The retinoblastoma (RB) pathway is a conserved eukaryotic cell cycle regulator that is thought to control cell cycle progression through periodic dissociation of the repressor protein, RB, from the activator proteins E2F and DP. This study shows that in the unicellular alga Chlamydomonas, the cell cycle is regulated by a constitutively chromatin-bound RB-E2F-DP ternary complex whose subunits do not undergo periodic dissociation.
Abstract
We examined the cell cycle dynamics of the retinoblastoma (RB) protein complex in the unicellular alga Chlamydomonas reinhardtii that has single homologs for each subunit—RB, E2F, and DP. We found that Chlamydomonas RB (encoded by MAT3) is a cell cycle–regulated phosphoprotein, that E2F1-DP1 can bind to a consensus E2F site, and that all three proteins interact in vivo to form a complex that can be quantitatively immunopurified. Yeast two-hybrid assays revealed the formation of a ternary complex between MAT3, DP1, and E2F1 that requires a C-terminal motif in E2F1 analogous to the RB binding domain of plant and animal E2Fs. We examined the abundance of MAT3/RB and E2F1-DP1 in highly synchronous cultures and found that they are synthesized and remain stably associated throughout the cell cycle with no detectable fraction of free E2F1-DP1. Consistent with their stable association, MAT3/RB and DP1 are constitutively nuclear, and MAT3/RB does not require DP1-E2F1 for nuclear localization. In the nucleus, MAT3/RB remains bound to chromatin throughout the cell cycle, and its chromatin binding is mediated through E2F1-DP1. Together, our data show that E2F-DP complexes can regulate the cell cycle without dissociation of their RB-related subunit and that other changes may be sufficient to convert RB-E2F-DP from a cell cycle repressor to an activator.
INTRODUCTION
A fundamental principle of cell cycle control involves regulatory switches that integrate internal and external information and drive coherent and unidirectional cell cycle progression (Morgan, 2007). A key regulator of the G1-S transition is the retinoblastoma (RB) tumor suppressor pathway. RB is part of a cell cycle network involving cyclin-dependent kinases (CDKs) that phosphorylate RB-related proteins (also referred to as pocket proteins) to regulate the initiation of S-phase (Gutierrez et al., 2002; Burkhart and Sage, 2008). Pocket proteins repress the cell cycle by binding to the E2F family of transcription factors that form a DNA binding protein by heterodimerizing with a related protein, DP. E2F-DP heterodimers bind specific response elements in the promoters of cell cycle genes and are thought to function primarily in transcriptional control (McClellan and Slack, 2007; van den Heuvel and Dyson, 2008). RB-related proteins and E2F-DP complexes are important for cell cycle regulation in diverse eukaryotic groups, including plants, animals, algae, and amoebae (Trimarchi and Lees, 2002; Robbens et al., 2005; Fang et al., 2006; MacWilliams et al., 2006; De Veylder et al., 2007; van den Heuvel and Dyson, 2008; Moulager et al., 2010).
Much of the detailed information on molecular mechanisms of pocket protein regulation has come from studies of mammalian cells (Burkhart and Sage, 2008). The model for cell cycle regulation in mammals involves sequential phosphorylation of pocket proteins during G1 and at the G1-S boundary by CDK complexes that include CDK3-Cyclin C, CDK4/6-Cyclin D, and CDK2-Cyclin E (Malumbres and Barbacid, 2005). Hyperphosphorylation of RB leads to its dissociation from E2F-DP, which then activates the transcription of S-phase genes. This regulated dissociation model is supported by in vitro and in vivo evidence (Cobrinik, 2005).
RB-related proteins (RBRs) have been identified in embyrophytes (vascular and nonvascular land plants) and green algae (Xie et al., 1996; Ach et al., 1997; Kong et al., 2000; Umen and Goodenough, 2001; Dewitte and Murray, 2003; Robbens et al., 2005; Sabelli et al., 2005; Inze and De Veylder, 2006; Ferris et al., 2010). Embryophyte RBRs are cell cycle–regulated phosphoproteins (Huntley et al., 1998; Boniotti and Gutierrez, 2001; Sabelli et al., 2005) that bind E2Fs (Huntley et al., 1998; Ramírez-Parra et al., 1999, 2003; Sekine et al., 1999; Sabelli et al., 2005). As in animals, embryophytes contain multiple E2F and DP isoforms that regulate cell division and differentiation (Huntley et al., 1998; Kosugi and Ohashi, 2002; Mariconti et al., 2002; Vandepoele et al., 2002; Sabelli et al., 2005; Desvoyes et al., 2006). Null mutations in Arabidopsis thaliana RBR1 are gametophyte lethal (Ebel et al., 2004) and cannot be propagated, while RBR knockdowns in Arabidopsis or tobacco have been used to show a role for RBRs in repressing proliferation and promoting differentiation (Menges et al., 2005; Wildwater et al., 2005; Jordan et al., 2007; Borghi et al., 2010). Interaction with chromatin-modifying proteins has also been shown as an important function of RBRs (Williams and Grafi, 2000; Shen, 2002; Rossi et al., 2003; Costa and Gutierrez-Marcos, 2008; Johnston et al., 2008; Jullien et al., 2008). However, the dynamics of RBR protein complex formation with E2F1 and DP1 during the plant cell cycle have not been closely examined.
Whereas regulated dissociation of RBRs from E2F-DP heterodimers is assumed to be required for cell cycle regulation, there is also evidence for S-phase RB-E2F-DP complexes (Wells et al., 2003; Ianari et al., 2009), whose relative importance and function in cell cycle control is still unclear. The investigation of RB-mediated cell cycle regulation in a simpler model system could shed light on the dynamics of RB-E2F-DP interactions in both plants and animals.
Chlamydomonas reinhardtii is a unicellular green alga whose sequenced genome encodes single homologs of RB, E2F, and DP (Bisova et al., 2005; Fang et al., 2006; Merchant et al., 2007). The Chlamydomonas RB homolog MAT3 has been shown to regulate the cell cycle and cell size homeostasis (Umen and Goodenough, 2001). Chlamydomonas uses a variation of the mitotic cell cycle, termed multiple fission, in which cells undergo a prolonged G1 period that is followed by multiple alternating rounds of S-phase and mitosis (S/M) to produce a uniform population of daughters (see Supplemental Figure 1 online) (Spudich and Sager, 1980; Craigie and Cavalier-Smith, 1982; Donnan and John, 1983; McAteer et al., 1985; John, 1987; Oldenhof et al., 2007). The extended G1 phase and rapid division cycles that characterize multiple fission are modifications that are critical for development and reproduction in plants and animals, making its investigation relevant for understanding both unicellular and multicellular cell cycles (Umen, 2005).
MAT3/RB regulates the cell cycle at two key points. An early/mid G1 control point, termed Commitment, is passed when cells attain a minimum size that allows them to divide at least once. Cells that have passed Commitment remain in G1 for an additional 5 to 8 h, during which time they can continue to grow if conditions permit. At the end of G1, mother cells undergo one or more rounds of S/M to produce 2n daughter cells. The S/M size checkpoint couples cell division number to mother cell size such that larger mother cells undergo more rounds of S/M than do smaller mother cells, thus ensuring a uniform-sized population of daughters (Craigie and Cavalier-Smith, 1982; John, 1987; Umen and Goodenough, 2001; Matsumura et al., 2003; Umen, 2005) (see Supplemental Figure 1 online). Under physiological conditions of alternating light and dark periods (e.g., 14 h light/10 h dark), the Chlamydomonas cell cycle becomes highly synchronized with G1 occurring in the light phase and S/M taking place early in the dark (Bisova et al., 2005), and this natural synchrony is advantageous for isolating homogenous populations of cells at defined positions in the cell cycle.
Cells that are missing the RB homolog MAT3 have a small cell phenotype that is caused by passage through Commitment at a prematurely small size and by supernumerary rounds of S/M that lead to formation of tiny daughters (Umen and Goodenough, 2001). A suppressor screen led to the identification of multiple alleles of DP1 and E2F1 that reversed the mat3 small cell phenotype and also could cause the opposite phenotype, large cells (Fang et al., 2006). The suppressor alleles dp1 and e2f1 were epistatic to mat3 and did not show any additive effects when combined. These genetic results suggest that the repressor MAT3/RB acts on a single DP1-E2F1 complex to regulate the cell cycle. Interestingly, cell cycle transcription defects were not observed in mat3, dp1, or e2f1 mutants despite their cell cycle defects (Fang et al., 2006). Whether this finding reflects an alternative means of cell cycle control by the Chlamydomonas RB pathway or the existence of cryptic transcriptional targets is under investigation. Additional suppressor loci were found in a subsequent screen that do not correspond to known transcriptional regulators but have cell cycle defects and may represent new targets of the RB pathway (Fang and Umen, 2008). Taken together, the data from Chlamydomonas indicate that the genetic architecture of the RB pathway is conserved, with MAT3/RB as a repressor and E2F1/DP1 a downstream cell cycle activator. Moreover, viable knockout mutations in MAT3/RB and DP1 indicated that neither protein is essential for proliferation but that the RB pathway is critical for coupling cell size to cell cycle progression. These properties make Chlamydomonas a highly tractable model for further investigation of the mechanism by which MAT3/RB and E2F1-DP1 regulate the cell cycle.
In this study, we extended our dissection of RB-mediated cell cycle control by examining the spatial and temporal interaction of MAT3/RB with E2F1 and DP1 during the cell cycle. Our findings suggest that regulated dissociation of the repressor MAT3/RB from the activators E2F1-DP1 need not occur for the RB pathway to control periodic cell cycle entry and exit. Instead, our data point to the existence of a stable chromatin-associated MAT3/RB-E2F1-DP1 ternary complex that may be modified in other ways to regulate the switch between cell cycle activation and repression.
RESULTS
Detection of MAT3/RB in Chlamydomonas
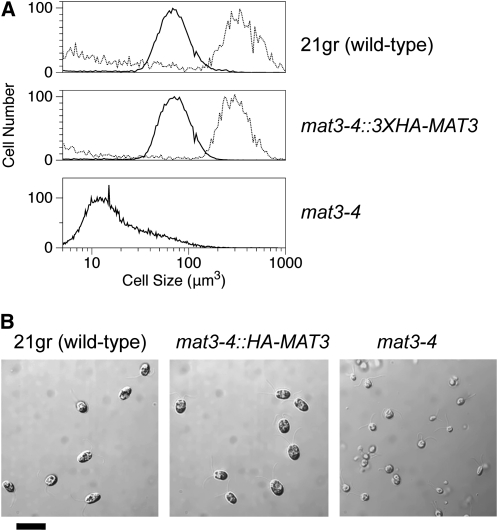
To detect and purify MAT3/RB, we constructed an N-terminal triple hemagglutinin (HA) epitope-tagged allele from a genomic MAT3 clone under the control of its native promoter and terminator (3XHA-MAT3) (see Methods; see Supplemental Figure 2 online). When transformed into a mat3-4 null mutant strain, we obtained complementation rates with 3XHA-MAT3 that were similar to those obtained with a wild-type genomic MAT3 construct that was used previously (Umen and Goodenough, 2001). The complemented mat3-4::3XHA-MAT3 transgenic lines had a wild-type size distribution and growth rate, could be synchronized with alternating light-dark cycles, and behaved in all respects like the wild-type control strain (Figures 1A and 1B)
Figure 1.
Complementation of mat3-4 with HA-Tagged MAT3.
(A) Size distribution of daughter cells (solid black line) or predivision mother cells (11 h light; dashed line) from synchronized wild type (21gr; top panel), mat3-4::3XHA-MAT3 (complemented strain; middle panel), or mat3-4 (bottom panel).
(B) Micrographs of daughter cells from (A). Bar = 20 μm.
We verified the expression of 3xHA-MAT3 by immunoblotting with anti-HA antibodies and observed a single ~130-kD band at the expected molecular mass for 3XHA-MAT3 that was not seen in untagged wild-type strains (see Supplemental Figure 3 online).
