Abstract
More than 25 loci have been linked to type 1 diabetes (T1D) in the nonobese diabetic (NOD) mouse, but identification of the underlying genes remains challenging. We describe here the positional cloning of a T1D susceptibility locus, Idd11, located on mouse chromosome 4. Sequence analysis of a series of congenic NOD mouse strains over a critical 6.9-kb interval in these mice and in 25 inbred strains identified several haplotypes, including a unique NOD haplotype, associated with varying levels of T1D susceptibility. Haplotype diversity within this interval between congenic NOD mouse strains was due to a recombination hotspot that generated four crossover breakpoints, including one with a complex conversion tract. The Idd11 haplotype and recombination hotspot are located within a predicted gene of unknown function, which exhibits decreased expression in relevant tissues of NOD mice. Notably, it was the recombination hotspot that aided our mapping of Idd11 and confirms that recombination hotspots can create genetic variation affecting a common polygenic disease. This finding has implications for human genetic association studies, which may be affected by the approximately 33,000 estimated hotspots in the genome.
Type 1 diabetes (T1D) is a polygenic autoimmune disease in which lymphocytes mediate the destruction of insulin-producing beta cells in the pancreas (Atkinson and Eisenbarth 2001). The events that trigger the pathogenic autoimmune response are still not clear, but recent genome-wide association studies indicate that there are more than 40 loci affecting susceptibility to T1D in humans (Hakonarson et al. 2007; Todd et al. 2007; The Wellcome Trust Case Control Consortium 2007; Barrett et al. 2009; Concannon et al. 2009a). These studies have detected association of T1D with the common variants for previously identified genes and new candidates. Except for the HLA locus, however, these loci have small effects upon disease risk (odds ratio < 2.5) and fail to adequately explain the genetic variance for T1D (Concannon et al. 2009b). Instead, it has been proposed that rare/private mutations with larger effects may account for the missing genetic variance in complex genetic diseases (Goldstein 2009). Whether rare or common, identifying the actual causative variants for T1D has proved challenging due to genetic heterogeneity among affected individuals.
A complementary approach to human studies is the use of inbred mouse strains, in which genetic heterogeneity is avoided, and selective mating can precisely map genes for which allelic variation affects disease susceptibility. Analysis of the nonobese diabetic (NOD) mouse strain, which spontaneously develops T1D similar to humans, has been widely used to better understand disease pathology and gain key insights into the genetics of T1D (Atkinson and Leiter 1999). In parallel to human studies, more than 25 loci (termed Idd) have been linked to T1D in the NOD mouse (Serreze and Leiter 2001; Ridgway et al. 2008). Confirmation of these loci is best achieved using congenic mouse strains (Rogner and Avner 2003), which are generated by controlled mating of NOD mice with diabetes-resistant strains to introduce a donor-derived chromosome interval carrying a resistant allele onto the susceptible NOD genetic background. By testing smaller donor-derived intervals for their effect upon diabetes onset, a region small enough to be sequenced for disease-causing variants can be identified. To date, congenic NOD strains have confirmed Idd loci on chromosomes (chr) 1–4, 6, 7, 11, 13, 17, and 18 (Serreze and Leiter 2001; Ridgway et al. 2008). Besides the MHC region on Chr 17, at least eight of these loci have been dissected into smaller intervals using congenic NOD strains, and accumulating evidence has identified B2m, Il2, Il21, Ctla4, Slc11a1, and Trpv1 as genes for which NOD mice harbor T1D susceptibility alleles (Hamilton-Williams et al. 2001; Kissler et al. 2006; Razavi et al. 2006; Yamanouchi et al. 2007; Araki et al. 2009; McGuire et al. 2009). Preliminary evidence also suggests that Arntl2 and Stat5b are potential T1D susceptibility genes (Hung et al. 2006; Laloraya et al. 2006).
Although few causative variants have been defined to date, the NOD alleles that increase T1D risk in this mouse strain can be either rare or common among inbred mouse strains. For example, the effect of the Idd1 locus is attributed to the relatively rare MHC class II variant (Abg7), together with a more common variant found in other inbred mouse strains that encodes a deletion in the I-E-α chain promoter (Serreze and Leiter 2001). This observation, along with human genetic studies, suggests that increased T1D risk in humans may also result from the combination of rare and common variants within the human population (Concannon et al. 2009b).
Despite the identification of several Idd genes to date, this limited collection does not fully explain T1D pathogenesis or the underlying genetic architecture for T1D risk. One of the many Idd loci still to be identified is Idd11, which is located on Chr 4 and originally linked to T1D in NOD backcrosses to the C57BL/6 (B6) and SJL strains (Morahan et al. 1994). As B6 mice carry a resistance allele for Idd11, congenic strains with different Chr 4-B6–derived intervals on the NOD genetic background were produced (NOD.B6Idd11A, NOD.B6Idd11B, NOD.B6Idd11C, NOD.B6Idd11D). Three of these congenic NOD strains demonstrated significant diabetes resistance, thus confirming and localizing Idd11 to an ∼8-Mb interval on Chr 4 (Brodnicki et al. 2000, 2005).
To localize Idd11 further and identify the underlying gene, we established new congenic NOD strains to dissect this ∼8-Mb interval and monitored them for diabetes onset. Remarkably, each of these smaller congenic intervals were derived from recombination breakpoints within the same 6.9-kb interval that resulted in varying levels of T1D susceptibility for the different congenic NOD strains. Here, we report the sequence analysis of these breakpoints and the identification of a recombination hotspot that led to the discovery of a novel candidate gene (GenBank mRNA: AK005651) for Idd11. This gene of unknown function exhibits decreased expression in the thymus and spleen of NOD mice. Furthermore, NOD mice carry a unique haplotype for Idd11 compared with 25 other inbred mouse strains analyzed. Our findings demonstrate that recombination hotspots, which have been relatively neglected in human association studies, can create unique DNA sequence variation that has relatively large effects upon the risk for a common polygenic disease.
