Abstract
Pooled-DNA sequencing strategies enable fast, accurate, and cost-effect detection of rare variants, but current approaches are not able to accurately identify short insertions and deletions (indels), despite their pivotal role in genetic disease. Furthermore, the sensitivity and specificity of these methods depend on arbitrary, user-selected significance thresholds, whose optimal values change from experiment to experiment. Here, we present a combined experimental and computational strategy that combines a synthetically engineered DNA library inserted in each run and a new computational approach named SPLINTER that detects and quantifies short indels and substitutions in large pools. SPLINTER integrates information from the synthetic library to select the optimal significance thresholds for every experiment. We show that SPLINTER detects indels (up to 4 bp) and substitutions in large pools with high sensitivity and specificity, accurately quantifies variant frequency (r = 0.999), and compares favorably with existing algorithms for the analysis of pooled sequencing data. We applied our approach to analyze a cohort of 1152 individuals, identifying 48 variants and validating 14 of 14 (100%) predictions by individual genotyping. Thus, our strategy provides a novel and sensitive method that will speed the discovery of novel disease-causing rare variants.
Understanding the genetic basis of common diseases is an important step toward the goal of personalized medicine (Ng et al. 2008). At present, two distinct hypotheses are under debate (Goldstein 2009; Manolio et al. 2009). The common variant, common disease (CVCD) hypothesis states that disease-causing alleles are common in the human population (frequency > 5%) (Reich and Lander 2001). In contrast, the rare variant, common disease (RVCD) hypothesis posits that multiple disease-causing alleles, which individually occur at low frequencies (<<1%), cumulatively explain a large portion of disease susceptibility (Cohen et al. 2004; Ji et al. 2008). Recent evidence favors the RVCD hypothesis, as common variants have failed to explain many complex traits (Manolio et al. 2009), while rare genetic variants have been successfully associated with HDL levels (Cohen et al. 2004), blood pressure (Ji et al. 2008), obesity (Ahituv et al. 2007), and colorectal cancer (Fearnhead et al. 2004, 2005).
Due to their low frequencies, identifying rare, disease-associated variants requires genotyping large cohorts in order to reach the appropriate statistical power (e.g., 5000 individuals are required to detect mutations present at 0.1% in the population with a probability of 96%). “Collapsing” methods in which rare variants are grouped together before association with disease have been shown to improve statistical power (Li and Leal 2008), but analysis of large cohorts is still required. One recent strategy for genotyping large cohorts consists of pooled-sample sequencing, where individual samples are pooled prior to analysis on a next-generation sequencing platform (Van Tassel et al. 2008; Druley et al. 2009; Erlich et al. 2009; Koboldt et al. 2009; Prabhu and Pe'er 2009). By leveraging the massively parallel output of second-generation DNA sequencing, pooled-sample sequencing allows fast and accurate detection of rare variants in thousands of samples at a fraction of time and cost of traditional methods. Individual sample identities can be recovered using a combinatorial pooling strategy (such as DNA Sudoku) (Erlich et al. 2009).
Despite the promise of this method for studying rare genetic variants, current computational approaches pose a bottleneck because they are focused either on single individual genotyping (Li et al. 2008) or on the detection of common variants in small-sized pools (Koboldt et al. 2009). Our previously developed SNPseeker algorithm allows the detection of single nucleotide substitutions in large pooled samples (Druley et al. 2009), but still fails to address two important key challenges in rare variant detection.
First, presently no algorithm has been able to detect indels in pools larger than 42 individuals without the presence of many false-positives (∼40%) (Koboldt et al. 2009), despite the fact that they account for one-quarter of the known mutations implicated in Mendelian diseases (Ng et al. 2008; Stenson et al. 2009). In particular, short indels represent the most common type of this class of variation (Ng et al. 2008) and have been reported to occur as rare germline variants associated with genetic diseases such as breast and ovarian cancer (King et al. 2003). Efforts to detect disease-associated genetic variants will therefore greatly benefit from the ability to accurately detect rare short indels.
Second, in order to accurately detect rare variants in a large pooled sample, an optimal significance cutoff for the accurate discrimination of true variants from false-positives must be chosen. This parameter is, in practice, affected by sequencing error rates and average coverage, which have been shown to change for every run (Druley et al. 2009). Failure to define an optimal cutoff results in lower sensitivity and increased false-positive rates. Since the rare variant hypothesis posits that individual disease-associated mutations will be extremely rare (but cumulatively common), it is absolutely critical to be able to specifically discriminate, in every experiment, a single heterozygous individual in a large cohort from the background noise. Until now this has not been reliably demonstrated.
