Abstract
TATA-binding protein (TBP) nucleates the assembly of the transcription preinitiation complex (PIC), and although TBP can bind promoters with high stability in vitro, recent results establish that virtually the entire TBP population is highly dynamic in yeast nuclei in vivo. This dynamic behavior is surprising in light of models that posit that a stable TBP-containing scaffold facilitates transcription reinitiation at active promoters. The dynamic behavior of TBP is a consequence of the enzymatic activity of the essential Snf2/Swi2 ATPase Mot1, suggesting that ensuring a highly mobile TBP population is critical for transcriptional regulation on a global scale. Here high-resolution tiling arrays were used to define how perturbed TBP dynamics impact the precision of RNA synthesis in Saccharomyces cerevisiae. We find that Mot1 plays a broad role in establishing the precision and efficiency of RNA synthesis: In mot1-42 cells, RNA length changes were observed for 713 genes, about twice the number observed in set2Δ cells, which display a previously reported propensity for spurious initiation within open reading frames. Loss of Mot1 led to both aberrant transcription initiation and termination, with prematurely terminated transcripts representing the largest class of events. Genetic and genomic analyses support the conclusion that these effects on RNA length are mechanistically tied to dynamic TBP occupancies at certain types of promoters. These results suggest a new model whereby dynamic disassembly of the PIC can influence productive RNA synthesis.
The RNA polymerase II (Pol II) transcription machinery consists of a collection of general transcription factors (GTFs) and the multisubunit Pol II enzyme itself (Reese 2003; Hahn 2004). Assembly of the Pol II preinitiation complex (PIC) on promoters is highly orchestrated by transcriptional regulators and coregulators that influence GTF recruitment by direct interaction with the transcription machinery and by modulating the promoter chromatin template (Hahn 2004). PIC assembly is nucleated by the TATA-binding protein (TBP), which physically interacts with multiple GTFs and DNA. TBP recruitment to promoters is often rate limiting for transcription in vivo (Pugh 2000).
Interaction of the TBP saddle with the TATA box results in severe bending and unwinding of the DNA (Burley and Roeder 1996). In vitro, the resultant complex forms a specialized, long-lived substrate for accrual of the other GTFs. Biochemical evidence indicates that a TBP-containing subcomplex remains on promoter DNA following the departure of Pol II (Hahn 2004). This complex, termed the scaffold, can facilitate transcription reinitiation in vitro (Hahn 2004). Although the in vitro evidence in support of a stable reinitiation intermediate is strong, PIC dynamics may be influenced by other factors in vivo. For example, stable TBP–DNA binding is antagonized by Mot1, a Snf2/Swi2-related ATPase that dissociates the TBP–DNA complex (Auble 2009). As another example, the NC2 heterodimer interacts with TBP to form an encircling clamp that allows TBP to diffuse along the DNA contour (Kamada et al. 2001; Schluesche et al. 2007). In fact, recent measurements of TBP mobility in living yeast cells demonstrate that all detectable TBP is highly mobile, displaying Mot1-dependent FRAP recovery times of <15 sec (Sprouse et al. 2008). Importantly, while the recovery times are rapid, they are markedly slower than can be explained by diffusion and are instead consistent with transient interaction with chromatin. This suggests that the entire (or nearly entire) TBP pool is rapidly recycled, leading to rapid redistribution of TBP among chromatin binding sites.
Several fundamental questions are raised by the observed high mobility of TBP in vivo. If TBP is rapidly recycled from sites on chromatin, what is the nature of these sites? Given the pervasive RNA synthesis in yeast cells under these conditions, it would appear that there may be active promoters for which PICs are rapidly recycled. If this is true, how and why are such dynamics important for promoter function? When TBP dynamics are compromised, are new or different types of RNA made, or is simply the quantity at the annotated genes changed? To begin to address these questions, we developed a general genomic strategy to identify aberrant RNA species in mutant strains of interest. Surprisingly, we find that compromising TBP dynamics via a conditional mutation of Mot1 gave rise to many hundreds of changes in RNA length, the largest category of which includes transcripts that were apparently initiated properly but failed to reach the end of the gene. In parallel, we determined how Mot1 affects TBP occupancy genome-wide for comparison with the RNA effects. The results support a model in which Mot1-mediated TBP dynamics at the promoter influence transcription elongation efficiency. These results argue that in contrast to prevailing views, at many promoters, PIC dynamics can play an important role in conferring efficiency and accuracy of transcription elongation.
Results
We first compared RNA from wild-type (WT) and mot1-42 yeast cells using Affymetrix genomic tiling arrays that interrogate the yeast genome at 5-bp resolution. mot1-42 is a temperature-sensitive allele that encodes a protein that is biochemically inactive in vitro (Darst et al. 2003), and prior work established that this allele induces changes in gene expression in vivo that closely parallel other severe, conditional mot1 alleles (Dasgupta et al. 2002). WT and mutant strains were grown at permissive temperature (30°C) and then shifted for 45 min to 35°C prior to harvesting RNA. This temperature shift did not impair growth of WT cells but did dramatically inhibit growth of the mot1-42 cells (Darst et al. 2003). We calculated the gene expression changes from the tiling array data using methods similar to those applied to conventional gene expression arrays (see Methods). These expression changes were well correlated with expression changes defined previously (Supplemental Fig. 1; Sprouse et al. 2009), thus validating the use of tiling arrays for quantitative RNA analysis.
