Abstract
It is a challenge to identify the molecular networks contributing to the neural basis of human speech. Mutations in transcription factor FOXP2 cause difficulties mastering fluent speech (developmental verbal dyspraxia, DVD), whereas mutations of sushi-repeat protein SRPX2 lead to epilepsy of the rolandic (sylvian) speech areas, with DVD or with bilateral perisylvian polymicrogyria. Pathophysiological mechanisms driven by SRPX2 involve modified interaction with the plasminogen activator receptor (uPAR). Independent chromatin-immunoprecipitation microarray screening has identified the uPAR gene promoter as a potential target site bound by FOXP2. Here, we directly tested for the existence of a transcriptional regulatory network between human FOXP2 and the SRPX2/uPAR complex. In silico searches followed by gel retardation assays identified specific efficient FOXP2-binding sites in each of the promoter regions of SRPX2 and uPAR. In FOXP2-transfected cells, significant decreases were observed in the amounts of both SRPX2 (43.6%) and uPAR (38.6%) native transcripts. Luciferase reporter assays demonstrated that FOXP2 expression yielded a marked inhibition of SRPX2 (80.2%) and uPAR (77.5%) promoter activity. A mutant FOXP2 that causes DVD (p.R553H) failed to bind to SRPX2 and uPAR target sites and showed impaired down-regulation of SRPX2 and uPAR promoter activity. In a patient with polymicrogyria of the left rolandic operculum, a novel FOXP2 mutation (p.M406T) was found in the leucine-zipper (dimerization) domain. p.M406T partially impaired the FOXP2 regulation of SRPX2 promoter activity, whereas that of the uPAR promoter remained unchanged. Together with recently described FOXP2-CNTNAP2 and SRPX2/uPAR links, the FOXP2-SRPX2/uPAR network provides exciting insights into molecular pathways underlying speech-related disorders.
INTRODUCTION
The development and functioning of the brain depend on very precise and complicated molecular networks that must be regulated both in time and in space. Animal models and human genetic analyses have identified a large number of proteins participating in crucial brain processes, but little is known about how these individual proteins organize in regulatory and interacting networks. Deciphering the neurogenetic pathways that are associated with speech-related circuits of the brain, while being of prime scientific importance, represents an even more challenging problem given that speech is a unique human trait. Nevertheless, in recent years, progress has been made by studying families where impairment of speech-associated processes is inherited in a monogenic fashion. It has thus been possible to pinpoint genes that constitute key entry points into the molecular networks underlying such processes (1,2). In particular, rare loss-of-function mutations in the FOXP2 gene (OMIM 605317) can cause a severe speech and language disorder (OMIM 602081) (3–5). FOXP2-related disorder is characterized by difficulties in mastering sequences of oral and facial movements, impairing speech articulation (developmental verbal dyspraxia, DVD), accompanied by additional problems with expressive and receptive language. FOXP2 belongs to a family of transcription factors that have crucial functions in a wide range of physiological processes and that are characterized by a forkhead-box (FOX) DNA-binding domain (6). In addition to the generation and characterization of various animal models (7–12), subsequent studies have focussed on the identification of FOXP2-targeted promoter regions (13,14). At least some putative FOXP2-regulated genes are likely to represent important players in speech-associated pathways. Indeed, a recent study found that FOXP2 directly regulates the CNTNAP2 gene (OMIM 604569) which encodes a contactin-associated protein of the neurexin superfamily and showed that polymorphisms in this target gene are associated with specific language impairment (15).
Human genetic analyses have also implicated mutations of the sushi-repeat protein SRPX2 (OMIM 300642) in a range of speech-related disorders. These include epileptic (rolandic/sylvian epilepsy), functional (DVD) and developmental (bilateral perisylvian polymicrogyria) disorders of the speech areas of the cortex (OMIM 300643, 300388) (16). Several proteins that physically interact with SRPX2 have then been identified, suggesting the involvement of the extracellular matrix proteolysis machinery in the pathology and physiology of speech-associated circuits. Most notably, SRPX2 was found to directly interact with uPAR (OMIM 173391), the plasminogen activator receptor of the urokinase type (also known as PLAUR), and the SRPX2/uPAR ligand–receptor interaction showed quantitative modification caused by a pathogenic mutation of SRPX2 (17).
Interestingly, the uPAR promoter region was identified among the top 100 potential FOXP2 targets in a high-throughput chromatin-immunoprecipitation microarray screen (ChIP-chip) using human neuronal models (14). Given that FOXP2 and SRPX2 mutations cause related disorders of speech processing and associated brain areas, this observation raised the exciting possibility of a functional link between the FOXP2 transcription factor on the one hand and the SRPX2/uPAR complex on the other hand. In the present study, we show that human FOXP2 down-regulates both the SRPX2 and the uPAR genes and that this transcriptional regulation is lost when FOXP2 bears a pathogenic p.R553H mutation that is known to cause DVD. Moreover, we report the discovery of a new FOXP2 coding mutation in a patient with polymicrogyria of the left rolandic operculum and demonstrate that it partially impairs the proper FOXP2 regulation of SRPX2 promoter activity. Our study thus identifies a novel genetic and regulatory network that is altered in disorders affecting speech processing and functioning/development of speech-related brain areas.
RESULTS
Bioinformatic detection of FOX, FOXP and FOXP2 consensus-binding sites in SRPX2 and uPAR promoters
FOX transcription factors bind to consensus DNA sites in the cis regulatory regions of their target genes. Consensus-binding sites with increased levels of specificity have been defined for transcription factors of the general FOX family (FOX sites: TRTTKRY, where R = A or G, K = T or G and Y = T or C), for members of the FOXP subfamily (FOXP sites: TATTTRT) and for FOXP2 itself (FOXP2 sites: AATTTG and ATTTGT). The FOXP2 consensus sequence ATTTGT is contained within the FOXP sites and the FOXP sites also conform to the more general FOX sites (18). The promoter regions of the human SRPX2 and of the human uPAR genes were screened in silico for the presence of FOX, FOXP and FOXP2 sites, including approximately 1.5 kb of DNA sequence 5′ to each corresponding and canonical transcription start site (TSS) (SRPX2: GenBank accession no. NM_014467; uPAR: GenBank accession no. NM_002659). In silico and 5′-RACE (rapid amplification of cDNA ends) polymerase chain reaction (PCR) experiments confirmed that the SRPX2 and uPAR brain transcripts detected so far did not display any alternative and more distal 5′ TSS (data not shown). Several consensus sequences of various types (FOX, FOXP, FOXP2) were identified in the promoter regions of both genes (Table 1, Supplementary Material, Fig. S1). Seven sites [SRP1–SRP7, from 3′ to 5′ of the (+) strand] were identified for SRPX2: SRP1, SRP2, SRP5 and SRP7 were of the FOX type and SRP3, SRP4 and SRP6 fitted the consensus for FOXP2. Six sites (UP1–UP6) were detected for uPAR: UP1–UP3 were of the FOX type and UP4–UP6 were of the FOXP type. Some of these sites (SRP1, UP2, UP5 and UP6) were composed of more than one consensus sequence. The presence of such consensus-binding sites in silico was thus consistent with a possible regulation of SRPX2 and of uPAR by transcription factors of the FOX family. Notably, comparative searches of those consensus-binding sites (SRP1-7 and UP1-6, respectively) in the Srpx2 and Upar promoter sequences from chimpanzee and mouse revealed subtle to dramatic differences with their human promoter counterparts, with the most obvious evolutionary modification being the lack of any of the UP1 to UP6 consensus forkhead binding site in the mouse Upar promoter region (Supplementary Material, Fig. S1).
Table 1.
