Leaf shape diversity relies on transient morphogenetic activity in leaf margins. This study identifies cytokinin as an important regulator of the extended morphogenetic activity that characterizes the leaf margin of compound tomato leaves.
Abstract
Leaf shape diversity relies on transient morphogenetic activity in leaf margins. However, how this morphogenetic capacity is maintained is still poorly understood. Here, we uncover a role for the hormone cytokinin (CK) in the regulation of morphogenetic activity of compound leaves in tomato (Solanum lycopersicum). Manipulation of CK levels led to alterations in leaf complexity and revealed a unique potential for prolonged growth and morphogenesis in tomato leaves. We further demonstrate that the effect of CK on leaf complexity depends on proper localization of auxin signaling. Genetic analysis showed that reduction of CK levels suppresses the effect of Knotted1 like homeobox (KNOXI) proteins on leaf shape and that CK can substitute for KNOXI activity at the leaf margin, suggesting that CK mediates the activity of KNOXI proteins in the regulation of leaf shape. These results imply that CK regulates flexible leaf patterning by dynamic interaction with additional hormones and transcription factors.
INTRODUCTION
Compound leaf development is a flexible process that responds sensitively to changes in genetic, hormonal, and environmental factors, making leaves an attractive system to study mechanisms of morphogenesis (Efroni et al., 2010). Leaf development can be roughly divided into three successive and overlapping stages. In the first stage, termed initiation (I), the leaf emerges from the flanks of the shoot apical meristem (SAM). During the second stage, primary morphogenesis (PM), leaves expand laterally, the basic leaf form is determined, and lateral structures such as leaflets, lobes, and serrations are elaborated from the leaf margin. In the final stage, secondary morphogenesis, the leaf grows substantially and differentiates by producing cell types typical of the mature leaf (Hagemann and Gleissberg, 1996; Poethig, 1997; Dengler and Tsukaya, 2001; Kaplan, 2001; Holtan and Hake, 2003; Efroni et al., 2008). Final leaf shape and size are affected by events that take place during all these stages. The developmental state of leaf primordia is followed by plastochrons, the interval between two successive leaves. Thus, the youngest leaf primordium is termed P1; it becomes P2 when the next primordium arises, and so on. Leaf growth is determinate, in that it lasts for a limited duration, but developing leaves retain transient organogenesis activity during PM in specific regions at their margins, termed marginal blastozones (Hagemann and Gleissberg, 1996). This results in the coexistence of tissues at different developmental stages and maturation states within the developing leaf.
Two basic leaf forms can be described with respect to the blade structure: simple leaves, in which there is a single undivided blade; and compound leaves, composed of multiple leaflets, each resembling a simple leaf. Classic and recent research has led to the hypothesis that compound-leaf development requires prolonged activity of the marginal blastozone during PM. Partially overlapping genetic mechanisms have been identified for leaf development from the SAM flanks and leaflet development from the leaf marginal blastozone (Hagemann and Gleissberg, 1996; Floyd and Bowman, 2010; Koenig and Sinha, 2010).
For example, Knotted1 like homeobox (KNOX1) genes are essential for the maintenance of indeterminate growth and morphogenetic activity of the SAM and also play essential roles in maintaining the transient indeterminacy and morphogenetic activity of the marginal blastozone of many species with compound leaves (Hareven et al., 1996; Janssen et al., 1998; Bharathan et al., 2002; Hake et al., 2004; Hay and Tsiantis, 2006; Kimura et al., 2008; Barth et al., 2009; Ramirez et al., 2009; Shani et al., 2009; Hay and Tsiantis, 2010). Conversely, TCP (teosinte branched-cycloidea-proliferating cell factor) domain proteins, such as LANCEOLATE (LA) in tomato (Solanum lycopersicum), negatively control SAM and marginal blastozone activity by promoting differentiation, and their downregulation during early leaf development is essential for compound-leaf development (Nath et al., 2003; Palatnik et al., 2003; Ori et al., 2007; Efroni et al., 2008). Thus, in the dominant La mutant, precocious LA activation leads to earlier differentiation and to a simplified leaf shape (Ori et al., 2007). In other species with compound leaves, such as pea (Pisum sativum) and Medicago truncatula, other factors play similar antagonistic roles in defining the window of morphogenetic activity (Hofer et al., 1997; Wang et al., 2008; Chen et al., 2010).
In species with either simple or compound leaves, CUC (cup-shaped cotyledon) transcription factors mark and define boundaries and maintain morphogenetic activities at both the SAM and the leaf’s marginal blastozone (Nikovics et al., 2006; Blein et al., 2008, 2010; Berger et al., 2009). Auxin maxima were shown to define both leaf and leaflet initiation sites, likely by a yet undefined interaction with CUC proteins (Reinhardt et al., 2000, 2003; Benková et al., 2003; Heisler et al., 2005; Barkoulas et al., 2008; Koenig et al., 2009; Blein et al., 2010; Canales et al., 2010).
The plant hormone cytokinin (CK) is involved in coordinating many developmental processes in the plant (Werner and Schmülling, 2009), including positive regulation of SAM size and activity (Werner et al., 2001, 2003; Giulini et al., 2004; Leibfried et al., 2005; Kurakawa et al., 2007; Sablowski, 2007; Galinha et al., 2009; Gordon et al., 2009; Veit, 2009; Perilli et al., 2010; Skylar et al., 2010; Zhao et al., 2010). CK biosynthesis was shown to be positively regulated by KNOXI proteins (Jasinski et al., 2005; Yanai et al., 2005; Sakamoto et al., 2006; Shani et al., 2006), and a mutual positive regulation was shown to exist between CK and WUSCHEL, an important regulator of SAM maintenance (Leibfried et al., 2005; Lindsay et al., 2006; Gordon et al., 2009). Conversely, SAM size was shown to be negatively regulated by type A response regulators (ARRs), which are induced by CK and negatively regulate CK signaling through a feedback loop (Giulini et al., 2004; Leibfried et al., 2005).
Here, we tested the possibility that CK is also involved in maintaining the activity of the leaf’s marginal blastozone. We demonstrate a role for CK in the regulation of compound-leaf development in tomato. Manipulation of CK levels in developing leaves led to alteration of the level of leaf complexity in tomato. We further show that CK acts downstream of KNOXI proteins in maintaining prolonged morphogenetic activity at the leaf margin but that the activities of KNOXI and CK only partially overlap.
RESULTS
CK Regulates Compound-Leaf Development
To test whether CK is involved in compound-leaf development, we manipulated CK levels in simple Arabidopsis thaliana leaves or compound tomato leaves by expressing either the CK biosynthesis gene Arabidopsis (At) ISOPENTENYL TRANSFERASE7 (At IPT7) or the CK degradation gene CYTOKININ OXIDASE3 (At CKX3) (Miyawaki et al., 2006; Werner and Schmülling, 2009). To distinguish between the roles of CK in the SAM and in developing leaves, we expressed these genes using the lateral organ–specific promoters ANTpro and FILpro in Arabidopsis and tomato, respectively (Figure 1A). The Arabidopsis ANTpro drives expression in the regions at the SAM flanks from which organs will initiate, and during early lateral organ development it is expressed throughout initiating organs. Later its expression becomes restricted to internal and basal regions (Elliott et al., 1996; Long and Barton, 1998; Schoof et al., 2000). In tomato, the FILpro from Arabidopsis drives expression throughout the primordia, including initiating leaflets and the intercalary region between them, until relatively late in leaf development (see Supplemental Figure 1 online; Lifschitz et al., 2006; Berger et al., 2009; Shani et al., 2009).
