Abstract
We have developed a novel biological semiconductor (BSC) based on electrical percolation through a multi-layer 3-D carbon nanotube-antibody network, which can measure biological interactions directly and electronically. In Electrical Percolation, the passage of current through the conductive network is dependent upon the continuity of the network. Molecular interactions, such as binding of antigens to the antibodies, disrupt the network continuity causing increased resistance of the network. A BSC is fabricated by immobilizing a pre-functionalized single-walled carbon nanotubes (SWNTs)-antibody complex directly on a Poly(methyl methacrylate) (PMMA) surface (also known as plexi-glass or Acrylic). We used the BSC for direct (label-free) electronic measurements of antibody-antigen binding, showing that, at slightly above the electrical percolation threshold of the network, binding of a specific antigen dramatically increases the electrical resistance. Using anti-Staphylococcal enterotoxin B (SEB) IgG as a “gate” and SEB as an “actuator”, we demonstrated that the BSC was able to detect SEB at concentrations of 1 ng/ml. The new BSCs may permit assembly of multiple sensors on the same chip to create “Biological Central Processing Units (CPUs)” with multiple biological elements, capable of processing and sorting out information on multiple analytes simultaneously.
Keywords: biosensor, semiconductor, carbon nanotubes, electrical percolation, antibody
Introduction
Biological semiconductors (BSC) are new electronic components that change their conductivity upon biological interactions such as protein-protein interactions, DNA-protein binding, DNA-DNA annealing, and hormone-receptor binding. The ability to measure such biological interactions directly and electronically has tremendous scientific, medical and industrial importance. One of the numerous possible applications of BSC technologies is for biodetection.
Nanomaterials are increasingly being adapted for direct sensing. For example, field effect transistor (FET) sensors 1, 2 based on single-walled carbon nanotubes (SWNTs) 3 were shown to be sensitive devices for directly detecting specific molecules without additional labeling. Such FETs rely on an electric field on the surface of individual carbon nanotube to control conductivity, and are highly sensitive to their environment. Conductance varies significantly with changes in electrostatic charges and surface adsorption of a variety of molecules 1, 4, 5. Using tubes grown directly on the chip by chemical vapor deposition (CVD), it was shown that a large conductance change can be achieved when individual tubes are utilized as gates for FETs in chemical sensors 1, 2, 6. In addition, a submonolayer of SWNTs fabricated by CVD 7 has been shown to exhibit semiconductor-like behavior (also based on an electric field on the surface of carbon nanotube to control conductivity) which can be gated to utilize surface interactions of biomolecules for biosensing 7, 8.
As an alternative to FET based electronic sensing, we propose here a novel approach for biosensing based on different physical principle: “electrical percolation,” in which the passage of current through a conductive network depends on the continuity of the network. Electrical percolation and carbon nanotube-based conductivity detectors have been reviewed recently 9, and several sensors that utilize electrical percolation for vapor 10 11 and solvents 12 sensing were described. In such sensors, changes in electrical conductivity were attributed to swelling of the polymer matrix and/or conductive modification due to the solvent absorption. However, the utilization of electrical percolation in the context of biological recognition element as a transistor gate has not been explored.
We hypothesize that when nanomaterial with a biological recognition element is used in a multilayer 3-D interconnected network, the number of contacts within the network can be varied by molecular interactions that change the resistance of the network. This change can be measured to determine the number of interactions and hence the concentration of the analyte. Such mechanism is different than that of the FETs used in biosensing. In FETs, the mobility of electrons within a single nanotube is dependent on surface interactions. In contrast, in this model, changes in electrical conductivity of the network are dependent the number of contacts of the elements within the network. Molecular interactions disrupt the network continuity resulting in increased resistance.
The use of electrical percolation for specific direct electronic gating requires a recognition element to bind with the biological target. Recognition elements can be ligands such as antibodies, DNA, receptors, aptamers, or hormones that control the electrical conductivity of the bio-nanocomposite containing the nanomaterial and recognition element. Our model suggests that using percolation principles, it will be possible to characterize changes in the connectivity of elements within the SWNT network by modeling electrical percolation as the flow of electricity through a randomly distributed network of conducting elements. In such a network, sites (vertices) or bonds (edges) are established by randomly placing resistors in a 3-D vector space with a statistically independent probability (p) of making contacts. At a critical threshold (pc), long-range connectivity within the vector space first appears (known as the “percolation threshold”)13. Beyond this threshold, the conducting elements increase precipitously and there is an onset of a sharp and very significant increase in the electrical conductivity of the material 14. Therefore, it is characteristic of the minimal concentration of conductive filler required to form a randomly distributed network that spans the whole material system. As previously stated, the concentration of conductive filler correlating to the percolation threshold will be affected not by the mobility of electrons within the filler, but rather by the characteristics that control the number of contacts and the contact resistance between filler elements. Thus, the principles governing the percolation threshold are not “electrochemical”, but rather “electrophysical” (e.g., morphology, scale, and orientation of the filler).