In silico detection of FOX, FOXP and FOXP2 consensus-binding sites
| Sequencea | Consensus-binding siteb |
Position (nt)c | |||
|---|---|---|---|---|---|
| FOX | FOXP | FOXP2 | |||
| SRPX2 | |||||
| SRP1d | TGTTTGTgagTGTTTAT | + | − | − | −110; −120 |
| SRP2 | GTAAACA | + | − | − | −305 |
| SRP3 | AATTTG | − | − | + | −664 |
| SRP4 | TATTTGT | + | + | + | −767 |
| SRP5 | TGTTGGC | + | − | − | −919 |
| SRP6 | TATTTGT | + | + | + | −1260 |
| SRP7 | TGTTTGAT | + | − | − | −1373 |
| uPAR | |||||
| UP1 | TATTTAC | + | − | − | −131 |
| UP2e | ACAAACAAACAAACA | + | − | − | −693; −697; −701 |
| UP3 | ATCAATA | + | − | − | −746 |
| UP4 | ATAAATA | + | + | − | −832 |
| UP5e | TATTTATTTATTTAT | + | + | − | −1160; −1164; −1168 |
| UP6f | ATAAATAAATAAATAAATA | + | + | − | −1319; −1323; −1327; −1331 |
The promoter regions of the human SRPX2 and uPAR genes were screened in silico for the presence of FOX, FOXP and FOXP2 consensus-binding sites (Supplementary Material, Fig. S1). nt, nucleotide.
aCore binding sites are represented in capital letters.
b(+) indicates the presence of a consensus-binding site and (−) indicates the absence of a consensus-binding site.
cPosition relative to the canonical TSS.
dTwo non-overlapping sites.
eThree overlapping sites.
fFour overlapping sites.
FOXP2 expression leads to a significant decrease in native SRPX2 and uPAR transcripts
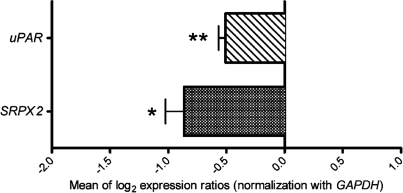
The possible regulation of SRPX2 and uPAR by FOXP2 was then addressed by measuring by quantitative RT–PCR (qRT–PCR) the expression of these genes in the presence of exogenous human FOXP2 protein encoded by the major transcript of FOXP2 (isoform 1; GenBank accession no. NM_014491). When HEK293T cells were transfected with a construct expressing recombinant FOXP2 protein, SRPX2 mRNA levels were significantly reduced (43.6% decrease, P < 0.05, two-tailed unpaired t-test) across replicate experiments when compared with control cells transfected with the corresponding non-recombinant vector (Fig. 1). Similarly, the amounts of uPAR transcripts also showed significant and replicated decrease (38.6% decrease, P < 0.01, two-tailed unpaired t-test) in FOXP2-transfected cells (Fig. 1). This indicated the existence of a functional link between FOXP2 and both the SRPX2 and the uPAR genes.
Figure 1.
qRT–PCR experiments. Wild-type FOXP2 protein down-regulates the native SRPX2 and uPAR genes. qRT–PCR experiments were performed from HEK293T cells transfected with pcDNA4/HisMax-FOXP2 construct. Expression changes are given as the mean of log2 expression ratios of cells transfected with pcDNA4/HisMax-FOXP2 compared with cells transfected with non-recombinant pcDNA4/HisMax vector and are normalized for equal expression of the GAPDH internal control. Values represent the mean of comparisons of five independent cDNA syntheses. P-values were calculated using two-tailed unpaired t-test. *P< 0.05 and **P < 0.01.
FOXP2 represses SRPX2 and uPAR promoters
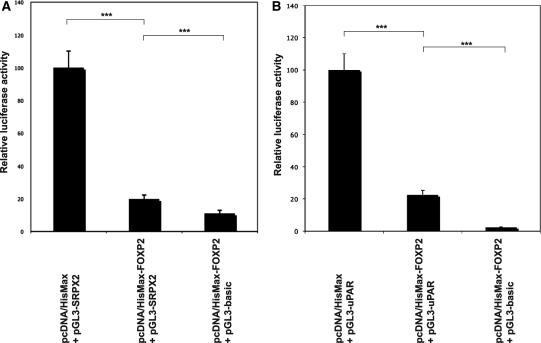
We next tested whether expression of FOXP2 could modify the activities of the SRPX2 and of the uPAR promoters by using a luciferase reporter assay. The promoter regions of each gene as defined above were subcloned into the appropriate vector (pGL3) 5′ to the coding sequence of the firefly luciferase reporter gene. Either construct (pGL3-SRPX2, pGL3-uPAR) was cotransfected with a FOXP2-containing vector (pcDNA4/HisMax-FOXP2) or with an empty (pcDNA4/HisMax) vector and with a β-galactosidase expression vector (pHSV-LacZ) for the normalization of transfection efficiencies. Significant and replicated repression of the SRPX2 promoter by FOXP2 protein was detected in pcDNA4/HisMax-FOXP2 transfected cells when compared with pcDNA4/HisMax-transfected cells (80.2% decrease in luciferase expression, P < 0.0005, two-tailed unpaired t-test) (Fig. 2A). As a positive control, FOXP2 was also found to repress the SV40 promoter (data not shown), as already described (18). Using the same experimental procedure, dramatic and reproducible repression (77.5% decrease in luciferase expression, P < 0.0005, two-tailed unpaired t-test) of the uPAR promoter by FOXP2 was shown (Fig. 2B). Together with the qRT–PCR data, the reporter assays shown here demonstrated the ability of the human FOXP2 protein to repress the activities of SRPX2 and uPAR promoters.
Figure 2.
Luciferase reporter assays. Wild-type FOXP2 protein down-regulates the activities of the SRPX2 (A) and of the uPAR (B) promoters. Either of the SRPX2 (pGL3-SRPX2) and uPAR (pGL3-uPAR) promoter constructs was co-transfected into HEK293T cells with FOXP2-containing (pcDNA4/HisMax-FOXP2) or with empty (pcDNA4/HisMax) vector and with a β-galactosidase expression vector (pHSV-LacZ) for the normalization of transfection efficiencies. In parallel, an empty (promoterless) vector (pGL3-basic) was transfected with pcDNA4/HisMax-FOXP2 and with pHSV-LacZ. Transcriptional activities were determined by quantifying the luciferase activity of cellular extracts prepared 48 h after transfection. Data show the mean ± SD relative activity from three experiments done in triplicate. Statistical significances were determined by two-tailed unpaired t-test. ***P < 0.0005.
FOXP2 directly binds to consensus sites in the SRPX2 and uPAR promoters
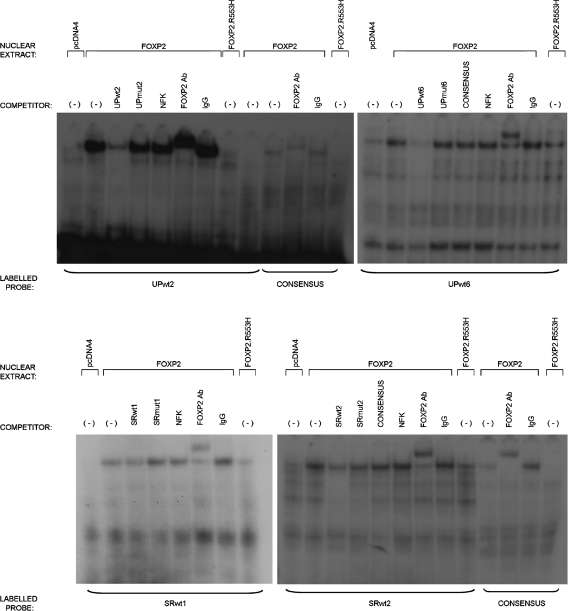
Repression of the SRPX2 and uPAR promoter activities and transcript expressions by FOXP2, and the presence of FOX, FOXP and FOXP2 consensus-binding sites within the promoter regions of SRPX2 and of uPAR, strongly suggested that FOXP2 could directly bind to these promoter regions. We tested this by carrying out electrophoretic mobility shift assays (EMSAs) for the 13 consensus-binding sites (SRP1-7 and UP1-6) that had been detected in silico in the SRPX2 and uPAR promoter sequences (Supplementary Material, Fig. S1 and Table S1). Nuclear extracts were prepared from HEK293T cells transfected with the FOXP2 expression construct (pcDNA4/HisMax-FOXP2). These extracts were able to bind to a positive control consensus FOXP2 site as previously shown (18) and to DNA probes corresponding to the UP2 and UP6 sites of the uPAR promoter region, as well as to the SRP1 and SRP2 sites of the SRPX2 promoter region (Fig. 3, Supplementary Material, Fig. S2). In contrast, no binding was observed when using nuclear extracts prepared from HEK293T cells transfected with an empty construct (pcDNA4/HisMax; line pcDNA4 in each panel of Fig. 3). The gel shifts that were observed with SRP1, SRP2, UP2 and UP6 probes were due to binding by FOXP2 protein, since addition of an N-terminal FOXP2 antibody caused larger complexes or ‘supershifts' to occur (line FOXP2Ab in each panel of Fig. 3). By comparison, no supershift was obtained when a non-specific IgG antibody was used. Binding to either of the SRP1, SRP2, UP2 and UP6 probes was specific, as demonstrated by competitive impairment with each corresponding unlabeled probe (lines UPwt2, UPwt6, SRwt1, SRwt2 in Fig. 3). Moreover, mutant (Supplementary Material, Table S1) and unlabeled forms of SRP1, SRP2, UP2 and UP6 probes (lines UPmut2, UPmut6, SRmut1, SRmut2 in Fig. 3) failed to compete with the FOXP2-binding abilities of their labeled and wild-type counterparts, as also shown for an irrelevant NFK unlabeled probe. Altogether, these data demonstrated efficient specific binding of FOXP2 to consensus sites situated within the promoter regions of both the SRPX2 and uPAR genes.