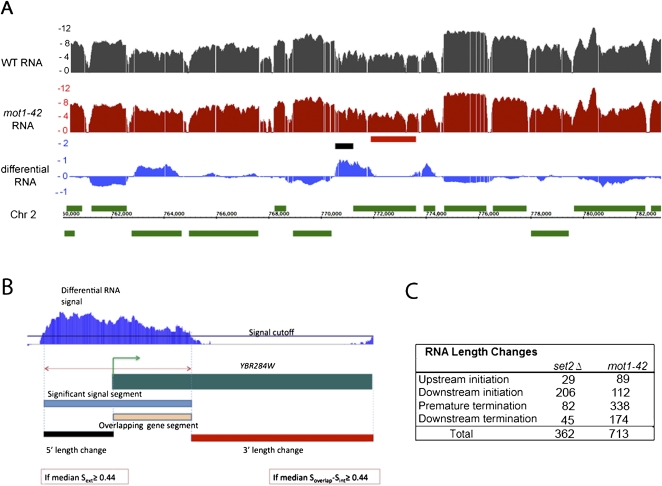
Although Mot1 exerts a global effect on transcription, most of these transcriptional effects are modest in magnitude. To better understand why Mot1-catalyzed TBP recycling is essential, we took advantage of the tiling arrays to determine whether Mot1-mediated TBP dynamics affect RNA precision as well as quantity. To test this idea, we developed a method to capture significant RNA hybridization signals that deviate from gene annotations (Fig. 1A,B). This approach was possible because in the overwhelming majority of cases, the WT RNA signals were closely aligned with annotated genes (an example is shown in Fig. 1A, and others are discussed below). By comparing the coordinates of gene annotations with differential RNA segments, a methodology was developed that can detect RNA segments that extend outside a gene annotation, or differential signals that overlap with only a portion of an open reading frame (ORF). The gene-dense nature of the budding yeast genome presented challenges for the analysis because differential RNA signals could potentially overlap with more than one gene annotation, giving rise to different types of apparent RNA length changes based on the relative orientation of the two genes (Supplemental Fig. S2). Nonetheless, it was possible to segregate the differential RNA signals into four categories in which the RNA segment overlap was defined provisionally with respect to the gene annotation (Fig. 1B). Thus, “upstream initiation” segments are RNAs that extended upstream of the normal transcription start site (TSS); “downstream initiation” events are RNAs apparently initiated within the ORF (also known as cryptic initiation events); “premature termination” segments correspond to RNAs within an ORF that do not include the normal 3′ end; and “downstream termination” events are those in which RNA extended beyond the termination site in WT cells. Although the arrays do not provide information about the strand specificity of the hybridized RNA, the strand-specific analyses described below confirm that most, if not all, of the detected events correspond to changes in RNA sense strand abundance.
Figure 1.
Global changes in RNA length in mot1-42 and set2Δ cells. (A) Integrated Genome Browser (Nicol et al. 2009) screen shot of log2 RNA profiles in WT (black) and mot1-42 (red) cells. The differential RNA profile (log2 mot1-42/WT) is shown in the blue track. Annotated genes are in green, with positions above or below the chromosomal coordinate indicating whether the genes are transcribed on the top or bottom strand. The short black and red horizontal bars denote RNA length changes captured by the method, as illustrated more clearly in panel B. (B) Overview of the method, illustrated with an enlargement of the chromosomal region in the center of the screen shot in A. In the case of the gene YBR284W, the significant differential RNA segment found using signal log2(mot1-42/WT) > 0.3 (blue segment) was compared with the annotation, and aberrant transcript length changes (denoted by the red and the black segments) were calculated. Thresholds requiring a ≥100 bp overlapping gene segment and length changes ≥150 bp long were applied. Soverlap and Sext refer to the differential RNA signal in the region where the differential segment overlaps (orange segment) and is external (black segment) to the annotation, respectively. Sint refers to the differential signal within the portion of the annotation that does not overlap the differential segment (red segment). By provisionally defining the differential RNA with respect to the overlapping gene, the aberrant RNAs were sorted among four groups (see text). In this example, a 5′ length change is classified as an upstream initiation event, whereas the partial overlapping gene segment and consequent change in 3′ length define a premature termination event. Although this example shows, for illustrative purposes, a gene with two types of RNA length changes, most of the genes with length changes in mot1-42 cells (92.4%) displayed only a single type as indicated in the table in panel C. (C) The table summarizes the number of aberrant RNAs identified from the differential RNA signal as in B in mot1-42 and set2Δ cells compared with WT cells. Of particular note is the large number of premature termination events in mot1-42 cells and the enrichment in downstream (cryptic) initiation events in set2Δ cells.
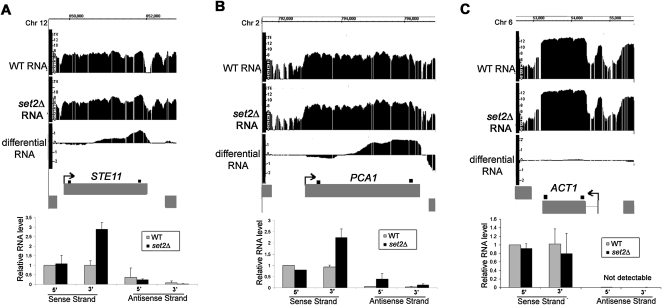
To validate the array-detected RNA length changes, RNA from set2Δ cells was compared with the WT using the same methodology. Loss of the H3K36 methyltransferase activity in set2Δ cells is well known to result in cryptic initiation within ORFs (Workman 2006). Consistent with the published data (Li et al. 2007), 206 instances of cryptic initiation were detected in set2Δ cells, and these “downstream initiation” events comprised the largest class of RNA length changes by far (Fig. 1C). Among the set2Δ-induced array-detected RNA length changes, we found and validated cryptic initiation in STE11 and PCA1, two genes previously shown to be susceptible to cryptic initiation in set2Δ cells (Fig. 2; Li et al. 2007).
Figure 2.
Confirmation of RNA length changes in set2Δ cells. (A–C) Integrated Genome Browser screenshots of log2 WT, set2Δ, and differential RNA levels (set2Δ/WT) for STE11, PCA1, and ACT1 genes. Bar graphs, relative RNA levels (arbitrary units) quantified by real-time PCR using sense- and antisense-specific 5′ and 3′ primer sets for each gene (shown as small black boxes above each gene). Average values are shown ±SE obtained by analysis of two independent RNA samples for each strain. Cryptic initiation was detected in STE11 and PCA1, whereas there was no significant change in ACT1 expression.
Using the same approach, we characterized RNA length changes from differential mot1-42 versus WT RNA. Strikingly, twice as many aberrant RNA species were detected in mot1-42 cells compared with set2Δ cells (Fig. 1C), and the mot1-42-induced length changes had a significantly different distribution among the four length change classes. Notably, a Mot1 defect led to 338 genes showing “premature termination,” the most prevalent class of events. The “premature termination” events fell into two categories: (1) “differential down” instances in which the RNA level was similar in the 5′ end of the ORF but diminished in the 3′ end of the ORF in mot1-42 cells compared with WT (77%) and (2) “differential up” instances in which a defect in Mot1 led to increased RNA in the 5′ portion of the ORF but the differential RNA failed to extend to the 3′ end of the ORF (23%). Although the mot1-42 and set2Δ data sets have different numbers of genes and different distributions among the length change categories, there are 12 genes that displayed premature termination in both mot1-42 and set2Δ cells. While few in number, the overlap is statistically significant (P = 0.02), suggesting that elongation efficiency in some genes may be under both Mot1 and Set2 control.