As a model system to demonstrate BSC, we used the detection of Staphylococcal enterotoxins (SEs). SEs are a group of twenty-one heat stable toxins implicated in foodborne diseases resulting from consumption of contaminated foods 15-19. Food poisoning by SEs causes gastrointestinal symptoms even at exposure levels as low as. 20-100 ng per person 20. In addition, SEs have been implicated in diseases such as atopic eczema 21-23, rheumatoid arthritis 24, 25, and toxic shock syndrome 26, and are recognized as potential bioweapons 27-30. SEs are traditionally assayed immunologically with enzyme-linked immunosorbent assays (ELISA) 31, which generally use optical detection. Other immunological assays for SE detection have also been described, including several biosensors 32-40. BSC-based assays for SEs (and for other microbial toxins) offer advantages such as speed and high-throughput.
Materials and methods
Materials and Reagents
Staphylococcal enterotoxin B (SEB), rabbit anti-SEB affinity purified IgG, and peroxidase (HRP) conjugated anti-SEB IgG were purchased from Toxin Technology (Sarasota, FL). Single-walled Carbon Nanotubes (CNTs) were obtained from Carbon Solutions Inc (Riverside, CA). Poly(diallyldimethylammonium chloride) polymer (PDDA) was purchased from Sigma-Aldrich (St. Louis, MO). Silver contact “Silver Liquid” was purchased from Electron Microscopy Sciences (Hatfield, PA). For ECL detection, Immun-Star HRP Chemiluminescence Kit was obtained from Bio-Rad (Hercutes, CA). All other reagents were of analytical grade and de-ionized water was used throughout.
Fabrication of BSC sensor
The BSC sensor used in this study was designed in CorelDraw11 (Corel Corp. Ontario, Canada) and micro-machined in 1.5 mm acrylic using a computer controlled laser cutter Epilog Legend CO2 65W cutter (Epilog, Golden, CO). Before engraving the common electrode for all sixteen BSCs, the connection well for the readout electrode and cutting the slots for the bio-nanocomposite material, the lower side of the PMMA sheet was coated with 3M 9770 adhesive transfer doublesided tape (Piedmont Plastics, Beltsville, MD) and the polycarbonate film was immobilized directly on the PMMA. The bio-nanocomposite was bonded to the polycarbonate film and the electrodes were filled with silver conducting paste.
Carbon nanotube preparation
The CNTs (30 mg) were first shortened and oxidized by mixing with concentrated sulfuric acid and nitric acid mixture (3:1 v/v) and sonicating with a Fisher (FS-14) sonicator for 6 h followed by extensive washing in water (100 ml) until neutralized (pH 7.0). Then the CNT were dispersed in 100 ml 1M NaOH solution for 5 min to achieve net negative charged carboxylic acid groups and washed with water (100 ml).
CNT functionalization
a linker molecule to the carbon nanotube was used 41. Poly(diallyldimethylammonium chloride (PDDA) is positively charged and SEB antibody is negatively charged, so the antibodies were electrostatically adsorbed onto carbon nanotube. The positively charged polycation was adsorbed by dispersing the CNT in 50 ml of 1 mg/mL PDDA containing 0.5 M NaCl for 30 min followed by centrifugation (10,000 RPM in Beckman centrifuge for 15 minutes) and washed with 100 ml of water.
CNT–antibody complex preparation
The CNT were functionalized by dispersing in a rabbit anti-SEB IgG phosphate buffer solution (20 mM, pH 8.0) at a concentration of 0.01 mg/mL for 1 h at room temperature, so that the antibody was adsorbed onto the CNT surface. After centrifugation (15 minutes) and washing extensively with water (10 ml), the modified CNT was stored at 4°C in pH 8.0 phosphate buffer at a concentration of approximately 1mg/mL for no more than two weeks before use.
BSC detection of SEB
The CNT–antibody complex described above is immobilized directly on PMMA or polycarbonate. The range of resistance tolerances for functional BSC is 30 to 100 ohm. Before applying SEB samples, the resistance of the BSC is first measured (R0) with an ohmmeter. Different concentrations of SEB samples in phosphate buffer are added to sample wells and incubated for 60 min at room temperature (25 °C). After washing, the BSC was dried at room temperature for 2 hours and the resistance measured again (R1). The difference between the two readings (R1 – R0) is used as signal corresponding to different concentration of SEB. The value obtained with concentration of 0 ng/mL SEB was defined as background. The ratio of the signal to background (S/B) was further used to quantify the SEB concentration. The raw data (in Ohms) depends on many factors including the initial reading of the sample matrix and variations among BSC. Therefore, the better measure is the ratio of S/B which accounts for such variations, and that is what is used to quantify the SEB concentration and to compare results from different experiments and different sample matrices.