MAT3/RB Is a Phosphoprotein
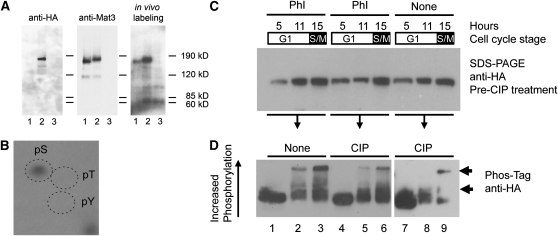
Site-specific phosphorylation of mammalian RB by CDKs occurs at the G1-S phase boundary and during S-, G2-, and M-phases (Buchkovich et al., 1989; DeCaprio et al., 1989; Mihara et al., 1989; Mittnacht and Weinberg, 1991; Mittnacht et al., 1994; Huntley et al., 1998; Boniotti and Gutierrez, 2001; Nakagami et al., 2002; Sabelli et al., 2005). As a first step to detect MAT3 phosphorylation, we used in vivo labeling and immunoprecipitation (IP), taking advantage of the fact that Chlamydomonas can rapidly and quantitatively incorporate 32PO4 under conditions where phosphate levels are still sufficient to support normal growth and cell cycle progression (see Methods). Labeling of asynchronous wild-type, mat3-4::3XHA-MAT3, and mat3-4 cultures for 30 to 60 min resulted in incorporation of >90% of added inorganic 32PO4. Lysates prepared from labeled cells were used for IPs with anti-MAT3 or anti-HA affinity matrices. IP pellets from all three labeled strains were fractionated by SDS-PAGE, transferred to polyvinylidene fluoride (PVDF) membranes, and subjected to both autoradiography and immunoblotting with anti-MAT3 or anti-HA antibodies (Figure 2A).32P-labeled bands migrating at the expected positions for MAT3 and 3XHA-MAT3 were detected in the wild-type and mat3-4::3XHA-MAT3 strains but not in the control mat3-4 strain (Figure 2A, right panel). The same bands were detected by immunoblotting using anti-MAT3 antibodies (Figure 2A, middle panel), whereas immunoblots using anti-HA antibodies detected a band only in the lane loaded with mat3-4::3XHA-MAT3 lysate (Figure 2A, left panel). The in vivo labeling experiments indicate that Chlamydomonas MAT3/RB, like metazoan and plant RB-related proteins, is a phosphoprotein.
Figure 2.
MAT3 Is a Cell Cycle–Regulated Phosphoprotein.
(A) In vivo 32PO4-labeled asynchronous cultures of the wild type (lane 1), mat3-4::3XHA-MAT3 (lane 2), and mat3-4 (lane 3) were immunoprecipitated using anti-MAT3 antibodies. IPs were separated by SDS-PAGE and blotted. The right panel is an autoradiograph of the blot, the middle panel is the same blot probed with anti-MAT3 antibodies, and the left panel is the blot stripped and reprobed with anti-HA antibodies.
(B) Autoradiograph from two-dimensional thin layer electrophoretic separation of hydrolyzed 3XHA-MAT3 immunoprecipitated from in vivo–labeled mitotic samples. The migration of unlabeled phosphoserine (pS), phosphothreonine (pT), and phosphotyrosine (pY) standards detected using ninhydrin is marked by dashed ovals.
(C) Anti-HA immunoblots to detect HA-MAT3 in mat3-4::3XHA-MAT3 whole-cell lysates from different cell cycle time points (5 h, pre-Commitment; 11 h, post-Commitment; 15 h, S/M). Each sample was split into three equal fractions. Phosphatase inhibitors (PhI) were added to two of the fractions prior to further processing. The blot shows starting protein levels fractionated by standard SDS-PAGE prior to incubation with CIP.
(D) Part of each sample described in (C) was treated with CIP as indicated and then fractionated by Phos-Tag SDS-PAGE and blotted to detect phosphorylated isoforms of HA-MAT3 (indicated by arrows). A longer exposure of the same blot is shown for lanes 7 to 9.
Previously, it has been reported that MAT3 has ~15 potential CDK phosphorylation sites, several of which are strong matches to the consensus S/T-P-X-K/R (Umen and Goodenough, 2001). IPs from S/M-phase cells were gel fractionated, and the radiolabeled MAT3 band was excised and subjected to partial acid hydrolysis. Labeled phosphoamino acids from MAT3/RB were mixed with unlabeled phosphoserine, phosphothreonine, and phosphotyrosine and then separated by two-dimensional thin layer electrophoresis followed by autoradiography (Blume-Jensen and Hunter, 2001). A single radioactive hydrolysis product was identified that comigrated with the phosphoserine standard, indicating that MAT3 phosphorylation occurs predominantly on Ser residues (Figure 2B). Candidate residues for CDK phosphorylation in MAT3 based on these data are shown in Supplemental Figure 4 online.
MAT3 Phosphorylation Is Cell Cycle Regulated
We used Phos-Tag SDS-PAGE and immunoblotting to examine the phosphorylation pattern of HA-MAT3 during the cell cycle (see Methods). Phos-Tag selectively retards the migration of phosphoproteins during electrophoresis, facilitating their detection (Kinoshita et al., 2006). Cells from synchronous cultures (>95% pure for each stage) were collected in early G1 (4 h, precommitment), late G1 (11 h, postcommitment), and S/M-phase (15 h). While we cannot separate S-phase and mitotic stages, their relative timing has been previously estimated (Coleman, 1982; Craigie and Cavalier-Smith, 1982; Harper and John, 1986; Harper, 1999). Based on these reports and the total time spent in S/M, at least 30% of cells from S/M samples are in S-phase, a value that is consistent with flow cytometric profiles of S/M cells (Fang et al., 2006) in which an S-phase population is evident.
MAT3 from precommitment cells migrated predominantly as a single band (Figure 2D, lane 1), suggesting that MAT3 is unphosphorylated or hypophosphorylated at this stage. MAT3 from postcommitment cells fractionated into additional slower-migrating species, indicating increased phosphorylation at this stage. These slower migrating species increased further in lysates from S/M cells (Figure 2D, lanes 2 and 3 indicated by arrows).
As previously noted (Kinoshita et al., 2006), the efficiency of membrane transfer from Phos-Tag gels decreases as phosphorylation increases (Figure 2C versus 2D), meaning that the ratio of hyper- to hypophosphorylated MAT3 could be even higher than what is seen in Figure 2D. We attempted to dephosphorylate HA-MAT3 using calf intestinal alkaline phosphatase (CIP) but found that CIP treatment made the protein highly unstable even in the presence of protease inhibitors (Figure 2D, lanes 7 to 9; see Methods). Loss of HA-MAT3 appears to be dependent on dephosphorylation as inclusion of phosphatase inhibitors prevented its disappearance (Figure 2D, lanes 4 to 6 compared with lanes 7 to 9). Although the CIP treatment was incomplete, we did observe a reduction in the apparent molecular mass of CIP-treated HA-MAT3 (Figure 2D, lanes 7 to 9), indicating the presence of protein phosphorylation.
E2F1 and DP1 Heterodimerize to Form a DNA Binding Protein
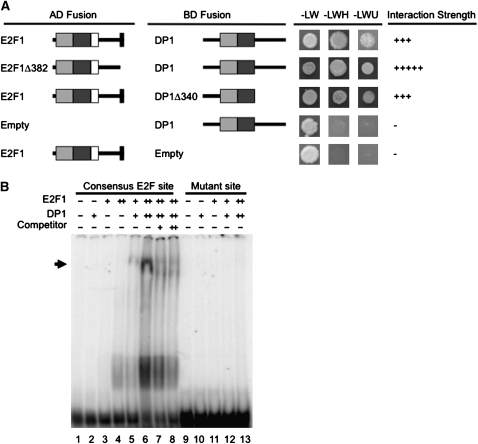
Chlamydomonas E2F1 and DP1 have N-terminal DNA binding and dimerization domains that are conserved with their homologs from metazoans and plants (see Supplemental Figures 5 and 6 online) (Bisova et al., 2005; Fang et al., 2006). We used a yeast two-hybrid (Y2H) assay (Miller and Stagljar, 2004) to determine whether Chlamydomonas E2F1 and DP1 preferentially heterodimerize as they do in other species. Each protein was fused to the yeast Gal4 DNA binding domain (Gal4-DB) as a bait construct or the Gal4 activation domain (Gal4-AD) as a prey construct. Plasmids were then transformed into a yeast strain that has the HIS3 and URA3 genes under the control of GAL4 binding sites as transcriptional reporters. The E2F1-DB bait construct showed autoactivation when expressed without a prey construct (data not shown), but the DP1-DB did not (Figure 3A). Neither E2F1-AD nor DP1-AD caused activation in the Y2H assay when coexpressed with a control construct expressing only the Gal4-DB (Figure 3A; see Supplemental Figure 7 online). By contrast, E2F1-AD coexpressed with the DP1-DB showed strong reporter activation in the Y2H assay, indicating a specific interaction between E2F1 and DP1 (Figure 3A). Glutathione S-transferase (GST) pull-down experiments using in vitro–translated E2F1 and DP1 confirmed their ability to bind each other directly (see Supplemental Figure 8 online).
Figure 3.
E2F1 and DP1 Bind Each Other and to a Consensus E2F Site.
(A) Y2H interaction assay between E2F1, DP1, and C-terminally truncated E2F1 and DP1. Light-gray boxes indicate the DNA binding domain, and dark-gray boxes indicate the dimerization domain from E2F1 and DP1 (see Supplemental Figures 5 and 6 online). White boxes indicate the marked box, and black boxes at the C terminus of E2F1 indicate the putative MAT3/RB binding domain (see Supplemental Figure 5 online). Gal4 activation domain (AD) or DNA binding domain (BD) fusions were tested for growth under nonselective conditions (−LW) or on media that select for HIS3 (−LWH + 30 mM 3-amino 1,2,3 triazole) or URA3 (−LWU) Y2H reporter expression. The “−” indicates no interaction, and the number of “+” symbols indicates relative strength of interaction. Additional controls are in Supplemental Figure 7 online.
(B) Electrophoretic mobility shift assay. As indicated above each lane, 5 (+) or 10 (++) pmol of recombinant E2F1 and/or DP1 was incubated with a 32P-labeled oligonucleotide consisting of an E2F consensus site or a mutant site. In lanes 7 and 8, 5× (+) or 10× (++) excess unlabeled specific competitor was mixed with the labeled oligonucleotides. The arrow indicates the position of the E2F1 and DP1-dependent gel shift band.
C-terminal truncations of both proteins were also tested in the Y2H assay. E2F1Δ382 is missing the last 55 amino acids that correspond to a region conserved with Volvox E2F1 (Prochnik et al., 2010) (see Supplemental Figure 5 online). However, this domain could not be unambiguously aligned to the RB binding motifs from animal or plant E2Fs that are also diverged from each other (Ramírez-Parra et al., 1999; Sekine et al., 1999). E2F1Δ209 is truncated just downstream of the conserved region containing its predicted DNA binding domain, dimerization domain, and marked box (see Supplemental Figure 5 online). DP1Δ340 is a C-terminal truncation that removes sequences downstream of the DNA binding, dimerization, and conserved domains (see Supplemental Figure 6 online) (Bisova et al., 2005; Fang et al., 2006). The truncation constructs did not diminish the interaction between E2F1 and DP1, and the E2F1Δ382 construct interacted with DP1 even more strongly than did full-length E2F1 (Figure 3A).
We next determined whether E2F1 and DP1 from Chlamydomonas could bind to a consensus E2F site (TTTCGCGC) that has been demonstrated to bind plant E2F-DP complexes (Ramírez-Parra and Gutierrez, 2000; Ramírez-Parra et al., 2003; Uemukai et al., 2005). Soluble recombinant hexahistidine-tagged E2F1 and DP1 (6His-E2F1 and 6His-DP1) were expressed in Escherichia coli and partially purified. These proteins were added individually and in combination to a 32P end–labeled double-stranded oligonucleotide that had a consensus E2F site or a mutated E2F site (TTTCGATC; Ramírez-Parra and Gutierrez, 2000; Ramírez-Parra et al., 2003). Protein-DNA complexes were detected using an electrophoretic mobility shift assay followed by autoradiography. Recombinant E2F1 alone showed some binding to the consensus site, whereas DP1 did not (Figure 3B, lanes 2 to 4), but when coincubated, the two proteins efficiently formed a new, slowly migrating complex (Figure 3B, lanes 5 and 6). A faster migrating complex bound to the oligonucleotide was also observed (Figure 3B, lanes 5 and 6) that is likely to contain monomeric E2F since it was absent in reactions containing a mutated E2F binding site (Figure 3B, lanes 2 and 9 to 13). Formation of the E2F1+DP1-dependent complex with the radiolabeled oligonucleotide was reduced by the presence of unlabeled competitor (Figure 3B, lanes 7 and 8), further supporting its sequence specificity. Together, our data are consistent with E2F1 and DP1 heterodimerizing to bind an E2F consensus site.
