Abstract
The complexity of the eukaryotic transcriptome is generated by the interplay of transcription initiation, termination, alternative splicing, and other forms of post-transcriptional modification. It was recently shown that RNA transcripts may also undergo cleavage and secondary 5′ capping. Here, we show that post-transcriptional cleavage of RNA contributes to the diversification of the transcriptome by generating a range of small RNAs and long coding and noncoding RNAs. Using genome-wide histone modification and RNA polymerase II occupancy data, we confirm that the vast majority of intraexonic CAGE tags are derived from post-transcriptional processing. By comparing exonic CAGE tags to tissue-matched PARE data, we show that the cleavage and subsequent secondary capping is regulated in a developmental-stage- and tissue-specific manner. Furthermore, we find evidence of prevalent RNA cleavage in numerous transcriptomic data sets, including SAGE, cDNA, small RNA libraries, and deep-sequenced size-fractionated pools of RNA. These cleavage products include mRNA variants that retain the potential to be translated into shortened functional protein isoforms. We conclude that post-transcriptional RNA cleavage is a key mechanism that expands the functional repertoire and scope for regulatory control of the eukaryotic transcriptome.
Recent technological advances in high-throughput sequencing have revealed unexpected complexity of the eukaryotic transcriptome. It is now evident that the majority of the mammalian genome is transcribed as interconnected networks of messenger RNAs (mRNAs) and long and short noncoding RNAs (Cheng et al. 2005; Carninci 2006; Carninci et al. 2006; Kapranov et al. 2007; Cloonan et al. 2008). This transcript diversity arises through several sequential steps comprising various combinations of alternative splicing, transcription initiation, and polyadenylation.
The enormous complexity of the mammalian transcriptome was first exposed by large-scale sequencing of full-length cDNA transcripts from mouse (Okazaki et al. 2002; Maeda et al. 2006) and chromosome tiling array analysis of RNAs expressed in human cell lines (Kapranov et al. 2002; Cheng et al. 2005; Kapranov et al. 2005). Capped analysis of gene expression (CAGE) analysis, a high-throughput technique for sequencing the 5′ end of capped RNAs, has revealed an unexpectedly wide range of alternative transcription initiation events (Carninci et al. 2005). One of the surprising findings from this study was the prevalence of CAGE tags that mapped within exonic coding sequences (Carninci et al. 2006). Initially, these intraexonic CAGE tags were thought to derive from transcripts initiated from internal promoters in these genes. However, a recent study by the CSHL/ENCODE Consortium showed that some exonic CAGE tags could be rescued by their mapping across exon–exon junctions (EEJs) (Fejes-Toth et al. 2009). Because the 5′ ends of these rescued CAGE tags are so proximal to EEJs, the minimal 5′ exons they define are considered too small for efficient splicing (Le Hir et al. 2000, 2001). Therefore, the authors concluded that CAGE tags across EEJs must have occurred as a consequence of post-splicing RNA cleavage and subsequent “secondary capping” of the cleaved transcript. This challenges the assumption that cleaved RNA transcripts are simply transitional intermediates in the degradation and recycling of mRNA, but could rather represent functional products generated from longer parental transcripts.
Here, we show that post-transcriptional cleavage events are widespread, conserved among eukaryotes, and generate a range of small RNAs and long coding and noncoding RNA (ncRNA) transcripts. By comparing exonic CAGE tags to tissue-matched parallel analysis of RNA ends (PARE) data, we show that the secondary capping of cleaved transcripts is a regulated process that is conserved between species and regulated in a developmental-stage- and tissue-specific manner. By deep-sequencing size-fractionated pools of human embryonic stem cell RNA, we show that the cleavage pathway has significant impact in remodeling the transcriptome. We conclude that post-transcriptional RNA cleavage is a common mechanism that, alongside transcription initiation, termination, alternative splicing, and editing, plays a significant part in the diversification of both the coding and noncoding transcriptional repertoire of the genome.
Results
Evidence of widespread intraexonic post-transcriptional cleavage
To identify likely sites of post-transcriptional RNA cleavage, we mapped CAGE tags against the mouse genome requiring exact matches and then mapped the remaining unmapped CAGE tags to all annotated EEJs as conducted previously (Fejes-Toth et al. 2009), finding 17,731 (0.2%) mouse and 6060 (0.4%) human tags mapped uniquely and exactly across coding EEJs (Supplemental Fig. S1; Supplemental Table S1; Carninci et al. 2006; Valen et al. 2009). Because the tags map overwhelmingly (99.9%) in the sense direction and only a tiny proportion (0.001%) can be mapped to shuffled EEJs, it is unlikely that EEJ-spanning CAGE tags are the result of a mapping artifact. As CAGE tags also frequently map within exons (Carninci et al. 2006), we considered the possibility that these tags may also arise as a consequence of cleavage. We identified 178,058 (∼13% of all mappable) CAGE tags in the FANTOM3 mouse CAGE library that mapped uniquely and exactly within the RefSeq-annotated coding sequence of 12,858 RefSeq mouse genes (∼61% of total) (Supplemental Table S2). Therefore, although cleavage is a relatively widespread event, specific cleavage events are relatively rare, particularly when considering the wide range of tissues from which CAGE libraries have been derived. We find that the ratio of CAGE tags mapping to annotated promoters compared to within coding exons is ∼5:1. A comparison of the incidence of CAGE tags mapping within exons with those crossing EEJs revealed that the distribution of mapped tags remained relatively constant throughout RefSeq genes (Supplemental Fig. S1). Although this observation supports the notion that intraexonic CAGE tags are also derived from post-transcriptional cleavage, it does not exclude the possibility that alternate promoters may drive the initiation of other transcripts in these regions.
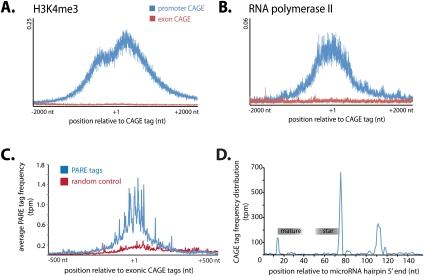
Therefore, we investigated whether epigenetic signatures of active gene promoters, including H3K4me1, H3K4me2, H3K4me3, H2AZ, H3K9ac, H3K18ac, and H2BK12ac, are associated with intraexonic CAGE tags (Fig. 1A; Supplemental Fig. S2; Barski et al. 2007; Wang et al. 2008). In contrast to the epigenetic signatures of active gene promoters, we did not observe enrichment for any of the examined chromatin modifications associated with intraexonic CAGE tags. We also did not observe a peak of RNA polymerase II (RNAPII) occupancy at exonic CAGE tag sites in contrast to that observed at promoters (Fig. 1A). Furthermore, we did not observe any significant association of TATA-box or CpG islands with intraexonic CAGE tags relative to gene promoters (Supplemental Fig. S3; Carninci et al. 2006). Together these results provide strong evidence that intraexonic CAGE tags are not indicative of conventional transcription initiation, and that the majority are likely generated as a consequence of post-transcriptional cleavage.
Figure 1.
Frequency distribution for modified chromatin immunoprecipitated tags relative to CAGE sites associated with gene promoters and exons. (A,B) The average frequency of DNA tags immunoprecipitated with antibodies to H3K4me3 (A) and RNA polymerase II (B) within a 4-kb window centered on CAGE tags mapping to promoters (blue) and coding exons (red) (Wang et al. 2008). (C) Frequency distribution of PARE tags (blue) within a 1000-nt window centered on exonic CAGE tags (indicated at +1). A random control showing PARE tags frequency distribution across random sites in matched exons shown (red). (D) Frequency of CAGE tags mapping across microRNA hairpin with mature and star sequences indicated.
Cleavage and secondary capping are distinct processes
Next, we considered the relationship between the cleavage and subsequent capping of an RNA transcript. PARE analysis permits the genome-wide sequencing of 5′-monophosphate-cleaved ends of polyadenylated RNA to provide an indication of uncapped transcripts (German et al. 2009). A comparison of PARE libraries to tissue-matched CAGE libraries (mouse brain, lung, and liver) allows us to compare capped and uncapped cleaved transcripts.