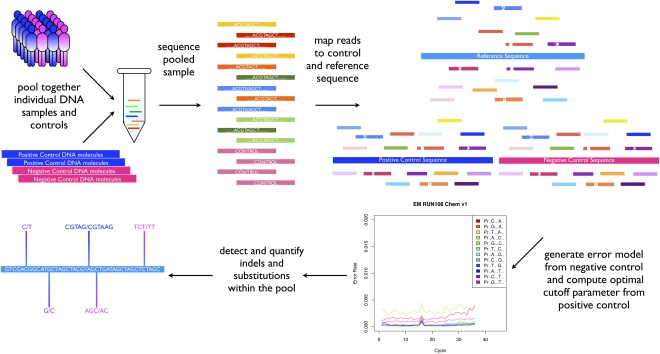
To address these important challenges, we have developed a novel experimental and computational strategy that combines a synthetically engineered DNA library inserted in each run and a new computational approach named SPLINTER (short indel prediction by large deviation inference and nonlinear true frequency estimation by recursion). This approach allows accurate detection and quantification of short insertions, deletions, and substitutions by integrating information from the synthetic DNA library to tune SPLINTER and quantify specificity and sensitivity for every experiment in order to accurately detect and quantify indels and substitutions (Fig. 1; Supplemental Fig. 1).
Figure 1.
Experimental and computational pipeline for detection of indels and substitutions in large pooled DNA samples: DNA samples from a selected group of patients are individually pooled in a complex mixture to be used as a template for PCR amplification of selected genomic loci. The pool PCR products are then combined in an equimolar mix containing a DNA fragment without variants (negative control) and a synthetic pool with engineered mutations present at the lowest expected variant frequency present in the sample (positive control). The mix is then sequenced on Illumina Genome Analyzer LIX, and sequencing reads are mapped back to the sample and the controls reference sequence by gapped alignment. The negative control reads are used to generate a second-order error model to be used in the variant calling phase. The positive control allows determination of the optimal cutoff for maximizing specificity and sensitivity of the analysis. SPLINTER will then be used to analyze the pooled sample, resulting in detection and quantification of indels and substitutions present in the pool. The SPLINTER algorithm detects true segregated variants by comparing the frequency vector of observed read bases to an expected frequency vector defined by the error model. If the observed vector is significantly different from the expected vector, then SPLINTER will call that position a sequence variant. For each identified variant, SPLINTER will then perform maximum likelihood fit in order to estimate its frequency in the pooled sample.
SPLINTER requires the presence of two components: a negative control (1–2 kb of cloned plasmid DNA) used to generate a run-specific error model, and a positive control consisting of a synthetic DNA library simulating an artificial pool with mutations engineered at a known position and frequency. We tested SPLINTER on synthetically engineered pooled samples containing different mutations at different frequencies in a variety of sequence-context backgrounds, obtaining 100% sensitivity with no false-positives in pools up to 500 individuals. SPLINTER was also able to accurately quantify allele frequencies—predicted and observed allele frequencies were correlated with a correlation of 0.999. We find that SPLINTER significantly outperforms all of the other algorithms for the analysis of pooled sequencing data by being the most sensitive approach, while also returning almost no false-positives. We then applied our strategy to multiple pooled samples, identifying novel and already described sequence variants, all of which were independently validated.
Results
Detection of rare insertions and deletions in synthetic libraries
For each experiment, we first pooled equimolar amounts of sample DNA together with the controls and generated a DNA library to be sequenced on the Illumina Genome Analyzer IIx sequencing platform. We then mapped back the sequencing reads to their reference and built a run-specific error model from the negative control reads. Next, we optimized our cutoff parameters on the positive control and then called SNPs and indels on our sample (see Supplemental material). We first sought to determine the upper limit of the number of samples that SPLINTER can analyze in a pool. To do so, we generated three synthetic DNA libraries, each containing 15 different indels and substitutions (Supplemental Tables 1, 2; Supplemental material) introduced at frequencies of 0.005, 0.002, and 0.001, respectively (corresponding to cohorts of 100, 250, and 500 diploid individuals). We sequenced these libraries using the workflow shown in Figure 1. In each instance, SPLINTER was able to correctly identify every variant (15/15 variants) without making false-positive calls (2254/2254 true-negatives) (Fig. 3A, below; Supplemental Table 4). We concluded that SPLINTER can accurately and reliably detect single heterozygous mutations in pools of up to 500 individuals.
Figure 3.
