Abstract
The copy number variation in beta-defensin genes on human chromosome 8 has been proposed to underlie susceptibility to inflammatory disorders, but presents considerable challenges for accurate typing on the scale required for adequately powered case–control studies. In this work, we have used accurate methods of copy number typing based on the paralogue ratio test (PRT) to assess beta-defensin copy number in more than 1500 UK DNA samples including more than 1000 cases of Crohn's disease. A subset of 625 samples was typed using both PRT-based methods and standard real-time PCR methods, from which direct comparisons highlight potentially serious shortcomings of a real-time PCR assay for typing this variant. Comparing our PRT-based results with two previous studies based only on real-time PCR, we find no evidence to support the reported association of Crohn's disease with either low or high beta-defensin copy number; furthermore, it is noteworthy that there are disagreements between different studies on the observed frequency distribution of copy number states among European controls. We suggest safeguards to be adopted in assessing and reporting the accuracy of copy number measurement, with particular emphasis on integer clustering of results, to avoid reporting of spurious associations in future case–control studies.
INTRODUCTION
Among the many human genes known to display variation in copy number, the beta-defensin genes mapping to human chromosome 8p23.1 have been well established to be extensively variable in human populations (1–4). In European populations, there is a common copy number variation (CNV) between two and seven copies per person for a segmental duplication containing seven beta-defensin genes, including DEFB4, DEFB103–107 and SPAG11, but excluding the neighbouring DEFB1 gene. There is a correlation between copy number and beta-defensin gene expression at the mRNA level in isolated cells (1), though a recent study found that beta-defensin 2 protein production from ex vivo-cultured colonic biopsies was correlated with local inflammation, but not with DEFB4 copy number (5), and case–control studies have provided the clearest indications to date of the functional consequences of this CNV.
Given the functions of beta-defensins as antimicrobials and cytokines (6,7), investigators have tested for association between beta-defensin variation and inflammatory disease, including psoriasis (8) and Crohn's disease (9,10). In 2006, Fellerman et al. reported an association between low beta-defensin copy number and Crohn's disease of the colon from cohorts of relatively modest sample size (71 colonic Crohn's patients and 169 controls); no association with ileal disease was demonstrated (9). In contrast, a more recent study by Bentley et al. (10) reported an association between Crohn's disease and higher beta-defensin copy number, using real-time PCR measurement in 466 cases and 329 controls.
Measuring copy number accurately poses particular challenges for case–control association studies, especially in multi-allelic CNVs (11), such that distinguishing (for example) five copies of the beta-defensin segmental duplication reproducibly from six copies is not easily achievable using standard methods. The unresolved influence of typing error on the outcome of different studies is evident in recent reports of discordant findings for the association between HIV infection and the CCL3L1 CNV, despite the CCL3L1 CNV varying across a relatively low copy number range (generally zero to four copies in European populations) (12–16). If just a small fraction of typing error applied differentially between case and control DNA samples, studies involving several hundred samples could easily generate spurious associations attributable either directly to differential error or to differential removal of samples failing to satisfy a typing quality threshold (17). Given that the highest copy numbers are intrinsically the most error-prone, even small differences of quality in the underlying CNV measurements could lead to an artefactual shift in the copy number distributions between cases and controls, mimicking a statistically significant biological association.
Even in the relatively low copy number range exemplified by the CCL3L1 CNV, those recent reports highlight the technical difficulty of obtaining convincingly accurate copy number measurements using methods based on real-time PCR. The study of Field et al. explicitly compared the data from the same samples (several thousand cases and controls in a study of type I diabetes), using either real-time PCR or assays using the paralogue ratio test (PRT) (14). PRT uses a single primer pair to amplify both test and reference amplicons simultaneously, so that the potential error arising from the different amplification efficiencies of distinct test and reference amplicons is reduced (18). Field et al. showed an apparently highly significant association between CCL3L1 copy number and type I diabetes using real-time PCR data. However, these same data showed a significant deviation from the expectations of the Hardy–Weinberg equilibrium; neither that deviation nor the association with type I diabetes was replicated after PRT-based measurement of copy number. The authors concluded that the apparently significant association was attributable to the effect of differential error of real-time PCR measurement methods between cases and controls, rather than to a true association (14).
In contrast with CCL3L1, the beta-defensin CNV varies across a higher copy number range (commonly 2–7), and even methods such as PRT, carefully designed and implemented to address the difficulty of the CNV-counting problem, are only capable of making the necessary fine distinctions of dosage with some level of unavoidable error (8,18,19). In this work, we combine PRT methods of CNV determination modified to reduce typing error with careful analysis of typing results to analyse beta-defensin copy number in more than 1000 cases of Crohn's disease from the UK. We find no support for the association previously reported by Fellerman et al. or for the association with higher copy number reported by Bentley et al. A direct comparison of results from the same samples typed either by real-time PCR or by our new implementation of PRT casts serious doubt on the suitability of this real-time PCR assay for accurate determination of beta-defensin CNV in large numbers of samples, as required by robust case–control association studies.
