Abstract
Point mutations result from errors made during DNA replication or repair, so they are usually expected to be homogeneous across all regions of a genome. However, we have found a region of chloroplast DNA in plants related to sweetpea (Lathyrus) whose local point mutation rate is at least 20 times higher than elsewhere in the same molecule. There are very few precedents for such heterogeneity in any genome, and we suspect that the hypermutable region may be subject to an unusual process such as repeated DNA breakage and repair. The region is 1.5 kb long and coincides with a gene, ycf4, whose rate of evolution has increased dramatically. The product of ycf4, a photosystem I assembly protein, is more divergent within the single genus Lathyrus than between cyanobacteria and other angiosperms. Moreover, ycf4 has been lost from the chloroplast genome in Lathyrus odoratus and separately in three other groups of legumes. Each of the four consecutive genes ycf4-psaI-accD-rps16 has been lost in at least one member of the legume “inverted repeat loss” clade, despite the rarity of chloroplast gene losses in angiosperms. We established that accD has relocated to the nucleus in Trifolium species, but were unable to find nuclear copies of ycf4 or psaI in Lathyrus. Our results suggest that, as well as accelerating sequence evolution, localized hypermutation has contributed to the phenomenon of gene loss or relocation to the nucleus.
The genome organization and gene content of chloroplast DNA (cpDNA) are highly conserved among most flowering plant species (Palmer 1985; Sugiura 1992; Jansen et al. 2007). The chloroplast genome of the most recent common ancestor of all angiosperms contained 113 different genes (four rRNA genes, 30 tRNA genes, and 79 protein genes), and this content has been retained in many angiosperms (Kim and Lee 2004). Rates of synonymous nucleotide substitution in chloroplast genes are generally low (a few fold lower than plant nuclear genes) and relatively homogeneous within a genome except for a threefold difference in rate between the large inverted repeat (IR) and single-copy regions (Wolfe et al. 1987; Drouin et al. 2008). Lineage-specific variation in chloroplast synonymous rates has been documented (Gaut et al. 1993; Guo et al. 2007) but is relatively modest compared to the vast differences seen among some plant mitochondrial lineages (Palmer et al. 2000; Mower et al. 2007; Sloan et al. 2009).
Some angiosperm cpDNAs have fewer than the 79 canonical protein genes due to gene losses. Most notable here are parasitic plants such as Cuscuta and Epifagus that have lost some or all photosynthetic ability (Wolfe et al. 1992; Funk et al. 2007; McNeal et al. 2007). Chloroplast gene losses are rarer in photosynthetic species, because in many cases the gene cannot simply be discarded and must instead be either functionally transferred to the nuclear genome or functionally replaced by a nuclear gene (“gene substitution”). Successful gene transfers from the chloroplast to the nuclear genome during angiosperm evolution have been reported for rpl22 in legumes (Gantt et al. 1991); for infA in several lineages, including almost all rosids (Millen et al. 2001); and for rpl32 in two families of Malpighiales (Cusack and Wolfe 2007; Ueda et al. 2007). In addition, Ueda et al. (2008) identified gene substitution as the mechanism of loss of the rps16 gene from cpDNA in Medicago and Populus. The loss of rps16 from cpDNA is compensated by dual targeting (to chloroplasts as well as mitochondria) of mitochondrial ribosomal protein S16, which is encoded by a nuclear gene. Several other examples of losses of genes from cpDNA in photosynthetic angiosperms have been reported, and it is striking that the few species in which gene losses have occurred tend also to be those whose chloroplast genomes are highly rearranged relative to the ancestral angiosperm organization (Jansen et al. 2007). As with angiosperm mitochondrial genomes (Adams and Palmer 2003), most of the genes that have been lost from chloroplast genomes during recent evolution have coded for ribosomal proteins (Jansen et al. 2007). There have been no published reports of the loss of genes coding for components of photosystems I or II (psa and psb genes), the electron transfer chain (pet genes), or the chloroplast ATP synthase (atp genes) from cpDNA in any angiosperms except parasitic species (Wolfe et al. 1992; Funk et al. 2007; McNeal et al. 2007).
One group of angiosperms that is known to be relatively prone to cpDNA rearrangement and gene loss is the legume family (Fabaceae) (Palmer et al. 1988). The large IR that is otherwise almost universally present in chloroplast genomes is absent from one large clade of legumes (the IR loss clade, or IRLC) (Wojciechowski et al. 2004), some of which also show other rearrangements of gene order. Chloroplast genomes in the IRLC species are also notable for having significant amounts of repetitive DNA, something not usually seen in angiosperm cpDNA (Milligan et al. 1989; Saski et al. 2005; Cai et al. 2008). Five instances of gene loss from the IRLC chloroplast genomes have been discovered. As well as the aforementioned gene transfers of rpl22 and infA and substitution of rps16 (Gantt et al. 1991; Millen et al. 2001; Ueda et al. 2008), it has been reported that that accD is completely absent from Trifolium subterraneum (subclover) cpDNA, and that ycf4 in Pisum sativum (pea) is either absent or a pseudogene (Nagano et al. 1991a; Smith et al. 1991; Cai et al. 2008). Slot-blot hybridization experiments suggested that ycf4 and rps16 may have been lost independently multiple times in different lineages of legumes (Doyle et al. 1995).
In the course of this study, we reviewed all reported instances (in published papers or in GenBank annotations) of gene loss among the 103 complete angiosperm chloroplast genome sequences that are publicly available, and found that 27 different protein-coding genes have been lost in at least one lineage (Supplemental Table S1). We found that some reported gene losses are simply due to annotation errors; because of this, the numbers of losses we describe here are slightly different from those in Jansen et al. (2007). In particular, we noticed that the gene ycf4, which was originally not identified in the genome sequences of the legumes Glycine max (soybean; Saski et al. 2005), T. subterraneum (subclover; Cai et al. 2008), Cicer arietinum (chickpea), and Medicago truncatula (Jansen et al. 2008), is in fact present in the cpDNAs of all these species but is so divergent that it was not recognized by the DOGMA software (Wyman et al. 2004) used to annotate them. This discovery prompted us to investigate the rapid evolution of ycf4 and its surrounding region in legumes.
Ycf4 is a thylakoid protein that has been shown to play a role in regulating photosystem I assembly in cyanobacteria (Wilde et al. 1995) and to be essential for photosystem I assembly in Chlamydomonas (Boudreau et al. 1997; Onishi and Takahashi 2009). Experiments in Chlamydomonas indicate that Ycf4 is the second of three scaffold proteins that act sequentially during the assembly process, with Ycf4's roles being to stabilize an intermediate subcomplex consisting of the PsaAB heterodimer and the three stromal subunits PsaCDE, and to add the PsaF subunit to this subcomplex (Ozawa et al. 2009). As well as the loss of ycf4 in P. sativum, several other previous studies have indicated that the evolution of ycf4 in legumes may be unusual. In soybean and Lotus japonicus, the Ycf4 protein, which is almost universally 184 or 185 amino acids long, has expanded to about 200 residues (Reverdatto et al. 1995; Kato et al. 2000). The gene also has a high rate of synonymous nucleotide substitution between the latter two species (Perry and Wolfe 2002). Phylogenetic trees for phaseoloid legumes constructed using ycf4 were incongruent with trees constructed using seven other genes, due to accelerated evolution of codon positions 1 and 2 in ycf4 (Stefanovic et al. 2009). In blot hybridizations to DNAs from 280 diverse angiosperms (as in Millen et al. 2001) using a ycf4 probe from tobacco, we observed (SS and JDP, unpubl.) strong hybridization to all DNAs except those from the only Papillionoid legumes surveyed: Medicago (no signal from five species) and Vigna (considerably diminished signal). We show here that ycf4 is situated in a local mutation hotspot, in Lathyrus, and possibly in other legume species, resulting in dramatic acceleration of sequence evolution in some species and evolutionary gene losses in others.
