Abstract
Antifolate resistance in Plasmodium vivax is caused by point mutations in genes encoding dihydrofolate reductase (pvdhfr) and dihydropteroate synthase (pvdhps). In this study, we used direct sequencing to survey pvdhfr and pvdhps mutations in 122 clinical P. vivax isolates from a central and a southern province of China. For pvdhfr, 36.9% were wild-type, whereas mutations were detected at four codons (57, 58, 61, and 117). The S117N/T mutation was the most prevalent (48.4%), followed by the T61M mutation (18.9%). Six pvdhfr mutant alleles were found, ranging from 37.7% to 0.8%. The dramatically different pvdhfr allele frequencies between the two P. vivax populations might be caused by different drug histories or intrinsic difference between temperate and subtropical strains. In contrast, except polymorphisms within a repeat region, no resistance-conferring mutations were detected in pvdhps. Our result suggests that P. vivax populations in China may be relatively susceptible to sulfadoxine-pyrimethamine.
Introduction
Plasmodium vivax is the most widespread cause of malaria outside Africa and causes 132–391 million clinical infections each year.1 Recent evidence of severe disease manifestations associated with P. vivax infections has dramatically changed the traditional view of P. vivax malaria as the benign tertian malaria.2,3 The ability of P. vivax to cause disease relapses adds significantly to the morbidity. In China, the malaria eradication campaign advocated by the World Health Organization in the mid-1950s was highly successful, and malaria was nearly eliminated from the central provinces. However, the predominant malaria parasite P. vivax in central China has demonstrated resilience to eradication and become increasingly prevalent in five central provinces for the past 10 years.4–6 With P. vivax malaria outbreaks occurring frequently in many counties of the five central provinces in recent years, P. vivax has become the predominant parasite species and is responsible for more than 90% of malaria cases in China.7
Although artemisinin-combination therapies show promise for treating patients with P. vivax malaria, first-line therapies for the radical cure of vivax malaria in China are still chloroquine (CQ) and primaquine. This drug combination has been used for more than 60 years, and there is increasing evidence showing the emergence and spread of CQ resistance in P. vivax, especially in Southeast Asia.8 Although the antifolate drug combination of sulfadoxine-pyrimethamine (SP) has replaced CQ for treating patients with P. falciparum malaria in many malaria-endemic regions, SP has not been recommended for treating patients with P. vivax malaria because earlier studies showed that P. vivax parasites seemed intrinsically resistant to SP.9,10 However, there is evidence indicating that SP resistance in P. vivax also results from drug selection pressure. Because mixed species infections with P. falciparum and P. vivax are common in many malarious regions,11 extensive use of SP to treat patients with P. falciparum malaria may have inadvertently exposed P. vivax to these drugs. Likewise, rapid emergence and spread of antifolate resistance observed in P. falciparum populations may be equally applicable to P. vivax populations.12,13
Drug resistance has always been a challenge for malaria control, and surveillance plays an essential role in resistance management. Resistance mechanisms to SP in both parasite species involve point mutations in the genes encoding dihydrofolate reductase (DHFR) and dihydropteroate synthase (DHPS), which are therapeutic targets of pyrimethamine and sulfadoxine, respectively.14,15 Because P. vivax cannot be cultured continuously, antifolate resistance studies have relied heavily on molecular genotyping and drug assays in heterologous systems expressing parasite DHFR and DHPS.16
Molecular genotyping of pvdhfr and pvdhps has been performed in many P. vivax-endemic areas.16 To date, some 20 mutations have been detected in pvdhfr; among them mutations at codons 57, 58, 61, 117, and 173, corresponding to positions 50, 51, 59, 108 and 164 in pfdhfr, respectively, are associated with pyrimethamine resistance.17–20 A structural study has confirmed that the most common mutations (S58R and S117N) cause structural changes in P. vivax DHFR and lead to decreased binding to pyrimethamine.21 A limited number of clinical studies have associated pvdhfr quadruple mutations with increased risks of SP treatment failure.19,22,23 For pvdhps, mutations at codons 382, 383, 512, 553, and 585, corresponding to codons 436, 437, 540, 581, and 613 in pfdhps, respectively, are predicted to confer increased resistance to sulfadoxine.24 The ubiquitous mutation V585 in global parasite isolates was proposed to be the key amino acid affecting the conformation of the drug binding domain,24 and other mutations may further reduce the sensitivity to sulfadoxine.20,24–27 Molecular surveys carried out in P. vivax-endemic areas have detected different geographic distributions and prevalences of pvdhfr and pvdhps mutations; some regions showed near fixation of some mutant alleles, which is correlated with regional difference in the use of SP.27–30 The extensive use of SP in malaria treatment in many malarious regions may be responsible to the multiple origins of resistance-conferring mutations in pvdhfr and pvdhps.31,32
Most of the studies on antifolate resistance in P. vivax were performed in regions where P. vivax and P. falciparum coexist. Because P. vivax malaria has not been directly treated with antifolate drugs, the evolution of mutations in the pvdhfr and pvdhps genes could only be inferred from possible selection by treatment of P. falciparum malaria with antifolate drugs. So far, mutations in pvdhfr and pvdhps have been studied from limited P. vivax samples from temperate zone countries, where P. vivax is the predominant malaria parasite species and has a dramatically different relapse phenotype compared with tropical strains.20,33 To study the evolution of antifolate resistance in P. vivax, we investigated genetic variations in pvdhfr and pvdhps genes of 122 malaria samples collected from two temperate zone provinces of central China (Anhui and Guizhou). Our study identified dramatically different patterns of selection for the pvdhfr and pvdhps genes.
