Abstract
Efficient sequencing of animal and plant genomes by next-generation technology should allow many neglected organisms of biological and medical importance to be better understood. As a test case, we have assembled a draft genome of Caenorhabditis sp. 3 PS1010 through a combination of direct sequencing and scaffolding with RNA-seq. We first sequenced genomic DNA and mixed-stage cDNA using paired 75-nt reads from an Illumina GAII. A set of 230 million genomic reads yielded an 80-Mb assembly, with a supercontig N50 of 5.0 kb, covering 90% of 429 kb from previously published genomic contigs. Mixed-stage poly(A)+ cDNA gave 47.3 million mappable 75-mers (including 5.1 million spliced reads), which separately assembled into 17.8 Mb of cDNA, with an N50 of 1.06 kb. By further scaffolding our genomic supercontigs with cDNA, we increased their N50 to 9.4 kb, nearly double the average gene size in C. elegans. We predicted 22,851 protein-coding genes, and detected expression in 78% of them. Multigenome alignment and data filtering identified 2672 DNA elements conserved between PS1010 and C. elegans that are likely to encode regulatory sequences or previously unknown ncRNAs. Genomic and cDNA sequencing followed by joint assembly is a rapid and useful strategy for biological analysis.
Sanger sequencing of eukaryotic genomes and transcriptomes has enabled large-scale gene discovery and evolutionary comparisons, but has also been a laborious process requiring multiple centers, millions of dollars, and years per genome. The human genome draft sequence was at first so fragmented that mRNAs and expressed sequence tags (ESTs) were used to scaffold it (Kent and Haussler 2001); three more years were needed to drive the human genome sequence to its near-finished state (International Human Genome Sequencing Consortium 2004). The search for variation in the human genome led to new DNA sequencing methods, producing short reads at much lower cost that can be aligned to a reference genome (Bentley et al. 2008). To allow de novo genome assembly from these short reads, programs that use de Bruijn graphs rather than classic overlapping have been developed and applied to microbial genomes (Zerbino and Birney 2008; Chaisson et al. 2009). Similarly, a transcriptome can be assembled into expressed sequence tags either by mapping reads onto a genome sequence or by de novo assembly (Haas and Zody 2010).
There are many organisms that have been studied by, at most, a small cohort of researchers, which are unlikely ever to be sequenced by a genome center, but which, nevertheless, could be useful to biology and medicine if their genome could be characterized. For instance, there are between 40,000 and 10 million nematode species (Blaxter 1998), many of which are important parasites and pests. As an instance of their possible analysis, we have generated a draft genome and transcriptome of the nematode Caenorhabditis sp. 3 PS1010 (NCBI taxonomy ID 96668; henceforth, “PS1010”). PS1010 is a nematode in the same genus as C. elegans, but is more distantly related to C. elegans and C. briggsae than they are to each other (Fig. 1; Kiontke and Fitch 2005; K Kiontke, pers. comm.). DNA sequence divergence between PS1010 and C. elegans is comparable to that between mammals and birds (Kiontke and Fitch 2005). Nevertheless, PS1010 still has identifiable, highly conserved noncoding DNA elements in common with C. elegans (Kuntz et al. 2008). We have sequenced the genome and transcriptome of PS1010 using Illumina paired and unpaired 75-nt reads, used Velvet (Zerbino and Birney 2008) to assemble supercontigs from both cDNA and genomic DNA, and then assembled both sequence sets into a gene-centric draft genome assembly of 79.8 Mb (Fig. 2). This approach both improved the assembly and produced better gene models over the entire expression range of the transcriptome. Our assembly has a supercontig N50 of 9.4 kb, nearly twice the average gene lengths of C. elegans and C. briggsae (Stein et al. 2003); it encodes a full Caenorhabditis proteome, and 2672 highly conserved DNA elements that may be regulatory.
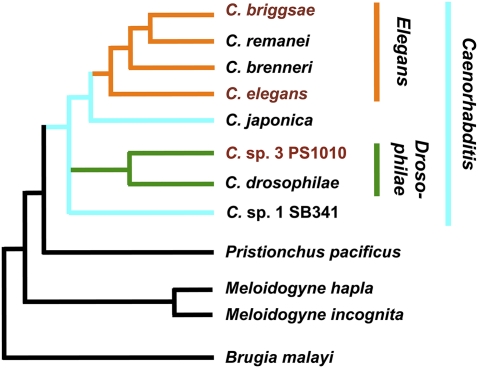
Figure 1.
Phylogenetic relationship of Caenorhabditis sp. 3 PS1010 to representative Caenorhabditis species and other nematode species discussed in this work, as determined by Kiontke et al. (2007), Meldal et al. (2007), and K Kiontke (pers. comm.). PS1010 is an outgroup to previously sequenced Caenorhabditis species, all of which closely resemble C. elegans, and most of which are formally considered part of an Elegans species group (Sudhaus and Kiontke 1996; Kiontke et al. 2007). PS1010 itself falls into a newly characterized Drosophilae species group, whose members substantially differ from Elegans species in morphology and mating behavior (K Kiontke, pers. comm.). However, PS1010 is still more closely related to C. elegans than non-Caenorhabditis nematodes such as Pristionchus, Meloidogyne, and Brugia, and hence, is more likely to show noncoding sequence conservation with C. elegans and C. briggsae. Phylogram not drawn to scale.
