Mutations in the maize thiamine biosynthesis gene bladekiller1 induce meristem abortion phenotypes that are completely rescued by exogenous thiamine. The data illustrate the essential role of metabolic cofactors during the proliferative growth of the undifferentiated stem cell populations that comprise plant meristems.
Abstract
Plant shoots undergo organogenesis throughout their life cycle via the perpetuation of stem cell pools called shoot apical meristems (SAMs). SAM maintenance requires the coordinated equilibrium between stem cell division and differentiation and is regulated by integrated networks of gene expression, hormonal signaling, and metabolite sensing. Here, we show that the maize (Zea mays) mutant bladekiller1-R (blk1-R) is defective in leaf blade development and meristem maintenance and exhibits a progressive reduction in SAM size that results in premature shoot abortion. Molecular markers for stem cell maintenance and organ initiation reveal that both of these meristematic functions are progressively compromised in blk1-R mutants, especially in the inflorescence and floral meristems. Positional cloning of blk1-R identified a predicted missense mutation in a highly conserved amino acid encoded by thiamine biosynthesis2 (thi2). Consistent with chromosome dosage studies suggesting that blk1-R is a null mutation, biochemical analyses confirm that the wild-type THI2 enzyme copurifies with a thiazole precursor to thiamine, whereas the mutant enzyme does not. Heterologous expression studies confirm that THI2 is targeted to chloroplasts. All blk1-R mutant phenotypes are rescued by exogenous thiamine supplementation, suggesting that blk1-R is a thiamine auxotroph. These results provide insight into the role of metabolic cofactors, such as thiamine, during the proliferation of stem and initial cell populations.
INTRODUCTION
Shoot apical meristems (SAMs) are responsible for the development of all the organs in plant shoots and require a complex signaling network wherein indeterminate stem cells are juxtaposed to initial cells of lateral organs such as leaves (reviewed in Barton, 2009). Formed during embryogenesis and maintained until reproductive stages of development, SAMs are self-organizing, self-renewing, and organogenic. Persistence of these pools of meristematic stem cells throughout the plant life cycle accounts for a strategic difference in plant and animal development, such that the majority of organs present in adult plants are formed postembryonically. SAMs must maintain a precise equilibrium during which cells lost to newly initiated organs are replenished by stem cells that divide to sustain the meristem. Failure to maintain the stem cell pool results in meristem abortion, whereas stem cell overproliferation can lead to fasciated meristems and abnormal organogenesis. Genetic analyses of meristem mutants have contributed to models where this essential balance between stem cell maintenance and stem cell differentiation is maintained via integrated networks of gene expression, hormonal signaling, and metabolism (reviewed in Fleming, 2006; Barton, 2009).
Continued stem cell activity depends on a high cytokinin to auxin and gibberellin hormone regime. Class-I KNOTTED1-LIKE HOMEOBOX (KNOX) transcription factors help maintain this ratio by simultaneously preventing cell differentiation through the repression of gibberellin signaling (Sakamoto et al., 2001; Bolduc and Hake, 2009) and promoting cell division via stimulation of cytokinin biosynthesis (Jasinski et al., 2005; Yanai et al., 2005). The founding member of the knox gene family, maize (Zea mays) knotted1 (kn1), is a useful marker of indeterminacy and is expressed throughout the meristem but downregulated at organ initiation sites (Jackson et al., 1994; Long et al., 1996). Loss of KN1 function leads to meristem consumption, underlining its importance in meristem maintenance (Vollbrecht et al., 2000).
WUSCHEL (WUS), a transcription factor required for meristem maintenance in Arabidopsis thaliana, refines cytokinin signaling by acting non-cell autonomously from a presumptive stem cell organizing center to repress negative regulators of cytokinin signaling called type-A Arabidopsis response regulators (arr5, arr6, arr7, and arr15) (Leibfried et al., 2005). Accordingly, overexpression of type-A arr’s results in a SAM consumption phenotype that mimics loss-of-function WUS mutants. The maize gene aberrant phyllotaxy1 (abph1) encodes a cytokinin-inducible type-A ARR homolog that represses meristem size via the regulation of cytokinin and auxin signaling (Giulini et al., 2004; Lee et al., 2009). Maize abph1 mutants have enlarged meristems and often generate a decussate leaf phyllotaxy (two leaves per node initiated perpendicular to leaves at adjoining nodes) rather than the distichous (one leaf per node in two ranks) pattern of wild-type maize (Jackson and Hake, 1999).
A shift in hormone signaling accompanies the onset of organ initiation at the SAM flanks. Concordantly, localized transport of auxin, as indicated by the auxin efflux protein PIN-FORMED1 (PIN1) and the auxin reporter DR5, is the earliest known molecular marker for leaf initiation and vascular development (Reinhardt et al., 2000, 2003; Heisler et al., 2005; Bayer et al., 2009). The accumulation of auxin at the SAM periphery is both necessary and sufficient for leaf initiation. Exogenous auxin application to the meristem periphery can induce new leaf formation (Reinhardt et al., 2000), while blocking auxin transport with N-1-naphthylphthalamic acid halts their initiation (Reinhardt et al., 2000; Scanlon, 2003).
Precise auxin regulation is also necessary for the initiation of axillary meristems from inflorescences. Mutations to the Arabidopsis pin1 or the PIN1 regulator pinoid (pid) lead to a pin-like inflorescence that lacks flowers (Okada et al., 1991; Bennett et al., 1995). Likewise, treatment of the maize inflorescence with N-1-naphthylphthalamic acid (Wu and McSteen, 2007) and mutations to the YUCCA-like auxin biosynthetic gene sparse inflorescence1 (Gallavotti et al., 2008) or the pid homolog barren inflorescence2 (McSteen et al., 2007) reduce axillary meristem initiation. Accumulation of the maize PIN1 ortholog, PIN1a, and DR5 expression precedes meristem initiation events within the maize inflorescence, further supporting a role for auxin accumulation in this process (Wu and McSteen, 2007).
Less research has focused on the interplay of metabolites and meristem function. As an active center of cell division, regulation of carbohydrate metabolism is vital to SAM function. This interdependence is highlighted by analyses of the Arabidopsis gene stimpy (Wu et al., 2005), which encodes a WOX transcription factor that is necessary for maintaining cell divisions within the SAM. Sucrose application fully rescues the seedling-lethal phenotype of stimpy mutants, illustrating the importance of carbohydrate regulation in SAM function. A role for sugar signaling is further demonstrated by analyses of meristem determinacy mutant ramosa3 (ra3), which causes abnormal branching of the female inflorescence (ear) (Satoh-Nagasawa et al., 2006). The ra3 gene encodes a trehalose-6-phosphate phosphatase, which is expressed in a small domain below axillary meristems and is purported to regulate branching via production of a mobile trehalose signal (Satoh-Nagasawa et al., 2006).
The B vitamin thiamine is synthesized in plants, bacteria, and fungi but not in animals; nutritional deficiency in humans leads to the neurological disease beriberi (Bates, 2007). Phosphorylated thiamine derivatives are essential cofactors used in the biosynthesis of terpenes and branched-chain amino acids and in a variety of carbohydrate metabolic pathways, including the Calvin cycle, nonoxidative pentose phosphate pathways, the citric acid cycle, and glycolysis. Thiamine auxotrophs in Arabidopsis illustrate that this cofactor is required for plant viability (Langridge, 1955; Feenstra, 1964; Redei, 1965; Li and Redei, 1969; Koornneef and Hanhart, 1981). Without thiamine supplementation, the Arabidopsis th1, py, and thi1 mutants die after the production of several albino leaves. The th2 and th3 auxotrophs are viable but have chlorotic and necrotic regions in leaves. Several of these loci are known to encode enzymes in the thiamine biosynthetic pathway. Arabidopsis THI1, in particular, catalyzes the biosynthesis of adenosine diphospho-5-(β-ethyl)-4-methylthiazole-2-carboxylic acid (ADT), a thiazole precursor to thiamine (Papini-Terzi et al., 2003).
