Abstract
DNA methylation is an essential epigenetic modification that plays a key role associated with the regulation of gene expression during differentiation, but in disease states such as cancer, the DNA methylation landscape is often deregulated. There are now numerous technologies available to interrogate the DNA methylation status of CpG sites in a targeted or genome-wide fashion, but each method, due to intrinsic biases, potentially interrogates different fractions of the genome. In this study, we compare the affinity-purification of methylated DNA between two popular genome-wide techniques, methylated DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain-based capture (MBDCap), and show that each technique operates in a different domain of the CpG density landscape. We explored the effect of whole-genome amplification and illustrate that it can reduce sensitivity for detecting DNA methylation in GC-rich regions of the genome. By using MBDCap, we compare and contrast microarray- and sequencing-based readouts and highlight the impact that copy number variation (CNV) can make in differential comparisons of methylomes. These studies reveal that the analysis of DNA methylation data and genome coverage is highly dependent on the method employed, and consideration must be made in light of the GC content, the extent of DNA amplification, and the copy number.
DNA methylation, which is one of the most studied epigenetic marks, involves the addition of a methyl group to the 5 position of the cytosine pyrimidine ring and occurs primarily at CpG dinucleotides in mammals (Jones 1999). DNA methylation patterns are established early in development and are associated with the regulation and maintenance of gene expression during differentiation (Sorensen et al. 2010). Methylation patterns can also be disrupted in many disease states, and in particular, changes in DNA methylation at CpG island-associated promoters can play a role in the development of cancer (Jones and Baylin 2002, 2007; Jaenisch and Bird 2003).
There are now numerous methods available for determining CpG methylation status (for review, see Laird 2010), including methods focused at the level of CpG islands (Ponzielli et al. 2008; Kaminsky et al. 2009), individual promoters (Weber et al. 2005, 2007; Novak et al. 2008), and, increasingly, genome-“scale” (Meissner et al. 2008; Gu et al. 2010) and genome-wide methods, either at high (Lister et al. 2009) or low resolution (Serre et al. 2009; Ruike et al. 2010). These later methods can be broadly classified into the following designations: reduced representation approaches that are based on methylation-sensitive (e.g., HELP) (Oda et al. 2009) or specific (e.g., CHARM) (Irizarry et al. 2008) restriction digestion (for review, see Jeddeloh et al. 2008), affinity-based methods such as methyl-DNA immunoprecipitation (MeDIP) (Weber et al. 2005, 2007; Novak et al. 2008) and methyl-CpG binding domain-based capture (MBDCap) (Rauch et al. 2006, 2008; Serre et al. 2009), and the more direct bisulphite treatment-based methods (Lister et al. 2009); coupling of reduced representation and bisulphite treatment has now been demonstrated (Meissner et al. 2008; Gu et al. 2010), and other combinations are also possible. However, there are still many challenges involved in interpreting data from DNA methylation-based assays, due to complex effects, both technical and biological that are introduced at various steps in the procedure, in addition to implicit biases from the methods employed. These include cellular purity and DNA quality, DNA amplification bias in GC-rich regions, and the effects of copy number aberrations. The focus of this study is on DNA methylation analyses using affinity-based approaches, in combination with promoter DNA microarrays or high-throughput DNA sequencing readouts.
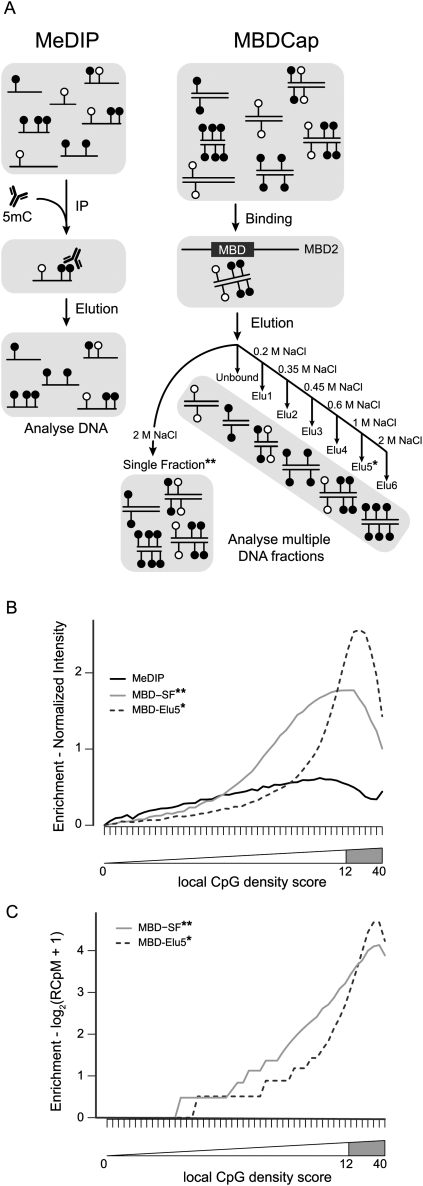
Chromatin immunoprecipitation (ChIP) has been used extensively to study protein–DNA interactions (Ren et al. 2000), and recently, an extensive benchmarking study has been conducted comparing microarray platforms and analysis methods (Johnson et al. 2008). Comparison studies for DNA methylation platforms are now starting to emerge (Li et al. 2010). MeDIP uses an antibody to 5-methyl-cytosine, targeting single-stranded DNA (Weng et al. 2009; Ruike et al. 2010), while the MBDCap approach uses the methyl-CpG binding domain of the MBD2 protein to capture double-stranded DNA (Serre et al. 2009). Furthermore, the MBDCap approach can use a series of salt fractionation steps that allows specific methylation density to be assessed (Fig. 1A). Methylation status of the immunoprecipitated DNA isolated from MeDIP or MBDCap is analyzed using tiling microarrays or high-throughput sequencing, and multiple platforms of each are available. Because DNA is often limiting and derived from mixed cell types, especially in studies with clinical samples, it can be difficult to isolate sufficient amounts of pure DNA to hybridize to microarrays directly or to sequence. DNA amplification techniques have been developed to address this problem (Paris 2009). Here, we show that DNA amplification can result in depletion of GC-rich regions and therefore may particularly impact on the interpretation of DNA methylation in CpG islands. Most importantly, copy number variation (CNV) can impact on the interpretation of DNA methylation levels, and in cancer, this can be critical within the regions harboring gene amplification and/or deletions. Furthermore, promoter tiling array data can be used to adjust for copy number changes, and we show that copy number aberrations can have a significant impact on genome-wide DNA methylation analysis.
Figure 1.
(A) Schematic showing the capture of methylated DNA into populations of single-stranded (MeDIP) or double-stranded (MBDCap) fragments. (B) Summarized probe intensities for enrichment of fully methylated DNA with MeDIP and two variations of MethylMiner-based enrichment. X-axis shows the local CpG density group (1–50). Y-axis shows the log2-scale input-normalized intensity. Each line shows the median intensity for the input-normalized intensities for the probes in the bin (here, for probes with GC content of 11 only). The intensities are further normalized such that the median in the lowest bin is 0. The location of probes within CpG islands is shown by the gray-shaded region, corresponding to a local CpG density score between 12 and 40. (C) Summarized read counts in bins of 1000 bases over the same genomic regions interrogated by the Affymetrix Promoter 1.0R array. Each line represents the median log2 read count (RCpM indicates read counts per million mapped); the summaries are normalized such that the median with the lowest bin is 0.