Results
Mapping Idd11 using congenic mouse strains
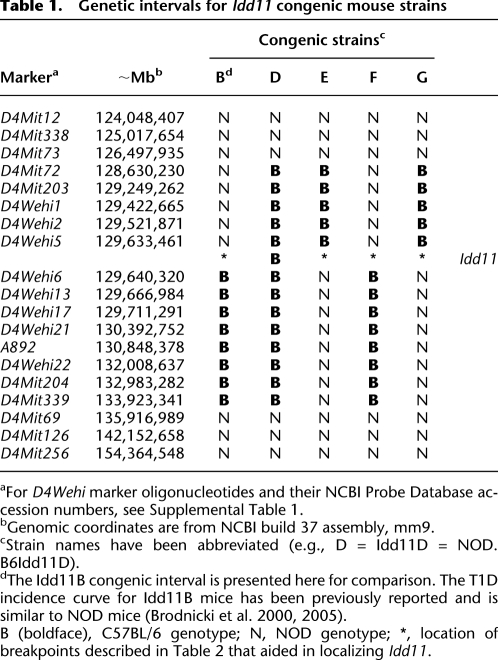
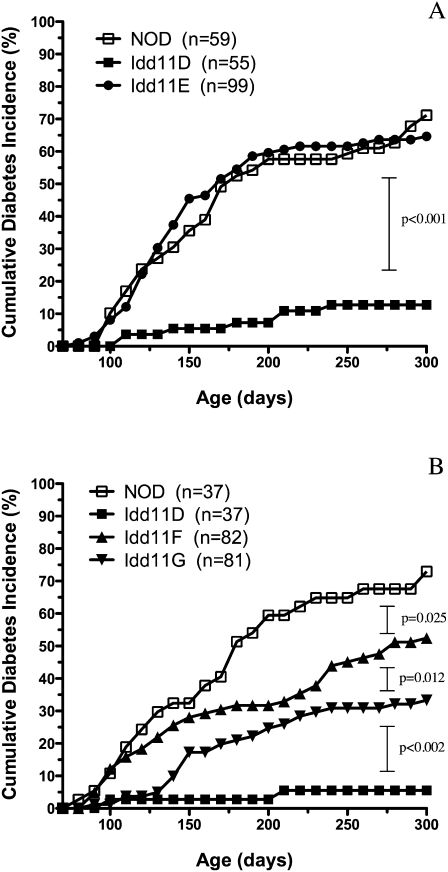
To localize Idd11 further, new congenic mouse strains were derived from NOD.B6Idd11D because this strain carried the smallest Chr 4-B6–derived interval providing diabetes protection among our panel of previously characterized congenic strains (Table 1; Brodnicki et al. 2000, 2005). Briefly, heterozygous NOD.B6Idd11D mice were intercrossed to generate F2 progeny that were screened for recombination events by genotyping novel markers we identified within this interval (Table 1; Supplemental Table 1; Supplemental Fig. 1). Three new congenic NOD strains (NOD.B6Idd11E, NOD.B6Idd11F, NOD.B6Idd11G) were established from recombinant F2 mice, and these were monitored for diabetes onset compared to NOD and Idd11D mice (note that henceforth congenic strain names are abbreviated, e.g., NOD.B6Idd11D = Idd11D) (Fig. 1).
Table 1.
Genetic intervals for Idd11 congenic mouse strains
aFor D4Wehi marker oligonucleotides and their NCBI Probe Database accession numbers, see Supplemental Table 1.
bGenomic coordinates are from NCBI build 37 assembly, mm9.
cStrain names have been abbreviated (e.g., D = Idd11D = NOD.B6Idd11D).
dThe Idd11B congenic interval is presented here for comparison. The T1D incidence curve for Idd11B mice has been previously reported and is similar to NOD mice (Brodnicki et al. 2000, 2005).
B (boldface), C57BL/6 genotype; N, NOD genotype; *, location of breakpoints described in Table 2 that aided in localizing Idd11.
Figure 1.
Cumulative diabetes incidence curves for Idd11 congenic strains. Female cohorts were monitored for diabetes by measuring urinary glucose levels. Pairwise comparisons of diabetes incidence curves were performed using the log-rank test. As a historical footnote, Idd11E was monitored first because it was presumed that this strain would be protected against diabetes given that Idd11B exhibited a similar diabetes incidence curve to NOD (Brodnicki et al. 2000). Once we observed that Idd11E was not protected against diabetes, we established cohorts for Idd11F and Idd11G to confirm this result using independent congenic strains, which initially appeared to have congenic intervals similar to Idd11B and Idd11E, respectively (Table 1). Subsequent sequence analysis of the interval between D4Wehi5 and D4Wehi6 (Table 2) identified genetic variation that explained the diabetes incidence observed for Idd11B (Brodnicki et al. 2000) and Idd11E (A).
Comparison of the resulting diabetes incidence curves indicated that Idd11 mapped to an ∼6.9-kb interval, between D4Wehi5 and D4Wehi6 (Table 1; Fig. 1A,B). However, the Idd11 locus appeared to be more complex than expected because the congenic strains exhibited varying levels of T1D incidence. For example, Idd11E and Idd11G seemed to have identical B6-derived intervals, but Idd11E was more susceptible to T1D than Idd11G (65% vs. 33% diabetic by 300 d). On the other hand, the Idd11G interval provided less diabetes protection compared with the larger Idd11D interval (33% vs. 6%) but provided greater protection compared with the Idd11F interval (33% vs. 52%). As Idd11E, Idd11F, and Idd11G were derived from the Idd11D strain, we postulated that sequencing the recombinant boundary between D4Wehi5 and D4Wehi6 would explain the variability in T1D susceptibility between the congenic strains.
Sequence analysis of the Idd11 critical interval
Sequence analysis of the NOD, B6, and congenic NOD strains identified several sequence variants within the ∼6.9-kb interval (Table 2; Supplemental Tables 2, 3). Remarkably, Idd11E and Idd11G were isogenic except at sequence variant 3, demonstrating that allelic variation at this position can significantly affect T1D susceptibility. However, the B6-derived allele at this position alone could not account for all of the Idd11 effect since it conferred different levels of diabetes protection to Idd11D (∼6% diabetic by 300 d), Idd11F (∼52%), and Idd11G (∼33%) (Fig. 1A,B; Table 2). This comparison of diabetes incidence curves indicated that Idd11 was likely due to a haplotype effect in which B6-derived sequences are required at more than one position to provide optimal T1D protection.
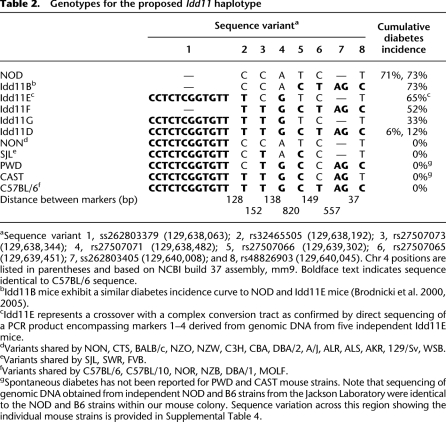
Table 2.
Genotypes for the proposed Idd11 haplotype
aSequence variant 1, ss262803379 (129,638,063); 2, rs32465505 (129,638,192); 3, rs27507073 (129,638,344); 4, rs27507071 (129,638,482); 5, rs27507066 (129,639,302); 6, rs27507065 (129,639,451); 7, ss262803405 (129,640,008); and 8, rs48826903 (129,640,045). Chr 4 positions are listed in parentheses and based on NCBI build 37 assembly, mm9. Boldface text indicates sequence identical to C57BL/6 sequence.
bIdd11B mice exhibit a similar diabetes incidence curve to NOD and Idd11E mice (Brodnicki et al. 2000, 2005).
cIdd11E represents a crossover with a complex conversion tract as confirmed by direct sequencing of a PCR product encompassing markers 1–4 derived from genomic DNA from five independent Idd11E mice.
dVariants shared by NON, CTS, BALB/c, NZO, NZW, C3H, CBA, DBA/2, A/J, ALR, ALS, AKR, 129/Sv, WSB.
eVariants shared by SJL, SWR, FVB.
fVariants shared by C57BL/6, C57BL/10, NOR, NZB, DBA/1, MOLF.
gSpontaneous diabetes has not been reported for PWD and CAST mouse strains. Note that sequencing of genomic DNA obtained from independent NOD and B6 strains from the Jackson Laboratory were identical to the NOD and B6 strains within our mouse colony. Sequence variation across this region showing the individual mouse strains is provided in Supplemental Table 4.
