The authors clone the rice FLOURY ENDOSPERM2 (FLO2) gene; flo2 mutants have aberrant endosperm, and FLO2 overexpressors have enlarged grains. Gene expression and protein interaction studies indicate that FLO2, a novel tetratricopeptide repeat containing protein, regulates storage starch and protein gene expression in rice endosperm development and may also play a role in heat tolerance.
Abstract
Rice (Oryza sativa) endosperm accumulates a massive amount of storage starch and storage proteins during seed development. However, little is known about the regulatory system involved in the production of storage substances. The rice flo2 mutation resulted in reduced grain size and starch quality. Map-based cloning identified FLOURY ENDOSPERM2 (FLO2), a member of a novel gene family conserved in plants, as the gene responsible for the rice flo2 mutation. FLO2 harbors a tetratricopeptide repeat motif, considered to mediate a protein–protein interactions. FLO2 was abundantly expressed in developing seeds coincident with production of storage starch and protein, as well as in leaves, while abundant expression of its homologs was observed only in leaves. The flo2 mutation decreased expression of genes involved in production of storage starch and storage proteins in the endosperm. Differences between cultivars in their responsiveness of FLO2 expression during high-temperature stress indicated that FLO2 may be involved in heat tolerance during seed development. Overexpression of FLO2 enlarged the size of grains significantly. These results suggest that FLO2 plays a pivotal regulatory role in rice grain size and starch quality by affecting storage substance accumulation in the endosperm.
INTRODUCTION
Endosperm is a storage organ in which a massive amount of storage starch and storage proteins are accumulated during seed development. Endosperm development is well characterized in maize (Zea mays). It has been reported that the expression of genes involved in starch and storage protein synthesis of maize endosperm is coordinated and that these genes act in a concerted manner (Giroux et al., 1994). Storage proteins are synthesized at high levels and accumulate in specific organelles, called protein bodies (Shewry et al., 1995). The maize floury mutants (fl1 and fl2) result in soft and starchy endosperm with a reduced amount of prolamin (zein) proteins, which are representative storage proteins in maize endosperm. The responsible gene for the maize fl1 mutation (FL1) encodes an endoplasmic reticulum protein, which participates in protein body formation by facilitating the localization of 22-kD α-zein (Holding et al., 2007). The maize fl2 and defective endosperm B30 mutants have small, misshapen protein bodies, resulting from single amino acid substitutions that cause an uncleaved signal peptide of 22-kD α-zein (Coleman et al., 1997) and 19-kD α-zein (Kim et al., 2004), respectively. The Mucronate mutant, which also has misshapen protein bodies, results from an abnormal 16-kD γ-zein (Kim et al., 2006). Maize opaque2, which encodes a leucine zipper protein, is a regulatory locus that plays an essential role in controlling the expression of genes encoding the 22-kD α-zein proteins (Schmidt et al., 1990, 1992).
Storage starch is synthesized in the amyloplasts, specialized plastids contained in the endosperm (Martin and Smith, 1995). Storage starch in seed endosperm is made up of amylose (linear α-1,4-polyglucans) and amylopectin (α-1,6-branched polyglucans). Starch is synthesized from sucrose after the latter is converted to ADP-glucose. Mutants showing abnormal features of storage starch in the endosperm, such as floury, white-core or opaque kernel phenotypes have been isolated (Satoh and Omura, 1981). In the case of starch accumulation in rice endosperm, key enzymes have been identified previously using mutants showing peculiar features of storage starch. Loss-of-function mutations of the genes encoding for granule-bound starch synthase (GBSS) and branching enzyme IIb (BEIIb) were found to be responsible for the glutinous mutation wx and the amylose extender mutation ae, respectively (Mizuno et al., 1993; Nishi et al., 2001). Other enzymes involved in starch biosynthesis, such as ADP-glucose pyrophosphorylase (AGPase), soluble starch synthase (SS), and branching enzyme I (BEI), have been determined through the similarity to their previously identified maize homologs (Kawasaki et al., 1993; Hirose and Terao, 2004; Kawagoe et al., 2005) and functionally characterized in detail (Nakamura, 2002; Satoh et al., 2003).