SEB detection using ECL
For the control experiment, after SEB binding the BSC was then blocked with 1% BSA in 15 μl buffer for 30 min. A HRP conjugated anti-rabbit IgG was added to the captured SEB and after 60 minutes incubation and washing, ECL was achieved by adding 7 uL of ECL buffer (formed by mixing the two solutions from the chemiluminescent kit in a 1:1 volume ratio) into each well and the ECL intensity was measured immediately with a custom-built point-of-care CCD detector 42-44. The CCD-based detector consists of an SXVF-M7.cooled CCD camera (Adirondack Video Astronomy, Hudson Falls, NY) equipped with a 5mm extension tube and a 12mmPentax f1.2 lens (Spytown, Utopia, NY). The luminescence was measured after 2 and 10 minutes of exposure. The CCD image intensities were analyzed using ImageJ software, developed and distributed freely by NIH (http://rsb.info.nih.gov/ij/download.html), and the data generated was then imported into Microsoft Excel (Microsoft, Redmond, WA) for further manipulation.
Results and Discussion
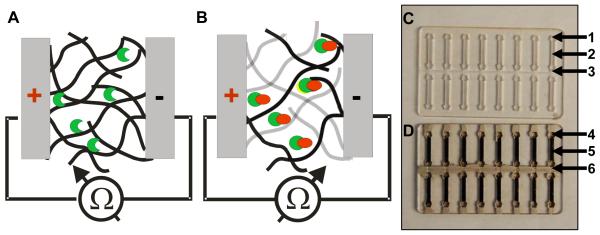
To demonstrate this new concept, we fabricated a detector with the bio-nanocomposite material by depositing pre-functionalized SWNTs with biological ligands to form a biological semiconductor (BSC) layer. A schematic of a sensor using the electrical percolation BSC is shown in figure 1. In the low resistance mode (figure 1 A), the SWNT-antibody network of the BSC (black lines) is shown with no antigens bound to antibodies (half-moon shape). In the high resistance mode (figure 1B), binding of antigens (circles) results in disruption of the network thus increasing electrical resistance. Non-contact SWNTs are shown in grey in the Figure.
Figure 1. Electrical percolation biological semiconductor (BSC).
A. schematic of the bio-nanocomposite layer of the BSC with no antigen bound to antibodies (shown as a half-moon shape); B. Binding of the SEB antigen (circles) results in disruption of the network (non-contact CNTs are shown in grey) thus increasing the resistance; C. a PMMA circuit board with 16 BSCs in two rows. Each BSC contains a connection well for the readout electrode (1), a detection channel (2) and a common electrode for all sixteen BSCs in the two rows (3). D. PMMA circuit board with the SWNT-antibody complex gate immobilized into the channel (5); readout electrode well (4) and the common electrode (6) are filled with silver conducting paste.
A simplified prototype of the BSC sensor is shown in figure 1C. The BSC is a unipolar device, with two electrodes painted with silver contact paste on both sides of the printed SWNT-antibody bio-nanocomposite. Several BSCs can be easily fabricated in a row on the same surface. At the circuit level, each BSC contains a connection well (figure 1C-1) for the silver electrode, a channel for the bio-nanocomposite (figure 1C-2) and a channel for the silver electrode. The silver electrode is common to all eight BSCs in a single row (figure 1C-3). The assembled chip is shown in figure 1D: the SWNT-antibody complex gate, which is immobilized into the channel to form the network (figure 1D-5), the connection well with the silver electrode (figure 1D-4), and the common connector filled with silver conducting solution (figure 1D-6).
The SWNTs are functionalized with rabbit anti-SEB IgG. A previously developed CNT functionalization scheme is employed for binding the SWNTs with the antibodies 42-44. The bio-nanocomposite is then immobilized by drying it directly on the surface of either Poly(methyl methacrylate) (PMMA) or polycarbonate wells (figure 1D) fabricated by laser micromachining42-44.
The electrical percolation BSC is operated simply by measuring the electrical resistance (figure 1D-4) between the silver paste electrodes (figure 1D-6). Binding of the specific antigen to the antibody disrupts the network (figure 1B) and increases the resistance. The amount of binding of the specific antigen to the antibody controls the overall resistance of the electrical percolation BSC network, which is measured by an ohmmeter via each BSC electrode and the common electrode.
A circuit board with sixteen electrical percolation BSCs was used in a conventional immunodetection assay by allowing binding of SEB to the antibody gate and washing off unbound material. The measurement value was calculated as the difference between the initial reading recorded (R0) with no SEB and the reading with SEB (R1). The difference between the two readings (R1 – R0) was measured as the signal, and is normalized by R0 to obtain the signal-to-baseline ratio. The use of a common electrode simplifies fabrication but it introduces a constant difference of resistance among the 16 BSC (based on their position relative to the measuring point). In our case, since we are measuring the difference between the two readings (R1 – R0), this constant difference will have no effect.