Results
MeDIP and MBDCap enrich different fractions of the genome based on CpG density
MeDIP and MBDCap are two capture methods commonly used to interrogate genome-wide DNA methylation patterns. These two techniques have inherent differences, namely, antibody versus MBD capture. We asked if each method was comparable in the detection of the same methylated genomic DNA sequences. Fully methylated human genomic DNA treated with SssI methyltransferase was used to benchmark the two affinity-based DNA methylation mapping platforms. For the MBDCap comparison, the MethylMiner protocol was used, where DNA can be eluted in a high-salt buffer (2 M NaCl) as a single fraction, here referred to as MBD-SF, or eluted as distinct subpopulations based on the degree of methylation, using an increasing concentration of NaCl from 200 mM to 2000 mM. MBD-Elu5 denotes the 1 M fraction of the elution series (Fig. 1A). After SssI treatment, essentially every CpG site in the genome is methylated, allowing a direct comparison of enrichment between MeDIP and MBDCap, which was interrogated using Affymetrix Human Promoter 1.0R arrays containing more than 4.5 million 25mer probes spanning 23,155 promoters. Figure 1B shows summarized input-subtracted promoter tiling array signals from MeDIP, MBD-Elu5, and MBD-SF of fully methylated DNA after stratifying probes according to the local genomic CpG density (for a formal definition, see Methods). Several key observations can be made: First, the overall degree of enrichment is higher for MBDCap-based procedures, especially for CpG-dense methylated DNA (probes with high local CpG density); second, due to the nature of the salt elution steps, MBD-Elu5 enriches primarily for CpG-dense material only, whereas MBD-SF enriches for a broader range of CpG densities, albeit at a lower average level. An attenuation of promoter microarray signal was observed at the highest CpG density regions, and notably, the attenuation seems to occur at a higher local CpG density for the MethylMiner-based procedure than MeDIP. As MeDIP recovers single-stranded DNA and MBDCap recovers double-stranded DNA, it is possible that topoisomerase activity from M.SssI may compromise amplification of DNA recovered from MeDIP relative to MBDCap (Matsuo et al. 1994). However, we found that enrichment levels of methylated LNCaP DNA were also favored by MBDCap in comparison to MeDIP for CpG dense regions, albeit at reduced levels (Supplemental Fig. 1), supporting the use of MBDCap-based procedures for favoring interrogation of CpG-rich regions.
Next we asked if the apparent CpG bias was also observed with sequencing data. Similar to the previous experiment, fully methylated DNA was used to perform the MBDCap enrichment, and we analyzed the eluted DNA using a high-throughput sequencing readout. Using one lane of Illumina Genome Analyzer sequencing with 36-base single end reads for each of MBD-Elu5 and MBD-SF, 8,616,022 and 11,557,035 uniquely mapped reads were obtained, respectively, to the reference human genome (see Methods). In order to compare the methylation readout against promoter arrays, the number of reads (in nonoverlapping 1000-bp bins) mapping to the subset of the genome interrogated by the Affymetrix tiling array was calculated. Figure 1C shows the read counts normalized to depth, similarly grouped by local CpG density. Supplemental Figure 2 shows enrichment profiles across the whole genome, highlighting the different profile of enrichment (and CpG density) of promoters compared to the entire genome. The CpG density bins in Figure 1C represent approximately the local CpG density bins in Figure 1B, but the enrichment levels are not directly comparable. Overall, a similar enrichment profile was observed for the sequencing data, where MBD-Elu5 enriches for densely methylated regions and MBD-SF enriches for a slightly broader range of CpG density. As before, a slight drop in the number of reads in very high CpG density regions was observed, but not to the same extent as observed in the tiling array data. It is not clear whether the decrease can be attributed to PCR-based amplification in the library preparation or cluster generation step, or whether there are other biases introduced in mapping reads to the genome, or whether this is an inherent property of the affinity-based techniques.
Whole-genome amplification can bias DNA methylation calls in CpG dense regions
Given the signal attenuation observed in the fully methylated enrichment experiments, we next examined if this is due to whole-genome amplification (WGA), a required step in the protocol to generate enough material for hybridization to the promoter arrays. It is well established that the GC content of a DNA template can affect the efficiency of amplification, often resulting in a bias against the GC-rich regions of the genome (Bredel et al. 2005; Pugh et al. 2008; Teo et al. 2008). Johnson et al. (2008) also report a significant drop in sensitivity, most notably for Affymetrix tiling arrays, when amplified DNA is hybridized. Amplification bias is perhaps even more of a potential concern in DNA methylation mapping, since CpG islands are GC-rich by their very nature and therefore more prone to any extant bias.
We were initially alerted to a potential problem involving GC bias when GSTP1, which is highly methylated at its CpG island-associated promoter in prostate cancer and is unmethylated in normal cells (Song et al. 2002; Nakayama et al. 2004), showed little differential enrichment at the probe-level on the Affymetrix promoter tiling arrays after MeDIP enrichment and WGA of prostate cancer (LNCaP) and normal prostate epithelial cells (PrECs) (see Supplemental Fig. 3A; Coolen et al. 2010). MeDIP-qPCR experiments, used as a control before hybridization, confirmed a strong affinity of methylated DNA at the GSTP1 locus, both before (77-fold) and after (76-fold) amplification (Supplemental Fig. 3B). Consequently, even though the degree of enrichment was maintained between LNCaP and PrECs, the absolute copy number of GSTP1 molecules in the population decreased proportionally after WGA, from 676 and 8.7 copies/ng beforehand to 32 and 0.43 copies/ng afterward, respectively. These data suggest that DNA amplification can result in an apparent loss of methylation detection for regions of the genome that are amplified less efficiently, such as GC-rich regions. To illustrate this further, Supplemental Figure 4 shows probe-level data for the CpG-rich promoters of WNT2 and CAV2 and the CpG-poor promoters of AGR2, PTN, and SOSTDC1, all of which are validated to be hypermethylated in prostate cancer cells. For the WNT2 and CAV2 promoters, the microarray signal representing differential methylation is observed only in regions flanking the CpG island. We hypothesize that WGA has ablated the absolute levels of these DNA molecules, similar to GSTP1. Notably, the AGR2, PTN, and SOSTDC1 promoters, which are of lower CpG content, exhibit a differential signal throughout the region validated to be differentially methylated. It is realistic to expect that many GC-rich regions, such as CpG islands, while differentially enriched between populations before amplification, become diluted to be below the lower detection limit on the promoter microarray after WGA.