Recently, it was reported that natural variation at the DEP1 locus, including a gain-of-function mutation causing truncation of the encoded phosphatidylethanolamine binding protein-like domain protein, enhances grain yield in rice (Oryza sativa; Huang et al., 2009). Mutation of the plastidial phosphorylase (Pho1) gene results in smaller starch granules and in modified amylopectin structure (Satoh et al., 2008). The rice protein kinase SPK phosphorylates sucrose synthase to enhance the degrading activity of sucrose in endosperm and strongly influences the strength of sink productivity (Asano et al., 2002). In addition, it has been shown that high-temperature stress during the grain-filling stage causes deleterious effects on the yield and quality of crop products. Several starch synthesis-related genes, such as GBSS, BEI, and BEIIb, are downregulated in rice grains developed under high-temperature stress, resulting in chalky grain appearance as well as reduction of grain weight (Yamakawa et al., 2007). These reports suggest there is a complex regulatory network working for storage substance accumulation.
Several transcription factors are involved in the expression of the genes for storage substances. A wheat (Triticum aestivum) NAC gene, NAM-B1, is associated with the accumulation of grain protein, zinc, and iron through acceleration of senescence with increases in nutrient remobilization from leaves to developing grains (Uauy et al., 2006). It has been shown that heterodimers of a bZIP transcription factor and a DOF transcriptional factor regulate storage protein gene expression by binding a conserved cis-element and work for the accumulation of storage proteins in the endosperm of many cereals (Pysh et al., 1993; Albani et al., 1997; Vicente-Carbajosa et al., 1997, 1998; Conlan et al., 1999; Onodera et al., 2001). The DOF transcription factor also binds to GAMYB, a key factor of gibberellin signaling, and regulates α-amylase expression in rice and barley (Hordeum vulgare; Diaz et al., 2002; Yamamoto et al., 2006; Kawakatsu et al., 2009). However, little is known about the regulatory system involved in the production of storage substances.
The rice floury endosperm mutants (flo1 to flo5), which have been isolated by N-methyl-N-nitrosourea (MNU) mutagenesis screening, show soft and chalky endosperm (Satoh and Omura, 1981; Kaushik and Khush, 1991). flo4 and flo5 have been identified as mutations of PPDKB (for pyruvate orthophosphate dikinase B) (Kang et al., 2005) and starch synthase IIIa (SSIIIa) (Fujita et al., 2007; Ryoo et al., 2007), respectively. The flo2 mutant exhibits extremely reduced levels of BEI expression in developing rice endosperm along with decreased levels of other starch-synthesizing enzymes, including AGPase, GBSS, SS, and BEIIb (Kawasaki et al., 1996).
Many features of rice grains are changed by high-temperature environment during seed development, and in some cases, the grains will display a chalky texture, milky appearance, and lower weight (Yamakawa et al., 2007). Recently, we found that the gene responsible for the rice flo2 mutant is also essential for the high-temperature resistant trait of a rice cultivar, suggesting that the character of the flo2 endosperm is a phenocopy of the effects of high temperature on rice grain development (She et al., 2010). In this article, we report the identification of the gene responsible for the flo2 mutation, which encodes a functionally unknown protein involved in a novel mechanism affecting grain size and quality control.
RESULTS
Rice flo2 Mutants Produce Aberrant Storage Substances
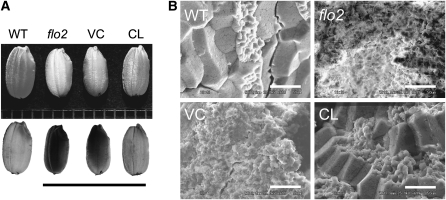
To determine the effects of the flo2 mutation, we first conducted a detailed examination of the mutant phenotype. The flo2 mutants showed no apparent differences in visible features during the vegetative stage; plant height, the number of leaves, the number of tillers, and the number of panicles were similar to those of wild-type plants. No significant difference was found in the timing of flowering. The number of panicles per plant, the number of spikelets per panicle, and the ratio of ripened grains did not significantly differ from those of the wild-type plant. The flo2 grains had white and floury endosperm (Figure 1A), and scanning electron microscopy images of transverse sections of flo2 grains indicated that this endosperm was filled with loosely packed, small, and spherical starch granules with large air spaces, while the wild-type endosperm consisted of densely packed, large, and irregularly polyhedral starch granules (Figure 1B).