Here, we describe the bladekiller1-R (blk1-R) mutant of maize, a thiamine auxotroph with defects in meristem maintenance and a novel leaf blade reduction phenotype. Positional cloning revealed that mutations to thiamine biosynthesis2 (thi2), a maize ortholog of Arabidopsis thi1 (Belanger et al., 1995), condition blk1 mutant phenotypes. Biochemical analyses suggest a conserved role for THI2 in thiamine biosynthesis and that the reference mutant allele, blk1-R, is a null. Importantly, all blk1-R mutant phenotypes are rescued by thiamine supplementation. These data provide a new perspective on the metabolic requirements of the SAM.
RESULTS
blk1-R Mutants Have Defects in Vegetative SAM Maintenance
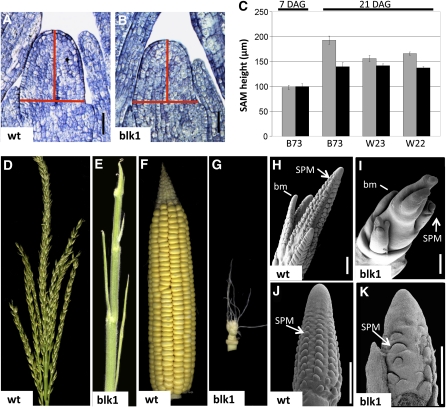
Plants homozygous for the blk1-R mutation were discovered as progeny with aborted shoot phenotypes segregating in the M2 generation of ethyl methanesulfonate (EMS)-treated maize. The blk1-R mutant allele was subsequently introgressed into the B73 inbred background for over nine generations. Longitudinal sections of seedling apices at 7 and 21 d after germination (DAG) reveal that while mutant and wild-type SAMs are of similar height early in vegetative development, blk1-R SAMs fail to increase in height at the rate of their wild-type siblings and are on average 53 μm shorter at 21 DAG (Figures 1A to 1C). Despite this effect on SAM height, SAM width is unaffected in the blk1-R mutant background (Figures 1A and 1B).
Figure 1.
blk1-R Mutants Exhibit Progressive Defects in Shoot Meristem Maintenance.
(A) and (B) 21-DAG SAMs in B73 background; red lines indicate reference points for measurement for height and width. Bars = 50 μm.
(A) Wild-type (wt) SAM.
(B) blk1-R SAM is reduced in height.
(C) blk1-R meristems (black bars) are shorter than wild-type SAMs (gray bars) in the B73, W22, and W23 inbred backgrounds at 21 DAG. Note that SAM height is essentially unaffected at 7 DAG. Error bars indicate ±se; n = 15.
(D) to (G) Mature maize inflorescences.
(D) Wild-type tassel.
(E) blk1-R mutant tassel has barren branched devoid of florets.
(F) Wild-type ear.
(G) blk1-R ear is greatly reduced in size and has few florets.
(H) to (K) Scanning electron microscopy of immature maize inflorescences, SPMs, and tassel branch meristems (bm). Bars = 500 μm.
(H) Wild-type tassel primordium.
(I) blk1-R tassel primordium has aberrant branches and fewer SPMs.
(J) Wild-type ear primordium.
(K) blk1-R ear primordium is truncated and has fewer SPMs.
To further investigate the meristem size phenotype, the blk1-R mutant allele was introgressed for five generations into W22 and W23, inbred maize lines with shorter SAMs than inbred B73 (Vollbrecht et al., 2000). In agreement with previously published data, wild-type SAMs in W22 and W23 were smaller than B73 shoot meristems (Figure 1C); introgression of the blk1-R mutation into W22 and W23 reduced the mean SAM height by 29 and 15 μm, respectively. Interestingly, the average height of blk1-R mutant SAMs after 3 weeks of growth (~140 μm) was not significantly altered by inbred background, suggesting the blk1-R mutation is epistatic to factors leading to the greater meristem height observed in the B73 inbreds.
blk1-R Mutants Have Highly Reduced Inflorescence Development
Defects in blk1-R mutant shoot meristems are especially apparent during reproductive development. The spatially separated male and female inflorescence meristems of maize give rise to the pollen-bearing tassel and the seed-bearing ear, respectively (Figures 1D and 1F; Kiesselbach, 1949). The tassel develops at the apex of the plant following transition of the SAM into an inflorescence meristem. Maize ears develop from lateral meristems located in the axils of leaves. Each inflorescence comprises several types of axillary meristems (Figures 1H and 1J; reviewed in Bortiri and Hake, 2007): branch meristems produce the long branches located at the base of the maize tassel (Figures 1D and 1H) and are absent from the ear (Figures 1F and 1J); spikelet-pair meristems (SPMs) each generate two spikelet meristems; the spikelet meristems then yield short floral-bearing spikelets and give rise to floral meristems, which produce the floral organs. Mature blk1-R mutant tassels are sterile, forming several shortened, barren branches (Figure 1E). Similarly, the mature mutant ear is severely truncated and develops only a few seed-bearing florets (Figure 1G). Scanning electron microscopy of inflorescence primordia reveal that blk1-R mutants are defective in axillary meristem initiation and/or maintenance. Although mutant tassels do form a reduced number of branch meristems, very few SPMs are initiated, and those that do form are subsequently aborted (Figure 1I). Occasional spikelet and floral meristems are formed near the bases of blk1-R mutant ear primordia (Figure 1K); however, growth of the mutant female inflorescence aborts prior to elongation. These observations highlight the progressive nature of the blk1-R mutant phenotype, in that the most severe shoot meristem abnormalities occur in the latest emerging structures of the shoot.
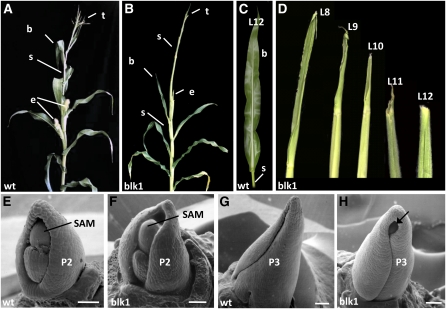
blk1-R Mutants Have Progressive Defects in Leaf Blade Development
Maize leaves develop from ~250 initial cells recruited from the flank of the SAM (Poethig, 1984). Asymmetric cell division, expansion, and differentiation within the leaf primordium give rise to a leaf with distinct morphological domains (reviewed in Hudson, 2000; Byrne et al., 2001). Mature leaves are subdivided along the longitudinal axis into a proximal sheath that encircles the stem and a distal blade; these two domains are separated by a hinge-like auricle and a fringe of epidermally derived, adaxial tissue called the ligule (Figures 2A and 2C). The blk1-R mutants exhibit a novel leaf reduction phenotype wherein the basal-most mature leaves (L1-L7) are morphologically normal and each consecutive leaf thereafter (L8-L18) exhibits an increasing degree of blade truncation, which is accompanied by shredding and necrosis of the blade margins and tip (Figures 2B and 2D). Although the extent of leaf reduction is variable from plant to plant and from leaf to leaf, the last two to four mutant leaves immediately below the tassel frequently contain no leaf blade and are composed of either sheath alone or of sheath topped by a ligule (Figure 2D).
Figure 2.
blk1-R Mutants Exhibit a Progressive Loss of Leaf Blade.
(A) Mature wild-type (wt) B73 plant. b, leaf blade; s, leaf sheath; e, ears; t, tassel.
