Abstract
Adenoviruses (AdV) broadly infect vertebrate hosts including a variety of primates. We identified a novel AdV in the feces of captive gorillas by isolation in cell culture, electron microscopy and PCR. From the supernatants of infected cultures we amplified DNA polymerase (DPOL), preterminal protein (pTP) and hexon gene sequences with generic pan primate AdV PCR assays. The sequences in-between were amplified by long-distance PCRs of 2 - 10 kb length, resulting in a final sequence of 15.6 kb. Phylogenetic analysis placed the novel gorilla AdV into a cluster of primate AdVs belonging to the species Human adenovirus B (HAdV-B). Depending on the analyzed gene, its position within the cluster was variable. To further elucidate its origin, feces samples of wild gorillas were analyzed. AdV hexon sequences were detected which are indicative for three distinct and novel gorilla HAdV-B viruses, among them a virus nearly identical to the novel AdV isolated from captive gorillas. This shows that the discovered virus is a member of a group of HAdV-B viruses that naturally infect gorillas. The mixed phylogenetic clusters of gorilla, chimpanzee, bonobo and human AdVs within the HAdV-B species indicate that host switches may have been a component of the evolution of human and non-human primate HAdV-B viruses.
Findings
Adenoviruses are non-enveloped icosahedral double-stranded DNA viruses that infect fish, amphibians, reptiles, birds and mammals [1]. Human adenoviruses (HAdV) are categorized into seven species (HAdV-A to HAdV-G) [2]. Each species includes a distinct number of serotypes [3]. In addition, intra-species shuffling of penton base, fiber and hexon genes by recombination has been frequently observed [4-6]. Simian adenoviruses have been discovered in monkeys and great apes [7-11]. They are very similar to HAdV, and most of them can be grouped into corresponding HAdV species or the newly established species Simian adenovirus A (SAdV-A).
In 2008, a group of Western lowland gorillas (Gorilla gorilla gorilla) suffered from prolonged diarrhea and self-limiting respiratory disease in the Zoological gardens of Münster, Germany. To isolate viral agents potentially responsible for the symptoms, fecal samples were suspended in phosphate-buffered saline, sterile filtered and cultured on MRC-5 cells and A549 cells. After eight days of culture, a cytopathogenic effect was observed. The culture supernatant was examined by electron microscopy, and virus-like structures were detected their size and general structure being consistent with that of adenoviruses (Additional Figure 1).
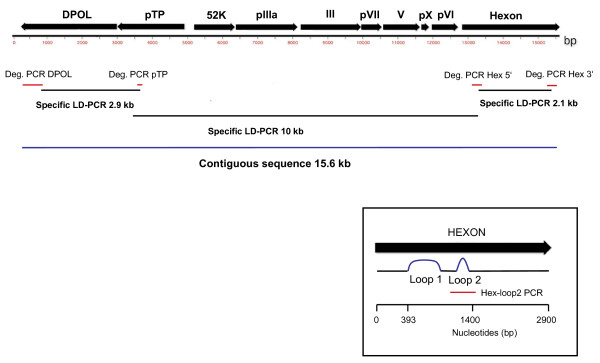
Figure 1.
PCR amplification strategy. Above the ruler, the amplified part of the AdV genome is depicted. Below, a schematic visualisation of the positions of the degenerated consensus PCRs (red lines), the long-distance PCRs (black lines) and the resulting contiguous sequence (blue line) is given. In the box, the hexon gene is magnified, and the positions of the hexon loops and the Hex-loop2 PCR (red line) are illustrated.
DNA was then extracted from culture supernatant using the Qiagen tissue kit according to the manufacturer's instructions (Qiagen, Hilden, Germany), and a generic primate adenovirus PCR was performed. For this purpose, a nested set of degenerate and deoxyinosine-substituted (deg/dI) primers was designed, targeting a highly conserved region of the DNA polymerase (DPOL) gene of primate mastadenoviruses (Figure 1; Table 1). PCR was performed in a total volume of 25 μl with 0.2 μl AmpliTaq Gold (Applied Biosystems), 20 pmol of each primer, 200 μM dNTPs, 2 mM MgCl2, and 5% DMSO. A T-Gradient thermocycler from Biometra was used with the following cycling conditions: 95°C for 12 min, and 45 cycles of 95°C for 30 sec, 45°C (1st round and 2nd round) for 30 sec and 72°C for 2 min, followed by a 15 min final extension step at 72°C. PCR products were purified using the Invisorb DNA clean up kit according to the instructions of the manufacturer (Invitek, Berlin, Germany), and directly sequenced with the Big Dye terminator cycle sequencing kit (Applied Biosystems, Warrington, UK) on a 377 automated DNA sequencer (Applied Biosystems). In BLAST analysis of GenBank, the sequence was most closely related to chimpanzee AdVs and human AdVs of the species HAdV-B. There was less similarity to the six gorilla HAdV-B viruses (SAdV-27.2; SAdV-28.2; SAdV-41.1; SAdV-41.2; SAdV-46; SAdV-47; Table 2) recently described [12]. Since the novel gorilla AdV was the seventh HAdV-B of this host, it was named for the purpose of this paper Gorilla gorilla adenovirus B7 (GgorAdV-B7).