Figure 1.
Phenotype of the flo2 Mutant.
(A) Representative seed of the wild type (Kinmaze), the flo2 mutant (EM37), vector control line (VC), and complemented line (CL). CL and VC are transformants with/without a wild-type FLO2 gene in the flo2 mutant EM37. The top panel shows the grain shapes of the seeds, and the bottom panel shows seeds illuminated with backlight. The floury phenotype (EM37 and VC) is indicated by a dark image. Wild-type and CL grains were more packed and transmitted light, whereas the flo2 mutant and VC grains were chalky and floury, resulting in dark images when illuminated with backlight. Bar = 1 cm.
(B) Scanning electron microscopy images of the transverse sections of the wild-type, flo2 mutant, VC, and CL grains. Bars = 50 μm.
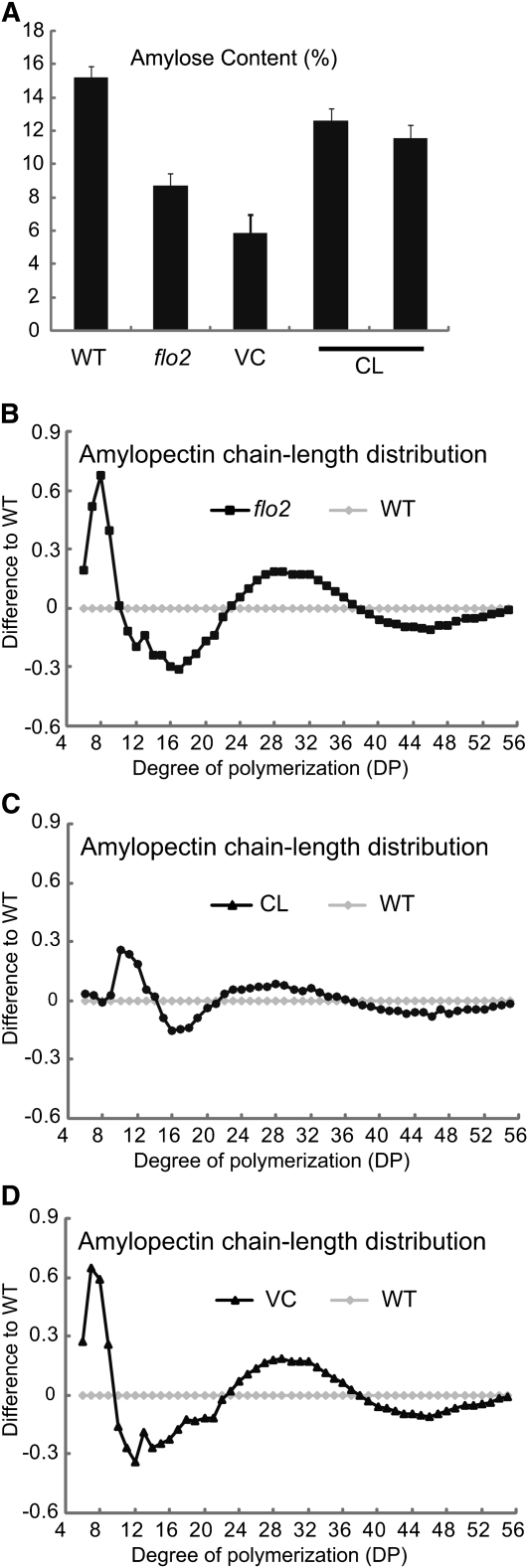
Grain size was significantly smaller than that of the wild type (89% on average, n > 40, P < 0.01), as were both grain weight and grain length. In the endosperm of the flo2 mutant, the amylose content was lower than that of the wild type (Figure 2A), and structural changes in amylopectin were also noted, with both the short and the long chains consisting of 9 to 21 degrees of polymerization (DP) and ≥38 DP, respectively, decreasing and the middle chains with 22 to 38 DP increasing (Figure 2B).