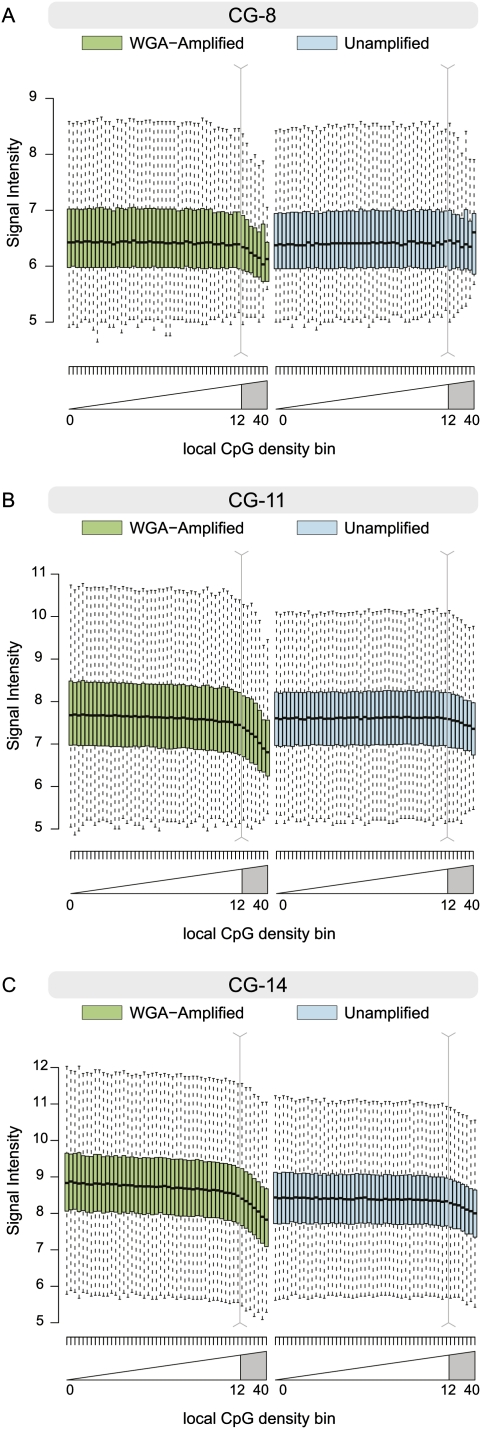
By using Affymetrix promoter tiling array data, the effect of WGA was studied directly by comparing the probe intensities from unamplified and amplified DNA from the same origin. For this experiment, genomic input DNA was used, without a methylated DNA affinity step. Figure 2 shows the distribution of the raw promoter array signal intensity for both an unamplified and WGA sample of the same input genomic DNA, across equally sized bins of local CpG density. Here, only probes with a GC content of 8, 11, and 14 (of the 25-mer probe) (Supplemental Fig. 5 displays the full range of probe GC contents) are displayed. Notably, a substantial drop in the promoter tiling microarray signal is observed for the probes in CpG-rich regions (local CpG density greater than 12 defines a CpG island). In addition, CpG-rich regions of the genome also show some attenuation in the hybridization signal from unamplified DNA when the probe GC content was greater than 11, suggesting that other effects, such as cross-hybridization and probe-specific temperature effects (Wei et al. 2008), may influence the signal observed on microarrays. It is also noted that the observed CpG density bias is not unique to the Affymetrix platform, since unamplified MeDIP-enriched fully methylated DNA samples analyzed on a NimbleGen platform (Gal-Yam et al. 2008) also exhibit attenuation over probes with a broad range of GC contents (Supplemental Fig. 6).
Figure 2.
Box-and-whisker plots of unnormalized log2-scale microarray intensities for unamplified and WGA-amplified genomic DNA. To control for the association between probe GC content and intensity, probes with GC content of 8, 11, and 14 (out of 25) are shown in A–C, respectively. Plots for the remaining probe GC contents (and further experimental samples) are shown in Supplemental Figure 5. Probes are grouped into 50 equally sized bins genome-wide-based on their local CpG density, as shown in Figure 1, B and C. Box-and-whisker plots show the 25th and 75th percentile as the bottom and top of the box, and the band represents the median; the whiskers show the lowest data point within 1.5 interquartile range (IQR) of the 25th percentile and the highest data point within 1.5 IQR of the 75th percentile.
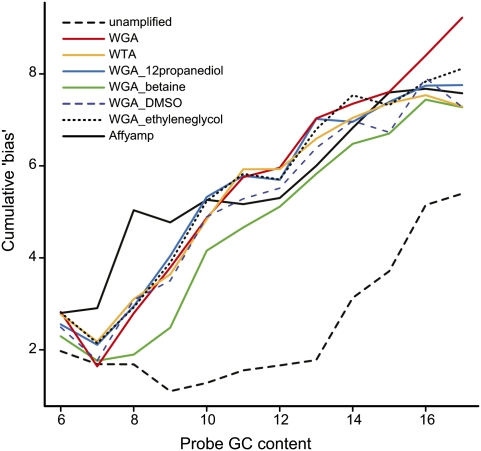
Since some form of genome-wide amplification is required to obtain sufficient DNA for array experiments, we next asked if recent variations to enhance the amplification of GC-rich regions (Zhang et al. 2009) could reduce the attenuation observed on the tiling arrays. Samples of unamplified genomic DNA with standard WGA, additives with WGA (Betaine, DMSO, ethylene glycol, 1,2-propanediol), and the different amplification conditions suggested by Affymetrix for ChIP-chip experiments (Fig. 3) are compared. To summarize the results, a cumulative bias score was calculated to quantify the signal attenuation in tiling array data across local CpG density bins for each group of probes at a GC content (for details of the bias score, see Methods; for explanation, see Supplemental Fig. 7). As shown previously in Figure 2, more CpG-rich attenuation (cumulative bias) at the higher probe GC content (greater than 11) occurs for unamplified DNA (Fig. 3). However, all the different amplification conditions tested show a much greater bias score than does unamplified DNA and did not reduce the signal attenuation (Fig. 3).
Figure 3.
Observed cumulative bias of various amplification methods. X-axis denotes the probe GC content. Y-axis denotes the cumulative bias score, which captures the cumulative signal attenuation over the 50 bins of local CpG density (for definition, see Methods). Each line represents a different amplification strategy.
Detection of differentially methylated regions
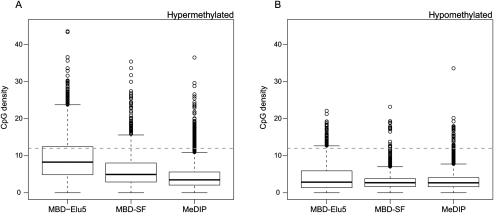
To explore the impact of these biases, we looked for differentially methylated regions (DMRs) across the promoter regions represented on the Affymetrix promoter tiling array, comparing the prostate cancer (LNCaP) and normal epithelial (PrEC) cell lines. Using a statistical procedure similar to model-based analysis of tiling arrays (MAT) (Johnson et al. 2006) at an estimated 5% false discovery rate (see Methods), 7384, 4398, and 3815 DMRs were detected between the two cell lines for MeDIP, MBD-Elu5, and MBD-SF, respectively. Given the enrichment profiles from Figure 1, it is not surprising that the detected DMRs showed CpG density distributions that reflect the enrichment profile, as shown in Figure 4. The differentially methylated promoters were split into hypermethylated (Fig. 4A) and hypomethylated in cancer (Fig. 4B) to illustrate the asymmetry in differential methylation between the cell lines. As expected, MBD-Elu5 discovers substantially more DMRs in CpG-rich regions and identifies the greatest proportion of CpG islands, and MBD-SF finds DMRs in a broader range of CpG density, while MeDIP identifies the lowest percentage of CpG-rich regions.