Table 1.
Primers for amplification of DPOL, pTP and hexon gene sequences
| Primer-set abbreviation | Targeted gene | Name of primer | sequence 5'-3' | PCR length | Annealing temp. |
|---|---|---|---|---|---|
| Degenerate primers | |||||
| DPOL-cons | DPOL | 4431-s$ | GTnTwyGAyAThTGyGGhATGTAyGC | ||
| 4428-as | GAGGCTGTCCGTrTC(n/i)CCGTA# | 956 bp | 45°C | ||
| 4428-s | CGGACGCCTCTGyTGGAC(n/i)AA | ||||
| 4429-as | GGCCAGCACrAA(n/i)GArGC | 650 bp | 45°C | ||
| Hex-5'-cons | Hexon | 4515-s | GTGGATGG(n/i)GA(r/i)GG(n/i)TACA | ||
| 4515-as | CGCACAACGTC(r/i)AA(n/i)AC(y/i)TC | 536 bp | 45°C | ||
| 4516-s | TGTAACATGAC(y/i)AA(r(i)GA(y/i)TGG | ||||
| 4516-as | CAGGGCCCCCAT(n/i)GACA | 381 bp | 45°C | ||
| Hex3'-cons | Hexon | 4517-s | CGCAATGGTC(n/i)TACATGCAC | ||
| 4517-as | CAGTGCCCGA(r/i)TA(k/i)GG(n/i)TT | 340 bp | 45°C | ||
| 4518-s | GCAGGACGC(y/i)TCGGAGTA | ||||
| 4518-as | CACCC(k/i)GTT(r/i)TC(n/i)CC | 230 bp | 45°C | ||
| pTP-cons | pTP | 4521-s | TGGCGACGT(n/i)GT(n/i)TACAG | ||
| 4521-as | CGGACT(y/i)(k/i)GA(r/i)CCTGAAA | 260 bp | 45°C | ||
| 4522-s | TACAGCCG(n/i)GTSTGGAAC | ||||
| 4522-as | CTGAAAGAGAGTTC(n/i)ACAGAATCA | 230 bp | 45°C | ||
| Specific primers for long-distance PCR | |||||
| pTP-DPOL-LD | pTP, DPOL | 4659-s | TCGCATCTCCAACGACCT | ||
| 4659-as | GCATCCATGGTGAAGATTCC | 2350 bp | 60°C | ||
| Hex-LD | Hexon | 4618-s | AGTTCGCTACACACTGGCTG | ||
| 4618-as | ATTGCGGTGATGATTGAATG | 1354 bp | 59°C | ||
| pTP-Hex-LD | pTP, Hexon | 4662-s | CTCGGTATCGTTGACGGC | ||
| 4662-as | GATCAACGGGCACAAAGC | 10044 bp | 60°C | ||
| Primers specific for AdV species B | |||||
| Hex-loop2 | Hexon | 5442s | GAACAAGATACTTTAGCATGTGGAA | ||
| 5442as | GATTGAATGGATTAACATTGTCC | 468 bp | 55°C | ||
| Hexon | 5443s | TAGAAAATCACGGGGTGGAAGA | |||
| 5443as | GGCATCCAAAGACCATCTG | 380 bp | 55°C | ||
$ s = sense, as = antisense # I = inosine
Table 2.