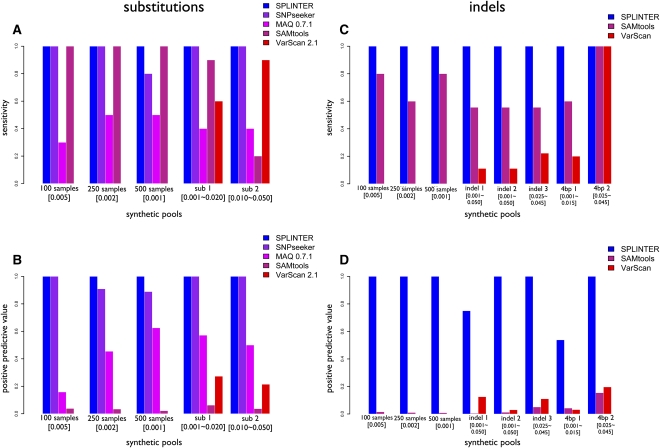
Comparison between SPLINTER and other variant calling algorithms: Substitutions (A,B) and indels (C,D) were analyzed independently. For each approach, performance was evaluated by assessing sensitivity (fraction of true-positive hits divided by total positives in the set) and positive predictive value (fraction of true-positive hits divided by total hits).
Estimation of required sequencing coverage for optimal indel and substitution detection
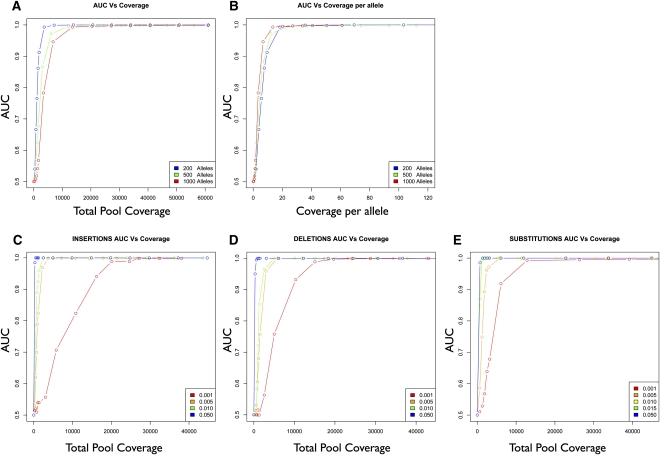
We next investigated how SPLINTER's accuracy changed as a function of average sequencing coverage. To do so, we sampled the sequencing data obtained for each of the three previous libraries at different fractions (Supplemental material) and then computed the accuracy of our predictions in the form of an area under a receiver-operator curve (AUC), a commonly used metric of accuracy ranging from 0.5 (random guessing) to 1 (100% sensitivity and specificity). By plotting AUC as a function of average sequencing coverage we found that accuracy increased with coverage, with high-frequency variants requiring less coverage than lower-frequency variants (Fig. 2A). By analyzing AUC as a function of coverage per allele, we observed a clear overlap of the curves for each pool, reaching AUC equal to 1 at ∼30-fold average coverage per haploid genome (Fig. 2B), indicating that accurate detection can be achieved given enough coverage independently of pool size.
Figure 2.
Relationship between variant detection accuracy and average sequencing coverage per base. (A) Accuracy expressed as AUC (area under the curve) (y-axis) plotted as a function of average sequencing coverage per base (x-axis) for synthetic pools with variants present at frequencies 1/200, 1/500, and 1/1000. (B) Same as in A, with average sequencing coverage per base per allele on the x-axis. (C–E) AUC (y-axis) as a function of average sequencing coverage per base (x-axis) for insertions (C), deletions (D), and substitutions (E). Variants are present at frequencies 1/1000, 5/1000, 10/1000, 15/1000, and 50/1000.
Recent resequencing efforts show that indel detection remains challenging, as their false-positive rate is 15-fold higher than substitutions (Pleasance et al. 2010). Our initial data suggested that indels can be detected as sensitively and accurately as substitutions. To test this hypothesis, we generated five additional DNA libraries with synthetic insertions, deletions, and substitutions included at a wide range of frequencies (from one to 50 variants in 1000 total alleles) (Supplemental Tables 2, 4). We achieved 100% sensitivity for all of the pools (9/9 indel variants and 10/10 substitution variants) with specificities between 99.91% and 100% (between 2263/2265 and 2259/2259 true-negatives). We then plotted the relationship between AUC and coverage for each set. Indels converged to AUC equal to 1 at a rate comparable to substitutions independently of the frequency of the mutation (Fig. 2A–C). Thus, we conclude that SPLINTER detects indels as accurately and as sensitively as it does substitutions.
Since many deleterious indels are 4 bp or shorter (King et al. 2003; Ng et al. 2008), we wanted to determine whether SPLINTER could accurately detect indels as large as 4 bp. We generated and sequenced two synthetic pools containing eight and 10 4-bp indels with frequencies ranging from 0.001 to 0.020 and from 0.025 to 0.045, respectively. SPLINTER achieved 100% sensitivity 10/10 variants and 100% specificity (2253/2253 true-negatives) for allele frequencies between 0.025 and 0.045 and 100% sensitivity (8/8 variants) and 99.5% specificity (2243/2253 true-negatives) between 0.001 and 0.020 (Supplemental Tables 3, 4). These results suggest that SPLINTER is sensitive and specific in detecting 4-bp indels.
