT, HSP60, COX4-IV, etc.) are often used to examine clearance of mitochondria by selective autophagy. Such protein analysis should also be supported by quantification of mitochondrial genome: nuclear genome ratio or by flow cytometry for mitochondrial dyes, and again it is important to verify that any changes in mitochondrial mass are indeed due to autophagy (eg by knocking down Beclin-1 or some other critical autophagy regulator) and not due to reduced mitochondrial biogenesis.
Recent evidence suggests that autophagosomes can form from late endosomes and trans-Golgi in cells that are deficient for Atg5 or Atg7 [32], and that this appears to be partly involved in eliminating mitochondria from reticulocytes [32,33]. However, Atg5/Atg7-independent autophagy requires Beclin1, suggesting that knockdown of Beclin1 may be a more generally robust approach to complete inhibition of macroautophagy, and to determining the importance of autophagy in an experimental system, than knockdown of either Atg5 or Atg7.
A brief comment on the use of chemical modulators of autophagy
The use of chemical modulators to assess autophagy in cells has been widely reported in the literature over the years, and while this may be valid when used in conjunction with genetic approaches, their use is now most commonly restricted to control experiments or, at the very least, comes with a lengthy description of the caveats involved. For example, one commonly used inhibitor of autophagy, 3-methyl adenine, clearly has additional effects on cell viability that cannot be fully explained in terms of current knowledge of autophagy. This is also true of other PI3K inhibitors, such as wortmannin (Figure 2). The lysosomotropic drug, chloroquine and the vacuolar ATPase inhibitor, Bafilomycin A1, are very useful to assess autophagic flux in cells but only if used at low doses and for short incubation periods (<4 h), since after this time Bafilomycin-A1, for example, will also inhibit the proteasome, endocytic trafficking and other cellular processes [20]. Similar criticisms pertaining to non-specific effects can also be levelled at agents that promote autophagy, such as rapamycin, lithium chloride, thapsigargin, tunicamycin, etc. For example, the use of rapamycin to induce autophagy (through inhibition of TORC1) can have differing effects, depending on cell type and relative activity of TORC2 and other signalling molecules in this pathway. Treatment of cells with lithium chloride will robustly induce autophagy by inhibiting inositol monophosphatase and increasing cellular levels of PI3P [34] but again is likely to have widespread effects on cell signalling, given that it also impacts Akt and Tor signalling. In short, such chemical approaches are useful but should be supported where possible by genetic approaches (Figure 2).
Figure 2.
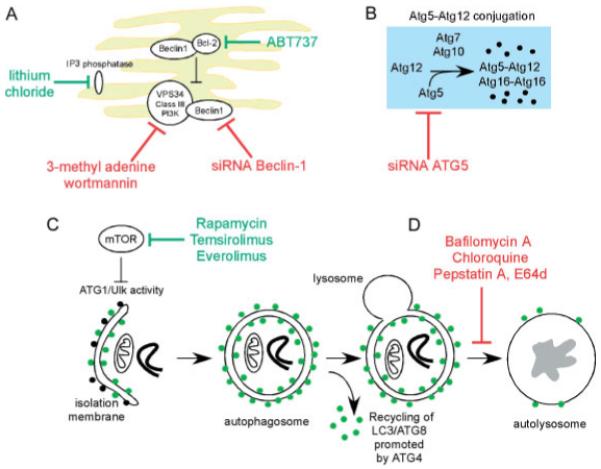
Diagram summarizing approaches to measure autophagy at different points during the autophagic process. Different agents and approaches can be used to induce or to block different steps in the autophagic process, from phagophore formation to lysosomal protease activity. (A) The activity of the Vps34-Beclin-1 complex at the ER is a focal point for artificial modulation of autophagy, with knockdown or knockout of Beclin1 being one of the most common genetic approaches to inhibiting autophagy. Treatment of cells with 3-methyl adenine or wortmannin, which inhibits the activity of Class III PI3 kinases such as Vps34, is a common chemical approach to inhibiting autophagy, although these drugs also likely have other off-target effects in the cell that can make interpretation of data difficult at times. Autophagy can be artificially induced in autophagy-competent cells by treatment with agents such as lithium chloride (which inhibits inositol phosphatase) that promote the levels of PI3P, the product of Vps34 activity and which is required for the recruitment of key factors to the expanding phagophore. Other agents, such as thapsigargin, act by inducing ER stress and mimicking physiological stress. Targeted peptides, such as ABT737, which block the interaction of Beclin-1 with Bcl-2 and other BH3-containing molecules, have also been validated as promoting autophagy [63]. (B) Akin to knockdown of Beclin-1, effective inhibition of autophagy may be achieved through knockdown of Atg5 [64]. With the discovery that there are Atg5-independent forms of autophagy, however, knockdown of Beclin-1 may be the preferred approach. (C) Rapamycin has been widely used to experimentally induce autophagy through its ability to block the inhibitory action of mTOR on Atg1 and autophagy. More efficacious derivatives of rapamycin, known as ‘rapalogues’ are in clinical trials for cancer therapy. Such rapalogues include temsirolimus and everolimus. (D) Various agents are commonly used to inhibit lysosomal turnover of autophagosome content, such as Bafilomycin A1, which inhibits the lysosomal Na+H+ ATPase, and chloroquine, which increases the pH of the lysosome, thereby preventing the activity of lysosomal acid proteases and causing autophagosomes to accumulate. Similar effects are induced by treatment with specific inhibitors of lysosomal proteases, such as pepstatin A or E64d. The increased accumulation of autophagosomes under conditions in which lysosomal proteases are inhibited is then used to assess the rate of autophagic flux in response to specific stresses.