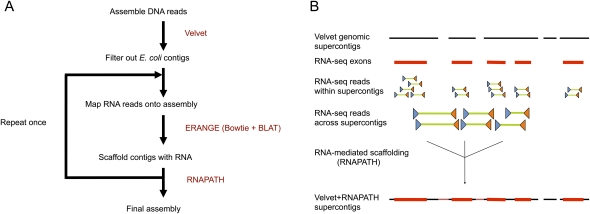
Figure 2.
Sequencing strategy. (A) Genomic reads are assembled using the Velvet short read assembler and then filtered for high similarity to E. coli. RNA-seq paired reads are mapped using a combination of Bowtie and BLAT onto this preliminary genomic assembly. The RNA-seq reads are then imported into ERANGE, where those reads with ends on separate supercontigs serve as input to the RNAPATH module. This process can be repeated with trimmed reads to increase mappable reads, if necessary. (B) Paired RNA reads (each pair represented by a blue and an orange triangle connected by a green line), where each read end maps to a separate genomic supercontig, can be used to scaffold (i.e., to join, order, and orient) those genomic supercontigs based on the two read ends pointing toward each other.
Results
Sequencing of the genome and RNA-seq-mediated scaffolding
We sequenced and assembled 200-bp fragment libraries at 100-fold nominal combined coverage (assuming a 100-Mb genome like C. elegans) to get an initial assembly of 79.8 Mb. This assembly's supercontig N50 improved from 1.5 kb to 5.0 kb with the addition of 375- and 450-bp fragment libraries, each having 35-fold coverage, for a final nominal coverage of 170-fold; however, coverage dropped significantly in regions of very low and very high GC content. These numbers do not include 497 supercontigs, totaling 4.6 Mb with an N50 of 64.7 kb and with ≥90% identity to genome sequences of Escherichia coli (PS1010’s laboratory food source); 10% of the raw reads mapped to E. coli sequences.
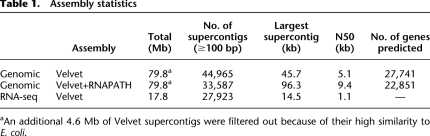
We also sequenced the mixed-stage larval transcriptome of PS1010 to a depth of 53.2 million 2 × 75 nt reads. Using pair-mates that mapped to different Velvet genomic supercontigs, we performed RNA-seq-mediated DNA scaffolding with the RNAPATH module in ERANGE 3.2 (Mortazavi et al. 2008) and generated a 79.8-Mb draft genome of PS1010 (Table 1); 15,450 Velvet supercontigs were placed into 4072 RNAPATH supercontigs.
Table 1.
Assembly statistics
aAn additional 4.6 Mb of Velvet supercontigs were filtered out because of their high similarity to E. coli.
To test the completeness of our assembly, we mapped its supercontigs onto 429 kb of PS1010 sequences already in GenBank, including 417 kb of pilot genome sequence (Kuntz et al. 2008). A total of 576 supercontigs generated by Velvet covered 90.7% of the previously known 429 kb. In contrast, the RNAPATH-based assembly covered the same sequence with only 402 supercontigs (Fig. 3). RNAPATH excluded 138 supercontigs that had more than 50% overlap with 81 PS1010 genomic repeats that amounted to an additional 1.1% of pilot sequence (or equivalently 10% of the gap sequence) along with 1% of standalone intronic supercontigs from the cDNA-mediated scaffolds (Fig. 3). The rest of the missing pilot sequence are in regions of low coverage with very low or very high GC content. Supercontigs composed of intergenic DNA or genes lacking RNA-seq data were left untouched. While still fragmented, this assembly is sufficient to analyze genes and should also contain a substantial fraction of noncoding elements conserved between PS1010 and C. elegans.
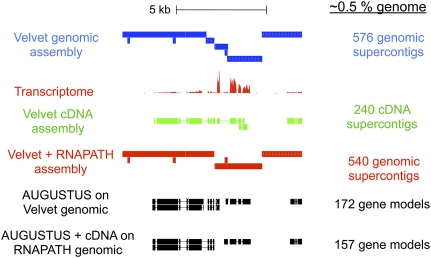
Figure 3.
An example of Velvet supercontigs (blue) and RNAPATH supercontigs (red) on 20 kb of Sanger-sequenced PS1010 fosmids (Kuntz et al. 2008) along with the ERANGE mapped coverage of the transcriptome reads (red), Velvet assembly of transcriptome (green), AUGUSTUS gene predictions on the original Velvet assembly and AUGUSTUS with Velvet-computed cDNA sequences assisted predictions on the final cDNA-scaffolded assembly. cDNA-mediated scaffolding combined with a genefinder improves the accuracy of gene models by allowing genes fragmented between genomic supercontigs to be on the same scaffold (broken line box). Summary statistics on the right are for the entire Sanger-sequenced 429 kb of sequence (corresponding to ∼0.5% of the genome).
Assembly of the transcriptome and gene annotation
To optimize our parameters for assembling PS1010 RNA-seq data into cDNA supercontigs, we first tested our parameters on a staged C. elegans L3 2 × 75 RNA-seq data set for which the correct outputs of assembly would be largely known. We found that Velvet typically made better assemblies of cDNA from moderately expressed genes than from strongly expressed ones. We thus assembled cDNA from such high-expression genes from a small subset of RNA-seq reads (one million), while doing a separate assembly with all of the reads for low-abundance cDNA and merging the resulting supercontigs by concatenating the FASTA sequences. Using this two-tiered strategy on our PS1010 RNA-seq data, we assembled 17.8 Mb of cDNA into 27,923 supercontigs with an N50 of 1.06 kb, 99% of which mapped back onto the genome. In contrast to genomic DNA, less than 0.02% of the RNA-seq reads mapped to E. coli. Velvet cDNA supercontigs were used as EST hints for the AUGUSTUS genefinder run with Caenorhabditis settings (Stanke et al. 2008) to predict 22,851 genes encoding 28,978 proteins. These gene models were used to evaluate expression levels with ERANGE (Supplemental Fig. S1); 63.2% of 161,032 predicted exons in 78.1% of the genes showed expression over 1 RPKM (read per kilobase per million reads) in our PS1010 RNA-seq data, corresponding to ≥6 reads for the median PS1010 exon length of 143.