RESULTS
Triplex PRT-based copy number typing
We adapted published PRT methods to provide a composite measure of beta-defensin copy number that would be sufficiently accurate and robust for application in case–control association studies, but remain simple and inexpensive in practice. Measurements of beta-defensin copy number applied to different parts of the copy variable repeat unit have clearly shown that the entire repeat unit varies coordinately, so that the measurement of any element in the repeat constitutes an effective copy number assay for the whole (1,19,20). Our ‘triplex’ assay contains three components: a ‘PRT' (PRT107A) and rs5889219 multi-allelic indel measure already described (19) together with a newly modified PRT assay (‘HSPD21’) based on the same heat shock protein pseudogene as used previously (18), but with alternative primers designed to use a reference locus on chromosome 21 rather than on chromosome 5 (Fig. 1). Internal comparisons between the results of these three independent assays allowed most samples to be assigned to integer copy number classes with high confidence; there was clear clustering of results from the two PRT components around maxima presumed to correspond to integer copy numbers (Fig. 2A and B). Empirical distributions of results for samples of known copy number were used to create a likelihood framework in which sets of triplex test results for individual samples could be used to assign the most likely copy number to a sample (see Materials and Methods). In 326 tests of reference samples of known copy number (copy numbers 3, 4, 5 or 6), an incorrect copy number was called on 11 occasions, all involving five- and six-copy samples and suggesting that the per-test error rate is of the order of 3–4%.
Figure 1.
The copy variable beta-defensin genomic region showing the locations in the repeat sampled by the PRT-based ‘triplex’ test. The approximate genomic extent of the seven copy variable beta-defensin genes DEFB4, DEFB103–DEFB107 and SPAG11 is shown, without the detail of intron–exon boundaries. Below this is shown an example result from ‘triplex’ PRT-based typing of copy number for beta-defensins. PRT assays were designed to specifically amplify DNA sequences that occur within the beta-defensin (DEFB) CNV repeat unit but have additional paralogues elsewhere in the genome. The primers are designed to amplify specifically from exactly two loci of the repeat sequence (one of which is at DEFB), with corresponding products distinguished by length. For example, HSPD21-PRT primers specifically amplify only the chr8 (DEFB) and chr21 copies of a repeat element, with products differing by 8 bp. The third assay (rs5889219) examines a triallelic indel polymorphism (123/126/128 bp) that can have different sizes in individual DEFB repeats. This trace shows a sample with a copy number of 4, reflected in the ratios between the ‘test’ (=DEFB) and ‘reference’ peaks in the two PRT systems HSPD21 and PRT107A, and in the approximate 1:1:2 ratios of alleles (consistent with a total of four copies) for the multi-allelic indel rs5889219. Each analysis is in turn performed in two parallel but independent replicate amplifications using different fluorescent labels which are combined before capillary electrophoresis. Data from the two different fluorescent labellings for each system are combined before further analysis. This combined analysis therefore returns six results useful in measurement of beta-defensin copy number: duplicates of two independent PRT-based estimates of copy number and duplicate ratios of allelic products for the multi-allelic indel.
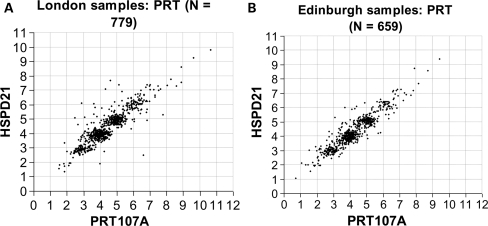
Figure 2.
Comparison of unrounded copy number measurements based on the HSPD21 and PRT107A PRT tests for (A) 779 cases and controls from the London collection and (B) 659 cases and controls from the Edinburgh collection, showing predominant clustering of measurements—most frequently centred around integer values 2, 3, 4, 5, 6 or 7. See also ‘Materials and Methods’ for details of procedures and data analysis.
Typing cases and controls
These methods were used to type DNA samples from Crohn's disease patients and appropriate control samples. From the London samples, we typed 666 Crohn's disease patients and 185 controls, from which 648 cases and 185 controls provided results sufficient for analysis; from the Edinburgh samples, we typed 384 cases and 350 controls, from which 358 and 314 samples (respectively) provided results acceptable for further analysis. We additionally typed 821 Edinburgh samples using real-time PCR methods (see Materials and Methods), including 625 (322 case and 303 control samples) for which we obtained CNV measurement data on the same DNA samples both from real-time PCR and the PRT-based triplex assay. In contrast with the results from PRT (Fig. 3A and B) which show clear grouping of measures around integer values, data from real-time PCR (analysed either by a ΔΔCT or standard-curve method) failed to show any such clustering around integer values (Fig. 3C). It is more parsimonious to propose that integer clusters have been dispersed by a high relative error in real-time PCR than to suggest that PRT-based measurements have artefactually created these integer clusters. Furthermore, although results from these two different methods of real-time PCR analysis are correlated (Fig. 4A), comparison of the same samples typed by PRT and real-time PCR (Fig. 4B) strongly suggests that for each copy number class indicated by clustering around integer values for PRT, real-time PCR delivers a wide range of values overlapping extensively with results from other integer classes. Overall, using either ΔΔCT or standard-curve analyses, more than 50% of integer copy number values from real-time PCR measurements of beta-defensin copy number disagree with corresponding integers deduced from the triplex assay in these 625 samples.