Monitoring autophagy by fluorescence microscopy
In addition to electron microscopy, earlier studies of autophagy relied heavily on cell staining and fluorescent microscopy. In particular, the over-expression of GFP–LC3, in which GFP (green fluorescent protein) is expressed as a fusion protein at the amino terminus of LC3, was widely used to measure autophagy [35]. These studies were limited, however, by several issues: (a) counting GFP-positive punctate structures in order to quantify relative levels of autophagy is a laborious and arguably subjective task, despite the use of computer software; (b) over-expressed GFP–LC3 can be incorporated into protein aggregates independent of autophagy; (c) transfection procedures used to introduce exogenous GFP–LC3 has been shown to induce autophagy; (d) GFP–LC3 is sensitive to acid pH and ceases to fluoresce once autophagosomes fuse with the lysosome, resulting in the inability to look at end-stages of autophagy [36,37]. For these reasons, the detection of the endogenous LC3B-II protein by immunofluorescence is preferred (Figure 1C). As with measurement of processed LC3B-II by western blot, this should be accompanied by controls that show levels of punctate LC3-positive autophagosomes in cells are dependent on critical autophagy regulators, such as Beclin-1, and accumulate following treatment with agents that block LC3 degradation and autophagosome turnover at the lysosome (eg Bafilomycin A1).
A novel tandem fluorescent tagged LC3 (tfLC3) expression vector has been developed that addresses the problem of GFP sensitivity to lysosomal proteases and allows analysis of late autolysosomes. In this vector, LC3 is fused to both GFP and RFP (red fluorescent protein), the latter being resistant to lysosomal proteolytic degradation. Thus, when RFP–GFP–LC3 is expressed in cells undergoing autophagy, at first both GFP and RFP are detected and autophagosomes appear as yellow puncta. Once the autophagosome fuses with the lysosome and matures to an autolysosome, GFP staining is lost and these structures now stain as red only [37]. This pattern of staining indicates a functional autophagic maturation process and thus changes in the dynamics of the yellow to red switch likely indicate altered autophagy. However, expression of tfLC3 in cells produces a background level of red fluorescence, probably due to basal autophagy and, as emphasized above, appropriate negative and positive controls should be included in the experiment.
WIPI-1, the human orthologue of Atg18 in yeast, binds PI3P (phosphatidylinositol 3-phosphate) and is required for recruitment of critical autophgy regulators to the phagophore. WIPI-1 also co-localizes with LC3-positive membrane structures in a manner that can be blocked by treatment with phosphoinositide 3-kinase inhibitors (eg wortmannin or LY294 002) or by expressing a mutant form of WIPI-1 that cannot bind PIP3 [38]. Thus, WIPI-1-positive puncta may also be used in conjunction with LC3B-II staining, and the aforementioned controls, as a functional readout of autophagy.
Monitoring autophagic flux
As discussed, measuring autophagy in a meaningful way requires an analysis of the rate of autophagic flux (the rate at which material is cleared from the cell by autophagy), as opposed to a snapshot look at autophagy at any one static point during that process from phagophore to autolysosome. One key approach to assess autophagic flux is to measure the rate of turnover of long-lived proteins that are normally turned over by autophagy [39]. This is performed by labelling intracellular proteins with either [14C]-leucine or [14C]-valine, followed by a long cold chase period and the time-dependent release of acid-soluble radioactivity is then measured by liquid scintillation counting. The rate of degradation is determined by calculating the ratio of acid-soluble radioactivity to the acid-precipitable cell fraction.