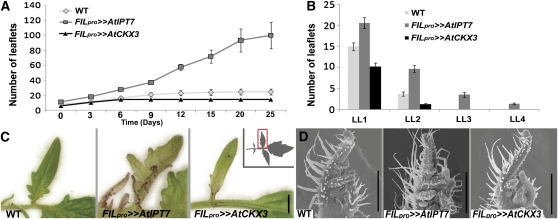
Figure 1.
CK Regulates Compound Leaf Development.
(A) A schematic illustration of the expression domains (shading) directed by the indicated promoters.
(B) Three-week-old Arabidopsis seedlings of the indicated genotypes.
(C) to (E) The sixth leaf (left) and a magnification of the second primary leaflet (right) of the indicated tomato genotypes.
(F) Successive leaves were removed from the plants and are shown in an acropetal sequence from left to right. L1 to L6, leaf 1 to leaf 6.
Xpro>>Y, plants expressing the Y gene under the control of the X promoter, using the LhG4 transactivation system. WT, wild type. Bars = 1 cm.
[See online article for color version of this figure.]
Leaves of ANTpro>>AtIPT7 Arabidopsis plants, expressing At IPT7 under the control of the ANTpro using the LhG4 transactivation system (Moore et al., 1998; see Methods), were similar to wild-type leaves (Figure 1B). ANTpro>>AtCKX3 Arabidopsis leaves were very small, round, and dark green, as described previously (Werner et al., 2003; Figure 1B). By contrast, At IPT7 expression in tomato leaves under the control of the FILpro led to the development of super-compound leaves with up to four orders of leaflets, compared with two orders in the wild-type leaf (Figures 1C, 1D, 1F, 2A, and 2B). In addition, leaflets were rounder than in the wild type (Figures 1C and 1D), and ectopic meristems and flowers were formed on the adaxial side of the leaf rachis (see Supplemental Figure 2 online). Conversely, expression of At CKX3 in tomato leaves led to the production of simplified leaves that made only primary leaflets (Figures 1E, 1F, 2A, and 2B). Furthermore, leaf margins of FILpro>>AtCKX3 plants were smooth, compared with the lobed margins of the wild-type leaf (Figure 2C). FILpro>>AtIPT7 leaves showed an opposite effect of enhanced morphogenetic activity at their margins, as manifested by an appearance of initiating primordia and the development of additional marginal structures (Figure 2C). In agreement, FILpro>>AtCKX3 leaves ceased leaflet initiation at a relatively early developmental stage relative to the wild type, while FILpro>>AtIPT7 continued to initiate leaflets much longer than the wild type (Figure 2A). Thus, tomato leaves have a potential for extended morphogenetic activity that lasts for weeks, and CK positively regulates this potential.
Figure 2.
CK Regulates a Prolonged Morphogenetic Activity of Tomato Leaf Margins.
(A) Total number of leaflets on the sixth leaf at successive time points during leaf growth. Time 0 refers to the day at which the leaf has emerged from the apex and expanded. The same leaves were analyzed every 3 to 5 d. Shown are averages ± se (n = 10).
(B) Number of leaflets in each order of leaflet reiteration, at time point 20 d in (A). LL1, number of primary and intercalary leaflets; LL2-LL4, number of secondary, tertiary, and quaternary leaflets, respectively, of the second primary leaflet. Shown are averages ± se (n = 10).
(C) A leaflet from a leaf at the P8 stage. The inset shows schematic illustration of the context (rectangle) of the images.
(D) Scanning electron micrographs of young leaf primordia at the P4 stage.
WT, wild type. Bars = 1 mm in (C) and 500 μm (D).
[See online article for color version of this figure.]
Examination of young primordia showed only mild effects during early leaf development of FILpro>>AtIPT7 and FILpro>>AtCKX3 leaves: Leaflet initiation was slightly accelerated in FILpro>>AtIPT7 primordia and slightly delayed in FILpro>>AtCKX3 primordia (Figure 2D; see Supplemental Figure 3 online). Constitutive expression of At CKX3 in tomato plants using the 35S promoter led to a strong phenotype of small plants with inhibited growth and small, simplified leaves (see Supplemental Figure 4 online), similar to Arabidopsis 35Spro:CKX3 (Werner et al., 2003).
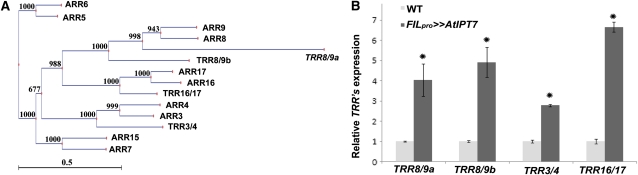
In Arabidopsis, ARRs have been shown to be rapidly induced by CK application (Imamura et al., 1998; D’Agostino et al., 2000; To and Kieber, 2008). A search in the tomato genomic and EST databases (http://solgenomics.net/) has identified seven ARR homologs. For further analysis, we used four of these genes that showed a relatively high degree of similarity. Interestingly, two pairs of closely related ARRs had a single close tomato homolog, and an additional pair had two close tomato homologs. The identified tomato genes were termed Tomato Response Regulator 3/4 (TRR3/4), TRR8/9a, TRR8/9b, and TRR16/17, according to the respective most similar Arabidopsis genes (Figure 3A; see Supplemental Figure 5 online).
Figure 3.
At IPT7 Overexpression Leads to Increased Expression of CK Response Markers.
(A) Unrooted tree showing the phylogenetic relationship among type-A response regulator proteins from tomato (TRR) and Arabidopsis (ARR). The scale bar indicates the distances in substitutions per amino acid.
(B) Quantification of TRR mRNA level in third leaves at the P5 stage of 17-d-old plants by quantitative RT-PCR. Shown are averages ± se (n = 3 biological repeats). *Significantly different relative to the respective wild type (WT) at P ≤ 0.05.
[See online article for color version of this figure.]
Quantitative RT-PCR analysis showed that mRNA levels of four of the identified TRRs were upregulated 2.5- to 6.5-fold in leaf primordia at the P5 stage of FILpro>>AtIPT7 plants (Figure 3B), suggesting that CK response was affected as expected in these plants. CK is a positive regulator of cell division and shoot regeneration (Miller et al., 1955). In Arabidopsis, the D-type cyclin (CYCD3) genes were shown to be induced by CK and to mediate its activity. CYCD3s were also shown to affect stage transition during lateral organ development in Arabidopsis (Riou-Khamlichi et al., 1999; Dewitte et al., 2007). We thus tested the effect of manipulating CK levels in developing leaves on the expression of tomato (Sl) CYCD3.3 (Kvarnheden et al., 2000). Sl CYCD3.3 was upregulated 2-fold in P5 and P7 primordia of FILpro>>AtIPT7 plants relative to the wild type but was unaffected in FILpro>>AtCKX3 (see Supplemental Figure 6 online).