To determine the relative specificity of CAGE- and PARE-tag methods, we compared their relative mapping densities at nuclear genes, which contain a 5′-G cap, and mitochondrial genes, that contain a 5′-monophosphate end (Scarpulla 2008). We found that in matched samples, CAGE tags mapped at an incidence of <3% of that of PARE tags to mitochondrial genes, while PARE tags map at an incidence of <0.3% to the nuclear gene transcription start site of genes, thereby indicating a high specificity to these two techniques (Supplemental Fig. S4). It also indicates that, unlike plants (Jiao et al. 2008), mammals are unlikely to contain prevalent uncapped mRNA transcripts.
In total, we identified 85,667 exonic cleavage sites by mapping PARE tags (see Methods). Like exonic CAGE tags, PARE tags map across EEJs, indicating that they are derived from post-transcriptional cleavage. Comparison of the frequency distribution of PARE tags at exonic CAGE tag sites indicates a distinct enrichment for PARE tags at exonic CAGE tag sites in all tissues (Fig. 1B). Furthermore, genes exhibit a correlated frequency for both CAGE and PARE tags, with some examples, such as Alb, being subject to similarly abundant cleavage according to both PARE and CAGE libraries (Supplemental Fig. S4). Although this closely links the cleavage and secondary capping processes, the low overlap between these data sets (5% of PARE tags overlap CAGE tags) suggests that these processes are mechanistically distinct.
Positional conservation of post-transcriptional cleavage sites
Given the prevalence of post-transcriptional cleavage in mouse and human, we next considered whether cleavage occurs at homologous positions in these species. We found 2713 sites to which the 5′ nucleotide of the intraexonic CAGE tags maps syntenically in both human and mouse. These homologous sites are also significantly enriched (3.8-fold, P < 0.01) in the frequency of exonic CAGE tags relative to nonconserved sites, and account for ∼10.6% and 9.6% of total mouse and human exonic CAGE tags, respectively (Supplemental Table S3). Furthermore, the human homologs of ∼50% of mouse genes similarly encompass exonic CAGE tags with a correlated exonic CAGE tag frequency of 0.32 (Supplemental Fig. S5; Supplemental Table S2).
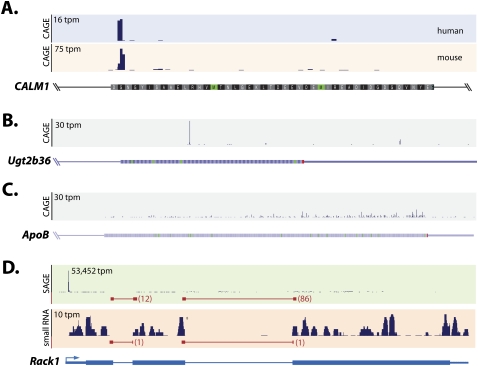
The analysis of conserved CAGE mapping sites revealed some striking examples, such as CALM1 (Fig. 2A), where a specific nucleotide position is subject to frequent post-transcriptional cleavage in both human and mouse. Therefore, we examined the distribution of CAGE tags across genes and found a considerable diversity in the organization of intraexonic CAGE tags, ranging from single specific events, as in the Ugt2b36 gene (Fig. 2B), to broad exonic CAGE clusters, as in the Apob gene (Fig. 2C). Unlike the specificity of single-peak cleavage events, the broad dispersed distribution of CAGE clusters through genes such as Apob seems unlikely to produce specific truncated long isoforms, but they may still produce a diversity of small RNAs. A comparison of the organization of CAGE tags within exons against CAGE tags at RefSeq-annotated promoters shows that intraexonic CAGE tags occur more frequently in single peaks than those at gene promoters (70% vs. 25%). The prevalence of single-peak CAGE tags for particular nucleotides raises the possibility for a sequence-specific contribution to post-transcriptional cleavage. However, motif analysis identified only a slight preference for guanine at the 5′-terminal nucleotides of exonic CAGE tags (Supplemental Fig. S3).
Figure 2.
Illustrative examples of post-transcriptional cleavage in human and mouse. (A) Exonic CAGE tags map to syntenic nucleotides in the human (blue panel) and mouse (beige panel) CALM1 gene. (B) Single peak distribution of exonic CAGE tags in the mouse Ugt2b36 gene. (C) Broad peak distribution of exonic CAGE tags in the mouse Apob gene. (D) Exonic SAGE tags (green panel) and small RNA tags (beige panel) map to the exon (black histogram) and across exon–exon junctions (red; tag count indicated) in the fly Rack1 gene (blue). (tpm) Tags per million.
Post-transcriptional cleavage occurs in lower eukaryotes
Given the prevalence of post-transcriptional cleavage in human and mouse, we next sought to identify whether this process occurs in other eukaryotic lineages. In Drosophila melanogaster, we analyzed deep-sequencing serial analysis of gene expression (SAGE) data sets that are derived from a modified protocol that, like CAGE, recognizes and sequences the 5′-capped end of full-length RNA transcripts (Hashimoto et al. 2004). We considered three libraries generated from different developmental stages (Ahsan et al. 2009), finding 27,515 (0.2%) SAGE tags that mapped across EEJs. Analysis of SAGE tags combined from all developmental stages revealed ∼14% mapped within FlyBase-annotated coding sequences, a similar proportion to that in human and mouse (Supplemental Table S4).
Post-transcriptional cleavage generates prevalent small RNAs
The CSHL/ENCODE Consortium found that 5′-modified small RNAs were enriched at post-transcriptional cleavage sites, and indicated that these small RNAs were likely to have been generated as a result of the cleavage process (Fejes-Toth et al. 2009). Using an independent small RNA deep-sequencing data set (Taft et al. 2009), we confirmed the previously described enrichment for small RNAs (15–30 nt) at exonic mapped CAGE sites in human THP1 cells (Supplemental Fig. S6). Based on the rationale that small RNAs are too small to be spliced by conventional machinery, we mapped small RNA tags across EEJs to determine whether small RNAs could be directly generated by cleavage. We found thousands of small RNAs that mapped exactly and uniquely across EEJs in 18 publicly available small RNA libraries derived from human, mouse, chicken, and fly (Supplemental Table S5; Ruby et al. 2006; Babiarz et al. 2008; Baek et al. 2008; Czech et al. 2008; Glazov et al. 2008). We also found that small RNAs derived from both nuclear and cytoplasmic preparations include spliced small RNAs (Supplemental Fig. S6; Taft et al. 2009). These EEJ-spanning small RNAs were orientated overwhelmingly in the sense direction, further supporting their derivation from a longer RNA precursor. The production of small RNAs by post-transcriptional cleavage is exemplified by the Drosophila Rack1 gene, which encompasses numerous small RNA tags and corresponding SAGE tags (Fig. 2D).
Consideration of CAGE tags at miRNA loci indicates an enrichment for CAGE tags corresponding to the sites of Drosha cleavage in numerous miRNA hairpins immediately downstream from the miRNA star sequence, suggesting that collateral transcripts produced during the processing of pri-miRNAs may be stable and subsequently capped (Fig. 1D). In some cases, such as mmu-mir-124-1, this Drosha-dependent cleavage may contribute to the post-transcriptional cleavage of longer host RNA transcripts (Supplemental Fig. S7). Therefore, we considered whether components of the small RNA processing enzymes are involved in the cleavage and generation of small exon-derived RNAs. However, we were unable to identify any common characteristics, such as sequence motifs, length, and prominent 5′-terminal nucleotides, that would define EEJ-spanning small RNAs. Furthermore, we did not observe ablation or reduction of EEJ-spanning small RNAs in libraries generated from Dicer1−/− or Dgcr8−/− mouse embryonic stem cells (Babiarz et al. 2008) or Dcr-1−/− fly ovaries (Supplemental Fig. S8; Czech et al. 2008). Similarly, EEJ-spanning small RNAs were not ablated in loqs−/− fly ovaries, as previously observed for endogenous siRNAs (Czech et al. 2008). Finally, EEJ-spanning RNAs were poorly represented in fly and human RNA sequencing libraries derived from RNA immunoprecipitation using antibodies targeting AGO1 and AGO2 (Supplemental Table S5; Czech et al. 2008; Ender et al. 2008). Together, these observations suggest that small exon-derived RNAs are distinct from known classes of small RNAs, such as endogenous siRNAs or miRNAs.