(B) Mature blk1-R mutant plants have truncated leaf blade development in apical nodes.
(C) Mature maize leaf (L12 = 12th leaf from base).
(D) Successive blk1-R leaves exhibit increasing degree of blade truncation; L12 is bladeless.
(E) to (H) Scanning electron micrograph of dissected 18-d-old SAM apices from blk1-R mutants and wild-type seedlings; plastochrons 2 and 3 (P2, P3); leaf numbers 15 and 16. Bars = 40 μm.
(E) Wild-type P2 has an apical hood covering the SAM.
(F) blk1-R P2 leaf is reduced.
(G) Wild-type P3 leaf envelops the underlying apex.
(H) blk1-R P3 leaf is reduced and exposes the shoot apex (arrow).
[See online article for color version of this figure.]
Scanning electron microscopy analysis of developing leaf primordia from 18-d-old seedlings revealed that the blk1-R blade truncation phenotype is evident as early as the second plastochron (P2). Wild-type P2 leaves form an apical hood that covers the SAM (Figure 2E); however, the blk1-R mutant P2 leaf fails to cover the SAM apex (Figure 2F). Whereas the margins of the P3 wild-type leaf primordium completely surround the shoot tip (Figure 2G), the mutant P3 primordium is truncated at the distal tip, exposing the underlying apex (Figure 2H). No evidence of tissue necrosis is observed in blk1-R mutant primordial leaves.
Molecular Markers Reveal blk1-R Mutants Are Progressively Deficient in Shoot Meristem Function
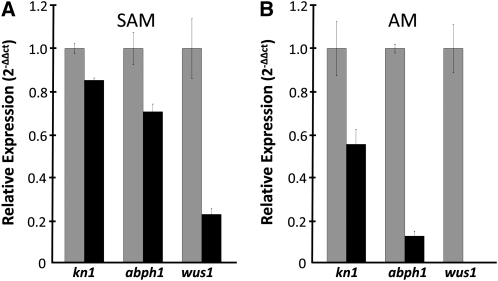
Morphological analyses indicate that blk1-R mutants exhibit a progressive decrease in SAM size (Figures 1A to 1C), which suggests a defect in stem cell maintenance. To explore this possibility, quantitative RT-PCR (qRT-PCR) analyses of laser-microdissected SAMs were used to compare the transcript accumulation of the several meristem maintenance marker genes, including the homeobox gene knotted1 (kn1; Jackson et al., 1994), the cytokinin response regulator abph1 (Giulini et al., 2004), and the putative stem cell organizer Zm wus1 (Nardmann and Werr, 2006). Mutant SAMs exhibit a significant decrease in the transcript abundance of these marker genes (Figure 3A), ranging from a 15% reduction in the meristem maintenance gene kn1 to a 78% reduction in the putative stem cell regulator wus1 and a 30% reduction in the cytokinin response regulator abph1. Likewise, transcript accumulation of kn1, abph1, and wus1 is significantly reduced in laser-microdissected (see Supplemental Figure 1 online) blk1-R mutant ear axillary meristems compared with wild-type siblings (Figure 3B). Surprisingly, no wus1 transcripts were amplified from blk1-R mutant lateral meristems. Notably, transcript accumulation of these meristems markers is more severely downregulated in mutant axillary meristems than in mutant vegetative SAMs (Figures 3A and 3B).
Figure 3.
Accumulation of Meristem Marker Transcripts Is Decreased in blk1-R Mutant Meristems.
qRT-PCR analyses of meristem marker gene transcript accumulation in wild type (gray bars) and blk1-R mutant (black bars) SAMs (A) and in ear axillary meristems (AM) (B). Each experiment used three biological replicates. Error bars denote 1 se.
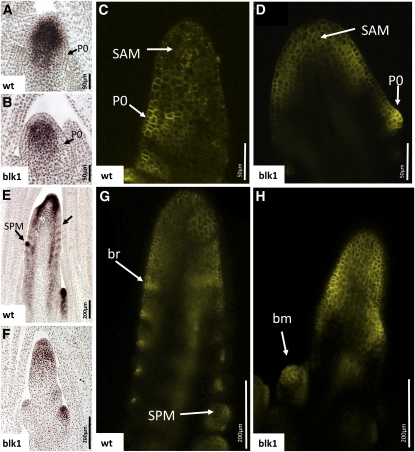
RNA in situ hybridizations were used to investigate whether reduced levels of kn1 in the blk1-R mutant SAM correlated with alterations in the cell/tissue specific accumulation pattern of this transcript. In wild-type apices, kn1 transcripts accumulate in the meristem proper but are absent at the site of leaf initiation (P0; Figure 4A). Mutant SAMs exhibit an equivalent kn1 accumulation pattern, although the signal intensity is consistently reduced (Figure 4B), corroborating the qRT-PCR data. As described previously (Gallavotti et al., 2008), accumulation of the auxin efflux reporter protein PIN1a is especially abundant at the P0 and serves as the earliest described marker of leaf initiation (Figure 4C). Although the leaf blade truncation phenotype of blk1-R mutants is evident in young leaf primordia (Figure 2), no overt differences in PIN1a~YFP (for yellow fluorescent protein) localization are observed in the blk1-R vegetative SAM (Figure 4D). Taken together, these data suggest that the earliest known events in maize leaf initiation are not affected in blk1-R mutants.
Figure 4.
kn1 and PIN1a~YFP Accumulation Patterns Are Normal in the blk1-R SAM but Are Aberrant in the Mutant Tassel Primordia.
(A), (B), (E), and (F) RNA in situ hybridizations with kn1 probe. wt, wild type.
(C), (D), (G), and (H) Confocal images of PIN1a~YFP fluorescence.
(A) kn1 mRNA accumulates in the indeterminate cells that make up the SAM proper but is downregulated at the P0 of wild-type 21-DAG seedlings.
(B) Mutant SAMs at 21-DAG have normal kn1 expression patterns, but mRNA levels are reduced.
(C) Wild-type SAMs at 21 DAG show PIN1a upregulation at the P0.
(D) blk1-R SAMs at 21 DAG have normal PIN1a upregulation at the P0.
(E) In wild-type tassel primordia, kn1 mRNA accumulates at the apices of branch primordia, at the apex of central tassel spike, at the presumptive sites of SPM initiation (arrow), and within developing SPMs.
(F) kn1 accumulation in blk1-R tassel primordia is reduced, and normal upregulation does not occur along the flanks of the inflorescence.
(G) PIN1a~YFP is upregulated in SPMs and along the flanks of the wild-type tassel primordium where it marks the initiation of bract primordia (br).
(H) PIN1a~YFP localization is aberrant in blk1-R mutant tassel primordia, which may account for the large reduction in SPM number. bm, branch meristem.
Initiation of lateral organ and axillary meristem primordia in the developing maize inflorescences is correlated with both kn1 expression and accumulation of PIN1 in a phyllotactic pattern along the inflorescence flank (Figures 4E and 4G; Vollbrecht et al., 2000; McSteen and Hake, 2001; Gallavotti et al., 2008). RNA in situ hybridizations of kn1 (Figure 4F) and confocal imaging of tassel primordia expressing PIN1a~YFP (Figure 4H) reveal that blk1-R mutant inflorescences do not accumulate kn1 or PIN1a~YFP in a banded pattern, which correlates with the failure to initiate normal whorls of tassel branches or rows of SPMs (Figures 1H and 1I). Whereas wild-type tassel primordia accumulate kn1 transcripts at the apices of branch primordia, at the apex of the central tassel spike, at the presumptive sites of SPM initiation, and within developing SPMs (Figure 4E), blk1-R mutant primordia exhibit an attenuated and dispersed kn1 accumulation pattern in the inflorescence and branch meristems (Figure 4F). Likewise, although some blk1-R mutant tassels accumulate PIN1a~YFP at the flank of the inflorescence in positions where an occasional branch meristem or SPM might form, no phyllotactic pattern of PIN1a accumulation is observed (Figure 4H). Thus, the phenotypic and molecular marker data demonstrate that the blk1-R mutation conditions a progressive defect in shoot meristem maintenance and function, in which the shoot meristem abortion phenotype becomes increasingly severe during later stages of development.