Materials and Methods
Sample collection and DNA extraction.
A total of 134 blood samples from patients with P. vivax malaria were obtained in 2006 and 2008 from symptomatic patients in two provinces of China. One hundred samples were obtained from Wuhe County in Anhui Province in the temperate zone, and 34 samples were obtained from Luodian County in Guizhou Province in the subtropical zone (Figure 1). All patients were diagnosed by microscopic examination of Giemsa-stained thin and thick blood smears. Fingerprick blood samples were collected on filter papers, dried, and stored at –20°C. DNA was extracted from blood spots by using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. The study protocol was reviewed and approved by the Ethics Committees of Guangxi and Anhui, China.
Figure 1.
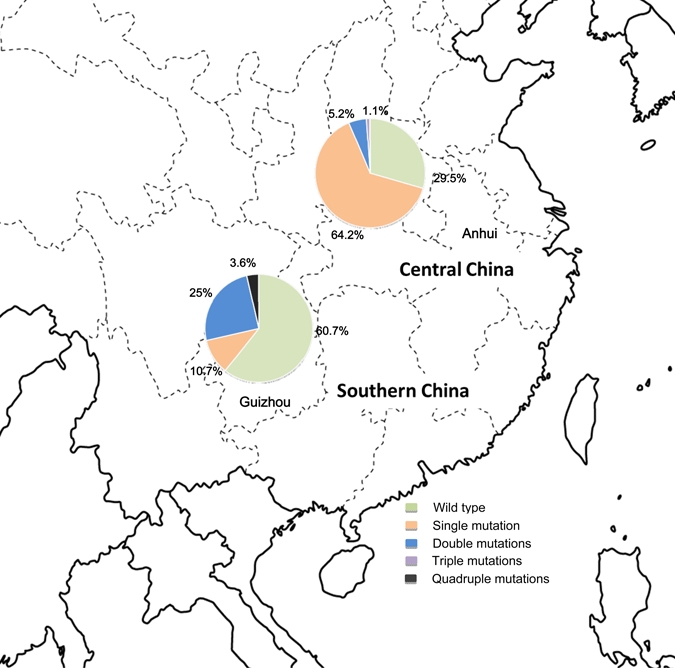
Sample collection sites in Anhui Province in central China and Guizhou Province in southern China. The pie charts show the frequency distribution of Plasmodium vivax dihydrofolate reductase synthase alleles. This figure appears in color at www.ajtmh.org.
Sequencing pvdhfr and pvdhps.
Genomic DNA was used as template for polymerase chain reaction (PCR) with the Advantage 2 polymerase mixture with proof-reading activity (Clontech, Mountain View, CA). The pvdhfr gene was amplified using the primer 5′-ATGGAGGACCTTTCAGATGTATTTGACATT-3′ and 5′-CTTGCTGTAAACCAAAAAGTCCAGAGTGGT-3′ as described.34 The PCR was performed with the following parameters: 94°C for 5 minutes; followed by 30 cycles at 94°C for 20 seconds, 60°C for 30 seconds, and 72°C for 1 minute; and a final elongation at 72°C 10 minutes. The pvdhps gene was amplified by nested PCR with the outer primers (forward 5′-TTGAAACACGCATTATGGTATCG-3′ and reverse 5′-AAGCGTAGCGACAGAAGAACG-3′) and inner primers (forward 5′-GAGATTACCCTAAGGTTGATGTATC-3′ and reverse 5′-GGTTTATTTGTCGATCCTGTG-3′).24,27 Another pair of primers (forward 5′-AATGGCAAGTGATGGGGCGAGCGTGATTGA-3′ and reverse 5′-CAGTCTGCACTCCCCGATGGCCGCGCCACC-3′) was used to amplify an approximately 700-basepair fragment of pvdhps in cases of amplification failure with the original primers. For pvdhps, PCR was performed using 35 cycles at 94°C for 1 minute, 60°C for 1 minute, and 72°C for 2 minutes. The PCR products were fractioned by electrophoresis on a 1.5% agarose gel, purified by PrepEase Gel Extraction Kit (United States Biochemicals, Cleveland, OH), and sequenced by using an ABI PRISM BigDye Terminator Cycle Sequencing Ready Reactions Kit on a 3730 xl Genetic Analyzer (Applied Biosystems, Foster City, CA).
Sequence analysis.