Figure 4.
Box-and-whisker plots of CpG density for putative DMRs (at estimated false discovery rate of 5%) between LNCaP and PrEC cells. Shown are hypermethylated (A) and hypomethylated (B) regions.
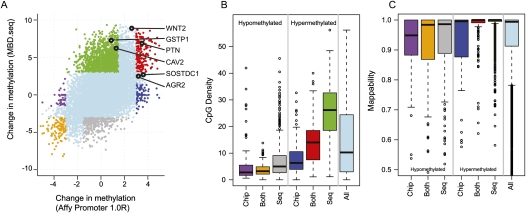
Next, MBD-SF enriched DNA was analyzed from the two cell lines using both promoter tiling array (MBDCap-chip) and high-throughput sequencing (MBDCap-seq), allowing us to compare directly the concordance of the two readouts. Since the tiling arrays only measure promoters, the sequencing data were summarized into bins of read counts at promoters so they could be directly compared. The statistical procedures used to detect differentially methylated promoters from the two platforms are fundamentally different, due to the nature of the data (probe intensities vs. read counts, see Methods). However, P-values should be on scales that are directly comparable. The P-values from each platform are back-transformed into Z-scores (i.e., quantiles of a standard normal distribution) to summarize the evidence for differential methylation, signed according to the direction of the change. Differential methylation Z-scores for both platforms are shown in Figure 5A. As expected, there is a general concordance (r = 0.46) since the two platforms are comparing enriched DNA from the same origin. One noticeable difference between the platforms is the range of Z-scores, with sequencing data giving a much larger range of evidence for differential methylation. Although it is not certain that every promoter-level difference above a threshold is indeed differentially methylated, the much wider range of Z-scores suggests the sequencing-based data have a higher sensitivity. Furthermore, comparison of MBDCap-seq data and quantitative Sequenom bisulphite-based DNA methylation data highlights a strong concordance (r = 0.81) (Supplemental Fig. 8). Figure 5A highlights that all six hypermethylated genes discussed previously are called differentially methylated by one of the readouts. Notably, the GSTP1 CpG island promoter shows a significantly higher number of reads in the region around the gene's transcription start site (TSS) for the cancer cells, but similar to the MeDIP-chip data (Fig. 5A), there is little evidence of differential methylation from the MBDCap-chip data (Supplemental Fig. 8). Similarly, the promoter of CAV2, a CpG-rich region, shows strong differential methylation for the sequencing but not for the microarray. The promoters of SOSTDC1 and AGR2, from low CpG density regions, are found by the microarray and only show moderate differential methylation in the sequencing data (Supplemental Fig. 9). Differential methylation calls for PTN and WNT2 are reasonably concordant. Furthermore, if a Z-score cutoff of three is set, the promoter array only detects 256 of the 2854 promoters detected by the sequencing data as differentially methylated, suggesting that its sensitivity is much lower. However, the tiling array also finds 246 regions differentially methylated that the sequencing data does not, suggesting there may also be inherent biases in the genomic regions that are suitable for sequencing.
Figure 5.
Comparison of MBD-SF tiling array and sequencing data. (A) Differential methylation Z-scores between LNCaP and PrEC cells using MBD-SF-seq (y-axis) and MBD-SF-chip (x-axis). The six validated genes that are shown in Supplemental Figures 3A and 4 are indicated with black dots. The remaining dot colors are chosen according to the differential methylation concordance between MBD-SF-seq and MBD-SF-chip Z-score as depicted in B. Note that some truly differentially methylated promoters, such as WNT2, are deemed “Indeterminate” by this concordance classification. (B) Box-and-whisker plots of CpG density for concordant and discordant differentially methylated promoters, with colors corresponding to the cutoffs shown in A. (C) Box-and-whisker plots of sequencing mapability of the concordant and discordant differentially methylated promoters, using the colors from A.
We next explored the CpG density of the concordant and discordant promoters (as defined by Z-score cutoffs; see Methods) that were detected using the two platforms (Fig. 5B), split into groups of hyper- and hypomethylated regions. The array-based readout detects a smaller percentage of CpG-rich regions in comparison with the sequencing-based data, supporting the observation that DNA amplification and other biases have a direct impact on the regions detected. The majority of regions detected by microarray and not by sequencing are in low CpG density regions. Conversely, sequencing-only detections are largely from CpG-rich regions. Furthermore, promoters deemed differentially methylated by microarray but not by sequencing, on average, have lower mapability (Fig. 5C). However, it should be noted that the probes in these same microarray-only detected regions exhibit a slightly higher probe copy number (see Supplemental Fig. 10).
The number of differentially methylated promoters identified by sequencing data is ultimately dependent on read depth. To estimate whether the saturation of promoter differential methylation has been achieved with the current sequencing depth, the MBD-SF-seq data set for LNCaP cells and PrECs was down-sampled at various fractions, and a curve was fitted to the number of differentially methylated promoters. The presented experiments capture an estimated 60%–68% of the differentially methylated promoters, while doubling the number of mapable reads will result in an estimated 78%–89% sensitivity (see Methods; Supplemental Fig. 11).
Copy number, input DNA, and DNA methylome data
Another important yet subtle aspect of identifying epigenetic changes, especially in a cancer setting, is the impact of copy number aberrations on the DNA methylation signal. Copy number aberrations can have a direct effect on transcript levels (Stranger et al. 2007). The effect is less clear in DNA methylome data, but the expectation is that genetically amplified (deleted) regions of the genome should be captured at a higher (lower) rate if the DNA is methylated. In the analysis of the ChIP-chip experiments, it is common practice to subtract the genomic DNA input signals from the immunoprecipitation signals in order to account for copy number, while indirectly this adjustment can also account for hybridization, sonication, or probe-specific effects. In two-color microarray experiments, this adjustment is done explicitly (Gal-Yam et al. 2008). Early sequencing-based DNA methylation mapping exercises have not included input DNA controls (Serre et al. 2009), and if included, biases are known to exist (Teytelman et al. 2009; Vega et al. 2009). It is highlighted here that signals from the input DNA can adequately identify copy number aberrations, and this knowledge will be critical to disentangling differential methylation from changes in copy number.