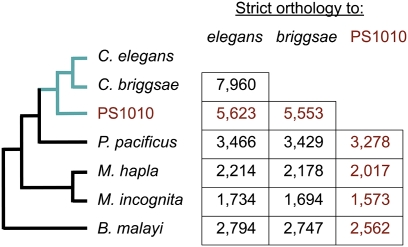
To characterize the completeness and content of PS1010's predicted genes, we used OrthoMCL (Li et al. 2003) and HMMER/PFAM-A (Finn et al. 2008; http://hmmer.janelia.org) to identify orthology groups and protein domains from PS1010, C. elegans, C. briggsae, Pristionchus pacificus (Dieterich et al. 2008), Meloidogyne hapla (Opperman et al. 2008), M. incognita (Abad et al. 2008), and Brugia malayi (Ghedin et al. 2007). All three Caenorhabditis proteomes had comparable similarity to those of the other nematodes (Fig. 4). A total of 5623 PS1010 genes (25% of 22,851) showed strict orthology (1:1 gene ratios in an orthology group) with C. elegans genes; this is ∼70% as many gene pairs as had strict orthology between C. briggsae and C. elegans. 11,633 (51%) displayed homology with at least one gene in another nematode genome; 11,630 (51%) encoded at least one of 3466 PFAM-A domains; 45 PFAM-A domains, encoded by 69 PS1010 genes, were found both in PS1010 and at least one non-Caenorhabditis nematode, yet were missing from both C. elegans and C. briggsae, suggesting that they have been specifically lost from the Elegans group (Supplemental Table S1). Two of these domains, encoded by one PS1010 gene apiece, are found in all four non-Caenorhabditis nematodes that we searched. Conversely, 85 PFAM-A domains were found in all nematode genomes except for PS1010 (Supplemental Table S2); at least some of these domains might exist within genes present in PS1010, but are missing from our assembly. (By comparison, 19 and 30 PFAM-A domains were found in all nematode genomes except for C. elegans and C. briggsae, respectively.) Some PFAM-A domains are represented in PS1010 by up to 370 genes, indicating that our genome assembly successfully captured extensively paralogous gene sets: for instance, 602 genes in PS1010 encode possible serpentine receptors, and 54 genes encode major sperm proteins (Supplemental Table S3). The number of predicted receptors is close to that for P. pacificus (613), though half that for C. elegans (1411) and C. briggsae (1120).
Figure 4.
The pattern of pairwise strict orthologies between the three Caenorhabditis species and the non-Caenorhabditis nematodes P. pacificus and M. hapla matches their known phylogeny. In particular, the numbers of strict orthologs between PS1010 and each of the outgroup species are comparable to those seen between C. elegans and C. briggsae and the same outgroups, suggesting that our protein-coding gene set is largely complete.
Identifying conserved noncoding elements
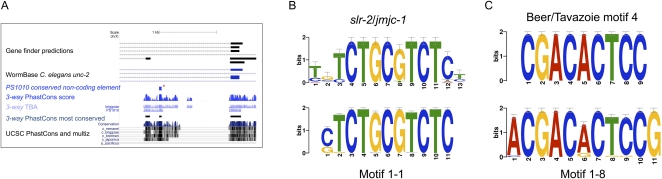
To identify noncoding sequences in the C. elegans genome that are highly conserved between C. elegans and PS1010, including ones likely to be regulatory, we used TBA/MULTIZ (Blanchette et al. 2004) to align PS1010 to the C. elegans and C. briggsae genomes and scanned the alignments with phastCons (Siepel et al. 2005) for three-species conservation. A total of 6.21% of the C. elegans genome showed conservation in 95,712 elements with an average size of 65 bp; 97.2% of these elements overlapped with repetitive DNA, known protein-coding or ncRNA exons, or alternative exon predictions (in some cases generated by us from ESTs and RNA-seq data; Supplemental Table S4). For example, the unc-2 gene had 53 unfiltered phastCons elements, but only three passed all of our filters, one of which marked a potential new promoter (Fig. 5A). Overlaps were conservatively defined as any match of ≥1 nt. We found 2672 filtered elements in all, comprising 0.08% of the C. elegans genome, ranging from 7 to 160 bp in size, with an average of 29 bp; 9.5% of these elements are ≥50 bp long (Supplemental Fig. S2). Two elements fall into a lin-39 enhancer conserved between C. elegans and PS1010 (Kuntz et al. 2008). In C. elegans, these elements disproportionately reside near genes annotated with 28 Gene Ontology (GO) terms (Supplemental Table S5). The most significantly enriched terms relate to reproduction, growth, and embryonic development.
Figure 5.