Interactions, Cell Cycle Accumulation, and Nuclear Localization of MAT3, DP1, and E2F1
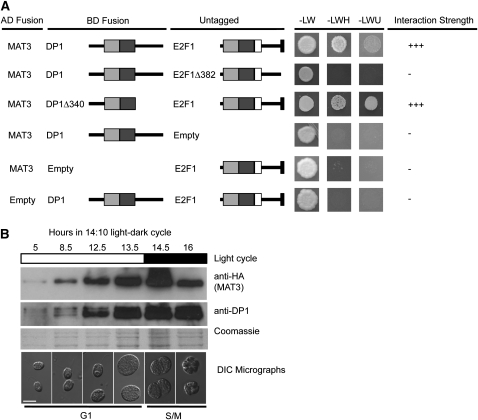
We used the Y2H assay to determine whether Chlamydomonas E2F1 and/or DP1 can interact with MAT3/RB. A MAT3-DB fusion was found to interact weakly with E2F1-AD but not with DP1-AD (see Supplemental Figure 7 online). To test whether MAT3/RB might require both E2F1 and DP1 for stable interaction, we used a modified Y2H bridging assay with MAT3-DB and DP1-AD fusions coexpressed with an unfused E2F1 construct. When coexpressed, the three proteins gave a strong Y2H activation signal, suggesting that MAT3 preferentially binds to E2F1-DP1 heterodimers (Figure 4A). However, when E2F1Δ382 or E2FΔ209 were used, both of which retain the ability to bind DP1 (Figure 3A), there was no detectable Y2H interaction with MAT3 (Figure 4A). These results indicate that the last 55 residues of E2F are required for MAT3 binding to E2F1-DP1 heterodimers and that this region of E2F1 may play an analogous role to the C-terminal RB binding domain of animal and plant E2Fs (Ramírez-Parra et al., 1999; Rubin et al., 2005). By contrast, the DP1Δ340 construct retained MAT3 binding (Figure 4A), suggesting that its C-terminal region is not required to recruit MAT3.
Figure 4.
MAT3/RB Interacts with E2F1-DP1 and Requires the C Terminus of E2F1 for Interaction.
(A) Untagged E2F1 or E2F1Δ382 was tested for their ability to bridge MAT3-AD and DP1-DBD or DP1Δ340-DBD in the Y2H assay. Y2H constructs are labeled the same as in Figure 3A. Additional controls are in Supplemental Figure 7 online.
(B) Immunoblots to detect 3XHA-MAT3 and DP1. Light-dark regime and cell cycle stage/time are depicted above and below each blot. Lysates prepared from equal numbers of mat3-4::3XHA-MAT3 cells were loaded in each lane. Top and middle panels are immunoblots probed with anti-HA and anti-DP1. Bottom panel is a Coomassie blue–stained gel to show total protein loaded on the basis of equal cell number per lane. Note that the 16-h time point was loaded based on mother cell number. Below each gel lane is a micrograph of cells from the corresponding time point. Bar = 10 μm.
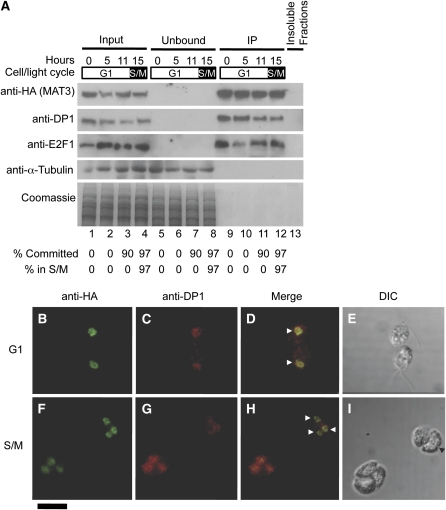
Antibodies raised against purified recombinant His-tagged DP1 (6His-DP1) or 6His-E2F1 could detect their respective antigens from whole-cell lysates (see Supplemental Figure 9 online). However, E2F1 was most easily detected in HA-MAT3 IP pellets, and the signal was absent in IPs from 3XHA-MAT3 e2f1-4 or 3XHA-MAT3 dp1-1 strains (see Supplemental Figures 9C and 9D online). The DP1 antibody was more sensitive than the E2F1 antibody and was used to detect E2F1-DP1 complexes in most subsequent experiments. In previous work, the abundance of MAT3, E2F1, and DP1 mRNAs during the cell cycle was found to be relatively constant, a pattern that was different from mRNAs corresponding to other cell cycle genes, such as those encoding mitotic and S-phase cyclins (Bisova et al., 2005). We used synchronous cultures of mat3-4::3XHA-MAT3 strains to prepare total lysates at different time points and then used immunoblotting of SDS-PAGE fractionated samples to compare the amount of MAT3 and DP1 proteins during the cell cycle. Both proteins accumulated approximately in proportion to cell size during G1 as shown when the gel was loaded by equal cell number per lane (Figure 4B; see Supplemental Figure 10 online). Because cells grow by up to 10-fold during G1, more total protein is loaded at later time points than early ones when equal cell numbers are loaded per lane. When equal protein was loaded per lane, it was clear that the concentration of HA-MAT3, DP1, and E2F1 remained constant with respect to total protein throughout the cell cycle (Figure 5A, lanes 1 to 4). This accumulation pattern suggests that MAT3, DP1, and E2F1 are expressed and function throughout the cell cycle, a finding that is consistent with their genetically defined roles in maintaining both the G1 Commitment size checkpoint and the S/M size checkpoint (Fang et al., 2006).
Figure 5.
MAT3/RB-DP1-E2F1 Relative Levels Are Constant, and the Proteins Remain Nuclear Localized throughout the Cell Cycle.
(A) Immunoblot detection of HA-MAT3, DP1, E2F1, and α-tubulin (top) and Coomassie staining of equivalently loaded gel run in parallel (bottom) from anti-HA immunoprecipitation of mat3-4::3XHA-MAT3 lysates. Cell cycle times from a 14 h:10 h light-dark synchronized culture are shown above each lane. Synchrony was determined from Commitment and mitotic index (S/M) assays shown for each fraction at the bottom. Equal total protein from each stage was used for the inputs. Lanes 1 to 4, 10% total input; lanes 5 to 8, 10% unbound fraction from IP; lanes 9 to 12, 10% IP pellet; lane 13, 10% of combined insoluble debris from all four lysates used for IPs.
(B) to (I) Immunofluorescence detection of 3XHA-MAT3 and DP1 during the cell cycle. G1 phase ([B] to [E]) or S/M phase ([F] to [I]) mat3-4::3XHA-MAT3 cells double-labeled with anti-HA (green signal; [B] and [F]) and anti-DP1 antibodies (red signal; [C] and [G]). (D) and (H) are merged signals from (B) and (C), and (F) and (G), respectively. The horizontal arrowheads mark examples of nuclei. (E) and (I) are differential interference contrast (DIC) images of cells from (B) to (D) and (F) to (I), respectively. The black arrowhead marks an incipient cleavage furrow in (I). Bar in bottom left corner = 10 μm.
MAT3/RB Remains Bound to E2F1 and DP1 throughout the Cell Cycle
In metazoans, CDK phosphorylation of pocket proteins is thought to cause their release from E2F-DP, thereby allowing E2F-DP–dependent cell cycle activation (reviewed in Trimarchi and Lees, 2002; Cobrinik, 2005; Dimova and Dyson, 2005; Burkhart and Sage, 2008). To determine the temporal binding pattern of MAT3/RB to E2F1-DP1 at different stages of the cell cycle, we quantified their association in synchronized cultures growing in a 14 h:10 h light:dark regime (Bisova et al., 2005). We prepared lysates from four stages: daughter cells (0 h, pre-Commitment), early G1 (5 h, pre-Commitment), mid G1 (11 h, post-Commitment), and S/M (15 h). Synchrony was assessed by microscopy to identify S/M phase cells and by Commitment status (Figure 5; see Methods). The cell cycle stage uniformity was >90% in each sample (Figure 5A, bottom). We prepared cell lysates from samples taken at each time point and used them for IP with anti-HA antibodies. We found that HA-MAT3 was able to co-IP DP1 and E2F1 equally and completely at each cell cycle stage (Figure 5A, compare input fractions, lanes 1 to 4, to IPs in lanes 9 to 12). We repeated this experiment using equal numbers of cells from each time point (see Supplemental Figure 10 online) and obtained similar results that match the overall accumulation patterns for each protein in total lysates (Figure 4B). Results from these experiments show that HA-MAT3, DP1, and E2F1 increase as cells grow and that their overall interaction remains stable throughout the cell cycle.
The presence of MAT3, E2F1, and DP1 in HA-MAT3 IP pellets was confirmed using tandem mass spectrometry (see Methods; see Supplemental Table 1 online). Importantly, these proteins were never detected in several parallel control IP experiments conducted using an untagged wild-type isogenic strain. Moreover, in cells with untagged MAT3, DP1 remained in the unbound fraction from anti-HA IPs (see Supplemental Figure 11 online).
MAT3/RB Is Constitutively Nuclear and Colocalizes with DP1 throughout the Cell Cycle
Human RB shows reduced affinity for nuclei during S-phase (Mittnacht and Weinberg, 1991; Szekely et al., 1991; Alberts et al., 1993; Stokke et al., 1993; Mancini et al., 1994; Mittnacht et al., 1994), though some RB has been reported to remain in the nucleus during S-phase (Kennedy et al., 2000). The unexpectedly stable association between MAT3/RB and E2F1-DP1 that we found prompted us to ask where they localize during the cell cycle and whether MAT3/RB cycles between the nucleus and cytoplasm. Like yeasts, Chlamydomonas undergoes closed mitoses with an intact nuclear envelope so nuclear proteins will not escape during division except through active export or exclusion (Johnson and Porter, 1968). We used indirect immunofluorescence (IF) to determine the localization of MAT3/RB in fixed synchronized cells from G1 and S/M. In G1, both proteins were detected exclusively in the nucleus (Figures 5B to 5I; see Supplemental Figure 12 online). Moreover, MAT3/RB and DP1 remained in the nucleus during S/M (Figures 5F to 5I). Among thousands of fixed S- and M-phase cells observed by IF, we always saw a strong signal for 3XHA-MAT3 that colocalized with the nuclear DNA and never saw any cytoplasmic staining above background (Figure 5; see Supplemental Figure 12 online; data not shown). Thus, there is no detectable MAT3/RB or DP1 outside the nucleus at any stage of the cell cycle.
MAT3/RB Can Localize to the Nucleus without DP1
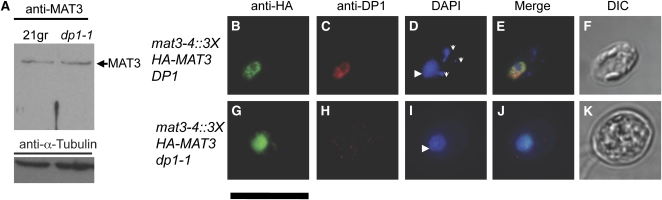
Human and mouse RB contain a bipartite nuclear localization signal in their C-terminal regions that is not conserved in plant and algal RB-related proteins (Zacksenhaus et al., 1993). Using automated prediction programs, we found no obvious nuclear localization signal in MAT3/RB, though such signals are poorly defined for algae and plants. We first tested whether the stability of MAT3/RB is affected by loss of E2F1-DP1 heterodimers and found that its levels did not change in a dp1-1 null mutant strain (Figure 6A). To test whether MAT3 requires the presence of an intact E2F1-DP1 complex for nuclear localization, we performed IF experiments with dp1-1 mat3-4::3XHA-MAT3 cells (Fang et al., 2006) (see Methods). 3XHA-MAT3 IF staining was unchanged in the mutant background, indicating that intact DP1-E2F1 is not required for nuclear localization of MAT3/RB (Figures 6B to 6K).
Figure 6.
MAT3/RB Stability and Nuclear Localization Are Unaffected by Loss of DP1
(A) Immunoblots of whole-cell lysates from the wild type or a dp1-1 strain using MAT3 polyclonal antibodies. The blots were reprobed with an α-tubulin antibody as a loading control (bottom).
(B) to (K) Indirect immunofluorescence microscopy with anti-HA and anti-DP1 antibodies in mat3-4::3XHA-MAT3 DP1 ([B] to [F]) or mat3-4::3XHA-MAT3 dp1-1 ([G] to [K]) cells. HA signal in green ([B] and [G]); DP1 signal in red ([C] and [H]).
(D) and (I) DAPI staining for DNA. Nucleus (larger, dimly staining structure; arrowheads) and chloroplast nucleoids (brighter puncta; vertical arrows) are marked.
(E) and (J) Merged signals.