Adenoviruses, accession numbers and hosts
| Adenovirus | Abbreviation | GenBank accession number | Host | Wild (Gabon) | Captive |
|---|---|---|---|---|---|
| HAdV-B of this study | |||||
| Gorilla gorilla adenovirus B7 | GgorAdV-B7 | HQ292614 | Western lowland gorilla | + | + |
| Gorilla gorilla adenovirus B8 | GgorAdV-B8 | HQ292615 | Western lowland gorilla | + | + |
| Gorilla gorilla adenovirus B9 | GgorAdV-B9 | HQ292616 | Western lowland gorilla | + | |
| Gorilla gorilla adenovirus B10 | GgorAdV-B10 | HQ292617 | Western lowland gorilla | + | |
| Published HAdV-B | |||||
| Simian adenovirus 21 | SAdV-21 | AC000010 | Chimpanzee | + | |
| Simian adenovirus 27.1 | SAdV-27.1 | FJ025909 | Chimpanzee | + | |
| Simian adenovirus 27.2 | SAdV-27.2 | FJ025928 | Gorilla | + | |
| Simian adenovirus 28.1 | SAdV-28.1 | FJ025914 | Chimpanzee | + | |
| Simian adenovirus 28.2 | SAdV-28.2 | FJ025915 | Gorilla | + | |
| Simian adenovirus 29 | SAdV-29 | FJ025916 | Chimpanzee | + | |
| Simian adenovirus 32 | SAdV-32 | FJ025911 | Chimpanzee | + | |
| Simian adenovirus 33 | SAdV-33 | JF025908 | Chimpanzee | + | |
| Simian adenovirus 35.1 | SAdV-35.1 | FJ025912 | Chimpanzee | + | |
| Simian adenovirus 35.2 | SAdV-35.2 | FJ025910 | Bonobo | + | |
| Simian adenovirus 41.1 | SAdV-41.1 | FJ025913 | Gorilla | + | |
| Simian adenovirus 41.2 | SAdV-41.2 | FJ025927 | Gorilla | + | |
| Simian adenovirus 46 | SAdV-46 | FJ025930 | Gorilla | + | |
| Simian adenovirus 47 | SAdV-47 | FJ025929 | Gorilla | + | |
| Human adenovirus B3 | HAdV-B3 | DQ086466 | Human | ||
| Human adenovirus B7 | HAdV-B7 | AC000018 | Human | ||
| Human adenovirus B11 | HAdV-B11 | AY163756 | Human | ||
| Human adenovirus B14 | HAdV-B14 | AY803294 | Human | ||
| Human adenovirus B16 | HAdV-B16 | AY601636 | Human | ||
| Human adenovirus B21 | HAdV-B21 | AY601633 | Human | ||
| Human adenovirus B34 | HAdV-B34 | AY737797 | Human | ||
| Human adenovirus B35 | HAdV-B35 | AY271307 | Human | ||
| Human adenovirus B50 | HAdV-B50 | AY737798 | Human |
To acquire extended sequence information of GgorAdV-B7, three additional nested PCR assays were designed (Table 1) targeting the preterminal protein (pTP) and two conserved regions at the 5'- and 3'-end of the hexon gene (Figure 1). PCRs were performed as described above, except that elongation at 72°C was for 1 min. With each primer set products of the expected size were obtained. BLAST analysis of their sequences also revealed a HAdV-B-like virus (not shown). To prove that the DPOL, pTP and hexon sequences originate from the same virus, we connected them with long-distance (LD) PCRs (Figure 1) using the TaKaRa-EX PCR system according to the instructions of the manufacturer (Takara Bio Inc., Otsu, Japan). The LD primer pairs are listed with their annealing temperatures in Table 1. Three overlapping PCR products were generated and sequenced by primer walking. A final contiguous sequence of 15637 bp was obtained spanning the genes DPOL, pTP and 52 k, the genes encoding the AdV proteins pIIIa, III (penton base), pVII, V, pX and pVI, and the hexon gene of GgorAdV-B7 (Figure 1).