Cumulatively, these observations suggest that CK regulates the level of leaf complexity by enabling extended morphogenetic activity at the leaf margin and regulating its duration but cannot cause the conversion of the Arabidopsis simple-leaf structure into a compound one.
Compound-Leaf Development Is Correlated with Extended CK Response at the Leaf Margin
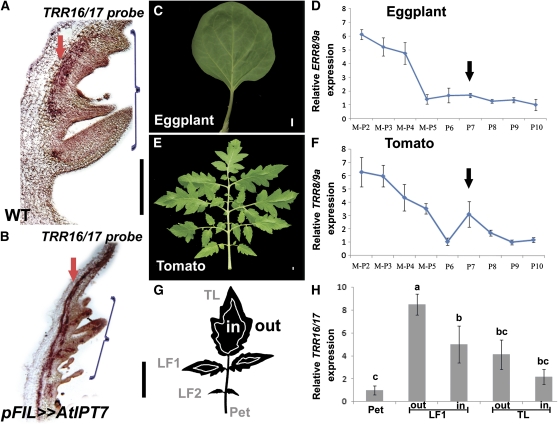
To further understand the role of CK in the development of the compound tomato leaf, we examined the spatial distribution of the CK response marker TRR16/17 in developing wild-type and FILpro>>AtIPT7 tomato leaves using in situ hybridization. In wild-type plants, TRR16/17 expression was observed at the margins of the leaf procambium (Figure 4A), as expected from the recently described role of CK in Arabidopsis root procambium development (Mähönen et al., 2006). Expression was also observed in leaf margins and leaflets, consistent with the involvement of CK in coordinating the morphogenetic activity of the tomato leaf margin (Figure 4A). In FILpro>>AtIPT7 leaves, TRR16/17 distribution was similar to that in wild-type leaves (Figure 4B), although expression was elevated, as revealed by a shorter exposure required to detect a signal.
Figure 4.
Enhanced CK Response in Leaf Margins of Compound Leaves.
(A) and (B) TRR16/17 expression in P4 leaf primordia of the indicated genotypes, assayed by mRNA in situ hybridization. Arrows point to procambial expression; brackets indicate the expression at the leaf margin. (A) was exposed for 36 h and (B) for 12 h. WT, wild type.
(C) and (E) Mature fifth leaf of eggplant (C) and tomato (E).
(D) and (F) Relative ERR8/9A (D) and TRR8/9A (F) mRNA expression at successive stages of leaf maturation of wild-type eggplant or tomato, respectively, as measured by quantitative RT-PCR. Averages ± se (n = 3 to 6 biological repeats) are indicated. M-P2, SAM and the two youngest leaf primordia (P1 and P2). Arrow points to the second peak of elevated expression at the P7 stage.
(G) A schematic illustration of the dissected domains of P8, used for the TRR16/17 quantification shown in (H).
(H) Relative TRR16/17 mRNA levels in different domains of a fifth wild-type tomato leaf at the P8 stage. The domains are illustrated in (G). mRNA levels were assayed by quantitative RT-PCR. Shown are averages ± se (n = 3). Bars with different letters are significantly different at P < 0.05. TL, terminal leaflet; LF1 and LF2, lateral leaflet 1 and 2, respectively; Pet, leaf petiole.
Bars = 200 μm in (A) and (B) and 1 cm in (C) and (E).
[See online article for color version of this figure.]
To examine whether CK response is prolonged during compound-leaf development, we compared the expression dynamics of representative CK response genes at successive developmental stages between tomato and eggplant (Solanum melongena), a Solanum species with simple leaves (Figure 4C). TRR8/9a and TRR16/17, as well as ERR8/9a (EGGPLANT RR8/9a, the closest homolog of TRR8/9a in eggplant), showed relatively high mRNA expression in SAM and young leaf primordia of tomato and eggplant, respectively, and a gradual reduction in expression was observed as leaves matured. However, ERR8/9a downregulation in eggplant leaves was earlier and steeper than in tomato (Figures 2D and 2F; see Supplemental Figure 7 online). Moreover, while eggplant primordia retained low ERR8/9a expression at later stages (Figure 4D), in tomato primordia, TRR8/9a and TRR16/17 expression was re-elevated at stage P7, around the developmental stage at which secondary leaflets develop (Figure 4F; see Supplemental Figure 7 online). This is consistent with the elimination of secondary leaflets and marginal lobing in FILpro>>AtCKX3 leaves and with the expression of TRR16/17 in leaf margins (Figures 1E, 4A, and 4B).
Different parts of the young tomato leaf primordium are at different developmental stages, and vascular CK response could account for much of the expression at later stages. Therefore, to test the relevance of the marginal morphogenetic activity to the extended TRR expression, we quantified TRR16/17 expression at different domains of dissected tomato fifth leaf primordia at the P8 stage (Figure 4G). TRR16/17 expression was relatively high in basal and marginal domains of the leaf (Figure 4H), regions that are considered younger and still meristematic at this developmental stage (Sun, 1957). Expression was relatively low in the petiole, implying that it does not reflect expression in mature vasculature (Figure 4H).
Together, these results imply that an extended CK activity in leaf margins is correlated with leaf complexity, confirming its role in controlling leaf-marginal morphogenetic activity.
The Control of Leaf Complexity by CK Requires Proper Localization of Auxin Response
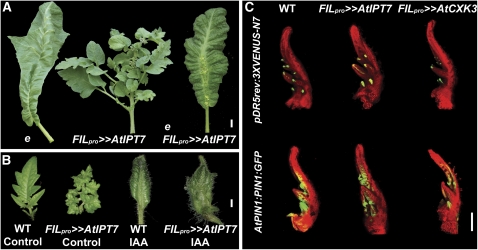
The plant hormone auxin has been recently implied in the regulation of leaflet initiation and growth (Barkoulas et al., 2008; Berger et al., 2009; Koenig et al., 2009). entire (e) tomato mutants have a simpler leaf form relative to the wild type (Figure 5A). The E gene was recently shown to encode Sl IAA9, a protein from the AUX/IAA (Auxin/indole-3-acetic acid) family of auxin response repressors, suggesting that auxin response is upregulated between leaflets in e mutant leaves (Wang et al., 2005; Berger et al., 2009). To examine the interaction between CK and auxin in the context of compound-leaf development, we expressed FIL pro >>AtIPT7 in the background of the e mutant. The e phenotype was epistatic to that of At IPT7 overexpression with respect to the level of leaf complexity, preventing the formation of super-compound leaves, although e FILpro>>AtIPT7 leaves still produced ectopic meristems from their rachises (Figure 5A). In agreement, microapplication of auxin (IAA; 1 mM) directly to both wild-type and FILpro>>AtIPT7 P2 leaf primordia resulted in simple mature leaves and a suppression of the super-compound phenotype caused by At IPT7 (Figure 5B). Auxin (IAA; 5 μM or 1 mM) suppressed the FILpro>>AtIPT7 phenotype also when applied to older primordia (see Supplemental Figure 8 online).
Figure 5.
CK Requires Proper Localization of Auxin Response to Regulate Leaf Shape.
(A) Tomato sixth leaves of the indicated genotypes. The e mutant phenotype is epistatic to that of pFIL>>AtIPT7.