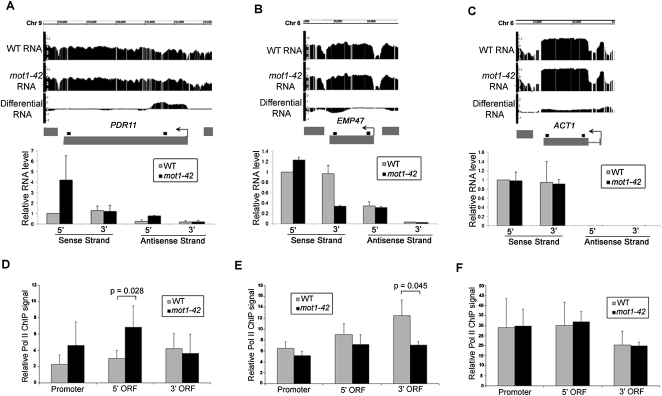
Selected RNA length changes were validated by real-time PCR, including two genes with premature termination defects (Fig. 3A–C). Validation data for four other array-detected RNA length changes are shown in Figure 4, A, B, E, and F. Note that strand-specific real-time PCR showed that the differential RNA effects are attributable largely, if not entirely, to changes in sense strand abundance. The results thus far support a role for Mot1 in maintaining RNA profiles that match annotated genes but do not address whether this effect is mediated through transcription or some other effect on RNA processing. To address this question, chromatin immunoprecipitation (ChIP) was performed to assess the Pol II density on genes for which the RNA length changes occurred. As shown in Figures 3, D through F, and 4, C, D, G, and H, differential changes in the RNA level correlated with changes in Pol II occupancy as expected if the differential RNA signals arose through changes in transcription.
Figure 3.
Validation of premature termination RNA length changes in mot1-42 cells and correlation with Pol II density. (A–C, top) Screen shots showing log2 WT, mot1-42, and differential RNA signals across PDR11, EMP47, and ACT1 genes. (Bottom) Relative RNA levels were quantified by real-time PCR for both the sense and antisense strands. Primers were specific for either the 5′ or 3′ end of each gene, and the amplified product is represented by the small black square above each gene. Results shown are from the means of two independent RNA samples with associated SE. PDR11 is an example of a gene with increased 5′ transcription in mot1-42 cells (classified as differentially up), whereas EMP47 had similar levels of 5′ RNA but less 3′ RNA in mot1-42 versus WT cells (classified as differentially down). No RNA length change was detected for ACT1, and the change in RNA level across the open reading frame was small (≤5%) in comparison to the total level of total ACT1 RNA in both strains. Antisense ACT1 RNA was not detectable. (D–F) Relative Pol II ChIP signals in the promoter, 5′ end, and 3′ end of each ORF. The results were obtained using the 8WG16 antibody and are shown as the mean of three independent biological replicates ± SD. The indicated P-values were determined using a one-tailed paired t-test of the log-transformed ChIP values. Note the correspondence between the changes in Pol II ChIP and RNA length changes: The 5′ ORF of PDR11 had an increased level of Pol II in mot1-42 cells that corresponded with increased 5′ ORF RNA level. Similarly, Pol II ChIP signal was decreased in the 3′ ORF of EMP47, consistent with the decrease in 3′ RNA in mot1-42 cells. As expected, there were no significant changes in Pol II ChIP for ACT1.
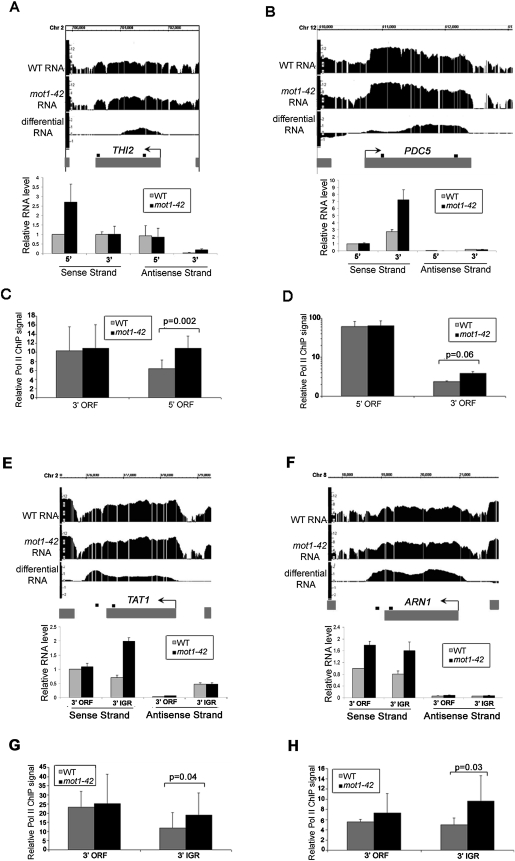
Figure 4.
Confirmation of cryptic initiation and 3′ transcript length changes in mot1-42 cells. (A,B,E,F) Screenshots of log2 WT, mot1-42, and differential RNA levels for four selected genes are shown in the upper part of each panel. Relative RNA levels quantified by real-time PCR using sense- and antisense-specific primers are shown in the bar graphs. The graphs show the average ± SE obtained by analysis of two independent RNA samples for each strain. The small black boxes above each gene indicate the locations of each primer set. (C,D,G,H) Relative RNA Pol II ChIP signals were obtained at the 5′ end and the 3′ end of the ORF (THI2 and PDC5) or the 3′ end of the ORF and the 3′ intergenic region (TAT1 and ARN1). The results were obtained using the 8WG16 antibody and are shown as the mean of three biological replicates ± SD. The indicated P-values were determined using a one-tailed paired t-test of the log-transformed ChIP values. The same primer sets were used as the RNA analysis. THI2 is an example of prematurely terminated RNA, and PDC5 is an example of downstream initiation. In contrast, TAT1 and ARN1 are examples of downstream termination RNAs. For all of the genes, there is an increase in Pol II ChIP levels that correspond with the RNA length change. Note that the Pol II ChIP data for PDC5 are shown on a log scale. The strong signal at the 5′ end of the PDC5 ORF is consistent with previously published data (Steinmetz et al. 2006).