Comparison of SPLINTER with other variant discovery approaches
We next compared SPLINTER with existing tools for variant calling. We used the synthetic DNA libraries previously described to benchmark the sensitivity and positive predictive value of each method. We compared SPLINTER with SNPseeker (Druley et al. 2009), MAQ (Li et al. 2008), SAMtools (Li et al. 2009), and VarScan (Koboldt et al. 2009) for the detection of substitutions (Fig. 3A,B) and with SAMtools and VarScan for the detection of indels (Fig. 3C,D). For each data set analyzed, SPLINTER significantly outperformed every other approach. In all of the synthetic libraries containing substitutions, SPLINTER detected all of the synthetic variants with no false-positives, thus achieving a 100% sensitivity and specificity. SNPseeker also achieved perfect accuracy in the pool simulating 100 individuals, but had a 20% positive predictive value in the libraries simulating 250 and 500 individuals, and had only an 80% sensitivity in the 500 individual library. The other approaches detected variants with substantially lower sensitivity and positive predictive values in all libraries. For each indel set, SPLINTER returned all of the true variants with no false-positives, except for the indel 1 set and the 4-bp 1 set (∼30% and ∼50% positive predictive values, respectively). In comparison, every other approach resulted in false-positive rates greater than 80%, while achieving low sensitivity, with the exception of the second 4-bp set. We also compared SPLINTER with a recently published new algorithm for pooled DNA variant detection called CRISP (Vikas 2010) for both substitution and indel detection (Supplemental Fig. 2). SPLINTER outperformed CRISP in both sensitivity (at most 40% increment) and positive predictive value (at most 80% increment).
In order to distinguish whether the improved accuracy in variant finding originated from improved alignments or improved variant calling, we also compared the performance of SPLINTER using our alignment algorithm versus using reads aligned with Novoalign (http://www.novocraft.com). Both aligners resulted in a comparable performance in finding true variants (Supplemental Fig. 3), although our aligner showed small increases in sensitivity and positive predictive value in several of the analyzed pools. This result suggests that improved variant calling accuracy mostly depended on the variant calling algorithm and not the underlying aligner. Taken together, there results demonstrate that SPLINTER outperforms other approaches at detecting single nucleotide substitutions and indels in large pools.
Estimation of the frequency of rare insertions and deletions in synthetic libraries
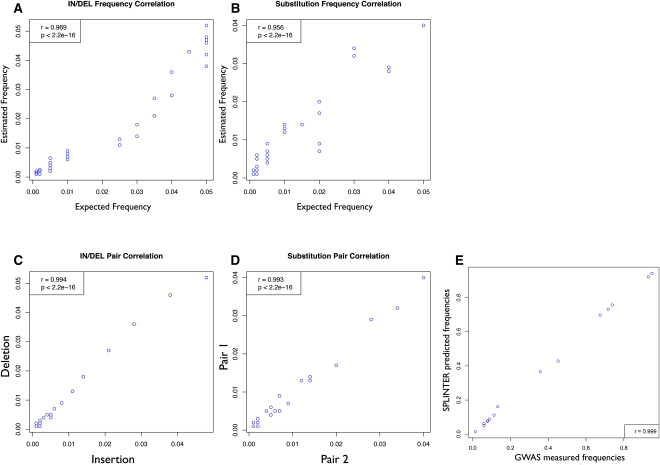
Having established that SPLINTER could detect rare variants in pooled samples, we next examined whether SPLINTER could also accurately determine the frequencies of the identified variants. We compared estimated and expected indel frequencies from all of our libraries (frequency range 0.001 ∼ 0.050) and found a very high correlation (r = 0.969, P < 2.2 × 10−16; Fig. 4A), indicating that SPLINTER was able to accurately estimate allele frequencies. We next sought to better understand the causes of the observed errors in our allele frequency estimates. Allele quantification can be affected by pipetting errors during DNA pooling and by preferential amplification of specific alleles in the pooled PCR. To distinguish between these two sources of error, we constructed all of our plasmids so that each contained two mutations spaced far enough apart to be analyzed independently (i.e., with no overlapping reads). If pipetting error and amplification bias are the major sources of error in allele quantification, then the estimated allele frequencies of mutations on the same plasmid will be highly correlated. This was indeed the case. Frequency estimates for mutations within the same molecule were very highly correlated (r = 0.995, P < 2.2 × 10−16; Fig. 4C), indicating that most of the noise in variant quantification was due to experimental error. We similarly observed very high correlations with substitutions (frequency correlation r = 0.956, P < 2.2 × 10−16; pair correlation r = 0.993, P < 2.2 × 10−16; Fig. 4D) and 4-bp indels (frequency correlation r = 0.962, P = 1.501 × 10−11; pair correlation r = 0.939, P = 5.599 × 10−5) (Supplemental Fig. 4). Based on these results, we reasoned that robotic pooling of samples might improve allelic quantification. Therefore, we robotically pooled and sequenced a large cohort of 974 people previously analyzed in a GWA study (see Methods). As expected, we observed an almost perfect correlation (r = 0.999, P < 2.2 × 10−16; Fig. 4E) between the GWA frequencies and the frequencies estimated by SPLINTER, indicating that inaccurate pipetting was indeed a primary source of error.