(B) Tomato leaves of the indicated genotypes, with or without microapplication of 1 mM IAA. Microapplication of IAA led to the development of simple leaves and suppressed the super-compound leaf phenotype of pFIL>>AtIPT7. WT, wild type.
(C) Distribution of the pDR5rev:3XVENUS-N7 and AtPIN1:PIN1-GFP markers. Shown are confocal microscope images of P4-staged leaves. The background leaf represents chlorophyll autofluorescence. Genotypes are as described in Figure 1.
Bars = 1 cm in (A), 1 mm in (B), and 200 μm in (C).
[See online article for color version of this figure.]
These findings imply that CK-regulated morphogenesis at the leaf margin depends on proper localization of the auxin signal. To further understand the interaction between auxin and CK, we examined the effect of manipulating CK levels on the distribution of auxin response and transport markers. A slight reduction in the distribution of the auxin response marker pDR5rev:3XVENUS-N7 and the auxin transporter AtPIN1:PIN1-GFP (Heisler et al., 2005; Bayer et al., 2009) was observed in FILpro>>AtCKX3 leaves (Figure 5C). However, these alterations were likely secondary to the developmental effects of the transgene, as the principal distribution of these markers remained similar to that in wild-type leaves. The distribution of these markers was not altered in FILpro>>AtIPT7 leaves (Figure 5C).
CK Acts Downstream of KNOXI Proteins in Compound-Leaf Development
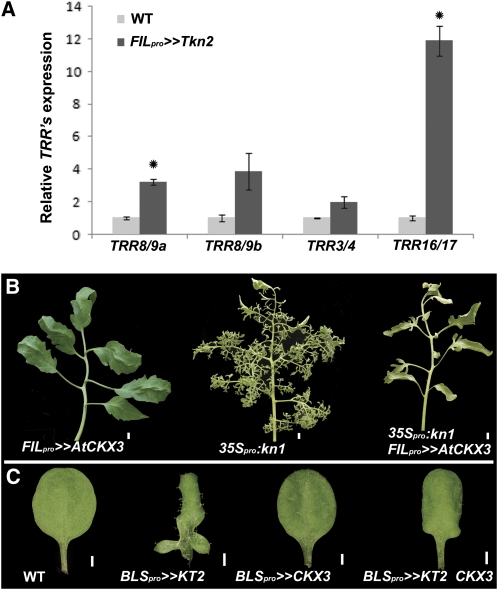
CK and KNOXI transcription factors both regulate compound-leaf development (Hareven et al., 1996; Janssen et al., 1998; Hay and Tsiantis, 2006; Shani et al., 2009; this study). In the SAM, KNOXI proteins were shown to act upstream of CK by positively regulating CK biosynthesis, and CK could partially compensate for loss of KNOXI function (Jasinski et al., 2005; Yanai et al., 2005; Sakamoto et al., 2006). This prompted us to ask whether CK mediates the activity of KNOXI proteins in the context of compound-leaf development. We thus tested the effect of KNOXI overexpression on the expression of the TRR genes. TRR expression was upregulated 2- to 12-fold in apices of tomato plants overexpressing the KNOXI gene Tkn2 specifically in lateral organs, relative to the wild type (Figure 6A).
Figure 6.
CK Acts Downstream of KNOXI in the Regulation of Leaf Complexity.
(A) Relative mRNA expression of the indicated TRR genes in 17-d-old tomato apices as measured by quantitative RT-PCR. Averages ± se (n = 3) are indicated for each genotype. *Significantly different relative to the respective wild type (WT) at P ≤ 0.05.
(B) Tomato fifth leaves. p35:kn1 plants express the maize KNOXI gene knotted1 by direct fusion to the 35S promoter, and all other transgenes are expressed using the transactivation system and are as in Figure 1.
(C) Arabidopsis fifth leaves of the indicated genotypes. At CKX3 coexpression suppresses the KT2 lobbed-leaf phenotype.
Bars = 1 cm in (B) and 1 mm in (C).
[See online article for color version of this figure.]
To test the biological relevance of this activation, we examined whether reducing CK levels will suppress the KNOXI overexpression phenotype. Leaf-specific expression of At CKX3 suppressed the super-compound leaf phenotype of 35Spro:Kn1 tomato plants, which ubiquitously overexpress the maize (Zea mays) KNOXI gene kn1 (Figure 6B). Similarly, coexpression of At CKX3 suppressed the phenotype of BLSpro>>Tkn2, expressing Tkn2 at later stages of leaf development (see Supplemental Figure 9 online). At CKX3 coexpression also suppressed the leaf lobing phenotype of Arabidopsis BLSpro>>KT2 plants, expressing the KNOXI gene KT2 in leaves (Figure 6C).
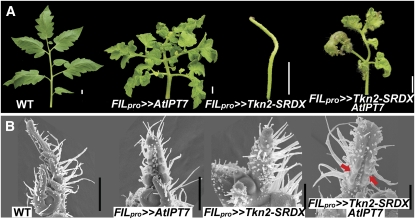
If CK acts downstream of KNOXI proteins in compound leaves, elevating CK levels is expected to compensate for downregulation of KNOXI activity. To test this prediction, we coexpressed At IPT7 and Tkn2-SRDX, a fusion of Tkn2 with a repression domain (Shani et al., 2009), in tomato leaves. At IPT7 overexpression was epistatic to the aberrant and simple leaf phenotype of Tkn2-SRDX, when coexpressed in leaves (Figure 7A). Examination of the mRNA expression levels of Tkn2, representing the combined expression of the endogenous Tkn2 gene and the Tkn2-SRDX transgene, verified that the Tkn2-SRDX transgene was not suppressed by the coexpression of At IPT7 (see Supplemental Figure 10 online). Examination of early stages of leaf development by scanning electron microscopy showed that At IPT7 coexpression specifically rescued the meristematic activity of the marginal blastozone, as manifested by the lack of trichome development and the initiation of leaflets (Figure 7B). In summary, these results imply that CK acts downstream of KNOXI genes in compound-leaf elaboration.
Figure 7.
Cytokinin Can Compensate for KNOXI Activity in Tomato Compound Leaves.
(A) Tomato fourth leaves of the indicated genotypes. Coexpression of At IPT7 suppresses the pFIL>>TKN2-SRDX simple-leaf phenotype.
(B) Scanning electron micrographs showing early leaf development. Arrows mark the sites of leaflets initiation in pFIL>>TKN2-SRDX AtIPT7 leaf margins.
WT, wild type. Bars = 1 cm in (A) and 500 μm in (B).
[See online article for color version of this figure.]
DISCUSSION
During leaf development, the leaf margin transiently retains morphogenetic potential that is responsible for the extremely flexible process of leaf morphogenesis. As a result, leaves show enormous size and shape diversity both within and among species. However, how this morphogenetic capacity is maintained is still poorly understood. Here, we show that the tomato leaf keeps initiating leaflets for weeks after its emergence and that CK regulates the extent of this prolonged morphogenetic activity. In agreement, CK response markers showed extended expression during the development of compound tomato leaves in comparison to simple-leaved eggplant. The activity of CK in promoting SAM activity was thus recruited for the transient indeterminate growth of developing leaves during their evolution from shoots.