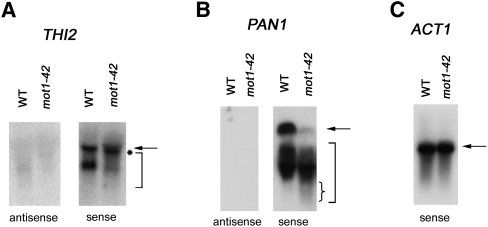
To further confirm the interpretations of the differential RNA tiling array data, we analyzed RNAs by Northern blotting using strand-specific probes. We chose genes associated with premature termination for which the full-length and predicted short RNAs were appropriately sized for detection on the blot, as well as being reasonably well resolved from each other. THI2 displayed premature termination and was in the differentially up class. As shown in Figure 5A, primarily two species of THI2 RNA were detected in WT cells, a full-length species and a discrete shorter RNA that has not been characterized. In mot1-42 cells, most of the full-length THI2 RNA was slightly shifted to a position of smaller size (marked by asterisk), and a heterogeneous smear of shorter RNAs was detected (marked by bracket). This is consistent with the tiling array and RT-PCR data showing an increase in prematurely terminated RNA in mot1-42 cells. As expected, all of the detectable RNA was sense RNA, confirming the premature termination designation. PAN1 was also detected as a gene associated with premature termination, but in the differentially down class. PAN1 is a Mot1-activated gene, and as expected, there was less full-length PAN1 RNA in mot1-42 cells compared with WT. A smear of shorter PAN1 RNA was readily detected, and as expected for a gene in the differentially down class, PAN1 short RNA was at least as abundant in mot1-42 cells as it was in WT cells. Moreover, we observed that the shortest RNA species detected were more abundant in mot1-42 cells compared with WT cells (small bracket). As expected, all of the detectable PAN1 RNA was sense RNA. In contrast, ACT1 RNA was a discrete species unaffected by mot1-42 (Fig. 5C). Interestingly, the presence of shorter THI2 and PAN1 RNA in WT cells suggests that these genes are prone to premature termination even in WT cells but that these effects are exaggerated by mutation of MOT1.
Figure 5.
Confirmation of RNA length changes by Northern blotting. Total RNA from WT or mot1-42 cells was resolved by gel electrophoresis, transferred to nylon membrane, and probed with radiolabeled single DNA strands to detect gene-specific sense or antisense RNAs, as indicated. (A) THI2. Full-length RNA is indicated by the arrow. Note the slight shift in mobility of full-length RNA (indicated by the asterisk) and the smear of smaller RNA species (bracket) in mot1-42 versus WT samples. Only sense RNA was detected. The probes spanned +240 to +930 with respect to the start of the open reading frame. (For map of probe location, see Supplemental Fig. S8.) (B) PAN1 was identified as a Mot1-activated gene displaying premature termination in the differentially down class. Consistent with this, note the decrease in full-length RNA (arrow) in mot1-42 cells, which was in contrast to the shorter RNAs (large bracket) whose abundance was comparable in mot1-42 versus WT cells. However, the small RNA species were distributed such that the shortest RNAs were more prominent in mot1-42 cells compared with WT (denoted by small brace). Only sense RNA was detected. The probes spanned +420 to +864 with respect to the start of the ORF. (For screen shot of PAN1 tiling RNA signals and map of probe location, see Supplemental Fig. S8.) (C) ACT1 control RNA was detected with a sense-strand specific probe. As expected, discrete full-length bands were detected in both WT and mot1-42 cells, with no quantitative difference between them. The ACT1 probe spanned +14 to +184 with respect to the start of the ORF.
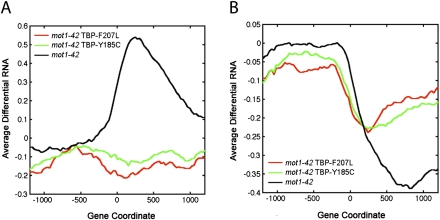
In a previous study (Sprouse et al. 2009), we identified two TBP alleles that bypass the requirement for Mot1 in vivo. These TBPs restored appropriate expression levels to many Mot1-regulated genes in vivo and allowed cells to grow without Mot1, which is otherwise essential (Sprouse et al. 2009). Biochemically, the bypass TBPs were defective for interaction with other GTFs or DNA, consistent with the critical activity Mot1 provides in destabilizing TBP-containing complexes in vivo (Sprouse et al. 2009). RNA tiling array analysis demonstrated that the two TBP bypass alleles suppressed the premature termination RNA length changes observed in mot1-42 cells (Fig. 6A,B). Suppression of the premature termination RNA synthesis from the differential up class was essentially complete, whereas the bypass TBPs partially restored efficient RNA synthesis to the differentially down gene class. Interestingly, the bypass TBPs were able to suppress each of the other RNA length change classes as well (Supplemental Fig. S7). We conclude that the mot1-42-mediated effects on RNA length can be explained by a direct effect of Mot1 on TBP dynamics.
Figure 6.
Suppression of the premature termination defects in mot1 cells by mutations in TBP. (A,B) Profiles of average differential RNA—defined as the average of the differential RNA signal as determined in Supplemental Methods (RNA Tiling Array Data)—for genes showing premature termination length changes in mot1-42 versus WT cells (black lines). The x-axis indicates the position along the chromosome in base pairs relative to the start of the gene annotation (zero). For comparison, the plots show the average differential RNA profiles for these same genes in mot1-42 cells harboring TBP-F207L versus WT (red lines) and mot1-42 TBP-Y185C versus WT (green lines). Premature termination length effects were subclassified depending on whether the differential signal was positive (A, 77 genes) or negative (B, 264 genes). Average signals were obtained by smoothing over a 30-bp window.
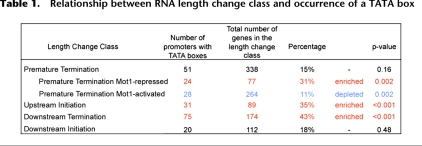
Computational approaches were employed to determine if there are promoter features or aspects of local genomic organization that correlate with the RNA length changes observed in mot1 cells. First, we found that upstream initiation and downstream termination classes of RNA length changes are enriched in genes whose promoters possess TATA boxes (Table 1; Basehoar et al. 2004). The TATA box association with premature termination genes was only detected for the differentially up class; the differentially down premature termination genes are significantly underrepresented in TATA-containing genes. In contrast, no significant TATA box enrichment or depletion was seen for the downstream initiation genes. These results argue that the nature of the core promoter is related to the propensity to generate a certain type of aberrant RNA. This observation provides further support for the notion that these RNA length changes result from direct, promoter-mediated effects on transcriptional elongation.