Figure 2.
Amylose Content and Amylopectin Composition of the flo2 and Wild-Type Grain.
(A) Amylose content of the wild type (Kinmaze), flo2 mutant (EM37), VC, and CL. Representative VC and CL were analyzed. Error bars show sd (n = 6).
(B) to (D) Comparison of the chain-length profile of amylopectins. Debranched amylopectin was compared with the wild type. Differences from the wild type in chain length distribution of amylopectins are shown. Amylopectin profile of the flo2 grain (B), CL (C), and VC (D).
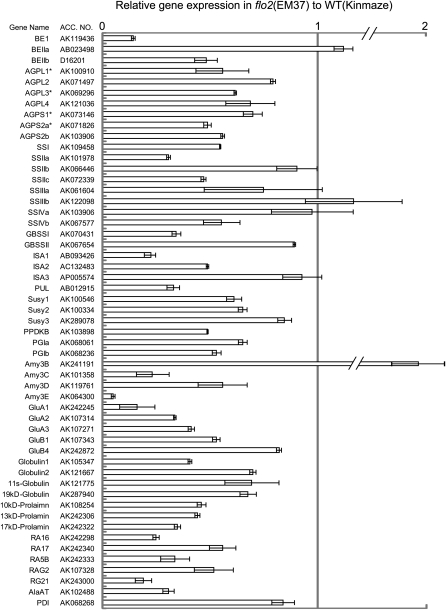
Real-time quantitative RT-PCR indicated that expression levels of many genes that participated in starch biosynthesis were significantly decreased, such as genes for BEI, BEIIb, AGPases (AGPL1, AGPL2, AGPL3, AGPL4, AGPS1, AGPS2a, and AGPS2b), soluble SSs (SSI, SSIIa, SSIIc, SSIIIa, and SSIVb), GBSSI, isoamylases (ISA1 and ISA2), and pullulanase (PUL). Expression of genes for SSIIb, SSIIIb, SSIVa, GBSSII, BEIIa, and ISA3 was little decreased (Figure 3). Gene expression for sucrose synthase (Susy1 and Susy2), PPDKB, glucose-6-phosphate isomerases (PGIa and PGIb), and α-amylase (Amy3C, Amy3D, and Amy3E) was also reduced, while expression of the genes for Susy3 and Amy3B was little decreased (Figure 3). The expression of storage protein genes encoding glutelins (GluA1, GluA2, GluA3, and GluB1), globulins (Globulin1, Globulin2, 11S-globulin, and 19-kD globulin), prolamins (10 kD, 13 kD, and 17 kD), major allergenic protein genes (14 to 16 kD; RA16, RA17, RAG5B, RAG2, and RG21), and the alanine aminotransferase gene was also significantly reduced, whereas expression of GluB4 and protein disulfate isomerase was little decreased (Figure 3). In short, the flo2 mutant showed reduced expression of a large portion of the genes participating in storage starch and storage protein biosynthesis in developing rice seeds.
Figure 3.
Expression Levels of the Genes Involved in Production of Storage Starch and Protein in the flo2 Mutant.