Additional assays that avoid the use of radioactivity have also been developed. For example, starvation-dependent accumulation of the 32 kDa form of betaine homocysteine methyltransferase (BHMT), which is inhibited by wortmannin and induced by rapamycin, has been shown to be a specific readout of autophagy in the liver [40]. However, BHMT is only expressed in the liver and kidney and, thus, a modified version of BHMT (GST-BHMT) has been developed that can be over-expressed in non-BHMT-expressing cells to measure autophagic flux [41,42]. Intracellular lactate dehydrogenase (LDH) is also specifically sequestered by autophagosomes and subsequently degraded in lysosomes [43]. Thus, LDH enzyme activity detected in isolated cellular membrane fractions can be used to quantify autophagy in a temporal manner. Additional autophagy substrates that may be used in such assays include transfected neomycin phosphotransferase and fatty acid synthase [44,45].
Recently, a luciferase-reporter based assay for protein aggregate degradation by autophagy was described. Firefly luciferase was fused at its aminoterminal end to polyglutamine repeats (either 19 or 80 repeats) to generate polyQ19 or polyQ80-luciferase [46]. PolyQ80-luciferase forms protein aggregates when expressed in cells that are specifically degraded by autophagy, whereas polyQ19-luciferase is soluble in the cytosol and thereby functions as a normalization control. Changes in the ratio of polyQ80-luciferase to polyQ19-luciferase were then reported to monitor autophagic flux in vivo and in vitro.
New assays similar to these discussed briefly here are being developed constantly, reflecting the increased need to measure autophagic flux in a reliable and quantitative manner. The journal Autophagy (www.landesbioscience.com/journals/autophagy/) has a regular ‘Toolbox’ section that discusses new approaches to measuring autophagy and also now has a blog site for experimentalists to ask technical questions of the readership.
Measuring autophagy in vivo
The increased interest in autophagy research is driven in part by the realization that the successful execution of many important physiological functions, such as reticulocyte maturation, antigen presentation, turnover of protein aggregates and elimination of pathogenic bacteria, is dependent on autophagy. Furthermore, mutation or inactivation of key autophagy genes has been linked to major human disease conditions, including neurodegenerative diseases, inflammatory bowel disease, cardiomyopathies and cancer [1]. Genetic targeting of key autophagy regulators in the mouse germline has further highlighted the importance of autophagy for normal mammalian development and in disease processes [1]. For example, Atg5-deficient mice were born, but died within 1 day of birth due to a dependence on autophagy of the heart and diaphragm for nutrients and energy during the immediate postnatal period [47]. Similar phenotypes were observed for mice lacking Atg7 or Atg3 [48]. Tissue-specific knockout of Atg5 or Atg7 in brain led to the early onset of neurodegeneration and progressive deficits in motor function [49,50], while loss of Beclin 1 (Atg6) resulted in early embryonic lethality characterized by widespread cell death, abnormal ectodermal layer with reduced cavitations and reduced embryo size [48,51]. The development of new genetically engineered mouse models targeting other autophagy regulators will undoubtedly continue to provide important information about the role of autophagy in vivo.
However, the generation of these mouse models does not just confirm the significance of autophagy for development but also provides important tools to further investigate the molecular mechanisms of autophagy in vivo and their relevance for disease aetiology and treatment. As with approaches to monitor autophagy in vitro, there are caveats attached to some of the current approaches to study autophagy in vivo, as discussed further below.
Analysis of autophagy in mouse models was initially driven by electron microscopy of fixed tissues or western blot for LC3 processing in tissue extracts. The development of a transgenic mouse line in which the autophagy indicator GFP–LC3 was expressed under the control of a strong ubiquitous promoter containing the cytomegalovirus immediate-early enhancer and the chicken β-actin promoter (CAG promoter) has provided a particularly useful tool to examine autophagy in mouse models [47,52,53]. GFP–LC3B transgenic mice develop normally and importantly exhibit punctate GFP–LC3 staining by fluorescence microscopy in key tissues undergoing autophagy, for example in the heart of perinatal mice, and importantly this staining pattern was has been shown to be dependent on functional autophagy [47]. Similar transgenic mouse lines expressing mCherry have now also been made and, like RFP-LC3 in cell culture, this line will likely have the benefit of allowing detection of late-stage autolysosomes that cannot be detected with GFP–LC3 [54]. Additional methods that can be used in conjunction with mouse genetics as positive and negative controls, respectively, include treating mice with rapamycin to induce autophagy or with hydroxychloroquine to inhibit autophagic flux.