Multiple sequence alignment and downstream analysis were carried out by using the CLUSTAL W program35 and the BioEdit package. Amino acid sequences were compared with wild-type sequences (GenBank accession nos. X98123 and AY186730 for pvdhfr and pvdhps, respectively).
Results
Mutant pvdhfr alleles and their frequencies.
Sequence polymorphisms were assessed in pvdhfr and pvdhps genes from 134 P. vivax samples obtained from two regions of China. Among these samples, the pvdhfr gene was successfully amplified and sequenced in 122 isolates. Compared with the wild-type sequence, pvdhfr genes from the 122 clinical samples had point mutations at 10 codons, among which four resulted in amino acid substitutions. The six synonymous point mutations were AAC to AAT (both coding for Asn) at codon 50, and GGT to GGG (both coding for Gly) at codons 89, 94, 95, 100, and 101, respectively. Point mutations at the latter five codons have not been reported16 and were found in 35 (28.7%) samples. Nonsynonymous mutations were detected at positions 57, 58, 61, and 117, and the point mutation at codon 173 was not observed in our samples (Table 1). Overall, 45 of the 122 samples (36.9%) were identified as containing wild-type pvdhfr, and the remaining 63.1% of isolates had at least one amino acid substitution. The most predominant pvdhfr S117N mutation was found in almost half of the parasite samples (48.4%), whereas the T61M, S58R, and F57L mutations were found in 23 (18.9%), 12 (9.8%), and 1 (0.8%) of the samples, respectively.
Table 1.
Point mutations in dihydrofolate reductase synthase among Plasmodium vivax isolates from China*
| Allele | Genotype* | Allele frequency: no. (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| 57 | 58 | 61 | 117 | 173 | Total | Anhui | Guizhou | |
| 1 | F | S | T | S | I | 45 (36.9) | 28 (29.5) | 17 (60.7) |
| 2 | F | S | T | N | I | 45 (36.9) | 42 (44.2) | 3 (10.7) |
| 3 | F | S | M | S | I | 19 (15.6) | 19 (20.0) | 0 |
| 4 | F | R | T | N | I | 10 (8.4) | 3 (3.2) | 7 (25.0) |
| 5 | F | S | M | N | I | 2 (1.6) | 2 (2.1) | 0 |
| 6 | F | R | M | N | I | 1 (0.8) | 1 (1.1) | 0 |
| 7 | L | R | M | T | I | 1 (0.8) | 0 | 1 (3.6) |
Mutated amino acids are shown in bold.
On the basis of amino acid changes, the pvdhfr sequences can be grouped into seven alleles; two alleles (5 and 6) were novel, and the remaining five have been previously identified (1, 2, 3, 4, and 7). Except for the wild-type allele, the most prevalent mutant allele was the one with a single mutation at codon 117 (36.9%). Triple (S58R/T61M/S117N) and quadruple mutations (F57L/S58R/T61M/S117T) were rare and accounted for less than 1% of the parasite isolates. Two types of double mutations (S58R/S117N and T61M/S117N) were found in 8.4% and 1.6% of parasite isolates, respectively. The two P. vivax parasite populations in China also differed in pvdhfr allele frequencies (Table 1 and Figure 1). The wild-type pvdhfr genotype was found in 60.7% of the parasite population from Guizhou and was found in 29.5% of the parasites from central China. Although each site had three mutant pvdhfr alleles (single, double, and triple mutations), a quadruple mutant allele (F57L/S58R/T61M/S117T) was only found in the population from southern China.
In addition to these point mutations, we observed variations at the central tandem repeat region between amino acid positions 88 and 105 of the pvdhfr gene.17,28,36 Size polymorphism in this region is resulted from variation in number of copies of the 18-basepair repeat, which is not essential for substrate binding.21 Most (77.0%) of the isolates sequenced contain three copies of the repeat, and 23.0% contained two 18-basepair repeats. None had a single copy of the 18-basepair repeat. Short indels and mutations were also found within the repeat unit of the sequence GGDN. A total of 22.1% of the samples showed a pattern in the repeat region similar to that in the wild type. The remaining samples can be divided into three types on the basis of the mutation at residue 99. We identified a deletion mutation, a previously reported H99S mutation,28 and a novel H99D mutation, which accounted for 23.0%, 49.2%, and 5.7% of the parasite samples, respectively.
Mutations in pvdhps alleles and their frequencies.
The pvdhps gene was successfully amplified and sequenced for 87 parasite isolates. In contrast to the pvdhfr gene, no single nucleotide polymorphism was present in pvdhps except for a novel tandem repeat variation. The amino acids at codons 382, 383, 512, 553, and 585 were all wild type. The 585V mutation has been suggested to be responsible for the intrinsic resistance of P. vivax to antifolates.24 We sequenced the repeat region of the pvdhps gene (spanning amino acids 603–666) of 63 parasite isolates from central China. Length polymorphism in this region is caused by a variable number of tandem repeat unit G(E/D)(A/G/S)KLTN.