Results
Rapid evolution of ycf4 in legumes
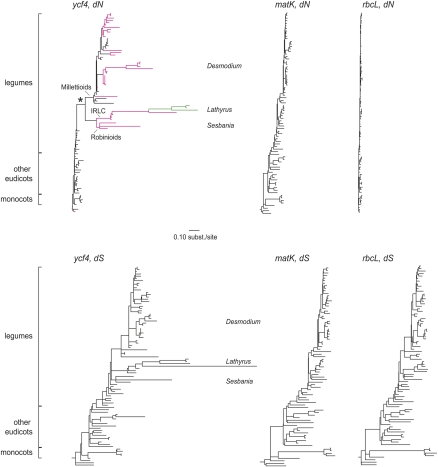
To investigate acceleration of the evolutionary rate of ycf4 in legumes, we compared its nonsynonymous and synonymous nucleotide substitution rates in different angiosperm lineages to the rates observed in two other, widely sequenced chloroplast genes, rbcL and matK. This analysis included new ycf4 sequence data from Lathyrus and other legumes, together with sequences from a previous phylogenetic study (GenBank [http://www.ncbi.nlm.nih.gov/Genbank/] accession nos. EU717431–EU717464; Stefanovic et al. 2009) and other database sequences. For each gene, we used a likelihood model to estimate the numbers of nonsynonymous (dN) and synonymous (dS) nucleotide substitutions that occurred on each branch of an angiosperm phylogenetic tree (see Methods). In the dN trees, ycf4 is seen to evolve much faster in most legumes than in other angiosperms (Fig. 1) but no similar acceleration is seen in legume rbcL or matK, which suggests that the acceleration is locus-specific, as well as lineage-specific. Within legumes, the first accelerated branch is the one leading to a large clade (Millettioids, Robinioids, and the IRLC; asterisk in Fig. 1), and the legumes that are outgroups to this branch do not show acceleration. This branch is also the first one on which the Ycf4 protein size expands above 200 amino acids (Fig. 1). Even faster periods of dN evolution are seen in the genera Desmodium and Lathyrus relative to other legumes. Ycf4 is a pseudogene in three of six Desmodium species we sequenced and in Clitoria ternatea (Supplemental Fig. S1C, left panel). In the dS trees, some acceleration is seen in ycf4 of legumes relative to other angiosperms, particularly in Lathyrus, but again no similar acceleration is seen in legume rbcL or matK (Fig. 1). The genus Lathyrus also shows by far the greatest increases in Ycf4 size, reaching 340 residues in Lathyrus latifolius and Lathyrus cirrhosus.
Figure 1.
Synonymous and nonsynonymous divergence in angiosperm chloroplast ycf4 sequences. Shown are dN (upper) and dS (lower) trees resulting from a codon-based likelihood analysis and a constrained topology, rooted using gymnosperm sequences (which are not included in the trees). All trees are drawn to the same scale. The species are in the same order from top to bottom in all trees, to the greatest extent possible, and are named in full in Supplemental Figure S1. Magenta branches in the dN tree for ycf4 indicate those on which the Ycf4 protein length is (or is inferred to have been) ≥200 amino acid residues; green branches indicate lengths ≥300 residues. The asterisk marks the branch (leading to Millettioids, Robinioids, and IRLC) in which rate acceleration is first seen. Trees for chloroplast rbcL and matK genes do not show comparable rate heterogeneity at either synonymous or nonsynonymous sites.
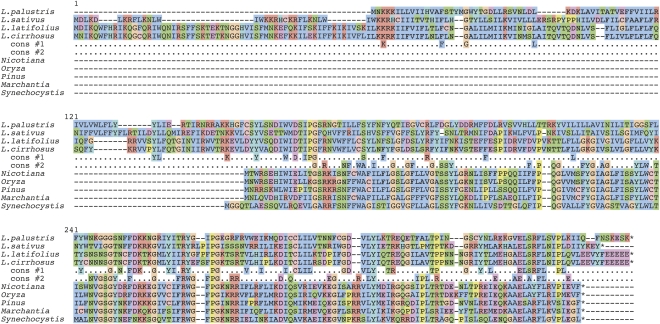
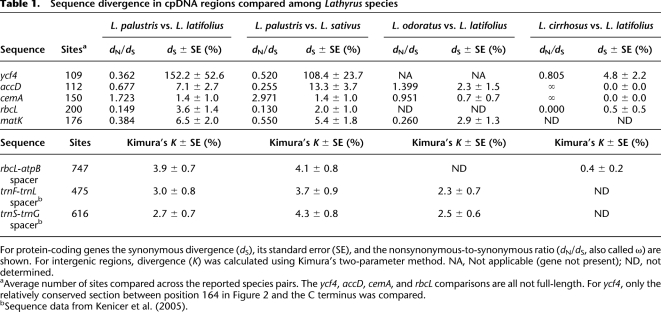
Remarkably, there is less amino acid sequence conservation between the Ycf4 proteins of two species within the genus Lathyrus (31% identity between Lathyrus palustris and L. cirrhosus), than between tobacco and the cyanobacterium Synechocystis (45% identity). Nevertheless, ycf4 can be inferred to be functional in the four Lathyrus species in which it is intact (Fig. 2), for two reasons. First, even though the level of amino acid sequence conservation among Lathyrus species is very low, many of the sites in the C-terminal part of the protein (beginning at position 248 in Fig. 2) that are conserved among other land plants and cyanobacteria are also conserved in Lathyrus. Second, comparing ycf4 sequences among Lathyrus species shows that they have lower levels of nonsynonymous than synonymous nucleotide substitutions (dN/dS < 1) (Table 1), which is a hallmark of sequences that are being constrained to code for proteins (Kimura 1977; Graur and Li 1999). We therefore infer that these long ycf4 genes in Lathyrus species are biologically functional. However, the level of constraint on Lathyrus ycf4 is lower than on other angiosperm ycf4s (e.g., dN/dS = 0.15 between tobacco and spinach ycf4, compared with dN/dS = 0.36–0.81 within the genus Lathyrus). Tests for positive (Darwinian) selection suggested that some Desmodium branches within the ycf4 tree have undergone adaptive evolution, and in separate analyses, site-specific tests for positive selection were significant for some codons in ycf4 when the whole legume tree was considered (data not shown). However, in view of the evidence that the whole region around ycf4 has a high mutation rate (see below), and because we also found some dN/dS values greater than 1 within the genus Lathyrus for two genes flanking ycf4 (cemA and accD) (Table 1), we suspect that the high dN/dS values are artifacts stemming from a combination of an increased mutation rate and lessened constraints on protein sequences, rather than being indicative of positive selection on multiple adjacent genes.
Figure 2.
Alignments of Ycf4 protein sequences from four Lathyrus species, four diverse land plants, and the cyanobacterium Synechocystis. Cons #1 shows residues that are absolutely conserved among the four Lathyrus species. Cons #2 shows residues that are absolutely conserved among Nicotiana tabacum, Oryza sativa, Pinus thunbergii, Marchantia polymorpha, and Synechocystis species PCC 6803. The alignment was made using MUSCLE as implemented in SeaView (Gouy et al. 2010) with default coloring of conservative amino acid substitution groups.
Table 1.
Sequence divergence in cpDNA regions compared among Lathyrus species
For protein-coding genes the synonymous divergence (dS), its standard error (SE), and the nonsynonymous-to-synonymous ratio (dN/dS, also called ω) are shown. For intergenic regions, divergence (K) was calculated using Kimura's two-parameter method. NA, Not applicable (gene not present); ND, not determined.
aAverage number of sites compared across the reported species pairs. The ycf4, accD, cemA, and rbcL comparisons are all not full-length. For ycf4, only the relatively conserved section between position 164 in Figure 2 and the C terminus was compared.
bSequence data from Kenicer et al. (2005).
We may have slightly overestimated the divergence of Lathyrus Ycf4 proteins because we inferred protein sequences from chloroplast DNA sequences, whereas some chloroplast transcripts are known to undergo mRNA editing (Stern et al. 2010). Editing in angiosperms involves C → U changes and typically occurs at 30–40 sites per genome (Tsudzuki et al. 2001; Inada et al. 2004). However, even extensive C → U editing could only marginally reduce the divergence in Lathyrus Ycf4. For example, if we assume that every possible C → U editing event that could increase the similarity between L. palustris and L. cirrhosus Ycf4 proteins actually occurs, their sequence identity only increases from 31% to 32%. Furthermore, no sites in ycf4 are known to undergo mRNA editing in other species (Tsudzuki et al. 2001; Chateigner-Boutin and Small 2007; and our analyses of EST data from M. truncatula, Lotus japonicus, and G. max).