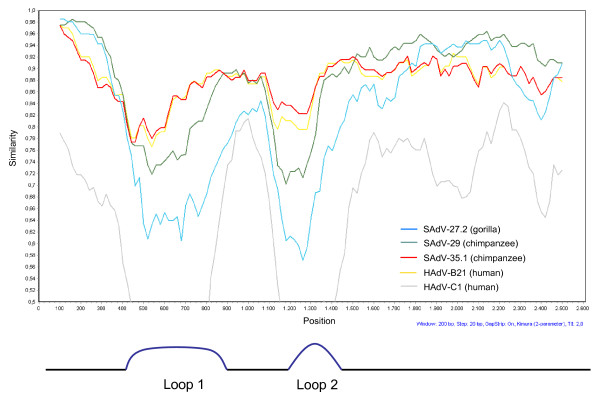
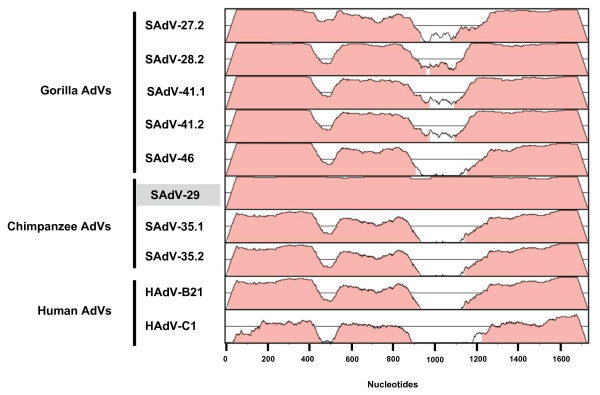
Since the AdV hexon gene is an important member of the core gene set used for AdV classification [2,3,13,14], we compared the available partial hexon gene of GgorAdV-B7 (2.7 kb) pair-wise with the corresponding hexon sequences of the most closely related human and chimpanzee HAdV-B viruses and all published gorilla HAdV-B viruses. The highest identity percentages were 96.6% for SAdV-35.1 (chimpanzee AdV) and 96% for HAdV-B21. The gorilla AdV SAdV-27.2, SAdV-28.2, SAdV-41.1, SAdV-41.2, SAdV-46 and SAdV-47 revealed only 87-91% identity. This closer relationship to HAdV-B21 and SAdV-35.1 was restricted to the loop-encoding regions 1 and 2 [15] as visible in an analysis with the software SIMPLOT http://sray.med.som.jhmi.edu (Figure 2). In pair-wise comparisons of DPOL and pTP genes, GgorAdV-B7 was equally closely related to chimpanzee and gorilla HAdV-B viruses (96-99.9%). However, the penton base gene of GgorAdV-B7 showed a striking similarity (99.7%) only to that of SAdV-29 (chimpanzee AdV). Using the program mVISTA http://genome.lbl.gov/vista/index.shtml, the near-perfect match of the GgorAdV-B7 and SAdV-29 penton base genes over the entire gene length is clearly visible (Figure 3).
Figure 2.
Simplot analysis of the hexon gene. 2.8 kb of the GgorAdV-B7 hexon gene were compared to the hexon genes of selected chimpanzee (green and red line), gorilla (blue line) and human (yellow and grey line) AdVs. Below the plot, the analysis parameters are listed in blue font. The hexon Loop 1 and Loop 2 regions are indicated at the bottom.
Figure 3.
Pairwise sequence alignment of the penton base open reading frame. The penton base gene of GgorAdV-B7 was compared with those of five gorilla-, three chimpanzee- and two human AdVs. The 50-100 percent sequence conservation is represented by the height of each data point along the y axis. The chimpanzee AdV SAdV-29 is highlighted in grey designating the exceptionally high similarity to GgorAdV-B7.
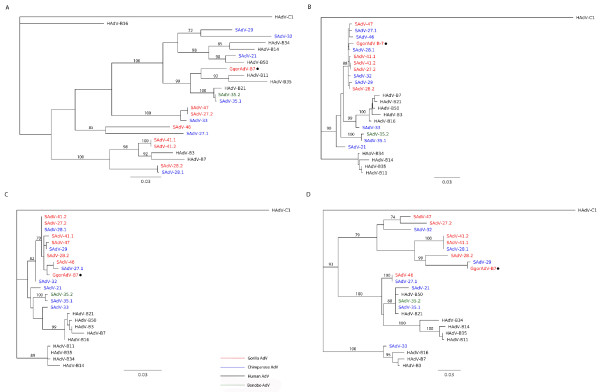
With the PhyML plug-in 2.0.1 of the Geneious Pro 5.0.4 software, phylogenetic trees were constructed on the basis of hexon, DPOL, pTP and penton base gene alignments (Figures 4). All published, completely sequenced HAdV-B viruses were included. In the hexon-based tree, GgorAdV-B7 clustered with HAdV-B21, SAdV-35.1 (chimpanzee AdV) and SAdV-35.2 (bonobo AdV) (Figure 4a). DPOL and pTP analyses placed GgorAdV-B7 into a tight cluster of gorilla and chimpanzee AdVs (Figure 4b and 4c). In the tree derived from the penton base, GgorAdV-B7 branched separately, nearly at the same position as SAdV-29 (Figure 4d). With the MrBayes 2.0.2 plug-in (Geneious Pro) or the Neighbor-Joining module of MacVector 10.6, trees with the same AdV clusters and similarly supported topology were obtained (data not shown).