Figure 4.
Precise quantification of rare genetic variants in synthetic and real samples. (A,B) Correlation between variant frequency measured by SPLINTER (y-axis) and expected variant frequency (x-axis) from eight synthetic pools for indels (A) and substitutions (B). (C,D) Pair correlation between mutation pairs present in the same DNA molecule for indels (C) and substitutions (D). (E) Correlation between variant frequency measured from GWA study (x-axis) and SPLINTER estimated frequency (y-axis).
High-throughput discovery of rare indels in large patient cohorts
Finally, we applied SPLINTER to a large human cohort as a “real-world” test of the algorithm. We sequenced 14 loci (2596 bp total) in 1152 individuals, which were divided into nine pools (94–178 individuals per pool) (see Methods). For every sequenced pool, we included a negative and positive control to tune SPLINTER. We identified, on average, 19 variants per pool (for a total of 151 variants, see Supplemental Table 6). To confirm SPLINTER's accuracy, we examined the overlap of our hits with variants listed in dbSNP. We observed large overlapping fractions—between 68.5% and 100% of the identified variants in each pool could be found in dbSNP (Supplemental Tables 5, 6). In all cases, statistical significance was reached (Fisher's exact test; Supplemental Table 5). We selected 14 variants (three novel variants and 11 from dbSNP) from the largest analyzed pool for independent validation by individual genotyping using the Sequenom iPLEX platform. All 14 variants were confirmed, resulting in 100% positive predictive value. Furthermore, allele frequencies were highly correlated with those estimated by SPLINTER (r = 0.985, P = 5.958 × 10−9; Supplemental Table 8; Supplemental Fig. 5). Together, these results demonstrate the utility of the SPLINTER methodology for the rapid analysis of large populations of individuals. All of the computational tools, source codes, and the experimental datasets presented in this study can be accessed at http://cgs.wustl.edu/∼fvallania/4_splinter_2010/5_splinter_webpage/SPLINTER_supporting_material.html.
Discussion
Rare genetic variation is likely to describe a substantial portion of heterogeneity in common and complex diseases. Identifying disease-associated rare variants requires the analysis of multiple loci in large cohorts. We have shown that a novel experimental design combined with SPLINTER can accurately identify genetic variants in large pools, leading to several advantages over other computational strategies.
First, we found that SPLINTER identified genetic variants with high sensitivity and precision, whereas the other methods were unable to detect a large fraction of the variation present in the samples. We found that a sequencing coverage of ∼30× per haploid genome was required to detect mutations with high sensitivity and specificity. In earlier work, we successfully analyzed pooled samples using SNPseeker at lower sequencing coverage (∼13.8-fold per haploid genome) (Druley et al. 2009). However, in that study most of the variants were present in many individuals in the pool, suggesting that in order to detect singleton alleles with ∼100% confidence in a variety of different sequence contexts a higher sequence coverage is required. This finding is confirmed by recent resequencing studies of single cancer genomes, where near-optimal accuracy of somatic SNP detection (3% false discovery rate) was achieved at ∼40-fold average haploid genome coverage (Pleasance et al. 2010), and by the lower performance of SNPseeker when compared with SPLINTER in detecting substitutions present at one in a 1000 in both sensitivity and precision.
Second, our strategy incorporates a synthetic positive control and a negative control, which allow estimation of sensitivity and specificity for each experiment. This is important because run-to-run variations in sequencing error rates can influence accuracy and perturb the optimal P-value cutoffs. The inclusion of the control DNA has a negligible impact on experiment cost. One single-end sequencing lane (∼30 million 36-bp-long reads per lane) can provide enough coverage to analyze ∼25 kb of genomic DNA in 500 patients, with the control sequences accounting for ∼4% of the total sequencing data.
Third, SPLINTER can accurately and sensitively detect indels with a high sensitivity and accuracy. Detection of indels, even in single genome resequencing studies, is indeed a challenging problem due to the difficulties in reducing the false-positive rate while retaining good sensitivity (Pleasance et al. 2010). In addition, previously published approaches cannot detect indels (Li et al. 2008; Druley et al. 2009), or can only be applied to small-sized cohorts (42 people) (Koboldt et al. 2009). Together, these issues have limited the application of pooled DNA sequencing. We have shown here that SPLINTER can accurately discriminate single indels in pools as large as 500 individuals with high sensitivity and specificity. By comparison, the best performing algorithm achieved at best an 80% false-positive rate.