Positional Cloning Identifies blk1-R as a Mutation in a Thiamine Biosynthetic Gene
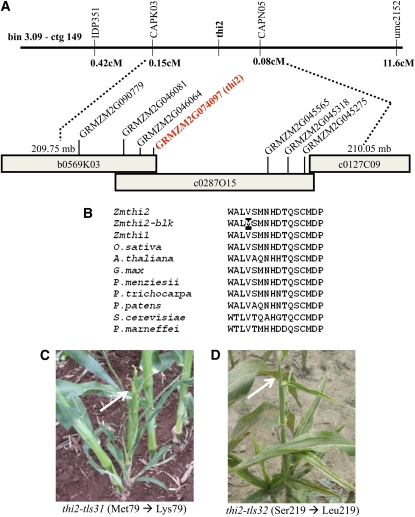
The blk1-R mutation was mapped to the long arm of chromosome 3 using the B-A translocation lines as described by Beckett (1978). F1 plants hypoploid for TB-3La displayed phenotypes that were equivalent to blk1-R homozygous plants, suggesting that the blk1-R mutation is a genetic null. The blk1-R mutation was mapped to contig 149 of the maize agarose fingerprinted contig map provided by the Arizona Genomics Institute (http://www2.genome.arizona.edu/genomes/maize), by linkage to simple sequence repeat marker umc2152 and the insertion-deletion marker IDP351 (Figure 5A). An F2 mapping population of 686 mutants was used to fine map the mutation to a 0.23-centimorgan interval delimited by the genetic markers CAPSK03 (two recombinants) and CAPSN05 (one recombinant), a region comprising ~300 kb (Figure 5A). Analysis of this sequenced region (Schnable et al., 2009; maizesequence.org [http://www.maizesequence.org/index.html]) revealed seven candidate open reading frames (ORFs); each ORF was evaluated for sequence polymorphisms disrupting the predicted gene product. A G-to-A point mutation within the blk1-R allele of the thi2 gene (GRMZM2G074097) was identified that is predicted to convert Val-211 to Met-211 within the translated protein. Amino acid residue Val-211 is conserved in comparisons of THI2 homologs from four angiosperms (Arabidopsis, Oryza sativa, Glycine max, and Populus trichocarpa), one gymnosperm (Pseudotsuga menziesii), one bryophyte (Physcomitrella patens), and two fungi (Saccharomyces cerevisiae and Penicillium marneffei) (Figure 5B). Two additional mutant thi2 alleles (thi2-tls31 and thi2-tls32), which condition equivalent blk1 phenotypes and also harbor missense mutations in conserved THI2 residues (Met-79 to Lys-79 and Ser-219 to Leu-219, respectively), confirm that mutations to thi2 render blk1 mutant phenotypes (Figures 5C and 5D). Thus, the blk1-R mutation is hereafter designated as the thi2-blk1 allele of thi2.
Figure 5.
Positional Cloning of the blk1-R Mutation Reveals a Missense Mutation in a Conserved Residue Encoded by the thi2 Gene.
(A) The blk1-R mutation was mapped to a small interval in Bin 3.09, ctg 149. The listed genetic markers (top) were used to delimit the interval. Recombination frequency is in centimorgans (cM). The region spans three BACs and contains seven genes with predicted ORFs (bottom).
(B) Amino acid sequence alignment of THI homologs in various organisms. Sixteen amino acids surrounding the blk1-R mutation (black) are shown.
(C) and (D) Two additional recessive mutant alleles of thi2, thi2-tlsl31 (C), and thi2-tls32 (D), condition equivalent shoot meristem maintenance, and leaf reduction phenotypes (arrows).
[See online article for color version of this figure.]
thi2 Transcripts Accumulate in Rapidly Dividing Tissues
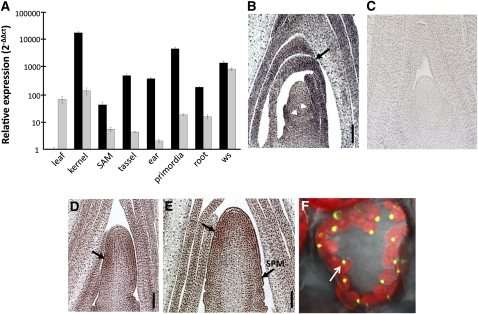
THI2 shares 95% amino acid similarity with the predicted gene product of thi1, a paralog located on maize chromosome 8 (Belanger et al., 1995). To decipher the tissue specificity and timing of expression of these nearly identical paralogs, qRT-PCR analyses were performed on cDNA derived from 14-DAG whole seedlings, seedling roots, 20 d after pollination kernels, laser-microdissected SAMs, developing ears and tassels (2 to 4 mm), immature leaf primordia (P2-P6), and mature adult leaves (L9-L10). Transcript accumulation of thi1 and thi2 was differentially regulated in all tissues tested (Figure 6A). Compared with thi1, thi2 transcripts preferentially accumulated in the rapidly dividing tissues comprising the kernel (145-fold), SAM (5-fold), immature tassel (130-fold) and ear (228-fold), and leaf primordia (182-fold) but were downregulated in mature leaves (74-fold). Substantial overlap of thi1 and thi2 transcript accumulation was noted in developing kernels and young seedlings.
Figure 6.
thi2 Transcript Accumulation and Protein Localization.
(A) Comparative transcript accumulation of thi1 (gray bars) and thi2 (black bars), normalized to levels of 18srRNA, within maize tissues relative to thi2 in the mature leaf = 1.0. Primordia, P2-P6 leaf primordia; leaf, mature L9-L10; ws, whole 14 DAG seedlings. Errors bars indicate ±se.
(B) RNA in situ hybridization detects thi transcripts in initiating leaf primordia (white arrowheads) and distal regions of older primordia (black arrows) of vegetative apices.
(C) Control in situ hybridization using a thi probe synthesized in the sense orientation.
(D) and (E) thi accumulates along the flanks of the 0.5-mm tassel primordium ([D]; arrow) and at the apex (arrow) and spikelet pair meristems (SPM) of the 1.5-mm tassel primordium (E).
(F) Confocal image of THI2-GFP fusion protein after transient expression in N. benthamiana leaves (arrow). Chlorophyll autoflorescence is shown in red, and GFP/chlorophyll fluorescence overlap is represented as yellow.
Bars = 100 μm.
A 1253-bp probe spanning the thi2 coding region was generated that is predicted to hybridize to both thi1 and thi2 transcripts, owing to the sequence similarity between thi1 and thi2. RNA in situ hybridizations of seedling apices revealed thi transcript accumulation in the distal regions of young leaf primordia (P2-P4) and at the SAM flank where leaf primordia initiate (Figure 6B). In support of qRT-PCR data (Figure 6A), thi transcripts were reduced in the SAM proper. Prior to the initiation of spikelet pair meristems, thi transcripts accumulated at the flank of the tassel inflorescence meristem (Figure 6C); later in tassel development, thi expression was upregulated in the developing spikelet pair meristems and in the inflorescence apex (Figure 6D).