We show that CK acts downstream to KNOXI proteins in the context of the leaf margin. The suppression of the simple leaf phenotype of FILpro>>Tkn2-SRDX by coexpression of At IPT7 is striking, as in most cases a simplified leaf phenotype is epistatic to a more compound leaf phenotype, in both tomato and other species (Hareven et al., 1996; Ori et al., 2007; Chen et al., 2010). Furthermore, both the e and the La mutant backgrounds, which have simplified leaves due to a defect in leaflet separation and precocious leaf maturation, respectively, were epistatic to the FILpro>>IPT7 phenotype. This further supports the conclusion that CK mediates KNOXI activity in compound-leaf development. It should be noted, however, that tomato leaves react differently to manipulations of CK levels and KNOXI activity in similar developmental contexts. For example, KNOXI genes were shown to dramatically extend both the I and PM stages of leaf development (Shani et al., 2009). By contrast, CK elevation throughout early leaf development using FILpro did not affect the transition to PM but only affected the morphogenetic activity during PM. Expression of KNOXI genes by BLSpro extended PM, while overexpressing At IPT7 by BLSpro caused uncontrolled growth between veins but did not affect marginal growth and did not delay the transition to secondary morphogenesis (see Supplemental Figure 11 online).
Therefore, while KNOXI proteins act through CK to promote the marginal morphogenetic activity, the context-specific response of the developing leaf to these factors only partially overlaps. The low sensitivity of the Arabidopsis leaves to At IPT7 expression may result from a combination of the relatively low sensitivity of the Arabidopsis leaf to KNOXI activity during the I stage (Hay et al., 2003; Shani et al., 2009) and the relatively short PM in these leaves; while KNOXI proteins can extend PM also in Arabidopsis, CK acts more locally within an extended PM. In agreement, while overexpression of At IPT7 at different stages of Arabidopsis leaf development did not affect leaf shape, coexpression of At IPT7 enhanced the BLSpro>>KT2 phenotype (see Supplemental Figure 12 online). We propose that KNOXI proteins act in two pathways: (1) activating factors that delay stage transition, partially by inhibiting gibberellins (Hay et al., 2002; Bolduc and Hake, 2009) and (2) activating CK to maintain the meristematic potential of the compound-leaf margins.
Proper localization of auxin maxima and response is shown here to be required for the regulation of leaf development by CK. The ratio between auxin and CK rather than their absolute amounts is known to affect many developmental processes (Skoog and Miller, 1957; Moubayidin et al., 2009). Recently, several mechanisms by which the plant assesses this ratio have been elucidated in embryos, roots, and shoots. These include repression of auxin response and transport by CK (Dello Ioio et al., 2008; Galinha et al., 2009; Lee et al., 2009; Ruzicka et al., 2009; Wolters and Jürgens, 2009; Moubayidin et al., 2010), as well as repression or activation of CK response by auxin (Müller and Sheen, 2008; Zhao et al., 2010). Here, the distribution of an auxin response marker and the auxin transporter PIN1 appeared to be essentially similar between the wild type and the genotypes with altered CK levels during early stages of leaf development, although the prolonged morphogenesis is likely accompanied by prolonged auxin signaling at the leaf margin. Thus, the reason that disruption of the auxin response gradients suppresses the effect of CK on leaf shape is still unclear.
We propose that CK is required for morphogenetic activity of the marginal blastozone but that differential auxin distribution is required in the context of this morphogenetic activity for leaflet initiation and separation. Alternatively, the suppression of the CK effect by uniformly high auxin levels or response could be a result of an earlier activity of auxin, leading to the elimination of the leaf domain that is responsive to CK. In conclusion, the genetic and environmental flexibility of compound leaves shape is regulated by the balance between the activities of CK, other hormones, and transcription factors.
METHODS
Plant Material
Tomato (Solanum lycopersicum cv M82, sp) and eggplant (Solanum melongena) plants were grown initially in a growth room at 16-h-day and 8-h-night conditions at 24 to 25°C. Four-week-old seedlings were transferred to greenhouse conditions at natural daylength (~14 and 10 h light in the summer and winter, respectively) 2°C at night and 25°C during the day. Arabidopsis thaliana plants were grown under long-day fluorescent light (16 h light/8 h dark, 21°C). The described Arabidopsis plants are all in the Landsberg erecta background. Most of the described transgenic plants were produced using the LhG4 transactivation system (Moore et al., 1998). In this system, a driver line (PRO:LhG4) that expresses the synthetic transcription factor LhG4 under a specific promoter is crossed to a responder line (OP:GENE), expressing a gene of interest downstream to several copies of the Escherichia coli Operator, recognized by LhG4. The resulting plants that harbor both constructs (PRO>>GENE) express the gene of interest in the expression domain of the specific promoter. The following Arabidopsis and tomato transgenic plants and mutants were described before: Arabidopsis BLSpro:LhG4 and ANTpro:LhG4 (Efroni et al., 2008), OP:KNAT2 (KT2), OP:Tkn2, OP:Tkn2-SRDX, tomato BLSpro:LhG4, and FILpro:LhG4 (Shani et al., 2009), 35Spro:LhG4 (Alvarez et al., 2006), 35Spro:Kn1 (Hareven et al., 1996), AtPIN1:PIN1-GFP (Moneymaker background) (Bayer et al., 2009), e (e2-n0741) (Berger et al., 2009), OP:AtIPT7, OP:AtCKX3, and pDR5rev:3XVENUS-N7 lines were generated during this research as described below.
Cloning and Plant Transformation
The Arabidopsis (Columbia) At IPT7 gene was amplified from genomic DNA, with annealing temperature of 58°C and 30 cycles, cloned downstream to an array of E. coli Operator (OP) sequences and transferred into the binary vector pART27 (Gleave, 1992). The At CKX3 construct, kindly provided by Thomas Schmülling and Tomáš Werner (Freie Universität Berlin), was cloned downstream to an OP array and subcloned into the binary pART27 vector. The pDR5rev:3XVENUS-N7 construct, kindly provided by Elliot Meyerowitz and Marcus Heisler (California Institute of Technology), was subcloned into pART27 vector. Kanamycin-resistant transformants for both species were selected as described (Shani et al., 2009). Primers used for At IPT7 cloning are described in Supplemental Table 1 online.
Plant Genetics
Phenotypic analyses were performed with selected operator:GENE (OP:GENE) responder lines that were crossed to promoter:LhG4 driver lines. Ten to twelve independent responder lines were crossed to the FILpro (tomato) or ANTpro (Arabidopsis) driver lines, and a representative line was selected for further analysis. FILpro>>AtIPT7 plants were sterile. Therefore, to generate FILpro>>AtIPT7 e plants, homozygous OP:AtIPT7 and FILpro:LhG4 plants were each crossed separately to plants homozygous for the e mutation, which causes leaves to have a simpler form. F2 individuals displaying the e phenotype and carrying the respective transgene (selected by their resistance to kanamycin) were crossed to each other (e OP:IPT7 × e FILpro:LhG4). Three-quarters of the F1 plants showed the e phenotype, while the remaining one-quarter possessed both the FILpro:LhG4 and op:AtIPT7 transgenes. A similar strategy was used to introduce FILpro>>AtIPT7 and FILpro>AtCKX into the pDR5rev:3XVENUS-N7, AtPIN1:PIN1-GFP, and op:Tkn2-SRDX backgrounds.