Total RNA extracted from 10 DAF developing seeds was used for real-time RT-PCR analysis. Expression of representative genes involved in storage starch production, storage proteins, and carbon metabolism in the flo2 mutant (EM37) are shown relative to the wild type (Kinmaze), which is set as 1. Each gene name is indicated by a simplified representation with the accession number of the corresponding full-length cDNA. BE1, BEIIa, and BEIIb, branching enzyme I, IIa, and IIb, respectively; AGPL (AGPL1, AGPL2, AGPL3, and AGPL4) and AGPS (AGPS1, AGPS2a, and AGPS2b), ADP glucose pyrophosphorylase large subunit and small subunit, respectively; SSI, SSIIa, SSIIb, SSIIc, SSIIIa, SSIIIb, SSIIIc, SSIVa, and SSIVb, soluble starch synthase I, IIa, IIb, IIc, IIIa, IIIb, IIIc, IVa, and IVb, respectively; GBSSI and GBSSII, granule-bound starch synthase I and II; ISA1, ISA2, and ISA3, corresponding isoamylase isozymes; PUL, pullulanase; Susy1, Susy2, and Susy3, sucrose synthase 1, 2, and 3, respectively; PPDKB, pyruvate phosphate dikinase B; PGI-a and PGI-b, glucose-6-phosphate isomerase a and b; Amy3B, Amy3C, Amy3D, and Amy3E, α-amylase 3B, 3C, 3D, and 3E, respectively; GluA1, GluA2, GluA3, GluB1, and GluB4, glutelin A1, A2, A3, B1, and B4, respectively; Globulin1, Globulin2, 11S-globulin, and 19-kD globulin, corresponding globulin species; 10-, 13-, and 17-kD prolamin, prolamins with each size; RA16, RA17, RAG5B, RAG2, and RG21, species of rice allergenic proteins; AlaAT, alanine aminotransferase; PDI, protein disulfate isomerase. Asterisks show subunits of the plastidic AGPase. Error bar shows sd (n = 3).
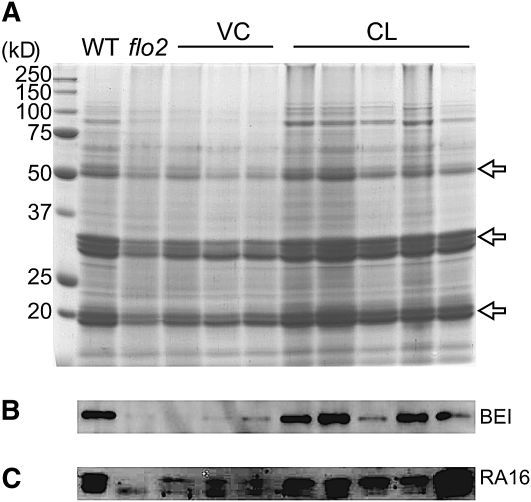
We next set out to determine whether there was also an alteration of protein accumulation in the flo2 mutants. In the flo2 grains, reduced accumulation of glutelin, a representative storage protein, was observed (Figure 4A), suggesting decreased production of storage proteins. Protein gel blot analyses also detected an extreme reduction in the amount of BEI and of major allergenic proteins, as well as of RA16 and homologs, in the grain of the flo2 mutant (Figures 4B and 4C). These results suggest that the flo2 mutation reduced the production of storage substances in total in the rice endosperm.
Figure 4.
Accumulation of Glutelins, BEI, and RA16 in Developing Seeds.
(A) Accumulation of glutelins in developing seeds of the wild type (Kinmaze), flo2 mutant (EM37), VC, and CL. Each lane contains proteins from one-hundredth of the total glutelin fraction, which was extracted from the mixture of 10 rice grains of each line. Proteins are shown by Coomassie blue staining after separation on SDS-PAGE. Arrows indicate 51-kD glutelin precursor, 30- to 36-kD α-subunits of glutelins, and 19- to 22-kD β-subunits of glutelins, respectively. Size markers are shown on the left.
(B) and (C) Accumulation of BEI (B) and RA16 (C) in developing seeds 10 DAF of the wild type, flo2 mutant, VC (three independent lines), and CL (five independent lines) is shown by protein gel blot analysis using antiserum raised against BEI and RA16, respectively. The RA16 antiserum detects highly conserved 14- to 16-kD allergenic proteins with slightly different molecular mass. Each lane contains 1 μg of total protein, which was separated by SDS-PAGE.