Gene losses and repetitive DNA in the region around ycf4 in legumes
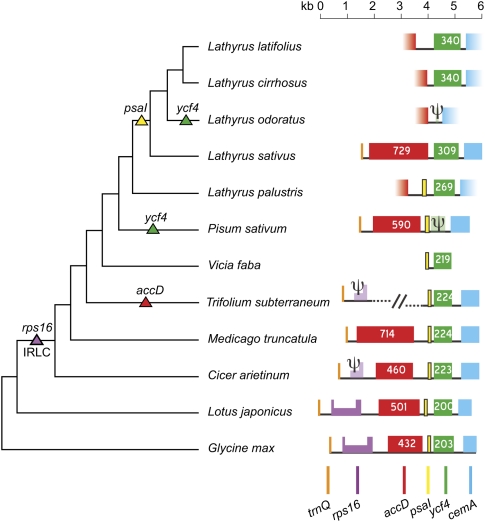
We sequenced the region flanking the ycf4 locus in five Lathyrus species, P. sativum (pea) and Vicia faba (broad bean) and compared it to the available data for other legumes (Fig. 3). This comparison reveals a history of multiple gene losses and gene length changes within a small region of cpDNA. We identified ycf4 pseudogenes in both P. sativum and Lathyrus odoratus (sweetpea), which must be the result of two separate losses of the gene (Fig. 3). The small photosystem I gene psaI, normally found immediately upstream of ycf4, is missing from a clade of four Lathyrus species but is present in L. palustris. Also in this region of the genome, the ribosomal protein gene rps16 was lost from cpDNA in the common ancestor of the IRLC clade (Doyle et al. 1995), and accD, coding for a subunit of acetyl-CoA carboxylase, is missing from T. subterraneum cpDNA, which has become rearranged in this region (Cai et al. 2008). Both ycf4 and accD show extensive length variation among the legume species that retain them (Fig. 3).
Figure 3.
Gene organization around the ycf4 locus in some legumes. Triangles indicate evolutionary losses of the indicated genes. Numbers indicate the numbers of codons in accD and ycf4 genes. Psi symbols denote pseudogenes. All genes are transcribed from left to right. Fading colors denote genes that were not completely sequenced. The half-height region in rps16 represents an intron. The slash marks indicate a genomic rearrangement in T. subterraneum (Cai et al. 2008). The topology of the phylogenetic tree (not drawn to scale) is from Asmussen and Liston (1998), Wojciechowski et al. (2004), and Kenicer et al. (2005).
The expansion of the accD open reading frame is partly explained by the presence of numerous tandemly repeated sequences in this region of legume cpDNA. As reported previously (Nagano et al. 1991a; Smith et al. 1991), and shown by a dot-matrix plot in Supplemental Figure S2A, P. sativum accD contains several in-frame internal repeats of up to 37 codons long. L. sativus accD has a similarly repetitive structure, but the sections of the gene that are repeated are different in the two species (Supplemental Fig. S2B,C). There are tandem repeats in the intergenic DNA between accD and ycf4 in L. latifolius (Supplemental Fig. S2D), and a tandem repeat of 15 codons is located within the 5′ end of L. sativus ycf4 (Supplemental Fig. S2B). All the repeats are species-specific, which suggests that these minisatellite-like sequences have a high turnover rate. However, some other species, such as L. odoratus, do not contain tandem repeats in this region, and the expanded size of ycf4 in most Lathyrus species is not primarily due to the accumulation of repeats.
Sequences of the P. sativum and L. sativus chloroplast genomes
To establish whether the patterns of evolution seen around the ycf4 locus are atypical of the rest of the genome, we sequenced the chloroplast genome of L. sativus (grasspea; 121,020 bp) and completed the genome sequence of P. sativum cpDNA (pea; 122,169 bp) (Supplemental Fig. S3). Both of these genomes lack the IR. They have rearrangements of gene order relative to the ancestral angiosperm order, as represented by tobacco, and also relative to each other. The gene order in P. sativum can be obtained from the tobacco order by eight inversion steps (Palmer et al. 1988), beginning with a 50-kb inversion that is shared by most legumes and that placed rps16 beside accD. The first three inversions occurred before the separation of the lineages giving rise to P. sativum and L. sativus, after which there were five more inversions specific to P. sativum, and three more inversions specific to L. sativus (Supplemental Fig. S3). None of the inversions in P. sativum or L. sativus is shared with the highly rearranged cpDNA of T. subterraneum (Cai et al. 2008), other than the initial 50-kb inversion (Supplemental Fig. S3E).
The L. sativus genome sequence shows that it shares four gene losses that have already been reported in P. sativum: infA, rps16, rpl22, and rpl23 (Gantt et al. 1991; Nagano et al. 1991a,b; Millen et al. 2001); whereas L. sativus ycf4 is intact. The status of rpl23 in P. sativum has been unclear because it contains a 190-bp frameshifting insert close to the normal 3′ end (Nagano et al. 1991b), but in L. sativus the same insert is present and more than half of rpl23 is missing, so we infer that rpl23 is a pseudogene in both species. In addition, these two species differ by the absence of psaI in L. sativus, and of ycf4 in P. sativum. As well as the gene losses, P. sativum and L. sativus both lack two of the 21 introns normally found in angiosperm cpDNA—the first intron of clpP, and the cis-intron of rps12. These intron losses were reported previously as part of a survey that showed their occurrence at about the time of origin of the IRLC clade (Jansen et al. 2008).
Legume chloroplast genomes have long been thought to contain more repetitive sequences than other cpDNAs (e.g., Saski et al. 2005), and this is confirmed by dot-matrix analysis. Using a cutoff of 28 matching bases per 30-bp window, there are very few repeated sequences of this size in the tobacco and spinach chloroplast genomes other than the IR and some similarities among group II introns and among iso-accepting tRNA genes (Supplemental Fig. S4). However in P. sativum, L. sativus, and Lotus japonicus (as a representative IR-containing legume), it is striking that there are many tandem or near-tandem repeats (i.e., dots near the main diagonal in Supplemental Fig. S4), and the region around ycf4 stands out as particularly repetitive.
Sites of gene loss coincide with a mutation hotspot
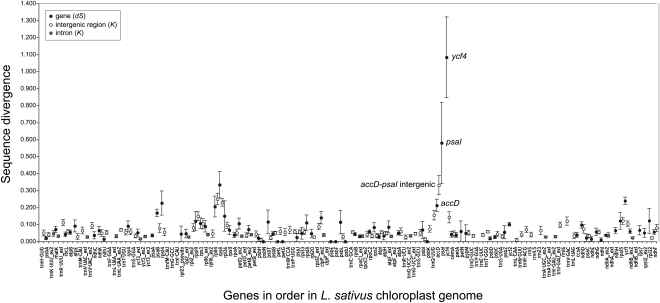
We measured synonymous divergence in each protein-coding gene between the P. sativum and L. sativus chloroplast genomes using dS (black circle symbols in Fig. 4), calculated by the yn00 program (Yang 2007). For most loci, the divergence between these species is less than 0.1 substitutions per site (median dS = 0.055 synonymous substitutions per site). Ycf4 cannot be included directly in this comparison because it is absent from P. sativum cpDNA, so instead, for ycf4 in Figure 4 we have plotted the dS value between L. palustris and L. sativus, which is 20-fold higher (1.084) than the median even though the comparison is over a shorter divergence time. We observed even higher dS values in comparing ycf4 between L. palustris and L. cirrhosus (1.481) or L. latifolius (1.522). Similarly, psaI is missing from L. sativus cpDNA so instead we compared P. sativum psaI to L. palustris psaI and found dS = 0.580, which again is much higher than the genome average for P. sativum versus L. sativus (0.055). For accD we compared only the regions of the gene that could be reliably aligned between P. sativum and L. sativus, and obtained a dS value (0.212) that is 3.8 times the genome average. This spike in local dS values is matched by a local increase in divergence in the intergenic regions near the ycf4 and accD loci (all compared between P. sativum and L. sativus using Kimura's K; open symbols in Fig. 4).
Figure 4.
Sequence divergence between the P. sativum and L. sativus chloroplast genomes. The x-axis lists genes or exons in the order in which they occur in the L. sativus genome. Black filled circles show dS (number of synonymous substitutions per synonymous site) for each orthologous protein gene pair, calculated using yn00 (Yang 2007). White and gray filled circles show divergence (K) for each intergenic region or intron, respectively, calculated by Kimura's two-parameter method (Kimura 1983). Vertical bars, dS or K ± 1 SE. Because ycf4 is a pseudogene in P. sativum and psaI is not present in L. sativus, the dS value plotted for ycf4 is for a comparison between L. sativus and L. palustris, and the dS value plotted for psaI is for a comparison between P. sativum and L. palustris (see text). No divergence values are plotted for intergenic regions that are not flanked by the same genes in the two species or that are shorter than 100 bp.