Figure 3.
Electrophoretic mobility shift assays. FOXP2 directly binds sites identified within the SRPX2 and uPAR promoters. EMSAs were used to determine whether direct interactions occurred between FOXP2 and the promoter regions of the SRPX2 and uPAR genes. DNA probes were designed based on the presence of predicted FOX, FOXP and FOXP2-binding sites (Table 1, Supplementary Material, Table S1). Radiolabeled probes that had shown binding in earlier experiments (Supplementary Material, Fig. S2) were incubated with nuclear extracts taken from HEK293T cells transfected with either an empty vector (pcDNA4), wild-type FOXP2 or FOXP2.R553H. The binding of FOXP2 to a known consensus binding site (CONSENSUS) is shown as a positive control. Binding assays were also performed in the presence of competition from unlabeled wild-type (wt), mutant (mut) and irrelevant (NFK) probes. In each case, FOXP2 binding to the labeled wild-type probe was efficiently impaired via competition with unlabeled wild-type competitor probe, but not by mutant or irrelevant competitor probes, displaying the specificity of the interaction. Addition of an antibody directed to FOXP2 (FOXP2 Ab) caused a supershift to occur in each case, an effect not observed when an irrelevant control antibody (IgG) was added. This confirmed that the identity of the protein causing gel retardation is indeed FOXP2.
Pathogenic mutation of FOXP2 disrupts its functional links with SRPX2 and uPAR
Rare mutations in transcription factor FOXP2 lead to DVD (3–5); hence, targets downstream to FOXP2 are obvious candidates for being involved in pathophysiological mechanisms. Based on the data presented above, SRPX2 and uPAR may be considered particularly strong candidates because of the demonstrated relationship between the SRPX2/uPAR complex and disorders of the speech cortex, including DVD itself (13,14). This question was addressed by studying the most well-characterized aetiological mutation of FOXP2 (p.R553H), assessing its functional effects on the SRPX2 and uPAR genes. p.R553H is a pathogenic substitution in the DNA-binding domain of the FOXP2 protein, found in all 15 affected members of an extensively studied multigenerational family segregating speech and language disorder (the KE family) (3).
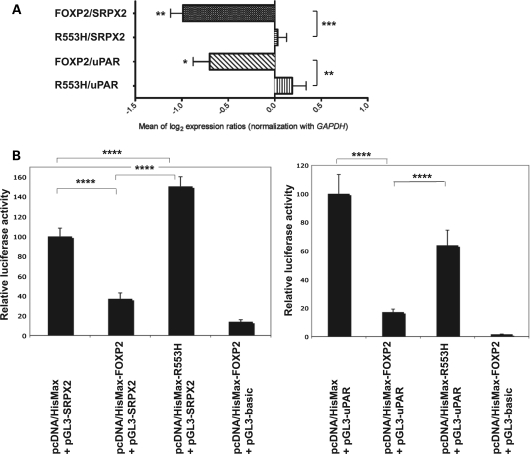
qRT–PCR experiments showed that, in contrast to wild-type FOXP2, p.R553H mutant FOXP2 showed a significant and complete loss of repression of native SRPX2 and uPAR expression (P < 0.001 and P < 0.01, respectively, two-tailed unpaired t-test) (Fig. 4A). Luciferase assays yielded consistent data; the p.R553H mutation led to significant losses of repression of SRPX2 and uPAR promoters when compared with wild-type FOXP2 (P < 0.0001 for each, two-tailed unpaired t-test) (Fig. 4B). There even was increased activity (P < 0.0001, two-tailed unpaired t-test) of the SRPX2 promoter in the presence of mutant p.R553H FOXP2 (Fig. 4B), when compared with cells transfected with a pcDNA4/HisMax empty vector. Overall, these experiments demonstrated a strong reduction in the capacity of mutant p.R553H FOXP2 protein to down-regulate SRPX2 and uPAR.
Figure 4.
Regulation of, and binding on, the SRPX2 and uPAR promoters is altered by a pathogenic p.R553H FOXP2 mutation. (A) qRT–PCR experiments showing loss of down-regulation of the native SRPX2 and uPAR expressions by mutant FOXP2. Expression changes are given as the mean of log2 expression ratios of HEK293T cells transfected with pcDNA4/HisMax-FOXP2 (wild-type FOXP2) or with pcDNA4/HisMax-R553H (mutant p.R553H FOXP2), when compared with HEK293T cells transfected with non-recombinant pcDNA4/HisMax vector, and are normalized for equal expression of the GAPDH internal control. Values represent the mean of comparisons of five independent cDNA syntheses. P-values were calculated using two-tailed unpaired t-test. *P < 0.05, **P < 0.01 and ***P < 0.001. (B) Luciferase reporter assays showing loss of down-regulation of the SRPX2 and uPAR promoter activities by mutant p.R553H FOXP2. Either of the SRPX2 (pGL3-SRPX2) and uPAR (pGL3-uPAR) promoter constructs was co-transfected into HEK293T cells with FOXP2-containing (pcDNA4/HisMax-FOXP2 or pcDNA4/HisMax-R553H) or with empty (pcDNA4/HisMax) vector and with a β-galactosidase expression vector (pHSV-LacZ) for the normalization of transfection efficiencies. In parallel, the empty (promoterless) vector (pGL3-basic) was transfected with pcDNA4/HisMax-FOXP2 and with pHSV-LacZ. Transcriptional activity was determined by quantifying the luciferase activity of cellular extracts prepared 48 h. after transfection. Data show the mean ± SD relative activity from three experiments done in triplicate. Statistical significances were determined by two-tailed unpaired t-test. ****P < 0.0001.
Loss of down-regulation of SRPX2 and of uPAR by mutant FOXP2 could be partly due to disrupted nuclear localization (18). Indeed, alteration of FOXP2 nuclear targeting was confirmed in the present study by immunocytochemistry with Xpress epitope antibody after transfection of HEK293T cells with tagged constructs allowing expression of wild-type or p.R553H FOXP2 (Fig. 5). While increased cytoplasmic localization was clearly detected when compared with wild-type FOXP2, a proportion of mutant p.R553H FOXP2 protein still showed nuclear localization, consistent with previously published data (18). FOXP2 DNA-binding capabilities can be affected by the p.R553H substitution (18) and this question was thus more specifically addressed with respect to the SRPX2 (SRP1 and SRP2) and uPAR (UP2 and UP6) binding sites using the same EMSA procedure as described above. Mutant p.R553H FOXP2 not only displayed reduced binding to a previously defined consensus site used as a control (18), but also showed unambiguous impairment (of variable degree) in its ability to bind to the consensus SRP1, SRP2, UP2 and UP6 target sites in the promoter regions of SRPX2 and of uPAR (Fig. 3).
Figure 5.