Conserved noncoding elements. (A) A highly conserved noncoding element (red star) in an intron of unc-2, as identified by phastCons and passing all of our filters, is a possible promoter element. Conserved PS1010 elements are typically subsets of the conserved elements shared between Elegans group genomes. (B) The most statistically significant predicted motif from the highly conserved noncoding elements (motif 1-1) (Table 2) is equivalent to the slr-2/jmjc-1-responsive motif of Kirienko and Fay (2010). (C) A functionally uncharacterized motif (1-8) closely resembles motif 4 of Beer and Tavazoie (2004).
One possible explanation of our persistent residue of longer elements might be that they overlap highly conserved noncoding RNA genes whose expression is rare enough to have eluded annotation. To test this idea, we checked our elements for overlaps with 3672 putative ncRNAs predicted in C. elegans by Missal et al. (2006) with RNAz, of which 1290 remained completely novel by January 2010 (i.e., they still did not overlap annotated protein- or nonprotein-coding exons in WormBase WS210). A total of 128 of our elements (4.8%) indeed overlapped RNAz predictions, and 72 elements (2.7%) had ≥80% overlap with the novel RNAz subset; the latter set of elements ranged in size from 10 to 120 nt, with a mean of 35 nt (Supplemental Table S4; Supplemental Fig. S2). However, 95% of our elements had no overlap with RNAz predictions, and this nonoverlapping majority ranged from 7 to 160 nt in size with a mean of 29 nt. This observation suggests that the filtered elements may identify novel, highly conserved ncRNAs, but that such cryptic ncRNAs do not currently account for either the bulk of elements or even most of the larger ones.
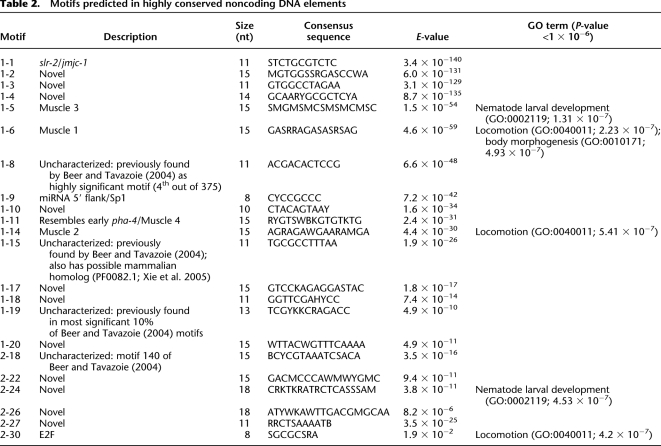
If these elements are genuinely regulatory, they should share recurrent motifs that at least partially match known regulatory sequences. We detected 22 motifs in 1193 elements with MEME (Bailey and Elkan 1994) and FIMO (Bailey et al. 2009). We then compared them with published motifs with TOMTOM (Gupta et al. 2007), finding significant similarities to motifs from C. elegans and the general literature (Table 2; Supplemental Table S6). Our most statistically significant predicted motif was equivalent to a slr-2/jmjc-1-dependent stress-response motif conserved between ecdysozoa and deuterostomes (Fig. 5B; Kirienko and Fay 2010). Other predicted motifs matched a miRNA promoter-associated motif (Ohler et al. 2004), the core promoter SP1 site (Li et al. 2004), muscle-specific motifs 1–4 of Zhao et al. (2007), early and late PHA-4 binding sites (Gaudet et al. 2004), the M-2 pharyngeal muscle motif (Ao et al. 2004), and the E2F binding site (van den Heuvel and Dyson, 2008). Subsets of those elements with matches to muscle-specific motifs were disproportionately found near genes annotated with GO terms for locomotion, body morphogenesis, and nematode larval development. The unc-2 element in Figure 5A contains a match to the E2B-like motif 2-30, which is also associated with locomotion; unc-2 itself encodes a voltage-gated calcium channel α1 subunit required for normal movement (Mathews et al. 2003). Other predicted motifs were of comparable statistical significance, but did not match sites with known functions. Four of them matched previous predictions by Beer and Tavazoie (2004), indicating that at least some of these novel motifs are likely to be real, uncharacterized regulatory sites conserved between PS1010 and C. elegans (Table 2; Fig. 5C).
Table 2.
Motifs predicted in highly conserved noncoding DNA elements
Discussion
We have carried out next-generation sequencing and analysis of the nematode Caenorhabditis sp. 3 PS1010, identifying approximately 18,000 expressed protein-coding genes in ∼90% of its genome, along with approximately 2700 noncoding DNA elements highly conserved between PS1010 and C. elegans. PS1010 is a member of the Caenorhabditis genus, but is not part of the Elegans group (Kiontke and Fitch 2005), which includes C. briggsae, three other recently sequenced nematodes (C. remanei, C. brenneri, and C. japonica), and an increasing number of unnamed elegans-like species. Instead, PS1010 belongs to a newly defined Drosophilae group within Caenorhabditis (Fig. 1; K Kiontke, pers. comm.). PS1010s conservation of genes and noncoding DNA therefore defines traits likely to be strongly required throughout the Caenorhabditis genus, despite overt differences in morphology and behavior between Elegans and Drosophilae group species (Sudhaus and Kiontke 1996; K Kiontke, pers. comm.) and despite sequence divergence comparable to that between humans and birds (Kiontke and Fitch 2005). In particular, the 2672 candidate DNA elements that passed our extensive filters probably encode either highly conserved regulatory elements or cryptic exons missed in the extensive annotation of C. elegans. While elements have an average size of 66 bp before being filtered with known exons, filtered elements average 29 bp in size (Supplemental Table S4). In addition, recurrent motifs found within the filtered elements include matches to several published regulatory motifs, and two elements mapped into a lin-39 enhancer previously shown to be conserved between C. elegans and PS1010 (Kuntz et al. 2008). These results are consistent with the hypothesis that many filtered elements are regulatory. Kuntz et al. (2008) found three other lin-39 enhancers conserved in PS1010 that our elements did not detect; we suspect that this arises from a limited ability of our gene–centric assembly to be aligned by TBA/MULTIZ in regions far from exons. This, in turn, suggests that a better PS1010 genome assembly might reveal significantly more than 2700 noncoding DNA elements to be conserved between C. elegans and PS1010.