In the absence of GFP–LC3 transgenic mice or other such indicator lines, one approach that can be useful is the in vivo labelling of autophagosomes with monodansylcadaverine (MDC), a compound that autofluoresces and specifically integrates into the lipid membranes of autophagosomes and autolysosomes [55]. Overlap between MDC-positive structure and punctate GFP–LC3 in tissues from mice injected with this compound has been used to validate the use of MDC and to examine levels of autophagy [54,56].
The role of autophagy in preventing human diseases [1], including cancer, neurodegenerative disorders and inflammatory bowel disease, has also made it important to examine autophagy in situ in human tissue sections. Immunohistochemical staining for levels of autophagy is now possible following the generation of an epitope specific antibody to processed LC3B (Novus NBP1-19 167) that can be used to detect autophagosome formation in tissue sections. Expression of Beclin-1 and other autophagy regulators has also been detected in situ by immunohistochemical staining, where its expression in colon tumours and hepatocellular carcinoma is prognostic of increased disease-free, long-term survival [57,58]. While PCR-based approaches have been used to measure the mRNA levels of autophagy regulators, such as Beclin-1, ATG1, DRAM and LC3 [59], which are transcriptionally up-regulated by important factors such as p53, E2F-1 and NF-κB [60-62], this approach has limited diagnostic value as a measure of autophagy, given the extent to which autophagy is regulated at the post-translational level, although may be informative if also supported by examination of protein levels and autophagosome formation in situ.
Conclusions
Given that autophagy is a highly dynamic and complex process that is tightly regulated at multiple steps, researchers need to carefully choose relevant methods and controls to assess autophagy and be aware of potential caveats in how such data is interpreted. More importantly, researchers should make use of several approaches (chemical and genetic) before making final conclusions about how autophagy is deregulated or functioning in their system. Overall, assays to detect endogenous LC3 processing in the presence or absence of inhibitors of lysosomal turnover of autophagosome content, complemented by analysis of effects of knockdown or knockout of autophagy regulators, such as Beclin-1, remains undoubtedly one of the most straight-forward and reliable assays to quantify autophagic flux in cell systems and in tissues. However, there are new methods being developed constantly to more reliably quantify autophagic flux, and the reader is advised to keep up with these advances since, as with most experimental approaches, there remain important caveats with the major assays most commonly in use and how such assays are interpreted.
Teaching materials.
PowerPoint slides of the figures from this review are supplied as supporting information in the online version of this article.
Acknowledgment
The authors acknowledge financial support from the National Cancer Institute (Grant No. RO1 CA131188; to KFM) and the Swiss National Foundation (Award No. PBZHP3-123296; to SB).
Footnotes
No conflicts of interest were declared.
References
- 1.Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010 doi: 10.1002/path.2697. DOI: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deter RL, De Duve C. Influence of glucagon, an inducer of cellular autophagy, on some physical properties of rat liver lysosomes. J Cell Biol. 1967;33:437–449. doi: 10.1083/jcb.33.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eskelinen EL. Fine structure of the autophagosome. Methods Mol Biol. 2008;445:11–28. doi: 10.1007/978-1-59745-157-4_2. [DOI] [PubMed] [Google Scholar]
- 5.Klionsky DJ, Agostinis P, Agrawal DK, Bamber BA, Bassham DC, Bergamini E, et al. Guidelines for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 7.Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 8.Eskelinen EL. To be or not to be? Examples of incorrect identification of autophagic compartments in conventional transmission electron microscopy of mammalian cells. Autophagy. 2008;4:257–260. doi: 10.4161/auto.5179. [DOI] [PubMed] [Google Scholar]
- 9.Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. Monitoring autophagy by electron microscopy in mammalian cells. Methods Enzymol. 2009;452:143–164. doi: 10.1016/S0076-6879(08)03610-0. [DOI] [PubMed] [Google Scholar]