These data raised a critical question: How big is the Idd11 haplotype? NOD crosses with either SJL, NON, 129, C57L, C57BL/10 (B10), or NOR mouse strains have previously demonstrated the linkage of T1D to chr 4, suggesting that these strains, similar to B6, may harbor a T1D-resistance allele for Idd11 (Morahan et al. 1994; Rodrigues et al. 1994; McAleer et al. 1995; McDuffie 2000; Reifsnyder et al. 2005; Leiter et al. 2009). The B10-identified and the NOR-identified loci, which overlap the B6-defined Idd11 locus, have also been confirmed by congenic strains (Lyons et al. 2000; Reifsnyder et al. 2005). Sequence analysis determined that the B10 and NOR strains were identical to B6 across the congenic breakpoint positions (Table 2; Supplemental Table 4). In contrast, the SJL strain was identical to NOD at these variants except at 1, 3, and 5, for which SJL was identical to B6, B10, and NOR (Table 2; Supplemental Table 4). Further sequence analysis for these variants within 20 other inbred mouse strains (including wild-derived strains) indicated that the Idd11 haplotype consisted of at least variants 1–5, with NOD mice representing a unique haplotype marked by a 12-bp deletion at variant 1 (Table 2; Supplemental Table 4).
Confirmation of a recombination hotspot
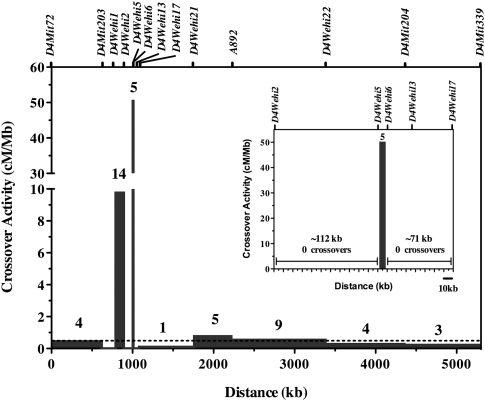
It was conspicuous that recombination breakpoints for four of our congenic strains occurred within an ∼1.2-kb interval and resulted in one instance of a crossover with a complex conversion tract (Idd11E) (Table 2). This dense clustering of crossover events and the presence of a complex conversion tract is characteristic of a recombination hotspot (Petes 2001; Jeffreys and May 2004; Bois 2007; Kauppi et al. 2007). To confirm and measure the frequency of the meiotic crossovers within the Idd11 haplotype, 723 F2 progeny were generated by intercrossing heterozygous (NOD × Idd11D)F1 mice and were screened for recombination events between D4Wehi5 and D4Wehi6. The calculated crossover activity was ∼50 cM Mb-1, which is 100-fold greater than the mouse genome average (∼0.5 cM Mb-1) (Shiroishi et al. 1995), and confirms that this interval harbors a recombination hotspot (Fig. 2). Genomic DNA available for 231 of these F2 progeny (representing 462 meiosis events) that did not exhibit a crossover between D4Wehi5 and D4Wehi6 was further genotyped for sequence variants 1–8 (Table 2). Only one noncrossover event (also termed gene conversion) was identified, at variant 4. This suggests a relatively low frequency (<0.5%) for noncrossover events in this hotspot, but gene conversion events can only be detected and estimated using the available sequence variants located within the hotspot interval. Hence, this conversion frequency is likely underestimated due to the relatively few variants within this interval between NOD and B6 mice. While other local recombination hotspots may reside elsewhere within the Idd11D interval, we observed no recombination events either ≤112 kb proximal to D4Wehi5 or ≤71 kb distal to D4Wehi6 (Fig. 2, inset). These flanking recombination “cold regions” have restricted our ability to generate smaller congenic intervals encompassing only the proposed Idd11 haplotype. It should be noted that while this recombination hotspot may generate noncrossover events, more than one variant within this haplotype would need to be converted to confer T1D protection in subsequent congenic NOD mice.
Figure 2.
Distribution of crossover breakpoints within the NOD.B6Idd11D congenic interval. Seven-hundred-twenty-three F2 pups (1446 meiosis events) were derived from intercrossing NOD.B6Idd11D F1 mice and were genotyped for markers within the NOD.B6Idd11D congenic interval. The crossover activity was calculated by dividing the frequency of crossovers in each interval by its length. For example, five pups had a recombination between D4Wehi5 and D4Wehi6, giving rise to a genetic distance of 0.345 cM across this 6859-bp interval, which is ∼50 cM Mb-1. Bars, crossover frequency in cM/Mb in each interval. Above each bar, the number of crossovers observed in that marker interval is shown. The dashed line indicates the average crossover activity for the mouse genome: ∼0.5cM/Mb (Shiroishi et al. 1995). Genetic markers are indicated by ticks above the plot. The inset displays an enlarged view of the interval between D4Wehi2 and D4Wehi17.
Bioinformatic and expression analysis of a novel candidate gene for Idd11
The Idd11 haplotype, including the recombination hotspot, is located within a predicted gene, termed AK005651 (i.e., GenBank mRNA accession number), supported by a collection of expressed sequence tags (ESTs) (Supplemental Fig. 2). The encoded transcript consists of five exons (Fig. 3A; Supplemental Table 5) and two splice isoforms (±exon 3) confirmed by RT-PCR and sequencing data (data not shown). AK005651 has a number of open reading frames, but none demonstrate clear protein domains or homology with known proteins nor does the transcript encode an obvious microRNA. Conservation between mammalian sequences across this region suggests the presence of a human ortholog, although no orthologous human spliced EST or transcript has yet been described (bioinformatic analysis is summarized in Supplemental Table 6).
Figure 3.
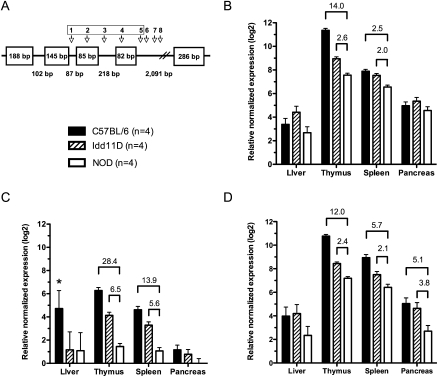
Schematic diagram and expression analysis of AK005651. (A) The Idd11 haplotype (variants 1–5) is located within a predicted gene, termed AK005651, consisting of five exons (Supplemental Table 5). Exon 3 is alternatively spliced giving rise to two different transcripts (±exon 3). Quantitative real-time PCR was performed to detect expression differences between mouse strains at 50 d of age for the spliced product derived from: (B) exons 4 and 5, which are present in both splice variants; (C) exons 2 and 3, (*) one of the four C57BL/6 samples was an extreme outlier (the mean relative normalized expression is 2.29 excluding this outlier); (D) exons 2 and 4. Approximate fold change (≥2) is shown only for significant pairwise comparisons between NOD and other mouse strains (P < 0.05, adjusted for multiple testing). Bars, mean expression level (±pooled SEM for each tissue).
If AK005651 does encode a protein, then variants 2 and 4 result in amino acid changes that depend on the open reading frame (as shown in Supplemental Table 7). For the three possible open reading frames, only one results in silent substitutions due to variants 2 and 4 (Supplemental Table 7, ORF 1), whereas these two variants produce amino acid changes in all other open reading frames between NOD and B6. The longest open reading frame for AK005651 (methionine to stop) is 53 amino acids and is located in exon 5. No sequence variation between NOD and B6 occurs in this open reading frame.