Figure 4.
Phylogenetic analysis. The Hexon (A), DPOL (B), pTP (C) and penton (D) sequences of GgorAdV-B7 and those of published NHP and human HAdV-B viruses were aligned and subjected to phylogenetic analysis. Human, chimpanzee, bonobo and gorilla AdVs are in black, blue, green and red font, respectively. AdVs discovered in this study are marked with black dots.
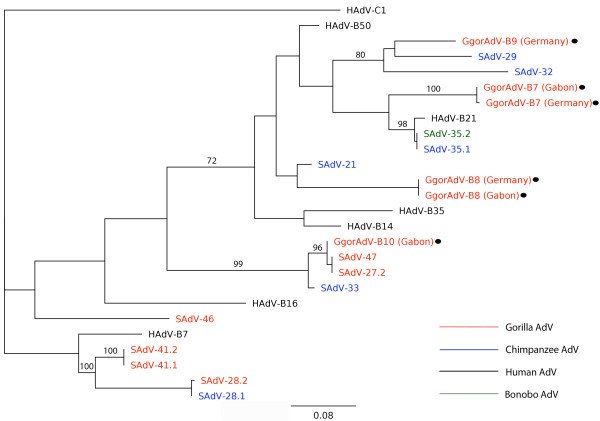
The remarkably close relatedness of GgorAdV-B7 to chimpanzee AdVs (SAdV-29 and 35.1) prompted us to investigate whether gorillas naturally host GgorAdV-B7. For this purpose, we examined wild Western lowland gorillas (Gorilla. g. gorilla) from Gabon and additional captive gorillas. Fecal samples were collected from 19 individuals in a remote area with little human presence in Loango National Park, Gabon. They originated from fresh nest sites or were freshly found on gorilla paths [16]. Samples were collected using single-use gloves and preserved by drying over silica. DNA was extracted following a previously described method [17]. In addition, ten necropsy samples (spleen, liver, pancreas, lymph node, tonsil, lung, kidney, urine) and one plasma sample were collected from four deceased captive gorillas in the Zoological gardens of Berlin as well as six fecal samples from three captive gorillas in the Zoological gardens of Münster, Germany. To test for the presence of HAdV-B viruses, we set up a nested PCR (PCR Hex-loop2; Table 1) which targets flanking sequences of a hyper variable region (loop 2) in the hexon gene (Figure 1) and amplifies 380 bp. The primers were deduced from HAdV-B sequences only and not degenerated. A total of 36 gorilla samples were screened. AdV DNA was only detected in feces (5/19 wild gorilla samples and 4/6 captive gorilla samples). In total, 9/25 fecal samples were PCR-positive (36%), and the products sequenced. Most importantly, a virus apparently identical to GgorAdV-B7 was identified in a wild gorilla from Gabon. Three additional HAdV-B viruses were also detected. Two were without close similarities to any published AdV sequence. The third one was nearly identical to the gorilla AdVs SAdV-27.2 and SAdV-47, which had been originally isolated from captive individuals [12]. They were tentatively named GgorAdV-B8, -B9 and -B10.
GgorAdV-B8 was detected in two fecal samples from wild gorillas in Gabon and in one sample from Münster. Its hexon sequence revealed the highest percentage of identity (86.5%) to SAdV-21 (chimpanzee HAdV-B). GgorAdV-B9 was only amplified from captive gorillas (two fecal samples from Münster) and showed 88% identity to SAdV-29 (chimpanzee HAdV-B). The GgorAdV-B7 to -B10 Hex-loop2 sequences and closely related sequences of published gorilla, chimpanzee, bonobo and human HAdV-B viruses were subjected to phylogenetic analysis as described above. In the tree, the HAdV-B viruses segregate into several subclades with members of two, three or four host species (human, chimpanzee, bonobo and gorilla) (Figure 5). This mixed clustering was also observed upon analysis of the nearly complete hexon gene (2.7 kb; Figure 4a) with GgorAdV-B7 only. It was partially visible in the penton base tree (Figure 4d) and entirely absent from the DPOL and pTP trees (Figures 4b and 4c). In addition, the phylogenetic position of a given primate AdV frequently differed, depending on the analyzed gene (compare Figures 4a to 4d).
Figure 5.