THI2 Is a Plastid-Localized Protein
Arabidopsis THI1 localizes to the mitochondria and chloroplasts (Chabregas et al., 2003). Previously, Belanger et al. (1995) used a THI1 antibody to show that the maize THI1 protein localizes to plastids. To verify the subcellular localization of THI2, a THI2-GFP (for green fluorescent protein) construct driven by the cauliflower mosaic virus 35S promoter was transiently expressed in Nicotiana benthamiana leaves. Confocal imaging of infiltrated leaf sections revealed that THI2-GFP accumulates in punctate spots within the chloroplast-containing mesophyll cells (Figure 6E). We speculate that the unusual punctate pattern of accumulation reflects the formation of inclusion bodies following overexpression of THI2. No THI2-GFP fluorescence was observed outside of the chloroplast or in epidermal cells. Although it is noted that overexpression from the cauliflower mosaic virus 35S promoter may bias targeting of THI2-GFP to the chloroplasts, no localization to the mitochondria was observed.
The thi2-blk Mutant Is Deficient in Thiamine Accumulation
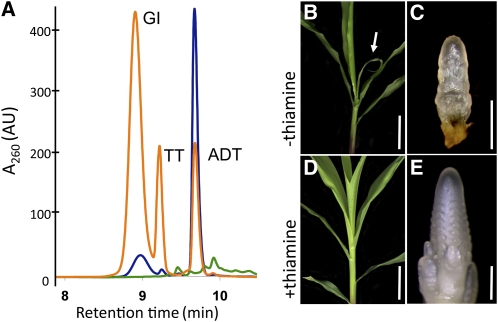
THI2 homologs from yeast (THI4) and Arabidopsis (THI1) copurify with ADT, a thiazole precursor to thiamine (Godoi et al., 2006; Jurgenson et al., 2006). Furthermore, the maize THI2 paralog, THI1, rescues the growth defects of yeast thi4 mutants, suggesting a conserved function during thiamine biosynthesis (Belanger et al., 1995). To verify the function of THI2, the protein was expressed in Escherichia coli, purified, and analyzed for bound substrate by HPLC. Similar to yeast THI4, the THI2 protein copurifies with ADT, indicating that THI2 also functions in thiamine biosynthesis (Figure 7A). Importantly, THI2-blk1 proteins expressed from the thi2-blk1 mutation did not purify with the ADT substrate (Figure 7A).
Figure 7.
thi2-blk1 Mutants Are Thiamine Auxotrophs
(A) HPLC analysis of THI2 (blue), THI2-blk1 (green), and yeast THI4 (orange) protein-bound metabolites. THI2, but not THI2-blk1 protein, copurifies with bound ADT substrate. GI, glycine-imine; TT, thiazole tautomer. AU, absorbance units.
(B) to (E) Thiamine treatment rescues blk1-R phenotypes. Untreated thi2-blk1 plant with reduced leaf blade (arrow) (B) and aborted shoot apex (C). Treated thi2-blk1 mutant plant showing rescued leaves (D) and tassel inflorescence (E).
Bars = 7 cm ([B] and [D]) and 500 μm ([C] and [E]).
HPLC analyses of thiamine content revealed that thi2-blk1 mutant seedlings accumulated ~50% less total thiamine than wild-type plants (wild type = 4.93 ± 0.10 nmol total thiamine/mg wet tissue; thi2-blk1 mutant = 2.27 ± 0.08 nmol total thiamine/mg wet tissue, n = 3). Furthermore, thi1-blk1 mutants accumulated ~30% less thiamine-diphosphate (a bioactive derivative of thiamine) than wild-type siblings (wild type = 1.56 ± 0.15 nmol thiamine-diphosphate/mg wet weight to thi2-blk1 mutant = 1.02 ± 0.03 nmol thiamine-diphosphate/mg wet weight). To test the hypothesis that the thi2-blk1 mutant phenotypes are caused by thiamine deficiency, mutant plants were treated with exogenous thiamine. Significantly, thiamine treatment rescued all thi2-blk1 mutant phenotypes, including defects in SAM size maintenance, leaf blade development, and axillary meristem initiation (Figures 7B to 7E; see Supplemental Figure 2 online). Furthermore, 100% of the progeny derived from thiamine-supplemented, self-pollinated thi2-blk1 plants exhibited thi2-blk1 mutant phenotypes, verifying that the rescued plants were indeed thi2-blk1 homozygotes.
DISCUSSION
thi2-blk1 Is a Null Allele of thi2
Positional cloning revealed that mutations to thi2 correlate with bladekiller phenotypes (Figure 5). The predicted THI2 amino acid sequence has homology to enzymes that generate the thiazole precursor to the B vitamin thiamine (Belanger et al., 1995). Accordingly, the THI2 protein copurifies with a thiazole intermediate (Figure 7A), suggesting a conserved role in thiamine biosynthesis. Both biochemical analyses and genetic dosage analyses suggested that thi2-blk1 is a null mutation. Plants homozygous for thi2-blk1 are phenotypically equivalent to hypoploid thi2-blk1/- individuals harboring a single dose of the thi2-blk1 mutation. Moreover, the THI2-blk1 protein does not purify with a thiazole intermediate (Figure 7A), indicating that the mutant enzyme is not functional. Loss of THI2 function conditions lower thiamine concentrations in thi2-blk1 mutant plants, and exogenous thiamine application complements all thi2-blk1 mutant phenotypes (Figures 7B to 7E). Taken together, these data reveal that the thi2-blk1 mutant is a thiamine auxotroph of maize.
Nonoverlapping Functions of the Maize THI2 and THI1 Paralogs
Belanger et al. (1995) confirmed that the nearly identical THI2 paralog THI1 can restore thiamine prototrophy in yeast. Despite their shared function in thiamine biosynthesis, THI1 and THI2 are not functionally redundant, as evidenced by the thi2-blk1 mutant phenotypes and by the distinct accumulation patterns of thi1 and thi2 transcripts (Figure 6A). Accumulation of thi2 transcripts is much higher than thi1 in developing kernels, whole SAMs, immature leaves, and tassel and ear primordia but relatively lower in mature leaves. These differences in transcript accumulation reflect the subfunctionalization of the maize THI paralogs, such that THI2 is heavily used in young, rapidly dividing tissues, and THI1 predominantly functions in mature green leaves.
The thi2-blk1 mutation conditions shoot meristem maintenance and leaf blade reduction phenotypes that are progressively more severe in later developmental stages (Figures 1 and 2). Shimamoto and Nelson (1981) proposed that thiamine stores transferred to developing maize kernels from maternal cob tissue are used by the germinating seedling. Extending this model, we propose that maternally derived thiamine combined with the THI1 function in developing kernels and young seedlings (Figure 6A) may account for the relatively weak thi2-blk1 mutant phenotypes during early developmental stages.
Development of the leaf blade is progressively diminished in thi2-blk1 mutants, whereas the leaf sheath is unaffected (Figures 2B and 2D). We speculate that the developing leaf blade may have higher thiamine demands than the sheath. In support of this model, in situ hybridization analyses reveal that thi transcript accumulation is higher in the distal portions of the P2-P4 leaf primordia than in the proximal regions (Figure 6B). In this view, the thiamine pool in thi2-blk1 mutant embryos may be sufficient for normal development of the basal-most leaves. However, as the thiamine pool is diminished in germinating seedlings due to the lack of THI2 activity, subsequent leaves would become increasingly thiamine deficient, leading to defects in blade development. Sheath development, on the other hand, may require less thiamine and thus is not affected by the loss of THI2 function. Identification of thi1 mutations and analyses of thi1 thi2 double mutant phenotypes will clarify the gene-specific functions of the maize thi paralogs.