- 10.Rubinsztein DC, Cuervo AM, Ravikumar B, Sarkar S, Korolchuk V, Kaushik S, et al. In search of an ‘autophagometer’. Autophagy. 2009;5:1–5. doi: 10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 11.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karim MR, Kanazawa T, Daigaku Y, Fujimura S, Miotto G, Kadowaki M. Cytosolic LC3 ratio as a sensitive index of macroautophagy in isolated rat hepatocytes and H4-II-E cells. Autophagy. 2007;3:553–560. doi: 10.4161/auto.4615. [DOI] [PubMed] [Google Scholar]
- 13.Tanida I, Ueno T, Kominami E. LC3 and Autophagy. In: Deretic V, editor. Methods in Molecular Biology: Autophagosome and Phagosome. Humana; Totowa, NJ: 2008. pp. 77–88. [DOI] [PubMed] [Google Scholar]
- 14.He H, Dang Y, Dai F, Guo Z, Wu J, She X, et al. Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J Biol Chem. 2003;278:29278–29287. doi: 10.1074/jbc.M303800200. [DOI] [PubMed] [Google Scholar]
- 15.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 16.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 17.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediated membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 18.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 19.Kimura S, Fujita N, Noda T, Yoshimori T. Monitoring autophagy in mammalian cultured cells through the dynamics of LC3. Methods Enzymol. 2009;452:1–12. doi: 10.1016/S0076-6879(08)03601-X. [DOI] [PubMed] [Google Scholar]
- 20.Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy. 2008;4:849–950. doi: 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- 21.Bjorkoy G, Lamark T, Pankiv S, Overvatn A, Brech A, Johansen T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 2009;452:181–197. doi: 10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- 22.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24121–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 23.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 24.Waguri S, Komatsu M. Biochemical and morphological detection of inclusion bodies in autophagy-deficient mice. Methods Enzymol. 2009;453:181–196. doi: 10.1016/S0076-6879(08)04009-3. [DOI] [PubMed] [Google Scholar]
- 25.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tolkovsky AM. Mitophagy. Biochim Biophys Acta. 2009;1793:1508–1515. doi: 10.1016/j.bbamcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 27.Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandoval H, Thiagarajan P, Dasgupta SK, Scumacker A, Prchal JT, Chen M, et al. Essential role for Nix in autophagic maturation of red cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, et al. Mitochondrial autophagy is a HIF-1 dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Tracy K, Macleod KF. Regulation of mitochondrial integrity, autophagy and cell survival by BNIP3. Autophagy. 2007;3:616–619. doi: 10.4161/auto.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–659. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Randall MS, Loyd MR, Dorsey FC, Kundu M, Cleveland JL, et al. Mitochondrial clearance is regulated by Atg7-dependent and -independent mechanisms during reticulocyte maturation. Blood. 2009;114:157–164. doi: 10.1182/blood-2008-04-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170:1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kadowaki M, Karim MR. Cytosolic LC3 ratio as a quantitative index of macroautophagy. Methods Enzymol. 2009;452:199–213. doi: 10.1016/S0076-6879(08)03613-6. [DOI] [PubMed] [Google Scholar]
- 36.Kuma A, Matsui M, Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 2007;3:323–328. doi: 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- 37.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 38.Proikas-Cezanne T, Ruckerbauer S, Stierhof YD, Berg C, Nordheim A. Human WIPI-1 puncta-formation: a novel assay to assess mammalian autophagy. FEBS Lett. 2007;581:3396–3404. doi: 10.1016/j.febslet.2007.06.040. [DOI] [PubMed] [Google Scholar]
- 39.Bauvy C, Meijer AJ, Codogno P. Assaying of autophagic protein degradation. Methods Enzymol. 2009;452:47–61. doi: 10.1016/S0076-6879(08)03604-5. [DOI] [PubMed] [Google Scholar]
- 40.Ueno T, Ishidoh K, Mineki R, Tanida I, Murayama K, Kadowaki M, et al. Autolysosomal membrane-associated betaine homocysteine methyltransferase. Limited degradation fragment of a sequestered cytosolic enzyme monitoring autophagy. J Biol Chem. 1999;274:15222–15229. doi: 10.1074/jbc.274.21.15222. [DOI] [PubMed] [Google Scholar]