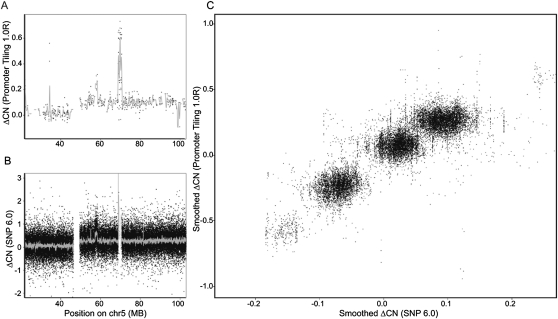
To validate the use of genomic input DNA tiling array data to account for copy number changes, Affymetrix SNP 6.0 array data were collected on the same two cell lines that have genome-wide DNA methylation data. To define copy number changes between the two cell lines, the input DNA tiling arrays were processed similarly to the gene expression data, resulting in a promoter-level summary of the change in copy number after accounting for probe-specific effects (see Methods) (Irizarry et al. 2003). Figure 6 shows the change in copy number between the two cell lines for chromosome 5 (for all other chromosomes, see Supplemental Fig. 12), emphasizing the strong association between promoter-level summaries of genomic input DNA on the promoter tiling array (Fig. 6A), and summarized SNP and copy number probes from the genotyping arrays (Fig. 6B). It also demonstrates the potential of using promoter-level summaries of input DNA signals for discovering copy number aberrations in the absence of directly collecting SNP array data or similar. Figure 6C shows the relationship between smoothed estimates of copy number (see Methods) for the two platforms, suggesting the genome-wide correspondence of copy number changes is quite high (r = 0.86).
Figure 6.
Using promoter tiling arrays to estimate changes in copy number. (A) Y-axis is the difference in copy number between the prostate cancer and normal epithelial cell line using the Affymetrix Promoter 1.0R array along human chromosome 5. The gray line represents kernel-smoothed differences over 200 kb. (B) Y-axis shows the difference in copy number using the Affymetrix SNP 6.0 array along the same region of chromosome 5. The gray line represents kernel-smoothed differences over 50 kb. (C) X-axis and y-axis represent the smoothed copy number changes between the prostate cancer and epithelial cell lines for the Promoter 1.0R and SNP 6.0 arrays, respectively, genome-wide over a common set of loci.
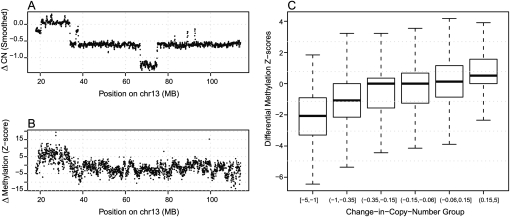
Next, we highlight that the effects of copy number aberrations are prominent in affinity-based epigenome data, affecting both DNA methylation and ChIP assays. The differential methylation detection exercise (using MBD-SF on prostate cancer LNCaP and PrEC lines) was extended to the entire genome, using read counts over 1500 base pair bins, and Z-scores were computed for each region. Figure 7A illustrates changes in MBDCap-seq read counts along chromosome 13. As expected, they are correlated with changes in copy number (Fig. 7B) for the same regions. Figure 7C shows the distribution of DMR Z-scores genome-wide, stratified by their corresponding change-in-copy-number status. Taken together, these observations underscore the need to integrate CNV explicitly with epigenome analyses. Interestingly, there is a large (∼10 M base pairs) region near 70 MB on chromosome 13 that shows deletion (SNP array data), but no corresponding change in methylation (MBDCap-seq). The MBDCap-seq data from the rest of this chromosome suggest that it is a region of hypermethylation, implying a potential link between regional hypermethylation and genome stability.
Figure 7.
Effects of copy number changes on differential methylation detection. (A) Differential methylation Z-score for between LNCaP and PrEC cells, using MBD-SF-seq, for human chromosome 13. (B) Smoothed Affymetrix SNP 6.0 array data showing corresponding changes in copy number. (C) Genome-wide distributions of Z-scores, stratified by the change-in-copy-number status of the corresponding regions.
Discussion
With the rapid growth of the different genome-wide technologies available for DNA methylation analysis (Lister et al. 2009; Oda et al. 2009; Serre et al. 2009; Ruike et al. 2010), it is timely to stop and reevaluate the limitations and benefits of the different techniques. We have evaluated the technical and data analysis aspects of promoter-level tiling microarray and genome-wide sequencing-based DNA methylation data and find that there are several hurdles that need to be overcome before a high sensitivity platform with genome-wide methylation coverage will emerge. Perhaps not unexpectedly, different enrichment techniques and readouts give different snapshots of the DNA methylome. Knowledge of inherent biases and limitations of each method should encourage protocol improvements and facilitate data integration from multiple platforms, as well as the development of improved bioinformatics tools to extract meaningful biological interpretation.
Comparisons of affinity-based methylome mapping techniques are now beginning to appear (Li et al. 2010). Here, a comparison of MBDCap- and MeDIP-based affinity capture strategies was performed, as well as a comparison of promoter tiling arrays and sequencing-based readouts. In addition, we highlight the technical limitations due to WGA, describe a novel method to assess amplification genome-wide with tiling arrays, and illustrate biological biases attributable to copy number that are relevant to the effective analysis of the DNA methylome. By using fully methylated DNA and LNCaP DNA, enrichment profiles were compared across the CpG density spectrum of MeDIP and two versions of MBDCap: MBD-Elu5, the 1000-mM fraction that elutes densely methylated DNA, and MBD-SF, a single elution encompassing all methylated fractions. Our data reveal higher overall enrichment using the methyl DNA binding domain protocol from MethylMiner, compared with immunoprecipitation with the 5-methylcytosine monoclonal antibody, especially in CpG-rich regions. MBD-Elu5 preferentially elutes CpG-rich DNA, while MBD-SF contains DNA molecules spanning a broader range of CpG densities. For all methods analyzed by promoter microarrays, a marked drop in signal intensity was observed at the CpG-dense regions, including at many of the CpG islands. This attenuation was found to be largely due to WGA but is also confounded by other effects, such as GC content of the microarray probes and cross-hybridization, which affects the signal and therefore sensitivity and dynamic range. Interestingly, CHARM has compared favorably in performance to MeDIP (Irizarry et al. 2008) and notably without an amplification step. However, CHARM does require large amounts of starting DNA and is typically used with a custom microarray.
Unfortunately for the MeDIP and MBDCap approaches, it is rarely feasible to get sufficient affinity-purified DNA for genome-wide analyses, thereby necessitating amplification before hybridization to microarrays. In some cases, pooling multiple samples may be a reasonable alternative, but this results in a loss of valuable replicate information. Furthermore, pooling affinity-purified DNA is often not practical when analyzing DNA from low cell numbers, such as formalin-fixed paraffin-embedded clinical samples. Our results suggest that amplification reduces sensitivity and permits only a subfraction of the genome to be interrogated. Specifically, tiling array probes representing CpG-rich regions, which are arguably of greatest interest for methylation mapping, appear to show a lower intensity and a compressed dynamic range. As a result, the ability to detect differential methylation using amplified DNA is compromised at many CpG islands.