Sequence variation within the proposed Idd11 haplotype may also affect gene expression. The sequence encompassing variants 1–4 contains predicted transcription factor binding sites (TFBSs) based upon conservative analyses using TRANSFAC matrices (Matys et al. 2006), but none of the Idd11 sequence variants occur within these predicted TFBSs (Supplemental Table 8). Variants 1 and 2 do occur in a promoter-associated regulatory feature (Supplemental Fig. 2) predicted by Ensembl based upon DNase I hypersensitivity and the enrichment of histone H3K4me3 in different cell types (Mikkelsen et al. 2007). Real-time PCR analysis indicated that AK005651 is differentially expressed between NOD and diabetes-resistant mouse strains (i.e., B6 and Idd11D) in tissues relevant to T1D pathogenesis. Exon 4 and 5 were present in both splice variants, and B6 and Idd11D mice had at least a twofold increase in thymic and splenic expression of these spliced exons (Fig. 3B). On the other hand, expression analysis of Spocd1 and Bai2, two genes flanking AK005651 (Supplemental Fig. 2), indicated that the sequence variation within AK005651, in particular variants 1–4 for the Idd11 haplotype, is unlikely to affect transcription levels of these two flanking genes (Supplemental Fig. 3).
Expression differences may also reflect deficient exon splicing. Sequence variants 2–5 do not disrupt canonical splice sites, although they may alter cryptic intron/exon splicing motifs (Chasin 2007; Wang and Burge 2008). In contrast, the unique NOD 12-bp deletion at position 1 shortens the intron between exons 2 and 3. Real-time PCR detected a significant decrease in a spliced product from these two exons in thymic and splenic RNA isolated from NOD mice compared with B6 and Idd11D mice (Fig. 3C). The spliced product from exons 2 and 4 was also significantly decreased in the NOD thymus and spleen, as well as the pancreas, compared with B6 and Idd11D (Fig. 3D). We note that the anomalous result for the liver (Fig. 3C) was due to one extreme outlier. This B6 sample did not result in an outlier using other real-time PCR probes and was the only such outlier observed within our expression data set (Figs. 3, 4; Supplemental Fig. 3).
Figure 4.
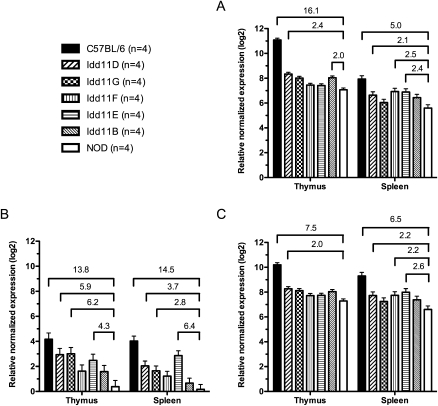
Expression analysis of AK005651 in congenic NOD mouse strains. Quantitative real-time PCR was performed to detect expression differences between mouse strains at 50 d of age for the spliced product derived from the following: (A) exons 4 and 5, which are present in both splice variants; (B) exons 2 and 3; (C) exons 2 and 4. Different cohorts of NOD, B6, and Idd11D mice to those in Figure 3 were generated and used in conjunction with Idd11B, Idd11E, Idd11F, and Idd11G mice for this experiment. Fold change (≥2) is shown only for significant pairwise comparisons between NOD and other mouse strains (P < 0.05, adjusted for multiple testing). Bars, mean expression level (±pooled SEM for each tissue).
To further dissect the effect of the Idd11 haplotype upon AK005651, expression analysis was performed using thymic and splenic RNA isolated from our series of congenic mouse strains (Fig. 4; note: NOD, B6, Idd11D represent different cohorts to those in Fig. 3). Variant 1 (12-bp ins/del) had the most distinct effect upon AK005651 expression. Congenic mice harboring the B6-derived insertion (Idd11D, Idd11G, Idd11E) exhibited significantly higher transcription levels of the spliced product for exons 2/3 compared with NOD mice (Fig. 4B). However, the effect of the Idd11 haplotype upon the transcription levels of spliced products for exons 2/4 and for exons 4/5 appears to be more complex. While the two spliced products had similar expression profiles in the respective tissues (Fig. 4A,C), the different Idd11 haplotypes led to varied expression between strains, as well as between tissues. For example, only Idd11D exhibited twofold or more increases for both splice products in both tissues; whereas Idd11E and Idd11F had more than twofold increases only in the spleen. These results indicate that different combinations of B6-derived sequences within the Idd11 haplotype have varying effects upon AK005651 expression, with increased expression for all three spliced products in both the thymus and spleen correlating with the greatest degree of T1D protection (i.e., Idd11D).
Discussion
Positional cloning using congenic mouse strains has been used to confirm and localize a number of T1D susceptibility loci (Serreze and Leiter 2001; Ridgway et al. 2008). Nonetheless, only a handful of genes for Idd loci have been identified to date, principally because it takes large mouse colonies and significant breeding time to generate a series of congenic mouse strains that maps a locus to within a genomic interval containing only one or a few candidate genes. Even when the candidate genes are identified (whether in mouse or human genetic studies), it is often difficult to determine which sequence variant(s) within the mapped interval are causal, unless they are obvious mutations that disrupt known coding, splicing, or regulatory motifs (Hindorff et al. 2009). In this study, a recombination hotspot generated a unique series of congenic mouse strains for Idd11 and enabled us to identify a novel candidate gene for which sequence variation affects T1D susceptibility in the NOD mouse.
Comparison of the sequence and diabetes incidence curves between our congenic NOD strains indicated that Idd11 is due to a haplotype, consisting of at least five variants, for which the composition has variable effects on susceptibility to T1D. For example, the Idd11D congenic strain has the complete B6-derived haplotype and the best protection against diabetes onset, whereas Idd11E and Id11B lack two or more B6-derived alleles within this haplotype and have similar diabetes incidence to NOD mice (Table 2; Fig. 1; Brodnicki et al. 2000). In contrast, Idd11F and Idd11G mice lack one B6-derived allele within the proposed Idd11 haplotype but do so at different positions and exhibited different T1D incidence curves (Table 2; Fig. 1). Thus these variable effects depend on the number and combination of B6-derived alleles within this haplotype. Similar associations of polymorphic haplotypes with autoimmune disease are well established for the major histocompatibility complex (Fernando et al. 2008), the human cytokine gene cluster on chr 5q31 (Rioux et al. 2001), and the mouse SLAM/CD2 gene cluster (Wandstrat et al. 2004). However, the Idd11 haplotype appears to be confined to a single gene, whereas these other haplotypes encompass multiple genes.
Sequence analysis determined that NOD mice have a unique Idd11 haplotype due to a 12-bp deletion. Notably, the NOD strain was generated from the same inbreeding program as the CTS and NON strains (Makino et al. 1980; Beck et al. 2000), but these two strains (as well as 18 other laboratory and four wild-derived inbred strains) do not have the 12-bp deletion, suggesting this microdeletion arose as a de novo mutation or was preferentially inherited from a different parental ancestor during the derivation of the NOD strain. In either case, further sequence comparison was required because this microdeletion did not fully explain the varying levels of T1D susceptibility observed in our congenic mouse strains. Two additional sequence variants (rs27507073 and rs27507066) were associated with T1D based on resistance phenotypes determined by segregation and congenic analyses (Brodnicki et al. 2000, 2005; Lyons et al. 2000; Reifsnyder et al. 2005). Our finding does not imply that the Idd11 haplotype is limited to these three variants, but only indicates the minimum size of the B6-derived haplotype providing protection against T1D in NOD mice.