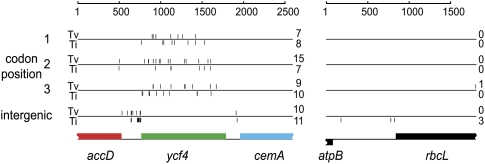
The very high level of synonymous substitution in ycf4 made us question whether the mutational process at this locus might somehow be different than elsewhere in the genome. We investigated this possibility by sequencing regions of cpDNA from L. latifolius and L. cirrhosus (Fig. 3), two species that are evidently very closely related because there is only 1 nucleotide (nt) substitution between their rbcL genes, and only three substitutions in the atpB-rbcL intergenic spacer (Fig. 5). There are only two differences out of 1256 bp in the combined partial accD and cemA sequences obtained from these species, compared with 56 differences in the 1023-bp-long ycf4 (dS = 0.048, dN = 0.039). Most strikingly, there are 19 differences (10% divergence) in the spacer between accD and ycf4. This spacer (from which psaI has been lost) is 238 bp in L. cirrhosus, most of which can be aligned to L. latifolius, but in L. latifolius the spacer has expanded to 648 bp due to the presence of multiple tandem repeat sequences comprising a 57-bp repeat unit (six complete and three partial copies) and a 67-bp repeat unit (two complete and one partial copy) (Supplemental Fig. S5). These results show that a region of ∼1500 bp in these Lathyrus genomes, extending through the accD-ycf4 spacer and most if not all of ycf4 itself, is a hotspot with a mutation rate that is dramatically higher than in the rest of the genome. Despite this high mutation rate, the types of nucleotide substitution occurring in the ycf4 region do not seem particularly biased, with an overall transition/transversion ratio of 0.9 (Fig. 5).
Figure 5.
Sequence divergence between L. latifolius and L. cirrhosus in the accD-ycf4-cemA region (left) and the atpB-rbcL region (right). Vertical tickmarks indicate the locations of each nucleotide substitution, categorized according to whether it occurs at codon position 1, 2, or 3; or in intergenic DNA; and as a transversion (Tv; tickmarks above the horizontal lines) or a transition (Ti; tickmarks below the horizontal line). The total numbers of each type of substitution are shown on the right. Supplemental Figure S5 shows the nucleotide sequence alignment summarized in the left panel.
There also appears to be a smaller second peak of divergence values in the region around the genes rpl14 and rps8 (Fig. 4), which is the site from which infA was lost in an early ancestor of Fabales and Cucurbitales (Millen et al. 2001). We found that sites of gene loss coincide with fast-evolving intergenic regions more often than expected by chance: Five of the six most divergent intergenic regions between P. sativum and L. sativus are also the sites of five of their six gene losses (ycf4, psaI, rps16, infA, rpl22; P = 2 × 10−6 by the hypergeometric test).
Mutation rate in ycf4 exceeds the rate in the nuclear genome
Early work on rates of nucleotide substitution in plant genomes concluded that the synonymous substitution rate (assumed to be equal to the mutation rate) is about four times higher in plant nuclear genomes than in the single-copy regions of chloroplast genomes (Wolfe et al. 1987, 1989; Gaut 1998; Muse 2000). Given the heterogeneity of synonymous rates seen within the Lathyrus chloroplast genomes, we wondered how these rates compared with the rate in the nuclear genome. Relatively few nuclear genes have been sequenced from Lathyrus species, so we generated new expressed sequence tag (EST) data from Lathyrus odoratus (sweetpea; see below) and identified putatively orthologous nuclear genes between these and database sequences from P. sativum. Among 56 putative orthologs, the median dS is 0.131 (Supplemental Table S2), which is 2.4 times higher than the median dS (0.055) for chloroplast genes compared between P. sativum and L. sativus. Thus in comparisons between Lathyrus and P. sativum, as in other flowering plant comparisons, the synonymous divergence in most parts of the chloroplast genome is lower than in the nuclear genome. The synonymous divergence in ycf4, however, is at least 10 times greater than in the nuclear genome (the ratios of the dS values given above, 1.084/0.131 = 8.3 and 1.522/0.131 = 11.6, are underestimates of the actual ratio because the numerators involve a shorter divergence time).
Transfer of Trifolium accD to the nucleus
We suspect that ycf4 and psaI have been transferred to the nuclear genome in the Lathyrus species that lack them in cpDNA, because these species are fully photosynthetic and must have a functional photosystem I. However, we were unable to find nuclear copies of these genes. We made numerous unsuccessful attempts (see Methods) to amplify ycf4 and psaI by PCR from genomic DNA of L. odoratus (which lacks both of them in its cpDNA and has a smaller ycf4 pseudogene than P. sativum). We then made cDNA from young green leaves of L. odoratus and sequenced 8702 ESTs. None of the ESTs were derived from a nuclear ycf4 or psaI, even though we did find ESTs corresponding to seven of the nine other nuclear-encoded subunits of photosystem I (Jolley et al. 2005), and to the older nuclear-transferred genes infA and rpl22. PsaI is a very small protein (34–40 amino acids) that is conserved between cyanobacteria and land plants and is physically located toward the exterior of photosystem I in P. sativum, where it interacts strongly with PsaH (Jolley et al. 2005; Amunts et al. 2007). It seems unlikely that photosystem I in Lathyrus could function efficiently without PsaI, although tobacco plants with a psaI knockout do not show a mutant phenotype under standard growth conditions (MA Schöttler and R Bock, pers. comm.). Most of the small membrane-spanning subunits of photosystem I appear to be nonessential, and knockout lines do not display visible mutant phenotypes (Varotto et al. 2002; Jensen et al. 2007; Schöttler et al. 2007). However, the loss of individual small membrane-spanning subunits usually affects the assembly of other subunits and results in lower efficiencies of excitation transfer and electron transfer (Varotto et al. 2002; Jensen et al. 2007; Schöttler et al. 2007), which would be evolutionarily deleterious. The only other known cases of loss of psaI from plastid DNA are in the parasitic species Cuscuta gronovii and Cuscuta obtusiflora which have reduced levels of photosynthesis but retain all other photosynthesis genes (Funk et al. 2007; McNeal et al. 2007), and in the nonphotosynthetic parasite Epifagus (Wolfe et al. 1992).
Although we have direct evidence for association between gene losses and a mutation hotspot only in the genus Lathryus, it is intriguing that other species in the IRLC legume clade show evolutionary losses of other genes that neighbor ycf4 and psaI (Fig. 3). The loss of rps16 in the common ancestor of the IRLC clade can be explained in terms of gene substitution by the nuclear gene for mitochondrial RPS16, as already demonstrated for Medicago (Ueda et al. 2008), and so does not necessitate a gene transfer to the nucleus. Rps16 has been lost on multiple independent occasions during land plant evolution (Supplemental Table S1; Ohyama et al. 1986; Tsudzuki et al. 1992; Ueda et al. 2008), so it is possible that its multiple losses in legumes are simply the result of relatively easy and/or early substitution by the mitochondrial gene.
The other IRLC legume gene loss in the neighborhood of ycf4 and psaI is the loss of accD in Trifolium (Fig. 3). AccD codes for a subunit of acetyl-CoA carboxylase, which functions in lipid synthesis and is an essential chloroplast gene in tobacco (Kode et al. 2005). The loss in Trifolium is one of five separate known instances of loss of accD in angiosperm cpDNAs (Supplemental Table S1). In grass species—the only case that has been studied in detail—the prokaryotic multisubunit carboxylase in the plastid has been completely replaced by a nuclear-encoded single-chain carboxylase of eukaryotic ancestry (Konishi et al. 1996; Gornicki et al. 1997). We identified an evolutionary transfer of accD to the nucleus in Trifolium. Using high-throughput EST sequence data from Trifolium repens (white clover), we found a cDNA structure consisting of a fusion between a gene for plastid lipoamide dehydrogenase (LPD2) and accD (Supplemental Fig. S6A–D). We confirmed the presence of a fused mRNA by reverse transcriptase PCR and Sanger sequencing (Supplemental Fig. S6E).