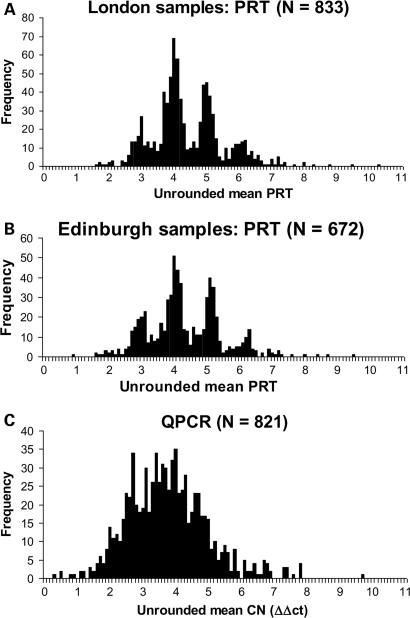
Figure 3.
Distribution of PRT-based results for (A) 833 London and (B) 672 Edinburgh cases and controls, showing the mean of HSPD21 and PRT107A measurements, or (if one PRT failed) the single PRT recorded, with clear peaks around integer values. (C) Real-time PCR data from 821 Edinburgh samples analysed by a ΔΔCT method shows no such clustering, suggesting a wider range of measurement error.
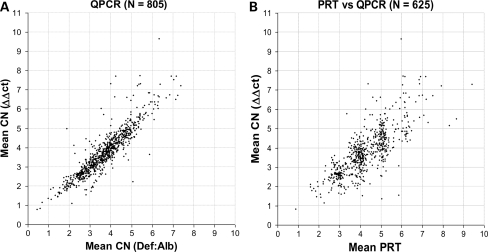
Figure 4.
Real-time PCR data (A) comparing two different methods of analysing real-time PCR data, indicating no serious discrepancy between analysis methods but no obvious clustering around integer values. (B) Real-time and PRT-based measurements on the same samples are compared. The PRT-based data show grouping around the integer values, but are spread vertically in this presentation by the much greater variation in measurements resulting from real-time PCR.
Case–control analysis
Using the triplex PRT-based assay, we were unable to find any significant association between beta-defensin copy number and Crohn's disease for the London samples (Fig. 5A, χ2 P = 0.381) or for the Edinburgh samples (Fig. 5B, P = 0.066). ΔΔCT analysis of real-time PCR data for Edinburgh samples (Fig. 5C) also failed to show a significant difference (P = 0.094). Exactly, the same 303 control DNA samples from Edinburgh have a mean copy number of 4.23 typed by the PRT-based triplex, but a mean of 3.79 by real-time PCR/ΔΔCT; the difference between the copy number measurements of the same samples by different methods is highly significant (P = 8.9 × 10−7). This suggests that there may be serious and pervasive typing error in the results of our real-time PCR assay.
Figure 5.
Frequency distributions of integer copy number classes for (A) 648 cases and 185 controls from the London collection using the PRT-based triplex test (P = 0.381), (B) 358 cases and 314 controls from the Edinburgh collection by the triplex test (P = 0.066) and (C) 407 cases and 411 controls from the Edinburgh collection using real-time PCR (analysed by a ΔΔCT method: P = 0.094).
Combining all available case and control samples typed in this study (1006 cases, 499 controls) showed a small increase in mean copy number in cases (4.45, compared with 4.305 for controls), which was apparently significant (χ2 P = 7 × 10−4). However, comparison of copy number distributions from PRT-based typing of control samples from Edinburgh and London/ECACC showed a weakly significant difference (P = 0.04). Given possible heterogeneity between populations for copy number frequencies, we examined the combined data in a Cochran–Mantel–Haenszel analysis, which attributed only marginal significance (P = 0.032) to the apparent excess of samples with five or more copies among Crohn's disease patients.
A specific comparison of copy numbers among Crohn's patients with colonic disease showed no significant differences in the mean copy number compared with controls, with either English (272 cases/185 controls) or Scottish (121 cases/314 controls) samples (χ2 P > 0.1). A combined analysis of 393 cases and 499 controls from the UK showed a difference which was not only of borderline significance (P = 0.047), but also showed that the mean beta-defensin copy number for the pooled UK colonic cases (4.41) was in fact higher than the mean copy number for UK controls (4.305). We therefore find no evidence to support the association reported by Fellerman et al. between low beta-defensin copy number and Crohn's disease of the colon.
The best analysis of the association of copy number data with disease would come from an analysis that explicitly accounted for the measurement error in the evaluation of evidence for association, such as that formulated in the CNVtools package of Barnes et al. (21). We attempted to apply these improved methods of analysis to our PRT data, but were unable to obtain convergence in the clustering phase of the analysis; we speculate that this failure comes from a combination of the requirement to extract a relatively large number of closely spaced copy number classes using only a small number of individual data points (two PRTs) per sample. Analyses like those implemented in CNV tools, however, remain most likely to be robust to the effects of measurement error.
We simulated the power of our study to detect the specific effect reported by Fellermann et al. of increased relative risk for samples with copy numbers lower than 4; we found >99% power to detect an odds ratio of 3.06 and 70% to detect an effect at the lower bound of the 95% confidence interval found by Fellermann et al. (an odds ratio of 1.46). A study of the size used in our work also has about 95% power to detect the specific finding of Bentley et al. (odds ratio of 1.54 for samples with five or more copies), and even 49% power to detect an effect of half the size (i.e. an odds ratio of 1.27). We do not therefore believe that our attempt to reproduce these reported associations with Crohn's disease has been seriously compromised by lack of power.