Idd11 is not the only T1D susceptibility locus on chr 4. Genetic studies have mapped at least three other Idd loci to this chromosome. An outcross between NOD and B10 originally detected a locus, termed Idd9, linked next to the telomere of chr 4 (Rodrigues et al. 1994). Congenic NOD strains, harboring B10-derived intervals, subsequently expanded and dissected this locus into three subloci (Idd9.1, Idd9.2, Idd9.3) (Lyons et al. 2000), with Idd9.1 mapped to the same region we had previously localized Idd11 (Morahan et al. 1994). The haplotype analysis reported here suggests that Idd11 and Idd9.1 are the same because B6 and B10 are identical by descent for this chromosome region. A NOD outcross with the NOR strain also detected linkage to the region encompassing Idd11, which was confirmed by NOD.NOR-chr 4 congenic mice (Reifsnyder et al. 2005). NOR is an inbred recombinant congenic strain derived from NOD and C57BLKS/J (Prochazka et al. 1992), and our sequence analysis indicated that the Idd11 haplotype for NOR is B6-derived. However, NOD.NOR-chr 4 mice harbor a larger congenic interval than the B6-derived intervals in our Idd11 congenic strains. Thus, resistant NOR-derived alleles at other chr 4 loci may also contribute to the overall diabetes protection observed for this congenic strain (Reifsnyder et al. 2005). Lastly, NOD outcrosses with the NON and 129/SvImJ mouse strains also detected linkage of T1D to chr 4 (McAleer et al. 1995; Leiter et al. 2009), but it is more likely that these linkage results were due to allelic variation at other loci because these two strains are NOD-like for the Idd11 haplotype—their allelic composition is not predicted to be protective based on comparison with the Idd11E haplotype for Idd11 (Table 2).
Idd11 appears to represent a “gene-based functional haplotype”: a defined sequence interval taken as a unit because individual variants are not sufficient to act as separate disease markers or completely account for the associated phenotypic effect (Hoehe 2003). The Idd11 haplotype, including the recombination hotspot, is located within AK005651, a predicted gene of unknown function. None of the open reading frames demonstrate homology with known proteins or evolutionary conservation, suggesting that AK005651 may encode a long noncoding RNA (Mercer et al. 2009; Ponting et al. 2009), although it was not detected in a recent large noncoding RNA screen of four mouse cell types (Guttman et al. 2009). As current bioinformatics approaches are limited in deciphering the function of AK005651, we investigated the effect of sequence variation on AK005651 expression.
The different Idd11 haplotypes, represented by our series of congenic strains, have variable effects on AK005651 expression, which is perhaps not surprising given their variable effects on susceptibility to T1D. Real-time PCR indicated that the NOD Idd11 haplotype is associated with decreased expression of AK005651 in the thymus and spleen. In particular, the unique NOD microdeletion was associated with the decreased expression of the exon 2/3 splice product, suggesting that this microdeletion may shorten the intron in which it resides, impeding the splicing lariat structure and altering expression of splice isoforms (Black 2003). However, increased splicing of these two exons was not sufficient to increase T1D protection (e.g., Idd11E mice develop T1D similar to NOD mice but do not harbor the NOD microdeletion and have increased expression of the exon 2/3 splice product). Further comparison of our congenic strains also indicated that only Idd11D, which has the greatest T1D protection, exhibited significantly increased expression of AK005651 for all three spliced products in both the thymus and spleen compared with NOD. Idd11G and Idd11F have the next best T1D protection, respectively, but lack a similar increase and correlation between AK005651 expression and T1D protection. This lack of correlation may reflect that sequence variation within the Idd11 haplotype also affects the function of the encoded gene product (whether a large noncoding RNA or protein) leading to increased T1D protection, especially in combination with altered gene expression. The low level of expression for AK005651, although seemingly technically challenging (e.g., Ct values > 30 using the maximum amount of RNA/cDNA), was reproducibly detected by three different real-time PCR assays in different mouse cohorts. Given the relatively small differences observed between congenic strains, further studies are required to determine the function of AK005651 and how sequence composition within the Idd11 haplotype affects gene function and expression to alter T1D susceptibility. The recent availability of efficient NOD ES cells (Nichols et al. 2009) should also enable replacement of the NOD variants with B6-derived protective variants to ultimately confirm the effect of this apparent gene-based functional haplotype. At present, AK005651 represents a novel disease susceptibility gene compared with the current set of identified Idd genes, which mainly encode proteins of the immune system (e.g., MHC molecules, B2M, IL2, IL21, CTLA4, SLC11A1) that were relatively well characterized before their discovery as T1D genes (Serreze and Leiter 2001; Ridgway et al. 2008).
Discovery of Idd11 relied on the presence of a recombination hotspot. The ability to map a disease locus using congenic mice depends on the occurrence of recombination events to generate successively smaller genomic intervals to identify the underlying gene. However, recombination during meiosis is not random (Petes 2001; Kauppi et al. 2004). Recombination hotspots and associated flanking coldspots ultimately restrict how small the congenic interval can become and dictate the number of candidate genes to be investigated. Paradoxically, Idd11 mapped to a recombination hotspot. Although this hotspot demonstrated lower crossover activity compared with other hotspots within the mouse genome (Guillon and de Massy 2002; Yauk et al. 2003; Kauppi et al. 2007; Paigen et al. 2008), it was highly localized (∼1.2 kb) and resulted in one instance of a crossover with a complex conversion tract. This recombination hotspot provided the molecular mechanism that generated the “hybrid haplotypes” in our congenic NOD strains. It seems likely it is also responsible for the underlying allelic variation at this locus within the Mus species.
Rather than restricting our ability to map Idd11, this recombination hotspot enabled us to identify specific variants accounting for the varying levels of T1D susceptibility observed in our congenic strains. For example, the noncrossover/conversion event at rs27507073 (variant 3 in the Idd11 haplotype) significantly increased the risk of T1D in Idd11E mice compared with Idd11G mice. Gene conversion, as well as genomic rearrangement, associated with recombination hotspots has been implicated in a number of Mendelian diseases (Lupski and Stankiewicz 2005; Chen et al. 2007; Turner et al. 2008). Our congenic strains demonstrate that recombination hotspots can generate unique hybrid haplotypes, due to shuffled haplotypes and/or complex conversion tracts, which increase the risk for a complex genetic disease if they arise on a genetic background of some liability. Up to 85% of T1D cases in the human population are sporadic (Karvonen et al. 2001). Undoubtedly these affected individuals inherited some combination of diabetogenic alleles from their parents, but common variants (including copy number variation) identified in recent genome-wide association studies have failed to explain the total T1D risk attributable to genetic factors (Concannon et al. 2009b; Conrad et al. 2009; The Wellcome Trust Case Control Consortium 2010). Instead, recombination hotspots may contribute to the generation of rare variants and/or hybrid haplotypes accounting for the significantly increased T1D risk in those individuals who have inherited a certain level of genetic liability due to common variants.