Phylogenetic analysis of the hexon loop 2 region. For details, see legend of Figure 4.
Taken together, these observations cannot be explained by co-speciation. Rather, they are in line with recombination and host switching. Such events have been previously discussed to be involved in the evolution of human AdV, because intra-species shuffling of penton base, fiber and hexon genes has been frequently observed [4-6]. In addition, a recombinant between viruses of the sub-clades HAdV-B1 and HAdV-B2 has been isolated from a captive chimpanzee [12]. Here, a close similarity of GgorAdV-B7 to SAdV-29 (complete sequence, excluding hexon loops) and to SAdV-35.1 (hexon loops) was observed (Figure 2). However, since in the loop region the nucleic acid identity between GgorAdV-B7 and SAdV-35.1 was well below 100%, it is unlikely that the existing AdVs are parent viruses in a recent recombination event giving rise to GgorAdV-B7. Rather, a more ancient one with subsequent genetic drift may have been involved or recombination with an unknown AdV, as suggested for HAdV-A18 [18].
Shuffling of genes by recombination between AdVs that naturally infect different host species (e.g., great ape and human AdVs) but under certain conditions co-infect the same host, may be an additional mechanism by which AdVs exchange genetic information. This could occur in places where contacts between humans and apes are frequent like in zoos and animal facilities. In addition, people who are involved in hunting primates and preparation of bush meat [19] are at risk to be infected. So far, infections of humans with non-human primate (NHP) AdVs have not been observed. Nevertheless, antibodies with specificity for chimpanzee HAdV-C viruses have been detected in humans from Sub-Saharan Africa and were significantly less frequent in people from the United States of America and Thailand [20]. In addition, the species HAdV-E comprises only one human serotype but more than 12 great ape serotypes. Therefore, the human HAdV-E was thought to be the result of a zoonotic transmission from chimpanzees to humans [21]. The gorilla AdV described in the present study (GgorAdV-B7) is highly similar to chimpanzee AdVs. Thus, a transmission event between chimpanzees and gorillas was possibly involved which is further indication for the potential of AdVs to jump between closely related hosts.
Very little is known about the pathogenic properties of NHP-AdV. GgorAdV-B7 was originally discovered in a group of gorillas suffering from prolonged diarrhea and self-limiting respiratory infection. Since human species B AdV have been linked to respiratory diseases [22-24], an etiological association of GgorAdV-B7 with the observed respiratory symptoms is possible. However, recent studies reported the frequent shedding of AdVs in the feces of healthy captive chimpanzees and gorillas [12,25]. Therefore, further investigations are needed.
Knowledge about the spectrum of AdV in wild great apes in general [25] is very limited. Specifically from wild gorillas, no information has been published. Although examining only a small set of samples, our findings show that AdV infecting captive gorillas can readily be found in wild animals (GgorAdV-B7; GgorAdV-B8). This is a good example of how humans may be brought into contact with new pathogens, not only locally through bushmeat hunting in regions where NHP live naturally, but also in other regions of the world where NHP are housed in zoos.
The high variety of known and novel HAdV-B viruses in great apes calls for larger studies to understand the diversity of AdVs currently circulating in African NHP as well as in local human populations. It is justified to assume that such studies will improve our insight into the zoonotic potential of adenoviruses and possibly answer the intriguing question whether AdVs of non-human primates have already contributed to the human "adeno-virosphere".
Accession numbers
The sequences reported in this study were deposited in GenBank under the accession numbers listed in Table 2.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
DW and JK performed the cell culture experiments. DW, NS, JK and JH performed the molecular genetic studies. FHL, CB and MMR coordinated field work and sample collection. JH and CL sampled gorilla feces. DW, NS, JK, FHL and BE conceived of the study, and participated in its design and coordination. DW, FHL, CB, JK and BE drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Negative stain electron micrograph of adenovirus-like particles isolated from a fecal sample of a captive gorilla. Negatively stained with 1% uranyl acetate. Virus particles are 70-90 nm in diameter with an icosahedral shape. Scale bar = 200 nm.
Contributor Information
Diana Wevers, Email: weversd@rki.de.
Fabian H Leendertz, Email: leendertzf@rki.de.
Nelly Scuda, Email: ScudaN@rki.de.
Christophe Boesch, Email: boesch@eva.mpg.de.
Martha M Robbins, Email: robbins@eva.mpg.de.
Josephine Head, Email: Josephine.Head@eva.mpg.de.