Table 1.
Relationship between RNA length change class and occurrence of a TATA box
Recent work has revealed two classes of noncoding RNA in yeast, termed SUTs (stable unannotated transcripts) and CUTs (cryptic unstable transcripts) (Wyers et al. 2005; Davis and Ares 2006; David et al. 2006; Neil et al. 2009; Xu et al. 2009). An extensive and detailed investigation uncovered no statistically significant relationship between the occurrence of a particular type of aberrant RNA in mot1-42 cells and the occurrence of a SUT or CUT flanking or within the annotated gene in which the RNA length change was detected (Supplemental Fig. S6; data not shown). As a second approach, we classified annotated genes as Mot1-activated, Mot1-repressed, or Mot1-unaffected, based on the overall change in RNA level quantified in the gene-centric analysis of the tiling data described above (Supplemental Fig. S1). Interestingly, in this case, significant relationships were discovered between Mot1-regulated genes and the presence of a SUT or CUT proximal or overlapping the annotation. As shown in Figure 7, Mot1-repressed genes (scored as differentially up) are enriched in genes with a SUT that overlaps their transcribed regions. These SUTs are transcribed in the opposite sense as the affected gene. On the other hand, Mot1-activated genes (scored as differentially down) tend to have an overlapping CUT near the 3′ end of the gene. Again, these CUTs are transcribed in the opposite sense as the affected gene. Although the underlying mechanisms are unknown, collectively, these results suggest that Mot1's effects on transcription are influenced by antisense transcription of SUTs or CUTs with a particular proximal relationship to the affected gene.
Figure 7.
Relationship between Mot1-regulated genes and CUT and SUT RNAs. The diagrams illustrate the two significant relationships detected between Mot1-mediated gene expression and noncoding transcription. Mot1-activated genes tend to have an overlapping CUT transcribed in the antisense direction. The CUT transcripts initiate within the transcribed region of the Mot1-activated gene, ∼40 bp on average upstream of the Mot1-activated gene termination site. In contrast, Mot1-repressed genes tend to have an overlapping antisense SUT. These SUTs initiate ∼40 bp on average downstream from the termination site for the Mot1-repressed gene.
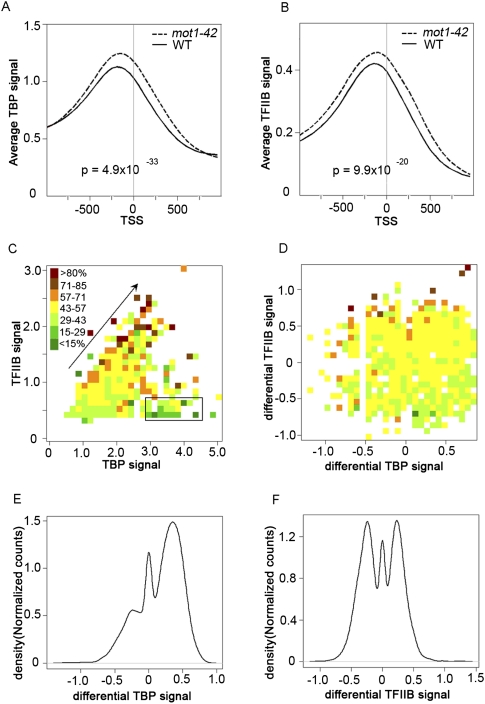
Next, we sought to correlate TBP chromatin localization with the global changes in the transcription observed in mot1 cells. Published work has documented TBP distribution genome-wide (Kim and Iyer 2004; Zanton and Pugh 2004; van Werven et al. 2008; Venters and Pugh 2009), as well as the Mot1 distribution and its correlation with TBP genome-wide (Geisberg and Struhl 2004; Zanton and Pugh 2004; van Werven et al. 2008). To determine the genome-wide dependence of TBP localization on Mot1 and to compare Mot1-mediated localization to changes in RNA length, ChIP with microarray hybridization (ChIP-chip) was performed using these same tiling arrays. These results allowed us to define the locations of TBP binding as well as the relative TBP occupancies genome-wide in both WT and mot1-42 cells. To distinguish productive TBP binding from nonproductive binding to Pol II promoters, ChIP-chip was performed in parallel for TFIIB. The results obtained with WT cells are in good agreement with published data (van Werven et al. 2009; Venters and Pugh 2009). For example, TBP and TFIIB were localized near each other and primarily in promoters, 126 and 123 bp, respectively, upstream from the TSS (Fig. 8A,B; Nagalakshmi et al. 2008). As expected, these locations correlate well with nucleosome-free regions (Supplemental Fig. S5; data not shown; Whitehouse et al. 2007). In addition, although there was a weak positive correlation between TBP promoter occupancy and gene expression, TFIIB promoter occupancy in both WT and mot1-42 cells tracked much more closely with RNA level (Fig. 8C).
Figure 8.
Global effects of Mot1 on TBP and TFIIB genomic distribution and correlations with transcription. Plots of average TBP (A) and average TFIIB chromatin (B) occupancies obtained by aligning all annotated Pol II genes with respect to the transcription start site (TSS). The signals were smoothed over a 50-bp sliding window (WT, solid curves; mot1-42, dashed curves). In both cases, the differences in the distributions are highly significant as determined by the Kolmogorov-Smirnov test (P-values as indicated). (C) The heat map displays the transcriptome-wide relationship between TBP occupancy (x-axis), TFIIB occupancy (y-axis), and RNA level (color) in WT cells. Each box represents one or more genes whose TBP and TFIIB occupancies fall within the box's x-axis/y-axis values. The box color is the relative median expression value on a scale defined by the range of all medians in the data set. Note the general trend that increasing TBP and TFIIB promoter occupancy is correlated with increasing RNA level (arrow). However, the correlation between TFIIB and RNA levels is better because there are genes with high TBP occupancy but low expression (black rectangle). (D) The heat map is similar to that in panel C but shows the transcriptome-wide relationship between the differential TBP signal (x-axis), the differential TFIIB signal (y-axis), and the change in RNA level (color) in mot1-42 versus WT cells. As in C, changes in TFIIB occupancy are reasonably well correlated with changes in transcription, whereas changes in TBP promoter occupancy do not correlate as well with changes in RNA level. (E,F) Density distributions of the differential TBP and differential TFIIB signals (as indicated), over the promoters for all the genes. The plots show that in mot1-42 cells compared with WT, TBP occupancies increased at the majority of promoters. In contrast, TFIIB occupancies increased or decreased at roughly equal numbers of promoters, consistent with changes in gene expression in both positive and negative directions.