One goal of our work was to devise an analytical tool kit for animal or plant genomes 70–300 Mb in size, usable by a small research group, with (in our case) a particular focus on nematode species. PS1010 was a good test case for this, because we had previously Sanger-sequenced fosmids representing roughly 0.5% of its genome (Kuntz et al. 2008) and, therefore, we could assess the quality of our genomic data through several rounds of sequencing and assembly. We were able to produce a genome assembly, which, though unsuitable for analyses of long-range regulation or multigene synteny, does support analyses of gene function, orthology, and short-range gene regulation. We also found that the PS1010 genome and transcriptome could be determined effectively with a single round of sequencing (e.g., one run of an Illumina flow cell). This finding opens the prospect of a wider survey of the nematode phylum at a reasonable cost.
Can this approach be extended to the vast number of uncharacterized nematodes, most of which probably cannot be cultured in the laboratory? Setting aside the daunting issue of chromosome diminution in some nematode clades such as Ascaris (Müller and Tobler 2000), this will depend on whether PS1010 is representative of other nematode genomes in its polymorphism and repeat structure. We did not inbreed PS1010 before sequencing, as was necessary for species such as C. brenneri (Barrière et al. 2009). However, PS1010 was isolated from a small sample of Caenorhabditis sp. 3 worms and underwent years of continuous culture before being frozen (K Kiontke, pers. comm.). Thus, PS1010 had probably already undergone a bottleneck that lowered its polymorphism and facilitated assembly. For new species that are difficult or impossible to inbreed, DNA from a single worm would allow reconstructing (at worst) two haplotypes at the cost of deeper sequencing. Moreover, current short-read assemblers are designed to deal with error-prone reads and so should tolerate higher levels of polymorphism than their predecessors.
When sequencing DNA from one or a few worms of an unculturable species, constructing jumping libraries from paired ends of larger genomic fragments with specified lengths will probably not be an option for scaffolding genome assemblies. In this case, transcriptomes could be a useful alternative for local scaffolding of genomes: RNA, after being reverse-transcribed, can be amplified from small samples in the same way as genomic DNA. Moreover, such RNA-seq samples would be from whole organisms, and so would be likely to express large fractions of all genes at some level. While cDNA-scaffolded assemblies will not match the quality of assemblies based on jumping libraries and run the risk of excluding intronic fragments, their scaffolding will improve as the depth and variety of RNA-seq samples from different developmental stages are added. Such assemblies will be best for genes most strongly expressed in biologically important life stages (e.g., infectious larvae). This is similar to the analysis of a cancer transcriptome in the context of its matching cancer genome. Both features could help decipher genomes replete with intronic and intergenic repeats, such as those of the nematode Panagrellus redivivus (de Chastonay et al. 1990) and of some plants. Velvet-assembled RNA-seq data makes gene predictions more reliable, and our overall strategy makes it feasible for individual laboratories to sequence the genomes of multicellular eukaryotes. cDNA-scaffolded assembly should thus enable draft genomes of many neglected organisms.
Methods
Worm culture
The strain PS1010 was obtained from the Caenorhabditis Genetics Center. These worms did not thrive on normal C. elegans growth media. We thus grew PS1010 on nutrient agar (Difco) supplemented with 0.1% v/v cholesterol and incubated with E. coli HB101 as food. Worms were grown to high density on five to ten 10-cm plates, collected with M9 buffer, cleaned of bacteria by sucrose centrifugation (Lewis and Fleming 1995), and bleached before growing cultures for DNA or RNA harvests.
Isolation of DNA and RNA
Recently bleached cultures of PS1010 were expanded on five to ten 10-cm nutrient agar/HB101 plates to starvation. After starving worms for 1–2 d to rid them of E. coli, they were collected with M9, sucrose-centrifuged, and snap-frozen with liquid nitrogen in ∼100-μL aliquots before storing at −80°C. Worms were thawed and refrozen three times to promote cuticle breakage before extracting either genomic DNA or bulk RNA. Genomic DNA was extracted by two rounds of proteinase K digestion and phenol-chloroform extraction, with an intermediate step of RNase A digestion in TE; bulk RNA was extracted with the Qiagen RNeasy mini kit.
Genome and transcriptome sequencing
Genomic DNA libraries were built using Illumina's standard paired-end protocol (Bentley et al. 2008). Four libraries were built using different size cuts ranging from 200- to 450-bp fragments and were sequenced as 75-mers (Supplemental Table S7). The 200-nt fragment RNA library was built largely as described (Mortazavi et al. 2008) with an added 12 rounds of column filtration following the last PCR steps and was sequenced as paired 75-mers. All libraries were sequenced on the Illumina Genome Analyzer II following the manufacturer's recommendations. Genome and RNA-seq reads were submitted to the Sequence Read Archive under accession number SRA023844.