Thiamine Is a Regulator of Stem/Initial Cell Activity
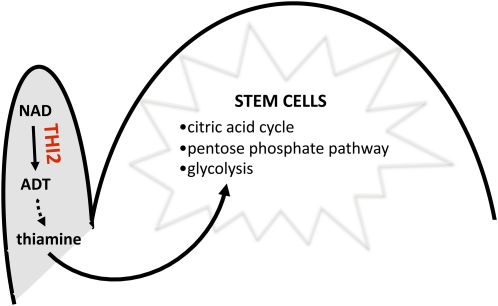
Our analysis of THI2 function reveals that actively dividing meristematic stem cells and organ initial cells are particularly sensitive to thiamine deprivation (Figures 1 and 2). The thi2-blk1 leaf phenotype is blade specific (Figures 2B and 2D) and correlated with thi transcript accumulation in the distal domains of wild-type leaf primordia (Figure 6B). Likewise, the axillary meristems that elaborate the tassel and ear inflorescences are also sensitive to thi2-blk1–induced thiamine deficiency (Figures 1D to 1K), and thi2 transcripts are upregulated at the sites of SPM formation (Figure 6D). Conversely, whereas thi2 transcripts accumulate in leaf initials comprising the P0, they are not detected in the SAM proper (Figure 6B), despite the fact that thi2-blk1 mutants exhibited a progressive defect in the maintenance of SAM stem cells (Figures 1A to 1C and 3A). These data suggest that relatively little thiamine is synthesized in the SAM proper, although previous SAM-specific microarray and RNA-seq analyses revealed an abundance of transcripts encoding thiamine-using metabolic enzymes (Emrich et al., 2007; Brooks et al., 2009). In light of these findings, we propose a model wherein thiamine synthesized in lateral organ primordia is transported into the SAM, where it is required for meristem maintenance (Figure 8).
Figure 8.
Model for THI2 Function during SAM Maintenance.
THI2 functions in leaves and leaf primordia during biosynthesis of ADT, the thiazole precursor to thiamine. Thiamine is then transported from leaves into the meristem and participates in various carbohydrate metabolic reactions required for stem and initial cell activities.
[See online article for color version of this figure.]
This model is supported by early work with plant tissue culture, which revealed that meristematic activity requires supplemental thiamine (Wightman and Brown, 1953). Thiamine function during the maintenance of proliferative stem/initial cell growth may be conserved in plants and animals. The growth of animal cancer cells, actively dividing populations of malignant stem cells, is suppressed by treatment with thiamine antagonists (Boros et al., 1998; Liu et al., 2010). Thiamine is an essential cofactor for transketolase function, and the proliferation of tumor cells requires the nonoxidative, transketolase-dependent pentose phosphate pathway for ribose synthesis (Boros et al., 1998). The use of transketolase inhibitors, such as the thiamine analog oxythiamine, leads to significant decreases in tumor proliferation (Comín-Anduix et al., 2001). Moreover, supplementing tumors with thiamine leads to transketolase activation and increases in tumor growth (Comín-Anduix et al., 2001). Although it is not known whether the effects of thiamine on meristem maintenance in maize are mediated by a transketolase-based mechanism, the inhibition of thiamine function blocks the proliferative growth of the undifferentiated stem cell populations that comprise both cancer cells in animals and meristematic cells in plants. Our data emphasize that in addition to traditional analyses of transcriptional regulation, phytohormones, and signal transduction, a complete understanding of the mechanisms regulating meristem and initial cell function in plants must also incorporate models describing the role of essential metabolites, such as thiamine.
METHODS
Plant Material and Treatments
The thi2-blk1 mutation was generated by EMS treatment of maize (Zea mays) pollen and was crossed onto wild-type individuals in an unknown genetic background. The tasselless (tls) mutant alleles, thi2-tls31 and thi2-tls32, were generated by EMS mutagenesis in the A632 background. Heterozygous thi2-blk1/+ individuals were backcrossed at least nine times into the B73 inbred background and five times into the W22 and W23 inbred lines. Genetic analyses were conducted in Ithaca, NY or in Juana Diaz, Puerto Rico. For molecular studies and phenotypic analysis, plants were grown under greenhouse conditions at Cornell University. Thiamine rescue experiments were completed by the twice weekly application of 100 mL of a 30 mM aqueous solution of thiamine-HCL (Sigma-Aldrich) to the soil of growing plants.
Histological Analysis and Scanning Electron Microscopy
Maize apices were hand-dissected from 7- or 21-DAG plants, fixed in 3.7% formalin/acetic acid/alcohol, and paraffin embedded, and 10-μm sections were stained with 0.05% toluidine blue O as described (Ruzin, 1999). All micrographs were imaged on a Zeiss Z1-Apotome microscope, and meristem measurements were made using Zeiss Axiovision software, release 4.6.
For scanning electron microscopy analysis, samples were hand-dissected and immediately frozen in a nitrogen slush. The samples were then moved to a BalTec cryopreparation chamber (BalTec, Technotrade) attached to a Hitachi S4500 scanning electron microscope, sputter coated with 30 nm of platinum at −150°C, and a vacuum pressure of 2 × 10−2 Torr under argon. The coated samples were then viewed at −163°C on the scanning electron microscope stage using 5 kV.
Mapping and Cloning of thi2-blk
B-A translocation stocks were obtained from the Maize Genetics COOP Stock Center. Plants hyperploid for TB-3La and heterozygous for the endosperm marker a1 were crossed onto thi2-blk1 heterozygous plants to generate individuals hypoploid for much of the long arm of chromosome 3 (described in Scanlon et al., 1994). An F2 mapping population was generated by crossing thi2-blk1 heterozygotes introgressed into B73 nine times onto the Mo17 inbred. Known molecular markers mapped to chromosome 3L were available at maizegdb.org and used to delimit the mapping interval. Sequence available at maizesequence.org and magi.plantgenomics.iastate.edu was used identify candidate genes and build primers for amplifying genomic regions. Sequence polymorphisms between Mo17 and the thi2-blk1 progenitor were used to design additional molecular markers that further narrowed the mapping interval (see Supplemental Table 1 online). A thi2 (GRMZM2G074097) product was amplified by PCR using thi2-cDNA primers, which flank the predicted coding region (see Supplemental Table 1 online).
THI2 Localization
A thi2 PCR product was generated using the thi2-gfp primers (see Supplemental Table 1 online) and was subsequently cloned into the pENTR vector (Invitrogen). Recombination with the pEarleyGate103 gateway vector (Earley et al., 2006) allowed a C-terminal fusion of THI2 with GFP. The resultant vector was electroporated into Agrobacterium tumefaciens strain C58C1, and cultures were grown overnight in Luria-Bertani media containing 50 μg/mL kanamycin and 50 μg/mL tetracycline. Pelleted cells were resuspended in 10 mM MgCl2 to an OD600 of 0.2. This suspension was then used to infiltrate 5-week-old Nicotiana benthamiana plants that were grown under fluorescent lights. THI2-GFP was visualized in leaf discs under water ~3 d after infiltration.