Fourth, SPLINTER can accurately quantify the frequency of the alleles present in the pool. Although high correlations between real and estimated frequencies were observed, small discrepancies may result in errors in variant association to a phenotype if the variant is rare and the effect of the variant is high. Our pair correlation analysis shows that the major source of errors in quantification does not come from SPLINTER, but rather from pipetting errors in pool construction as indicated by the improved correlations after robotic pipetting of the pools. This issue can, in fact, be resolved by performing orthogonal validation of the samples, which will be highly facilitated by the overall performance of SPLINTER in detecting rare variants as opposed to other methods. In contrast, the major source of error in array-based pooled DNA analysis is array variation, being seven times higher than pool construction variation (Macgregor 2007). This observation argues that our approach shows even higher accuracy compared with other experimental platforms.
Finally, our approach can be applied to any pooled cohort or any heterogeneous sample of any size and can be easily scaled up to whole-exome and whole-genome analysis. Given the presence of a positive control to infer the optimal parameters, pooled samples can accurately be analyzed without limitations on experimental design or achieved coverage. In this study, we used PCR to amplify the various genomic regions, but our strategy is also compatible with solid and liquid-phase genomic capture approaches (Mamanova et al. 2010).
We found that alignment errors decreased our ability to detect large indels. This explains why SPLINTER performed slightly worse in the analysis of the 4-bp indel libraries relative to the 1–2-bp indel libraries. To detect the longer indels, it was necessary to allow larger gaps in our read alignments, which increased the overall alignment noise. We believe this was due to potential sequencing artifacts or sample contaminants aligning back to the reference sequence, thereby reducing the signal coming from true variants. This limitation can be overcome with longer sequencing read lengths, which should reduce the ambiguity in aligning reads while allowing larger gaps (in this work, all sequencing reads were 36 bp in length). Similarly, while whole-genome analysis may present additional challenges due to increased sequence complexity, compared with the analyzed synthetic controls we expect it to mostly impact the read alignment step in the analysis pipeline, which can be overcome by generating paired-end and/or longer sequencing reads. In addition, with reduced error rate, fewer observations at a given variant position will be needed to provide confidence in the variant call. Nevertheless, our approach is the first one to accurately call short indels in large pooled samples.
One departure of our algorithm from other variant calling programs is that SPLINTER does not incorporate quality scores in any step of the analysis. We have found that our error model captures essentially the same information that is contained in quality scores (see Supplemental material; Druley et al. 2009 and so including quality score information does not improve SPLINTER's performance. The high performance of our method compared with others that use quality scores (Li et al. 2008; Koboldt et al. 2009) suggests that this viewpoint is likely correct. Additionally, analyzing reads aligned with quality scores resulted in equal or lower performance when compared with reads aligned using our aligner (see Supplemental Fig. 5).
To obtain a complete understanding of the molecular causes of common diseases, it is critical to be able to detect and analyze rare variants (Van Tassel et al. 2008; Druley et al. 2009; Erlich et al. 2009; Koboldt et al. 2009; Prabhu and Pe'er 2009). Pooled DNA sequencing is an important method for rare variant analysis, since it enables the rapid and cost-effective analysis of thousand or tens of thousands of individuals. SPLINTER will also be useful for analyzing samples that are naturally heterogeneous—e.g., for the detection and quantification of rare somatic mutations in tumor samples (Stingl and Caldas 2007). A second promising application is detection of induced mutations in in vitro evolution experiments (Barrick at al. 2009; Beaumont et al. 2009). Thus, we expect SPLINTER will become a useful tool for the analysis of data generated by next-generation sequencing methods.