In plastids, lipoamide dehydrogenase is a component of pyruvate dehydrogenase, a complex that makes acetyl-CoA (Lutziger and Oliver 2000; Drea et al. 2001). The T. repens nuclear transcript codes for a predicted protein of 805 amino acids, with residues 1–512 (including a transit peptide) derived from LPD2 and residues 513–805 derived from accD. By comparison to the known genomic structures of LPD genes in M. truncatula, we infer that in T. repens the accD sequence has replaced the final two exons (exons 14 and 15) of its LPD2 gene, with the point of fusion occurring at the third codon of exon 14. We did not find any evidence for alternative splicing of the LPD2–accD fusion to form two products, as occurs with the SOD–rpl32 fusion in mangrove trees (Cusack and Wolfe 2007). The fusion to accD probably rendered LPD2 unable to code for functional lipoamide dehydrogenase, because the fusion protein lacks some conserved residues normally provided by exons 14 and 15, but T. repens retains and expresses a paralogous gene LPD1 that also codes for plastid lipoamide dehydrogenase (Supplemental Fig. S6A). We found the transferred gene in T. repens, but we presume that the transfer is shared by other Trifolium species, including the two that have been demonstrated to have no accD in their cpDNAs (T. subterraneum and Trifolium pratense) (Doyle et al. 1995; Cai et al. 2008). We also found database ESTs for a nuclear accD in T. pratense (red clover), but they are too short to confirm that this species also has the LPD2–accD fusion. Phylogenetic analysis indicates that the T. repens and T. pratense nuclear sequences have a monophyletic origin and that the transfer of accD to the nucleus occurred within the IRLC clade (Supplemental Fig. S6F), consistent with the change in LPD2 gene structure that occurred after Trifolium diverged from Medicago (Supplemental Fig. S6A). The Trifolium nuclear accD gene is transcribed in both T. repens and T. pratense, is predicted to have a functional transit peptide in T. repens (TargetP cTP score 0.976) (Emanuelsson et al. 2000), and shows evidence of selection to maintain its AccD-coding function (dN/dS = 0.26 between T. repens and T. pratense in the accD region of the transcript). Moreover, the Trifolium nuclear mRNAs code for a leucine residue at a site that undergoes an essential Ser → Leu mRNA edit in P. sativum plastids (Supplemental Fig. S6D; Sasaki et al. 2001; Inada et al. 2004).
Discussion
The genomic region around ycf4 in Lathyrus is a dramatic hotspot for point mutations. It is difficult to quantify the factor by which its mutation rate is increased relative to the rest of the genome, but comparisons of synonymous site divergence indicate an increase of at least 20-fold, both in comparisons between P. sativum and L. sativus (Fig. 4) and among Lathyrus species (Table 1). Between L. latifolius and L. cirrhosus, the increase may be even greater (Fig. 5; Table 1). Even a 20-fold mutation rate increase only goes partway toward explaining how the protein sequence divergence between L. palustris and L. cirrhosus (with a divergence time of <10 Myr) (Kenicer et al. 2005) exceeds that between other angiosperms and cyanobacteria (separated by >1000 Myr); a relaxation of selective constraints on the Ycf4 protein in legumes must be involved too. Although there have been previous reports that the variance of synonymous substitution rates among genes in many eukaryotic genomes is greater than expected by chance (e.g., Baer et al. 2007; Fox et al. 2008), there are few if any precedents for the phenomenon that we describe here—a sharply localized mutation rate acceleration of great magnitude in one specific region of a genome. The existence of the hotspot violates the common assumption that the point mutation rate is approximately constant in all regions of the same genome (Kimura 1983), which underpins the silent molecular clock hypothesis (Ochman and Wilson 1987). Our results bear some similarities to the “mutation showers” (transient localized hypermutation events) that have been found in some studies on the genomic distribution of spontaneous mutations (Drake 2007; Wang et al. 2007; Nishant et al. 2009). As well as being a mutation hotspot, ycf4 and its neighbors also appear to be a hotspot for the formation and turnover of minisatellite sequences in Lathyrus.
The previous study most relevant to our findings is that of Erixon and Oxelman (2008), who reported somewhat similar results for the chloroplast clpP gene in Silene and Oenothera species. For some interspecies comparisons in their study, both dN and dS were elevated in clpP compared with other chloroplast genes, although the dS elevations were at most fivefold for clpP, compared with at least 20-fold for ycf4 in Lathyrus. Also, insertions of repetitive amino acid sequence regions occurred in some of the fast-evolving taxa. Locus-specific rate accelerations affecting both dN and dS were reported in cpDNA of Geraniaceae, but in this case, the accelerations occurred in numerous genes (Guisinger et al. 2008). In all IR-containing cpDNAs, the synonymous rate is higher in single-copy genes than in IR-located genes, probably due to a copy-number effect during DNA repair (Wolfe et al. 1987; Birky and Walsh 1992; Perry and Wolfe 2002). Dramatic accelerations of synonymous rates have been found in the mitochondrial genomes of some plants, such as Plantago, Pelargonium, and certain Silene species (Cho et al. 2004; Parkinson et al. 2005; Mower et al. 2007; Sloan et al. 2009). Most of these mitochondrial accelerations appear to affect all genes in the genome similarly, but among-gene rate heterogeneity was found within the mtDNAs of a few species (Mower et al. 2007), including a 40-fold difference in synonymous rates between atp9 and three other mitochondrial genes in Silene (Sloan et al. 2009). Because plant mitochondrial genomes are relatively large and do not show much gene order conservation, most studies have only examined individual genes so the sizes of the genomic regions affected by rate acceleration are not known.
Apart from these organellar examples, there are very few precedents for a mutation rate change that is so pronounced over such a short physical distance. One early study (Martin and Meyerowitz 1986) reported a 2-kb region of noncoding DNA near the glue gene cluster of three Drosophila species, which contained an abrupt boundary between a conserved region and a nonconserved region with a 10-fold elevated substitution rate, but this report has not been followed up with more extensive analyses based on complete genome sequence data. An abrupt boundary of evolutionary rates also occurs on the mammalian X chromosome at the junction between the pseudoautosomal region and the X-specific region. The pseudoautosomal part of the gene Fxy, which spans this junction in laboratory mice, has a synonymous rate about 60 times faster than the X-specific part of the gene, probably because the high recombination rate in the pseudoautosomal part leads to high levels of biased gene conversion (Perry and Ashworth 1999; Duret and Galtier 2009).
Is the chloroplast hypermutation phenomenon unique to Lathyrus? At present, Lathyrus is the only legume genus for which we have extensive sequence data from more than one species, so we are unable to say whether the same hotspot is present in legumes outside this genus. Therefore the only gene losses we can potentially attribute directly to hypermutation are those of ycf4 in L. odoratus and of psaI in the ancestor of four Lathyrus species. Ycf4 is also evolving fast in Desmodium and has been lost in three species of that genus. The losses of ycf4 in P. sativum, of accD in Trifolium, and the older loss of rps16 in the ancestor of the IRLC clade are suggestive, but we have no direct evidence that these loci were fast-evolving prior to the gene losses. It is possible that a hotspot has existed throughout legume evolution and was the cause of the ycf4 acceleration seen in the common ancestor of Millettioids, Robinioids, and the IRLC (Fig. 1) but that the exact location of the hotspot (and its associated tandem repeat sequences) has varied somewhat among lineages, affecting ycf4 in some taxa, but accD or psaI in others. We do not know the molecular basis for the increases in either the point mutation rate or the length mutation rate, but we speculate that they might be connected. We suggest that a correlation between the two rates could develop if, for some reason, the genomic region around ycf4 was subject to repeated DNA breakage and repair (cf. Guisinger et al. 2008; Yang et al. 2008). In this regard, it is interesting to note that only a few angiosperm species have cpDNAs that are highly rearranged relative to the canonical gene order, but among these, there are several independent lineages that are both highly rearranged and contain rapidly-evolving protein genes (Jansen et al. 2007). These lineages include Jasminum (acceleration of accD; Lee et al. 2007), Silene (acceleration of clpP; Erixon and Oxelman 2008), and now Lathyrus (acceleration of ycf4). The phylogenetic diversity of these lineages suggests that hypermutable regions may exist in other angiosperm cpDNAs, and our findings may go some way toward explaining the apparent bursts of organelle-to-nucleus gene transfer seen in some angiosperms.