From 63 parasite samples, seven repeat haplotypes (RH1–6, and D) were recognized and only one (type D) was previously reported26,32 (Figure 2). The number of repeats in our samples ranged from four to nine. The most common haplotype is RH1, which represented (49.2%) of the analyzed samples and appeared to be different from SalI strain by only two mutational steps. Three other haplotypes (RH2, RH3, and RH4) were present in the samples at similar frequencies (11.1–14.3%). Within the repeat region, seven novel synonymous mutations were observed: position 615 (ACG to ACC) in 47 sequences (74.6%), position 616 (AAT to AAC) in 47 sequences (74.6%), position 631 (GGG to GGT) in 16 sequences (25.4%), position 634 (AAA to AAG) in 7 sequences (11.1%), position 638 (GGG to GGA) in 7 sequences (11.11%), position 662 (AAG to AAA) in 9 sequences (14.3%), and position 664 (ACC to ACT) in 9 sequences (14.3%). Mutation 615 is always linked to mutation 616 and mutation 634 is always linked to the mutation 638. The repeat region is not predicted to directly bind sulfa drugs, and is therefore unlikely involved in resistance to sulfa drugs in P. vivax.
Figure 2.

Polymorphisms in the repetitive domain of Plasmodium vivax dihydropteroate synthase. Deletions are denoted by dashes. RH1–RH6 are from this study. A*–H* correspond to the alleles described by Hawkins and others,32 and A′–D′ represent the alleles from Menegons and others. 26 Sequences were aligned with the Sal I reference sequence (www.tigr.org), and the repeat units are labeled consecutively as R1 through R13. The frequency of each allele identified in this study is shown as number (N) and percentage (%).
Discussion
Chloroquine-primaquine is still the first-line treatment for P. vivax malaria. With the emergence of CQ-resistant P. vivax strains in many malarious regions, it is also becoming a high priority to monitor drug resistance and develop drugs for future treatment of P. vivax malaria. Although antifolate drugs have not been recommended for treating P. vivax malaria, recent studies suggest that this family of drugs may find a role in future treatment of P. vivax malaria in certain regions.16,37 In this study, we have analyzed pvdhfr and pvdhps mutations in two P. vivax populations from both temperate zone and subtropical provinces of China. Our study showed that mutant pvdhfr genotypes were present at relatively high levels, but the mutations mostly occurred at single amino acids. In addition, no resistance-conferring mutations were found in pvdhps, suggesting that the P. vivax parasites in China may be relatively sensitive to SP.
It is well established that in P. falciparum accumulation of multiple mutations in pfdhfr and pfdhps is associated with in vitro resistance and clinical treatment failures with SP.14,15 A resistance mechanism to antifolates in P. vivax has been proposed that is similar to that in P. falciparum and linked to mutations at homologous positions in pvdhfr and pvdhps. For example, mutations at amino acids 58 and 117 of pvdhfr correspond to mutations at positions 59 and 108 in pfdhfr, respectively, which are associated with pyrimethamine resistance. Results from in vitro drug assays with limited numbers of field samples and tests using a yeast expression system are generally agreeable with this assumption.18,20,38 Furthermore, limited clinical assessments of the efficacy of SP have associated pvdhfr quadruple mutations and increased risks of clinical resistance to SP,19,22,23,25 suggesting that molecular genotyping data for pvdhfr and pvdhps should provide useful information about SP resistance in P. vivax.
Molecular epidemiologic studies in different areas have showed dramatically different mutations rates in pvdhfr and pvdhps.16 In Thailand where antifolates have been used heavily since the 1970s as a replacement of CQ for treating P. falciparum malaria, quadruple pvdhfr mutations and double pvdhps mutations have reached prevalences of approximately 60% and 72%, respectively, especially in P. vivax populations along Thailand–Myanmar border areas.27 The prevalence of highly mutated pvdhfr and pvdhps is correlated with the poor therapeutic efficacy of SP against P. vivax malaria in this region.23,39 Similar pvdhfr quadruple mutations are also prevalent in certain areas of Indonesia and Papua New Guinea.18 In comparison, in southern Asia areas such as India, Pakistan, and Sri Lanka, parasites harboring multiple mutations in pvdhfr and pvdhps are relatively rare.28,30,40 In such regions where P. falciparum and P. vivax coexist, mutations in pvdhfr and pvdhps have been attributed to selective pressure of antifolate drugs specific for controlling P. falciparum. Accordingly, pvdhfr and pvdhps mutation rates are expected to be much lower in temperate zone regions where P. vivax is the only or predominant malaria parasite. Limited studies on some temperate zone P. vivax samples support this prediction.33,41 Although an earlier study that used seven P. vivax isolates collected in 1994 from China showed that 71% of the parasites were wild-type at the pvdhfr locus, the small sample size may not be representative.20 In this study, we found that mutant pvdhfr genotypes were present in approximately 63% of the clinical isolates. However, most pvdhfr mutations occurred at single amino acids, and triple and quadruple mutations were only found in two parasite isolates. In addition, no additional amino acid changes were found in pvdhps.