Determination of the Gene Responsible for the flo2 Mutation
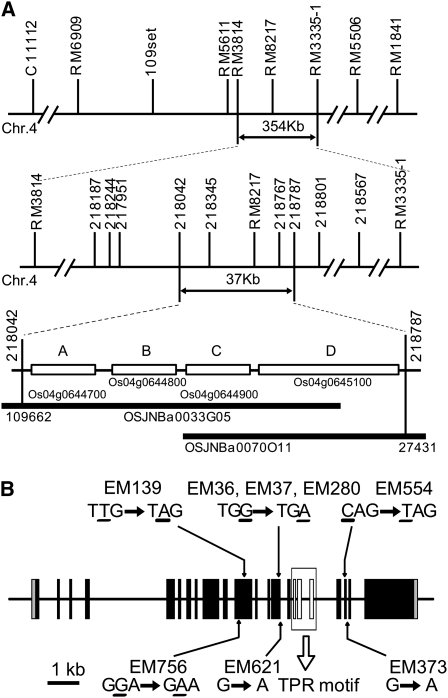
We then set out to identify the gene responsible for the flo2 mutation by map-based cloning using F2 progenies of a cross between an flo2 mutant, EM37, which was made by treatment with MNU, and an Indica cultivar, Kasalath. Approximately 4000 F2 lines were developed and analyzed. The flo2 phenotype segregated 3:1 in the F2 generations, suggesting that the gene responsible for the flo2 phenotype was a single locus in the genome. Using 634 individual F2 lines, a coarse map of the locus was created. The candidate gene was initially mapped to a location of ~354 kb in a 111-centimorgan region of chromosome 4 and then narrowed down to a 37-kb region by fine mapping using genetic markers that were newly established based upon the polymorphic DNA sequences between the parental lines (Figure 5A).
Figure 5.
Positional Cloning and Structure of Rice FLO2.
(A) Fine mapping of the FLO2 locus. The region of the FLO2 locus was mapped to a 354-kb region by published EST/SSR markers (RM3814 and RM3335-1), then narrowed to a 37-kb region by newly created markers (218042 and 218787) in chromosome 4 (Chr. 4), which contained four predicted genes shown by the gene names registered in the database. Markers used for the mapping are shown in the figure and are detailed in Supplemental Table 2 online.
(B) Exon/intron structure of FLO2. The FLO2 gene is composed of 23 exons (filled box) with 22 introns, including three TPR motifs (open box) in the middle. Eight mutants showing the flo2 phenotype contained one base substitution in the FLO2 gene that generated a premature stop codon (EM36, EM37, EM139, EM280, and EM554), a presumable splicing site (EM373 and EM624), or an amino acid change (EM756).
This region contained four predicted genes, and subsequent nucleotide sequence analysis of the 37-kb region revealed a G-to-A nucleotide substitution in the flo2 mutant in one of the candidate genes, Os04g0645100, while no mutation was found in other regions. This mutation generated a stop codon in the predicted reading frame of the corresponding gene (Figure 5B) and was detected in all tested F2 lines homozygous for the recessive flo2 phenotype.
To verify that this locus was responsible for the flo2 phenotype, we complemented flo2 mutants with transgenic lines harboring a complete wild-type copy of the Os04g0645100 gene. These complemented lines (CL) set seeds of normal size and shape. Scanning electron microscopy images of transverse sections of CL grains show rugged surfaces as observed in those of the wild-type endosperm, which consisted of densely packed polyhedral-shaped starch granules (Figure 1). The amylose content was also mostly restored to wild type in CL (Figure 2A), the chain length distribution profile of amylopectin shifted toward that of the wild type (Figure 2). CL grains accumulated levels of glutelins, BEI, and RA16 in the endosperm that were similar to the wild type (Figure 3). These results indicate that the loss-of-function mutation of this gene causes the phenotype of the flo2 mutation. Consequently, we identified the Os04g0645100 gene as the gene responsible for the flo2 mutation, FLOURY ENDOSPERM2 (FLO2).