It is likely that many factors dictate whether a gene can be lost from an organelle genome. One property that is common to the gene transfer and gene substitution processes is that they both involve a phase during which the organelle gene and the nuclear gene coexist in the same species (Timmis et al. 2004). Analogous to a gene duplication, this two-gene phase can be resolved either by losing the organelle copy (resulting in a successful transfer of function) or by losing the nuclear copy (restoring the status quo). Intermediates in this process, and sister lineages where the two-gene phase was resolved in opposite ways, have been identified (Adams et al. 1999). Brandvain and Wade (2009) have shown theoretically that the ratio between the point mutation rates in the organelle and nuclear copies has a profound influence on the direction in which the two-gene phase is resolved. If the organelle mutation rate is lower than the nuclear mutation rate, as is true for most plant mitochondrial and chloroplast genes, then gene transfer will not occur unless there is a benefit to relocating the gene. By contrast, if the organelle rate exceeds the nuclear rate, then gene transfer is predicted to occur even in the absence of any benefit (Brandvain and Wade 2009). Therefore, in a genome such as Lathyrus cpDNA, in which the mutation rate exceeds the nuclear rate only in one hypermutable region, we should expect to see more transfers, substitutions, or losses of genes from the hypermutable region than from the rest of the genome. This argument provides a plausible explanation for the losses of ycf4 and psaI seen in some Lathyrus species and, perhaps more generally, for the cluster of losses from the rps16-accD-psaI-ycf4 region seen in other legume cpDNAs.
Methods
Plant material
Seeds of Lathyrus sativus (cv. Cicerchia Marchigiana) were purchased from B&T World Seeds. Seeds of L. cirrhosus (accession no. LAT17) were obtained from the Leibniz Institute of Plant Genetics and Crop Plant Research. Other Lathyrus species were purchased from Thompson & Morgan. Additional sequencing of P. sativum cpDNA was done using cv. Feltham First.
Nucleotide sequencing
The P. sativum (pea) chloroplast genome sequence was completed by S.A. and J.C.G. using the chain termination method (Sanger et al. 1977) with fluorescent dideoxynucleotides on PCR products amplified from cloned PstI fragments (Palmer and Thompson 1981), from cpDNA extracted from isolated chloroplasts, or from total DNA extracted from shoots of 8-d-old seedlings. Chloroplasts were isolated by the high-salt method (Bookjans et al. 1984), and DNA was extracted by the CTAB (hexadecyltrimethylammonium bromide) method (Milligan 1989). Previously published regions were not resequenced except at the borders or where there were discrepancies between publications. Newly sequenced regions were completed on both strands, and all the PstI sites used for cloning were confirmed by sequencing spanning PCR fragments. At the ycf4 locus, there is a 2-bp deletion in the sequence reported by Nagano et al. (1991a), relative to the sequence reported by Smith et al. (1991), both of which were obtained from the same cloned 17.3-kb PstI fragment from P. sativum cv. Alaska. We confirmed that this 2-bp deletion exists, in both cv. Alaska and cv. Feltham First. This correction means that the reported ORF157 (Smith et al. 1991) does not exist.
The L. sativus (grasspea) chloroplast genome sequence was determined by A.M.M., T.A.K., and K.H.W. Approximately 150 seeds were grown on soil in the greenhouse. Seedling shoots were harvested at 7 d post-germination, and cpDNA was prepared according to the method described by Milligan (1989) except that chloroplast lysis and cpDNA recovery procedures were modified. Chloroplasts were lysed by adding a 1/5 volume of 10% CTAB (Sigma-Aldrich) and heating for 20 min at 70°C. This was followed by a chloroform extraction, treatment with RNaseA (10 μg/mL), and isopropanol precipitation of cpDNA. A plasmid library of nebulized fragments was constructed from 5 μg of cpDNA by GATC-Biotech. The genome sequence was assembled from 1536 Sanger shotgun sequence reads with primer-walking to close gaps.
We tried unsuccessfully to amplify ycf4 and psaI by PCR from L. odoratus genomic DNA using 16 and 9 primer combinations, respectively, and a range of amplification conditions. These primers were designed based on amino acid residues conserved among known Fabaceae Ycf4 and PsaI proteins, but primer design for these genes is difficult due to the fast rate of ycf4 evolution and the short length of psaI, as well as the high A+T content of the region. To obtain EST data from L. odoratus, we isolated poly(A) mRNA from leaves of 3-d-old seedlings. A normalized cDNA library was constructed by GATC-Biotech, and the 3′ ends of 8702 cDNAs were sequenced by Agencourt Biosciences. ESTs were assembled into contigs, and putative orthologs between these contigs and P. sativum sequence data from GenBank were identified according the method of Sémon and Wolfe (2008).
The other new sequence data indicated in Figures 3 and 5 were generated by PCR amplification and sequencing (by primer walking) of at least three independent cloned products for each region. The cpDNA region that normally contains ycf4 was PCR amplified from L. latifolius, L. cirrhosus, L. odoratus, and L. palustris using primers designed from the P. sativum accD (5′-AAACAGGCACAGGTCAASTAAATGG-3′) and cemA (5′-GACGGAGATACACGATTTAAATAACG-3′) genes. The atpB–rbcL region from L. latifolius, L. cirrhosus, and L. palustris was amplified with primers 5′-TGRAAAARCTACATCGAGTACCGGAGG-3′ and 5′-TATGATCTCCACCAGACATACG-3′. T. repens mRNA sequences coding for LPD1 and the LPD2–accD fusion gene were identified among 700,000 ESTs obtained by high-throughput pyrosequencing of flower, leaf, and stolon mRNA from the inbred line S (7S.4.6.3.3.4.4.10) (DM and SB, unpubl.) and assembled manually. The structure of the LPD2-accD junction was confirmed by reverse transcriptase-PCR from T. repens leaf mRNA (commercial variety Nusiral) and Sanger sequencing.
Computational methods
Sequence divergence for most analyses was calculated using yn00 from the PAML package (Yang 2007) for coding regions and Kimura's two-parameter method (Kimura 1983) for noncoding regions. Gene sequences were aligned by reverse-translation of ClustalW alignments of the corresponding protein sequences. Noncoding sequences were aligned using ClustalW with manual adjustment for regions around ycf4. For the analysis in Figure 1 and Supplemental Figure S1, we first constructed a maximum likelihood phylogeny (in PAUP) from matK sequences using the HKY substitution model with a four category gamma rate distribution. The transition/transversion ratio and shape parameter were estimated iteratively until the topology converged. This analysis included legume ycf4 sequences from Stefanovic et al. (2009; GenBank [http://www.ncbi.nlm.nih.gov/Genbank/] accession nos. EU717431–EU717464). The dN and dS branch lengths for the matK, ycf4, and rbcL trees were estimated based on the PAML/codeML free-ratio model, using the fixed topology obtained from the above matK ML analysis. Dot-matrix plots were made using DNAMAN (http://www.lynnon.com) (Huang and Zhang 2004).
Acknowledgments
We thank Gavin Conant for help with Figure S3, Shusei Sato for Trifolium cDNA clones, Greg Kenicer for prepublication access to sequence data, and Ralph Bock for discussion. S.A. and J.C.G. thank Chris Maddren (Department of Genetics, University of Cambridge) for DNA sequencing. This study was supported by Science Foundation Ireland (K.H.W., T.A.K.), European Commission FP5 Plastid Factory (J.C.G., T.A.K.), US National Institutes of Health (J.D.P.), and the METACyt Initiative of Indiana University, funded in part through a major grant from the Lilly Endowment, Inc. (J.D.P., D.W.R.).
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession nos. HM029359–HM029371, HM048906–HM048910, and GO313838–GO322539.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.111955.110.