Carsten Ludwig, Email: Ludwig@allwetterzoo.de.
Joachim Kühn, Email: kuehnj@uni-muenster.de.
Bernhard Ehlers, Email: ehlersb@rki.de.
Acknowledgements
We thank Michael Laue for electron microscopic analysis. The technical assistance of Sonja Liebmann and Cornelia Walter is kindly acknowledged. We thank the Agence Nationale des Parcs Nationaux (ANPN) and the Centre National de la Recherche Scientifique et Technique (CENAREST) of Gabon for permission to conduct research in Loango National Park. We also thank the Société pour la Conservation et le Développement (SCD) and Wildlife Conservation Society (WCS) for financial and logistical support. For sample collection from wild gorilla, we thank L. Rabanal, L. Mackaga, E. R. Guizard, N. Tagg, B. Graw, E. Fairet, M. Gregoire, L. Rankin and the other field assistants of the Loango Ape project. For DNA extraction and identification of individual samples from wild gorilla we thank M. Arandjelovic and L. Vigilant from the Max-Planck-Institute for Evolutionary Anthropology, Leipzig.
References
- Davison AJ, Benko M, Harrach B. Genetic content and evolution of adenoviruses. J Gen Virol. 2003;84:2895–2908. doi: 10.1099/vir.0.19497-0. [DOI] [PubMed] [Google Scholar]
- Benkö M, Harrach B, Both G, Russel W, Adair BM, Adam E, de Jong JC, Hess M, Johnson M, Kajon A, In: Virus Taxonomy: VIIIth Report of the International Committee on Taxonomy of Viruses. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editor. Elsevier; 2005. Family Adenoviridae; pp. 213–228. [Google Scholar]
- Bailey A, Mautner V. Phylogenetic relationships among adenovirus serotypes. Virology. 1994;205:438–452. doi: 10.1006/viro.1994.1664. [DOI] [PubMed] [Google Scholar]
- Lukashev AN, Ivanova OE, Eremeeva TP, Iggo RD. Evidence of frequent recombination among human adenoviruses. Journal of General Virology. 2008;89:380–388. doi: 10.1099/vir.0.83057-0. [DOI] [PubMed] [Google Scholar]
- Robinson CM, Rajaiya J, Walsh MP, Seto D, Dyer DW, Jones MS, Chodosh J. Computational analysis of human adenovirus type 22 provides evidence for recombination among species D human adenoviruses in the penton base gene. Journal of Virology. 2009;83:8980–8985. doi: 10.1128/JVI.00786-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh MP, Chintakuntlawar A, Robinson CM, Madisch I, Harrach B, Hudson NR, Schnurr D, Heim A, Chodosh J, Seto D, Jones MS. Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs G, Harrach B, Zakhartchouk AN, Davison AJ. Complete genome sequence of simian adenovirus 1: An Old World monkey adenovirus with two fiber genes. Journal of General Virology. 2005;86:1681–1686. doi: 10.1099/vir.0.80757-0. [DOI] [PubMed] [Google Scholar]
- Basnight M Jr, Rogers NG, Gibbs CJ Jr, Gajdusek DC. Characterization of four new adenovirus serotypes isolated from chimpanzee tissue explants. American Journal of Epidemiology. 1971;94:166–171. doi: 10.1093/oxfordjournals.aje.a121308. [DOI] [PubMed] [Google Scholar]
- Kovacs GM, Davison AJ, Zakhartchouk AN, Harrach B. Analysis of the first complete genome sequence of an Old World monkey adenovirus reveals a lineage distinct from the six human adenovirus species. Journal of General Virology. 2004;85:2799–2807. doi: 10.1099/vir.0.80225-0. [DOI] [PubMed] [Google Scholar]
- Bányai K, Esona MD, Liu A, Wang Y, Tu X, Jiang B. Molecular detection of novel adenoviruses in fecal specimens of captive monkeys with diarrhea in China. Veterinary Microbiology. 2010;142:416–419. doi: 10.1016/j.vetmic.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Kidd AH, Garwicz D, Oberg M. Human and simian adenoviruses: Phylogenetic inferences from analysis of VA RNA genes. Virology. 1995;207:32–45. doi: 10.1006/viro.1995.1049. [DOI] [PubMed] [Google Scholar]