DISCUSSION
Our results do not support the findings of Bentley et al. (10) of a significantly increased beta-defensin copy number among Crohn's disease patients. Bentley et al. (10) found a mean increase in copy number of 0.41 repeats among affected individuals of European origin from New Zealand (mean copy number 4.36 in Crohn's patients, 3.95 in controls); in contrast, we find no clear increase in mean copy number (4.48 for cases and 4.42 for controls) for London samples, and the elevation seen among the Edinburgh samples (mean copy number for cases = 4.40 and mean for controls = 4.24) is not significant.
It is of particular concern that the most striking difference between our results and those of Bentley et al. is that the New Zealand group of controls has a substantially lower mean copy number than our UK controls (3.95 for European New Zealanders versus 4.305 for combined UK control samples). We analysed the frequency distribution of beta-defensin copy number classes between the controls of European origin typed in this study, by Fellermann et al., by Bentley et al. and of the European controls typed by McCarroll et al. (22). The distribution of copy number classes found in this study, or by Fellermann et al., shows no significant discrepancy with that found by McCarroll et al.; however, although it remains possible that there are genuine differences of copy number frequencies between New Zealanders and other populations of European origin, the copy number distribution in controls of European origin typed by Bentley et al. is significantly different from that determined by McCarroll et al. (P = 3.4 × 10−4) and from the distribution in this study (P = 1.9 × 10−7).
In the recent report from the Welcome Trust Case–Control Consortium (WTCCC) on CNV associations in eight common diseases, no significant association was observed between beta-defensin copy number and Crohn's disease (23). Array-CGH was used to interrogate more than 3000 CNVs, including high-quality analysis of the beta-defensin CNV. The report included a useful and detailed discussion of artefactual influences on copy number determination using their custom array-CGH platform; although the detailed experimental and biological factors influencing test outcome will differ between their customized genome-wide platform and methods (such as PRT and real-time PCR) for the analysis of individual candidate CNVs, their evidence resonates with our own findings in demonstrating the clear potential for reporting spurious associations with CNVs, especially multi-allelic CNVs. The WTCCC study also failed to replicate the reported association of rheumatoid arthritis with CCL3L1/CCL4L1 copy number (24), which (like the reported associations of Crohn's disease with beta-defensin copy number) relied on copy number determination by real-time PCR, unsupported by independent methods.
The studies of both Bentley et al. and Fellerman et al. depended exclusively on real-time PCR methods for the measurement of beta-defensin copy number (9,10). Both used a two-colour real-time PCR assay relative to ALB as a control locus. The study of Fellermann et al. assigned samples to copy number classes using both a standard curve and a ΔΔCT analysis, but their report does not show the distribution of unrounded measurements, but only the data as assigned to integer classes. Bentley et al. used a standard-curve method of analysis, and although unrounded data are shown in their Supplementary Material, there is no clear indication of clustering around integer values. Although mosaicism for beta-defensin copy number among the cells sampled to provide a DNA sample from a patient or control is in principle possible, there is no good evidence justifying this interpretation of a non-integer copy number measurement for beta-defensins. Indeed, the clear assignment of most samples to values close to an integer using our PRT-based methods, alongside the comparison with real-time PCR results from the same samples (Figs 3 and 4), strongly suggests that even when carefully applied, some real-time PCR measurements may be too prone to influences from factors other than the true copy number to be valuable in making the fine distinctions of DNA representation necessary to type samples accurately. It is of particular concern that measurements applying two analyses of the real-time PCR data (Fig. 4A) show good mutual correlation, despite apparently being subject to measurement error of sufficient magnitude to obscure the resolution of results into integer-centred copy number classes. Similarly, the data of Bentley et al. do not show any clear tendency to cluster around integer values, even at the relatively low copy numbers of 2, 3 or 4. This in turn suggests that in this case the real-time PCR measurements of beta-defensin copy number may be influenced by additional factors (such as the physicochemical state of the DNA) to a sufficient extent to result in a frequently incorrect integer call (Fig. 4B), but may nevertheless return data reproducible enough to create a false impression of robustly accurate measurement (Fig. 4A).
Our experiences in side-by-side typing of this CNV by two methods on the same samples reinforce the view that great care should be taken in case–control association studies using multi-allelic copy number variants (11). Although measurement error that applies equally to cases and controls will largely act to obscure any true association that exists, even a very small differential bias—in which cases and controls are subject to different biases in typing results, perhaps deriving from different methods of DNA preparation, storage or handling (17)—applied systematically to large case–control comparisons can result in spurious but apparently highly significant associations with copy number. Our experience in this work leads us to the conclusion that copy number measurement using real-time PCR appears particularly prone to measurement error, as is also apparent from the general failure of real-time PCR methods to resolve copy number clusters clearly for n > 2 at the CCL3L1 copy number variant (13,14,16). This observation is consistent with the experience of Perne et al. (25), who concluded that real-time PCR was the least consistent of the three methods they tested for beta-defensin copy number measurement.
Very generally, whatever copy number measurement is used, in order to avoid misinterpreting differential measurement bias as a true biological association, we would recommend some simple steps in the design and analysis of copy number-based case–control association studies.