Isolation of TRR Genes
The BLASTN and TBLASTX search programs and the SOL genomics network (SGN; www.sgn.cornell.edu) were used to identify sequences of ARR homologs from tomato. Eggplant ERR8/9a was amplified from eggplant cDNA using tomato TRR8/9a primers.
Phylogenetic Analysis
Multiple sequence alignment (see Supplemental Data Set 1) and phylogenetic tree calculation were conducted using CLC Main Workbench 5.6.1 program, using the neighbor-joining algorithm (www.clcbio.com). Bootstrap values are from 1000 trials.
Tissue Collection, RNA Analysis, and Statistical Analysis
Each leaf is characterized by its position on the plant (for example, L1 is the first leaf produced by the plant and L5 the fifth) and by its developmental stage. Thus, L5 P1 is the fifth leaf when it has just initiated from the SAM, and it becomes L5 P2 after the next primordium initiates. For analysis of gene expression during leaf development, tissue was collected at successive stages of plant development, such that the fifth leaf of the plant (L5) was at the corresponding developmental stage in all samples. For very young leaf primordia at the P1-P3 stages, the leaf was at the respective developmental stage and was collected with younger leaf primordia and the SAM. At least three biological repeats, each consisting of pooled material from three different plants, were collected for each developmental stage. Quantitative real-time RT-PCR analysis was performed using the TaKaRa SYBR Premix Ex Taq II (RR081Q) kit and a Corbett Research Rotor-Gene 6000 cycler. For each gene, a standard curve was obtained using dilutions of a cDNA sample, and only reactions in which the standard curve was linear and the sample values fell within the standard curve (efficiency of 0.96 to 1.02 and R2 > 0.99) were used. Quantification of each gene was performed using the Corbett Research Rotor-Gene software. The levels of each gene and of the TUBULIN (TUB) reference gene were separately calculated at the logarithmic phase relative to the respective standard curve. At least three independent technical repeats were performed for each cDNA sample, and at least three biological repeats were used for each genotype. Relative expression of each sample was calculated by dividing the expression level by that of TUB (in arbitrary units). Gene/TUB ratios were then averaged and presented as a ratio of a control treatment, the value of which was set to 1. Primer sequences are detailed in Supplemental Table 1 online.
Statistical analysis was performed using JMP software (SAS Institute). Student’s t test was used for comparison of means, which were deemed significantly different at P < 0.05.
In Situ Hybridization
Antisense and sense probes were produced by in vitro transcription with digoxigenin-11-UTP (Roche) using T7 or SP6 RNA polymerase (Promega). Tissue samples were fixed overnight in 4% paraformaldehyde, gradually transferred to ethanol and then to Histoclear (Gadot), and embedded in Paraplast Plus (McCormick Scientific). Eight-micrometer-thick tissue sections were produced and mounted on ProbeOnPlus slides (Fisher Biotech). Slides were treated successively with Histoclear, an ethanol series, water, Proteinase K (1 μg/mL), PBS, 4% paraformaldehyde, PBS, triethanolamine (0.1 M, with stirring), PBS, and increasing ethanol series up to 100% ethanol. Hybridization was performed overnight at 55°C. After hybridization, slides were washed successively several times with 0.2× SSC at 55°C, several times with NTE (0.5 M NaCl, 10 mM Tris-HCl, pH 7.5, and 1 mM EDTA) at 37°C, and blocked with 1% fresh Boehringer block and then with 1% BSA solution (1% BSA, 100 mM Tris, pH 7.5, 150 mM NaCl, and 0.3% Triton X-100). Slides were incubated in pairs with antidigoxigenin antibodies (Roche) for 2 h at room temperature and then washed three times with 1% BSA solution and two times with 100 mM Tris, pH 9.5, and 100 mM NaCl. Slides were incubated with NBP/BCIP substrate solution (Roche) for 1 to 3 d and then washed with water, mounted, and analyzed. The expression pattern detected by the TRR16/17 antisense probe was compared with a control TRR16/17 sense probe, which showed only background signal. Primer sequences are detailed in Supplemental Table 1 online.
Imaging, Microscopy, and Microapplication of IAA
The pattern of GFP and VENUS expression was detected by a confocal laser scanning microscope (model LSM510; Zeiss) with the argon laser set at 488 nm for excitation, a long-pass 560-nm filter for chlorophyll emission, and 505- to 530-nm filter for GFP and VENUS emission, as described (Shani et al., 2009). Scanning electron microscopy was performed using a JEOL 5410 LV microscope as described (Shani et al., 2009). Microapplication of 1 mM IAA (Duchefa Brand I0901) was performed as described (Reinhardt et al., 2000).
Accession Numbers
Sequence data for Arabidopsis (At) or tomato (Sl) genes used in this study can be found in the Arabidopsis Genome Initiative or Sol Genomics Network under the following accession numbers: At IPT7 (AT3G23630), At CKX3 (AT3G23630), At KT2 (AT1G70510), Sl TRR3,4 (SGN-U577676), Sl TRR8/9a (SGN-U572841), Sl TRR8/9b (SGN-U572839), Sl TRR16,17 (SGN-U601012), Sl Tkn2 (SGN-U321206), Sl E (AF022020), and Sl CYCD3.3 (SGN-U586344).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. FILpro Drives Expression throughout the Leaf until Relatively Late Stages of Leaf Development.
Supplemental Figure 2. FILpro>>AtIPT7 Plants Grow Indeterminately.
Supplemental Figure 3. Early Leaf Development in Genotypes with Altered CK Levels.
Supplemental Figure 4. Constitutive Overexpression of At CKX3 in Tomato Results in Stunted Plants with Simple Leaves.
Supplemental Figure 5. Sequence Comparison of Tomato and Arabidopsis Type A RR Genes.
Supplemental Figure 6. CYCD3.3 Is Upregulated in FILpro>>AtIPT7 Plants.
Supplemental Figure 7. A Second Peak of a CK Response Marker in P7 Tomato Primordia.
Supplemental Figure 8. Exogenous Auxin Application Suppresses the FILpro>>AtIPT7 Super-Compound Leaf Phenotype.
Supplemental Figure 9. Coexpression of At CKX3 Suppresses the Super-Compound Leaf Phenotype Caused by Tkn2 Expression in Leaf Margins.
Supplemental Figure 10. Tkn2-SRDX Expression Remains High When Coexpressed with At IPT7.
Supplemental Figure 11. Late Expression of At IPT7 Does Not Affect Leaf Complexity but Causes Ectopic Growth between Veins.
Supplemental Figure 12. Coexpression of At IPT7 Enhances the Leaf Phenotype Caused by KT2 Overexpression at Late Stages of Arabidopsis Leaf Development by BLSpro.
Supplemental Table 1. Primers Used in This Study.
Supplemental Data Set 1. Text File of the Sequences and Alignment Used for the Phylogenetic Analysis Shown in Figure 3A.