Sequencing-based assays require less starting material. However, amplification during the sequencing protocol also has the potential to introduce some sequence bias in CpG-rich regions, albeit to a lesser extent. Promoter tiling array and high-throughput sequencing readouts of the same populations of MBDCap-enriched methylated DNA were compared. Even though the concordance is strong, it is the discordance that highlights the differences in the platform-specific snapshots of the methylome. DMRs that are detected by sequencing and not by microarray are commonly located in the CpG-rich regions of the genome, validating the loss in sensitivity on microarrays that is partly attributable to WGA. Furthermore, sequencing-based assays are strongly affected by enrichment levels, such that highly enriched regions are sequenced to a greater depth, resulting in higher power to detect changes. Therefore, microarrays may be better suited to interrogating regions of lower enrichment, such as those in lower CpG density areas, where the cost of sequencing to obtain sufficient coverage may become limiting. To a smaller extent, some of the DMRs detected by microarrays and not by sequencing are in regions of lower “mapability,” suggesting microarrays may have improved sensitivity in these regions, if unique probes can be designed. Furthermore, the extent to which the genomic repeat elements are present in affinity-captured methylated DNA is largely unknown. Longer or paired-end reads will result in higher mapability, while paired-end reads will be essential to studying the effects of repeat elements. Overall, sequencing data appear to be more sensitive for the discovery of DMRs, in terms of total detections, and they carry the obvious advantage that the entire genome can be interrogated. However, the complexities introduced by the enrichment levels, the amount of sequencing used, amplification, methylated repeat elements, CpG density, and mapability are cumulatively significant, suggesting that array and sequencing platforms may be complementary for cost-effective and comprehensive analysis of differential methylation.
Last, we studied the subtle effects that CNV may introduce into DNA methylation data sets. The use of input DNA hybridized to tiling arrays for economical copy number aberration detection was validated, and we highlighted that genetic changes can significantly confound the identification of epigenetic differences, if not explicitly integrated into the analysis. This will be of particular importance when analyzing cancer methylomes, for example, where copy number aberrations are widespread. Extrapolation of the information within existing copy number databases or from existing genomic DNA microarray or arrayCGH data should be straightforward. However, the implication of our results is that, in some cases, additional resources will need to be dedicated to collect CNV information (e.g., CNV-seq) (Xie and Tammi 2009). Copy number biases will not only confound genome-wide methylation analyses but will also be present in other affinity-based epigenome mapping exercises, such as chromatin immunopreciptation experiments studying histone modifications.
Many exciting new approaches have recently emerged to study genome-wide DNA methylation (Clarke et al. 2009; Lister et al. 2009; Flusberg et al. 2010), and along with these novel approaches have come an abundance of challenges, mainly associated with the interpretation of the growing masses of data. A better understanding of these technologies and the impact of current laboratory protocols, such as MeDIP and MBDCap, will go a long way toward the development of suitable and sensitive protocols for the genome-wide analysis of the methylome.
Methods
Cell lines and culture conditions
LNCaP prostrate cancer cells were cultured as described previously (Song et al. 2002). Normal PrECs (Cambrex Bio Science catalog no. CC-2555) were cultured according to the manufacturer's instructions in Prostate Epithelial Growth Media (PrEGM; Cambrex Bio Science catalog no. CC-3166).
Methylation profiling by MeDIP
DNA was extracted from the cell lines using the Puragene extraction kit (Gentra Systems). For fully methylated positive control DNA, CpG genome universal methylated DNA was obtained from Millipore (catalog no. 57821). The MeDIP assay was performed on 4 μg of sonicated genomic DNA (300–500 bp) in 1× IP buffer (10 mM sodium phosphate at pH 7.0, 140 mM NaCl and 0.05% Triton X-100). Ten micrograms anti-5-methylcytosine mouse monoclonal antibody (Calbiochem clone 162 33 D3 catalog no. NA81) was incubated overnight in 500 μL 1× IP buffer, and the DNA/antibody complexes were collected with 80 μL Protein A/G PLUS agarose beads (Santa Cruz sc-2003). The beads were washed three times with 1× IP buffer at 4°C and twice with 1 mL TE buffer at room temperature. The immune complexes were eluted with freshly prepared 1% SDS and 0.1 M NaHCO3, and the DNA was purified by phenol/chloroform extraction and ethanol precipitation and resuspended in 30 μL H2O. Input samples were processed in parallel.
Isolation of methylated DNA by MBDCap
The MethylMiner Methylated DNA Enrichment Kit (Invitrogen) was used to isolate the methylated DNA. One microgram of genomic DNA was sonicated to 100–500 bp. Then 3.5 μg (7 μL) of MBD-Biotin Protein was coupled to 10 μL of Dynabeads M-280 Streptavidin according to the manufacturer's instructions. The MBD-magnetic beads conjugates were washed three times and resuspended in 1 volume of 1× bind/wash buffer. The capture reaction was performed by adding 1 μg sonicated DNA to the MBD-magnetic beads on a rotating mixer for 1 h at room temperature. All capture reactions were done in duplicate. The beads were washed three times with 1× bind/wash buffer. The methylated DNA was eluted in one of two ways: (1) as a single fraction with a high-salt elution buffer (2000 mM NaCl), denoted MBD-SF; or (2) as distinct subpopulations based on the degree of methylation using an increasing NaCl concentration of the elution buffer, from 200 mM to 2000 mM in a stepwise gradient (elution 1, 200 mM; elution 2, 350 mM; elution 3, 450 mM; elution 4, 600 mM; elution 5, 1000 mM; and elution 6, 2000 mM). Each fraction was concentrated by ethanol precipitation using 1 μL glycogen (20 μg/μL), 1/10th volume of 3 M sodium acetate (pH 5.2), and two sample volumes of 100% ethanol, and was resuspended in 60 μL H20.
WGA and promoter array analyses
Immunoprecipitated DNA and input DNA from MeDIP immunoprecipitations and MBD-Capture reactions were amplified with GenomePlex Complete WGA Kit (Sigma catalog no. WGA2), according to the manufacturer's instructions. Fifty nanograms of DNA was used in each amplification reaction. The reactions were cleaned up using cDNA cleanup columns (Affymetrix no. 900371), and 7.5 μg of amplified DNA was fragmented and labeled according to Affymetrix Chromatin Immunoprecipitation Assay Protocol P/N 702238 Rev. 3. Affymetrix GeneChip Human Promoter 1.0R arrays (P/N. 900777) were hybridized using the GeneChip Hybridization wash and stain kit (P/N 900720).
Amplification bias experiments
WGA reactions were performed in the presence of reagents known to enhance the amplification of GC-rich DNA (Zhang et al. 2009). Betaine (Sigma B0300-IVL; final 2.2 M), ethylene glycol (Sigma E-9129; lot 23H00252; final 1.075 M), 1,2 propanediol (Sigma 398039; final 0.816M), and DMSO (Stratagene catalog no. 600260-53; final 4%) were used in separate 100 μL WGA reactions. Following the amplification, the DNA was purified, labeled, fragmented, and hybridized to the Affymetrix GeneChip Human Promoter 1.0R arrays as described above.