Copy number should be determined by more than one independent measurement per sample.
Case and control DNA samples should be analysed together, preferably interspersing case and control samples in the same plates, to reduce the impact of any ‘batch’ effects on typing.
Reference samples of known copy number should be included in all typing experiments, and data from these samples should be used to assess the true accuracy of the methods used.
In assessing the accuracy of typing for non-reference samples, a general assessment of the quality of the data can be made by examining the degree of clustering around integer values, unless there is strong evidence for true mosaicism, the absence of clear peaks around integer values in the frequency distribution suggests that error is sufficiently frequent and extensive to obscure these clusters.
A sufficiently large data set will include many measurements of apparently the same copy number (for example, n = 4 for beta-defensins), and the distributions for unrounded measurements around a central value can be compared between cases and controls within these integer classes (see Supplementary Material) to test for evidence of differential bias.
Where the data allow, a package such as CNVtools (21) should be used to model and account for measurement error in the evaluation of disease associations.
Finally, in order to allow a full assessment of the overall quality of the copy number typing, there should ideally be full disclosure of all measurements made on all samples in published work (see Supplementary Material). Of particular importance in this respect is the full reporting of the real (unrounded) copy number estimates, rather than of the frequencies of results already rounded to the closest integer.
MATERIALS AND METHODS
DNA samples: Scotland (‘Edinburgh’)
Patients with Crohn's disease attended the inflammatory bowel disease (IBD) clinic at the Western General Hospital, Edinburgh, Scotland, which is a tertiary referral centre for IBD in South-East Scotland. The diagnosis of Crohn's disease adhered to the criteria of Lennard-Jones (26). Age at diagnosis and disease phenotype (location and behaviour) were classified according to the Montreal classification (27). Written informed consent was obtained from all patients. Healthy controls were either unrelated spouses/friends of IBD patients or anonymized blood samples obtained from the Scottish Blood Transfusion Service. The Medicine and Oncology Subcommittee of the Lothian Local Research Ethics Committee approved the study protocol (LREC 2000/4/192). DNA was extracted by either a modified salting out technique as used previously (28) or by using a Nucleon blood DNA extraction kit according to the manufacturer's protocol. Of the 435 (total) Crohn's disease samples typed in this study, 418 were also included in the WTCCC (29).
DNA samples: England (‘London’)
DNA samples designated as extracted at the ‘London’ Centre were white European patients with Crohn's disease recruited from specialist IBD clinics in London and Newcastle as reported elsewhere (30) after informed consent and ethical review (REC 05/Q0502/127). DNA was extracted from 10 ml of venous blood by salt/chloroform extraction (30). Of the 666 samples used in this study, 491 were also included in the WTCCC. Human random control (HRC) samples from the UK (from the ECACC collection: http://www.hpacultures.org.uk/) were used as control samples for the London Crohn's disease cohort.
Copy number typing using real-time PCR
Copy number estimation of the human DEFB4 and human serum albumin (ALB) genes was carried out using a QuantiTect Multiplex PCR kit (Qiagen, UK) and assayed on a Rotorgene (Corbett Research, now Qiagen, UK). Taqman primers and probes were obtained (Applied Biosystems, UK) as 20× mixes. ALB was used as a reference gene as it has a single copy per haploid genome (31). Cycling conditions were: initial hold at 95°C for 3 min followed by 45 cycles of: 95°C for 20 s, 60°C for 30 s, 72°C for 30 s. Primer sequences used: DEFB4 forward primer: TGTGGCTGATGCGGATTCA, DEFB4 reverse primer: CGTATCTTTGGACACCATAGTTTAATTTGG; DEFB4 FAM-labelled probe CATGTCGCACGTCTCTG; ALB forward primer: GGCTCAGCAGTGGAATACTCT, ALB reverse primer: TTGCCCTATAAAGCACTCTTTCATGT; ALB JOE-labelled probe TTCAGCCTAATTCCC. Patient DNA samples were quantified and 5 ng of DNA was used for each duplicate reaction. A DNA sample of known copy number was used to construct a DNA concentration standard curve (from 50 ng to 0.1 ng DNA per reaction). DNA samples of known copy numbers (assayed by Ed Hollox, University of Leicester), ranging from 2 to 7, were used as known controls in all assays. The raw data were analysed by comparing the DEFB4:ALB threshold cycle ratios (ΔΔCt method) with samples of known copy number. In addition, the same data were analysed using a standard curve method comparing the DEFB4:ALB concentration ratios with samples of known copy number. These methods gave similar results for each sample for the copy number estimation.
Copy number typing using PRT-based methods
In general, the PRT seeks to avoid the problems inherent in comparative PCR methods (including real-time PCR) caused by the comparison between the yield of two dissimilar amplicons, for which the amplification process may be differentially susceptible to minor perturbations in experimental conditions (18). To achieve this, PRT primers are designed to exploit sequence similarities between elements (often dispersed repeats) present both in the copy variable unit and at another genomic location. A single pair of primers is designed to amplify both the region under study (the ‘test’ locus) and exactly one other example from a region of constant copy number (the ‘reference’ locus). If the (very similar) test and reference products can be distinguished and quantified separately, for example, by length, the ratio of test:reference product yields provides an accurate measure of the copy number.