Genome and cDNA assembly using Velvet
Raw reads were first mapped using Bowtie 0.12.1 (Langmead et al. 2009) onto the existing 439 kb of PS1010 sequence in GenBank to determine optimal insert sizes for paired mates using ERANGE 3.2 (Mortazavi et al. 2008). Raw reads were assembled with Velvet v.0.7.56 (Zerbino and Birney 2008) using k = 47 nt, expected coverage of 200, minimum coverage of four, minimum pair count of two, supercontigs ≥100 nt, and specified insert sizes for the two longest fragment libraries, where the settings were optimized for the highest N50. Supercontigs showing ≥90% matches to any E. coli assembly in GenBank with BLAT (Kent 2002) were filtered out. The remaining supercontigs were used for the transcriptome-mediated scaffolding of the genome (Fig. 2). Ungapped transcriptome mate-ends were mapped with Bowtie 0.12.1 using the settings “-v 2 -e 240 -k 11 -m 10 --strata --best”, allowing matches of ≥70/75. Reads that did not map with Bowtie were then mapped at 70/75 using BLAT, filtered with pslReps, and imported as 3.6 million splice reads (with at least 6 nt on the short end of the splice) with ERANGE. We mapped RNA-seq reads first with Bowtie onto the cDNA-scaffolded genomic assembly; ERANGE extracted 43.7 million uniquely mappable reads and 0.6 million multireads from the Bowtie mappings. The remaining reads did not map primarily because of poor sequence quality.
RNA-seq reads were assembled into cDNA with Velvet using a two-tiered strategy. One million paired reads were used to assemble cDNA from highly expressed genes with the settings “-exp_cov 100 -ins_length 200 -cov_cutoff 4. -min_contig_lgth 100”. In parallel, all of the reads were used to assemble cDNA from moderately expressed genes with the settings “-exp_cov 1000 -ins_length 200 -cov_cutoff 4. -min_contig_lgth 100”. The resulting cDNA supercontigs were collectively mapped to the genome with BLAT and used as hints to the AUGUSTUS 2.3 genefinder (Stanke et al. 2008).
Transcriptome scaffolding of the genome using RNAPATH
The mapped RNA-seq reads were imported into ERANGE sqlite datasets, and paired-mates with both uniquely mappable (ungapped or spliced) ends on different supercontigs were exported out for the genomic scaffolding by the new RNAPATH module within ERANGE 3.2; while we could have exported these reads instead to a general scaffolding program such as Bambus (Pop et al. 2004), we opted to have the code more tightly integrated within ERANGE. The read-mates were used to build an edge-weighted adjacency matrix of the supercontigs; only the top two edges per supercontigs with weights greater than two were kept. Scaffolding proceeded by starting at leaves and following the highest-weighted edges, reverse-complementing supercontigs as necessary to keep read-mates oriented toward each other; supercontigs and edges that were included in a scaffold could not be reused in any subsequent scaffold. We repeated the scaffolding a second time to obtain the final assembly, which is available in GenBank (accession no. AEHI01000000) and WormBase.
Annotating genomic DNA and protein-coding genes
AUGUSTUS was run on the PS1010 Velvet+RNAPATH assembly with C. elegans parameters. We also ran AUGUSTUS on the C. elegans genome using either de novo parameters, or the following data sources for hints: ∼355,000 C. elegans ESTs from GenBank; public RNA-seq data from GenBank; or our own Velvet-assembled cDNA supercontigs, from our own C. elegans RNA-seq data.
To calculate RPKM expression levels on a per-exon and per-gene basis with ERANGE using the AUGUSTUS gene models, RNA reads were mapped onto the Velvet+RNAPATH assembly. Bowtie and BLAT were run at the same settings used for genomic assembly, but using the first 50 bp of each read and allowing up to five mismatches.
To find repetitive elements in our genome assembly, we ran RepeatModeler, which itself runs both RECON (Bao and Eddy 2002) and RepeatScout (Price et al. 2005) before merging their predictions. We identified 422 repetitive elements in the PS1010 genome, which we mapped to 429 kb of Sanger-sequenced PS1010 genomic DNA with BLAT.
Protein sequence analyses were done on the PS1010 predicted proteome itself, the WormBase WS210 predicted proteomes of C. elegans, C. briggsae, and P. pacificus, the WormBase WS207 predicted proteome of M. hapla, the predicted proteome of M. incognita from http://www.inra.fr/meloidogyne_incognita/genomic_resources/downloads, and a hybrid predicted proteome for B. malayi from both WormBase WS209, and our own de novo AUGUSTUS predictions using B. malayi parameters. We determined orthologies with OrthoMCL 1.3 (Li et al. 2003), run with standard settings. Since OrthoMCL outputs are protein based (overcounting genes with multiple products), groups were mapped from proteins to genes with Perl, and “orthology groups” with one gene were discarded. Genes were considered to belong to strict orthology groups if there was no more than one gene from each species in that group (i.e., 1:1, 1:1:1, etc.). Less stringently, a PS1010 gene was considered to have homology of some sort if it fell into any orthology group that had non-PS1010 genes as members. Protein domains with an E-value of ≤10−6 were found in all seven proteomes with hmmscan from HMMER 3.0b3 (http://hmmer.janelia.org) and release 24.0 of PFAM-A (Finn et al. 2008).