Interestingly, in mot1-42 cells, TBP promoter occupancies were increased genome-wide compared with WT cells (Fig. 8A,B). More detailed analyses supported the conclusion that overall, TBP occupancies increased in promoters across the board regardless of whether Mot1 exerted a detectable effect on expression of the gene (Fig. 8; data not shown). The implications of these observations are discussed below. Figure 8C shows the global relationship between TBP promoter occupancy (x-axis), TFIIB promoter occupancy (y-axis), and RNA level (color). The diagonal arrow shows that for a significant number of genes, increasing TBP and TFIIB promoter occupancy was correlated with increasing RNA level, as indicated by a transition in the color along the diagonal from green (lower RNA level) to brown/red (higher RNA level). However, the plot also shows that TFIIB promoter occupancy was much better correlated with RNA level than TBP, as indicated by the substantial number of genes with low associated RNA levels (colored green) but with high TBP occupancy (demarcated by the rectangle). The global effects of Mot1 on gene expression, as well as TBP and TFIIB occupancy, are shown in Figure 8D. Differential TFIIB promoter occupancy was reasonably well correlated with differential expression mediated by loss of Mot1. Note that an increase in gene expression in mot1-42 cells (brown/red) was generally associated with an increase in TFIIB promoter occupancy (higher y-axis value), whereas decreases in gene expression (green) generally displayed a decrease in TFIIB promoter occupancy (lower y-axis value). In contrast, there was no obvious correlation between the change in gene expression (color) and the change in TBP promoter occupancy (x-axis value). Figure 8, E and F, supports these general conclusions. These histograms show how the global distributions of TBP and TFIIB changed in mot1-42 versus WT cells. Most promoters (60%) had increased TBP occupancies in mot1-42 versus WT cells, as illustrated by the large peak of positive differential TBP in Figure 8E. The small peak centered over zero indicates that there were some genes (20%) whose TBP occupancies did not change, and the left-hand shoulder reflects promoters whose TBP occupancies decreased in mot1-42 cells. In contrast, as shown in Figure 8F, the two large peaks indicate that TFIIB occupancies were increased or decreased at roughly equal numbers of promoters (39% increased and 42% decreased). The correlation of the differential TFIIB signal with differential RNA (Fig. 8D) indicates that the bimodal distribution of differential TFIIB in Figure 8F was a consequence of the nature of the Mot1-mediated transcriptional effect. To further explore the nature of Mot1-mediated effects on TBP, a peak finding algorithm was employed to map more precisely the loci of protein binding (Supplemental Fig. S3). Most promoters possessed single TBP peaks (67.4%); about a third (32.7%) possessed TFIIB peaks; and close to half of the detected TBP peaks (47.3%) were associated with a TATA motif (Supplemental Fig. S4). Thus, in many instances the effects of Mot1 on TBP and TFIIB promoter occupancy appear to reflect changes in the occupancies of single, discrete complexes formed on promoters.
Finally, using several computational approaches, we investigated the relationship between TBP occupancy and the RNA length changes that occurred in mot1-42 cells (see Supplemental materials and Methods). For the altered initiation events in particular, the relatively small number of affected genes made statistical analysis difficult. Nonetheless, we observed that the “downstream initiation” events appear to have a relatively straightforward origin: In mot1-42 cells, the shift in TBP localization parallels the apparent site of initiation of the new RNA (Pearson correlation = 0.51; data not shown). This suggests that changes in initiation occurred because Mot1 failed to clear TBP from cryptic sites that can nucleate the assembly of functional transcription complexes. It is unclear how a defect in Mot1 could give rise to RNAs with extended 3′ ends, but the correlation of these different aberrant RNA species with promoter type (Table 1) suggests that termination or 3′ processing events can somehow be influenced by TBP dynamics, depending on the type of promoter.
Discussion
The results presented here reveal several new and unanticipated relationships between TBP dynamics and transcription. Notably, we find that impaired turnover of TBP at promoters correlates with the production of a predominant class of “premature termination” RNAs. The Pol II ChIP and TBP allele suppression results argue that Mot1-mediated effects on RNA length distribution are attributable to changes in transcription of the affected genes, and that these occur as a consequence of altered TBP dynamic behavior at promoters. Moreover, the genes that display RNA length changes tend to have certain promoter attributes (TATA versus TATA-less). The stochastic failure of Pol II elongation that ensues when TBP dynamics are perturbed suggests that Mot1-catalyzed clearance of TBP may be important for promoter recruitment or activity of the accessory factors that subsequently promote Pol II elongation. Another attractive model is that there may be communication between factors associated with the promoter and the elongating Pol II. A possible explanation is that a physical connection persists between the promoter and elongating Pol II, which can stall elongation some distance from the promoter. The role of Mot1 then would be to facilitate the dissolution of the TBP complex at the promoter to release a restrained Pol II. Alternatively, there may be communication between the promoter and an elongating Pol II to facilitate transit through chromatin or to ensure that downstream RNA processing events are coupled to transcription. Regardless of the mechanism, it is striking that a large number of yeast genes rely to some extent on TBP dynamics to ensure accurate and efficient RNA elongation.
The genome-wide increase in TBP occupancy observed in mot1-42 cells compared with WT reported here fits well with the rapid mobility of essentially the whole TBP pool assessed by live cell imaging (Sprouse et al. 2008) and shows that TBP occupancy is generally limiting at promoters in vivo. On the other hand, although Mot1 regulates a substantial proportion of the transcriptome, not all yeast genes are affected in mot1-42 cells. Presumably, the Mot1-affected genes are especially sensitive to TBP occupancy, whereas other genes are rate-limited in some other critical step of the transcription cycle. A recent study concluded that TBP turnover rates are different at different classes of promoters (van Werven et al. 2009). This study relied on a replacement strategy in which expression of a differentially tagged form of TBP was induced and the rate of its association with promoters was tracked. As the production of the new TBP pool required on the order of 30 min, this approach would not capture the very rapid Mot1-catalyzed dynamics (occurring on the order of seconds) that our previous work has shown accounts for the behavior of the majority of the TBP pool (Sprouse et al. 2008). Although no genomic scale method exists yet for detection of such rapid, locus-specific dynamic behavior, the analysis of the TBP occupancy profile in mot1-42 cells reported here significantly extends prior work by showing that Mot1 does target virtually the entire pool of promoter bound TBP in vivo. This observation provides support for a recent study that modeled the dynamic behavior of GTFs based on previously published ChIP-chip data, which found that interactions between TBP and chromatin are best described by models in which the interactions are rather transient (Samorodnitsky and Pugh 2010). Our suspicion is that GTF–promoter interactions span a wide range of lifetimes. Because the long-lived interactions detected by kinetic ChIP experiments appear to represent a relatively small (albeit very important) proportion of the total number of chromatin interactions, they may well be below the limit of detection in the live cell imaging experiments that capture the behavior of the overall GTF pool.