Based on the assumption that EEJ-spanning small RNAs can serve as a proxy for post-transcriptional cleavage, we examined small RNA sequencing libraries from Caenorhabditis elegans, where CAGE or SAGE data are unavailable, to ascertain the extent of this phenomenon in other metazoa. We found abundant EEJ-spanning small RNAs (Supplemental Table S5) that, given their predominantly sense orientation and lack of 5′-terminal guanine, are unlikely to correspond to endogenous siRNAs (Ruby et al. 2006), and thereby suggest that post-transcriptional cleavage is a relatively ancient and evolutionarily conserved process.
Post-transcriptional cleavage produces long RNA transcripts
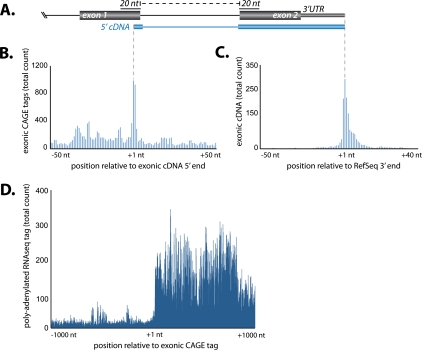
Because CAGE libraries are constructed from polyadenylated RNA >200 nt (Carninci et al. 2006), a major product of post-transcriptional cleavage is likely to be long RNA transcripts. Therefore, we examined cDNA libraries for the presence of long RNA transcripts generated by post-transcriptional cleavage by identifying cDNA transcripts whose 5′ termini map no more than 20 nt upstream of an EEJ using a strategy similar to that used for CAGE tags (see above; Fig. 3A). To minimize the incidence of false positives due to incomplete reverse transcription, we restricted our analysis to FANTOM3 cDNA libraries, which had been curated for full-length transcripts by a cap-trapping step prior to cDNA sequencing (Carninci et al. 2003, 2005). We found 887 transcripts that initiated within 20 nt 5′ of an EEJ, indicative of genesis by post-transcriptional cleavage, as well as a further 3303 cDNAs that initiate within RefSeq-annotated coding sequences (Supplemental Table S6). We find that the ratio of 5′ termini of cDNAs mapping to annotated ends compared to within coding exons is ∼6.7:1. Furthermore, 27% of 5′ termini mapping to exons are supported by both CAGE and cDNA evidence. At the 3′ end, we found 35% of cDNAs originating within exons share the same polyadenylation site as the host RefSeq gene (Fig. 3D). Given that exonic CAGE tags are derived from polyadenylated RNA >200 nt (Carninci et al. 2006) and numerous full-length cDNA transcripts are derived from post-transcriptional cleavage, we suggest that, in addition to small RNAs, long RNAs are a major product of the post-transcriptional cleavage process.
Figure 3.
Evidence of post-transcriptional cleavage from full-length cDNA transcripts. (A) Schematic diagram illustrating the mapping of 5′-cDNA termini that span exon–exon junctions (EEJs) and 3′-cDNA termini to the annotated 3′ end of the host RefSeq gene. (B) Cumulative frequency distribution of mouse exonic CAGE tags within a 100-nt window cantered on the 5′ termini of exonic cDNAs. (C) Cumulative frequency distribution of 3′-cDNA termini distribution centered on a 100-nt window centered on the annotated 3′ end of the host RefSeq gene. (D) Cumulative frequency distribution of polyadenylated events within a 2-kb window centered on exonic CAGE tags. RNA sequencing library derived from mouse brain (Parkhomchuk et al. 2009).
The process of post-transcriptional cleavage infers the existence of an upstream collateral transcript. However, the stability and polyadenylation status of the putative upstream transcript are uncertain. To identify any stable polyadenylated upstream transcripts, we compared the mapping positions of mouse deep-sequencing tags after the removal of 3′ nongenomic poly(A) additions with putative cleavage sites. The analysis did not reveal enrichment for polyadenylation sites coincident to or upstream of exonic mouse brain CAGE tags or polyadenylated tags that crossed EEJs (Fig. 3D), suggesting that any upstream collateral transcript is nonpolyadenylated. Furthermore, we used real-time PCR with poly(T) primers at four noted cleavage sites within the Calm1, Bptf, Pnpla2, and Polr2a genes. However, we were unable to identify the expression of any stable upstream polyadenylated cleaved transcripts (Supplemental Fig. S9).
Sequencing of size-fractionated RNA resolves cleaved transcripts from spliced isoforms
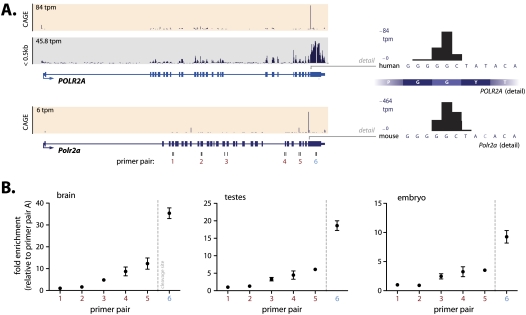
Prevalent post-transcriptional cleavage anticipates the biogenesis of smaller transcript isoforms of differing sizes. Although such isoforms should be detectable by Northern analysis, it is not possible to distinguish isoforms produced by cleavage from those produced by alternative splicing. To overcome this problem, we used deep sequencing of size-fractionated polyadenylated RNA from human embryonic stem cells (see Methods). This approach simultaneously resolves the size and splicing status of different isoforms, thereby discriminating between alternatively spliced and cleaved transcripts. To identify genes that had potentially undergone post-transcriptional cleavage, we used the smallest RefSeq-annotated isoform to predict the smallest size fraction where the resulting mRNA should appear; omitting any genes where deep-sequencing tags indicated alternative unannotated splice junctions. Using this approach, we found the mean relative expression of genes in smaller size fractions was 27% and identified a number of candidate cleaved mRNAs (Supplemental Fig. S10; Supplemental Table S7). Although we could not discount a contribution of degradation to this enrichment, we noted that the number of exonic CAGE tags correlated with this enrichment (R2 = 0.37, P < 0.01). Indeed, the cumulative sum of RNA-seq tags is enriched 50 nt downstream relative to 50 nt upstream of such exonic CAGE tags in lower size fractions (P < 0.01) (Supplemental Fig. S10). This is illustrated by examples such as POLR2A that exhibit enriched RNA-seq coverage downstream from a cleavage site that is conserved between human and mouse, suggesting prevalent cleavage of this transcript (Fig. 4A). To confirm the independent expression of a truncated isoform from the POLR2A loci, we performed RT-PCR across the cleavage site. RT-PCR was performed with cDNA libraries generated with random hexamer priming (Fig. 4B). We show that the sequence downstream from the identified cleavage site displayed a significantly higher expression than the upstream sequence in mouse brain, embryo, and testes tissue, consistent with the generation of a downstream truncated transcript.
Figure 4.
Detailed analysis of Polr2a cleavage. (A) Genome Browser view showing human (top panel) and mouse (bottom panel) Polr2a gene. Frequency distribution of CAGE tags (beige panels) indicates prevalent cleavage in the terminal Polr2a exon in both human and mouse (note that promoter CAGE tags are omitted). Nucleotide-level resolution of the Polr2a cleavage site (right panels) indicates conservation of the cleavage event between human and mouse. Size-fractionated RNA sequencing tags from human embryonic stem cells (gray panel) shows enriched expression immediately downstream from the cleavage event. (B) RT-PCR using primer pairs in mouse brain, testes, and embryo shows enriched expression downstream from the cleavage site. Positions of the RT-PCR primers (1–6) are indicated in A.