The new ‘triplex’ composite system for typing beta-defensin copy number incorporates two PRT systems [PRT107A (19) and HSPD21 PRT (see below)] together with an allelic ratio test based on the multi-allelic indel rs5889219 (19). HSPD21 PRT uses the same heat-shock pseudogene HSPDP3 as described previously (18) but uses an alternative placement of primers to amplify beta-defensin-linked DNA in conjunction with reference products from chromosome 21 using primers HSPD21F (GAGGTCACTGTGATCAAAGAT) and HSPD21R (AACCTTCAGCACAGCTACTC). Unlike the original HSPDP3 PRT described (18), the test and reference products for HSPD21 PRT and PRT107A can be resolved by length without restriction digestion, and so can be analysed without further processing. The rs5889219 primers used (5DELF AAACCAATACCCTTTCCAAG with 5DELR4 TTCTCTTTTGTTTCAGATTCAGATG) give products in the range 123–128 bp. HSPD21 and PRT107A products were amplified from 10 ng of genomic DNA in duplex in 10 μl of PCRs using an initial denaturation step at 95°C for 5 min followed by 22 cycles of 95°C 30 s/58°C 30 s/70°C 1 min, followed by a single step of 58°C 30 s/70°C 40 min. rs5889219 was amplified separately in 10 μl reactions with 25 cycles of 95°C 30 s/50°C 30 s/70°C 1 min, followed by a single step of 50°C 1 min/70°C 20 min. To add to the measurement accuracy without greatly increasing the cost, each PCR was carried out twice using different fluorescently labelled primers (FAM- or HEX-labelled PRT107AF, FAM- or NED-labelled HSPD21R, HEX- or NED-labelled 5DELF), and all the products mixed before electrophoresis on a single ABI3100 capillary. In all cases, PRT measurements were carried out on mixed plates containing randomly interspersed case, control and reference DNA samples, as well as no-DNA blanks.
Likelihood analysis of triplex copy number measurements
PRT data were initially calibrated as described (18,19) relative to standard reference samples of known copy number obtained from the ECACC-HRC collection using the mean ratio from the two fluorescent labels for each PRT, resulting in a single (unrounded) copy number estimate for each sample from each PRT. In cases from both cohorts and controls from the Edinburgh collection, maxima in the initially calibrated distributions for PRT107A were not centred precisely on integer values; in contrast, the ECACC-HRC samples used as controls for the London-based cases showed no such shift in the centres of PRT107A clusters, implying that copy number values measured by PRT107A can be sensitive to differences in DNA preparation methods. To avoid propagating error into further analyses, a linear transformation was applied to all PRT107A values from non-ECACC samples to improve the consistency with integer clustering (Supplementary Material) before further analysis. In addition to two copy number estimates derived from PRT, there were also (in two fluorescent labels) measurements of rs5889219 allele representations for each sample.
Although both PRT107A and HSPD21 PRT return numerical estimates of copy number, rs5889219 ratios only indicate consistency with particular integer values. We therefore developed a likelihood framework in which data from both PRT and rs5889219 measurements could be combined to indicate the probable copy number state for each sample. To allow simple placement of most samples in integer copy number classes, but to simultaneously allow some evaluation of the statistical confidence that could be placed in these allocations, the likelihood method evaluated the joint probability of all the data observed for a given sample across a range of copy numbers from n = 1 to n = 9. The distribution of PRT measurements for each given true copy number was modelled using a set of Gaussian distributions with means and standard deviations derived for each PRT from extensive empirical data. rs5889219 ratios were modelled using a similarity score, again derived from extensive empirical observations. For each set of data from the triplex test, a copy number maximizing the likelihood of all the observations was derived, together with a ‘minimum ratio’, the likelihood ratio between the most likely and the next most likely integer copy numbers, used as a crude index of the confidence of the copy number call. Further analysis of some samples in which the support for the maximum-likelihood copy number was relatively low, or in which there was evidence of discordancy between different components of the assay (n = 48 for London samples, n = 51 for Edinburgh samples), was undertaken. Microsatellite typing (19) was carried out to clarify the integer copy number for 35 of these, and the reported copy number altered in 20 cases (see annotations to full data set in Supplementary Material). Further details of methods and programs used are available on request.
Tests of association with disease status were carried out using χ2 tests on 3 × 2 contingency tables, classifying samples into low (3 or fewer), central (n = 4) or high (5 or more) copy number categories.