(F) and (K) DIC images.
MAT3/RB Remains Chromatin Bound throughout the Cell Cycle
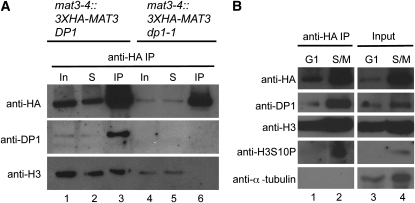
The finding that MAT3/RB is bound to E2F1-DP1 throughout the cell cycle, including during S-phase and mitosis, suggests that the MAT3/RB-E2F-DP complex may also be stably associated with chromatin throughout the cell cycle. To test this possibility, HA-MAT3 was immunoprecipitated from extracts of cells prepared from synchronized cultures at different points in the cell cycle. Prior to IP, the lysates were sonicated to shear DNA into fragments of <500 bp (see Methods). Following IP, we tested for chromatin association by immunoblotting with antibodies that recognize histone H3 (Casas-Mollano et al., 2007) and a conserved mitotic phosphorylation of histone H3 on Ser-10 (S10P) (Crosio et al., 2002). Histone H3 coimmunoprecipitated with HA-MAT3/RB in wild-type cells (Figure 7A, lane 3), but not in dp1 mutants (Figure 7A, lane 6), indicating that MAT3/RB associates with chromatin and requires intact E2F1-DP1 complexes to do so (Figure 7A). The assay for chromatin association of MAT3/RB was repeated with synchronized samples taken from G1 or S/M cultures (Figure 7B). Total Histone H3 coimmunoprecipitated with MAT3/RB at both cell cycle phases (Figure 7B, lanes 1 and 2), whereas H3S10P was detected only in S/M lysates, where it coimmunoprecipitated with MAT3/RB (Figure 7B, lanes 2 and 4). These data indicate that MAT3/RB-E2F1-DP1 complexes remain associated with chromatin during the entire cell cycle.
Figure 7.
MAT3/RB Binds to Chromatin throughout the Cell Cycle and Requires DP1 for Chromatin Binding.
(A) Immunoblot of IP inputs (In), unbound supernatant fraction (S), and anti-HA IPs (IP) from mat3-4::3XHA-MAT3 DP1 and mat3-4::3X-HA-MAT3 dp1-1 probed with antibodies raised to HA (HA-MAT3), DP1, and Histone H3 (H3).
(B) Immunoblot of anti-HA IPs (lanes 1 and 2) and IP inputs (lanes 3 and 4) from G1- and S/M-phase synchronized mat3-4::3XHA-MAT3 lysates probed with antibodies against HA (to detect HA-MAT3), DP1, Histone H3 (H3), Histone H3S10P (H3S10P), and α-tubulin.
DISCUSSION
Identification of an RB-E2F-DP Protein Complex in Chlamydomonas
In previous work, we genetically defined a canonical RB pathway in Chlamydomonas where MAT3/RB acts as a cell cycle repressor and DP1 and E2F1 are downstream cell cycle activators (Fang et al., 2006). Here, we extended this work and found a protein complex in vivo containing MAT3/RB, E2F1, and DP1 that was detected by coimmunoprecipitation and immunoblotting, mass spectrometry, and immunofluorescence. Interactions between the subunits of this complex were confirmed using Y2H and in vitro pull-down assays. MAT3/RB was found to be a nuclear phosphoprotein and was constitutively bound to chromatin, an interaction that required at least the DP1 subunit of E2F1-DP1. Additionally, E2F1 and DP1 from Chlamydomonas have the ability to bind a consensus E2F site as a heterodimer. The pocket protein binding consensus found in the C terminus of animal and some plant E2Fs was not found in Chlamydomonas E2F1 (Mariconti et al., 2002; Vandepoele et al., 2002; Bisova et al., 2005; Fang et al., 2006). However, we found that the C-terminal residues of E2F1 are conserved between Chlamydomonas and Volvox and that this region has some compositional similarity to the RB binding motif in animal and plant E2Fs, being enriched for acidic and aromatic residues (see Supplemental Figure 5 online) (Helin et al., 1992; Ramírez-Parra et al., 1999; Ramírez-Parra et al., 2003). Indeed, this region of Chlamydomonas E2F1 appears be critical for mediating the interaction between MAT3 and E2F1-DP1 complexes as shown by Y2H assays (Figure 4A). Moreover, its partial deletion in the e2f1-4 allele (Fang et al., 2006) (see Supplemental Figure 5 online) appears to abolish the ability of e2f1-4 to coIP with HA-MAT3 (see Supplemental Figure 9 online). The C-terminal region of DP1 did not appear to be important for MAT3 binding (Figure 4A), a finding that is consistent with data from animal and plant RB, E2F, and DP (Ramírez-Parra et al., 1999, 2003; Rubin et al., 2005).
In animals, a second interaction was identified between the C-terminal domain of RB (RB-C) and the extended interface formed by E2F-DP heterodimers that is conserved in Chlamydomonas E2F1 and DP1 (Bisova et al., 2005; Rubin et al., 2005; Fang et al., 2006). Our Y2H results showing a weak binary interaction between MAT3/RB and E2F1 (see Supplemental Figure 7B online), and a stronger interaction when DP1 was present (Figure 4A) is consistent with a two-interface binding mode as previously reported (Rubin et al., 2005). The results from our experiments suggest analogous sets of binding interactions between RB-E2F-DP from Chlamydomonas, animals, and plants and underscore the fundamental similarities between the RB pathways in diverse eukaryotes.
The abundance of the MAT3/RB-E2F1-DP1 increases gradually as cells grow during G1, a property that is found for many proteins in Chlamydomonas that must increase during G1-phase to maintain uniform concentrations (Figure 4B). Therefore, this pattern is a default state that ensures protein homeostasis in cells that can grow in mass by >10-fold during a single cell cycle. On the other hand, MAT3/RB-DP1-E2F1 are nuclear, and their concentration within the nucleus may increase during G1 depending on whether the nuclear volume also increases in proportion to cytoplasmic volume as it does in yeasts (Jorgensen et al., 2007; Neumann and Nurse, 2007). It is possible that nuclear concentration of MAT3/RB-E2F1-DP1 has some role in sensing cell size through the nucleocytoplasmic ratio (Umen, 2005), an idea that merits further testing.
E2F1 and DP1 Form a Conserved DNA Binding Complex
The conservation of DNA binding by E2F1 and DP1 from Chlamydomonas is important in light of previous experiments in which we were unable to identify transcriptional defects in dp1, e2f1, and mat3 mutants, despite their having cell size checkpoint defects (Fang et al., 2006). The question of whether E2F1 and DP1 act primarily as transcription factors in Chlamydomonas remains open, but our finding in this study of conserved DNA and chromatin binding indicate that the function of E2F1-DP1 in Chlamydomonas is likely mediated through association with chromatin (Figure 7).
Moreover, our previous work showing that DP1-E2F1 activity is not essential for cell cycle progression or for transcription of cell cycle genes is now supported by similar findings in mice where cells lacking activator E2Fs (E2F1-E2F3) did not have serious proliferation defects or transcriptional defects for E2F1 targets (Chong et al., 2009). Additional experiments are under way to identify genomic binding sites for MAT3/RB and E2F1-DP1 that may provide information on how these proteins regulate the cell cycle.
Dissociation of MAT3/RB from E2F1-DP1 Is Not Required for Cell Cycle Regulation
In animals, pocket protein complexes have been investigated and appear to lose affinity for E2F-DP heterodimers when phosphorylated by CDKs (Lundberg and Weinberg, 1998; Rubin et al., 2005; Burke et al., 2010). However, there have been reports of intact S-phase RB-E2F complexes (Wells et al., 2003), and DNA damage–induced E2F-dependent transcription can also be mediated by intact RB-E2F complexes (Ianari et al., 2009). The relative contribution of free E2F-DP complexes versus RB-E2F-DP complexes in driving normal cell cycle progression is not completely clear. In plants, the model for RB-E2F-DP interactions has been partly inferred from animal studies (reviewed in Sabelli and Larkins, 2009), but RB-E2F-DP complexes have not been isolated from plants to compare their composition at different cell cycle stages. Here, we present evidence for a stable MAT3/RB-E2F1-DP1 complex that remains intact and bound to chromatin throughout the cell cycle, including S- and M-phases.
Four lines of evidence from this work demonstrate that intact MAT3/RB-E2F-DP complexes regulate the cell cycle without dissociation of the MAT3/RB subunit. First, quantitative coimmunoprecipitation demonstrated that MAT3/RB is bound to E2F1-DP1 at all stages of the cell cycle from highly synchronous cultures (Figure 5A). The amount of E2F1-DP1 that was present in 3XHA-MAT3 IP pellets remained constant, and the relative abundance of DP1 and MAT3/RB at different cell cycle stages was similar, meaning that our IP results were not skewed by an excess of DP1-E2F1 during S/M (Figure 5A). Although S- and M-phase cells cannot be physically separated, S-phase cells are clearly detectable in S/M populations and are not a minor fraction (Coleman, 1982; Craigie and Cavalier-Smith, 1982; Harper and John, 1986; John, 1987; Harper, 1999). Second, our localization experiments indicated that MAT3/RB and DP1 (and by inference, E2F1) colocalize to the nucleus throughout the cell cycle, even in S-phase and mitotic cells (Figures 5B to 5I). Third, we found that MAT3 is increasingly phosphoryated as the cell cycle progresses, with maximum phosphorylation occurring during S/M-phase (Figure 2D). Were E2F1-DP1 released from hyperphosphorylated MAT3, it would have been easily detectable as an increase in unbound/decrease in bound E2F1-DP1 in our IPs from post-Commitment or S/M samples. Instead, our results indicate that phosphorylated MAT3 remains bound to E2F1/DP1 heterodimers with equal stoichiometry at all cell cycle stages (Figure 5). Fourth, we found that MAT3/RB associates with chromatin at all stages of the cell cycle (Figure 7B). These chromatin binding experiments rule out the possibility that MAT3/RB regulates E2F1-DP1 by removing them from chromatin. On the contrary, intact DP1-E2F1 was found to be essential for recruiting MAT3/RB to chromatin as revealed by our inability to coimmunoprecipitate chromatin-associated HA-MAT3 in dp1 mutants (Figure 7A). Whereas it is possible that a small fraction of free E2F1-DP1 escaped detection and is responsible for driving cell cycle progression, a more parsimonious explanation for our data is that the MAT3/RB-E2F1-DP1 ternary complex remains intact and stably bound to chromatin throughout the cell cycle.
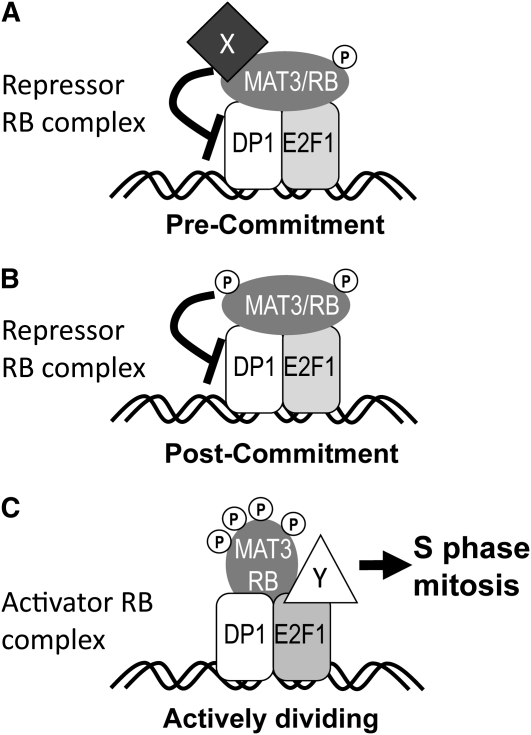
These findings then raise the question of how a stably associated RB-E2F-DP complex might regulate the cell cycle. We propose that it is periodic modification of the complex bound to chromatin that allows cell cycle repression or activation. In this model, phosphorylation or other changes in MAT3/RB, E2F1, and DP1 would change their interactions with effectors that either repress or activate the cell cycle (Figure 8). Our data on cell cycle–regulated phosphorylation of MAT3/RB support this model. Whereas no phospho-MAT3 was detected in pre-Commitment samples, we did see phospho-isoforms in post-Commitment G1 cells several hours prior to the initiation of S-phase, and we detected further phosphorylation during S/M (Figure 2). This model for a stable ternary complex does not contradict the notion that the relative affinities of the subunits change during the cell cycle; indeed, such changes in affinity have been seen in vitro upon CDK phosphorylation of mammalian RB (Zheng et al., 1999; Burke et al., 2010), but altered affinity in vitro does not necessarily imply dissociation in vivo. Our model is consistent with the known association of RB-related proteins with dozens of effectors, including chromatin modifying proteins, replication proteins, transcription factors, and others that may change in their temporal associations with RB-related proteins to regulate either cell cycle progression or differentiation (Williams and Grafi, 2000; Morris and Dyson, 2001; Egelkrout et al., 2002; Shen, 2002; Ausín et al., 2004; Mosquna et al., 2004; Magyar et al., 2005; Ramírez-Parra and Gutierrez, 2007; Burkhart and Sage, 2008; Jullien et al., 2008; van den Heuvel and Dyson, 2008; Chen et al., 2009; Sabelli et al., 2009). Our findings provide a basis for further investigation of RB-E2F-DP complexes and how a stable ternary complex may be modified as a switch to drive cell cycle progression. Of special interest are the post-Commitment and S/M MAT3/RB-E2F1-DP1 complexes whose differences from early G1 complexes should reflect the mechanism by which this regulator is converted from a repressor to a cell cycle activator.