SWNT-antibody bio-nanocomposite percolation
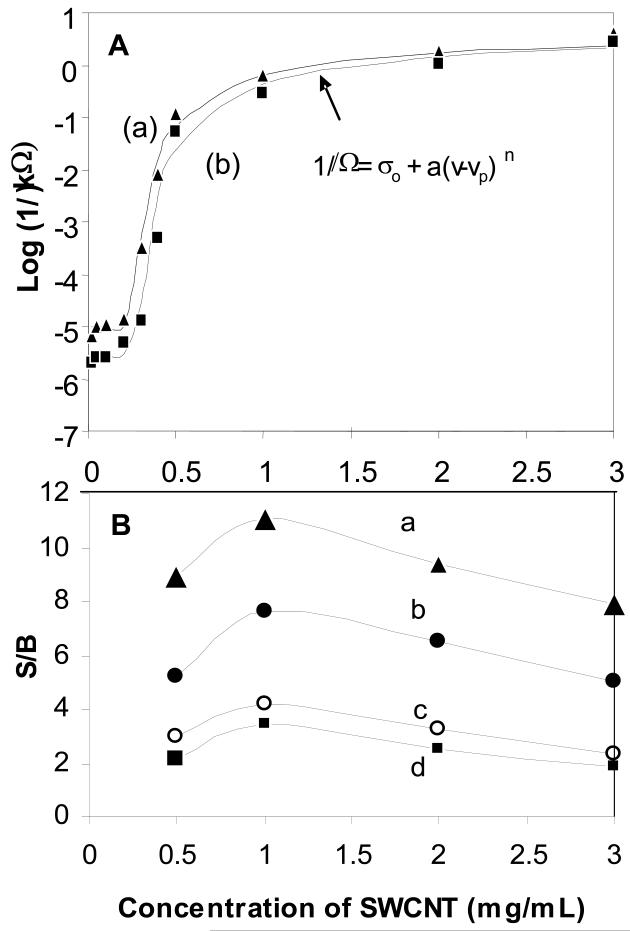
The percolation of the SWNT-antibody network was established using various concentrations, v, of SWNT immobilized onto a PMMA surface without (Figure 2A-a) and with anti-SEB antibody (Figure 2A-b). Their resistance, Ω, was measured to determine the percolation threshold, vp, using a conventional power law equation from percolation theory (see form of equation in Figure 2A and arrow pointing to data fit) with a baseline resistance, σo Using the power law fit, it was possible to determine that the percolation threshold for the SWNT-antibody network is between 0.2 to 0.3 mg/mL, and does not change significantly after antibody immobilization in Figure 2A. The rate of change in resistance is directly related to the power-law exponent, n, which was 8 and the power-law coefficient, a, which was 5.×10−6. There are three characteristic regimes in SWNT concentration associated with these values: (1) between ~0.2 to 0.5 mg/mL the percolation threshold is characterized by a steep change (approximately four orders of magnitude) in resistance due to the onset of percolation, (2) between ~0.5 to 1 mg/mL the change levels off and the increase is approximately one order of magnitude, (3) over ~1 mg/ml the resistance levels off and does not change significantly with higher concentrations of SWNT resulting in complete percolation. Over the entire range, the total change in resistance is approximately five orders of magnitude. The percolation threshold of the SWNT-antibody bio-nanocomposite network also indicates that its typical resistance (figure 2A-b) will be higher than the resistance that is attributed to the SWNT only (figure 2A-a), presumably due to the contacts between the antibody and the functionalized SWNT.
Figure 2. Percolation of the SWNT bio-nanocomposite.
(A).The measured resistance of various concentrations, v, of SWNTs immobilized onto a PMMA surface without (a-triangle) and with (b-rectangle) anti-SEB antibody. (B) The effect of SWNT concentration on BSC responce to SEB binding. Various concentrations of SWNT were immobilized with anti-SEB antibody and reacted with SEB: (a) 100 ng/ml SEB, (b) 10 ng/ml, (c) 1 ng/ml and (d) 0.5 ng/ml. For all SEB concentrations, the highest S/B was detected at the point of complete percolation corresponding to 1 mg/ml SWNT.
We propose the following model for the electrical percolation BSC. At the percolation transition point, the point above the percolation threshold where the change in resistance begins to level off, there is a still relatively low statistical distribution of “contacts” between the CNT-antibody complexes in the network. Therefore, small changes in the CNT-antibody complexes can lead to dramatic changes in conductivity. Based on this model, we predict that bio-nanocomposite prepared with 1 mg/mL of SWNT will be the most sensitive to molecular interactions for immunodetection, since this is the concentration at which the change in resistance begins to level off, consistent with the complete percolation of the SWNTs.
To validate the prediction that the point where complete percolation occurs (1 mg/mL) will be the most sensitive to molecular interactions, we analyzed the response of the BSC over a range of SWNTs concentrations (0.5-3 mg/ml) in response to binding of broad range of SEB concentration (0.5 - 100 ng/ml). At the transition point of 1 mg/ml, the BSC exhibited peak sensitivity to all SEB concentrations (Figure 2B). This result confirmed our predictions and clearly suggests that the mechanism of the BSC sensor is electrical percolation. Moreover, for all concentrations of SWNTs bio-nanocomposite, the S/B increases with increasing SEB concentration, suggesting that the new BSC can be used for direct biosensing and bioactuation.