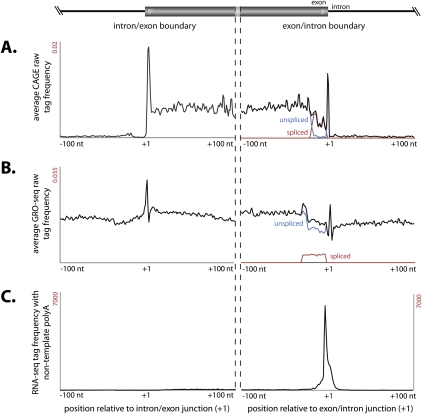
CAGE tags are enriched at the 5′ end of exons
Although we did not observe any significant difference between the cleavage of spliced compared to nonspliced genes (Supplemental Fig. S11), when we considered the distribution of CAGE tags across individual exons, we observed an enrichment for exonic CAGE tags immediately downstream from the 5′ intron–exon junction (Fig. 5A). The close association of this subset of CAGE tags with the 5′ splice junction prompted us to analyze the 3′ junction, where we also observed a similar enrichment for intronic CAGE tags immediately downstream from the 3′ exon junction (Fig. 5B). Because the intronic location of these CAGE tags suggests that they are not a consequence of the mature mRNA cleavage process, we considered both 5′ and 3′ intron–exon-junction-associated CAGE subsets with recently published deep sequencing of nuclear run-on transcription (Core et al. 2008) and observed an enrichment for nascent transcripts generated by elongating RNAPII whose 5′ termini aligned with both CAGE subsets (Fig. 5B). Furthermore, unlike transcription initiation at gene promoters, there is no evidence of divergent antisense transcription at these CAGE sites (Supplemental Fig. S11). In addition, we identified a distinct enrichment for polyadenylation events at exonic 3′ borders (Supplemental Fig. 5C; Parkhomchuk et al. 2009). Together, we propose a model whereby CAGE sites arise through a cotranscriptional but transcription initiation-independent mechanism to account for these patterns (see Discussion).
Figure 5.
Distinct class of exonic CAGE tags associated with splice junctions. (A) Average CAGE tags frequency distribution within a 200-nt window centered on splice junctions (indicated at +1) shows a distinct enriched peak immediately downstream from the intron–exon junction (first column) and exon–intron junction (second column). (B) Distribution of GRO-seq tags derived from nuclear run-on (Core et al. 2008) across splice junctions peak immediately downstream from the intron–exon junction and exon–intron junction. (C) Cumulative frequency distribution of polyadenylated RNA deep-sequencing tags from mouse brain (Parkhomchuk et al. 2009) across exon–intron splice junctions.
Post-transcriptional cleavage is tissue- and developmental-stage-specific
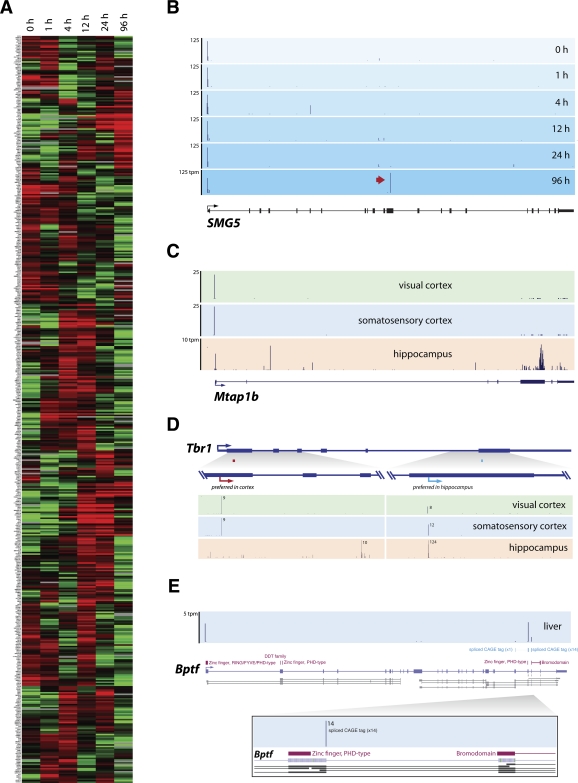
To determine whether post-transcriptional cleavage occurs in a tissue- and developmental-stage-specific manner that would be indicative of a regulated process, we analyzed CAGE libraries derived from a differentiating human myelomonocytic leukemia cell line (THP1) (Suzuki et al. 2009) and a range of mouse tissues that had sufficient depth for comparing relative CAGE frequency (Valen et al. 2009). First, we compared the intraexonic CAGE frequency of human genes across six time points in human THP1 cells following phorbal myristate acetate (PMA) stimulation. To permit robust statistical analysis, we then selected the top 500 genes ranked by CAGE frequency, which corresponded to a minimum frequency of 250 CAGE tags per million (tpm) across the time course, for further study (Fig. 6A). Within this set, we found the frequency of CAGE tags in 164 (32%) genes varied by at least fourfold between at least two points. Consistent with a relevant biological role in the experimental system examined, a number of genes with well-established functions in macrophage biology, such as CSF1R, MYB, and CD109, were identified in this subset (Suzuki et al. 2009).
Figure 6.
Tissue-specific cleavage events. (A) Cluster analysis of the expression of the 500 genes containing the highest exonic CAGE tag frequency shows the up-regulation (red) and down-regulation (green) of mRNA cleavage in human THP-1 cell differentiation. Expression level was determined as the normalized CAGE tag frequency mapping to coding exons. (B) Genome Browser view of human SMG5 gene with CAGE tag mapping (blue panels) across different THP1 time points shows specific cleavage event (red arrow) at 96 h post-PMA-stimulation. (C) Mouse Mtap1b gene showing CAGE tags mapping in the visual (green panel), somatosensory (blue panel) cortex, and hippocampus (beige panel). (D) Mouse Tbr1 gene shows alternative exonic CAGE tag locations preferred between the visual and somatosensory cortex (red arrow) and the hippocampus (blue arrow). (E) CAGE tags mapping (blue panel) to Bptf (violet bars) bisect distinct encoded protein domains (purple bars), potentially generating alternative protein-coding isoforms. (Tpm) Tags per million.
To determine whether the observed changes in intraexonic CAGE tag frequency exhibit independent profiles to the rate of host gene transcription initiation, we compared the ratio of CAGE tags mapping to exons with those at the promoter of the host gene. Although a weak correlation between promoter strength and intraexonic CAGE frequency was observed (R2 = 0.13) (Supplemental Fig. S12), we found that, rather than exonic CAGE frequency being proportional to transcription initiation as would be expected by systematic RNA degradation, there was considerable variance (Supplemental Fig. S12). For example, intraexonic CAGE tags within the SMG5 gene undergo a dramatic induction at 96 h, despite a relatively constant rate of transcription initiation (Fig. 6B). In addition, there was almost no overlap of RNAPII occupancy with intraexonic CAGE tags (0.7%), in contrast to the overlap of RNAPII binding sites at CAGE tags at gene promoters (13%).
We next extended this analysis to examine the tissue-specific changes in the frequency of intraexonic CAGE tags. The results similarly showed dynamic changes in intraexonic CAGE frequency across eight mouse tissues in 92% of the genes analyzed (Supplemental Fig. S13). This is exemplified by the Mtap1b gene, which, despite showing expression in all tissues sampled, only contains intraexonic CAGE tags in the hippocampus (Fig. 6C), and the Tbr1 gene, where alternative intraexonic CAGE clusters are preferred in different regions of the brain (Fig. 6D). In contrast, we also observe considerable conservation of cleavage sites across tissues, such as within the Foxb2 gene, where the same site was preferred in all eight tissues (Supplemental Fig. S14). Together, these results suggest that at least a subset of intraexonic CAGE tags is produced via a regulated process (see Discussion).