Confocal Microscopy
To generate thi2-blk1 mutants expressing PIN1a-YFP, thi2-blk1 heterozygotes were crossed with PIN1a-YFP transgenic individuals (kindly provided by D. Jackson, Cold Spring Harbor Laboratory) and then self-fertilized (Gallavotti et al., 2008). SAMs and immature tassels were uncovered by hand-dissection and visualized in water. Images of fluorescent proteins were collected on a Leica TCS-SP5 confocal microscope (Leica Microsystems) using a ×10 or ×20 objective (numerical aperture 0.4 or 0.7, respectively). YFP was excited with an argon-ion laser (514 nm), and emitted light was collected from 517 to 564 nm. GFP was excited with the argon laser (488 nm), and emitted light was collected between 498 and 510 nm. Chloroplasts were excited with the argon laser (488 nm), and emitted light was collected from 610 to 669 nm. The GFP and chloroplast signals were collected separately and later superimposed. Light images were collected simultaneously with the fluorescence images using the transmitted light detector. Images were processed using Leica LAS-AF software (version 1.8.2) and Adobe Photoshop CS2 version 9.0.2 (Adobe Systems).
qRT-PCR and in Situ Hybridizations
All transcript accumulation experiments were performed in the B73 inbred background or with thi2-blk1 mutants introgressed into B73 at least nine times. Laser microdissection and subsequent RNA preparation were completed as outlined by Zhang et al. (2007) and Ohtsu et al. (2007). In brief, dissected apices from 21-DAG seedlings and lateral meristems that were still producing husk leaves at presumptive ear nodes were acetone fixed, paraffin embedded, sectioned at 10 μm, and laser microdissected. Five meristems were captured, and pooled per biological replicate before RNA extraction and T7-based RNA amplification. Total RNA was extracted from 20 d after pollination endosperm and embryo, 21-DAG roots, immature nongreen leaves (P2-P6), 2- to 4-mm ear and tassel primordia, and fully expanded adult leaves (L12) using TRIzol (Invitrogen). For all replicates, 1 μg of RNA was treated with DNase and used in a SuperscriptIII-based cDNA synthesis (Invitrogen). The qRT-PCR analyses were performed with SYBR-green methodology and gene-specific primers (see Supplemental Table 1 online) as described by Zhang et al. (2007). Three biological replicates for each genotype or tissue type were used during every experiment. Data are presented using the 2−ΔΔct method (Livak and Schmittgen, 2001) with threshold values normalized to levels of β-6-tubulin (Figure 3) or 18srRNA (Figure 6).
Mutant and wild-type 21-DAG or 28- to 35-DAG seedlings were processed for in situ hybridizations as described (Jackson, 1991) and with modifications (Long et al., 1996). A minimum of five replicates for each probe was used to get a consensus pattern of expression. Primers for probe construction are listed in Supplemental Table 1 online.
Biochemical Analyses
Thiamine compounds were extracted in 15% trichloroacetic acid from 14-DAG wild-type and thi2-blk1 seedlings and HPLC analyzed according to published protocols (Rapala-Kozik et al., 2008). Truncated Zm thi2 and Zm thi2-blk1 clone missing the 47 N-terminal residues of the encoded proteins were prepared by the Cornell Protein Facility using standard methods. Protein expression and purification and HPLC analysis of bound metabolites were completed as described (Chatterjee et al., 2006), with some modifications. THI2 proteins were first purified using a Ni-NTA column (Qiagen) (equilibrated with 50 mM Tris, pH 8.0, 300 mM NaCl, and 10 mM imidazole) and then by gel filtration (Superdex 200 column [26/60, GE Healthsciences], equilibrated with 50 mM Tris buffer, pH 8.0, and 100 mM NaCl).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Z. mays thi1 (NM_001112226), Z. mays thi2 (NM_001112227), Z. mays 18s rRNA (AF168884.1), Z. mays β-6-tubulin (L10633), kn1 (NM_001111966), abph1 (NM_001111384), Z. mays wus1 (NM_001112490), IDP351 (AY111998), BE_NO5 (CG797330); Arabidopsis THI1 (NP_200288), Oryza sativa THI1 (AAZ93636), Glycine max THI1 (BAA88228), Populus trichocarpa THI1 (XP_002305603), Pseudotsuga menziesii THI1 (AAV92531), Physcomitrella patens THI1 (XP_001755171), Saccharomyces cerevisiae THI1 (NP_011660), and Penicillium marneffei THI1 (XP_002146762).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Vegetative Axillary Meristems from Presumptive Ear Nodes Were Laser Microdissected for Tissue-Specific RNA Extraction.
Supplemental Figure 2. Thiamine Treatment Rescues Defects in SAM Height at 21 DAG.
Supplemental Table 1. Primers Used in This Study.
Acknowledgments
We thank C. Daugherty for assistance with electron microscopy, M. Srivastava at the Boyce Thompson Institute Plant Cell Imaging Facility for assistance with confocal microscopy, J. Jantz for help with plant propagation, Todd Pontius and Jessica Levy for assistance with fine-mapping thi2-tls31, and the Maize Inflorescence Project for generating the thi2-tls31 and thi2-tls32 alleles. Funding from the USDA National Research Initiative and the National Science Foundation is also acknowledged. We thank D. Jackson at the Cold Spring Harbor Laboratory for creating and sharing the Zm PIN1a-YFP transgenic line.
References
- Barton M.K. (2010). Twenty years on: The inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 341: 95–113 [DOI] [PubMed] [Google Scholar]
- Bates C. (2007). Thiamine. Handbook of Vitamins, Zempleni J., Rucker R., McCormick D., Suttie J., (Boca Raton, FL: CRC Press; ), pp. 252–288 [Google Scholar]
- Bayer E.M., Smith R.S., Mandel T., Nakayama N., Sauer M., Prusinkiewicz P., Kuhlemeier C. (2009). Integration of transport-based models for phyllotaxis and midvein formation. Genes Dev. 23: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett J.B. (1978). B-A translocations in maize: I. Use in locating genes to chromosome arms. J. Hered. 69: 27–36 [Google Scholar]
- Belanger F.C., Leustek T., Chu B., Kriz A.L. (1995). Evidence for the thiamine biosynthetic pathway in higher-plant plastids and its developmental regulation. Plant Mol. Biol. 29: 809–821 [DOI] [PubMed] [Google Scholar]
- Bennett S.R.M., Alvarez J., Bossinger G., Smyth D. (1995). Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 8: 505–520 [Google Scholar]
- Bolduc N., Hake S. (2009). The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21: 1647–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros L.G., Brandes J.L., Lee W.N., Cascante M., Puigjaner J., Revesz E., Bray T.M., Schirmer W.J., Melvin W.S. (1998). Thiamine supplementation to cancer patients: a double edged sword. Anticancer Res. 18(1B): 595–602 [PubMed] [Google Scholar]
- Bortiri E., Hake S. (2007). Flowering and determinacy in maize. J. Exp. Bot. 58: 909–916 [DOI] [PubMed] [Google Scholar]
- Brooks L., III, et al. (2009). Microdissection of shoot meristem functional domains. PLoS Genet. 5: e1000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M., Timmermans M., Kidner C., Martienssen R. (2001). Development of leaf shape. Curr. Opin. Plant Biol. 4: 38–43 [DOI] [PubMed] [Google Scholar]
- Chabregas S.M., Luche D.D., Van Sluys M.A., Menck C.F., Silva-Filho M.C. (2003). Differential usage of two in-frame translational start codons regulates subcellular localization of Arabidopsis thaliana THI1. J. Cell Sci. 116: 285–291 [DOI] [PubMed] [Google Scholar]