Typing success and missingness
From 666 Crohn's disease DNA samples from the London collection, PRT-based triplex data suitable for analysis were produced by 648 samples and by all of 185 control samples (P = 0.049). Among the Edinburgh samples, 358 out of 384 Crohn's disease DNA samples produced suitable triplex data and 314 out of 350 control samples (P = 0.11). The overall call rate for typing was therefore approximately 95%. In CNV studies at multi-allelic loci such as the beta-defensins, there is a particular danger that application of a quality threshold will lead to differential rejection of higher copy number samples (because it will be harder for these samples to meet a quality criterion). More seriously, if this affects cases and controls differentially, there is the potential for a spurious association to be generated. We therefore tested our data for the potential effects of applying a quality control filter, accepting only data resulting in a ‘minimum ratio’ (see above) greater than 20. This led overall to the rejection of just under 10% of samples and to a highly significant overrepresentation of higher copy numbers among the rejected samples (mean copy number of rejected samples 5.06, compared with 4.35 for accepted samples, χ2 P = 9.7 × 10−8). Nevertheless, we found no evidence that this affected cases and controls differentially among either the Edinburgh or London samples (P > 0.05).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from the Chief Scientist's Office, Scottish Government (to M.C.A.), a PhD scholarship from the Ministry of Higher Education, Malaysia (to S.A.B.), a Studentship from the Portuguese FCT (to R.P.), grant funding from the Wellcome Trust (programme grant funding to J.S., grant 081808/CGM to C.G.M.), the National Institutes of Health Research Biomedical Research Centre at Guy's and St Thomas’ NHS Foundation Trust in partnership with King's College London (to N.J.P. and C.G.M.) and the Guy's and St Thomas' Charity (to C.G.M.).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Ed Hollox (University of Leicester) for helpful discussions and interactions in this work and to Dr Tom Reader (University of Nottingham) for help with analytical methods.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Hollox E.J., Armour J.A.L., Barber J.C.K. Extensive normal copy number variation of a beta-defensin antimicrobial-gene cluster. Am. J. Hum. Genet. 2003;73:591–600. doi: 10.1086/378157. doi:10.1086/378157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hollox E.J., Davies J., Griesenbach U., Burgess J., Alton E.W.F.W., Armour J.A.L. Beta-defensin genomic copy number is not a modifier locus for cystic fibrosis. J. Negat. Results Biomed. 2005;4:9. doi: 10.1186/1477-5751-4-9. doi:10.1186/1477-5751-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linzmeier R.M., Ganz T. Human defensin gene copy number polymorphisms: comprehensive analysis of independent variation in alpha- and beta-defensin regions at 8p22–p23. Genomics. 2005;86:423–30. doi: 10.1016/j.ygeno.2005.06.003. doi:10.1016/j.ygeno.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Hollox E.J., Barber J.C.K., Brookes A.J., Armour J.A.L. Defensins and the dynamic genome: what we can learn from structural variation at human chromosome band 8p23.1. Genome Res. 2008;18:1686–1697. doi: 10.1101/gr.080945.108. doi:10.1101/gr.080945.108. [DOI] [PubMed] [Google Scholar]
- 5.Aldhous M., Noble C., Satsangi J. Dysregulation of human beta-defensin-2 protein in inflammatory bowel disease. PLoS ONE. 2009;4:e6285. doi: 10.1371/journal.pone.0006285. doi:10.1371/journal.pone.0006285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003;3:710–20. doi: 10.1038/nri1180. doi:10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 7.Niyonsaba F., Ogawa H., Nagaoka I. Human beta-defensin-2 functions as a chemotactic agent for tumour necrosis factor-alpha-treated human neutrophils. Immunology. 2004;111:273–81. doi: 10.1111/j.0019-2805.2004.01816.x. doi:10.1111/j.0019-2805.2004.01816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hollox E.J., Huffmeier U., Zeeuwen P.L.J.M., Palla R., Lascorz J., Rodijk-Olthuis D., van de Kerkhof P.C.M., Traupe H., de Jongh G., Heijer M.d., et al. Psoriasis is associated with increased [beta]-defensin genomic copy number. Nat. Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. doi:10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fellermann K., Stange D.E., Schaeffeler E., Schmalzl H., Wehkamp J., Bevins C.L., Reinisch W., Teml A., Schwab M., Lichter P., et al. A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am. J. Hum. Genet. 2006;79:439–448. doi: 10.1086/505915. doi:10.1086/505915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bentley R., Pearson J., Gearry R., Barclay M., McKinney C., Merriman T., Roberts R. Association of higher DEFB4 genomic copy number with Crohn's disease. Am. J. Gastroenterol. 2010;105:354–359. doi: 10.1038/ajg.2009.582. doi:10.1038/ajg.2009.582. [DOI] [PubMed] [Google Scholar]
- 11.McCarroll S.A. Copy-number analysis goes more than skin deep. Nat. Genet. 2008;40:5–6. doi: 10.1038/ng0108-5. doi:10.1038/ng0108-5. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez E., Kulkarni H., Bolivar H., Mangano A., Sanchez R., Catano G., Nibbs R.J., Freedman B.I., Quinones M.P., Bamshad M.J., et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. doi:10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 13.Bhattacharya T., Stanton J., Kim E.Y., Kunstman K.J., Phair J.P., Jacobson L.P., Wolinsky S.M. CCL3L1 and HIV/AIDS susceptibility. Nat. Med. 2009;15:1112–1115. doi: 10.1038/nm1009-1112. doi:10.1038/nm1009-1112. [DOI] [PubMed] [Google Scholar]