BSC-based analysis of SEB
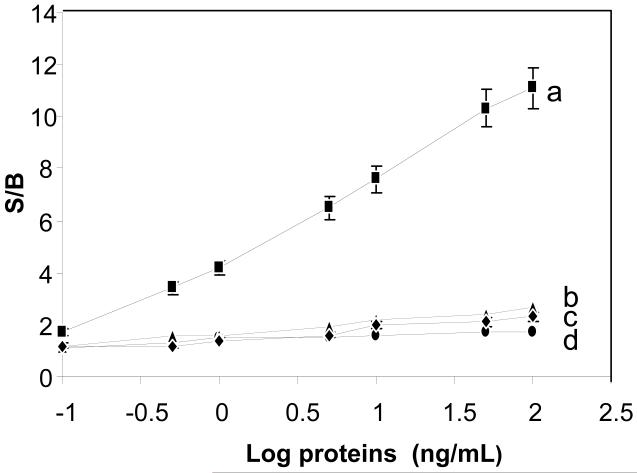
To show the specificity of the BSC response, various amounts of SEB (from 0.1-100 ng/mL) in buffer were added to the chip with 1 mg/ml of SWNT (figure 3a). The resistance increased proportionally to the amount of SEB. Non-specific antigens were used to study the BSC leak rate, which is the change in resistance with non-specific binding and is an indication of the specificity and the selectivity of BSC actuation. Various non-specific antigens were used, including a smaller molecular weight (14 kDa) protein, lysozyme (figure 3b), and a higher molecular weight (150 kDa) protein, human IgG (figure 3c). As shown in figure 3, the level of non-specific binding in these semiconductors is relatively small regardless of concentration, which is similar to the S/B for SEB concentrations when there is no antibody on the SWNTs (figure 3d). The measurement is rapid. The exact time to get a stable reading depends on the SEB concentration, but is not longer than a few minutes. If sample preparation is needed, the analysis may be longer depending on the protocol for sample preparation used.
Figure 3. Electrical characteristics of SEB actuation of BSC.
Various concentrations of SEB (0, 0.1, 1, 5, 10, and 50 ng/mL) were applied to the sensor composed of 1 mg/ml SWNT with immobilized anti-SEB IgG (a rectangle). Nonspecific binding of similar concentrations of lysozyme (b-diamond) and human IgG (c-triangle) to the chip is shown, along with SWNT without antibody (d-circle). Error bar = SD (n=8).
To determine the limit of detection (LOD) for SEB, the S/B ratio from eight replicas of various concentrations of SEB was compared to buffer. A T-test demonstrated that at 1 ng/ml, the S/B ratio is significantly different (P<0.00017) from the value using buffer only. Thus, the current configuration has a LOD of 1 ng/mL for SEB.
In comparison, the ELISA LOD, using sandwich assays combined with optical detection for SE, ranges from 0.5 to 2 ng/g of food 45-50.
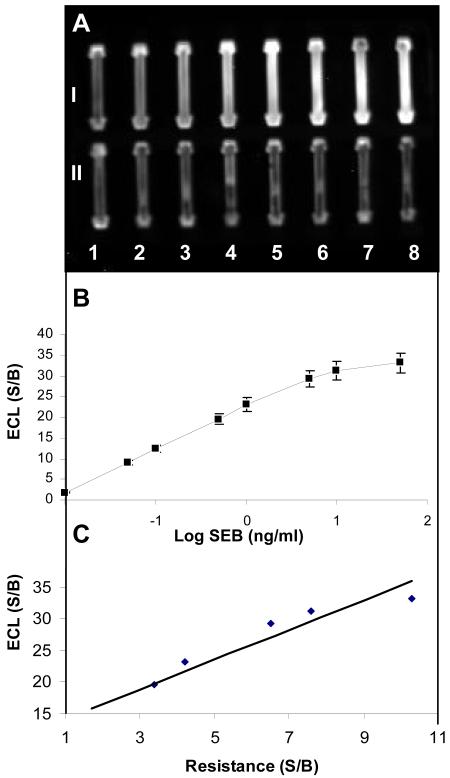
To confirm that the percolation of the SWNT-antibody and the antibody gate mechanisms shown in figures 2 and 3 depend on SEB binding, an independent measurement of bound SEB to the SWNTs bio-nanocomposite was carried out using a sandwich immunoassay detected by Enhanced Chemiluminescence (ECL). As shown in Figure 4A, the intensity of the signal from the captured SEB on the BSC chip is proportional to the amount of SEB. Quantitative analysis of the data (Figure 4C) suggests a high correlation between the amount of SEB and the ECL signal and that there is a very high correlation (R2= 0.9942) between the electrical measurements (Figure 3A) and the ECL measurements (Figure 4B). The linear regression is also highly significant (p<0.0056), suggesting that the anti-SEB antibody on the BSC chip did indeed capture SEB, and that the direct electrical measurements are in agreement with the indirect sandwich immunoassay detected by ECL.