It has been proposed that point mutations in pvdhfr, such as those in pfdhfr, are acquired sequentially in response to drug pressure and mutations at residues 117 and 58 arise first.17 Consistent with this proposal, mutation S117N was the most prevalent point mutation in pvdhfr in the two P. vivax populations in China. In regions of extensive SP use, two mutant genotypes at residue 117 (S117T and S117N) have been observed; the former has been associated with highly mutated pvdhfr and may be a key mutation for subsequent acquisition of additional mutations and development of high resistance to SP.19,22 In Thailand, for example, the pvdhfr T61M mutation, which has reached a high level of prevalence (> 60%), is mostly linked with the S117T mutation.17,27 Consistent with the scarcity of the S117T mutation in China, highly mutated pvdhfr (triple or quadruple mutations) was also rare. However, the 61M mutation was relatively frequent (20%), especially in the temperate zone P. vivax population. Also noteworthy is that most of the 61M mutations were found as a single mutation in the central China P. vivax population, suggesting that this mutation could arise independently.
Although the presence of the pvdhfr 57L58R61M117T quadruple mutant associated with clinical resistance to SP was found in an isolate from southern China, it is unknown whether this mutant genotype was generated locally or imported from neighboring subtropical provinces, where P. falciparum was also present. It is obvious that the two P. vivax populations in China have drastically different pvdhfr haplotype frequencies, which may be attributed to different drug selection pressures or the intrinsic differences between the temperate and tropical strains of P. vivax. Furthermore, highly polymorphic repeat motifs in pvdhfr and pvdhps may indicate a lack of SP-selective sweep in these parasite populations. The different resistance-conferring pvdhfr mutation haplotypes may have evolved independently in different P. vivax populations in China.31 Because in malarious regions where P. falciparum and P. vivax coexist mutant pvdhfr and pvdhps alleles are presumably the result of exposure to antifolate drugs that are used for treatment of P. falciparum malaria, it is thus counterintuitive that the parasites carrying pvdhfr mutations are more prevalent in the temperate population from Anhui province, where P. vivax is the only malaria parasite species, than in the subtropical population from Guizhou Province. However, in the temperate regions in which P. vivax is endemic, antifolate drugs have been used mostly as prophylaxis. In contrast, other antimalarial drugs such as piperaquine and artemisinins, have been used to treat P. falciparum malaria in the subtropical regions. Therefore, different pvdhfr allele prevalences in the two P. vivax populations in China might be attributed to different drug histories in these areas.
It appears that in P. falciparum SP poses asymmetric selection of pfdhfr and pfdhps and mutations in pfdhfr usually occur first.14 This selection seems to be a similar in P. vivax because higher frequencies of pvdhps mutations were only observed in regions such as Thailand where highly mutated pvdhfr haplotypes were present.17,27 This finding has been thought to be caused by the intrinsic difference in mutation rates associated with these two genes, which reflects the relative fitness of the resulting enzymes in the absence of the drug pressure.14 Similar to P. vivax populations from other regions with low antifolate use, the P. vivax populations from China contain resistance-conferring mutations in pvdhfr but not in pvdhps.
When compared with other malaria-endemic regions of world, the antifolate drug use history in China is different. From late 1950s to early 1960s, only pyrimethamine was used heavily for malaria prophylaxis in all malaria-endemic areas of China.42 In P. vivax-endemic areas, use of pyrimethamine plus primaquine for eight weeks has been a practice for malaria prophylaxis during the transmission season.43 From 1966 to early 1970s, it was replaced with antimalarial drug combination no. 1 (pyrimethamine-dapsone) and later no. 2 (sulfadoxine and pyrimethamine) for prophylaxis. Drug combination no. 2 contains only half the amount of sulfadoxine (250 mg) as Fansidar (500 mg).42 Therefore, it is plausible that differential use of pyrimethamine and sulfa drugs in China may be partially responsible for the asymmetric selection of pvdhfr and pvdhps.
The pvdhfr and pvdhps mutation rates generally coincide with the extent of SP use and may be a useful indicator for predicting clinical efficacy of SP. In China, extensive use of pyrimethamine for P. vivax malaria prophylaxis and treatment may have led to evolution of pvdhfr mutations. Nevertheless, the rarity of highly mutated pvdhfr and absence of mutations in pvdhps suggest that these parasite populations might be relatively susceptible to SP.22 A recent in vitro study on in vitro sensitivity of P. vivax isolates from this region to a number of antimalarial drugs supported this assumption (Feng L and others, unpublished data). In addition, an earlier clinical study in southern China indicated that eight-week use of pyrimethamine-primaquine prophylaxis during the P. vivax transmission season was highly effective in preventing P. vivax malaria.44 Similarly, a clinical study found pyrimethamine monotherapy was effective in 65.5% of P. vivax malaria cases in central China, whereas the 34.5% of the cases displayed grade I and II resistance.45 These data indicate that P. vivax populations in China may be relatively susceptible to antifolate drugs.
Acknowledgments
We thank Ziyi Miao for extracting DNA from malaria-infected samples.