Figure 8.
Model for Cell Cycle Regulation by a Stable Nuclear MAT3/RB-E2F1-DP1 Complex.
(A) MAT3/RB actively represses the cell cycle in pre-Commitment cells while bound to E2F1-DP1, potentially through the recruitment of other repressor proteins signified by protein X. Phosphorylation of MAT3/RB is indicated by gray P symbols.
(B) MAT3/RB-E2F1-DP1 become modified or primed after Commitment, potentially by phosphorylation and/or loss of corepressors.
(C) A conformational change in MAT3/RB-DP1-E2F1 occurs during S/M that allows recruitment of cell cycle activators signified by protein Y binding to E2F1. The conformational change in MAT3/RB may be caused by further phosphorylation during S/M.
METHODS
Chlamydomonas reinhardtii Strains and Culture Conditions
Strains 21gr (wild-type, MT+), 6145 (wild-type, MT-), dp1-1, e2f1-4, and mat3-4 have been described previously (Umen and Goodenough, 2001; Fang et al., 2006). mat3-4::3XHA-MAT3 was constructed as described below and was also crossed to dp1-1 and e2f1-4 to generate dp1-1 mat3-4::3XHA-MAT3 and mat3-4::3XHA-MAT3 e2f1-4. All cultures were grown in high salt medium (Harris, 1989) under ~250 μE light at 24°C in a 14 h:10 h light:dark cycle unless otherwise noted.
Synchronous cultures were monitored for cell size and cell concentration using a Multisizer 3 Coulter Counter (Beckman) and for mitotic progression by phase contrast microscopy. Commitment assays were done as previously described (Umen and Goodenough, 2001; Fang et al., 2006)
Plasmids
The 3XHA-MAT3 plasmid was created as follows. An XbaI recognition site was inserted just after the start codon of plasmid pMAT3.1, a derivative of pMAT3 that contains 439 bp of additional 3′ untranslated region sequence at its 3′ end compared with pMAT3 (Umen and Goodenough, 2001). Three tandem copies of an HA epitope tag (from plasmid mTn-3xHA/GFP; GenBank U54830) were amplified with primers containing SpeI sites at each end, and the SpeI-cut product was ligated into the compatible XbaI site that had been engineered into pMAT3.1. A clone with the 3XHA tag inserted in the correct orientation (p3XHA-MAT3.1, predicted amino acid sequence depicted in Supplemental Figure 2 online) was used to cotransform mat3-4 along with pSI103 that has the paromomycin resistance marker gene aphVIII (Sizova et al., 2001) using glass beads as described previously with selection on TAP agar + 15 μg/mL paromomycin (Umen and Goodenough, 2001). Paromomycin-resistant mat3-4 transformants that were dark green and grew as fast as wild-type colonies were picked and tested for their size distribution and for expression of HA-tagged MAT3. Four independent mat3-4::3XHA-MAT3 complemented lines behaved indistinguishably from wild-type strains with respect to cell size, growth, and cell cycle kinetics and were used for further experiments.
Yeast two-hybrid vectors were created by ligating cDNAs amplified by PCR with oligonucleotides listed in Supplemental Table 2 online, into the NdeI and EcoRI sites of either pGAD-T7 or pGBKT7 (Clontech). The MAT3 cDNA was a gift from Kazusa DNA Research Institute, Japan (Asamizu et al., 2004), and E2F1 and DP1 were previously described (Fang et al., 2006). pBridge-DP1(BD)-E2F1 was created by amplifying the DP1 cDNA (see Supplemental Table 2 online) and cloning into the EcoRI and BamHI sites of pBridge (Clontech) to make pBridge-DP1(BD). E2F1 was amplified (see Supplemental Table 2 online), partially digested with NotI and BglII, and the full-length fragment was gel extracted and ligated into the corresponding sites in pBridge-DP1(BD) to make pBridge-DP1(BD)-E2F1.
pBridge-DP1(BD)-E2F1Δ382 and pBridge-DP1(BD)-E2F1Δ209 were created by amplifying the E2F1 CDS (see Supplemental Table 2 online), digesting with NdeI and BglII, and ligating into NdeI- and BglII-digested pBridge-DP1(BD). pBridge-DP1Δ340(BD)-E2F1 was created by amplifying the DP1 CDS (see Supplemental Table 2 online), digesting with BamHI and EcoRI and ligating to EcoRI- and BamHI-digested pBridge to create pBridge-DP1Δ340(BD). pBridge-DP1(BD)-E2F1 and pBridge-DP1Δ340(BD) were digested with BglII and XbaI, and fragments corresponding to the pBridge with E2F1 and DP1Δ340 were gel purified and ligated together to generate pBridge-DP1Δ340(BD)-E2F1.
pGST-MAT3 was created by ligating MAT3 cDNA from pGAD-MAT3 into EcoRI- and NotI-digested pGEX-4T2 (GE Healthcare). GST-DP1 and GST-E2F were created by a similar procedure with ligation of DP1 and E2F1 to EcoRI- and NotI-digested pGEX-4T2. pETDuet-DP1 was constructed by ligating EcoRI- and XhoI-digested DP1 cDNA from pGEM-T-easy (Fang et al., 2006) to EcoRI- and SalI-digested pETDuet-1 (Novagen). pETDuet-E2F1 was constructed by ligating EcoRI-and HindIII-digested E2F1 from a pGEM-T-easy cDNA construct (Fang et al., 2006) to the EcoRI and HindIII sites in pETDuet-1.
In Vivo Labeling
Strains 6145 (wild-type), mat3-4, and mat3-4::3XHA-MAT3 were grown in TP, pH 7.4 (TAP without acetate; Harris, 1989), that contained 10% of the normal amount of phosphate (0.1 mM) that supported normal growth. Just prior to labeling, cells were resuspended in the same medium containing no additional phosphate and labeled for 30 to 60 min with the addition of 1 mCi of 32PO4H2. Uptake was monitored by comparing input counts to those from supernatants and pellets and was consistently >90%. After labeling, unlabeled phosphate was added to 1 mM for 30 min and the cells pelleted and processed for IP as described below, but without the addition of dithiobis(succinimidyl) propionate (DSP). Radioactive bands corresponding to HA-MAT3 were identified by autoradiography of dried SDS-PAGE gels and in parallel by immunoblots probed with anti-HA antibodies. Gel slices were excised, protein eluted from the gel, and then partially acid hydrolyzed. Hydrolysates were mixed with phosphoamino acid standards and separated by two-dimensional thin layer electrophoresis and stained with ninhydrin before exposing the plate to film (Blume-Jensen and Hunter, 2001). Samples of all labeled cultures were monitored for growth and subsequent cell cycle progression after labeling and were unchanged compared with unlabeled controls. Unsynchronized cultures used for IPs (Figure 2A) were grown in continuous light and had cell cycle distributions of ~20% G1 pre-Commitment, 75% G1 post-Commitment, and 5% S/M. Phosphoamino acid analysis (Figure 2B) was performed on IPs from synchronized mat3-4::3XHA-MAT3 cells that were >85% in S/M.
Recombinant Proteins
Overnight cultures of either GST, GST-MAT3, GST-DP1 and GST-E2F1, pDuet-E2F1, or pDuet-DP1 in Rosetta(DE3) Escherichia coli cells were diluted 1000-fold into fresh Luria-Bertani with 50 μg/mL carbenecillin and grown at 37°C. When cultures reached an OD600 of 0.5, 1 mM isopropyl β-d thiolgalactopyranoside was added to induce recombinant protein expression. After 4 h with isopropyl β-d thiolgalactopyranoside, cells were harvested by centrifugation and dry cell pellets stored at −80°C. Frozen cells were thawed on ice and resuspended in one-tenth original culture volume of EB (100 mM Tris-HCl, pH 8.0, 500 mM NaCl, and 10 mM imidazole) and sonicated eight times for 2 min each on ice with a Branson sonicator at 50% power with a duty cycle of 0.5 s on and 0.5 s off. His-tagged DP1 and E2F1 were purified from inclusion bodies. Lysates were collected by centrifugation at 14,000g for 20 min at 4°C. Pellets from the centrifugation were resuspended in EB and sonicated briefly, after which urea was added to 6 M to solubilize the proteins. Solubilized inclusion bodies were centrifuged at 14,000g for 20 min at 4°C, and the supernatant was applied directly to 1 mL of Ni-NTA agarose (Qiagen) for 1 h at 4°C with continuous inversion. Ni-NTA beads were collected by centrifugation at 5000g for 10 min at 4°C and were washed with WB (100 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10 mM imidazole, and 6 M urea) six times. His-tagged proteins were then eluted in WB with 1 M imidazole.
GST, GST-MAT3, GST-DP1, and GST-E2F1 were purified from the soluble fraction (above) after lysis and centrifugation by binding the supernatant fraction to 1 mL glutathione-Sepharose (GE Healthcare), washing three times with WB lacking urea, and eluting with WB lacking urea and supplemented with 100 mM reduced glutathione.
Prior to electrophoretic mobility shift assay experiments, eluted 6xHis-E2F1 or 6xHis-DP1 were refolded by dialysis into 20 mM Tris-HCl, pH 7.4, and 150 mM NaCl at 4°C. Very little aggregated protein was observed after extensive dialysis, and the refolded proteins bound to each other but not to purified GST (data not shown). Refolded protein was purified by chromatography on either a 10-mL Bio-Rad S10 (6xHis-E2F1) or 10-mL Bio-Rad Q10 (6xHis-DP1) ion exchange column, and protein was eluted by a linear gradient of 0 to 1 M NaCl in 20 mM Tris-HCl, pH 8.0. Protein purity was >95% as assessed by Coomassie Brilliant Blue staining of SDS-PAGE gels from purified fractions.
Electromobility Shift Assay
The DNA oligonucleotide 5′-ATTTAAGTTTCGCGCCCTTTCTCAA-3′ that has a mammalian consensus E2F binding site (underlined) and a mutated version 5′-ATTTAAGTTTCGATCCCTTTCTCAA-3′ (bold residues) were each end labeled and annealed to reverse cDNA oligonucleotides to generate double-stranded probes for binding and mobility shift assays (Carey and Smale, 2000). End labeling was done with 200 pmol of each oligonucleotide using polynucleotide kinase (NEB) and 15 μCi of ATP[γ-32P] at 37°C for 30 min according to the manufacturer’s instructions. Unincorporated label was removed by spin-column gel filtration (Sigma-Aldrich S0185)
Either 5 or 10 pmol of 6xHis-DP1 and 6xHis-E2F1 were mixed in various combinations in 5 mM Tris-HCl, pH 8.0, 150 mM KCl, 25 mM EDTA, 0.1% Triton X-100, 12.5% glycerol, and 1 mM DTT (Carey and Smale, 2000). One microgram of sheared salmon sperm DNA or 5 to 10 pmol unlabeled double-stranded oligonucleotide competitor was mixed with 5 or 10 pmol of labeled double-stranded oligonucleotides and added to the proteins, incubated at 4°C for 30 min, and then loaded directly on a 4% acrylamide TBE gel run at 18 mA for 2 h at 4°C (Carey and Smale, 2000). The gel was then vacuum dried and the radiolabeled probe detected using a phosphor imager (Molecular Dynamics).