Conserved genomic DNA elements and motifs
The PS1010 draft assembly was aligned to the genomes of C. elegans and C. briggsae with TBA/MULTIZ (Blanchette et al. 2004) at BLASTZ settings T = 0, W = 8, K = 2200 and then analyzed for conservation using phastCons (Siepel et al. 2005) with standard settings. We trained phastCons on alignments of C. elegans chromosome IV, and then used it with otherwise standard settings to generate a list of elements in the C. elegans genome conserved in both C. briggsae and PS1010. We required that they be at least 10 nt long, with 80% overlap in all three genomes, and that they not overlap any of the following datasets in C. elegans: complex or simple repetitive elements; known exons from protein-coding or noncoding RNA (ncRNA) genes annotated in WormBase WS210; BLASTN hits (E ≤ 10−3) against C. elegans ncRNA sequences from WS210; alternative exon predictions by genefinders, such as mGene; which are provided in WormBase as supplementary data rather than official gene models (Schweikert et al. 2009); and our own exon predictions with AUGUSTUS. Statistically overrepresented GO terms of neighboring C. elegans genes were found with the Cistematic module of ERANGE (Mortazavi et al. 2006).
We extracted motifs of 6–18 nt in size from the filtered elements with MEME (-minw 6 -maxw 18) (Bailey and Elkan 1994), allowing any number of motifs per sequence (-mod anr), setting a minimum significance of P ≤ 0.05, allowing up to 50 instances of a motif (-nmotifs 50), and using a Markov-1 background dinucleotide frequency model that we generated from 47.7 MB of filtered C. elegans genome sequence. This consisted of sequence from which we had first removed repeats and exons (known or predicted), and then had removed any sequence fragments under 30 nt. We compared motifs with TOMTOM (Gupta et al. 2007) using Euclidean distances to measure their similarity, Q-values (Storey and Tibshirani 2003) to define highly significant matches, and P-values to qualify weak ones. Previously published motifs were extracted from JASPAR (Portales-Casamar et al. 2010), Drosophila Flyreg v2 (http://www.danielpollard.com/bergman2004_matrices.html; Bergman et al. 2005), TRANSFAC (Matys et al. 2006) via the TOMTOM Web portal (Bailey et al. 2009), and WormBase (Harris et al. 2010). To find subsets of conserved DNA elements containing instances of particular motifs, we ran FIMO with default parameters (Bailey et al. 2009); these subsets were, in turn, scanned for overrepresented GO terms in neighboring genes with ERANGE/Cistematic.
More details of some procedures above are given in the Supplemental Methods.
Acknowledgments
We thank Robin Giblin-Davis for providing PS1010 in 1991, Oren Schaedel for use of his C. elegans L3 RNA-seq data, Todd Ciche and Karin Kiontke for advice on worm culture and RNA extractions, Henry Amrhein and Diane Trout for computational support, and Adler Dillman, Karin Kiontke, Adrienne Roeder, Hillel Schwartz, and Allyson Whittaker for comments on the manuscript. Sequencing was performed in the Millard and Muriel Jacobs Genetics and Genomics Laboratory at Caltech (I.A., L.S.). This work was supported by the Howard Hughes Medical Institute, with which P.W.S. is an Investigator, the Beckman Institute Functional Genomics Center, the Caltech Moore Cell Center, grants HG02223 and HG003162 from the National Human Genome Research Institute, and grant GM084389 from the National Institute of General Medical Sciences.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession no. AEHI01000000 and to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA023844.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.111021.110.
References
- Abad P, Gouzy J, Aury JM, Castagnone-Sereno P, Danchin EG, Deleury E, Perfus-Barbeoch L, Anthouard V, Artiguenave F, Blok VC, et al. 2008. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat Biotechnol 26: 909–915 [DOI] [PubMed] [Google Scholar]
- Ao W, Gaudet J, Kent WJ, Muttumu S, Mango SE 2004. Environmentally induced foregut remodeling by PHA-4/FoxA and DAF-12/NHR. Science 305: 1743–1746 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS 2009. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z, Eddy SR 2002. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res 12: 1269–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrière A, Yang SY, Pekarek E, Thomas CG, Haag ES, Ruvinsky I 2009. Detecting heterozygosity in shotgun genome assemblies: Lessons from obligately outcrossing nematodes. Genome Res 19: 470–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer MA, Tavazoie S 2004. Predicting gene expression from sequence. Cell 117: 185–198 [DOI] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman CM, Carlson JW, Celniker SE 2005. Drosophila DNase I footprint database: A systematic genome annotation of transcription factor binding sites in the fruitfly, Drosophila melanogaster. Bioinformatics 21: 1747–1749 [DOI] [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res 14: 708–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter M 1998. Caenorhabditis elegans is a nematode. Science 282: 2041–2046 [DOI] [PubMed] [Google Scholar]
- Chaisson MJ, Brinza D, Pevzner PA 2009. De novo fragment assembly with short mate-paired reads: Does the read length matter? Genome Res 19: 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Chastonay Y, Muller F, Tobler H 1990. Two highly reiterated nucleotide sequences in the low C-value genome of Panagrellus redivivus. Gene 93: 199–204 [DOI] [PubMed] [Google Scholar]