Supplementary Material
Acknowledgments
We thank Thomas Schmülling and Tomáš Werner for the At CKX3 construct, Elliot Meyerowitz and Marcus Heisler for the pDR5rev:3XVENUS construct, Cris Kuhlemeier and Emmanuelle Bayer for the AtPIN1:PIN1-GFP tomato seeds, Yuval Eshed for driver constructs and seeds, Shai Fleishon for help with the characterization of TRRs, Roni Shalev and Ido Shwartz for help with cloning and tomato transformation, and Leor Eshed-Williams for critical reading of the manuscript. This work was supported by grants from the Israel Science Foundation (689/05 and 60/10), U.S.–Israel Binational Agricultural Research and Development Fund (IS 04140-08C), and the Israeli Ministry of Agriculture (837-0010-06) to N.O. and from the Israel Science Foundation (253/06) to D.W. and N.O.
References
- Alvarez J.P., Pekker I., Goldshmidt A., Blum E., Amsellem Z., Eshed Y. (2006). Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkoulas M., Hay A., Kougioumoutzi E., Tsiantis M. (2008). A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat. Genet. 40: 1136–1141 [DOI] [PubMed] [Google Scholar]
- Barth S., Geier T., Eimert K., Watillon B., Sangwan R.S., Gleissberg S. (2009). KNOX overexpression in transgenic Kohleria (Gesneriaceae) prolongs the activity of proximal leaf blastozones and drastically alters segment fate. Planta 230: 1081–1091 [DOI] [PubMed] [Google Scholar]
- Bayer E.M., Smith R.S., Mandel T., Nakayama N., Sauer M., Prusinkiewicz P., Kuhlemeier C. (2009). Integration of transport-based models for phyllotaxis and midvein formation. Genes Dev. 23: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Berger Y., Harpaz-Saad S., Brand A., Melnik H., Sirding N., Alvarez J.P., Zinder M., Samach A., Eshed Y., Ori N. (2009). The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 136: 823–832 [DOI] [PubMed] [Google Scholar]
- Bharathan G., Goliber T.E., Moore C., Kessler S., Pham T., Sinha N.R. (2002). Homologies in leaf form inferred from KNOXI gene expression during development. Science 296: 1858–1860 [DOI] [PubMed] [Google Scholar]
- Blein T., Hasson A., Laufs P. (2010). Leaf development: What it needs to be complex. Curr. Opin. Plant Biol. 13: 75–82 [DOI] [PubMed] [Google Scholar]
- Blein T., Pulido A., Vialette-Guiraud A., Nikovics K., Morin H., Hay A., Johansen I.E., Tsiantis M., Laufs P. (2008). A conserved molecular framework for compound leaf development. Science 322: 1835–1839 [DOI] [PubMed] [Google Scholar]
- Bolduc N., Hake S. (2009). The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21: 1647–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales C., Barkoulas M., Galinha C., Tsiantis M. (2010). Weeds of change: Cardamine hirsuta as a new model system for studying dissected leaf development. J. Plant Res. 123: 25–33 [DOI] [PubMed] [Google Scholar]
- Chen J., et al. (2010). Control of dissected leaf morphology by a Cys(2)His(2) zinc finger transcription factor in the model legume Medicago truncatula. Proc. Natl. Acad. Sci. USA 107: 10754–10759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino I.B., Deruère J., Kieber J.J. (2000). Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R., Nakamura K., Moubayidin L., Perilli S., Taniguchi M., Morita M.T., Aoyama T., Costantino P., Sabatini S. (2008). A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384 [DOI] [PubMed] [Google Scholar]
- Dengler N.G., Tsukaya H. (2001). Leaf morphogenesis in dicotyledons: current issues. Int. J. Plant Sci. 162: 459–464 [Google Scholar]
- Dewitte W., Scofield S., Alcasabas A.A., Maughan S.C., Menges M., Braun N., Collins C., Nieuwland J., Prinsen E., Sundaresan V., Murray J.A. (2007). Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. USA 104: 14537–14542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I., Blum E., Goldshmidt A., Eshed Y. (2008). A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I., Eshed Y., Lifschitz E. (2010). Morphogenesis of simple and compound leaves: A critical review. Plant Cell 22: 1019–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott R.C., Betzner A.S., Huttner E., Oakes M.P., Tucker W.Q., Gerentes D., Perez P., Smyth D.R. (1996). AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8: 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd S.K., Bowman J.L. (2010). Gene expression patterns in seed plant shoot meristems and leaves: Homoplasy or homology? J. Plant Res. 123: 43–55 [DOI] [PubMed] [Google Scholar]
- Galinha C., Bilsborough G., Tsiantis M. (2009). Hormonal input in plant meristems: A balancing act. Semin. Cell Dev. Biol. 20: 1149–1156 [DOI] [PubMed] [Google Scholar]
- Giulini A., Wang J., Jackson D. (2004). Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 430: 1031–1034 [DOI] [PubMed] [Google Scholar]
- Gleave A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Gordon S.P., Chickarmane V.S., Ohno C., Meyerowitz E.M. (2009). Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 106: 16529–16534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann W., Gleissberg S. (1996). Organogenetic capacity of leaves: The significance of marginal blastozones in angiosperms. Plant Syst. Evol. 199: 121–152 [Google Scholar]
- Hake S., Smith H.M., Holtan H., Magnani E., Mele G., Ramirez J. (2004). The role of knox genes in plant development. Annu. Rev. Cell Dev. Biol. 20: 125–151 [DOI] [PubMed] [Google Scholar]
- Hareven D., Gutfinger T., Parnis A., Eshed Y., Lifschitz E. (1996). The making of a compound leaf: genetic manipulation of leaf architecture in tomato. Cell 84: 735–744 [DOI] [PubMed] [Google Scholar]
- Hay A., Jackson D., Ori N., Hake S. (2003). Analysis of the competence to respond to KNOTTED1 activity in Arabidopsis leaves using a steroid induction system. Plant Physiol. 131: 1671–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A., Kaur H., Phillips A., Hedden P., Hake S., Tsiantis M. (2002). The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. 12: 1557–1565 [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2006). The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 38: 942–947 [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2010). KNOX genes: Versatile regulators of plant development and diversity. Development 137: 3153–3165 [DOI] [PubMed] [Google Scholar]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Hofer J., Turner L., Hellens R., Ambrose M., Matthews P., Michael A., Ellis N. (1997). UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr. Biol. 7: 581–587 [DOI] [PubMed] [Google Scholar]
- Holtan H.E., Hake S. (2003). Quantitative trait locus analysis of leaf dissection in tomato using Lycopersicon pennellii segmental introgression lines. Genetics 165: 1541–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura A., Hanaki N., Umeda H., Nakamura A., Suzuki T., Ueguchi C., Mizuno T. (1998). Response regulators implicated in His-to-Asp phosphotransfer signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 2691–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen B.J., Lund L., Sinha N. (1998). Overexpression of a homeobox gene, LeT6, reveals indeterminate features in the tomato compound leaf. Plant Physiol. 117: 771–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., Phillips A., Hedden P., Tsiantis M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Kaplan D.R. (2001). Fundamental concepts of leaf morphology and morphogenesis: A contribution to the interpretation of molecular genetic mutants. Int. J. Plant Sci. 162: 465–474 [Google Scholar]
- Kimura S., Koenig D., Kang J., Yoong F.Y., Sinha N. (2008). Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr. Biol. 18: 672–677 [DOI] [PubMed] [Google Scholar]