Our study represents a primary example of a recombination hotspot associated with a complex genetic disease. Ng et al. (2010) have also recently described a recombination hotspot in a region of GABRB2 for which haplotypes are associated with schizophrenia. Similar to our study in congenic mice, their human study suggests that recombination hotspots are likely to contribute to the etiology of complex genetic diseases (Ng et al. 2010). However, sequence variation for the region on the human chr 1p35 encompassing the homologous Idd11 locus has not been associated with T1D (Barrett et al. 2009). This is not necessarily unexpected as the NOD mouse represents a “single case study” with a collection of diabetogenic alleles, some of which are unique to the NOD mouse strain, while others are common within the Mus species (Atkinson and Leiter 1999; Serreze and Leiter 2001). Given that the NOD Idd11 haplotype is unique, the equivalent susceptibility locus in humans might not exist or the equivalent human allele(s) may not be detectable in recent association studies because it is rare and/or resides in a recombination hotspot. HapMap data phase III (release 2, February 2009) shows that the homologous interval on the human chr 1p35 (between rs16834708 and rs13426) exhibits weak to no linkage disequilibrium (r2 < 0.1) (The International HapMap Consortium 2007). Such potential recombination hotspots (∼33,000 estimated in the human genome) (The International HapMap Consortium 2007) have so far been neglected in genetic studies due to the statistical methods employed for detecting association (Kauppi et al. 2004; The International HapMap Consortium 2007), but their ability to give rise to gene conversion and unique hybrid haplotypes, as observed for the Idd11 locus, demonstrates their capacity for producing sequence variation with demonstrable effects upon complex traits.
Methods
Mice
NOD/Lt (NOD) and C57BL/6 (B6) mouse strains were obtained from The Walter & Eliza Hall Institute specific pathogen-free (SPF) facilities. NOD.B6Idd11A, NOD.B6Idd11B, NOD.B6Idd11C, and NOD.B6Idd11D were established after 10 backcross generations or more using a conventional breeding approach for congenic mouse strains as previously described (Rogner and Avner 2003; Brodnicki et al. 2005). NOD.B6Idd11E, NOD.B6Idd11F, and NOD.B6Idd11G mouse strains were generated from (NOD × NOD.B6Idd11D)F2 progeny (Supplemental Fig. 1), which were screened for recombination events between D4Mit72 and D4Mit204. New congenic intervals, which dissected the Idd11D interval (Fig. 1), were fixed to homozygosity by brother–sister mating.
Genotyping and sequencing
DNA samples were extracted from tail biopsies by standard methods and genotyped with polymorphic markers by PCR (Supplemental Table 1; Brodnicki et al. 2000). NOD.B6Idd11D was genotyped using a 10-cM averaged genome-wide marker panel, and no B6-derived alleles were found outside the congenic interval (note that all other congenic strains described were derived from NOD.B6Idd11D). To fine-map the recombination sites, new genetic markers (i.e., nucleotide repeats) were identified using the publicly available mouse genome sequence (NCBI Build 37 assembly; mm9), the UCSC Genome Browser (http://genome.ucsc.edu/) (Kuhn et al. 2009), and the Tandem Repeats Finder program (Benson 1999). These markers (D4Wehi1–D4Wehi22) were shown to be polymorphic between the NOD and B6 mouse strains by PCR and gel electrophoresis of genomic DNA using sequence-specific oligonucleotides (Supplemental Table 1). Sequence within the defined Idd11 critical interval was determined by direct sequencing of overlapping PCR products (Supplemental Table 2) and the BigDye Terminator v3.1 sequence kit (Applied Biosystems). Sequence contigs for each strain were aligned to determine sequence variation between inbred strains (Megalign, DNAstar, Inc.). Genotyping of sequence variants 1–8 within the Idd11 haplotype (Table 2) was performed on a Roche LightCycler 480 using TaqMan probes (Applied Biosystems) (Supplemental Table 3) and Roche Probe Master Mix (Roche Applied Science) according to manufacturer's instructions.
Diabetes monitoring
Cohorts of female mice were housed in an SPF facility and tested once a week for elevated urinary glucose (>110 mmol/L) using Diastix reagent strips (Bayer Australia, Ltd.) over a 300-d time course. Three consecutive elevated readings indicated the onset of diabetes. Pairwise comparisons of the diabetes incidence between mouse strains were done using the log-rank test.
Bioinformatic analysis of AK005651
Genomic sequence and transcript sequences were aligned to the mouse genome (NCBI Build 37 assembly; mm9) using BLAT via the UCSC Genome Browser (http://genome.ucsc.edu/) (Kuhn et al. 2009). Sequence was compared with annotated ESTs, multispecies alignments, conserved elements, and the gene prediction algorithms available through the UCSC Genome Browser. Transcript sequences were also aligned to the NCBI EST database using BLASTN and to the NR protein database using BLASTX (http://blast.ncbi.nlm.nih.gov/Blast.cgi; Altschul et al. 1990). All potential transcripts were translated in six frames and searched for known domains represented by profile hidden Markov models from the PFAM database using hmmpfam (http://hmmer.janelia.org) (Finn et al. 2008). The RFAM database was used to evaluate the presence of a noncoding gene (http://rfam.janelia.org/; Gardner et al. 2009), and BLASTN was used to align transcript sequence to mature miRNAs and stem-loop sequences in miRBase (Griffiths-Jones et al. 2006). To determine if variants are within predicted TFBSs, 71 nucleotides (nt) of sequence centered on the individual variants were extracted from the mouse genome, and TFBSs were predicted in these sequences using only high-quality TRANSFAC matrices (Matys et al. 2006) representing vertebrate transcription factors and match score thresholds selected to give the minimum false-positive rate (Kel et al. 2003).
Quantitative real-time PCR
Tissues were taken from female mice (∼50 d old) and RNA isolated using TRIzol reagent (Invitrogen). cDNA was synthesized using Superscript III reverse transcriptase (Invitrogen). Quantitative real-time PCR was performed on a Roche LightCycler 480 using LightCycler Probe Master Reagent (Roche Diagnostics) or TaqMan Gene Expression Master Mix (Applied Biosystems) according to the manufacturer's instructions. Technical replicates were done in triplicate to calculate the average Ct value for each biological sample. Oligonucleotide primers and fluorescent probes were synthesized by Sigma Genosys, Roche Applied Sciences, or Applied Biosystems (Supplemental Table 9). The fluorescent probes used to detect the spliced products between AK005651 exon 2 and exon 3, as well as exon 2 and exon 4, are located across the spliced exon boundaries, whereas the fluorescent probe used to detect the spliced product between exon 4 and exon 5 is located entirely within exon 5 (Fig. 3) or across the spliced exon boundary (Fig. 4). The fluorescent probe used to detect the spliced product for Spocd1 was located in exon 9. The fluorescent probe used to detect the spliced product for Bai2 was located in exon 29. Thermal cycling consisted of a denaturation step (10 min at 95°C) and 45 amplification cycles (10 sec at 95°C, 15–30 sec at 60°C–66°C, 30 sec at 40°C). Products observed for each primer pair were confirmed by sequencing. Standard curves were generated for all primer sets to ensure exponential increase of targeted transcripts during amplification (efficiency = 10(−1/slope) = ∼2). ΔCt for each tissue was calculated as CtAK005651 − Ref, where Ref is the average Ct value of the reference genes Hmbs and Hprt1. Relative normalized log2 expression values were calculated for graphing purposes as 41 − mean(Ref) − ΔCt, where mean(Ref) is the grand mean of the reference genes Ct for each tissue. Here, 40 represents the practical Ct detection limit for real-time PCR, and 41 establishes the 0 point (i.e., no detectable gene expression) for the y-axis. Subtracting mean(Ref) converts the scores to log2-expression relative to the detection threshold scale from 0–41. Statistical significance for the difference in expression was obtained using pairwise t-tests with pooled standard deviations for each tissue. P-values were adjusted for multiple testing using Holm's method (Holm 1979).