Local CpG density
We use the definition of local CpG density given by Pelizzola et al. (2008), with a window of 600 bp (Pelizzola et al. 2008) since we hybridize genomic DNA fragments with an average length of 600 bases, and individual probes are measuring signal from adjacent genomic regions and are thus affected by the number of CpG sites in this region. Briefly, the local CpG density is a weighted count of CpG sites in the genome upstream and downstream 600 bases from a given point of interest (e.g., microarray probe location). Weight decreases linearly from 1 at the center of the point of interest to 0 at 600 bases up- or downstream. The score is a reflection of the number of CpG sites in close proximity to the point of interest.
Cumulative signal attenuation
The cumulative bias score captures the degree of attenuation at high local CpG density for a set of probes with given probe GC content. Using the statistics (median, 25th percentile, and 75th percentile) from bins 3–17 for each combination of probe GC content and sample, a median and variance for the combination were calculated. The cumulative bias is the sum of absolute deviations from the calculated median and variance among the 50 bins for the combination. A pictorial description is given in Supplemental Figure 7.
Untargeted promoter-level analysis of promoter array data
A probe-level score for the difference of interest was calculated (LNCaP signal − PrEC signal) and smoothed using a trimmed mean (600-bp window) and searched for a significant and persistent difference. To calculate a false discovery rate, the order of the probes is randomized and the same procedure is followed. The method is implemented in the regionStats function of the Repitools package (Statham et al. 2010).
Targeted promoter-level analysis of promoter array and sequencing data
For tiling array data, a probe-level score for the difference of interest was calculated (LNCaP signal − PrEC signal) using all the probes within 750 bases of every TSS. A one-sample t-statistic was then calculated to determine whether the average probe-level score for each TSS is significantly different from zero, as implemented in the blockStats function of the Repitools package (Statham et al. 2010). P-values are calculated from t-statistics. For sequencing data, the number of reads that mapped to within 750 bases of every TSS was counted. Then, an exact test for the difference in counts between LNCaP and PrEC was calculated using the Bioconductor edgeR package (Robinson et al. 2010).
Data normalization
The normalization for Affymetrix Human Promoter 1.0R arrays follows the adjustment proposed from the model-based analysis of tiling arrays (MAT) (Johnson et al. 2006), which compensates for the global effects of base composition and probe copy number.
By the nature of high-throughput sequencing experiments, each sample is sequenced to a different depth. The compensation for total read depth occurs at the following stages: (1) in the analysis of MBDCap-enriched SssI-treated DNA, signal levels are presented as read counts per million uniquely mapped, as shown in Figure 1C (promoters) and Supplemental Figure 2 (genome); and (2) the differential analysis of read counts at promoters, for comparing LNCaP-MBDCap versus PrEC-MBDCap, explicitly compensates for read depth (i.e., library size) in the edgeR software (Robinson et al. 2010).
Back-transformed Z-scores
To put observed differences between LNCaP and PrEC cells on a common scale, for both the tiling array and sequencing platforms, P-values were back-transformed into signed Z-scores. For each P-value, the Z-score is the value, z, of the standard normal distribution such that Pr(Z > z) = p/2, where p is the P-value. Regions with higher signal (or higher relative count) in LNCaP cells will have positive Z-scores, otherwise they will be negative.
Mapping Genome Analyzer sequencing reads
We mapped 36 base pair reads to the hg18 reference genome using Bowtie (Langmead et al. 2009), with up to three mismatches. Reads that mapped more than once (i.e., identical start sites) to a single genomic location were excluded.
Concordance of DMRs between promoter arrays and sequencing
Promoters were deemed to be concordant and hypermethylated if both platforms give a Z-score greater than 3 and to be concordant and hypomethylated if both Z-scores are less than −3. Hypermethylated discordant promoters were defined as one platform having a Z-score greater than 3 and the other platform with a Z-score less than 1.5. Similarly, cutoffs of −3 and −1.5 were used to define discordant hypomethylated promoters. Note that promoters deemed as differentially methylated by one platform and not by the other (i.e., between 1 and 3 or between −3 and −1) are considered indeterminate.
Down-sampling analysis
The counts were down-sampled for each gene promoter to accumulate total read counts between 20% and 100% of the original data set (10 data sets are sampled for each level of down-sampling). For each down-sampled data set, the number of DMRs (using an absolute Z-score cutoff of 3) was calculated. Using the median number of DMRs for each level of subsampling, a nonlinear curve of the form axc/(b + xc) was fitted (using the R nls function) in order to estimate the total number of DMRs (i.e., parameter a). The 95% confidence interval for the total number of DMRs is (5555, 6323). The data sets sampled at 100% reveal 3777 DMRs.
Smoothed copy number estimates
The change in copy number has been smoothed using a truncated Gaussian kernel smoother using a bandwidth of 50/200 kb (promoter/SNP array), using the implementation in the aroma.core R package (Bengtsson et al. 2008).
Mapability
Using Bowtie, all possible 36-bp reads from the entire human genome were mapped back to the genome. At every base, a read can either be unambiguously mapped starting at a given position or not. Mapability is the proportion of such reads that can be mapped for a given genomic region.
Acknowledgments
We thank Kate Patterson for help with preparation of the figures and critical reading of the manuscript and Oleg Mayba for mappability calculation code. This work is supported by National Health and Medical Research Council (NH&MRC) project (427614, 481347) (M.D.R., C.S., D.S.) and Fellowship (S.J.C.), Cancer Institute NSW grants (CINSW: S.J.C., A.L.S.), and NBCF Program Grant (S.J.C.) and ACRF. We also thank the Ramaciotti Centre, University of New South Wales (Sydney, Australia) for array hybridizations and Illumina GAII sequencing of MBDCap DNA.
Footnotes
[Supplemental material is available online at http://www.genome.org. The data from this study have been submitted to Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under SuperSeries accession no. GSE24546.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.110601.110.