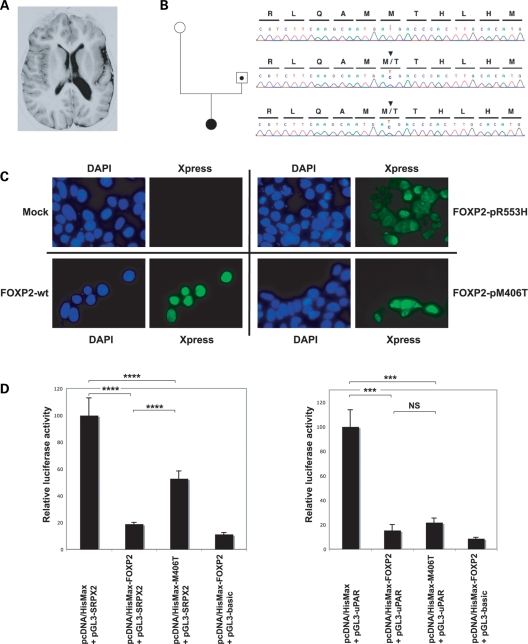
Novel p.M406T FOXP2 mutation in a patient with polymicrogyria of the left rolandic operculum partially impairs proper regulation of SRPX2. (A) MRI (axial inversion recovery section) of the patient with left opercular polymicrogyria. (B) c.T1591C mutation of FOXP2. The nucleotide sequences and translations are shown above the direct sequencing trace from PCR-amplified fragment. Top: Section of a wild-type sequence in the unaffected mother (white circle). Middle: Section of a sequence with the mutation in the carrier father (dotted square). Bottom: Section of a sequence with the mutation in the affected proband (full-blackened circle). Two more siblings were carriers, but did not show obvious neurological problems, although no MRI was performed (sequence traces not shown). (C) p.R553H and p.M406T mutant FOXP2 proteins display altered intracellular localizations. HEK293T cells were transfected with pcDNA4/HisMax (mock) vector (top, left), with pcDNA4/HisMax-FOXP2 (wt) vector (bottom, left), with pcDNA4/HisMax-R553H (p.R553H) vector (top, right) or with pcDNA4/HisMax-M406T (p.M406T) vector (bottom, right). Wild-type FOXP2 displayed unambiguous nuclear localization. Mutant p.R553H FOXP2 showed both nuclear and cytoplasmic localizations, as already described (18), and p.M406T FOXP2 also showed altered nuclear targeting. HisMax-tag fusion protein FOXP2 was detected using an antibody to the N-terminal Xpress™ tag (green). Blue: nuclear DAPI staining. (D) Luciferase reporter assays showing partial loss of down-regulation of the SRPX2 promoter activity by mutant p.M406T FOXP2. Activity of the uPAR promoter remained unchanged. Either of the SRPX2 (pGL3-SRPX2) and uPAR (pGL3-uPAR) promoter constructs was co-transfected into HEK293T cells with FOXP2-containing (pcDNA4/HisMax-FOXP2 or pcDNA4/HisMax-M406T) or with empty (pcDNA4/HisMax) vector and with a β-galactosidase expression vector (pHSV-LacZ) for the normalization of transfection efficiencies. In parallel, an empty (promoterless) vector (pGL3-basic) was transfected with pcDNA4/HisMax-FOXP2 and with pHSV-LacZ. Transcriptional activity was determined by quantifying the luciferase activity of cellular extracts prepared 48 h after transfection. Data show the mean ± SD relative activity from three experiments done in triplicate. Statistical significances were determined by two-tailed unpaired t-test. NS, not significant (P > 0.05), ***P < 0.001 and ****P < 0.0001.
A novel FOXP2 missense mutation in a patient with left opercular polymicrogyria partially impairs regulation of SRPX2
The regulatory link between FOXP2 and SRPX2 prompted us to screen for FOXP2 mutations in a series of 32 patients presenting with disorders of the speech cortex that can be caused by SRPX2 mutations [rolandic epilepsy (RE) with speech impairment, perisylvian polymicrogyria] or with other disorders of the same clinical spectrum [continuous spike-and-waves during sleep (CSWS) and Landau–Kleffner syndromes] (19). One heterozygous missense mutation (c.T1591C) of FOXP2 was found in a girl of Turkish origin presenting with focal epilepsy with CSWS and cognitive and language deficits. Magnetic resonance imaging (MRI) showed polymicrogyria of the left rolandic operculum (Fig. 5A and B). Her father also carried the mutation (Fig. 5B) but did not have any neurological problem and had normal MRI (data not shown), as verified by three independent and experienced radiologists and neuropaediatricians. The proband's sister and brother also had the mutation (data not shown). Their clinical phenotype could not be fully assessed and no MRI was performed, but no obvious neurological abnormality was noted. No compound heterozygous mutation was found in the patient and her mother and their karyotypes were normal; comparative genomic hybridization analysis of the patient's DNA using Agilent 60K oligonucleotides array did not detect any abnormality in the FOXP2 genomic area (data not shown). The mutation replaced a highly conserved methionine residue with a threonine (p.M406T) within the leucine-zipper domain of FOXP2 and was neither found in the SNP database (http://www.ncbi.nlm.nih.gov/snp at NCBI) nor in 222 unrelated control individuals (33 of Turkish origin + 189 Caucasians). In analyses of HEK293T cells transfected with tagged mutant constructs, p.M406T led to partial alteration in the nuclear localization of FOXP2 (Fig. 5C). Luciferase reporter assays were then conducted and showed that the p.M406T mutation led to significant loss of repression of the SRPX2 promoter, when compared with wild-type FOXP2 (P < 0.0001, two-tailed unpaired t-test) (Fig. 5D). In contrast to p.R553H (Fig. 4B), p.M406T FOXP2 partially retained the ability to repress SRPX2 promoter activity and had no effect on the repression of uPAR promoter activity (Fig. 5D).
DISCUSSION
The present study characterized a novel molecular regulatory network associated with the disorders of cortical speech areas and of speech processing. In particular, we demonstrated the direct down-regulation of SRPX2, a gene implicated in X-linked RE associated with DVD (RESDX syndrome) or with bilateral perisylvian polymicrogyria (BPP) (16), by the FOXP2 transcription factor, mutated in people with autosomal dominant DVD (3–5). Similarly, FOXP2 also down-regulated the uPAR gene, which encodes a receptor of SRPX2, which in turn displays a modified interaction with the mutant SRPX2 protein (17). Hence, our study shows that FOXP2 co-regulates the expression of the ligand SRPX2 and of its receptor, uPAR. That there is connectivity between FOXP2 targets had already been suggested (14); as far as we are aware, this is the first clear demonstration of regulated targets with a direct interaction between them. Our findings are also in good agreement with the usual down-regulating effects of FOXP2 on its direct targets as identified by ChIP-chip experiments (13,14); furthermore, whereas the SRPX2 promoter region was not present on the microarrays used in these previous studies, uPAR was detected among the 100 most significant FOXP2 targets in human neuronal models (14).
Wild-type FOXP2 was able to bind to several consensus sites in the promoter regions of SRPX2 (SRP1 and SRP2) and uPAR (UP2 and UP6). None of these four sites corresponded to the canonical and specific FOXP2 consensus sequences; instead, SRP1, SRP2 and UP2 were of the FOX type and UP6 of the FOXP type. The binding of FOXP2 to less stringent FOX or FOXP sites has already been observed (14). Among the four FOXP2 binding sites studied here, three (SRP1, UP2 and UP6) consisted of more than one forkhead consensus sequence; this situation would be consistent with the binding of FOXP2 through homo- or heterodimerization, as already proposed (20), and may also favor cross-binding to FOX and FOXP consensus sites. Both the regulations of SRPX2 and of uPAR were abolished by a p.R553H FOXP2 mutation that is known to cause DVD (3); the expression of p.R553H FOXP2 even led to significant increases in the activity of the SRPX2 promoter when compared with the control situation. This latter result is in line with the previous investigations of the p.R553H mutant form using the SV40 promoter as a reporter (18) and may be due to the interference of the mutant protein with the transactivation capabilities of endogenous FOXP proteins expressed in HEK293T cells. Consistent with the losses of regulation of SRPX2 and of uPAR, mutant FOXP2 also showed partial alteration in nuclear localization and disrupted binding to the SRPX2 and uPAR target sites. Altogether, these data argued in favor of a pathogenic involvement of the FOXP2-SRPX2/uPAR regulatory link.