Footnotes
Financial support: This study was supported by grants R21 AI078263-01 and U19AI089672 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Authors' addresses: Miao Miao, Long Cui, Jessica Ahlum, and Liwang Cui, Department of Entomology, Pennsylvania State University, 501 ASI Building, University Park, PA, E-mails: doublemiao@gmail.com, lxc31@psu.edu, jha127@psu.edu, and luc2@psu.edu. Zhaoqing Yang, Parasitology Department, Kunming Medical College, Kunming, Yunnan, China, E-mail: zhaoqingy92@hotmail.com. Yaming Huang, Malaria Department, Guangxi Centers foe Disease Control, Nanning, Guangxi, China, E-mail: hym9992003@yahoo.com.
References
- 1.Price RN, Tjitra E, Guerra CA, Yeung S, White NJ, Anstey NM. Vivax malaria: neglected and not benign. Am J Trop Med Hyg. 2007;77:79–87. [PMC free article] [PubMed] [Google Scholar]
- 2.Anstey NM, Russell B, Yeo TW, Price RN. The pathophysiology of vivax malaria. Trends Parasitol. 2009;25:220–227. doi: 10.1016/j.pt.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Mueller I, Galinski MR, Baird JK, Carlton JM, Kochar DK, Alonso PL, del Portillo HA. Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect Dis. 2009;9:555–566. doi: 10.1016/S1473-3099(09)70177-X. [DOI] [PubMed] [Google Scholar]
- 4.Sleigh AC, Liu XL, Jackson S, Li P, Shang LY. Resurgence of vivax malaria in Henan Province, China. Bull World Health Organ. 1998;76:265–270. [PMC free article] [PubMed] [Google Scholar]
- 5.Xu BL, Su YP, Shang LY, Zhang HW. Malaria control in Henan Province, People's Republic of China. Am J Trop Med Hyg. 2006;74:564–567. [PubMed] [Google Scholar]
- 6.Zhang W, Wang L, Fang L, Ma J, Xu Y, Jiang J, Hui F, Wang J, Liang S, Yang H, Cao W. Spatial analysis of malaria in Anhui province, China. Malar J. 2008;7:206. doi: 10.1186/1475-2875-7-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou SS, Wang Y, Tang LH. Malaria situation in the People's Republic of China in 2006. Chin J Parasitol Parasitic Dis. 2007;25:439–441. [PubMed] [Google Scholar]
- 8.Baird JK. Chloroquine resistance in Plasmodium vivax. Antimicrob Agents Chemother. 2004;48:4075–4083. doi: 10.1128/AAC.48.11.4075-4083.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doberstyn EB, Teerakiartkamjorn C, Andre RG, Phintuyothin P, Noeypatimanondh S. Treatment of vivax malaria with sulfadoxine-pyrimethamine and with pyrimethamine alone. Trans R Soc Trop Med Hyg. 1979;73:15–17. doi: 10.1016/0035-9203(79)90121-4. [DOI] [PubMed] [Google Scholar]
- 10.Young MD, Burgess RW. Pyrimethamine resistance in Plasmodium vivax malaria. Bull World Health Organ. 1959;20:27–36. [PMC free article] [PubMed] [Google Scholar]
- 11.Mayxay M, Pukrittayakamee S, Newton PN, White NJ. Mixed-species malaria infections in humans. Trends Parasitol. 2004;20:233–240. doi: 10.1016/j.pt.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Nair S, Williams JT, Brockman A, Paiphun L, Mayxay M, Newton PN, Guthmann JP, Smithuis FM, Hien TT, White NJ, Nosten F, Anderson TJ. A selective sweep driven by pyrimethamine treatment in southeast Asian malaria parasites. Mol Biol Evol. 2003;20:1526–1536. doi: 10.1093/molbev/msg162. [DOI] [PubMed] [Google Scholar]
- 13.Pearce RJ, Pota H, Evehe MS, Ba el H, Mombo-Ngoma G, Malisa AL, Ord R, Inojosa W, Matondo A, Diallo DA, Mbacham W, van den Broek IV, Swarthout TD, Getachew A, Dejene S, Grobusch MP, Njie F, Dunyo S, Kweku M, Owusu-Agyei S, Chandramohan D, Bonnet M, Guthmann JP, Clarke S, Barnes KI, Streat E, Katokele ST, Uusiku P, Agboghoroma CO, Elegba OY, Cisse B, A-Elbasit IE, Giha HA, Kachur SP, Lynch C, Rwakimari JB, Chanda P, Hawela M, Sharp B, Naidoo I, Roper C. Multiple origins and regional dispersal of resistant dhps in African Plasmodium falciparum malaria. PLoS Med. 2009;6:e1000055. doi: 10.1371/journal.pmed.1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sibley CH, Hyde JE, Sims PF, Plowe CV, Kublin JG, Mberu EK, Cowman AF, Winstanley PA, Watkins WM, Nzila AM. Pyrimethamine-sulfadoxine resistance in Plasmodium falciparum: what next? Trends Parasitol. 2001;17:582–588. doi: 10.1016/s1471-4922(01)02085-2. [DOI] [PubMed] [Google Scholar]
- 15.Gregson A, Plowe CV. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol Rev. 2005;57:117–145. doi: 10.1124/pr.57.1.4. [DOI] [PubMed] [Google Scholar]