References
- Bengtsson H, Simpson K, Bullard J, Hansen K 2008. aroma.affymetrix: A generic framework in R for analyzing small to very large Affymetrix data sets in bounded memory, tech report no. 745. Department of Statistics, University of California, Berkeley [Google Scholar]
- Bredel M, Bredel C, Juric D, Kim Y, Vogel H, Harsh GR, Recht LD, Pollack JR, Sikic BI 2005. Amplification of whole tumor genomes and gene-by-gene mapping of genomic aberrations from limited sources of fresh-frozen and paraffin-embedded DNA. J Mol Diagn 7: 171–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H 2009. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol 4: 265–270 [DOI] [PubMed] [Google Scholar]
- Coolen MW, Stirzaker C, Song JZ, Statham AL, Kassir Z, Moreno CS, Young AN, Varma V, Speed TP, Cowley M, et al. 2010. Consolidation of the cancer genome into domains of repressive chromatin by long-range epigenetic silencing (LRES) reduces transcriptional plasticity. Nat Cell Biol 12: 235–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW 2010. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods 7: 461–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X, Lin JC, Liang G, Jones PA, Tanay A 2008. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci 105: 12979–12984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Bock C, Mikkelsen TS, Jager N, Smith ZD, Tomazou E, Gnirke A, Lander ES, Meissner A 2010. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods 7: 133–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15 doi: 10.1093/nar/gng015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Carvalho B, Wu H, Brandenburg SA, Jeddeloh JA, Wen B, Feinberg AP 2008. Comprehensive high-throughput arrays for relative methylation (CHARM). Genome Res 18: 780–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A 2003. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet 33: 245–254 [DOI] [PubMed] [Google Scholar]
- Jeddeloh JA, Greally JM, Rando OJ 2008. Reduced-representation methylation mapping. Genome Biol 9: 231 doi: 10.1186/gb-2008-9-8-231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS 2006. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci 103: 12457–12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Li W, Gordon DB, Bhattacharjee A, Curry B, Ghosh J, Brizuela L, Carroll JS, Brown M, Flicek P, et al. 2008. Systematic evaluation of variability in ChIP-chip experiments using predefined DNA targets. Genome Res 18: 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA 1999. The DNA methylation paradox. Trends Genet 15: 34–37 [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB 2002. The fundamental role of epigenetic events in cancer. Natl Rev 3: 415–428 [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB 2007. The epigenomics of cancer. Cell 128: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky ZA, Tang T, Wang SC, Ptak C, Oh GH, Wong AH, Feldcamp LA, Virtanen C, Halfvarson J, Tysk C, et al. 2009. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet 41: 240–245 [DOI] [PubMed] [Google Scholar]
- Laird PW 2010. Principles and challenges of genome-wide DNA methylation analysis. Natl Rev 11: 191–203 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Ye M, Li Y, Yan Z, Butcher LM, Sun J, Han X, Chen Q, Zhang X, Wang J 2010. Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods (in press). doi: 10.1016/j.ymeth.2010.04.009 [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K, Silke J, Gramatikoff K, Schaffner W 1994. The CpG-specific methylase SssI has topoisomerase activity in the presence of Mg2+. Nucleic Acids Res 22: 5354–5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M, Gonzalgo ML, Yegnasubramanian S, Lin X, De Marzo AM, Nelson WG 2004. GSTP1 CpG island hypermethylation as a molecular biomarker for prostate cancer. J Cell Biochem 91: 540–552 [DOI] [PubMed] [Google Scholar]
- Novak P, Jensen T, Oshiro MM, Watts GS, Kim CJ, Futscher BW 2008. Agglomerative epigenetic aberrations are a common event in human breast cancer. Cancer Res 68: 8616–8625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda M, Glass JL, Thompson RF, Mo Y, Olivier EN, Figueroa ME, Selzer RR, Richmond TA, Zhang X, Dannenberg L, et al. 2009. High-resolution genome-wide cytosine methylation profiling with simultaneous copy number analysis and optimization for limited cell numbers. Nucleic Acids Res 37: 3829–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris PL 2009. A whole-genome amplification protocol for a wide variety of DNAs, including those from formalin-fixed and paraffin-embedded tissue. Methods Mol Biol 556: 89–98 [DOI] [PubMed] [Google Scholar]
- Pelizzola M, Koga Y, Urban AE, Krauthammer M, Weissman S, Halaban R, Molinaro AM 2008. MEDME: An experimental and analytical methodology for the estimation of DNA methylation levels based on microarray derived MeDIP-enrichment. Genome Res 18: 1652–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponzielli R, Boutros PC, Katz S, Stojanova A, Hanley AP, Khosravi F, Bros C, Jurisica I, Penn LZ 2008. Optimization of experimental design parameters for high-throughput chromatin immunoprecipitation studies. Nucleic Acids Res 36: e144 doi: 10.1093/nar/gkn735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh TJ, Delaney AD, Farnoud N, Flibotte S, Griffith M, Li HI, Qian H, Farinha P, Gascoyne RD, Marra MA 2008. Impact of whole genome amplification on analysis of copy number variants. Nucleic Acids Res 36: e80 doi: 10.1093/nar/gkn378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch T, Li H, Wu X, Pfeifer GP 2006. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res 66: 7939–7947 [DOI] [PubMed] [Google Scholar]
- Rauch TA, Zhong X, Wu X, Wang M, Kernstine KH, Wang Z, Riggs AD, Pfeifer GP 2008. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci 105: 252–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. 2000. Genome-wide location and function of DNA binding proteins. Science 290: 2306–2309 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK 2010. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruike Y, Imanaka Y, Sato F, Shimizu K, Tsujimoto G 2010. Genome-wide analysis of aberrant methylation in human breast cancer cells using methyl-DNA immunoprecipitation combined with high-throughput sequencing. BMC Genomics 11: 137 doi: 10.1186/1471-2164-11-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre D, Lee BH, Ting AH 2009. MBD-isolated genome sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res 38: 391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JZ, Stirzaker C, Harrison J, Melki JR, Clark SJ 2002. Hypermethylation trigger of the glutathione-S-transferase gene (GSTP1) in prostate cancer cells. Oncogene 21: 1048–1061 [DOI] [PubMed] [Google Scholar]
- Sorensen AL, Jacobsen BM, Reiner AH, Andersen IS, Collas P 2010. Promoter DNA methylation patterns of differentiated cells are largely programmed at the progenitor stage. Mol Biol Cell 21: 2066–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statham AL, Strbenac D, Coolen MW, Stirzaker C, Clark SJ, Robinson MD 2010. Repitools: An R package for the analysis of enrichment-based epigenomic data. Bioinformatics 26: 1662–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, et al. 2007. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315: 848–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo YY, Inouye M, Small KS, Fry AE, Potter SC, Dunstan SJ, Seielstad M, Barroso I, Wareham NJ, Rockett KA, et al. 2008. Whole genome-amplified DNA: Insights and imputation. Nat Methods 5: 279–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teytelman L, Ozaydin B, Zill O, Lefrancois P, Snyder M, Rine J, Eisen MB 2009. Impact of chromatin structures on DNA processing for genomic analyses. PLoS ONE 4: e6700 doi: 10.1371/journal.pone.0006700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega VB, Cheung E, Palanisamy N, Sung WK 2009. Inherent signals in sequencing-based chromatin-immunoprecipitation control libraries. PLoS ONE 4: e5241 doi: 10.1371/journal.pone.0005241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D 2005. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37: 853–862 [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466 [DOI] [PubMed] [Google Scholar]
- Wei H, Kuan PF, Tian S, Yang C, Nie J, Sengupta S, Ruotti V, Jonsdottir GA, Keles S, Thomson JA, et al. 2008. A study of the relationships between oligonucleotide properties and hybridization signal intensities from NimbleGen microarray datasets. Nucleic Acids Res 36: 2926–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng YI, Huang TH, Yan P.S 2009. Methylated DNA immunoprecipitation and microarray-based analysis: Detection of DNA methylation in breast cancer cell lines. Methods Mol Biol 590: 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Tammi MT 2009. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics 10: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yang X, Meng L, Liu F, Shen C, Yang W 2009. Enhanced amplification of GC-rich DNA with two organic reagents. Biotechniques 47: 775–779 [DOI] [PubMed] [Google Scholar]