Acknowledgments
We thank S. Foote, J. Stankovich, Y. Hu, and S. Mannering for useful discussions; V. Marshall, M. Martyn, and G. Brammar for technical assistance; E.H. Leiter for providing genomic DNA for various inbred mouse strains; and the mouse care facility staff at the Walter & Eliza Hall Institute and Department of Medicine at The University of Melbourne. This work was supported by the Juvenile Diabetes Research Foundation (1-2005-925), the Cooperative Research Centre for Discovery of Genes for Common Human Diseases, the Australian NHMRC (575552), and the NIH/NIDDK (1R01 DK062882-01A1). I.K.L.T. is supported by a Melbourne International Research Scholarship. N.W. and M.P.A. are supported by Australian Postgraduate Awards. M.P.A. is also supported by a St. Vincent's Institute Foundation Scholarship. C.E. is supported by a Peter Doherty Fellowship. M.B. is supported by an NHMRC Career Development Award. G.S. is supported by a NHMRC Senior Research Fellowship. G.M. is supported by NHMRC Program Grant 516700 and by the Diabetes Research Foundation of Western Australia. T.S. is supported by an Australia Fellowship.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) under accession nos. ss262803370, ss262803372, ss262803374, ss262803376, ss262803379, ss262803382, ss262803385, ss262803388, ss262803390, ss262803391, ss262803392, ss262803394, ss262803397, ss262803400, ss262803402, ss262803403, ss262803404, and ss262803405, and to the NCBI Probe Database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=probe) under accession nos. 10544425–10544446.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.101881.109.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ 1990. Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Araki M, Chung D, Liu S, Rainbow DB, Chamberlain G, Garner V, Hunter KM, Vijayakrishnan L, Peterson LB, Oukka M, et al. 2009. Genetic evidence that the differential expression of the ligand-independent isoform of CTLA-4 is the molecular basis of the Idd5.1 type 1 diabetes region in nonobese diabetic mice. J Immunol 183: 5146–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson MA, Eisenbarth GS 2001. Type 1 diabetes: New perspectives on disease pathogenesis and treatment. Lancet 358: 221–229 [DOI] [PubMed] [Google Scholar]
- Atkinson MA, Leiter EH 1999. The NOD mouse model of type 1 diabetes: As good as it gets? Nat Med 5: 601–604 [DOI] [PubMed] [Google Scholar]
- Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, et al. 2009. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 41: 703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck JA, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig JT, Festing MF, Fisher EM 2000. Genealogies of mouse inbred strains. Nat Genet 24: 23–25 [DOI] [PubMed] [Google Scholar]
- Benson G 1999. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res 27: 573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336 [DOI] [PubMed] [Google Scholar]
- Bois PR 2007. A highly polymorphic meiotic recombination mouse hot spot exhibits incomplete repair. Mol Cell Biol 27: 7053–7062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodnicki TC, McClive P, Couper S, Morahan G 2000. Localization of Idd11 using NOD congenic mouse strains: Elimination of Slc9a1 as a candidate gene. Immunogenetics 51: 37–41 [DOI] [PubMed] [Google Scholar]
- Brodnicki TC, Fletcher AL, Pellicci DG, Berzins SP, McClive P, Quirk F, Webster KE, Scott HS, Boyd RL, Godfrey DI, et al. 2005. Localization of Idd11 is not associated with thymus and NKT cell abnormalities in NOD mice. Diabetes 54: 3453–3457 [DOI] [PubMed] [Google Scholar]
- Chasin LA 2007. Searching for splicing motifs. Adv Exp Med Biol 623: 85–106 [DOI] [PubMed] [Google Scholar]
- Chen JM, Cooper DN, Chuzhanova N, Ferec C, Patrinos GP 2007. Gene conversion: Mechanisms, evolution and human disease. Nat Rev Genet 8: 762–775 [DOI] [PubMed] [Google Scholar]
- Concannon P, Chen WM, Julier C, Morahan G, Akolkar B, Erlich HA, Hilner JE, Nerup J, Nierras C, Pociot F et al. 2009a. Genome-wide scan for linkage to type 1 diabetes in 2,496 multiplex families from the Type 1 Diabetes Genetics Consortium. Diabetes 58: 1018–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concannon P, Rich SS, Nepom GT 2009b. Genetics of type 1A diabetes. N Engl J Med 360: 1646–1654 [DOI] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, et al. 2009. Origins and functional impact of copy number variation in the human genome. Nature 464: 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando MM, Stevens CR, Walsh EC, De Jager PL, Goyette P, Plenge RM, Vyse TJ, Rioux JD 2008. Defining the role of the MHC in autoimmunity: A review and pooled analysis. PLoS Genet 4: e1000024 doi: 10.1371/journal.pgen.1000024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. 2008. The Pfam protein families database. Nucleic Acids Res 36: D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. 2009. Rfam: Updates to the RNA families database. Nucleic Acids Res 37: D136–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DB 2009. Common genetic variation and human traits. N Engl J Med 360: 1696–1698 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ 2006. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon H, de Massy B 2002. An initiation site for meiotic crossing-over and gene conversion in the mouse. Nat Genet 32: 296–299 [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakonarson H, Grant SF, Bradfield JP, Marchand L, Kim CE, Glessner JT, Grabs R, Casalunovo T, Taback SP, Frackelton EC, et al. 2007. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature 448: 591–594 [DOI] [PubMed] [Google Scholar]
- Hamilton-Williams EE, Serreze DV, Charlton B, Johnson EA, Marron MP, Mullbacher A, Slattery RM 2001. Transgenic rescue implicates β2-microglobulin as a diabetes susceptibility gene in nonobese diabetic (NOD) mice. Proc Natl Acad Sci 98: 11533–11538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci 106: 9362–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehe MR 2003. Haplotypes and the systematic analysis of genetic variation in genes and genomes. Pharmacogenomics 4: 547–570 [DOI] [PubMed] [Google Scholar]
- Holm S 1979. A simple sequentially rejective multiple test procedure. Scand J Stat 6: 65–70 [Google Scholar]
- Hung MS, Avner P, Rogner UC 2006. Identification of the transcription factor Arntl2 as a candidate gene for the type 1 diabetes locus Idd6. Hum Mol Genet 15: 2732–2742 [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys AJ, May CA 2004. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet 36: 151–156 [DOI] [PubMed] [Google Scholar]
- Karvonen M, Sekikawa A, LaPorte R, Tuomilehto J, Tuomilehto-Wolf E 2001. Type 1 diabetes: Global epidemiology. In The epidemiology of diabetes mellitus (ed. Ekoe JM et al. ), pp. 71–102 John Wiley & Sons, West Sussex [Google Scholar]
- Kauppi L, Jeffreys AJ, Keeney S 2004. Where the crossovers are: Recombination distributions in mammals. Nat Rev Genet 5: 413–424 [DOI] [PubMed] [Google Scholar]