The data presented here demonstrated the existence of a novel regulatory and functional link, FOXP2-SRPX2/uPAR, displaying impairment in various disorders involving speech-related areas and networks. These findings are reminiscent of the recent functional link that was found between FOXP2 and the gene coding for the contactin associated protein-like 2, CNTNAP2 (15). Whereas FOXP2 mutations cause DVD accompanied by expressive and receptive language deficits, CNTNAP2 shows genetic association not only with specific language impairment (15) but also with autism (21–23) and with epilepsy and schizophrenia (24). Moreover, a homozygous CNTNAP2 mutation in Amish children has been shown to cause focal epilepsy followed by language regression, behavioral problems and mental retardation (25). Hence, both the FOXP2-CNTNAP2 and the FOXP2-SRPX2/uPAR regulatory pathways make direct and functional molecular links between various disorders of the brain that are diagnostically distinct, but are well known to share clinical, epidemiological and neurobiological features. Epilepsy, autism, DVD and specific language impairment may well rely on partially overlapping molecular networks. More precisely, there is long known and increasing evidence for a connection between idiopathic focal epilepsies—RE particularly—on the one hand and cognitive, attention, speech and reading disabilities on the other hand (26–29). FOXP2 regulates the expression of both components of the SRPX2/uPAR complex involved in epileptic seizures of the rolandic area; interestingly, the centrotemporal electroencephalographic hallmark of RE has shown increased frequency in patients with DVD (30). To the best of our knowledge, no rolandic seizure has ever been reported in people with FOXP2 mutations. When compared with mutations in SRPX2, mutations in FOXP2 very likely disrupt the regulation of many more pathways that may have modulatory effects on the susceptibility to epileptic seizures.
A novel p.M406T FOXP2 heterozygous mutation was found in a girl with developmental defects of the speech cortex, i.e. polymicrogyria of the left rolandic operculum, hence recalling an SRPX2-related phenotype. The proband's father carried the mutation but did not manifest the disease and had normal MRI; this obviously argues against a simple causal role. Two other sibs also carried the mutation but their phenotype could not be ascertained and no MRI was done. This situation is reminiscent of that observed in other genetic cases: for instance, SHANK3 mutations can be found in patients with autistic syndrome disorder and their unaffected relatives (31). Nevertheless, several arguments account for a likely participation of the p.M406T FOXP2 mutation as a susceptibility factor to the polymicrogyria reported here. First, functional MRI abnormalities of various regions, including Broca's area in the inferior frontal gyrus, have already been detected in people carrying FOXP2 mutations (32). The aforementioned FOXP2 target, CNTNAP2, shows enriched expression in frontal gray matter of human fetal brain tissue (33), and FOXP2 itself displays high levels of expression in the developing perisylvian cortex (34). Secondly, p.M406T was neither found in 222 control individuals nor in available databases of human variation. Furthermore, the methionine residue that is mutated is highly conserved across evolution, and p.M406T occurred in a leucine zipper domain that is crucial for the dimerization and subsequent DNA binding of FOXP2 (20). Thirdly, the present study showed that there is a direct functional link between FOXP2 and the SRPX2/uPAR complex that in turn has been implicated in bilateral perisylvian polymicrogyria that predominated on the left side (16,17). More importantly, p.M406T was shown here to have functional consequences in vitro: it not only led to altered pattern of nuclear localization of FOXP2, but also to significantly reduced, albeit still active, regulation of the SRPX2 promoter activity; interestingly also, the promoter activity of uPAR remained unchanged. Hence, the effect of p.M406T was more subtle than that of p.R553H. This is consistent with the difference in the genetic influences associated with those two mutant FOXP2 proteins. p.M406T retained partial functionality and the effects of this risk factor are likely to be modulated by genomic background, environmental influences and stochastic developmental events, hence leading to incomplete penetrance in the pedigree reported here. Although the existence of compound heterozygous mutation cannot be firmly excluded, no other FOXP2 mutation was found in the patient and her mother and conventional as well as molecular cytogenetics did not reveal any gene rearrangement. Moreover, homozygous and compound heterozygous mice lacking functional Foxp2 not only have severe developmental delays and general motor dysfunction but also do not survive beyond 3 weeks after birth (10).
FOXP2 shows evidence of recent positive selection (35). FOXP2 underwent two non-synonymous changes since the human–chimpanzee split and cell-based studies suggest that several FOXP2 target genes may be differentially regulated by human and chimpanzee versions of the protein (36). SRPX2 also exhibited an accelerated rate of non-synonymous substitutions in the human lineage; although there was insufficient statistical power to demonstrate the action of positive selection (37), the human-specific SRPX2 evolutionary change was associated with modified interaction with uPAR (17). From this evolutionary viewpoint, differences in the regulation of expression of SRPX2 and of uPAR by FOXP2 in different species might have been important. The SRP2 consensus site is conserved in the mouse and chimpanzee Srpx2 promoters. In contrast, SRP1, while being slightly different between the human and chimpanzee promoters, was not detected in the mouse Srpx2 promoter. Interestingly, the UP2 and UP6 FOXP2-binding sites of the human uPAR promoter region were conserved between human and chimpanzee but did not exist at all in the promoter region of murine Upar. Although preliminary, these data may suggest that the regulations by FOXP2 of uPAR and of SRPX2 have appeared or have been modified during evolution since the rodent–primate split.
Generally, the functioning and development of brain circuits underlying human speech result from complex sequential processes that must be tightly regulated. The emerging picture arising from the study of various speech-related disorders (RE, DVD, BPP) is that of a complicated and intertwined network of regulation and interaction comprising FOXP2, uPAR, SRPX2 as well as CNTNAP2. Moreover, the situation is likely to be considerably more complex: FOXP2 certainly has many more functional targets (13,14), and uPAR (38,39) as well as SRPX2 (40,41) expression can be modulated by other transcription factors. In addition to uPAR, SRPX2 itself has many more possible partners (17), some of which such as FBN1 (Fibrillin-1) or PCSK6 (Subtilisin-like proprotein convertase 4, PACE4) also showed significant enrichment after ChIP-chip FOXP2 experiments (14). It is thus expected that alterations of several other molecular pathways will be found in future studies of speech-related syndromes. As such, it will be crucial to identify and study each of them, and then try to integrate how and when they may interfere with each other and with the development/functioning of speech-related areas and networks. From this viewpoint, the identification of the FOXP2-SRPX2/uPAR functional and genetic link, and its alteration by a FOXP2 pathogenic mutation, represent important entry points for deciphering the complicated regulatory networks of molecules that go awry in speech-related disorders. Together with recently described FOXP2-CNTNPA2 genetic and SRPX2/uPAR proteomic links, the present findings make novel genetic and molecular links between distinct phenotypes that share clinical, epidemiological and neurobiological features, including autism, epilepsy of speech-related areas and developmental speech and language disorders.
MATERIALS AND METHODS
In silico analyses and 5′-RACE experiments
The promoter regions of the human SRPX2 (GenBank accession no. NM_014467) and uPAR (GenBank accession no. NM_002659) genes were taken from the UCSC human genome browser web site (March 2006 human reference sequence, NCBI Build 36.1; http://genome.ucsc.edu/) and were screened in silico for the presence of FOX, FOXP and FOXP2 consensus-binding sites within 1.5 kb of DNA sequence 5′ to each canonical TSS and using the DNA Pattern Find program of the Sequence Manipulation Suite (42) at http://bioinfo.unice.fr/softwares/sms2/dna_pattern.html. The possible existence of alternative transcripts with more distal 5′ TSS was tested in silico and by RACE performed from human whole-brain total RNA using FirstChoice® RLM-RACE Kit (Ambion) according to the manufacturer's instructions and using the following primers: 5′-RACE Outer Primer and SRPX2-Race1 (5′-tagaacgagtggctcctggtctgt) or uPAR-Race1 (5′-gacgcaggtgtggagcagca) for the first PCR and 5′-RACE Inner Primer and SRPX2-Race2 (5′-cggcctccgggattctgttaacactgc) or uPAR-Race2 (5′-tgcgcggggtccctgcacgtcttctctcctt) for the nested PCR. Consensus-binding sites were as defined previously (14): for the general FOX family, TRTTKRY, where R = A or G, K = T or G and Y = T or C; for members of the FOXP subfamily, TATTTRT; for FOXP2, AATTTG and ATTTGT. For evolutionary comparisons, the promoter regions of Pan troglodytes Srpx2 (GenBank accession no. NM_001135656) and Upar (GenBank accession no. NM_001009031) genes and of Mus musculus Srpx2 (GenBank accession no. NM_026838) and Upar (GenBank accession no. NM_011113) genes were taken from the corresponding UCSC chimpanzee [Chimp Mar. 2006 (panTro2) assembly, Build 2 Version 1, October 2005] and mouse [Mouse July 2007 (mm9) assembly, Build 37] genome browsers at http://genome.ucsc.edu/, respectively.