- 16.Hawkins VN, Joshi H, Rungsihirunrat K, Na-Bangchang K, Sibley CH. Antifolates can have a role in the treatment of Plasmodium vivax. Trends Parasitol. 2007;23:213–222. doi: 10.1016/j.pt.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Imwong M, Pukrittayakamee S, Renia L, Letourneur F, Charlieu JP, Leartsakulpanich U, Looareesuwan S, White NJ, Snounou G. Novel point mutations in the dihydrofolate reductase gene of Plasmodium vivax: evidence for sequential selection by drug pressure. Antimicrob Agents Chemother. 2003;47:1514–1521. doi: 10.1128/AAC.47.5.1514-1521.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hastings MD, Maguire JD, Bangs MJ, Zimmerman PA, Reeder JC, Baird JK, Sibley CH. Novel Plasmodium vivax dhfr alleles from the Indonesian Archipelago and Papua New Guinea: association with pyrimethamine resistance determined by a Saccharomyces cerevisiae expression system. Antimicrob Agents Chemother. 2005;49:733–740. doi: 10.1128/AAC.49.2.733-740.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tjitra E, Baker J, Suprianto S, Cheng Q, Anstey NM. Therapeutic efficacies of artesunate-sulfadoxine-pyrimethamine and chloroquine-sulfadoxine-pyrimethamine in vivax malaria pilot studies: relationship to Plasmodium vivax dhfr mutations. Antimicrob Agents Chemother. 2002;46:3947–3953. doi: 10.1128/AAC.46.12.3947-3953.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Auliff A, Wilson DW, Russell B, Gao Q, Chen N, Anh le N, Maguire J, Bell D, O'Neil MT, Cheng Q. Amino acid mutations in Plasmodium vivax DHFR and DHPS from several geographical regions and susceptibility to antifolate drugs. Am J Trop Med Hyg. 2006;75:617–621. [PubMed] [Google Scholar]
- 21.Kongsaeree P, Khongsuk P, Leartsakulpanich U, Chitnumsub P, Tarnchompoo B, Walkinshaw MD, Yuthavong Y. Crystal structure of dihydrofolate reductase from Plasmodium vivax: pyrimethamine displacement linked with mutation-induced resistance. Proc Natl Acad Sci USA. 2005;102:13046–13051. doi: 10.1073/pnas.0501747102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hastings MD, Porter KM, Maguire JD, Susanti I, Kania W, Bangs MJ, Sibley CH, Baird JK. Dihydrofolate reductase mutations in Plasmodium vivax from Indonesia and therapeutic response to sulfadoxine plus pyrimethamine. J Infect Dis. 2004;189:744–750. doi: 10.1086/381397. [DOI] [PubMed] [Google Scholar]
- 23.Imwong M, Pukrittakayamee S, Looareesuwan S, Pasvol G, Poirreiz J, White NJ, Snounou G. Association of genetic mutations in Plasmodium vivax dhfr with resistance to sulfadoxine-pyrimethamine: geographical and clinical correlates. Antimicrob Agents Chemother. 2001;45:3122–3127. doi: 10.1128/AAC.45.11.3122-3127.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korsinczky M, Fischer K, Chen N, Baker J, Rieckmann K, Cheng Q. Sulfadoxine resistance in Plasmodium vivax is associated with a specific amino acid in dihydropteroate synthase at the putative sulfadoxine-binding site. Antimicrob Agents Chemother. 2004;48:2214–2222. doi: 10.1128/AAC.48.6.2214-2222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imwong M, Pukrittayakamee S, Cheng Q, Moore C, Looareesuwan S, Snounou G, White NJ, Day NP. Limited polymorphism in the dihydropteroate synthetase gene (dhps) of Plasmodium vivax isolates from Thailand. Antimicrob Agents Chemother. 2005;49:4393–4395. doi: 10.1128/AAC.49.10.4393-4395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menegon M, Majori G, Severini C. Genetic variations of the Plasmodium vivax dihydropteroate synthase gene. Acta Trop. 2006;98:196–199. doi: 10.1016/j.actatropica.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Rungsihirunrat K, Sibley CH, Mungthin M, Na-Bangchang K. Geographical distribution of amino acid mutations in Plasmodium vivax DHFR and DHPS from malaria endemic areas of Thailand. Am J Trop Med Hyg. 2008;78:462–467. [PubMed] [Google Scholar]
- 28.Kaur S, Prajapati SK, Kalyanaraman K, Mohmmed A, Joshi H, Chauhan VS. Plasmodium vivax dihydrofolate reductase point mutations from the Indian subcontinent. Acta Trop. 2006;97:174–180. doi: 10.1016/j.actatropica.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Barnadas C, Musset L, Legrand E, Tichit M, Briolant S, Fusai T, Rogier C, Bouchier C, Picot S, Menard D. High prevalence and fixation of Plasmodium vivax dhfr/dhps mutations related to sulfadoxine/pyrimethamine resistance in French Guiana. Am J Trop Med Hyg. 2009;81:19–22. [PubMed] [Google Scholar]