- Kauppi L, Jasin M, Keeney S 2007. Meiotic crossover hotspots contained in haplotype block boundaries of the mouse genome. Proc Natl Acad Sci 104: 13396–13401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E 2003. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res 31: 3576–3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissler S, Stern P, Takahashi K, Hunter K, Peterson LB, Wicker L 2006. In vivo RNA interference demonstrates a role for Nramp1 in modifying susceptibility to type 1 diabetes. Nat Genet 38: 479–483 [DOI] [PubMed] [Google Scholar]
- Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. 2009. The UCSC Genome Browser Database: Update 2009. Nucleic Acids Res 37: D755–D761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloraya M, Davoodi-Semiromi A, Kumar GP, McDuffie M, She JX 2006. Impaired Crkl expression contributes to the defective DNA binding of Stat5b in nonobese diabetic mice. Diabetes 55: 734–741 [DOI] [PubMed] [Google Scholar]
- Leiter EH, Reifsnyder PC, Wallace R, Li R, King B, Churchill GC 2009. NOD × 129.H2g7 backcross delineates 129S1/SvImJ-derived genomic regions modulating type 1 diabetes (T1D) development in mice. Diabetes 58: 1700–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Stankiewicz P 2005. Genomic disorders: Molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet 1: e49 doi: 10.1371/journal.pgen.0010049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons PA, Hancock WW, Denny P, Lord CJ, Hill NJ, Armitage N, Siegmund T, Todd JA, Phillips MS, Hess JF, et al. 2000. The NOD Idd9 genetic interval influences the pathogenicity of insulitis and contains molecular variants of Cd30, Tnfr2, and Cd137. Immunity 13: 107–115 [DOI] [PubMed] [Google Scholar]
- Makino S, Kunimoto K, Muraoka Y, Mizushima Y, Katagiri K, Tochino Y 1980. Breeding of a non-obese, diabetic strain of mice. Jikken Dobutsu 29: 1–13 [DOI] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. 2006. TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34: D108–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAleer MA, Reifsnyder P, Palmer SM, Prochazka M, Love JM, Copeman JB, Powell EE, Rodrigues NR, Prins JB, Serreze DV, et al. 1995. Crosses of NOD mice with the related NON strain. A polygenic model for IDDM. Diabetes 44: 1186–1195 [DOI] [PubMed] [Google Scholar]
- McDuffie M 2000. Derivation of diabetes-resistant congenic lines from the nonobese diabetic mouse. Clin Immunol 96: 119–130 [DOI] [PubMed] [Google Scholar]
- McGuire HM, Vogelzang A, Hill N, Flodstrom-Tullberg M, Sprent J, King C 2009. Loss of parity between IL-2 and IL-21 in the NOD Idd3 locus. Proc Natl Acad Sci 106: 19438–19443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS 2009. Long non-coding RNAs: Insights into functions. Nat Rev Genet 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morahan G, McClive P, Huang D, Little P, Baxter A 1994. Genetic and physiological association of diabetes susceptibility with raised Na+/H+ exchange activity. Proc Natl Acad Sci 91: 5898–5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SK, Lo WS, Pun FW, Zhao C, Yu Z, Chen J, Tong KL, Xu Z, Tsang SY, Yang Q, et al. 2010. A recombination hotspot in a schizophrenia-associated region of GABRB2. PLoS ONE 5: e9547 doi: 10.1371/journal.pone.0009547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Jones K, Phillips JM, Newland SA, Roode M, Mansfield W, Smith A, Cooke A 2009. Validated germline-competent embryonic stem cell lines from nonobese diabetic mice. Nat Med 15: 814–818 [DOI] [PubMed] [Google Scholar]
- Paigen K, Szatkiewicz JP, Sawyer K, Leahy N, Parvanov ED, Ng SH, Graber JH, Broman KW, Petkov PM 2008. The recombinational anatomy of a mouse chromosome. PLoS Genet 4: e1000119 doi: 10.1371/journal.pgen.1000119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes TD 2001. Meiotic recombination hot spots and cold spots. Nat Rev Genet 2: 360–369 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W 2009. Evolution and functions of long noncoding RNAs. Cell 136: 629–641 [DOI] [PubMed] [Google Scholar]
- Prochazka M, Serreze DV, Frankel WN, Leiter EH 1992. NOR/Lt mice: MHC-matched diabetes-resistant control strain for NOD mice. Diabetes 41: 98–106 [DOI] [PubMed] [Google Scholar]
- Razavi R, Chan Y, Afifiyan FN, Liu XJ, Wan X, Yantha J, Tsui H, Tang L, Tsai S, Santamaria P, et al. 2006. TRPV1+ sensory neurons control β cell stress and islet inflammation in autoimmune disease. Cell 127: 1123–1135 [DOI] [PubMed] [Google Scholar]
- Reifsnyder PC, Li R, Silveira PA, Churchill G, Serreze DV, Leiter EH 2005. Conditioning the genome identifies additional diabetes resistance loci in Type I diabetes resistant NOR/Lt mice. Genes Immun 6: 528–538 [DOI] [PubMed] [Google Scholar]
- Ridgway WM, Peterson LB, Todd JA, Rainbow DB, Healy B, Burren OS, Wicker LS 2008. Gene-gene interactions in the NOD mouse model of type 1 diabetes. Adv Immunol 100: 151–175 [DOI] [PubMed] [Google Scholar]
- Rioux JD, Daly MJ, Silverberg MS, Lindblad K, Steinhart H, Cohen Z, Delmonte T, Kocher K, Miller K, Guschwan S, et al. 2001. Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat Genet 29: 223–228 [DOI] [PubMed] [Google Scholar]
- Rodrigues NR, Cornall RJ, Chandler P, Simpson E, Wicker LS, Peterson LB, Todd JA 1994. Mapping of an insulin-dependent diabetes locus, Idd9, in NOD mice to chromosome 4. Mamm Genome 5: 167–170 [DOI] [PubMed] [Google Scholar]
- Rogner UC, Avner P 2003. Congenic mice: Cutting tools for complex immune disorders. Nat Rev Immunol 3: 243–252 [DOI] [PubMed] [Google Scholar]
- Serreze DV, Leiter EH 2001. Genes and cellular requirements for autoimmune diabetes susceptibility in nonobese diabetic mice. Curr Dir Autoimmun 4: 31–67 [DOI] [PubMed] [Google Scholar]
- Shiroishi T, Koide T, Yoshino M, Sagai T, Moriwaki K 1995. Hotspots of homologous recombination in mouse meiosis. Adv Biophys 31: 119–132 [DOI] [PubMed] [Google Scholar]
- Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, et al. 2007. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet 39: 857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DJ, Miretti M, Rajan D, Fiegler H, Carter NP, Blayney ML, Beck S, Hurles ME 2008. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat Genet 40: 90–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandstrat AE, Nguyen C, Limaye N, Chan AY, Subramanian S, Tian XH, Yim YS, Pertsemlidis A, Garner HR Jr, Morel L, et al. 2004. Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus. Immunity 21: 769–780 [DOI] [PubMed] [Google Scholar]
- Wang Z, Burge CB 2008. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium 2010. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 464: 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanouchi J, Rainbow D, Serra P, Howlett S, Hunter K, Garner VE, Gonzalez-Munoz A, Clark J, Veijola R, Cubbon R, et al. 2007. Interleukin-2 gene variation impairs regulatory T cell function and causes autoimmunity. Nat Genet 39: 329–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauk CL, Bois PR, Jeffreys AJ 2003. High-resolution sperm typing of meiotic recombination in the mouse MHC Eβ gene. EMBO J 22: 1389–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]