- Roy S, Vandenberghe LH, Kryazhimskiy S, Grant R, Calcedo R, Yuan X, Keough M, Sandhu A, Wang Q, Medina-Jaszek CA. et al. Isolation and characterization of adenoviruses persistently shed from the gastrointestinal tract of non-human primates. PLoS Pathogens. 2009;5 doi: 10.1371/journal.ppat.1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas I, Avellon A, Mosquera M, Jabado O, Echevarria JE, Campos RH, Rewers M, Perez-Brena P, Lipkin WI, Palacios G. Molecular identification of adenoviruses in clinical samples by analyzing a partial hexon genomic region. J Clin Microbiol. 2005;43:6176–6182. doi: 10.1128/JCM.43.12.6176-6182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford-Miksza LK, Schnurr DP. Adenovirus serotype evolution is driven by illegitimate recombination in the hypervariable regions of the hexon protein. Virology. 1996;224:357–367. doi: 10.1006/viro.1996.0543. [DOI] [PubMed] [Google Scholar]
- Torres S, Chodosh J, Seto D, Jones MS. The revolution in viral genomics as exemplified by the bioinformatic analysis of human adenoviruses. Viruses. 2010;2:1367–1381. doi: 10.3390/v2071367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arandjelovic M, Head J, Kühl H, Boesch C, Robbins MM, Maisels F, Vigilant L. Effective non-invasive genetic monitoring of multiple wild western gorilla groups. Biological Conservation. 2010;143:1780–1791. doi: 10.1016/j.biocon.2010.04.030. [DOI] [Google Scholar]
- Nsubuga AM, Robbins MM, Roeder AD, Morin PA, Boesch C, Vigilant L. Factors affecting the amount of genomic DNA extracted from ape faeces and the identification of an improved sample storage method. Mol Ecol. 2004;13:2089–2094. doi: 10.1111/j.1365-294X.2004.02207.x. [DOI] [PubMed] [Google Scholar]
- Walsh MP, Seto J, Tirado D, Chodosh J, Schnurr D, Seto D, Jones MS. Computational analysis of human adenovirus serotype 18. Virology. 2010;404:284–292. doi: 10.1016/j.virol.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EL, Blencowe E, Brandon K, Brown D, Burn RW, Cowlishaw G, Davies G, Dublin H, Fa JE, Milner-Gulland EJ. et al. Hunting for consensus: Reconciling bushmeat harvest, conservation, and development policy in West and Central Africa. Conservation Biology. 2007;21:884–887. doi: 10.1111/j.1523-1739.2006.00595.x. [DOI] [PubMed] [Google Scholar]
- Xiang Z, Li Y, Cun A, Yang W, Ellenberg S, Switzer WM, Kalish ML, Ertl HCJ. Chimpanzee adenovirus antibodies in humans, sub-Saharan Africa. Emerging Infectious Diseases. 2006;12:1596–1599. doi: 10.3201/eid1210.060078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkayastha A, Ditty SE, Su J, McGraw J, Hadfield TL, Tibbetts C, Seto D. Genomic and bioinformatics analysis of HAdV-4, a human adenovirus causing acute respiratory disease: Implications for gene therapy and vaccine vector development. Journal of Virology. 2005;79:2559–2572. doi: 10.1128/JVI.79.4.2559-2572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz H, Wigand R, Heinrich W. Worldwide epidemiology of human adenovirus infections. American Journal of Epidemiology. 1983;117:455–466. doi: 10.1093/oxfordjournals.aje.a113563. [DOI] [PubMed] [Google Scholar]
- Russell WC. In: Topley and Wilson's Virology. 10. Mahy BWJ, Ter Meulen V, Hodder Arnold, editor. 2005. Adenoviruses; pp. 439–443. [Google Scholar]
- Gray GC, Callahan JD, Hawksworth AW, Fisher CA, Gaydos JC. Respiratory diseases among U.S. Military personnel: Countering emerging threats. Emerging Infectious Diseases. 1999;5:379–387. doi: 10.3201/eid0503.990308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Singh J, Ruone S, Humphrey C, Yip CCY, Lau SKP, Anderson LJ, Kaur T. Short report: Identification of adenoviruses in fecal specimens from wild chimpanzees (Pan trogylodytes schweinfurthii) in Western Tanzania. American Journal of Tropical Medicine and Hygiene. 2010;82:967–970. doi: 10.4269/ajtmh.2010.09-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Negative stain electron micrograph of adenovirus-like particles isolated from a fecal sample of a captive gorilla. Negatively stained with 1% uranyl acetate. Virus particles are 70-90 nm in diameter with an icosahedral shape. Scale bar = 200 nm.