Constructs and site-directed mutagenesis
For luciferase gene reporter experiments, the promoter regions of SRPX2 and of uPAR as defined above were amplified by PCR from genomic DNA and subcloned into the pGL3 vector (Promega), 5′ to the coding sequence of the firefly luciferase reporter gene. Primers used were as follows: PROM_SRPX2.F: 5′-aaaaaggtacctaaactctgggagctggaga; PROM_SRPX2.R: 5′-aaaaaaagctttttatggtattttgtgcccttt; PROM_UPAR.F: 5′-aaaaactcgagctggcgtgcccctgtaa; PROM_UPAR.R: 5′-aaaaaaagcttggtccctgcacgtcttctct. Direct sequencing was used to confirm the integrity of each construction. The pcDNA4/HisMax-FOXP2 vector has been used previously (18). Quick-Change Site-Directed Mutagenesis kit (Stratagene) was used to generate the pcDNA4/HisMax-R553H and pcDNA4/HisMax-M406T vectors allowing the expression of the mutant forms (p.R553H and p.M406T) of FOXP2. The following primers were used: FOXP2.R553H-F: 5′-gcaacttggaagaatgcagtacatcataatcttagcctgc and FOXP2.R553H-R: 5′-gcaggctaagattatgatgtactgcattcttccaagttgc; FOXP2.M406T-F: 5′-cgtcttcaagcaatgacgacccacttgcacatgc and FOXP2.M406T-R: 5′-gcatgtgcaagtgggtcgtcattgcttgaagacg. The presence of the desired mutations (c.G2032A and c.T1591C, corresponding to p.R553H and to p.M406T, respectively, with nucleotide positions as in NM_014491) and the absence of any other unwanted mutation were confirmed by direct sequencing.
Cell cultures and transfections
HEK293T cells were grown in 5% CO2 at 37°C in Dulbecco's modified Eagle's medium (Lonza) supplemented with 2 mm l-of glutamine, 100 U/ml of penicillin, 100 mg/ml of streptomycin and 10% fetal calf serum. One day before transfection, cells were seeded in six-well plates with a concentration of 105 cells/well. When cells reached 70–80% confluence, they were transiently transfected using the Lipofectamine PlusTM reagent (Invitrogen) according to the manufacturer's instructions.
Real-time RT–PCR
HEK293T cells were transfected with expression vectors for either wild-type FOXP2 (pcDNA4/HisMax-FOXP2) or mutant FOXP2 (pcDNA4/HisMax-R553H), or with an empty vector (pcDNA4/HisMax). Cells were harvested 48 h after transfection and total RNAs were extracted in TRIzol reagent. One microgram of total RNA was reverse-transcribed using random hexamers and the Superscript(R) II RNase H reverse transcriptase (Invitrogen), according to the manufacturer's instructions.
PCR primers were designed for the SRPX2 and uPAR genes as well as for the GAPDH (glyceraldehyde-3-phosphate deshydrogenase) and B2M (beta-2-microglobulin) control genes, using the Universal Probe Library Assay Design Center at http://www.roche-applied-science.com/. Primers were: for SRPX2: forward 5′-agggcacaaaataccataaaaca and reverse 5′-gaagtcagccatcaattaacttctaa; for uPAR: forward 5′-cctctgcaggaccacgat and reverse 5′-tggtcttctctgagtgggtaca; for GAPDH: forward 5′-tccactggcgtcttcacc and reverse 5′-ggcagagatgatgaccctttt; for B2M: forward 5′-taggagggctggcaacttag, and reverse 5′-cttatgcacgcttaactatcttaacaa. Quantitative PCR was carried out using LightCycler® 480 SYBR Green I Master (Roche) in the LightCycler® 480 system (Roche). All primer pairs were optimized to ensure specific amplification of the PCR product and the absence of any primer dimer. Quantitative PCR standard curves were set up for all. Quantification was calculated using the comparative CT method. Fold changes were reported for cells transfected with either of wild-type (pcDNA4/HisMax-FOXP2) or mutant (pcDNA4/HisMax-R553H) FOXP2-containing expression vector and when compared with cells transfected with empty (pcDNA4/HisMax) vector. Relative quantification was performed using GAPDH as reference gene; the other control gene B2M did not show any significant variation (data not shown). For each gene, fold changes were reported as the mean of comparisons between five cDNA preparations. Data were expressed as mean ± SEM. Statistical significance was assessed using unpaired t-tests (two-tailed).
Luciferase reporter assays
HEK293T cells were cotransfected in six-well plates with (i) 150 ng of the appropriate reporter construct (pGL3-basic for negative control, pGL3-promoter for positive control, pGL3-SRPX2 or pGL3-uPAR construct for the analysis of each corresponding promoter activity), (ii) 150 ng of pHSV-LacZ for normalization and (iii) 1.2 µg of a construct for either the expression of FOXP2 wild-type (pcDNA4/HisMax-FOXP2), or the expression of mutant FOXP2 proteins (pcDNA4/HisMax-R553H or pcDNA4/HisMax-M406T), or no FOXP2 expression (empty pcDNA4/HisMax). Forty-eight hours after transfection, cells were lysed with 150 µl of Reporter Lysis Buffer (Promega). Firefly luciferase activity was quantified using the Luciferase Assay System (Promega) on a LB9507 Luminometer (Lumat). LacZ activity was measured using the β-Galactosidase Enzyme Assay System (Promega) on a NanoDrop™ 1000 (Thermo Fisher Scientific). All transfections were performed in triplicate and repeated in three independent experiments (nine biological replicates in total). The relative luciferase activities were then calculated with correction for transfection by β-galactosidase activity. Data were expressed as mean ± SEM. Statistical significance was assessed using unpaired t-tests (two-tailed).
EMSA experiments
HEK293T nuclear extracts (transfected with FOXP2, FOXP2.R553H or the pcDNA4 empty vector control) were prepared as described previously (18). Probes were designed as 24–30 nucleotide oligomers (Supplementary Material, Table S1) based around the predicted binding sites in the SRPX2 and uPAR promoters (Table 1). Mutant promoters were the same, however, with the core predicted binding site replaced (Supplementary Material, Table S1). A consensus probe previously shown to be efficiently bound by FOXP2 was used as a positive control (5′-agcttaaacaagacaacacaaataa) and an irrelevant promoter sequence (corresponding to an NFK binding site) was used as a negative control (5′-agctccgggggtgatttcactccccg). Oligonucleotide labeling and DNA-binding reactions were performed as described previously (18). Where unlabeled competitor probes were used to confirm specificity of DNA binding, they were added in 10-fold excess and pre-incubated at room temperature for 15 min, before the addition of labeled probe. Supershift assays were performed via pre-incubation of an N-terminal FOXP2 antibody (N-16; Santa Cruz) or a normal goat IgG negative control antibody (Santa Cruz) with nuclear lysates for 15 min at room temperature prior to the binding reaction. Protein–DNA interactions were resolved on a 5% polyacrylamide Tris/borate/EDTA gel.
Immunocytochemistry experiments
HEK293T cells were cultured on cover slips (LabTek I, Dutcher) and transfected with either a wild-type FOXP2-containing vector (pcDNA4/HisMax-FOXP2) or with a mutant FOXP2-containing vector (pcDNA4/HisMax-R553H or pcDNA4/HisMax-M406T) or with an empty vector (pcDNA4/HisMax). Forty-eight hours after transfection, cells were fixed using 4% paraformaldehyde solution at room temperature. HisMax-tag fusion protein FOXP2 was detected using an antibody to the N-terminal Xpress™ tag and nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole). Images were captured with a fluorescence microscope (Leica, DMR) equipped with a CoolSnap camera.