Figure 4. Sandwich Immunoassay of captured SEB on electrical percolation BSC detected using ECL.
Different concentrations of captured SEB incubated with Horseradish Peroxidase (HRP) conjugated anti-SEB IgG and assayed with ECL: A. ECL intensity measured with a custom-built Point-of-Care CCD detector after exposure time of 20 min at SEB concentrations of 0.01, 0.05, 0.1, 0.5, 1, 5, 10, and 50 ng/mL. B. plot of SEB concentration vs. ECL signal. C. correlation between the S/B of ECL and resistance measurements at various SEB concentrations.
Our interpretation of the data is that antigen binding leads to rearrangement of the SWNT-antibody network, resulting in physical depletion of electron carriers in the bulk of the SWNT-antibody bio-nanocomposite through changes in contact between the SWNTs. Such contacts are analogous to the physical edge of the conduction band. We suggest that being at this point, the antibody gate mechanism initiated by binding the antigen to the antibody shifts the complex closer to the band gap, which is an energy range where statistically few electron states exist so fewer electrons can jump between SWNTs. This is analogous to decreasing an electric field in a classical semiconductor, and therefore increases the electrical resistance of the SWNT-antibody network. The percolated SWNT-antibody network can therefore be considered the “conduction band”, and the number of electrons in the conduction band (i.e., the band gap) is physically determined by the number of SWNT-antibody complexes in the conduction band rather than by the conventional electronic band gap at the surface of the SWNT that is responsible for electrochemical detection principles.
Unlike field-effect transistors (FETs) based sensors, which rely on an electric field at the surface of the SWNTs to control conductivity, the response of the electrical percolation BSC can be attributed to the number of contacts of CNTs within the network. Since the number of contacts can be varied by molecular interactions (i.e., by antibody-antigen binding), changes in the resistance of the network can be used to determine the number of interactions and hence the concentration of the analyte.
One of the most attractive features of electrical percolation BSCs based on SWNTs is the simplicity of the preparation (screen printing). In contrast, FETs are often fabricated using on chip chemical vapor deposition (CVD) and require a high-tech infrastructure for microfabrication of solid-state semiconductor components. Furthermore, unlike FETs which are constructed with SWNTs as a single wire or sub-monolayer network, BSC do not need to be oriented, can be fabricated and functionalized in bulk. Electrical percolation BSCs can simply be printed on any non-conductive material to create biosensors capable of detecting a variety of molecules. Selectivity is achieved by printing different specific biological “gates”, such as antibodies, DNA, receptors, or aptamers, taking advantage of the natural selectivity of these biological molecules. Moreover, electrical percolation BSC production can be readily scaled to perform multi-analyte detection, unlike single CNT devices that are challenging to fabricate and functionalize.
Having simple biosensor technology may permit wider use of biosensors. Existing technologies are relatively complex, have limited capability for multi-analyte detection, and are costly. The BSC proposed here overcome each of these limitations. BSCs are very simple to fabricate and to operate and are capable of multi-analyte detection. Using BSCs, it may be possible to fabricate miniaturized “Biological Central Processing Units (CPUs)” with multiple biological elements, capable of processing and sorting out information on multiple analytes simultaneously. By combining them with computer algorithms, it is possible to automatically perform multi-analyte detection and make decisions analogous to the way a silicon chip processes digital information to make decisions important for direct biodetection of multiple microbial pathogens and their toxins, numerous cancer biomarkers, cardiovascular or kidney biomarkers.
Conclusion
In conclusion, we have demonstrated the fabrication of a novel biological semiconductor (BSC) based on electrical percolation through a multi-layer 3-D carbon nanotube-antibody network which can measure biological interactions directly and electronically. In this system, molecular interactions, such as binding of antigens to the antibodies, disrupt the network continuity causing increased resistance of the network. We demonstrated the use of BSC for direct electronic measurements of Staphylococcal enterotoxin B (SEB).
In the future, the new BSCs could be used to assemble multiple sensors on the same chip, creating “Biological Central Processing Units (CPUs)” with multiple biological elements that are capable of processing and sorting out information on multiple analytes simultaneously.
Beyond biosensing, a “Biological CPU” could be a transformational technology, providing users with immediate decision-making capability that is useful for many biomedical applications including regulation and actuation of implantable biomedical devices such as insulin pumps, cardiac assist devices or other theranostics.
Unlike FET-based sensors with oriented SWNTs synthesized on the chip, percolation based sensor rely on the conductivity of network. Various ligands can be used to functionalize pre-made SWNTs gates in bulk and the pre-made gates can be simply printed or deposited on non-conductive materials. These factors make BSC a practical approach for direct electronic sensing.