- 30.Schousboe ML, Rajakaruna RS, Salanti A, Hapuarachchi HC, Galappaththy GN, Bygbjerg IC, Amerasinghe PH, Konradsen F, Alifrangis M. Island-wide diversity in single nucleotide polymorphisms of the Plasmodium vivax dihydrofolate reductase and dihydropteroate synthetase genes in Sri Lanka. Malar J. 2007;6:28. doi: 10.1186/1475-2875-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hawkins VN, Auliff A, Prajapati SK, Rungsihirunrat K, Hapuarachchi HC, Maestre A, O'Neil MT, Cheng Q, Joshi H, Na-Bangchang K, Sibley CH. Multiple origins of resistance-conferring mutations in Plasmodium vivax dihydrofolate reductase. Malar J. 2008;7:72. doi: 10.1186/1475-2875-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hawkins VN, Suzuki SM, Rungsihirunrat K, Hapuarachchi HC, Maestre A, Na-Bangchang K, Sibley CH. Assessment of the origins and spread of putative resistance-conferring mutations in Plasmodium vivax dihydropteroate synthase. Am J Trop Med Hyg. 2009;81:348–355. [PubMed] [Google Scholar]
- 33.Brega S, de Monbrison F, Severini C, Udomsangpetch R, Sutanto I, Ruckert P, Peyron F, Picot S. Real-time PCR for dihydrofolate reductase gene single-nucleotide polymorphisms in Plasmodium vivax isolates. Antimicrob Agents Chemother. 2004;48:2581–2587. doi: 10.1128/AAC.48.7.2581-2587.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Na BK, Lee HW, Moon SU, In TS, Lin K, Maung M, Chung GT, Lee JK, Kim TS, Kong Y. Genetic variations of the dihydrofolate reductase gene of Plasmodium vivax in Mandalay Division, Myanmar. Parasitol Res. 2005;96:321–325. doi: 10.1007/s00436-005-1364-0. [DOI] [PubMed] [Google Scholar]
- 35.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Pecoulas PE, Tahar R, Ouatas T, Mazabraud A, Basco LK. Sequence variations in the Plasmodium vivax dihydrofolate reductase-thymidylate synthase gene and their relationship with pyrimethamine resistance. Mol Biochem Parasitol. 1998;92:265–273. doi: 10.1016/s0166-6851(97)00247-8. [DOI] [PubMed] [Google Scholar]
- 37.Hastings MD, Sibley CH. Pyrimethamine and WR99210 exert opposing selection on dihydrofolate reductase from Plasmodium vivax. Proc Natl Acad Sci USA. 2002;99:13137–13141. doi: 10.1073/pnas.182295999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rungsihirunrat K, Na-Bangchang K, Hawkins VN, Mungthin M, Sibley CH. Sensitivity to antifolates and genetic analysis of Plasmodium vivax isolates from Thailand. Am J Trop Med Hyg. 2007;76:1057–1065. [PubMed] [Google Scholar]
- 39.Pukrittayakamee S, Chantra A, Simpson JA, Vanijanonta S, Clemens R, Looareesuwan S, White NJ. Therapeutic responses to different antimalarial drugs in vivax malaria. Antimicrob Agents Chemother. 2000;44:1680–1685. doi: 10.1128/aac.44.6.1680-1685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khatoon L, Baliraine FN, Bonizzoni M, Malik SA, Yan G. Prevalence of antimalarial drug resistance mutations in Plasmodium vivax and P. falciparum from a malaria-endemic area of Pakistan. Am J Trop Med Hyg. 2009;81:525–528. [PMC free article] [PubMed] [Google Scholar]
- 41.Zakeri S, Motmaen SR, Afsharpad M, Djadid ND. Molecular characterization of antifolates resistance-associated genes, (dhfr and dhps) in Plasmodium vivax isolates from the Middle East. Malar J. 2009;8:20. doi: 10.1186/1475-2875-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Z. Malaria Control and Research in China. Beijing: People's Hygiene Press; 1991. [Google Scholar]
- 43.Shen J, Hu X. Comparison of the antimalarial effect of long-term pyrimethamine prophylaxis versus change to “antimalarial drug combination no. 3”. Railway Med J. 1978;6:90–91. [Google Scholar]
- 44.Su BY, Huang HM. Observation on the preventive effect of eight-week pyrimethamine and primaquine treatment on vivax malaria. Guangxi Med J. 1982;4:12–13. [Google Scholar]
- 45.Chen JS, Zhang KR, Geng ZW. Observation on clinical efficacy of pyrimethamine on Plasmodium vivax in Kaifeng District. Henan J Prev Med. 1977;9:51–56. [Google Scholar]