Primary Antibodies
The rat monclonal antibody 3F10 (Roche) was used for detection of the HA epitope on immunoblots and for immunofluorescence. Histone H3 and its phospho-Ser10 isoform were detected with antibodies purchased from Cell Signaling Technology (catalog numbers 9715 and 9706, respectively). Tubulin was detected with an anti-α-tubulin antibody purchased from Sigma-Aldrich (clone B-5-1-2, catalog number T6074). Other antibodies were generated using partially purified proteins (GST-MAT3, 6xHis-DP1, or 6xHis-E2F1) that were separated by SDS-PAGE, stained with Coomassie Brilliant Blue R 250, excised from the gel, and eluted. The identity of each recombinant protein was confirmed by tandem mass spectrometry performed by the Salk Institute Proteomics Core Facility. Approximately 2 mg of each protein was submitted for polyclonal antibody production in New Zealand white rabbits (Cocalico).
For DP1 antibody purification, ~20 mg of a Chlamydomonas dp1-1 (Fang et al., 2006) whole-cell lysate was prepared in pH 10.0 coupling buffer (Pierce AminoLink) by bead beating with 0.7-mm zirconium beads (Biospec) at 4°C for 20 min. The lysate was clarified by centrifugation at 20,000g for 30 min at 4°C, and the cleared supernatant was mixed with 1 mL AminoLink agarose beads (Pierce) overnight with constant agitation at 4°C. The reaction was terminated by adding 100 mM ethanolamine. The coupled proteins were washed to remove unbound material using two changes of buffer (100 mM glycine, pH 2.5, 20 mM Tris-HCl, pH 8, 500 mM NaCl, and 5% SDS). Total sera raised against recombinant DP1 was preabsorbed to the dp1-1 matrix overnight at 4°C in PBS, and the unbound fraction was used for affinity purification.
For affinity purification, ~5 mg of purified GST-MAT3, 6xHis-E2F1, or 6xHis-DP1 were coupled to 1 mL of CNBr-activated agarose beads using the manufacturer’s instructions (GE Healthcare) and washed extensively with 1× PBS and 50 mM glycine, pH 2.5, buffer to remove uncoupled protein. Approximately 20 mL of crude DP1 antisera or interference-purified DP1 antisera were precipitated with 40% ammonium sulfate and pelleted by centrifugation for 30 min at 14,000g at 4°C. Precipitated protein was resuspended in 5 mL of 1× PBS and dialyzed overnight against several changes of 1× PBS. Insoluble material was removed by centrifugation for 30 min at 4°C at 14,000g, and the supernatant was then incubated overnight with affinity beads. Unbound material was washed from the affinity beads with 30 column volumes of PBS, and bound IgG was eluted in 50 mM Gly, pH 2.5, and immediately neutralized and concentrated to between 0.3 and 1 mg/mL total protein in PBS. Antibody specificity and titer were tested by immunoblotting to detect recombinant proteins and proteins from whole-cell lysates as described in Results. E2F1 antibody specificity was demonstrated by immunobloting anti-HA IPs from mat3-4::3XHA-MAT3 e2f1-4 and mat3-4::3XHA-MAT3 dp1-1 (see Supplemental Figures 9C and 9D online). Blots were probed with anti-E2F1 antibody, stripped, and reprobed with anti-HA antibody to verify immunoprecipitation of HA-MAT3.
SDS-PAGE and Immunoblotting
SDS-PAGE was performed using standard procedures (Sambrook and Russell, 2001). Phos-Tag SDS-PAGE was performed as described (Kinoshita et al., 2006) by adding 100 μM each of Phos-Tag acrylamide and MnCl2 to standard SDS-PAGE gels. SDS-PAGE gels were blotted to Immobilon-P PVDF membranes (Millipore) in 10 mM Tris, 100 mM Gly, and 10% methanol for 45 min at 400 mA with an Xcel-Blot apparatus (Invitrogen). Phos-Tag gels were incubated for 10 min in transfer buffer with 1 mM EDTA, washed twice for 5 min in water, and then transferred to PVDF (Kinoshita et al., 2006). Membranes were blocked for 30 min in PBS and 5% nonfat dry milk. Blots were probed with primary antibodies diluted in TBS, 5% nonfat dry milk, 0.2% Tween 20, and 0.01% thimerasol for 4 to 12 h at room temperature. Antibodies were used at the following dilutions: 250 μg/mL anti-HA, 1:1000; 1 mg/mL anti-DP1, 1:100; 1 mg/mL anti-MAT3, 1:500; 1 mg/mL anti-E2F1, 1:100; and 2 mg/mL anti-α-tubulin, 1:20,000. Goat anti-rat or anti-rabbit secondary antibodies coupled to horseradish peroxidase (Pierce) were used at 1:2500 dilution and incubated with blots at room temperature in PBS, 5% nonfat dry milk, 0.2% Tween 20, and 0.01% thimerasol for 1 h. Blots were then washed three times with PBS and 0.2% Tween 20 at room temperature for 10 min. Some experiments (Figures 2C, 2D, and 5A; see Supplemental Figures 9C and 9D online) used the SNAP i.d. system for immunoblot processing (Millipore). Blocking was performed with 50 mL of 0.25% nonfat dry milk in TBS with 0.25% Tween 20. The same antibody amount as described above was used, but in a final volume of 3 mL of TBS, 0.25% nonfat dry milk, and 0.2% Tween 20. Blots were incubated for 10 min (anti-HA and secondary antibodies) or 1 h (anti-E2F1 or anti-DP1). After each antibody incubation step, the blots were washed three times with 30 mL of TBS and 0.2% Tween 20 under constant vacuum. Before chemiluminescence, the blots were briefly rinsed with deionized water. Antigen was detected by chemiluminescece (Pierce SuperSignal Pico or Dura for overnight detection) using autoradiographic film (HyperFilm; GE Healthcare).
Phosphatase Experiments
CIP treatment of whole-cell lysates was performed with cells resuspended in 1× NEB buffer 3 (50 mM TrisCl, pH 7.9, 100 NaCl, 10 mM MgCl2, and 1 mM DTT), including protease inhibitors as described above. Cells were divided into three equal parts, and then phosphatase inhibitors (PhI) (50 mM NaF and 50 mM Na2VO4) were added as indicated in Figure 2C. An aliquot of each sample was reserved in 1× SDS-PAGE buffer prior to CIP treatment so that input protein could be assessed (Figure 2C). Dephosphorylation was initiated by adding 200 units of CIP (NEB) to lysates as indicated. The lysates were incubated at 4°C for 30 min and then 37°C for 5 min; SDS-PAGE sample buffer was then added (1× final concentration), and the samples were boiled and fractionated by SDS-PAGE and then immunoblotted.
Immunofluorescence Microscopy
Chlamydomonas cells were harvested by centrifugation at 2500g for 5 min, washed once with PBS, and concentrated 10-fold. Cells were adhered to poly-l-lysine–coated cover slips for 10 min at room temperature, excess liquid was blotted away, and the cells were fixed by immersion into two to three changes of −20°C methanol and 10 mM DTT in prechilled coplin jars. Most experiments were done both with and without fixation of cells in 1% paraformaldehyde for 1 h at 4°C prior to adhesion to cover slips. The results with and without paraformaldehyde fixation were the same. Cover slips were rehydrated in PBS and 0.01% thimerasol overnight at 4°C. Cover slips were then blocked for 30 min in blocking buffer (5% BSA and 1% cold-water fish gelatin) and incubated for 30 min in the same buffer with 10% (v/v) normal goat sera (Antibodies Incorporated). Cover slips were incubated for 1 h in anti-HA and/or anti-DP1 antibodies at 1:750 and 1:50 dilutions for 1 h at room temperature and washed three times with 20% blocking buffer for 10 min each wash. Goat anti-rabbit or anti-rat secondary antibodies conjugated to AlexaFluor 488 or AlexaFluor 568 (Invitrogen) were then diluted in 20% blocking buffer and incubated with the cover slips for 1 h at room temperature in the dark. Following three washes with 20% blocking buffer diluted in PBS, the cover slips were incubated for 5 min in 2 nM 4',6-diamidino-2-phenylindole (DAPI) or a 1:10,000 dilution of Sytox Green (Invitrogen) and then washed for 5 min in PBS. Excess liquid was removed, and the cover slips were mounted with VectaShield (Vector Labs). Microscopy was performed with a DeltaVision Dv3000 deconvolution microscope (Applied Precision) with a ×100 objective, using the following filter sets: DAPI, excitation, 350/40, emission, 457/50; FITC, excitation, 490/20, emission, 528/38; rhodamine, excitation, 555/28, emission, 617/73. Image stacks were acquired with Softworx and subjected to deconvolution. Images in Figures 5 and 6 are average intensity Z-projections produced in ImageJ64 version 1.42 (http://rsb.info.nih.gov/ij/) and are representative of at least three experimental replicates.
Immunoprecipitation
At each time point, an equal number of cells or an equal mass of cells was centrifuged at 5000g in the presence of 0.005% Tween 20. Cell pellets were resuspended in PBS with protease and phosphatase inhibitors (P9599, Sigma-Aldrich; 10 mM PMSF, 10 mM benazmidine, 5 mM EDTA, 5 mM EGTA, 50 μM MG-132, 1 mM NaF, and 1 mM Na2VO4) at a concentration of 109 cells/mL and then cross-linked on ice for 30 min with 1 mM freshly prepared DSP (Pierce 22585). After cross-linking for 5 min on ice, the reactions were quenched by adding Tris-HCl, pH 7.5, to 100 mM. Cells were then immediately flash frozen in liquid nitrogen and stored at −80°C.
Frozen cell pellets were thawed on ice and lysed by sonication on ice with 10% power, and a 0.5 s on, 0.5 s off cycle for 2 min. This sonication procedure was repeated six to eight times. DNA from the lysates was precipitated and gel fractionated to assess shearing efficiency. Prior to immunoprecipitation, protein content was determined with the BCA or Bradford assay (Olson and Markwell, 2007). Immunoprecipitations were performed with either of two methods that produced similar results. Method 1: Lysates were centrifuged at 20,000g for 30 min at 4°C to remove insoluble material, and the supernatant was mixed with 50 μL of Sephadex G-25 (Sigma-Aldrich) for 30 min at 4°C to remove nonspecific agarose binding proteins. The beads were removed by centrifugation at 500g for 5 min at 4°C, and the supernatant was mixed with 10 μL of anti-HA affinity matrix (Roche 3F10 matrix, catalog number 11815016001) for 2 h at 4°C with constant agitation. The beads were collected by centrifugation at 500g at 4°C for 5 min and washed six times with modified RIPA buffer (50 mM HEPES-KOH, 50 mM NaCl, 5 mM EDTA, 5 mM EGTA, 1% Nonidet P-40, 10% glycerol, 1 mM Na2VO4, 100 mM benzamidine, 1 mM NaF, and 1 mM PMSF) for 5 min per wash, and washes were combined with unbound fractions for immunoblotting. After washing, bound proteins were eluted with 50 mM Gly, pH 2.5, for 20 min at 4°C or eluted by directly mixing with SDS-PAGE loading buffer with 100 mM DTT (to reverse the DSP cross-links) at 85°C for 10 min. Method 2: Quantitative IP experiments (Figure 5A; see Supplemental Figure 10 online) were repeated using magnetic beads. In this approach, we omitted cross-absorption of the whole-cell lysate against Sephadex G-25. One microgram of anti-HA antibody (Roche clone 3F10, catalog number 11867423001) was added directly to the lysate and incubated for 2 h at 4°C and then 10 μL of Magna ChIP particles (Millipore, catalog number 16-661) were added and incubated a further 2 h. Beads were collected magnetically in a tube holder for 5 min at 4°C and washed three times. Washes of the magnetic beads were combined with the unbound fractions. IP beads were directly eluted with 1× SDS-PAGE sample buffer as described above for the anti-HA agarose resin. The insoluble fraction created during lysate preparation was resuspended in RIPA with protease and phosphatase inhibitors and sonicated for 30 s at 10% power. Washing was repeated three times, and the supernatant from washing the insoluble fraction was combined with the total/input fraction prior to IP. By the third wash, we found the insoluble fraction to be depleted in protein content (determined by SDS-PAGE and gel staining; Figure 5A, lane 13). For immunoblotting, insoluble fractions from all four cell cycle time points were combined, boiled in SDS-PAGE sample buffer, and loaded proportionally.