- Dieterich C, Clifton SW, Schuster LN, Chinwalla A, Delehaunty K, Dinkelacker I, Fulton L, Fulton R, Godfrey J, Minx P, et al. 2008. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat Genet 40: 1193–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. 2008. The Pfam protein families database. Nucleic Acids Res 36: D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet J, Muttumu S, Horner M, Mango SE 2004. Whole-genome analysis of temporal gene expression during foregut development. PLoS Biol 2: e352 doi: 10.1371/journal.pbio.0020352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghedin E, Wang S, Spiro D, Caler E, Zhao Q, Crabtree J, Allen JE, Delcher AL, Guiliano DB, Miranda-Saavedra D, et al. 2007. Draft genome of the filarial nematode parasite Brugia malayi. Science 317: 1756–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS 2007. Quantifying similarity between motifs. Genome Biol 8: R24 doi: 10.1186/gb-2007-8-2-r24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Zody MC 2010. Advancing RNA-Seq analysis. Nat Biotechnol 28: 421–423 [DOI] [PubMed] [Google Scholar]
- Harris TW, Antoshechkin I, Bieri T, Blasiar D, Chan J, Chen WJ, De La Cruz N, Davis P, Duesbury M, Fang R, et al. 2010. WormBase: A comprehensive resource for nematode research. Nucleic Acids Res 38: D463–D467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium 2004. Finishing the euchromatic sequence of the human genome. Nature 431: 931–945 [DOI] [PubMed] [Google Scholar]
- Kent WJ 2002. BLAT—the BLAST-like aligment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Haussler D 2001. Assembly of the working draft of the human genome with GigAssembler. Genome Res 11: 1541–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke K, Fitch DHA 2005. The phylogenetic relationships of Caenorhabditis and other rhabditids. WormBook (ed. The C. elegans Research Community), pp 1–11 doi: 10.1895/wormbook.1.11.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke K, Barrière A, Kolotuev I, Podbilewicz B, Sommer R, Fitch DH, Félix MA 2007. Trends, stasis, and drift in the evolution of nematode vulva development. Curr Biol 17: 1925–1937 [DOI] [PubMed] [Google Scholar]
- Kirienko NV, Fay DS 2010. SLR-2 and JMJC-1 regulate an evolutionarily conserved stress-response network. EMBO J 29: 727–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz SG, Schwarz EM, DeModena JA, De Buysscher T, Trout D, Shizuya H, Sternberg PW, Wold BJ 2008. Multigenome DNA sequence conservation identifies Hox cis-regulatory elements. Genome Res 18: 1955–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JA, Fleming JT 1995. Basic culture methods. Methods Cell Biol 48: 3–29 [PubMed] [Google Scholar]
- Li L, Stoeckert CJ Jr, Roos DS 2003. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, He S, Sun JM, Davie JR 2004. Gene regulation by Sp1 and Sp3. Biochem Cell Biol 82: 460–471 [DOI] [PubMed] [Google Scholar]
- Mathews EA, García E, Santi CM, Mullen GP, Thacker C, Moerman DG, Snutch TP 2003. Critical residues of the Caenorhabditis elegans unc-2 voltage-gated calcium channel that affect behavioral and physiological properties. J Neurosci 23: 6537–6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. 2006. TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34: D108–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldal BH, Debenham NJ, De Ley P, De Ley IT, Vanfleteren JR, Vierstraete AR, Bert W, Borgonie G, Moens T, Tyler PA, et al. 2007. An improved molecular phylogeny of the Nematoda with special emphasis on marine taxa. Mol Phylogenet Evol 42: 622–636 [DOI] [PubMed] [Google Scholar]
- Missal K, Zhu X, Rose D, Deng W, Skogerbø G, Chen R, Stadler PF 2006. Prediction of structured non-coding RNAs in the genomes of the nematodes Caenorhabditis elegans and Caenorhabditis briggsae. J Exp Zool B Mol Dev Evol 306: 379–392 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Leeper Thompson EC, Garcia ST, Myers RM, Wold B 2006. Comparative genomics modeling of the NRSF/REST repressor network: From single conserved sites to genome-wide repertoire. Genome Res 16: 1208–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Müller F, Tobler H 2000. Chromatin diminution in the parasitic nematodes Ascaris suum and Parascaris univalens. Int J Parasitol 30: 391–399 [DOI] [PubMed] [Google Scholar]
- Ohler U, Yekta S, Lim LP, Bartel DP, Burge CB 2004. Patterns of flanking sequence conservation and a characteristic upstream motif for microRNA gene identification. RNA 10: 1309–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman CH, Bird DM, Williamson VM, Rokhsar DS, Burke M, Cohn J, Cromer J, Diener S, Gajan J, Graham S, et al. 2008. Sequence and genetic map of Meloidogyne hapla: A compact nematode genome for plant parasitism. Proc Natl Acad Sci 105: 14802–14807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop M, Kosak DS, Salzberg SL 2004. Hierarchical scaffolding with Bambus. Genome Res 14: 149–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A 2010. JASPAR 2010: The greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res 38: D105–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Jones NC, Pevzner PA 2005. De novo identification of repeat families in large genomes. Bioinformatics 21: i351–i358 [DOI] [PubMed] [Google Scholar]
- Schweikert G, Zien A, Zeller G, Behr J, Dieterich C, Ong CS, Philips P, De Bona F, Hartmann L, Bohlen A, et al. 2009. mGene: Accurate SVM-based gene finding with an application to nematode genomes. Genome Res 19: 2133–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24, 637–644 [DOI] [PubMed] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, et al. 2003. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS Biol 1: E45 doi: 10.1371/journal.pbio.0000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhaus W, Kiontke K 1996. Phylogeny of Rhabditis subgenus Caenorhabditis (Rhabditidae, Nematoda). J Zoo Syst Evol Res 34: 217–233 [Google Scholar]
- van den Heuvel S, Dyson NJ 2008. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol 9: 713–724 [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M 2005. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature 434: 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E 2008. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Schriefer LA, Stormo GD 2007. Identification of muscle-specific regulatory modules in Caenorhabditis elegans. Genome Res 17: 348–357 [DOI] [PMC free article] [PubMed] [Google Scholar]