References
- Adams KL, Palmer JD 2003. Evolution of mitochondrial gene content: Gene loss and transfer to the nucleus. Mol Phylogenet Evol 29: 380–395 [DOI] [PubMed] [Google Scholar]
- Adams KL, Song K, Roessler PG, Nugent JM, Doyle JL, Doyle JJ, Palmer JD 1999. Intracellular gene transfer in action: Dual transcription and multiple silencings of nuclear and mitochondrial cox2 genes in legumes. Proc Natl Acad Sci 96: 13863–13868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts A, Drory O, Nelson N 2007. The structure of a plant photosystem I supercomplex at 3.4 Å resolution. Nature 447: 58–63 [DOI] [PubMed] [Google Scholar]
- Asmussen CB, Liston A 1998. Chloroplast DNA characters, phylogeny, and classification of Lathyrus (Fabaceae). Am J Bot 85: 387–401 [PubMed] [Google Scholar]
- Baer CF, Miyamoto MM, Denver DR 2007. Mutation rate variation in multicellular eukaryotes: Causes and consequences. Nat Rev Genet 8: 619–631 [DOI] [PubMed] [Google Scholar]
- Birky CW Jr, Walsh JB 1992. Biased gene conversion, copy number, and apparent mutation rate differences within chloroplast and bacterial genomes. Genetics 130: 677–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookjans G, Stummann BM, Henningsen KW 1984. Preparation of chloroplast DNA from pea plastids isolated in a medium of high ionic strength. Anal Biochem 141: 244–247 [DOI] [PubMed] [Google Scholar]
- Boudreau E, Takahashi Y, Lemieux C, Turmel M, Rochaix JD 1997. The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J 16: 6095–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandvain Y, Wade MJ 2009. The functional transfer of genes from the mitochondria to the nucleus: The effects of selection, mutation, population size and rate of self-fertilization. Genetics 182: 1129–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Guisinger M, Kim HG, Ruck E, Blazier JC, McMurtry V, Kuehl JV, Boore J, Jansen RK 2008. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J Mol Evol 67: 696–704 [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Small I 2007. A rapid high-throughput method for the detection and quantification of RNA editing based on high-resolution melting of amplicons. Nucleic Acids Res 35: e114 doi: 10.1093/nar/gkm640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Mower JP, Qiu YL, Palmer JD 2004. Mitochondrial substitution rates are extraordinarily elevated and variable in a genus of flowering plants. Proc Natl Acad Sci 101: 17741–17746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack BP, Wolfe KH 2007. When gene marriages don't work out: Divorce by subfunctionalization. Trends Genet 23: 270–272 [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL, Palmer JD 1995. Multiple independent losses of two genes and one intron from legume chloroplast genomes. Syst Bot 20: 272–294 [Google Scholar]
- Drake JW 2007. Too many mutants with multiple mutations. Crit Rev Biochem Mol Biol 42: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drea SC, Mould RM, Hibberd JM, Gray JC, Kavanagh TA 2001. Tissue-specific and developmental-specific expression of an Arabidopsis thaliana gene encoding the lipoamide dehydrogenase component of the plastid pyruvate dehydrogenase complex. Plant Mol Biol 46: 705–715 [DOI] [PubMed] [Google Scholar]
- Drouin G, Daoud H, Xia J 2008. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol Phylogenet Evol 49: 827–831 [DOI] [PubMed] [Google Scholar]
- Duret L, Galtier N 2009. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet 10: 285–311 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Erixon P, Oxelman B 2008. Whole-gene positive selection, elevated synonymous substitution rates, duplication, and indel evolution of the chloroplast clpP1 gene. PLoS ONE 3: e1386 doi: 10.1371/journal.pone.0001386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AK, Tuch BB, Chuang JH 2008. Measuring the prevalence of regional mutation rates: An analysis of silent substitutions in mammals, fungi, and insects. BMC Evol Biol 8: 186 doi: 10.1186/1471-2148-8-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk HT, Berg S, Krupinska K, Maier UG, Krause K 2007. Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol 7: 45 doi: 10.1186/1471-2229-7-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantt JS, Baldauf SL, Calie PJ, Weeden NF, Palmer JD 1991. Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J 10: 3073–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS 1998. Molecular clocks and nucleotide substitution rates in higher plants. Evol Biol 30: 93–120 [Google Scholar]
- Gaut BS, Muse SV, Clegg MT 1993. Relative rates of nucleotide substitution in the chloroplast genome. Mol Phylogenet Evol 2: 89–96 [DOI] [PubMed] [Google Scholar]
- Gornicki P, Faris J, King I, Podkowinski J, Gill B, Haselkorn R 1997. Plastid-localized acetyl-CoA carboxylase of bread wheat is encoded by a single gene on each of the three ancestral chromosome sets. Proc Natl Acad Sci 94: 14179–14184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O 2010. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27: 221–224 [DOI] [PubMed] [Google Scholar]
- Graur D, Li W-H 1999. Fundamentals of molecular evolution. Sinauer, Sunderland, MA [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK 2008. Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc Natl Acad Sci 105: 18424–18429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Castillo-Ramirez S, Gonzales V, Bustos P, Fernandez-Vazquez JL, Santamaria RI, Arellano J, Cevallos MA, Davila G 2007. Rapid evolutionary change of common bean (Phaseolus vulgaris L.) plastome and genomic diversification of legume chloroplasts. BMC Genomics 8: 228 doi: 10.1186/1471-2164-8-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Zhang L 2004. Rapid and sensitive dot-matrix methods for genome analysis. Bioinformatics 20: 460–466 [DOI] [PubMed] [Google Scholar]
- Inada M, Sasaki T, Yukawa M, Tsudzuki T, Sugiura M 2004. A systematic search for RNA editing sites in pea chloroplasts: An editing event causes diversification from the evolutionarily conserved amino acid sequence. Plant Cell Physiol 45: 1615–1622 [DOI] [PubMed] [Google Scholar]
- Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Muller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci 104: 19369–19374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Wojciechowski MF, Sanniyasi E, Lee SB, Daniell H 2008. Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae). Mol Phylogenet Evol 48: 1204–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PE, Bassi R, Boekema EJ, Dekker JP, Jansson S, Leister D, Robinson C, Scheller HV 2007. Structure, function and regulation of plant photosystem I. Biochim Biophys Acta 1767: 335–352 [DOI] [PubMed] [Google Scholar]
- Jolley C, Ben-Shem A, Nelson N, Fromme P 2005. Structure of plant photosystem I revealed by theoretical modeling. J Biol Chem 280: 33627–33636 [DOI] [PubMed] [Google Scholar]
- Kato T, Kaneko T, Sato S, Nakamura Y, Tabata S 2000. Complete structure of the chloroplast genome of a legume, Lotus japonicus. DNA Res 7: 323–330 [DOI] [PubMed] [Google Scholar]
- Kenicer GJ, Kajita T, Pennington RT, Murata J 2005. Systematics and biogeography of Lathyrus (Leguminosae) based on internal transcribed spacer and cpDNA sequence data. Am J Bot 92: 1199–1209 [DOI] [PubMed] [Google Scholar]
- Kim K-J, Lee H-L 2004. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees). Comparative analysis of sequence evolution among 17 vascular plants. DNA Res 11: 247–261 [DOI] [PubMed] [Google Scholar]
- Kimura M 1977. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature 267: 275–276 [DOI] [PubMed] [Google Scholar]
- Kimura M 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, UK [Google Scholar]
- Kode V, Mudd EA, Iamtham S, Day A 2005. The tobacco plastid accD gene is essential and is required for leaf development. Plant J 44: 237–244 [DOI] [PubMed] [Google Scholar]
- Konishi T, Shinohara K, Yamada K, Sasaki Y 1996. Acetyl-CoA carboxylase in higher plants: Most plants other than gramineae have both the prokaryotic and the eukaryotic forms of this enzyme. Plant Cell Physiol 37: 117–122 [DOI] [PubMed] [Google Scholar]
- Lee HL, Jansen RK, Chumley TW, Kim KJ 2007. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol Biol Evol 24: 1161–1180 [DOI] [PubMed] [Google Scholar]
- Lutziger I, Oliver DJ 2000. Molecular evidence of a unique lipoamide dehydrogenase in plastids: Analysis of plastidic lipoamide dehydrogenase from Arabidopsis thaliana. FEBS Lett 484: 12–16 [DOI] [PubMed] [Google Scholar]