Mass Spectrometry
MAT3 IPs were fractionated by SDS-PAGE as described above, and the gel was silver stained (Shevchenko et al., 1996). Gel slices were excised and submitted for mass spectrometric identification of proteins by The Salk Institute proteomics facility. Proteins in gel slices were digested with trypsin and analyzed by liquid chromatography–electrospray ionization tandem mass spectrometry (Shevchenko et al., 1996) in a Bruker Esquire 3000 Plus quadrupole ion trap mass spectrometer (Bruker Daltonics). Tandem mass spectra were searched using the Mascot algorithm (Matrix Science) against the nonredundant National Center for Biotechnology Information database filtered for sequences from plants and version 3 of the predicted Chlamydomonas proteome (archived at http://genome.jgi-psf.org/Chlre4/Chlre4.download.ftp.html). HA-MAT3, E2F1, and DP1 peptides that were identified are reported in Supplemental Table 1 online.
GST Pull-Down Assays
Untagged E2F1 and DP1 were translated in vitro at 30°C for 90 min in the presence of 35S-Met (TNT with T7 RNA polymerase; Promega). Translations were then incubated with purified recombinant GST, GST-E2F1, or GST-DP1 (see above). Reactions were precentrifuged for 10 min at room temperature prior to pull down with glutathione-Sepharose beads (GE Healthcare). Beads were washed six times with PBS 0.2% Tween 20 and eluted by addition of PBS with 100 mM glutathione. Eluted proteins were separated by SDS-PAGE and subjected to autoradiography.
Yeast Two-Hybrid Experiments
Yeast strain Mav203 (Invitrogen) was cultured in synthetic medium and transformed using standard methods as recommended for Y2H assays (Clontech Yeast Protocols Manual). Cotransformed strains were maintained in synthetic dropout medium (SD) lacking Leu and Trp (-LW) supplemented with 2% glucose and yeast nitrogen base without amino acids. To test for His auxotrophy, cells were grown overnight in liquid SD-LW with 2% glucose, normalized to OD 1.0, and serially diluted to OD 0.1 and 0.01 in fresh SD-LW. Two microliters of cells were then spotted onto a series of SD plates lacking Leu, Trp, His (−LWH) or with 10, 20, 25, 30, 35, and 40 mM 3-amino 1,2,3 triazole. Bridged interactions with free E2F1, MAT3-AD, and DP1-DBD were detected with the Clontech pBridge system. Transformants were plated on −LWHM medium (lacking Leu, Trp, His, and Met). To test for uracil auxotrophy, the strains were also spotted on SD plates lacking Leu, Trp, and uracil (−LWU). All yeast strains were maintained and tested at 30°C.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: mTn-3xHA/GFP, GenBank U54830; Chlamydomonas MAT3, GenBank AAK68064; Chlamydomonas incerta MAT3, GenBank AAV41810; Volvox carteri (female) MAT3, GenBank GU784915; Chlorella sp NC64A RBR, Joint Genome Initiative protein ID 57746; Micromonas pusilla CCMP1545 RBR, Joint Genome Initiative protein ID 48829 (http://genome.jgi-psf.org/MicpuC2/MicpuC2.home.html); Arabidopsis thaliana RBR1, GenBank NP_566417; Ostreococcus tauri RB, GenBank AAV68604; Cyanidioschyzon merolae RBR, http://merolae.biol.s.u-tokyo.ac.jp/ ID CMT038C; Chlamydomonas E2F1, DQ417491; Volvox carteri E2F1, XP_002948442; Chlamydomonas DP1, DQ417492; and V. carteri DP1, XP_002955099.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Multiple Fission Cell Cycle of Chlamydomonas.
Supplemental Figure 2. Protein Sequence of 3XHA-MAT3.
Supplemental Figure 3. Detection of 3XHA-MAT3.
Supplemental Figure 4. Multiple Sequence Alignment and CDK Phosphorylation Site Conservation in RB-Related Proteins from Chlamydomonas, Other Algae, and Plants.
Supplemental Figure 5. E2F1 Protein Alignment.
Supplemental Figure 6. DP1 Protein Alignment.
Supplemental Figure 7. Additional Yeast Two-Hybrid Experiments and Controls.
Supplemental Figure 8. GST Pull-Down Using in Vitro–Translated E2F and DP1.
Supplemental Figure 9. DP1 and E2F1 Antibody Specificity.
Supplemental Figure 10. Coimmunoprecipitation of HA-MAT3, E2F1, and DP1 from Equal Numbers of Cells at Different Stages of the Cell Cycle.
Supplemental Figure 11. Specificity of HA Antibody.
Supplemental Figure 12. Nuclear Localization of DP1 and HA-MAT3.
Supplemental Table 1. Tandem Mass Spectrometry Data for HA-MAT3 Immunoprecipitations.
Supplemental Table 2. Oligonucleotides Used to Create Plasmids.
Supplementary Material
Acknowledgments
We thank Alice Kim for laboratory assistance, Cristina Lopez for helpful discussions, and Nancy Benson for administrative support. This work was supported by American Cancer Society Research Scholar Grant RSG-05-196-01-CCG to J.G.U., National Research Service Award Fellowship GM086037 to B.J.S.C.O., a Swiss National Science Foundation fellowship to M.O., National Science Foundation Integrative Graduate Education and Research Traineeship Fellowship 0504645 to J.M.Z., and United States Public Health Service Grant CA-832683 to T.H. T.H. is a Frank and Else Schilling American Cancer Society Professor.
References
- Ach R.A., Taranto P., Gruissem W. (1997). A conserved family of WD-40 proteins binds to the retinoblastoma protein in both plants and animals. Plant Cell 9: 1595–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts A.S., Thorburn A.M., Shenolikar S., Mumby M.C., Feramisco J.R. (1993). Regulation of cell cycle progression and nuclear affinity of the retinoblastoma protein by protein phosphatases. Proc. Natl. Acad. Sci. USA 90: 388–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamizu E., Nakamura Y., Miura K., Fukuzawa H., Fujiwara S., Hirono M., Iwamoto K., Matsuda Y., Minagawa J., Shimogawara K., Takahashi Y., Tabata S. (2004). Establishment of publicly available cDNA material and information resource of Chlamydomonas reinhardtii (Chlorophyta), to facilitate gene function analysis. Phycologia 43: 722–726 [Google Scholar]
- Ausín I., Alonso-Blanco C., Jarillo J.A., Ruiz-García L., Martínez-Zapater J.M. (2004). Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat. Genet. 36: 162–166 [DOI] [PubMed] [Google Scholar]
- Bisova K., Krylov D.M., Umen J.G. (2005). Genome-wide annotation and expression profiling of cell cycle regulatory genes in Chlamydomonas reinhardtii. Plant Physiol. 137: 475–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume-Jensen P., Hunter T. (2001). Two-dimensional phosphoamino acid analysis. Methods Mol. Biol. 124: 49–65 [DOI] [PubMed] [Google Scholar]
- Boniotti M.B., Gutierrez C. (2001). A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J. 28: 341–350 [DOI] [PubMed] [Google Scholar]
- Borghi L., Gutzat R., Fütterer J., Laizet Y., Hennig L., Gruissem W. (2010). Arabidopsis RETINOBLASTOMA-RELATED is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell 22: 1792–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchkovich K., Duffy L.A., Harlow E. (1989). The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell 58: 1097–1105 [DOI] [PubMed] [Google Scholar]
- Burke J.R., Deshong A.J., Pelton J.G., Rubin S.M. (2010). Phosphorylation-induced conformational changes in the retinoblastoma protein inhibit E2F transactivation domain binding. J. Biol. Chem. 285: 16286–16293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart D.L., Sage J. (2008). Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer 8: 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M., Smale S.T. (2000). Electrophoretic mobility shift assays. Transcriptional Regulation in Eukaryotes: Concepts, Strategies and Techniques. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ), pp. 493–496 [Google Scholar]
- Casas-Mollano J.A., van Dijk K., Eisenhart J., Cerutti H. (2007). SET3p monomethylates histone H3 on lysine 9 and is required for the silencing of tandemly repeated transgenes in Chlamydomonas. Nucleic Acids Res. 35: 939–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Hafidh S., Poh S.H., Twell D., Berger F. (2009). Proliferation and cell fate establishment during Arabidopsis male gametogenesis depends on the Retinoblastoma protein. Proc. Natl. Acad. Sci. USA 106: 7257–7262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J.-L., et al. (2009). E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature 462: 930–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobrinik D. (2005). Pocket proteins and cell cycle control. Oncogene 24: 2796–2809 [DOI] [PubMed] [Google Scholar]
- Coleman A. (1982). The nuclear cell cycle in the Chlamydomonas (Chlorophyceae). J. Phycol. 18: 192–195 [Google Scholar]
- Costa L.M., Gutierrez-Marcos J.F. (2008). Retinoblastoma makes its mark on imprinting in plants. PLoS Biol. 6: e212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R.A., Cavalier-Smith T. (1982). Cell volume and the control of the Chlamydomonas cell cycle. J. Cell Sci. 54: 173–191 [Google Scholar]
- Crosio C., Fimia G.M., Loury R., Kimura M., Okano Y., Zhou H., Sen S., Allis C.D., Sassone-Corsi P. (2002). Mitotic phosphorylation of histone H3: Spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 22: 874–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaprio J.A., Ludlow J.W., Lynch D., Furukawa Y., Griffin J., Piwnica-Worms H., Huang C.M., Livingston D.M. (1989). The product of the retinoblastoma susceptibility gene has properties of a cell cycle regulatory element. Cell 58: 1085–1095 [DOI] [PubMed] [Google Scholar]
- Desvoyes B., Ramírez-Parra E., Xie Q., Chua N. (2006). Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol. 140: 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T., Inzé D. (2007). The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 8: 655–665 [DOI] [PubMed] [Google Scholar]
- Dewitte W., Murray J.A. (2003). The plant cell cycle. Annu. Rev. Plant Biol. 54: 235–264 [DOI] [PubMed] [Google Scholar]
- Dimova D.K., Dyson N.J. (2005). The E2F transcriptional network: Old acquaintances with new faces. Oncogene 24: 2810–2826 [DOI] [PubMed] [Google Scholar]
- Donnan L., John P.C. (1983). Cell cycle control by timer and sizer in Chlamydomonas. Nature 304: 630–633 [DOI] [PubMed] [Google Scholar]
- Ebel C., Mariconti L., Gruissem W. (2004). Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429: 776–780 [DOI] [PubMed] [Google Scholar]
- Egelkrout E.M., Mariconti L., Settlage S.B., Cella R., Robertson D., Hanley-Bowdoin L. (2002). Two E2F elements regulate the proliferating cell nuclear antigen promoter differently during leaf development. Plant Cell 14: 3225–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S.-C., de los Reyes C., Umen J.G. (2006). Cell size checkpoint control by the retinoblastoma tumor suppressor pathway. PLoS Genet. 2: e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S.-C., Umen J.G. (2008). A suppressor screen in Chlamydomonas identifies novel components of the retinoblastoma tumor suppressor pathway. Genetics 178: 1295–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P., et al. (2010). Evolution of an expanded sex-determining locus in Volvox. Science 328: 351–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C., Ramírez-Parra E., Castellano M.M., del Pozo J.C. (2002). G(1) to S transition: More than a cell cycle engine switch. Curr. Opin. Plant Biol. 5: 480–486 [DOI] [PubMed] [Google Scholar]
- Harper J. (1999). Chlamydomonas cell cycle mutants. Int. Rev. Cytol. 189: 131–176 [Google Scholar]
- Harper J., John P. (1986). Coordination of division events in the Chlamydomonas cell cycle. Protoplasma. 131: 118–130 [Google Scholar]
- Harris E.H. (1989). The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. (San Diego, CA: Academic Press; ). [DOI] [PubMed] [Google Scholar]
- Helin K., Lees J.A., Vidal M., Dyson N., Harlow E., Fattaey A. (1992). A cDNA encoding a pRB-binding protein with properties of the transcription factor E2F. Cell 70: 337–350 [DOI] [PubMed] [Google Scholar]
- Huntley R., et al. (1998). The maize retinoblastoma protein homologue ZmRb-1 is regulated during leaf development and displays conserved interactions with G1/S regulators and plant cyclin D (CycD) proteins. Plant Mol. Biol. 37: 155–169 [DOI] [PubMed] [Google Scholar]
- Ianari A., Natale T., Calo E., Ferretti E., Alesse E., Screpanti I., Haigis K., Gulino A., Lees J.A. (2009). Proapoptotic function of the retinoblastoma tumor suppressor protein. Cancer Cell 15: 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inze D., De Veylder L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40: 77–105 [DOI] [PubMed] [Google Scholar]