Post-transcriptional cleavage generates a diversity of long coding isoforms
The tissue- and developmental-stage-specific RNA cleavage, combined with conservation of cleavage sites between human and mouse, suggest that post-transcriptional cleavage represents a regulated mechanism to diversify the transcriptome. The ability of this process to generate functional small RNAs has been previously discussed (see above; Fejes-Toth et al. 2009). For example, Alb1 and the well-characterized ncRNA XIST are subject to prevalent cleavage that generates large numbers of small RNAs (Supplemental Fig. S15). Furthermore, we also note that a number of well-characterized ncRNAs, such as the two neighboring ncRNAs NEAT1 and NEAT2, the latter of which is prevalent in all sequenced hES size fractions, are a source of numerous exonic CAGE tags and small RNAs in human embryonic stem cells (Supplemental Fig. S10). However, the specific and regulated cleavage of mRNAs described within this study (see above) appears to produce distinct long 5′-capped RNA transcripts that retain, albeit truncated, coding potential, in a manner similar to the recently reported case of the DUX4 gene (Snider et al. 2009). Indeed, the secondary 5′ capping of such cleaved transcripts may permit their correct localization and translation (Gu and Lima 2005), raising the possibility that a major role of RNA cleavage is to generate alternative mRNA isoforms.
To investigate the propensity of post-transcriptional cleavage to generate alternative mRNA isoforms, we examined the coding potential of cleaved transcripts. Using the 882 FANTOM3 cDNA transcripts whose 5′ end mapped across an EEJ, we found that ∼87% retain an open reading frame greater than 100 amino acids, initiating from an alternative downstream initiation codon (Supplemental Table S8). In some cases, this would produce an isoform with a different domain structure, such as in the case of Bptf, which is specifically cleaved between the regions encoding the bromodomain and the PHD finger domain in both human and mouse (Fig. 6E). The ability of similarly cleaved transcripts to be translated as functionally distinct proteins has been previously reported (Thoma et al. 2001; Hasselblatt et al. 2005; Snider et al. 2009). Although an N-terminal proteomics study, analogous to the 5′-terminal transcriptomic analysis conducted here, is required to determine the full contribution of this process to protein diversity, examination of publicly available N-terminal peptide sequences reveals an enrichment of peptides with N-terminal methionine downstream from cleavage sites relative to random positions in the coding sequence (Supplemental Fig. S16).
Discussion
Recent years have shown that the eukaryotic transcriptome comprises a remarkably complex network of long and short, coding and noncoding RNAs. In this study, we show that the process of post-transcriptional RNA cleavage, alongside differential transcription initiation, termination, alternative splicing, and other forms of post-transcriptional modification, expands the repertoire of coding and noncoding RNAs in the eukaryotic transcriptome. We show that post-transcriptional RNA cleavage is prevalent throughout eukaryotes and occurs in a developmental-stage- and tissue-specific manner in human and mouse. Together, these signatures indicate that, rather than simply being a transitory intermediate of RNA degradation, cleavage diversifies the transcriptome in a regulated manner.
A major product of post-transcriptional cleavage is long RNA transcripts. Although such long RNAs may ultimately be cleaved into smaller RNAs, we propose that truncated mRNA isoforms may also potentially be translated to C- or N-terminal truncated proteins. There are numerous precedents to support this proposal (Thoma et al. 2001; Hasselblatt et al. 2005; Snider et al. 2009). For example, a recent study found that a range of endogenous small and long RNA transcripts are generated from the DUX4 gene, including a 3′-cleaved isoform that undergoes translation from an internal methionine to generate a functionally distinct truncated protein that lacks the N-terminal domain (Snider et al. 2009). Moreover, the authors note that full-length DUX4 mRNA or protein has been rarely detected in vivo. Such examples provide proof-of-principle that post-transcriptional cleavage of mRNA transcripts has the ability to modulate the functional repertoire of the proteome in a manner analogous to mechanisms of proteolytic cleavage that contribute to proteomic diversity (Walsh et al. 2005).
In addition to protein-coding transcripts, we also show that post-transcriptional cleavage can yield an array of noncoding transcripts. Indeed, we note that a number of transcripts, such as Alb and Apob, are cleaved with such prevalence as to be unlikely to produce specific truncated isoforms. However, such transcripts may alternatively generate a wide range of small ncRNAs. As well as the generation of small RNAs generated by cleavage, there are several precedents for ncRNAs being processed from an mRNA precursor, such as mirtrons from splice junctions (Ruby et al. 2007) and endogenous siRNAs from mRNA duplexes (Okamura and Lai 2008). The ability to generate a range of noncoding transcripts from a coding precursor provides an intersecting avenue between the protein-coding and noncoding transcriptomes, thereby contributing to an increased blurring of the dichotomy between coding and regulatory transcripts (Kawashima et al. 2003; Chooniedass-Kothari et al. 2004; Dinger et al. 2008).
An important focus of future research is the identification of the factors responsible for mediating the specificity and process of post-transcriptional cleavage. Suites of RNases are responsible for the cleavage and processing of many nascent RNAs, including miRNAs, snoRNAs, rRNAs, and tRNAs, to their mature forms (Saida and Odaert 2007). Also, small antisense RNAs may direct cleavage by annealing with complementary RNA and forming a double-stranded substrate for RNase H cleavage (Cerritelli and Crouch 2009). Although the cleavage of these transcripts results in rapid degradation of the upstream capped, non-polyadenylated fragment, the 3′-uncapped polyadenylated fragment can exhibit remarkable stability, being efficiently translated even when the site of cleavage occurs within a few nucleotides upstream of the initiation codon (Thoma et al. 2001; Hasselblatt et al. 2005). Furthermore, the resultant N-terminal-truncated proteins have distinct functional differences from the full-length form. While this manuscript was under review, a study of transcriptome-wide 5′-capped mRNA cleavage products revealed several classes of endonucleolytic cleavage that were dependent on various nucleases, including Ago2 (Karginov et al. 2010). A subset of these mRNA cleavage events was shown to be Drosha-dependent and guided by miRNAs. This mechanism of cleavage is likely to underlie the post-transcriptional cleavage phenomenon described here.
The tissue-specific incidence of intraexonic CAGE tags is suggestive of a tightly regulated process. There are three main levels at which this regulation may occur: (1) the cleavage process, (2) secondary capping, or (3) degradation. Our PARE data analysis suggests that although cleavage and secondary capping are linked, they are nevertheless distinct processes with only a subset of cleaved processes undergoing secondary capping. Therefore, the selection of cleaved transcripts for secondary capping may be a contributing factor to the observed differential frequencies of intraexonic CAGE tags. Because differential secondary capping frequencies alone cannot explain the tissue-specific incidence of CAGE tags in many cases, we conclude that additional regulation is also occurring at either or both the degradation of rates of cleaved transcripts and the cleavage process itself.
The stability and ability of a transcript to be translated has been previously considered to be dependent on the presence of a 5′ cap and 3′ poly(A) tail. However, these studies demonstrate uncapped transcripts can be efficiently translated (Steiger and Decker 2001). Therefore, it is likely that the prevalent post-transcriptional cleavage documented in this study represents only a subset of the complement of cleaved RNA isoforms. Indeed, a recent transcriptomic analysis of uncapped mRNAs in Arabidopsis revealed that almost all mRNAs were simultaneously present as uncapped variants (Jiao et al. 2008). Nevertheless, the transcripts considered within this study and that of the CSHL/ENCODE Consortium show a transcript can be subject to “secondary” capping after cleavage (Fejes-Toth et al. 2009), a conclusion supported by a range of other studies (Schoenberg and Maquat 2009). The “secondary” cleavage may be catalyzed by a population of cytoplasmically located enzymes recently ascribed the ability to convert and cap the 5′ termini of RNA transcripts (Otsuka et al. 2009). Conversely, enzymes capable of adding and extending the polyadenylation tail at the 3′ termini of transcripts have also been well described (Mangus et al. 2003). Therefore, it seems there are multiple enzymes that can generate a range of capped and uncapped, polyadenylated and non-polyadenylated transcripts.