Genetic screening
Thirty-two patients presenting with various related disorders, such as RE with DVD, perisylvian polymicrogyria, CSWS and Landau–Kleffner syndromes, were subjected to careful clinical, neuropsychological and electroencephalogram recordings, as well as to neuroimaging examinations whenever appropriate. All patients had given informed consent for the collection of blood samples and DNA extractions. In particular, one girl with the p.M406T FOXP2 mutation had polymicrogyria of the left rolandic operculum as shown by MRI; she had focal epilepsy with CSWS and cognitive and language impairments. Pregnancy, birth and initial development were normal. Language acquisition was delayed with no clear regression. She had mental retardation with verbal, performance and full-scale IQs around 40 that remained unchanged from 7 to 16 years old. CSWS were detected at 6 years old and disappeared at the age of 10 years old. She also had right hemiparesia. At 25 years old, the patient was under 50 mg/day of phenobarbital and had rare nocturnal focal seizures; she still had cognitive and language deficits. The patient was born from unaffected parents. As her father also carried the mutation, he was submitted to MRI examination which did not reveal any abnormality (data not shown). The proband's sister and brother also carried the mutation; they did not show any obvious neurological disorder upon preliminary examination and no MRI was done. The whole coding sequence of the FOXP2 gene was screened for mutations by direct sequencing of each exon and its surrounding intronic sequences. Both strands of each corresponding PCR product (list of primers upon request) were sequenced with an ABI3700 DNA Analyzer (Applied Biosystems) and the data were analyzed with the Genalys 3.0 software (43).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by INSERM (Institut National de la Santé et de la Recherche Médicale) and by grants from ANR (Agence Nationale de la Recherche), FRC (Fédération pour la Recherche sur le Cerveau), the Wellcome Trust (project grant 080971 and core award 075491) and Autism Speaks. S.E.F. is a Royal Society Research Fellow. S.C.V. was funded by a Wellcome Trust VIP award and J.C. is a recipient of a French MRT (Ministry of Research and Technology) PhD fellowship.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all the family members and patients who participated in this study. Each participant gave informed consent prior to the study, according to the appropriate ethical committees. We thank H. Gara-Ksouri and F. Schaller-Marly at INMED for technical help and R. Leventer at Royal Children's Hospital, Victoria, Australia, for expertise in MRI interpretation.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Fisher S.E. On genes, speech, and language. N. Engl. J. Med. 2009;353:1655–1657. doi: 10.1056/NEJMp058207. doi:10.1056/NEJMp058207. [DOI] [PubMed] [Google Scholar]
- 2.Fisher S.E., Scharff C. FOXP2 as a molecular window into speech and language. Trends Genet. 2009;25:166–177. doi: 10.1016/j.tig.2009.03.002. doi:10.1016/j.tig.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Lai C.S., Fisher S.E., Hurst J.A., Vargha-Khadem F., Monaco A.P. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. doi:10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 4.MacDermot K.D., Bonora E., Sykes N., Coupe A.M., Lai C.S., Vernes S.C., Vargha-Khadem F., McKenzie F., Smith R.L., Monaco A.P., et al. Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am. J. Hum. Genet. 2005;76:1074–1080. doi: 10.1086/430841. doi:10.1086/430841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeesman S., Nowaczyk M.J., Teshima I., Roberts W., Cardy J.O., Brian J., Senman L., Feuk L., Osborne L.R., Scherer S.W. Speech and language impairment and oromotor dyspraxia due to deletion of 7q31 that involves FOXP2. Am. J. Med. Genet. 2006;140:509–514. doi: 10.1002/ajmg.a.31110. doi:10.1002/ajmg.a.31110. [DOI] [PubMed] [Google Scholar]
- 6.Lehmann O.J., Sowden J.C., Carlsson P., Jordan T., Bhattacharya S.S. Fox's in development and disease. Trends Genet. 2003;19:339–344. doi: 10.1016/S0168-9525(03)00111-2. doi:10.1016/S0168-9525(03)00111-2. [DOI] [PubMed] [Google Scholar]
- 7.Shu W., Cho J.Y., Jiang Y., Zhang M., Weisz D., Elder G.A., Schmeidler J., De Gasperi R., Sosa M.A., Rabidou D., et al. Altered ultrasonic vocalization in mice with a disruption in the Foxp2 gene. Proc. Natl Acad. Sci. USA. 2005;102:9643–9648. doi: 10.1073/pnas.0503739102. doi:10.1073/pnas.0503739102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.French C.A., Groszer M., Preece C., Coupe A.M., Rajewsky K., Fisher S.E. Generation of mice with a conditional Foxp2 null allele. Genesis. 2007;45:440–446. doi: 10.1002/dvg.20305. doi:10.1002/dvg.20305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haesler S., Rochefort C., Georgi B., Licznerski P., Osten P., Scharff C. Incomplete and inaccurate vocal imitation after knockdown of FoxP2 in songbird basal ganglia nucleus Area X. PLoS Biol. 2007;5:e321. doi: 10.1371/journal.pbio.0050321. doi:10.1371/journal.pbio.0050321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groszer M., Keays D.A., Deacon R.M., de Bono J.P., Prasad-Mulcare S., Gaub S., Baum M.G., French C.A., Nicod J., Coventry J.A., et al. Impaired synaptic plasticity and motor learning in mice with a point mutation implicated in human speech deficits. Curr. Biol. 2008;18:354–362. doi: 10.1016/j.cub.2008.01.060. doi:10.1016/j.cub.2008.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller J.E., Spiteri E., Condro M.C., Dosumu-Johnson R.T., Geschwind D.H., White S.A. Birdsong decreases protein levels of FoxP2, a molecule required for human speech. J. Neurophysiol. 2008;100:2015–2025. doi: 10.1152/jn.90415.2008. doi:10.1152/jn.90415.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enard W., Gehre S., Hammerschmidt K., Hölter M., Blass T., Somel M., Brückner M.K., Schreiweis C., Winter C., Sohr R., et al. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell. 2009;137:961–971. doi: 10.1016/j.cell.2009.03.041. doi:10.1016/j.cell.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 13.Spiteri E., Konopka G., Coppola G., Bomar J., Oldham M., Ou J., Vernes S.C., Fisher S.E., Ren B., Geschwind D.H., et al. Identification of the transcriptional targets of FOXP2, a gene linked to speech and language, in developing human brain. Am. J. Hum. Genet. 2007;81:1144–1157. doi: 10.1086/522237. doi:10.1086/522237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vernes S.C., Spiteri E., Nicod J., Groszer M., Taylor J.M., Davies K.E., Geschwind D.H., Fisher S.E. High-throughput analysis of promoter occupancy reveals direct neural targets of FOXP2, a gene mutated in speech and language disorders. Am. J. Hum. Genet. 2007;81:1232–1250. doi: 10.1086/522238. doi:10.1086/522238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vernes S.C., Newbury D.F., Abrahams B.S., Winchester L., Nicod J., Groszer M., Alarcón M., Oliver P.L., Davies K.E., Geschwind D.H., et al. A functional genetic link between distinct developmental language disorders. N. Engl. J. Med. 2008;359:2337–2345. doi: 10.1056/NEJMoa0802828. doi:10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roll P., Rudolf G., Pereira S., Royer B., Scheffer I.E., Massacrier A., Valenti M.P., Roeckel-Trevisiol N., Jamali S., Beclin C., et al. SRPX2 mutations in disorders of language cortex and cognition. Hum. Mol. Genet. 2006;15:1195–1207. doi: 10.1093/hmg/ddl035. doi:10.1093/hmg/ddl035. [DOI] [PubMed] [Google Scholar]