References
- (1).Kong J, Franklin NR, Zhou C, Chapline MG, Peng S, Cho K, Dai H. Science. 2000;287:622–625. doi: 10.1126/science.287.5453.622. [DOI] [PubMed] [Google Scholar]
- (2).Kong J, Dai HJ. Journal of Physical Chemistry B. 2001;105:2890–2893. [Google Scholar]
- (3).Iijima S. Nature. 1991;354:56–58. [Google Scholar]
- (4).Collins PG, Bradley K, Ishigami M, Zettl A. Science. 2000;287:1801–1804. doi: 10.1126/science.287.5459.1801. [DOI] [PubMed] [Google Scholar]
- (5).Shim M, Javey A, Kam NW, Dai H. J Am Chem Soc. 2001;123:11512–11513. doi: 10.1021/ja0169670. [DOI] [PubMed] [Google Scholar]
- (6).Tans SJ, Devoret MH, Dai HJ, Thess A, Smalley RE, Geerligs LJ, Dekker C. Nature. 1997;386:474–477. [Google Scholar]
- (7).Chen RJ, Bangsaruntip S, Drouvalakis KA, Kam NW, Shim M, Li Y, Kim W, Utz PJ, Dai H. Proc Natl Acad Sci U S A. 2003;100:4984–4989. doi: 10.1073/pnas.0837064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Chen RJ, Zhang Y, Wang D, Dai H. J Am Chem Soc. 2001;123:3838–3839. doi: 10.1021/ja010172b. [DOI] [PubMed] [Google Scholar]
- (9).Li C, Thostenson ET, Chou TW. Composites Science and Technology. 2008;68:1227–1249. [Google Scholar]
- (10).Zhang B, Fu RW, Zhang MQ, Dong XM, Wang LC, Pittman CU. Materials Research Bulletin. 2006;41:553–562. [Google Scholar]
- (11).Zhang B, Fu RW, Zhang MQ, Dong XM, Lan PL, Qiu JS. Sensors and Actuators B-Chemical. 2005;109:323–328. [Google Scholar]
- (12).Kobashi K, Villmow T, Andres T, Potschke P. Sensors and Actuators B-Chemical. 2008;134:787–795. [Google Scholar]
- (13).Grimmett G. Percolation. 2nd ed. Springer-Verlag; New York: 1999. [Google Scholar]
- (14).MacDonald RA, Voge CM, Kariolis M, Stegemann JP. Acta Biomater. 2008;4:1583–1592. doi: 10.1016/j.actbio.2008.07.005. [DOI] [PubMed] [Google Scholar]
- (15).Archer DL, Young FE. Clin Microbiol Rev. 1988;1:377–398. doi: 10.1128/cmr.1.4.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Olsen SJ, MacKinnon LC, Goulding JS, Bean NH, Slutsker L. Mor Mortal Wkly Rep CDC Surveill Summ. 2000;49:1–62. [PubMed] [Google Scholar]
- (17).Bean NH, Goulding JS, Lao C, Angulo FJ. Mor Mortal Wkly Rep CDC Surveill Summ. 1996;45:1–66. [PubMed] [Google Scholar]
- (18).Bunning VK, Lindsay JA, Archer DL. World Health Stat Q. 1997;50:51–56. [PubMed] [Google Scholar]
- (19).Garthright WE, Archer DL, Kvenberg JE. Public Health Rep. 1988;103:107–115. [PMC free article] [PubMed] [Google Scholar]
- (20).Asao T, Kumeda Y, Kawai T, Shibata T, Oda H, Haruki K, Nakazawa H, Kozaki S. Epidemiol Infect. 2003;130:33–40. doi: 10.1017/s0950268802007951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Breuer K, Wittmann M, Bosche B, Kapp A, Werfel T. Allergy. 2000;55:551–555. doi: 10.1034/j.1398-9995.2000.00432.x. [DOI] [PubMed] [Google Scholar]
- (22).Bunikowski R, Mielke M, Skarabis H, Herz U, Bergmann RL, Wahn U, Renz H. J Allergy Clin Immunol. 1999;103:119–124. doi: 10.1016/s0091-6749(99)70535-x. [DOI] [PubMed] [Google Scholar]
- (23).Mempel M, Lina G, Hojka M, Schnopp C, Seidl HP, Schafer T, Ring J, Vandenesch F, Abeck D. Eur J Clin Microbiol Infect Dis. 2003;22:306–309. doi: 10.1007/s10096-003-0928-0. [DOI] [PubMed] [Google Scholar]
- (24).Howell MD, Diveley JP, Lundeen KA, Esty A, Winters ST, Carlo DJ, Brostoff SW. Proc Natl Acad Sci U S A. 1991;88:10921–10925. doi: 10.1073/pnas.88.23.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Uematsu Y, Wege H, Straus A, Ott M, Bannwarth W, Lanchbury J, Panayi G, Steinmetz M. Proc Natl Acad Sci U S A. 1991;88:8534–8538. doi: 10.1073/pnas.88.19.8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Herz U, Bunikowski R, Mielke M, Renz H. Int Arch Allergy Immunol. 1999;118:240–241. doi: 10.1159/000024085. [DOI] [PubMed] [Google Scholar]