During this analysis, we observed a distinct enrichment for CAGE tags at splice junctions, suggesting cleavage and capping may occur at these sites. The cleavage of nascent transcription at these sites is also supported by RNAPII run-on analysis, where the profile of these nascent transcripts is similar to that observed at the 3′ termini of 3′ untranslated regions where cotranscriptional cleavage occurs. We suggest a model in which CAGE tags at splice junctions are generated by the co-transcriptional cleavage and capping of nascent transcripts, and this is similar to the recruitment of cleavage and capping enzymes at the 5′ and 3′ ends of genes by phosphorylated RNAPII (Moore and Proudfoot 2009). Indeed, we observe enrichment for phosphorylated RNAPII at these sites (Supplemental Fig. S11), and the preferential positioning of nucleosomes at exon borders, reminiscent of 5′ and 3′ gene termini, has been recently reported (Andersson et al. 2009; Nahkuri et al. 2009; Schwartz et al. 2009; Spies et al. 2009; Tilgner et al. 2009).
The data presented here show that post-transcriptional cleavage is a significant step in RNA processing, contributing to the functional range of the eukaryotic transcriptome. Given that regulation of RNA cleavage is likely to be more economical than the assembly of alternative transcription initiation complexes, the contribution of post-transcriptional cleavage to the diversity of the transcriptome may even exceed that of alternative promoters. However, it should be noted that despite its contribution to diversity, the contribution of cleavage to transcriptomic output as a whole is relatively small (estimated at ∼10% from CAGE tags). Nevertheless, despite being relatively rare, this process does seem widespread. Furthermore, changing cellular conditions may modulate global regulation of post-transcriptional processing in a manner similar to viral assault or heat-shock-inducing global changes to the degradation of RNA processing.
These models are reminiscent of a polycistronic system that is exploited in prokaryotes and integral to the gene expression of some eukaryotes such as trypanosomes (Kozak 1999; Clayton and Shapira 2007). Indeed, it seems likely that post-transcriptional cleavage is an ancient mechanism that occurs (in at least some form) throughout both prokaryotic and eukaryotic lineages. In support of this, it was recently reported that the 5′ termini of ∼7% of full-length cDNAs initiate from within the coding sequence of Saccharomyces cerevisiae genes (Miura et al. 2006), raising the possibility that post-transcriptional cleavage is an ancient mechanism of RNA processing that is conserved throughout the eukaryotic lineage. Although we were unable to identify any compelling instances of full-length cDNA within this subset that mapped proximally across EEJs, this was perhaps not surprising given the relative scarcity of splice junctions in yeast.
Given the potential significance of post-transcriptional cleavage in defining the cellular transcriptomic profile, it is likely that deregulation of this mechanism can contribute toward disease etiology. Indeed, the DUX4 truncated transcripts discussed above are considered candidates for involvement in facioscapulohumeral muscular dystrophy (Snider et al. 2009). In this study, we noted many disease candidate genes that are subject to cleavage, such as the Alzheimer's disease candidate gene BACE1 (Cole and Vassar 2008), which is subject to a conserved and high degree of cleavage in the mouse hippocampus and human. The dysregulation of post-transcriptional cleavage may result in the expression of aberrant protein isoforms, and even silent nucleotide polymorphisms may abolish cleavage sites that affect expression of the subsequent protein isoform. Therefore, a consideration of post-transcriptional cleavage may provide novel insights in the future analysis of diseases arising from the transcriptome.
Methods
CAGE and SAGE analysis
Human and mouse CAGE tags retrieved from RIKEN (http://fantom3.gsc.riken.jp/) and Drosophila SAGE tags retrieved from MachiBase (Ahsan et al. 2009) were mapped to the human (hg18), mouse (mm8), or Drosophila (dm3) genomes as appropriate with ZOOM requiring exact and unique matches (Lin et al. 2008). To identify EEJs spanning CAGE or SAGE tags, those tags that did not map to the genome were mapped to 60-nt regions centered on the splice sites defined by RefSeq gene annotations (Pruitt et al. 2007). CAGE or SAGE tags were defined as originating from a protein-coding sequence if they intersected entirely within a RefSeq-annotated coding region. The frequency distribution of CAGE tags mapping within coding exons and across EEJs was determined by summing the number of CAGE tags mapping within a 100-nt window centered on all RefSeq-annotated EEJs within coding sequences.
To identify conserved cleavage sites, syntenic locations of mouse CAGE tags in the human genome were identified using the LiftOver utility (Kuhn et al. 2009). Mouse CAGE tags that mapped to the same site as human CAGE tags were defined as conserved.
Immunoprecipitation deep-sequencing data analysis
Deep-sequencing tags derived from RNA polymerase II, H3K4me1, H3K4me2, H3K4me3, H2AZ, H3K9ac, H3K18ac, and H2BK12ac immunoprecipitation for resting CD4+ cells (Barski et al. 2007; Wang et al. 2008) were obtained from the NCBI short read archive (accession ID SRA000234, SRA000287) and mapped to the human genome (hg18) with ZOOM requiring exact and unique matches (Lin et al. 2008). To determine enrichment of chromatin marks with intraexonic or mRNA initiation sites, the relative mapping position of sequencing tags to the nucleotide associated with the highest CAGE tag frequency within the coding exon or promoter was plotted over a ±50-nt window.
PARE analysis
RNA was extracted from adult mouse tissues using TRIzol and PARE libraries prepared as described previously (German et al. 2009). Polyadenylated RNA was isolated from 200 μg of total RNA (from adult mouse brain, liver, or lung) using oligo(dT) dynabeads (Invitrogen) and an RNA adapter (5′-GUUCAGAGUUCUACAGUCCGAC-3′) ligated using T4 RNA ligase (Ambion). RNA was extracted with phenol/chloroform, ethanol-precipitated, and re-purified with oligo(dT) dynabeads. RNA was then reverse-transcribed (SuperscriptIII; Invitrogen) using the primer [5′-CGAGCACAGAATTAATACGACT(18)V-3′] and amplified by PCR (Phusion DNA polymerase; Finnzymes) using the primers (5′-GTTCAGAGTTCTACAGTCCGAC-3′ and 5′-CGAGCACAGAATTAATACGAC-3′). PCR conditions were seven cycles of 94°C for 30 sec, 60°C for 20 sec, and 72°C for 3 min. Products were gel-purified, cleaved with MmeI (New England Biolabs), and dephosphorylated (shrimp alkaline phosphatase; New England Biolabs). Samples were run on a 12% polyacrylamide gel, and a 42-nt band was excised. DNA was eluted from the gel overnight with 0.3 M NaCl, filtered through a Millex 0.45 μM column, and ethanol-precipitated. Products were then ligated using T4 DNA ligase (Ambion) to one of six double-stranded DNA adapters (top, 5′-P-TCGTATGCCGTCTTCTGCTTG-3′; bottom, NN: 3′-NNAGCATACGGCAGAAGACGAAC-5′) that varied in the composition of an additional first 6 nt (not in the given sequence) to enable barcoding of the separate tissue samples. Another 12% polyacrylamide gel was run, and a 92-nt band was excised and purified as above, followed by PCR amplification using the following conditions: 25 cycles of 94°C for 20 sec, 60°C for 20 sec, and 72°C for 20 sec. The product was again run on a polyacrylamide gel and purified prior to high-throughput sequencing using the Illumina GA platform. The data are available via the NCBI Gene Expression Omnibus (accession ID GSE22627).
Sequence motif detection
To search for possible motifs associated with post-transcriptional cleavage, 60-nt sequences centered on CAGE tag mapping sites either within exons or at promoters were obtained and analyzed with MEME (Bailey et al. 2009). Motifs were visualized with WebLogo (Crooks et al. 2004).