Methods
Preparation of the synthetic pools
Every synthetic pool library consists of a mixture of different oligonucleotides, where one is referred to as the wild-type allele and the others are mutants with respect to the wild type. We used the consensus sequence of the 72-bp exon 9 from TP53 (RefSeq accession no. NM_000546) as the “wild-type” insert into a pGEM-T Easy vector (Promega). We then designed a panel of different variations of this consensus sequence (see Supplemental Tables 1–3) containing single, double, and 4-bp indels, as well as single nucleotide substitutions. These vectors could then be pooled such that each mutation was present at different frequencies. Once pooled, a single PCR reaction was performed using primers that flanked the insertion site and generated a 335-bp amplicon. To facilitate ligation into the vector, each oligonucleotide was ordered with 5′ phosphorylation and an overhanging 3′ A from Integrated DNA Technologies. Complimentary oligonucleotide pairs were annealed as follows: 1 μL of sense and antisense oligonucleotide at 100 μM were mixed with 5 μL of 10× PCR buffer (Sigma-Aldrich) and brought to a final volume of 50 μL. The annealing mix was then warmed up to 95°C for 5 min, followed by 20 min at 25°C. Each annealed sequence was then ligated into the pGEM-T Easy Vector (Promega) according to the manufacturer's protocol and reagents. The final ligation product was then transformed into GC-10 competent cells (GeneChoice) using standard cloning protocol. Colonies were screened using “Blue/White” selection induced by Xgal and IPTG, White colonies were picked and grown on Luria broth agar with ampicillin for 12–16 h. Plasmid was then recovered from the transformed bacteria suspension using Qiaprep Spin Miniprep kit according to the manufacturer's protocol (Qiagen). Following insert validation by Sanger sequencing, plasmid pools were prepared by pooling each plasmid at the appropriate number of molecules in order to introduce the desired mutations at the desired frequency with respect to the wild-type background. Each pool was generated with a total number of 1011 plasmid molecules. This was chosen in order to mimic the best conditions described in the original pooled-DNA sequencing protocol11 to maximize the number of molecules available for analysis, while keeping fluid volumes tractable. Each pool was then PCR amplified using primer sequences flanking the plasmid insertion site (see Supplemental Table 4). Each PCR reaction was performed as follows: (1) 93°C for 2 min; (2) 93°C for 30 sec; (3) 56°C for 30 sec; (4) 65°C for 2 min; (5) repeat steps 2–4 for 18 cycles; (6) 65°C for 10 min. Each PCR mix contained 2.5 uL of 10× PfuUltra buffer, 10 μM forward and reverse primers, 1 M betaine (Sigma-Aldrich/Fluka), 1.25 U PfuUltra DNA polymerase, and between 30 and 50 ng of template DNA in a final volume of 25 uL. Each pool was then sequenced using a single lane of the Illumina Genome Analyzer II platform.
DNA library preparation and sequencing for pooled samples
After PCR amplification of target loci, a second pool was created by adding PCR products to the positive and negative controls for the analyzed pooled sample. In order to generate uniform sequencing coverage, every PCR product and control was pooled at the same number of molecules (chosen to be at least 1011 molecules [∼1 μg] in order to have enough material for the sequencing library preparation). Random ligation, sonication, and sequencing library preparation were performed as previously described with a few changes. DNA ligation was performed in a final volume of 50 μL. Prior to sonication, ligation products were diluted 1:10 using Qiagen PBI buffer from the QIAquick PCR Purification Kit (Qiagen). Fragmentation was then performed using the Bioruptor XL sonicator (Diagenode). Samples were sonicated in parallel with the following settings: 25 min of total sonication time, 40 sec of pulse followed by 20 sec without pulse, high power pulse setting. This resulted in each pool of large concatemers being randomly fragmented between 500 and 4000 bp (data not shown). Following sonication, DNA samples were then purified via the QIAquick PCR purification kit (Qiagen) and sequencing libraries were prepared according to the standard protocol for genomic sample preparation by Illumina (Illumina). Each library was then sequenced on a single lane of an Illumina Genome Analyzer II, generating 36-bp read lengths.
Variant calling in pooled samples
For each pooled sample, reads were compressed in order to reduce computational run-time and then aligned to their reference using a dynamic programming algorithm (see Supplemental material). Aligned reads were used to generate a run-specific error model from the incorporated negative control. The aligned file and the error model are then used by SPLINTER in input to detect the presence of a sequence variant in the pool at any analyzed position. Optimal detection of true variants was achieved by calibrating the P-value cutoff used by SPLINTER with information generated from the included positive control (see Supplemental material).
Acknowledgments
This work was partly supported by the Children's Discovery Institute grant MC-II-2006-1 (R.D.M., T.E.D.), the NIH Epigenetics Roadmap grant (1R01DA025744-01 and 3R01DA025744-02S1; R.D.M., F.L.M.V.), the Saigh Foundation (F.L.M.V., T.E.D.), and Hope Street Kids and Alex's Lemonade Stand “A” Award support (T.E.D.). We thank Michael DeBaun at Washington University in Saint Louis, School of Medicine, for providing the samples from the SIT cohort; Lee Tessler, David Mayhew, and the other members of the Mitra lab for helpful discussion on the SPLINTER algorithm; the Sequenom Core facility at the Human Genetics Division at Washington University in Saint Louis, School of Medicine for the Sequenom validation; and Jessica Hoisington-Lopez at the Center for Genome Science, Washington University in Saint Louis, School of Medicine for running the samples on the Illumina GAIIx platform. This work is dedicated to Natalina Vallania.