- 14.Field S.F., Howson J.M.M., Maier L.M., Walker S., Walker N.M., Smyth D.J., Armour J.A.L., Clayton D.G., Todd J.A. Experimental aspects of copy number variant assays at CCL3L1. Nat. Med. 2009;15:1115–1117. doi: 10.1038/nm1009-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He W.J., Kulkarni H., Castiblanco J., Shimizu C., Aluyen U., Maldonado R., Carrillo A., Griffin M., Lipsitt A., Beachy L., et al. Experimental aspects of copy number variant assays at CCL3L1 Reply. Nat. Med. 2009;15:1117–1120. doi: 10.1038/nm1009-1117. [DOI] [PubMed] [Google Scholar]
- 16.Urban T.J., Weintrob A.C., Fellay J., Colombo S., Shianna K.V., Gumbs C., Rotger M., Pelak K., Dang K.K., Detels R., et al. CCL3L1 and HIV/AIDS susceptibility. Nat. Med. 2009;15:1110–1112. doi: 10.1038/nm1009-1110. doi:10.1038/nm1009-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clayton D.G., Walker N.M., Smyth D.J., Pask R., Cooper J.D., Maier L.M., Smink L.J., Lam A.C., Ovington N.R., Stevens H.E., et al. Population structure, differential bias and genomic control in a large-scale, case–control association study. Nat. Genet. 2005;37:1243–1246. doi: 10.1038/ng1653. doi:10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 18.Armour J.A.L., Palla R., Zeeuwen P.L.J.M., den Heijer M., Schalkwijk J., Hollox E.J. Accurate, high-throughput typing of copy number variation using paralogue ratios from dispersed repeats. Nucleic Acids Res. 2007;35:e19. doi: 10.1093/nar/gkl1089. doi:10.1093/nar/gkl1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abu Bakar S., Hollox E.J., Armour J.A.L. Allelic crossover between distinct genomic locations generates copy number diversity in human beta-defensins Proc. Natl Acad. Sci. USA. 2009;106:853–858. doi: 10.1073/pnas.0809073106. doi:10.1073/pnas.0809073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groth M., Szafranski K., Taudien S., Huse K., Mueller O., Rosenstiel P., Nygren A.O.H., Schreiber S., Birkenmeier G., Platzer M. High-resolution mapping of the 8p23.1 beta-defensin cluster reveals strictly concordant copy number variation of all genes. Hum. Mutat. 2008;29:1247–1254. doi: 10.1002/humu.20751. doi:10.1002/humu.20751. [DOI] [PubMed] [Google Scholar]
- 21.Barnes C., Plagnol V., Fitzgerald T., Redon R., Marchini J., Clayton D., Hurles M.E. A robust statistical method for case–control association testing with copy number variation. Nat. Genet. 2008;40:1245–1252. doi: 10.1038/ng.206. doi:10.1038/ng.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCarroll S.A., Kuruvilla F.G., Korn J.M., Cawley S., Nemesh J., Wysoker A., Shapero M.H., de Bakker P.I.W., Maller J.B., Kirby A., et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. doi:10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 23.Wellcome Trust Case Control Consortium. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. doi:10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKinney C., Merriman M.E., Chapman P.T., Gow P.J., Harrison A.A., Highton J., Jones P.B.B., McLean L., O'Donnell J.L., Pokorny V., et al. Evidence for an influence of chemokine ligand 3-like 1 (CCL3L1) gene copy number on susceptibility to rheumatoid arthritis. Ann Rheum Dis. 2008;67:409–413. doi: 10.1136/ard.2007.075028. [DOI] [PubMed] [Google Scholar]
- 25.Perne A., Zhang X.H., Lehmann L.E., Groth M., Stuber F., Book M. Comparison of multiplex ligation-dependent probe amplification and real-time PCR accuracy for gene copy number quantification using the beta-defensin locus. BioTechniques. 2009;47:1023–1028. doi: 10.2144/000113300. doi:10.2144/000113300. [DOI] [PubMed] [Google Scholar]
- 26.Lennard-Jones J.E. Classification of inflammatory bowel disease. Scand. J. Gastroenterol. 1989;24(suppl.):2–6. doi: 10.3109/00365528909091339. doi:10.3109/00365528909091339. [DOI] [PubMed] [Google Scholar]
- 27.Silverberg M.S., Satsangi J., Ahmad T., Arnott I.D., Bernstein C.N., Brant S.R., Caprilli R., Colombel J.F., Gasche C., Geboes K., et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can. J. Gastroenterol. 2005;19(Suppl. A):5–36. doi: 10.1155/2005/269076. [DOI] [PubMed] [Google Scholar]
- 28.Arnott I.D.R., Nimmo E.R., Drummond H.E., Fennell J., Smith B.R.K., MacKinlay E., Morecroft J., Anderson N., Kelleher D., O'Sullivan M., et al. NOD2/CARD15, TLR4 and CD14 mutations in Scottish and Irish Crohn's disease patients: evidence for genetic heterogeneity within Europe. Genes Immun. 2004;5:417–425. doi: 10.1038/sj.gene.6364111. doi:10.1038/sj.gene.6364111. [DOI] [PubMed] [Google Scholar]
- 29.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. doi:10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prescott N.J., Fisher S.A., Franke A., Hampe J., Onnie C.M., Soars D., Bagnall R., Mirza M.M., Sanderson J., Forbes A., et al. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn's disease and is independent of CARD15 and IBD5. Gastroenterology. 2007;132:1665–1671. doi: 10.1053/j.gastro.2007.03.034. doi:10.1053/j.gastro.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 31.Chen Q.X., Book M., Fang X.M., Hoeft A., Stuber F. Screening of copy number polymorphisms in human beta-defensin genes using modified real-time quantitative PCR. J. Immunol. Methods. 2005;308:231–240. doi: 10.1016/j.jim.2005.11.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.