Full-length cDNA analysis
Full-length human and mouse cDNA sequences were obtained from RIKEN (http://fantom3.gsc.riken.jp/). Putative cleavage products were identified by intersecting 5′ cDNA coordinates with RefSeq-annotated internal coding exons or across EEJs as described above for CAGE/SAGE tag mapping. The CRITICA algorithm (Badger and Olsen 1999) was used to identify non-protein-coding from the RIKEN FANTOM3 full-length mouse cDNA library as described previously (Mercer et al. 2008).
Identification of polyadenylation sites
Deep-sequencing tags (accession ID SRA008290) (Parkhomchuk et al. 2009) that derived from polyadenylated mouse brain RNA were used to indentify polyadenylation sites using the following approach: Initially using ZOOM, tags from the sequencing library were aligned to the mouse genome with exact matching. Those tags that did not map exactly to the genome were then trimmed of two or more A nucleotides from the 3′ end, and then aligned again using ZOOM. Those tags that mapped uniquely and exactly following 3′ poly(A) trimming were considered to indicate polyadenylated RNA and therefore mark putative 3′ ends of transcripts on the genome.
Small RNA analysis
Small RNA data sets were obtained from the NCBI Sequence Read Archive (GEO accession series ID GSE10686, GSE11086, GSE11624, GSE7448, GSE9389, GSE12521, GSE9306, GSE10829). Post-transcriptionally derived small RNAs were identified using the same approach described above for CAGE and SAGE tags.
Post-transcriptional cleavage profiling
To determine whether post-transcriptional cleavage is tissue- and developmental-stage-specific, we compared frequencies of exonic and EEJ-spanning CAGE tags across eight mouse tissues (embryo, lung, liver, embryo, visual cortex, somatosensory cortex, cerebellum, and hippocampus) (Valen et al. 2009) and six time points during the differentiation of the human THP1 myelomonocytic leukemia cell line (Suzuki et al. 2009) for each gene. CAGE tags were normalized and represented as tags per million. The 500 genes that contained the highest frequency of exonic CAGE tags were clustered using the Cluster utility (Eisen et al. 1998). CAGE tag frequencies were log-transformed and visualized as a heat map. Exonic CAGE tag frequencies were compared to the CAGE tag frequency in the promoter for the gene subset. Promoter expression levels were defined as the sum of CAGE tags within the promoter region (±50-nt window around RefSeq-annotated transcription start site). The ratios of promoter to exonic CAGE tag frequency were calculated and visualized as a heat map.
RNA size fractionation and deep sequencing
RNA libraries for size fractionation and polysome enrichment for deep sequencing were generated from human embryonic stem cells (G Kolle and SM Grimmond, in prep.). Briefly, HES2 was grown as previously described (Laslett et al. 2007), and nuclei and cellular debris were removed by centrifugation. Cytoplasmic polyadenylated RNA was separated on a 1.2% agarose, and gel slices were excised corresponding to the following sizes: 0–0.5 kb, 0.5–2 kb, 2–3.5 kb, 3.5–6.5 kb, and 6.5–20+ kb. Slices were dissolved, and extracted RNA (1% of each fraction) was run on the Agilent Bioanalyser (Agilent) to confirm the correct size distribution and yield. Library molecules were clonally amplified onto 1-μm magnetic beads according to the SOLiD Template Bead Preparation protocol and sequenced using a SOLiD Analyzer as per the manufacturer's instructions (Applied Biosystems). Mapping of SOLiD sequencing tags was performed using a recursive mapping strategy to the human Genome version hg18 and a library of exon–exon junctions as described previously using RNAmate V.1.1 (Cloonan et al. 2009). For analysis of gene expression in size-fractionated RNA, the smallest RefSeq-annotated isoform was used to bin each gene according to size, with any gene where RNA-seq data showed evidence of unannotated RefSeq splice junctions being omitted. The relative expression in each size fraction was then summed for the smallest annotated RefSeq gene. The data are available at the NCBI Gene Expression Omnibus (accession ID GSE24355).
Coding potential analysis of post-transcriptional cleavage products
Open reading frames were computed from cDNA sequences in all three possible sense frames, with ATG signifying the start codon and an in-frame TAA or TGA signifying a stop codon. Optimal and suboptimal start codons were determined by the presence or absence of an upstream Kozak sequence (Kozak 2005).
Quantitative PCR
Total cellular RNA from mouse lung was purified using TRIzol (Invitrogen) according to the manufacturer's instructions, and any contaminating genomic DNA was removed by treatment with DNase I (Invitrogen) for 30 min at 37°C. To assess the yield and quantity of RNA produced, samples were run on an Agilent 2100 Bioanalyzer using the RNA 6000 Nano Chip kit (Agilent). The ratio of optical density at 260 and 280 nm was ≥1.8 in all cases. RNA was oligo(dT) reverse-transcribed with SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Quantitative RT-PCR was performed using the following primers crossing exonic CAGE tag sites: Pnpla2: GCATCTCCCTGACTCGTGTT (forward), TGGCAAGTTGTCTGAAATGC (reverse); Bptf: TTGGCATCTTGCAAAGTGAG (forward), GGTGCATCATTGGGGTCTAC (reverse); Calm1: ACTGGGTCAGAACCCAACAG (forward), GTTCTGCCGCACTGATGTAA (reverse); Pol2ra: TTACTCCCCTGCATGGTCTC (forward), AGGGGCTCTGGGGTGTATAG (reverse), with tubulin delta used as the internal standard. The targeted RNA and tubulin delta RNA was quantified on an ABI Prism 7000 Sequence Detection System with ABI Prism 7000 SDS software (v1.0; Applied Biosystems). Reactions contained SYBR Green PCR master mix (Applied Biosystems), primers, and template diluted appropriately in distilled water. The cycling conditions were 10 min (95°C), followed by 45 cycles of 15 sec (95°C), and 1 min (58°C). The comparative delta Ct method (Applied Biosystems) was used to determine relative RNA expression. To identify the presence of an upstream polyadenylated cleaved transcript, quantitative PCR was performed exactly as described above, except the reverse primers were replaced with poly(T) primers.
Acknowledgments
This work was supported by grants and fellowships from the Australian Research Council/University of Queensland cosponsored Federation Fellowship (FF0561986; J.S.M.), a National Health and Medical Research Council of Australia Career Development Award (CDA631542; M.E.D.), a Queensland Government Department of Employment, Economic Development and Innovation Smart Futures Fellowship (M.E.D.), and the Australian Stem Cell Centre.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession nos. GSE22627 and GSE24355.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.112128.110.