- 17.Royer-Zemmour B., Ponsole-Lenfant M., Gara H., Roll P., Lévêque C., Massacrier A., Ferracci G., Cillario J., Robaglia-Schlupp A., Vincentelli R., et al. Epileptic and developmental disorders of the speech cortex: ligand/receptor interaction of wild-type and mutant SRPX2 with the plasminogen activator receptor uPAR. Hum. Mol. Genet. 2008;17:3617–3630. doi: 10.1093/hmg/ddn256. doi:10.1093/hmg/ddn256. [DOI] [PubMed] [Google Scholar]
- 18.Vernes S.C., Nicod J., Elahi F.M., Coventry J.A., Kenny N., Coupe A.M., Bird L.E., Davies K.E., Fisher S.E. Functional genetic analysis of mutations implicated in a human speech and language disorder. Hum. Mol. Genet. 2006;15:3154–3167. doi: 10.1093/hmg/ddl392. doi:10.1093/hmg/ddl392. [DOI] [PubMed] [Google Scholar]
- 19.Rudolf G., Valenti M.P., Hirsch E., Szepetowski P. From rolandic epilepsy to continuous spike-and-waves during sleep and Landau–Kleffner syndromes: insights into possible genetic factors. Epilepsia. 2009;50(Suppl. 7):25–28. doi: 10.1111/j.1528-1167.2009.02214.x. doi:10.1111/j.1528-1167.2009.02214.x. [DOI] [PubMed] [Google Scholar]
- 20.Li S., Weidenfeld J., Morrisey E.E. Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol. Cell. Biol. 2004;24:809–822. doi: 10.1128/MCB.24.2.809-822.2004. doi:10.1128/MCB.24.2.809-822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alarcón M., Abrahams B.S., Stone J.L., Duvall J.A., Perederiy J.V., Bomar J.M., Sebat J., Wigler M., Martin C.L., Ledbetter D.H., et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am. J. Hum. Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. doi:10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arking D.E., Cutler D.J., Brune C.W., Teslovich T.M., West K., Ikeda M., Rea A., Guy M., Lin S., Cook E.H., et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am. J. Hum. Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. doi:10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakkaloglu B., O'Roak B.J., Louvi A., Gupta A.R., Abelson J.F., Morgan T.M., Chawarska K., Klin A., Ercan-Sencicek A.G., Stillman A.A., et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am. J. Hum. Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. doi:10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman J.I., Vrijenhoek T., Markx S., Janssen I.M., van der Vliet W.A., Faas B.H., Knoers N.V., Cahn W., Kahn R.S., Edelmann L., et al. CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol. Psychiatry. 2008;13:261–266. doi: 10.1038/sj.mp.4002049. doi:10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]
- 25.Strauss K.A., Puffenberger E.G., Huentelman M.J., Gottlieb S., Dobrin S.E., Parod J.M., Stephan D.A., Morton DH. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N. Engl. J. Med. 2006;354:1370–1377. doi: 10.1056/NEJMoa052773. doi:10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 26.Deonna T.W., Roulet E., Fontan D., Marcoz JP. Speech and oromotor deficits of epileptic origin in benign partial epilepsy of childhood with rolandic spikes (BPERS). Relationship to the acquired aphasia-epilepsy syndrome. Neuropediatrics. 1993;24:83–87. doi: 10.1055/s-2008-1071519. doi:10.1055/s-2008-1071519. [DOI] [PubMed] [Google Scholar]
- 27.Staden U., Isaacs E., Boyd S.G., Brandl U., Neville B.G. Language dysfunction in children with rolandic epilepsy. Neuropediatrics. 1998;29:242–248. doi: 10.1055/s-2007-973569. doi:10.1055/s-2007-973569. [DOI] [PubMed] [Google Scholar]
- 28.Metz-Lutz M.N., Kleitz C., de Saint Martin A., Massa R., Hirsch E., Marescaux C. Cognitive development in benign focal epilepsies of childhood. Dev. Neurosci. 1999;21:182–190. doi: 10.1159/000017397. doi:10.1159/000017397. [DOI] [PubMed] [Google Scholar]
- 29.Monjauze C., Tuller L., Hommet C., Barthez M.A., Khomsi A. Language in benign childhood epilepsy with centro-temporal spikes abbreviated form: rolandic epilepsy and language. Brain Lang. 2005;92:300–308. doi: 10.1016/j.bandl.2004.07.001. doi:10.1016/j.bandl.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 30.Echenne B., Cheminal R., Rivier F., Negre C., Touchon J., Billiard M. Epileptic electroencephalographic abnormalities and developmental dysphasias: a study of 32 patients. Brain Dev. 1992;14:216–225. doi: 10.1016/s0387-7604(12)80233-6. [DOI] [PubMed] [Google Scholar]
- 31.Durand C.M., Betancur C., Boeckers T.M., Bockmann J., Chaste P., Fauchereau F., Nygren G., Rastam M., Gillberg I.C., Anckarsäter H., et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 2007;39:25–27. doi: 10.1038/ng1933. doi:10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liégeois F., Baldeweg T., Connelly A., Gadian D.G., Mishkin M., Vargha-Khadem F. Language fMRI abnormalities associated with FOXP2 gene mutation. Nat. Neurosci. 2003;6:1230–1237. doi: 10.1038/nn1138. doi:10.1038/nn1138. [DOI] [PubMed] [Google Scholar]
- 33.Abrahams B.S., Tentler D., Perederiy J.V., Oldham M.C., Coppola G., Geschwind D.H. Genome-wide analyses of human perisylvian cerebral cortical patterning. Proc. Natl Acad. Sci. USA. 2007;104:17849–17854. doi: 10.1073/pnas.0706128104. doi:10.1073/pnas.0706128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson M.B., Kawasawa Y.I., Mason C.E., Krsnik Z., Coppola G., Bogdanovic D., Geschwind D.H., Mane S.M., State M.W., Sestan N. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. doi:10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Enard W., Przeworski M., Fisher S.E., Lai C.S., Wiebe V., Kitano T., Monaco A.P., Pääbo S. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. doi:10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- 36.Konopka G., Bomar J.M., Winden K., Coppola G., Jonsson Z.O., Gao F., Peng S., Preuss T.M., Wohlschlegel J.A., Geschwind D.H. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462:213–217. doi: 10.1038/nature08549. doi:10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Royer B., Soares D.C., Barlow P.N., Bontrop R.E., Roll P., Robaglia-Schlupp A., Blancher A., Levasseur A., Cau P., Pontarotti P., et al. Molecular evolution of the human SRPX2 gene that causes brain disorders of the rolandic and Sylvian speech areas. BMC Genet. 2007;8:72. doi: 10.1186/1471-2156-8-72. doi:10.1186/1471-2156-8-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H., Yang L., Jamaluddin M.S., Boyd D.D. The Kruppel-like KLF4 transcription factor, a novel regulator of urokinase receptor expression, drives synthesis of this binding site in colonic crypt luminal surface epithelial cells. J. Biol. Chem. 2004;279:22674–22683. doi: 10.1074/jbc.M401257200. doi:10.1074/jbc.M401257200. [DOI] [PubMed] [Google Scholar]
- 39.Büchler P., Reber H.A., Tomlinson J.S., Hankinson O., Kallifatidis G., Friess H., Herr I., Hines O.J. Transcriptional regulation of urokinase-type plasminogen activator receptor by hypoxia-inducible factor 1 is crucial for invasion of pancreatic and liver cancer. Neoplasia. 2009;11:196–206. doi: 10.1593/neo.08734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurosawa H., Goi K., Inukai T., Inaba T., Chang K.S., Shinjyo T., Rakestraw K.M., Naeve C.W., Look A.T. Two candidate downstream target genes for E2A-HLF. Blood. 1999;93:321–332. [PubMed] [Google Scholar]
- 41.Fledderus J.O., Boon R.A., Volger O.L., Hurttila H., Ylä-Herttuala S., Pannekoek H., Levonen A.L., Horrevoets A.J. KLF2 primes the antioxidant transcription factor Nrf2 for activation in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2008;28:1339–1346. doi: 10.1161/ATVBAHA.108.165811. doi:10.1161/ATVBAHA.108.165811. [DOI] [PubMed] [Google Scholar]
- 42.Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28:1102–1114. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi M., Matsuda F., Margetic N., Lathrop M. Automated identification of single nucleotide polymorphisms from sequencing data. J. Bioinform. Comput. Biol. 2003;1:253–265. doi: 10.1142/s021972000300006x. doi:10.1142/S021972000300006X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.