Author contributions: F.L.M.V. designed and implemented the SPLINTER algorithm. F.L.M.V., T.E.D., and R.D.M. designed the experiments. F.L.M.V. performed the sequencing experiments on the synthetic and real samples. FL.M.V. performed the data analysis and variant validation. J.W. and E.R. performed sequencing on the GWA sample and analyzed its frequency correlation. I.B. and M.P. provided reagents. F.L.M.V., R.D.M., and T.E.D. wrote the manuscript.
Footnotes
[Supplemental material is available online at www.genome.org. Sequencing data is available at http://cgs.wustl.edu/∼fvallania/4_splinter_2010/5_splinter_webpage/SPLINTER_supporting_material.html. Novel SNP data have been submitted to the NCBI dbSNP (http://www.ncbi.nlm.nih.gov/snp) under accession nos. rs113740468, rs78985299, and rs113225202. SPLINTER is available at http://www.ibridgenetwork.org/wustl/splinter.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.109157.110.
References
- Ahituv N, Kavaslar N, Schackwitz W, Ustaszewska A, Martin J, Hebert S, Doelle H, Ersoy B, Kryukov G, Schmidt S, et al. 2007. Medical sequencing at the extremes of human body mass. Am J Hum Genet 80: 779–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461: 1243–1247 [DOI] [PubMed] [Google Scholar]
- Beaumont HJ, Gallie J, Kost C, Ferguson GC, Rainey PB 2009. Experimental evolution of bet hedging. Nature 462: 90–93 [DOI] [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH 2004. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science 305: 869–872 [DOI] [PubMed] [Google Scholar]
- Druley TE, Vallania FL, Wegner DJ, Varley KE, Knowles OL, Bonds JA, Robison SW, Doniger SW, Hamvas A, Cole FS, et al. 2009. Quantification of rare allelic variants from pooled genomic DNA. Nat Methods 6: 263–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich Y, Chang K, Gordon A, Ronen R, Navon O, Rooks M, Hannon GJ 2009. DNA Sudoku–harnessing high-throughput sequencing for multiplexed specimen analysis. Genome Res 19: 1243–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearnhead NS, Wilding JL, Winney B, Tonks S, Bartlett S, Bicknell DC, Tomlinson IP, Mortensen NJ, Bodmer WF 2004. Multiple rare variants in different genes account for multifactorial inherited susceptibility to colorectal adenomas. Proc Natl Acad Sci 101: 15992–15997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearnhead NS, Winney B, Bodmer WF 2005. Rare variant hypothesis for multifactorial inheritance: Susceptibility to colorectal adenomas as a model. Cell Cycle 4: 521–525 [DOI] [PubMed] [Google Scholar]
- Goldstein DB 2009. Common genetic variation and human traits. N Engl J Med 360: 1696–1698 [DOI] [PubMed] [Google Scholar]
- Ji W, Foo JN, O'Roak BJ, Zhao H, Larson MG, Simon DB, Newton-Cheh C, State MW, Levy D, Lifton RP 2008. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet 40: 592–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Marks JH, Mandell JB, New York Breast Cancer Study Group 2003. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 302: 643–646 [DOI] [PubMed] [Google Scholar]
- Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, Ding L 2009. VarScan: Variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25: 2283–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM 2008. Methods for detecting associations with rare variants for common diseases: Application to analysis of sequence data. Am J Hum Genet 83: 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor S 2007. Pooling sources of error. Eur J Hum Genet 15: 501–504 [DOI] [PubMed] [Google Scholar]
- Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, Shendure J, Turner DJ 2010. Target-enrichment strategies for next-generation sequencing. Nat Methods 7: 111–118 [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Levy S, Huang J, Stockwell TB, Walenz BP, Li K, Axelrod N, Busam DA, Strausberg RL, Venter JC 2008. Genetic variation in an individual human exome. PLoS Genet 4: e1000160 doi: 10.1371/journal.pgen.1000160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. 2010. A small-cell lung cancer genome with complex signature of tobacco exposure. Nature 463: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu S, Pe'er I 2009. Overlapping pools for high-throughput targeted resequencing. Genome Res 19: 1254–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, Lander ES 2001. On the allelic spectrum of human disease. Trends Genet 17: 502–510 [DOI] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, Cooper DN 2009. The Human Gene Mutation Database: 2008 Update. Genome Med 1: 13 doi: 10.1186/gm13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J, Caldas C 2007. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat Rev Cancer 7: 791–799 [DOI] [PubMed] [Google Scholar]
- Van Tassell CP, Smith TP, Matukumalli LK, Taylor JF, Schnabel RD, Lawley CT, Haudenschild CD, Moore SS, Warren WC, Sonstegard TS 2008. SNP discovery and allele frequency estimation by deep sequencing of reduced representation libraries. Nat Methods 5: 247–252 [DOI] [PubMed] [Google Scholar]
- Vikas B 2010. A statistical method for the detection of variants from next-generation resequencing of DNA pools. Bioinformatics 26: 318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]