References
- Ahsan B, Saito TL, Hashimoto S, Muramatsu K, Tsuda M, Sasaki A, Matsushima K, Aigaki T, Morishita S 2009. MachiBase: A Drosophila melanogaster 5′-end mRNA transcription database. Nucleic Acids Res 37: D49–D53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J 2009. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res 19: 1732–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22: 2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger JH, Olsen GJ 1999. CRITICA: Coding region identification tool invoking comparative analysis. Mol Biol Evol 16: 512–524 [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP 2008. The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS 2009. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Carninci P 2006. Tagging mammalian transcription complexity. Trends Genet 22: 501–510 [DOI] [PubMed] [Google Scholar]
- Carninci P, Waki K, Shiraki T, Konno H, Shibata K, Itoh M, Aizawa K, Arakawa T, Ishii Y, Sasaki D, et al. 2003. Targeting a complex transcriptome: The construction of the mouse full-length cDNA encyclopedia. Genome Res 13: 1273–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38: 626–635 [DOI] [PubMed] [Google Scholar]
- Cerritelli SM, Crouch RJ 2009. Ribonuclease H: The enzymes in eukaryotes. FEBS J 276: 1494–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308: 1149–1154 [DOI] [PubMed] [Google Scholar]
- Chooniedass-Kothari S, Emberley E, Hamedani MK, Troup S, Wang X, Czosnek A, Hube F, Mutawe M, Watson PH, Leygue E 2004. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett 566: 43–47 [DOI] [PubMed] [Google Scholar]
- Clayton C, Shapira M 2007. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol Biochem Parasitol 156: 93–101 [DOI] [PubMed] [Google Scholar]
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. 2008. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods 5: 613–619 [DOI] [PubMed] [Google Scholar]
- Cloonan N, Xu Q, Faulkner GJ, Taylor DF, Tang DT, Kolle G, Grimmond SM 2009. RNA-MATE: A recursive mapping strategy for high-throughput RNA-sequencing data. Bioinformatics 25: 2615–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SL, Vassar R 2008. BACE1 structure and function in health and Alzheimer's disease. Curr Alzheimer Res 5: 100–120 [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE 2004. WebLogo: A sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453: 798–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Mattick JS 2008. Differentiating protein-coding and noncoding RNA: Challenges and ambiguities. PLoS Comput Biol 4: e1000176 doi: 10.1371/journal.pcbi.1000176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G 2008. A human snoRNA with microRNA-like functions. Mol Cell 32: 519–528 [DOI] [PubMed] [Google Scholar]
- Fejes-Toth K, Sotirova V, Sachidanandam R, Assaf G, Hannon G, Kapranov P, Foissac S, Willingham A, Duttagupta R, Dumais E, et al. 2009. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457: 1028–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- German MA, Luo S, Schroth G, Meyers BC, Green PJ 2009. Construction of parallel analysis of RNA ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat Protoc 4: 356–362 [DOI] [PubMed] [Google Scholar]
- Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, Tizard ML 2008. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res 18: 957–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Lima CD 2005. Processing the message: Structural insights into capping and decapping mRNA. Curr Opin Struct Biol 15: 99–106 [DOI] [PubMed] [Google Scholar]
- Hashimoto S, Suzuki Y, Kasai Y, Morohoshi K, Yamada T, Sese J, Morishita S, Sugano S, Matsushima K 2004. 5′-End SAGE for the analysis of transcriptional start sites. Nat Biotechnol 22: 1146–1149 [DOI] [PubMed] [Google Scholar]
- Hasselblatt P, Hockenjos B, Thoma C, Blum HE, Offensperger WB 2005. Translation of stable hepadnaviral mRNA cleavage fragments induced by the action of phosphorothioate-modified antisense oligodeoxynucleotides. Nucleic Acids Res 33: 114–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Riechmann JL, Meyerowitz EM 2008. Transcriptome-wide analysis of uncapped mRNAs in Arabidopsis reveals regulation of mRNA degradation. Plant Cell 20: 2571–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP, Gingeras TR 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296: 916–919 [DOI] [PubMed] [Google Scholar]
- Kapranov P, Drenkow J, Cheng J, Long J, Helt G, Dike S, Gingeras TR 2005. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res 15: 987–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Karginov FV, Cheloufl S, Chong MMW, Stark A, Smith AD, Hannon GJ 2010. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol Cell 38: 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima H, Takano H, Sugita S, Takahara Y, Sugimura K, Nakatani T 2003. A novel steroid receptor co-activator protein (SRAP) as an alternative form of steroid receptor RNA-activator gene: Expression in prostate cancer cells and enhancement of androgen receptor activity. Biochem J 369: 163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234: 187–208 [DOI] [PubMed] [Google Scholar]
- Kozak M 2005. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361: 13–37 [DOI] [PubMed] [Google Scholar]
- Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. 2009. The UCSC Genome Browser Database: Update 2009. Nucleic Acids Res 37: D755–D761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett AL, Grimmond S, Gardiner B, Stamp L, Lin A, Hawes SM, Wormald S, Nikolic-Paterson D, Haylock D, Pera MF 2007. Transcriptional analysis of early lineage commitment in human embryonic stem cells. BMC Dev Biol 7: 12 doi: 10.1186/1471-213X-7-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ 2000. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J 19: 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ 2001. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 20: 4987–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Zhang Z, Zhang MQ, Ma B, Li M 2008. ZOOM! Zillions of oligos mapped. Bioinformatics 24: 2431–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda N, Kasukawa T, Oyama R, Gough J, Frith M, Engstrom PG, Lenhard B, Aturaliya RN, Batalov S, Beisel KW, et al. 2006. Transcript annotation in FANTOM3: Mouse gene catalog based on physical cDNAs. PLoS Genet 2: e62 doi: 10.1371/journal.pgen.0020062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A 2003. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4: 223 doi: 10.1186/gb-2003-4-7-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS 2008. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci 105: 716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura F, Kawaguchi N, Sese J, Toyoda A, Hattori M, Morishita S, Ito T 2006. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc Natl Acad Sci 103: 17846–17851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136: 688–700 [DOI] [PubMed] [Google Scholar]
- Nahkuri S, Taft RJ, Mattick JS 2009. Nucleosomes are preferentially positioned at exons in somatic and sperm cells. Cell Cycle 8: 3420–3424 [DOI] [PubMed] [Google Scholar]
- Okamura K, Lai EC 2008. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol 9: 673–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563–573 [DOI] [PubMed] [Google Scholar]
- Otsuka Y, Kedersha NL, Schoenberg DR 2009. Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol Cell Biol 29: 2155–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, Soldatov A 2009. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res 37: e123 doi: 10.1093/nar/gkp596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR 2007. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35: D61–D65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127: 1193–1207 [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP 2007. Intronic microRNA precursors that bypass Drosha processing. Nature 448: 83–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saida F, Odaert B 2007. RNA recognition and cleavage by sequence-specific endoribonucleases. Protein Pept Lett 14: 103–111 [DOI] [PubMed] [Google Scholar]
- Scarpulla RC 2008. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev 88: 611–638 [DOI] [PubMed] [Google Scholar]
- Schoenberg DR, Maquat LE 2009. Re-capping the message. Trends Biochem Sci 34: 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, Ast G 2009. Chromatin organization marks exon–intron structure. Nat Struct Mol Biol 16: 990–995 [DOI] [PubMed] [Google Scholar]
- Snider L, Asawachaicharn A, Tyler AE, Geng LN, Petek LM, Maves L, Miller DG, Lemmers RJ, Winokur ST, Tawil R, et al. 2009. RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: New candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum Mol Genet 18: 2414–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Nielsen CB, Padgett RA, Burge CB 2009. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell 36: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger MA, Decker CJ 2001. New twists in understanding the fate of antisense oligodeoxynucleotide mRNA targets. Mol Cell 8: 732–733 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Forrest AR, van Nimwegen E, Daub CO, Balwierz PJ, Irvine KM, Lassmann T, Ravasi T, Hasegawa Y, de Hoon MJ, et al. 2009. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat Genet 41: 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest AR, Grimmond SM, Schroder K, et al. 2009. Tiny RNAs associated with transcription start sites in animals. Nat Genet 41: 572–578 [DOI] [PubMed] [Google Scholar]
- Thoma C, Hasselblatt P, Kock J, Chang SF, Hockenjos B, Will H, Hentze MW, Blum HE, von Weizsacker F, Offensperger WB 2001. Generation of stable mRNA fragments and translation of N-truncated proteins induced by antisense oligodeoxynucleotides. Mol Cell 8: 865–872 [DOI] [PubMed] [Google Scholar]
- Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R 2009. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol 16: 996–1001 [DOI] [PubMed] [Google Scholar]
- Valen E, Pascarella G, Chalk A, Maeda N, Kojima M, Kawazu C, Murata M, Nishiyori H, Lazarevic D, Motti D, et al. 2009. Genome-wide detection and analysis of hippocampus core promoters using DeepCAGE. Genome Res 19: 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CT, Garneau-Tsodikova S, Gatto GJ Jr 2005. Protein post-translational modifications: The chemistry of proteome diversifications. Angew Chem Int Ed Engl 44: 7342–7372 [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]