- (27).Wiener SL. Mil Med. 1996;161:251–256. [PubMed] [Google Scholar]
- (28).Ler SG, Lee FK, Gopalakrishnakone P. J Chromatogr A. 2006;1133:1–12. doi: 10.1016/j.chroma.2006.08.078. [DOI] [PubMed] [Google Scholar]
- (29).Henghold WB., 2nd Dermatol Clin. 2004;22:257–262. doi: 10.1016/j.det.2004.03.004. v. [DOI] [PubMed] [Google Scholar]
- (30).Rosenbloom M, Leikin JB, Vogel SN, Chaudry ZA. Am J Ther. 2002;9:5–14. doi: 10.1097/00045391-200201000-00003. [DOI] [PubMed] [Google Scholar]
- (31).Bennett RW. J Food Prot. 2005;68:1264–1270. doi: 10.4315/0362-028x-68.6.1264. [DOI] [PubMed] [Google Scholar]
- (32).Yu X, Kim SN, Papadimitrakopoulos F, Rusling JF. Mol Biosyst. 2005;1:70–78. doi: 10.1039/b502124c. [DOI] [PubMed] [Google Scholar]
- (33).Soelberg SD, Chinowsky T, Geiss G, Spinelli CB, Stevens R, Near S, Kauffman P, Yee S, Furlong CE. J Ind Microbiol Biotechnol. 2005;32:669–674. doi: 10.1007/s10295-005-0044-5. [DOI] [PubMed] [Google Scholar]
- (34).Shriver-Lake LC, Shubin YS, Ligler FS. J Food Prot. 2003;66:1851–1856. doi: 10.4315/0362-028x-66.10.1851. [DOI] [PubMed] [Google Scholar]
- (35).Sapsford KE, Taitt CR, Loo N, Ligler FS. Appl Environ Microbiol. 2005;71:5590–5592. doi: 10.1128/AEM.71.9.5590-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Rasooly L, Rasooly A. Int J Food Microbiol. 1999;49:119–127. doi: 10.1016/s0168-1605(99)00053-7. [DOI] [PubMed] [Google Scholar]
- (37).Rasooly A, Herold KE. J AOAC Int. 2006;89:873–883. [PubMed] [Google Scholar]
- (38).Rasooly A. J Food Prot. 2001;64:37–43. doi: 10.4315/0362-028x-64.1.37. [DOI] [PubMed] [Google Scholar]
- (39).Nedelkov D, Rasooly A, Nelson RW. Int J Food Microbiol. 2000;60:1–13. doi: 10.1016/s0168-1605(00)00328-7. [DOI] [PubMed] [Google Scholar]
- (40).Homola J, Dostalek J, Chen S, Rasooly A, Jiang S, Yee SS. Int J Food Microbiol. 2002;75:61–69. doi: 10.1016/s0168-1605(02)00010-7. [DOI] [PubMed] [Google Scholar]
- (41).Liu G, Lin Y. Anal Chem. 2006;78:835–843. doi: 10.1021/ac051559q. [DOI] [PubMed] [Google Scholar]
- (42).Yang M, Kostov Y, Rasooly A. Int J Food Microbiol. 2008;127:78–83. doi: 10.1016/j.ijfoodmicro.2008.06.012. [DOI] [PubMed] [Google Scholar]
- (43).Yang M, Kostov Y, Bruck HA, Rasooly A. Anal Chem. 2008;80:8532–8537. doi: 10.1021/ac801418n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Sapsford KE, Sun S, Francis J, Sharma S, Kostov Y, Rasooly A. Biosens Bioelectron. 2008;24:618–625. doi: 10.1016/j.bios.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Miyamoto T, Kamikado H, Kobayashi H, Honjoh K, Iio M. J Food Prot. 2003;66:1222–1226. doi: 10.4315/0362-028x-66.7.1222. [DOI] [PubMed] [Google Scholar]
- (46).Pan TM, Yu YL, Chiu SI, Lin SS. Zhonghua Min Guo Wei Sheng Wu Ji Mian Yi Xue Za Zhi. 1996;29:100–107. [PubMed] [Google Scholar]
- (47).Park CE, Akhtar M, Rayman MK. Appl Environ Microbiol. 1994;60:677–681. doi: 10.1128/aem.60.2.677-681.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Park CE, Warburton D, Laffey PJ. Int J Food Microbiol. 1996;29:281–295. doi: 10.1016/0168-1605(95)00046-1. [DOI] [PubMed] [Google Scholar]
- (49).Vernozy-Rozand C, Mazuy-Cruchaudet C, Bavai C, Richard Y. Lett Appl Microbiol. 2004;39:490–494. doi: 10.1111/j.1472-765X.2004.01602.x. [DOI] [PubMed] [Google Scholar]
- (50).Wieneke AA. Int J Food Microbiol. 1991;14:305–312. doi: 10.1016/0168-1605(91)90122-6. [DOI] [PubMed] [Google Scholar]