- Koenig D., Bayer E., Kang J., Kuhlemeier C., Sinha N. (2009). Auxin patterns Solanum lycopersicum leaf morphogenesis. Development 136: 2997–3006 [DOI] [PubMed] [Google Scholar]
- Koenig D., Sinha N. (2010). Evolution of leaf shape a pattern emerges. Curr. Top. Dev. Biol. 91: 169–183 [DOI] [PubMed] [Google Scholar]
- Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., Sakakibara H., Kyozuka J. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655 [DOI] [PubMed] [Google Scholar]
- Kvarnheden A., Yao J.L., Zhan X., O’Brien I., Morris B.A. (2000). Isolation of three distinct CycD3 genes expressed during fruit development in tomato. J. Exp. Bot. 51: 1789–1797 [DOI] [PubMed] [Google Scholar]
- Lee B.H., Johnston R., Yang Y., Gallavotti A., Kojima M., Travençolo B.A., Costa Lda.F., Sakakibara H., Jackson D. (2009). Studies of aberrant phyllotaxy1 mutants of maize indicate complex interactions between auxin and cytokinin signaling in the shoot apical meristem. Plant Physiol. 150: 205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A., To J.P., Busch W., Stehling S., Kehle A., Demar M., Kieber J.J., Lohmann J.U. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Lifschitz E., Eviatar T., Rozman A., Shalit A., Goldshmidt A., Amsellem Z., Alvarez J.P., Eshed Y. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 103: 6398–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay D.L., Sawhney V.K., Bonham-Smith P.C. (2006). Cytokinin-induced changes in CLAVATA1 and WUSCHEL expression temporally coincide with altered floral development in Arabidopsis. Plant Sci. 170: 1111–1117 [Google Scholar]
- Long J.A., Barton M.K. (1998). The development of apical embryonic pattern in Arabidopsis. Development 125: 3027–3035 [DOI] [PubMed] [Google Scholar]
- Mähönen A.P., Bishopp A., Higuchi M., Nieminen K.M., Kinoshita K., Törmäkangas K., Ikeda Y., Oka A., Kakimoto T., Helariutta Y. (2006). Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311: 94–98 [DOI] [PubMed] [Google Scholar]
- Miller C.O., Skoog F., von Saltza M.H., Strong F.M. (1955). Kinetin, a cell division factor from deoxyribonucleic acid. J. Am. Chem. Soc. 77: 1392–1393 [Google Scholar]
- Miyawaki K., Tarkowski P., Matsumoto-Kitano M., Kato T., Sato S., Tarkowska D., Tabata S., Sandberg G., Kakimoto T. (2006). Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. USA 103: 16598–16603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore I., Gälweiler L., Grosskopf D., Schell J., Palme K. (1998). A transcription activation system for regulated gene expression in transgenic plants. Proc. Natl. Acad. Sci. USA 95: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L., Di Mambro R., Sabatini S. (2009). Cytokinin-auxin crosstalk. Trends Plant Sci. 14: 557–562 [DOI] [PubMed] [Google Scholar]
- Moubayidin L., Perilli S., Dello Ioio R., Di Mambro R., Costantino P., Sabatini S. (2010). The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr. Biol. 20: 1138–1143 [DOI] [PubMed] [Google Scholar]
- Müller B., Sheen J. (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath U., Crawford B.C., Carpenter R., Coen E. (2003). Genetic control of surface curvature. Science 299: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Nikovics K., Blein T., Peaucelle A., Ishida T., Morin H., Aida M., Laufs P. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18: 2929–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39: 787–791 [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Allen E., Wu X., Schommer C., Schwab R., Carrington J.C., Weigel D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Perilli S., Moubayidin L., Sabatini S. (2010). The molecular basis of cytokinin function. Curr. Opin. Plant Biol. 13: 21–26 [DOI] [PubMed] [Google Scholar]
- Poethig R.S. (1997). Leaf morphogenesis in flowering plants. Plant Cell 9: 1077–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez J., Bolduc N., Lisch D., Hake S. (2009). Distal expression of knotted1 in maize leaves leads to reestablishment of proximal/distal patterning and leaf dissection. Plant Physiol. 151: 1878–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Mandel T., Kuhlemeier C. (2000). Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C., Huntley R., Jacqmard A., Murray J.A. (1999). Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Ruzicka K., Simásková M., Duclercq J., Petrásek J., Zazímalová E., Simon S., Friml J., Van Montagu M.C., Benková E. (2009). Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc. Natl. Acad. Sci. USA 106: 4284–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R. (2007). The dynamic plant stem cell niches. Curr. Opin. Plant Biol. 10: 639–644 [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Sakakibara H., Kojima M., Yamamoto Y., Nagasaki H., Inukai Y., Sato Y., Matsuoka M. (2006). Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol. 142: 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K.F.X., Jürgens G., Laux T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Shani E., Burko Y., Ben-Yaakov L., Berger Y., Amsellem Z., Goldshmidt A., Sharon E., Ori N. (2009). Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell 21: 3078–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E., Yanai O., Ori N. (2006). The role of hormones in shoot apical meristem function. Curr. Opin. Plant Biol. 9: 484–489 [DOI] [PubMed] [Google Scholar]
- Skoog F., Miller C.O. (1957). Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 54: 118–130 [PubMed] [Google Scholar]
- Skylar A., Hong F., Chory J., Weigel D., Wu X. (2010). STIMPY mediates cytokinin signaling during shoot meristem establishment in Arabidopsis seedlings. Development 137: 541–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C.N. (1957). Histogenesis of the leaf and structure of the shoot apex in Glycine max (L.) Merrill. Bull. Torrey Bot. Club 84: 163–174 [Google Scholar]
- To J.P., Kieber J.J. (2008). Cytokinin signaling: Two-components and more. Trends Plant Sci. 13: 85–92 [DOI] [PubMed] [Google Scholar]
- Veit B. (2009). Hormone mediated regulation of the shoot apical meristem. Plant Mol. Biol. 69: 397–408 [DOI] [PubMed] [Google Scholar]
- Wang H., Chen J., Wen J., Tadege M., Li G., Liu Y., Mysore K.S., Ratet P., Chen R. (2008). Control of compound leaf development by FLORICAULA/LEAFY ortholog SINGLE LEAFLET1 in Medicago truncatula. Plant Physiol. 146: 1759–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Jones B., Li Z., Frasse P., Delalande C., Regad F., Chaabouni S., Latché A., Pech J.C., Bouzayen M. (2005). The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17: 2676–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Motyka V., Strnad M., Schmülling T. (2001). Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 98: 10487–10492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Motyka V., Laucou V., Smets R., Van Onckelen H., Schmülling T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Schmülling T. (2009). Cytokinin action in plant development. Curr. Opin. Plant Biol. 12: 527–538 [DOI] [PubMed] [Google Scholar]
- Wolters H., Jürgens G. (2009). Survival of the flexible: hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 10: 305–317 [DOI] [PubMed] [Google Scholar]
- Yanai O., Shani E., Dolezal K., Tarkowski P., Sablowski R., Sandberg G., Samach A., Ori N. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15: 1566–1571 [DOI] [PubMed] [Google Scholar]
- Zhao Z., Andersen S.U., Ljung K., Dolezal K., Miotk A., Schultheiss S.J., Lohmann J.U. (2010). Hormonal control of the shoot stem-cell niche. Nature 465: 1089–1092 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.