Recent results show that constitutive yeast genes are expressed by infrequent initiation events clearly separated in time (Zenklusen et al. 2008). These observations also fit well with our measurements of highly dynamic TBP in vivo (Sprouse et al. 2008) and suggest that, in contrast to establishing a stable scaffold, many active promoters are subject to occupancy by transcription complexes that undergo rapid cycles of assembly and disassembly. Such dynamic behavior has been speculated to be important for ensuring appropriate start site selection and timely transcriptional regulatory responses (Auble 2009). However, the results presented here support the notion of a fundamental role for PIC dynamics in the process of RNA synthesis itself. More generally, the ability to rapidly characterize the spectrum of aberrant RNAs present in a particular mutant background using the approach outlined here will likely be of use in unraveling the molecular mechanisms responsible for these effects.
Methods
Yeast strains and growth conditions
Saccharomyces cerevisiae WT and mot1-42 strains used for RNA and ChIP analyses are derivatives of YPH499 (Sikorski and Hieter 1989) and were previously described (Darst et al. 2003; Dasgupta et al. 2005). SET2 was replaced with the kanMX cassette in YPH499 cells, and proper integration was confirmed by PCR. For TBP ChIP-chip, the C-terminally myc-tagged TBP strains were derived by transformation of PCR-generated DNA fragments into the same background containing the mutant alleles (YAD155, WT; YAD156, mot1-42). For the RNA analysis of mutant TBPs, previously described pRS314-borne alleles of TBP (WT, Y185C, or F207L) (Sprouse et al. 2009) were transformed into AY51 (WT) or AY87 (Y185C, F207L). Yeast strains were grown in YPD at 30°C to an OD600 ∼ 1.0. Then cells were shifted immediately for 45 min to 35°C and were harvested for isolation of total RNA or treated for ChIP as previously described (Dasgupta et al. 2005).
Expression analysis
Total RNA was isolated using hot-acid phenol extraction (Schmitt et al. 1990). For the tiling arrays, cDNA was synthesized from 7 μg of total RNA using the Affymetrix GeneChip WT Double-Stranded cDNA Synthesis Kit as recommended by the manufacturer, but with the addition 0.4 mM dUTP for subsequent fragmentation and biotin end-labeling. dsDNA was purified using the GeneChip Sample Cleanup Module (Affymetrix, Inc.). Fragmentation and labeling were performed using the GeneChip WT Double-Stranded DNA Terminal Labeling Kit. Fragmented DNA was confirmed to be between 25 and 100 bp using the Agilent RNA 6000 Nano Kit and an Agilent 2100 Bioanalyzer. Efficient labeling was confirmed by gel shift assay using NeutrAvidin. Samples were hybridized to S. cerevisiae Tiling 1.0R Arrays (Affymetrix), and raw data were generated by the Microarray Core Facility at the University of Virginia (Charlottesville). WT, mot1-42, and set2Δ RNA analyses were performed using two independent biological replicates for each. For the experiment in Figure 5, A and B, WT (TBP), mot1-42 (TBP-Y185C), and mot1-42 (TBP-F207L) RNA was analyzed as a single replicate. For quantitative real-time PCR validation, RT-PCR was first performed using the iScript Select cDNA Synthesis Kit (Bio-Rad) according to manufacturer's instructions. cDNA was then quantitated by real-time PCR using iQ SYBR Green Supermix (Bio-Rad) and the Bio-Rad MyiQ Single Color Real-Time PCR Detection System. Each experiment includes two independent biological replicates. Northern blotting was performed as previously described (Dasgupta et al. 2005) using strand-specific probes obtained by single-primer PCR in reactions that included [α-32P]dATP. In each case, the template for probe synthesis was a gel-purified PCR fragment spanning the differentially affected transcribed region. Primer sequences are shown in Supplemental Table 1.
Chromatin immunoprecipitation
ChIP assays were performed exactly as previously described (Dasgupta et al. 2005) with the following antibodies: for TBP ChIP, anti-myc (9E10) (Dasgupta et al. 2005); for TFIIB ChIP, a TFIIB rabbit polyclonal antibody (Dasgupta et al. 2005); and for RNA Pol II ChIP, the Pol II monoclonal antibody 8WG16 (Thompson et al. 1989; Bhaumik and Green 2001). ChIP material was then used for hybridization to tiling arrays or for quantitation by real-time qPCR. For the tiling arrays, library preparation and amplification of DNA for both ChIP and mock IP samples were performed using the GenomePlex Complete Whole Genome Amplification Kit (Sigma) as described (O'Green et al. 2006) with several modifications: dUTP was added in equimolar concentration to the dNTP mix (0.4 mM), and a second amplification was performed for both the ChIP and mock samples using 10 ng of the previously amplified material. Samples were purified with the QIAquick PCR purification kit (Qiagen) prior to re-amplification and fragmentation. Duplicate samples were combined to obtain 7.5 μg of material for fragmentation and labeling. Samples were hybridized to S. cerevisiae Tiling 1.0R arrays (Affymetrix), and raw data were generated by the Microarray Core Facility at the University of Virginia (Charlottesville). Three independent biological replicates were analyzed for TBP, and two independent biological replicates were analyzed for TFIIB. Quantitative real-time PCR was performed on ChIP, mock IP, and total samples as described for the RNA analysis. ChIP signals were obtained by subtracting mock IP signal from ChIP signal and normalizing against the input. ORF primers were identical to those used in the expression analysis. Three independent biological replicates were performed for each ChIP analysis. P-values were obtained by log transforming the calculated ChIP signals and a one-tailed, paired, t-test was conducted.