- 41.Mercer CA, Kaliappan A, Dennis PB. Macroautophagy-dependent, intralysosomal cleavage of a betaine homocysteine methyltransferase fusion protein requires stable multimerization. Autophagy. 2008;4:185–194. doi: 10.4161/auto.5275. [DOI] [PubMed] [Google Scholar]
- 42.Dennis PB, Mercer CA. The GST-BHMT assay and related assays for autophagy. Methods Enzymol. 2009;452:97–118. doi: 10.1016/S0076-6879(08)03607-0. [DOI] [PubMed] [Google Scholar]
- 43.Kopitz J, Kisen GO, Gordon PB, Bohley P, Seglen PO. Nonselective autophagy of cytosolic enzymes by isolated rat hepatocytes. J Cell Biol. 1990;111:941–953. doi: 10.1083/jcb.111.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nimmerjahn F, Milosevic S, Behrends U, Jaffee EM, Pardoll DM, Bornkamm GW, et al. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur J Immunol. 2003;33:1250–1259. doi: 10.1002/eji.200323730. [DOI] [PubMed] [Google Scholar]
- 45.Egner R, Thumm M, Straub M, Simeon A, Schuller HJ, Wolf DH. Tracing intracellular proteolytic pathways. Proteolysis of fatty acid synthase and other cytoplasmic proteins in the yeast Saccharomyces cerevisiae. J Biol Chem. 1993;268:27269–27276. [PubMed] [Google Scholar]
- 46.Ju JS, Miller SE, Jackson E, Cadwell K, Piwnica-Worms D, Weihl CC. Quantitation of selective autophagic protein aggregate degradation in vitro and in vivo using luciferase reporters. Autophagy. 2009;5:511–519. doi: 10.4161/auto.5.4.7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 48.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell. 2008;15:344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 50.Komatsu M, Waguri S, Chiba T, Murata S, Iwata S, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 51.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 52.Mizushima N, Kuma A. Autophagosomes in GFP-LC3 transgenic mice. Methods Mol Biol. 2008;445:119–124. doi: 10.1007/978-1-59745-157-4_7. [DOI] [PubMed] [Google Scholar]
- 53.Kuma A, Mizushima N. Chromosomal mapping of the GFP-LC3 transgene in GFP-LC3 mice. Autophagy. 2008;4:61–62. doi: 10.4161/auto.4846. [DOI] [PubMed] [Google Scholar]
- 54.Perry CN, Kyoi S, Hariharan N, Takagi H, Sadoshima J, Gottlieb RA. Novel methods for measuring cardiac autophagy in vivo. Methods Enzymol. 2009;453:325–342. doi: 10.1016/S0076-6879(08)04016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Munafo DB, Colombo MI. A novel assay to study autophagy: regulation of autophagosome vacuole size by amino acid deprivation. J Cell Sci. 2001;114:3619–3629. doi: 10.1242/jcs.114.20.3619. [DOI] [PubMed] [Google Scholar]
- 56.Vazquez CL, Colombo MI. Assays to assess autophagy induction and fusion of autophagic vacuoles with a degradative compartment, using monodansylcadaverine (MDC) and DQ-BSA. Methods Enzymol. 2009;452:85–95. doi: 10.1016/S0076-6879(08)03606-9. [DOI] [PubMed] [Google Scholar]
- 57.Ding ZB, Shi YH, Zhou J, Qiu SJ, Xu Y, Dai Z, et al. Association of autophagy defects with a malignant phenotype and poor prognosis of hepatocellular carcinoma. Cancer Res. 2008;68:9167–9175. doi: 10.1158/0008-5472.CAN-08-1573. [DOI] [PubMed] [Google Scholar]
- 58.Li B, Li CY, Peng RQ, Wu XJ, Wang HY, Wan DS, et al. The expression of beclin1 is associated with favorable prognosis in stage IIIB colon cancers. Autophagy. 2009;5:303–306. doi: 10.4161/auto.5.3.7491. [DOI] [PubMed] [Google Scholar]
- 59.Miracco C, Cevenini G, Franchi A, Luzi P, Cosci E, Mourmouras V, et al. Beclin1 and LC3 autophagic gene expression in cutaneous melanocytic lesions. Hum Pathol. 2009 doi: 10.1016/j.humpath.2009.09.004. ePub ahead of print; DOI: 10.1016/j.humpath.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 60.Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison PR, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 61.Polager S, Ofir M, Ginsberg D. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene. 2008;27:4860–4864. doi: 10.1038/onc.2008.117. [DOI] [PubMed] [Google Scholar]
- 62.Copetti T, Bertoli C, Dalla E, Demarchi F, Schneider C. p65/RelA modulates BECN1 transcription and autophagy. Mol Cell Biol. 2009;29:2594–2608. doi: 10.1128/MCB.01396-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, et al. Functional and physical interaction between Bcl-XL and a BH3-like domain in Beclin1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lum JJ, Bauer DE, Kong M, Harris MH, Li CY, Lindsten T, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–249. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]