- Martin CH, Meyerowitz EM 1986. Characterization of the boundaries between adjacent rapidly and slowly evolving genomic regions in Drosophila. Proc Natl Acad Sci 83: 8654–8658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal JR, Kuehl JV, Boore JL, Depamphilis CW 2007. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol 7: 57 doi: 10.1186/1471-2229-7-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen RS, Olmstead RG, Adams KL, Palmer JD, Lao NT, Heggie L, Kavanagh TA, Hibberd JM, Gray JC, Morden CW, et al. 2001. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13: 645–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan BG 1989. Purification of chloroplast DNA using hexadecyltrimethylammonium bromide. Plant Mol Biol Rep 7: 144–149 [Google Scholar]
- Milligan BG, Hampton JN, Palmer JD 1989. Dispersed repeats and structural reorganization in subclover chloroplast DNA. Mol Biol Evol 6: 355–368 [DOI] [PubMed] [Google Scholar]
- Mower JP, Touzet P, Gummow JS, Delph LF, Palmer JD 2007. Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol Biol 7: 135 doi: 10.1186/1471-2148-7-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse SV 2000. Examining rates and patterns of nucleotide substitution in plants. Plant Mol Biol 42: 25–43 [PubMed] [Google Scholar]
- Nagano Y, Matsuno R, Sasaki Y 1991a. Sequence and transcriptional analysis of the gene cluster trnQ-zfpA-psaI-ORF231-petA in pea chloroplasts. Curr Genet 20: 431–436 [DOI] [PubMed] [Google Scholar]
- Nagano Y, Ishikawa H, Matsuno R, Sasaki Y 1991b. Nucleotide sequence and expression of the ribosomal protein L2 gene in pea chloroplasts. Plant Mol Biol 17: 541–545 [DOI] [PubMed] [Google Scholar]
- Nishant KT, Singh ND, Alani E 2009. Genomic mutation rates: What high-throughput methods can tell us. BioEssays 31: 912–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Wilson AC 1987. Evolution in bacteria: Evidence for a universal substitution rate in cellular genomes. J Mol Evol 26: 74–86 [DOI] [PubMed] [Google Scholar]
- Ohyama K, Fukuzawa H, Kohchi T, Shirai H, Sano T, Sano S, Umesono K, Shiki Y, Takeuchi M, Chang Z, et al. 1986. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 322: 572–574 [Google Scholar]
- Onishi T, Takahashi Y 2009. Effects of site-directed mutations in the chloroplast-encoded ycf4 gene on photosystem I complex assembly in the green alga Chlamydomonas reinhardtii. Plant Cell Physiol 50: 1750–1760 [DOI] [PubMed] [Google Scholar]
- Ozawa SI, Nield J, Terao A, Stauber EJ, Hippler M, Koike H, Rochaix JD, Takahashi Y 2009. Biochemical and structural studies of the large Ycf4-photosystem I assembly complex of the green alga Chlamydomonas reinhardtii. Plant Cell 21: 2424–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD 1985. Comparative organization of chloroplast genomes. Annu Rev Genet 19: 325–354 [DOI] [PubMed] [Google Scholar]
- Palmer JD, Thompson WF 1981. Clone banks of the mung bean, pea and spinach chloroplast genomes. Gene 15: 21–26 [DOI] [PubMed] [Google Scholar]
- Palmer JD, Osorio B, Thompson WF 1988. Evolutionary significance of inversions in legume chloroplast DNAs. Curr Genet 14: 65–74 [Google Scholar]
- Palmer JD, Adams KL, Cho Y, Parkinson CL, Qiu YL, Song K 2000. Dynamic evolution of plant mitochondrial genomes: Mobile genes and introns and highly variable mutation rates. Proc Natl Acad Sci 97: 6960–6966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson CL, Mower JP, Qiu YL, Shirk AJ, Song K, Young ND, DePamphilis CW, Palmer JD 2005. Multiple major increases and decreases in mitochondrial substitution rates in the plant family Geraniaceae. BMC Evol Biol 5: 73 doi: 10.1186/1471-2148-5-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J, Ashworth A 1999. Evolutionary rate of a gene affected by chromosomal position. Curr Biol 9: 987–989 [DOI] [PubMed] [Google Scholar]
- Perry AS, Wolfe KH 2002. Nucleotide substitution rates in legume chloroplast DNA depend on the presence of the inverted repeat. J Mol Evol 55: 501–508 [DOI] [PubMed] [Google Scholar]
- Reverdatto SV, Beilinson V, Nielsen NC 1995. The rps16, accD, psaI, ORF 203, ORF 151, ORF 103, ORF 229 and petA gene cluster in the chloroplast genome of soybean (PGR95-051). Plant Physiol 109: 338 [Google Scholar]
- Sanger F, Nicklen S, Coulson AR 1977. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci 74: 5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Kozaki A, Ohmori A, Iguchi H, Nagano Y 2001. Chloroplast RNA editing required for functional acetyl-CoA carboxylase in plants. J Biol Chem 276: 3937–3940 [DOI] [PubMed] [Google Scholar]
- Saski C, Lee SB, Daniell H, Wood TC, Tomkins J, Kim HG, Jansen RK 2005. Complete chloroplast genome sequence of Glycine max and comparative analyses with other legume genomes. Plant Mol Biol 59: 309–322 [DOI] [PubMed] [Google Scholar]
- Schöttler MA, Flugel C, Thiele W, Stegemann S, Bock R 2007. The plastome-encoded PsaJ subunit is required for efficient photosystem I excitation, but not for plastocyanin oxidation in tobacco. Biochem J 403: 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sémon M, Wolfe KH 2008. Preferential subfunctionalization of slow-evolving genes in Xenopus laevis. Proc Natl Acad Sci 105: 8333–8338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Oxelman B, Rautenberg A, Taylor DR 2009. Phylogenetic analysis of mitochondrial substitution rate variation in the angiosperm tribe Sileneae. BMC Evol Biol 9: 260 doi: 10.1186/1471-2148-9-260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AG, Wilson RM, Kaethner TM, Willey DL, Gray JC 1991. Pea chloroplast genes encoding a 4 kDa polypeptide of photosystem I and a putative enzyme of C1 metabolism. Curr Genet 19: 403–410 [DOI] [PubMed] [Google Scholar]
- Stefanovic S, Pfeil BE, Palmer JD, Doyle JJ 2009. Relationships among phaseolid legumes based on sequences from eight chloroplast regions. Syst Bot 34: 115–128 [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR 2010. Chloroplast RNA metabolism. Annu Rev Plant Biol 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Sugiura M 1992. The chloroplast genome. Plant Mol Biol 19: 149–168 [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W 2004. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5: 123–135 [DOI] [PubMed] [Google Scholar]
- Tsudzuki J, Nakashima K, Tsudzuki T, Hiratsuka J, Shibata M, Wakasugi T, Sugiura M 1992. Chloroplast DNA of black pine retains a residual inverted repeat lacking rRNA genes: nucleotide sequences of trnQ, trnK, psbA, trnI and trnH and the absence of rps16. Mol Gen Genet 232: 206–214 [DOI] [PubMed] [Google Scholar]
- Tsudzuki T, Wakasugi T, Sugiura M 2001. Comparative analysis of RNA editing sites in higher plant chloroplasts. J Mol Evol 53: 327–332 [DOI] [PubMed] [Google Scholar]
- Ueda M, Fujimoto M, Arimura SI, Murata J, Tsutsumi N, Kadowaki KI 2007. Loss of the rpl32 gene from the chloroplast genome and subsequent acquisition of a preexisting transit peptide within the nuclear gene in Populus. Gene 402: 51–56 [DOI] [PubMed] [Google Scholar]
- Ueda M, Nishikawa T, Fujimoto M, Takanashi H, Arimura SI, Tsutsumi N, Kadowaki KI 2008. Substitution of the gene for chloroplast RPS16 was assisted by generation of a dual targeting signal. Mol Biol Evol 25: 1566–1575 [DOI] [PubMed] [Google Scholar]
- Varotto C, Pesaresi P, Jahns P, Lessnick A, Tizzano M, Schiavon F, Salamini F, Leister D 2002. Single and double knockouts of the genes for photosystem I subunits G, K, and H of Arabidopsis. Effects on photosystem I composition, photosynthetic electron flow, and state transitions. Plant Physiol 129: 616–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gonzalez KD, Scaringe WA, Tsai K, Liu N, Gu D, Li W, Hill KA, Sommer SS 2007. Evidence for mutation showers. Proc Natl Acad Sci 104: 8403–8408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde A, Hartel H, Hubschmann T, Hoffmann P, Shestakov SV, Borner T 1995. Inactivation of a Synechocystis sp strain PCC 6803 gene with homology to conserved chloroplast open reading frame 184 increases the photosystem II-to-photosystem I ratio. Plant Cell 7: 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski MF, Lavin M, Sanderson MJ 2004. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am J Bot 91: 1846–1862 [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Li WH, Sharp PM 1987. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci 84: 9054–9058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Sharp PM, Li W-H 1989. Rates of synonymous substitution in plant nuclear genes. J Mol Evol 29: 208–211 [Google Scholar]
- Wolfe KH, Morden CW, Palmer JD 1992. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci 89: 10648–10652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20: 3252–3255 [DOI] [PubMed] [Google Scholar]
- Yang Z 2007. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA 2008. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet 4: e1000264 doi: 10.1371/journal.pgen.1000264 [DOI] [PMC free article] [PubMed] [Google Scholar]