- John P.C.L. (1987). Control points in the Chlamydomonas cell cycle. Algal Development, Wiessner W., Robinson D.G., Starr R.C., (Berlin: Springer-Verlag; ), pp. 9–16 [Google Scholar]
- Johnson U.G., Porter K.R. (1968). Fine structure of cell division in Chlamydomonas reinhardi. Basal bodies and microtubules. J. Cell Biol. 38: 403–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston A.J., Matveeva E., Kirioukhova O., Grossniklaus U., Gruissem W. (2008). A dynamic reciprocal RBR-PRC2 regulatory circuit controls Arabidopsis gametophyte development. Curr. Biol. 18: 1680–1686 [DOI] [PubMed] [Google Scholar]
- Jordan C.V., Shen W., Hanley-Bowdoin L.K., Robertson D.N. (2007). Geminivirus-induced gene silencing of the tobacco retinoblastoma-related gene results in cell death and altered development. Plant Mol. Biol. 65: 163–175 [DOI] [PubMed] [Google Scholar]
- Jorgensen P., Edgington N.P., Schneider B.L., Rupes I., Tyers M., Futcher B. (2007). The size of the nucleus increases as yeast cells grow. Mol. Biol. Cell 18: 3523–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien P.E., Mosquna A., Ingouff M., Sakata T., Ohad N., Berger F. (2008). Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol. 6: e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy B.K., Barbie D.A., Classon M., Dyson N., Harlow E. (2000). Nuclear organization of DNA replication in primary mammalian cells. Genes Dev. 14: 2855–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E., Kinoshita-Kikuta E., Takiyama K., Koike T. (2006). Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteomics 5: 749–757 [DOI] [PubMed] [Google Scholar]
- Kong L.J., Orozco B.M., Roe J.L., Nagar S., Ou S., Feiler H.S., Durfee T., Miller A.B., Gruissem W., Robertson D., Hanley-Bowdoin L. (2000). A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 19: 3485–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (2002). Interaction of the Arabidopsis E2F and DP proteins confers their concomitant nuclear translocation and transactivation. Plant Physiol. 128: 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg A.S., Weinberg R.A. (1998). Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol. Cell. Biol. 18: 753–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacWilliams H., Doquang K., Pedrola R., Dollman G., Grassi D., Peis T., Tsang A., Ceccarelli A. (2006). A retinoblastoma ortholog controls stalk/spore preference in Dictyostelium. Development 133: 1287–1297 [DOI] [PubMed] [Google Scholar]
- Magyar Z., De Veylder L., Atanassova A., Bakó L., Inzé D., Bögre L. (2005). The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 17: 2527–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M., Barbacid M. (2005). Mammalian cyclin-dependent kinases. Trends Biochem. Sci. 30: 630–641 [DOI] [PubMed] [Google Scholar]
- Mancini M.A., Shan B., Nickerson J.A., Penman S., Lee W.H. (1994). The retinoblastoma gene product is a cell cycle-dependent, nuclear matrix-associated protein. Proc. Natl. Acad. Sci. USA 91: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariconti L., Pellegrini B., Cantoni R., Stevens R., Bergounioux C., Cella R., Albani D. (2002). The E2F family of transcription factors from Arabidopsis thaliana. Novel and conserved components of the retinoblastoma/E2F pathway in plants. J. Biol. Chem. 277: 9911–9919 [DOI] [PubMed] [Google Scholar]
- Matsumura K., Yagi T., Yasuda K. (2003). Role of timer and sizer in regulation of Chlamydomonas cell cycle. Biochem. Biophys. Res. Commun. 306: 1042–1049 [DOI] [PubMed] [Google Scholar]
- McAteer M., Donnan L., John P. (1985). The timing of division in Chlamydomonas. New Phytol. 99: 41–56 [Google Scholar]
- McClellan K.A., Slack R.S. (2007). Specific in vivo roles for E2Fs in differentiation and development. Cell Cycle 6: 2917–2927 [DOI] [PubMed] [Google Scholar]
- Menges M., de Jager S.M., Gruissem W., Murray J.A.H. (2005). Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41: 546–566 [DOI] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara K., Cao X.R., Yen A., Chandler S., Driscoll B., Murphree A.L., T’Ang A., Fung Y.K. (1989). Cell cycle-dependent regulation of phosphorylation of the human retinoblastoma gene product. Science 246: 1300–1303 [DOI] [PubMed] [Google Scholar]
- Miller J., Stagljar I. (2004). Using the yeast two-hybrid system to identify interacting proteins. Methods Mol. Biol. 261: 247–262 [DOI] [PubMed] [Google Scholar]
- Mittnacht S., Lees J.A., Desai D., Harlow E., Morgan D.O., Weinberg R.A. (1994). Distinct sub-populations of the retinoblastoma protein show a distinct pattern of phosphorylation. EMBO J. 13: 118–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittnacht S., Weinberg R.A. (1991). G1/S phosphorylation of the retinoblastoma protein is associated with an altered affinity for the nuclear compartment. Cell 65: 381–393 [DOI] [PubMed] [Google Scholar]
- Morgan D.O. (2007). The Cell Cycle: Principles of Control. (London: New Science Press; ). [Google Scholar]
- Morris E.J., Dyson N.J. (2001). Retinoblastoma protein partners. Adv. Cancer Res. 82: 1–54 [DOI] [PubMed] [Google Scholar]
- Mosquna A., Katz A., Shochat S., Grafi G., Ohad N. (2004). Interaction of FIE, a polycomb protein, with pRb: A possible mechanism regulating endosperm development. Mol. Genet. Genomics 271: 651–657 [DOI] [PubMed] [Google Scholar]
- Moulager M., Corellou F., Vergé V., Escande M.-L., Bouget F.-Y. (2010). Integration of light signals by the retinoblastoma pathway in the control of S phase entry in the picophytoplanktonic cell Ostreococcus. PLoS Genet. 6: e1000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H., Kawamura K., Sugisaka K., Sekine M., Shinmyo A. (2002). Phosphorylation of retinoblastoma-related protein by the cyclin D/cyclin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. Plant Cell 14: 1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann F.R., Nurse P. (2007). Nuclear size control in fission yeast. J. Cell Biol. 179: 593–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenhof H., Zachleder V., Van den Ende H. (2007). The cell cycle of Chlamydomonas reinhardtii: The role of the commitment point. Folia Microbiol. (Praha) 52: 23–60 [DOI] [PubMed] [Google Scholar]
- Olson B.J., Markwell J. (2007). Assays for determination of protein concentration. Current Protocols in Protein Science. (Hoboken, NJ: John Wiley & Sons; ), pp. 3.4.1-3.4.29 [DOI] [PubMed] [Google Scholar]
- Prochnik S.E., et al. (2010). Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329: 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Parra E., Fründt C., Gutierrez C. (2003). A genome-wide identification of E2F-regulated genes in Arabidopsis. Plant J. 33: 801–811 [DOI] [PubMed] [Google Scholar]
- Ramírez-Parra E., Gutierrez C. (2000). Characterization of wheat DP, a heterodimerization partner of the plant E2F transcription factor which stimulates E2F-DNA binding. FEBS Lett. 486: 73–78 [DOI] [PubMed] [Google Scholar]
- Ramírez-Parra E., Gutierrez C. (2007). E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol. 144: 105–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Parra E., Xie Q., Boniotti M.B., Gutierrez C. (1999). The cloning of plant E2F, a retinoblastoma-binding protein, reveals unique and conserved features with animal G(1)/S regulators. Nucleic Acids Res. 27: 3527–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbens S., Khadaroo B., Camasses A., Derelle E., Ferraz C., Inzé D., Van de Peer Y., Moreau H. (2005). Genome-wide analysis of core cell cycle genes in the unicellular green alga Ostreococcus tauri. Mol. Biol. Evol. 22: 589–597 [DOI] [PubMed] [Google Scholar]
- Rossi V., Locatelli S., Lanzanova C., Boniotti M.B., Varotto S., Pipal A., Goralik-Schramel M., Lusser A., Gatz C., Gutierrez C., Motto M. (2003). A maize histone deacetylase and retinoblastoma-related protein physically interact and cooperate in repressing gene transcription. Plant Mol. Biol. 51: 401–413 [DOI] [PubMed] [Google Scholar]
- Rubin S.M., Gall A.L., Zheng N., Pavletich N.P. (2005). Structure of the Rb C-terminal domain bound to E2F1-DP1: A mechanism for phosphorylation-induced E2F release. Cell 123: 1093–1106 [DOI] [PubMed] [Google Scholar]
- Sabelli P.A., Dante R.A., Leiva-Neto J.T., Jung R., Gordon-Kamm W.J., Larkins B.A. (2005). RBR3, a member of the retinoblastoma-related family from maize, is regulated by the RBR1/E2F pathway. Proc. Natl. Acad. Sci. USA 102: 13005–13012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli P.A., Hoerster G., Lizarraga L.E., Brown S.W., Gordon-Kamm W.J., Larkins B.A. (2009). Positive regulation of minichromosome maintenance gene expression, DNA replication, and cell transformation by a plant retinoblastoma gene. Proc. Natl. Acad. Sci. USA 106: 4042–4047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli P.A., Larkins B.A. (2009). Regulation and function of retinoblastoma-related plant genes. Plant Sci. 177: 540–548 [Google Scholar]
- Sambrook J., Russell D. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Sekine M., Ito M., Uemukai K., Maeda Y., Nakagami H., Shinmyo A. (1999). Isolation and characterization of the E2F-like gene in plants. FEBS Lett. 460: 117–122 [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Shen W.H. (2002). The plant E2F-Rb pathway and epigenetic control. Trends Plant Sci. 7: 505–511 [DOI] [PubMed] [Google Scholar]
- Sizova I., Fuhrmann M., Hegemann P. (2001). A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277: 221–229 [DOI] [PubMed] [Google Scholar]
- Spudich J.L., Sager R. (1980). Regulation of the Chlamydomonas cell cycle by light and dark. J. Cell Biol. 85: 136–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokke T., Erikstein B.K., Smedshammer L., Boye E., Steen H.B. (1993). The retinoblastoma gene product is bound in the nucleus in early G1 phase. Exp. Cell Res. 204: 147–155 [DOI] [PubMed] [Google Scholar]
- Szekely L., Uzvolgyi E., Jiang W.Q., Durko M., Wiman K.G., Klein G., Sumegi J. (1991). Subcellular localization of the retinoblastoma protein. Cell Growth Differ. 2: 287–295 [PubMed] [Google Scholar]
- Trimarchi J.M., Lees J.A. (2002). Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3: 11–20 [DOI] [PubMed] [Google Scholar]
- Uemukai K., Iwakawa H., Kosugi S., de Uemukai S., Kato K., Kondorosi E., Murray J.A.H., Ito M., Shinmyo A., Sekine M. (2005). Transcriptional activation of tobacco E2F is repressed by co-transfection with the retinoblastoma-related protein: Cyclin D expression overcomes this repressor activity. Plant Mol. Biol. 57: 83–100 [DOI] [PubMed] [Google Scholar]
- Umen J.G. (2005). The elusive sizer. Curr. Opin. Cell Biol. 17: 435–441 [DOI] [PubMed] [Google Scholar]
- Umen J.G., Goodenough U.W. (2001). Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev. 15: 1652–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S., Dyson N.J. (2008). Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell Biol. 9: 713–724 [DOI] [PubMed] [Google Scholar]
- Vandepoele K., Raes J., De Veylder L., Rouzé P., Rombauts S., Inzé D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J., Yan P.S., Cechvala M., Huang T., Farnham P.J. (2003). Identification of novel pRb binding sites using CpG microarrays suggests that E2F recruits pRb to specific genomic sites during S phase. Oncogene 22: 1445–1460 [DOI] [PubMed] [Google Scholar]
- Wildwater M., Campilho A., Perez-Perez J.M., Heidstra R., Blilou I., Korthout H., Chatterjee J., Mariconti L., Gruissem W., Scheres B. (2005). The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123: 1337–1349 [DOI] [PubMed] [Google Scholar]
- Williams L., Grafi G. (2000). The retinoblastoma protein - A bridge to heterochromatin. Trends Plant Sci. 5: 239–240 [DOI] [PubMed] [Google Scholar]
- Xie Q., Sanz-Burgos A.P., Hannon G.J., Gutiérrez C. (1996). Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J. 15: 4900–4908 [PMC free article] [PubMed] [Google Scholar]
- Zacksenhaus E., Bremner R., Phillips R.A., Gallie B.L. (1993). A bipartite nuclear localization signal in the retinoblastoma gene product and its importance for biological activity. Mol. Cell. Biol. 13: 4588–4599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N., Fraenkel E., Pabo C.O., Pavletich N.P. (1999). Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev. 13: 666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.