Tiling array data analysis
We performed both gene-biased and -unbiased analyses of the tiling array data. For unbiased analysis, we used the Affymetrix Tiling Array Software (TAS), which quantile normalizes replicate arrays, scales their median intensity to a user-defined value, and calculates the −10log10(P-value) and log2(pseudo-median) (or signal strength) associated with a one- or two-sample Wilcoxon signed rank test over a sliding window (Cawley et al. 2004). For the ChIP-chip analysis of TBP and TFIIB for mot1-42 and WT strains, we applied the two-sample test of the ChIP sample compared with mock IP using a window size of 500 bp. For the total RNA analysis, we applied the one-sample test using a window size of 50 bp. For the biased analysis, a gene-centric library (CDF) file was generated from yeast gene annotations (Fisk et al. 2006) and the Affymetrix tiling array library (BPMAP) file, which contained a probe set for every annotated yeast gene composed of all probes whose central position fell within the annotated start and stop of the gene. Normalized gene expression estimates were obtained by quantile normalizing the arrays and applying GCRMA. Lists of differentially expressed genes were obtained using the limma package in Bioconductor and applying a 5% false discovery rate cutoff. Additional methods are described in the Supplement. Tiling array data have been deposited in the NCBI Gene Expression Omnibus under accession number GSE18283.
Acknowledgments
We thank Mitch Smith and Jeff Smith for comments on the manuscript. This work was supported by the NIH grant GM55763 to D.T.A.
Footnotes
[Supplemental material is available online at http://www.genome.org. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE18283.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.109504.110.
References
- Auble DT 2009. The dynamic personality of TATA-binding protein. Trends Biochem Sci 34: 49–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116: 699–709 [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Green MR 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev 15: 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Roeder RG 1996. Biochemistry and structural biology of transcription factor IID (TFIID). Annu Rev Biochem 65: 769–799 [DOI] [PubMed] [Google Scholar]
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116: 499–509 [DOI] [PubMed] [Google Scholar]
- Darst RP, Dasgupta A, Zhu C, Hsu J-Y, Vroom A, Muldrow TA, Auble DT 2003. Mot1 regulates the DNA binding activity of free TATA-binding protein in an ATP-dependent manner. J Biol Chem 278: 13216–13226 [DOI] [PubMed] [Google Scholar]
- Dasgupta A, Darst RP, Martin KJ, Afshari CA, Auble DT 2002. Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms. Proc Natl Acad Sci 99: 2666–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A, Juedes SA, Sprouse RO, Auble DT 2005. Mot1-mediated control of transcription complex assembly and activity. EMBO J 24: 1717–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM 2006. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci 103: 5320–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Ares M Jr 2006. Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc Natl Acad Sci 103: 3262–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk DG, Ball CA, Dolinski K, Engel SR, Hong EL, Issel-Tarver L, Schwartz K, Sethuraman A, Botstein D, Cherry JM, et al. 2006. Saccharomyces cerevisiae S288C genome annotation: A working hypothesis. Yeast 23: 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg JV, Struhl K 2004. Cellular stress alters the transcriptional properties of promoter-bound Mot1–TBP complexes. Mol Cell 14: 479–489 [DOI] [PubMed] [Google Scholar]
- Hahn S 2004. Structure and mechanism of the RNA polymerase II transcription machinery. Nat Struct Mol Biol 11: 394–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada K, Shu F, Chen H, Malik S, Stelzer G, Roeder RG, Meisterernst M, Burley SK 2001. Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex. Cell 106: 71–81 [DOI] [PubMed] [Google Scholar]
- Kim J, Iyer VR 2004. Global role of TATA box-binding protein recruitment to promoters in mediating gene expression profiles. Mol Cell Biol 24: 8104–8112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL 2007. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev 21: 1422–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A 2009. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457: 1038–1042 [DOI] [PubMed] [Google Scholar]
- Nicol JW, Helt GA, Blanchard SG Jr, Raja A, Loraine AA 2009. The Integrated Genome Browser: Free software for distribution and exploration of genome-scale datasets. Bioinformatics 25: 2730–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Green H, Nicolet CM, Blahnik K, Green RD, Farnham PJ 2006. Comparison of sample preparation methods for ChIP-chip assays. Biotechniques 41: 577–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh BF 2000. Control of gene expression through regulation of the TATA-binding protein. Gene 255: 1–14 [DOI] [PubMed] [Google Scholar]
- Reese JC 2003. Basal transcription factors. Curr Opin Genet Dev 13: 114–118 [DOI] [PubMed] [Google Scholar]
- Samorodnitsky E, Pugh BF 2010. Genome-wide modeling of transcription preinitiation complex disassembly mechanisms using ChIP-chip data. PLoS Comput Biol 6: e1000733 doi: 10.1371/journal.pcbi.1000733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluesche P, Stelzer G, Piaia E, Lamb DC, Meisterernst M 2007. NC2 mobilizes TBP on core promoter TATA boxes. Nat Struct Mol Biol 14: 1196–1201 [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18: 3091–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprouse RO, Karpova TS, Mueller F, Dasgupta A, McNally JG, Auble DT 2008. Regulation of TATA binding protein dynamics in living yeast cells. Proc Natl Acad Sci 105: 13304–13308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprouse RO, Wells MN, Auble DT 2009. TATA-binding protein variants that bypass the requirement for Mot1 in vivo. J Biol Chem 284: 4525–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA 2006. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell 24: 735–746 [DOI] [PubMed] [Google Scholar]
- Thompson NE, Steinberg TH, Aronson DB, Burgess RR 1989. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J Biol Chem 264: 11511–11520 [PubMed] [Google Scholar]
- van Werven FJ, van Bakel H, van Teeffelen HAAM, Altelaar AFM, Koerkamp MG, Heck AJR, Holstege FCP, Timmers HTM 2008. Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes Dev 22: 2359–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven FJ, van Teeffelen HAAM, Holstege FCP, Timmers HTM 2009. Distinct promoter dynamics of the basal transcription factor TBP across the yeast genome. Nat Struct Mol Biol 16: 1043–1048 [DOI] [PubMed] [Google Scholar]
- Venters BJ, Pugh BF 2009. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res 19: 360–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T 2007. Chromatin remodelling at promoters suppresses antisense transcription. Nature 450: 1031–1035 [DOI] [PubMed] [Google Scholar]
- Workman JL 2006. Nucleosome displacement in transcription. Genes Dev 20: 2009–2017 [DOI] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle J-C, Dufour M-E, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. 2005. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanton SJ, Pugh BF 2004. Changes in genomewide occupancy of core transcriptional regulators during heat stress. Proc Natl Acad Sci 101: 16843–16848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH 2008. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol 15: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]