- Chatterjee A., Jurgenson C.T., Schroeder F.C., Ealick S.E., Begley T.P. (2006). Thiamin biosynthesis in eukaryotes: Characterization of the enzyme-bound product of thiazole synthase from Saccharomyces cerevisiae and its implications in thiazole biosynthesis. J. Am. Chem. Soc. 128: 7158–7159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comín-Anduix B., Boren J., Martinez S., Moro C., Centelles J.J., Trebukhina R., Petushok N., Lee W.N., Boros L.G., Cascante M. (2001). The effect of thiamine supplementation on tumour proliferation. A metabolic control analysis study. Eur. J. Biochem. 268: 4177–4182 [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Emrich S.J., Barbazuk W.B., Li L., Schnable P.S. (2007). Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res. 17: 69–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feenstra W.J. (1964). Isolation of nutritional mutants in Arabidopsis thaliana. Genetica 35: 259–269 [DOI] [PubMed] [Google Scholar]
- Fleming A. (2006). Metabolic aspects of organogenesis in the shoot apical meristem. J. Exp. Bot. 57: 1863–1870 [DOI] [PubMed] [Google Scholar]
- Gallavotti A., Yang Y., Schmidt R.J., Jackson D. (2008). The relationship between auxin transport and maize branching. Plant Physiol. 147: 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulini A., Wang J., Jackson D. (2004). Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 430: 1031–1034 [DOI] [PubMed] [Google Scholar]
- Godoi P.H., Galhardo R.S., Luche D.D., Van Sluys M.A., Menck C.F., Oliva G. (2006). Structure of the thiazole biosynthetic enzyme THI1 from Arabidopsis thaliana. J. Biol. Chem. 281: 30957–30966 [DOI] [PubMed] [Google Scholar]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Hudson A. (2000). Development of symmetry in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51: 349–370 [DOI] [PubMed] [Google Scholar]
- Jackson D. (1991). In situ hybridization in plants. Molecular Plant Pathology: A Practical Approach, Bowles D.J., Gurr S.J., McPherson M., (Oxford, UK: Oxford University Press; ), pp. 163–174 [Google Scholar]
- Jackson D., Hake S. (1999). Control of phyllotaxy in maize by the abphyl1 gene. Development 126: 315–323 [DOI] [PubMed] [Google Scholar]
- Jackson D., Veit B., Hake S. (1994). Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120: 405–413 [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., Phillips A., Hedden P., Tsiantis M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Jurgenson C.T., Chatterjee A., Begley T.P., Ealick S.E. (2006). Structural insights into the function of the thiamin biosynthetic enzyme Thi4 from Saccharomyces cerevisiae. Biochemistry 45: 11061–11070 [DOI] [PubMed] [Google Scholar]
- Kiesselbach T. (1949). The structure and reproduction of corn. Research Bulletin 161: University of Nebraska College of Agriculture [Google Scholar]
- Koornneef M., Hanhart C. (1981). A new thiamine locus in Arabidopsis. Arabidopsis Inf. Serv. 18: 52–58 [Google Scholar]
- Langridge J. (1955). Biochemical mutations in the crucifer Arabidopsis thaliana (L.) Heynh. Nature 176: 260–261 [DOI] [PubMed] [Google Scholar]
- Lee B.H., Johnston R., Yang Y., Gallavotti A., Kojima M., Travençolo B.A., Costa Lda.F., Sakakibara H., Jackson D. (2009). Studies of aberrant phyllotaxy1 mutants of maize indicate complex interactions between auxin and cytokinin signaling in the shoot apical meristem. Plant Physiol. 150: 205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A., To J.P., Busch W., Stehling S., Kehle A., Demar M., Kieber J.J., Lohmann J.U. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Li S.L., Rédei G.P. (1969). Thiamine mutants of the crucifer, Arabidopsis. Biochem. Genet. 3: 163–170 [DOI] [PubMed] [Google Scholar]
- Liu S., Monks N.R., Hanes J.W., Begley T.P., Yu H., Moscow J.A. (2010). Sensitivity of breast cancer cell lines to recombinant thiaminase I. Cancer Chemother. Pharmacol. 66: 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Long J.A., Moan E.I., Medford J.I., Barton M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- McSteen P., Hake S. (2001). barren inflorescence2 regulates axillary meristem development in the maize inflorescence. Development 128: 2881–2891 [DOI] [PubMed] [Google Scholar]
- McSteen P., Malcomber S., Skirpan A., Lunde C., Wu X., Kellogg E., Hake S. (2007). barren inflorescence2 encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol. 144: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardmann J., Werr W. (2006). The shoot stem cell niche in angiosperms: expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono- and dicot evolution. Mol. Biol. Evol. 23: 2492–2504 [DOI] [PubMed] [Google Scholar]
- Ohtsu K., et al. (2007). Global gene expression analysis of the shoot apical meristem of maize (Zea mays L.). Plant J. 52: 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Ueda J., Komaki M.K., Bell C.J., Shimura Y. (1991). Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3: 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papini-Terzi F.S., Galhardo R.S., Farias L.P., Menck C.F., Van Sluys M.A. (2003). Point mutation is responsible for Arabidopsis tz-201 mutant phenotype affecting thiamin biosynthesis. Plant Cell Physiol. 44: 856–860 [DOI] [PubMed] [Google Scholar]
- Poethig S. (1984). Cellular parameters of leaf morphogenesis in maize and tobacco. Contemporary Problems in Plant Anatomy, White R.A., Dickison W.C., (Orlando, FL: Academic Press; ), pp. 235–259 [Google Scholar]
- Rapala-Kozik M., Kowalska E., Ostrowska K. (2008). Modulation of thiamine metabolism in Zea mays seedlings under conditions of abiotic stress. J. Exp. Bot. 59: 4133–4143 [DOI] [PubMed] [Google Scholar]
- Redei G.P. (1965). Genetic blocks in the thiamine synthesis of the angiosperm Arabidopsis. Am. J. Bot. 52: 834–841 [Google Scholar]
- Reinhardt D., Mandel T., Kuhlemeier C. (2000). Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Ruzin S. (1999). Plant Microtechniques and Microscopy. (New York: Oxford University Press; ). [Google Scholar]
- Sakamoto T., Kamiya N., Ueguchi-Tanaka M., Iwahori S., Matsuoka M. (2001). KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 15: 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Nagasawa N., Nagasawa N., Malcomber S., Sakai H., Jackson D. (2006). A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441: 227–230 [DOI] [PubMed] [Google Scholar]
- Scanlon M.J. (2003). The polar auxin transport inhibitor N-1-naphthylphthalamic acid disrupts leaf initiation, KNOX protein regulation, and formation of leaf margins in maize. Plant Physiol. 133: 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon M.J., Stinard P.S., James M.G., Myers A.M., Robertson D.S. (1994). Genetic analysis of 63 mutations affecting maize kernel development isolated from Mutator stocks. Genetics 136: 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Shimamoto K., Nelson O.E. (1981). Movement of C-compounds from maternal tissue into maize seeds grown in vitro. Plant Physiol. 67: 429–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E., Reiser L., Hake S. (2000). Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127: 3161–3172 [DOI] [PubMed] [Google Scholar]
- Wightman F., Brown R. (1953). The effects of thiamin and nicotinic acid on meristematic activity in pea roots. J. Exp. Bot. 4: 184–196 [Google Scholar]
- Wu X., Dabi T., Weigel D. (2005). Requirement of homeobox gene STIMPY/WOX9 for Arabidopsis meristem growth and maintenance. Curr. Biol. 15: 436–440 [DOI] [PubMed] [Google Scholar]
- Wu X., McSteen P. (2007). The role of auxin transport during inflorescence development in maize, Zea mays (Poaceae). Am. J. Bot. 11: 1745–1755 [DOI] [PubMed] [Google Scholar]
- Yanai O., Shani E., Dolezal K., Tarkowski P., Sablowski R., Sandberg G., Samach A., Ori N. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15: 1566–1571 [DOI] [PubMed] [Google Scholar]
- Zhang X., Madi S., Borsuk L., Nettleton D., Elshire R.J., Buckner B., Janick-Buckner D., Beck J., Timmermans M., Schnable P.S., Scanlon M.J. (2007). Laser microdissection of narrow sheath mutant maize uncovers novel gene expression in the shoot apical meristem. PLoS Genet. 3: e101. [DOI] [PMC free article] [PubMed] [Google Scholar]