We determined the extent of the FLO2 transcript. FLO2 was predicted to contain 23 exons with 22 introns. Although the full-length mRNA from the FLO2 gene was predicted to have more than 5163 nucleotides, current databases only contained partial sequences for the FLO2 mRNA (AK072140 and AK100454, corresponding to the N-terminal region of the middle region and the middle region to the C-terminal region of the predicted open reading frame [ORF], respectively). We found that the partial end sequences of four clones in the unidentified rice full-length cDNA pool, which had been constructed by the Rice Genome Resource Center (Tsukuba, Japan), showed the corresponding nucleotide sequence of the predicted full-length ORF. Subsequently, we determined the entire nucleotide sequence of these cDNAs and confirmed that all four clones encoded the full-length predicted amino acid sequence deduced from the FLO2 gene.
The FLO2 gene was predicted to encode a protein composed of 1720 amino acid residues, in the middle region of which were three repeats of a tetratricopeptide repeat (TPR) motif, lying at the positions of 933 to 966, 975 to 1008, and 1017 to 1050 amino acid residues (Figure 5B). In other parts, no homology was found to any proteins with known function.
In addition, seven flo2 alleles isolated from the MNU mutagenesis screen also contained mutations in the FLO2 gene (Figure 5B). Four alleles were found with point mutations that generated a stop codon in the ORF: EM139, EM36, EM280, and EM554. In EM373 and EM621, a mutation was found at the junction of a splicing site, suggesting that it may interfere with the precise removal of the intron. EM756 had a point mutation causing a Gly (GGA) to Glu (GAA) substitution. All of these flo2 alleles produced similar phenotypes of floury grains with chalky endosperm, as observed in the EM37 grains, suggesting that these nucleotide substitutions caused the flo2 phenotype (Figure 5B).
FLO2 and Its Homologs Constitute a Novel Gene Family Unique to Plants
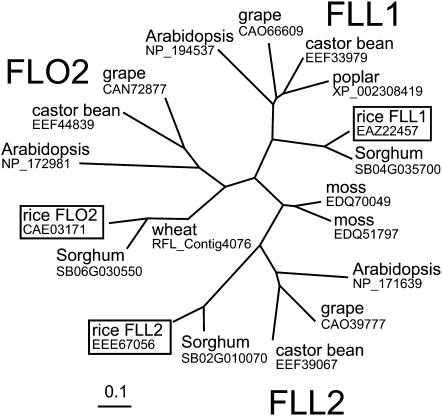
To determine whether FLO2 was part of a gene family, we looked for similar genes in the rice genome. We found two homologous genes, designated FLO2-LIKE1 (FLL1) (Os02g0255700) and FLL2 (Os07g0422000). The predicted proteins encoded by FLL1 and FLL2 showed 59.8 and 52.7% similarity in amino acid sequence, respectively, to FLO2. Orthologous genes to FLO2 and its homologs were also found in other plant genomes, such as in wheat, sorghum (Sorghum bicolor), poplar (Populus spp), grape (Vitis vinifera), castor bean (Ricinus communis), Arabidopsis thaliana, and moss (Physcomitrella patens) (Figure 6), but not in animals or yeast. These homologs in higher plants were classified into three groups, which contained FLO2, FLL1, and FLL2, respectively, whereas homologs of moss diverged from this group. These results suggest that FLO2 constitutes a novel conserved gene family in plants.
Figure 6.
Phylogenetic Tree of the Representative FLO2 Homologs.
Proteins are shown in the figure as the names of plant species with the corresponding accession numbers registered in protein databases (http://www.ncbi.nlm.nih.gov/ and http://www.ddbj.nig.ac.jp/index-j.html). Subfamilies including FLO2, FLL1, and FLL2 are indicated. Rice genes are boxed. Scale represents the number of differences between sequences.
FLO2 Interacts with Late Embryogenesis Abundant and Basic Helix-Loop-Helix Proteins
To identify the interaction partners of FLO2, we screened a cDNA library from developing rice seeds at 10 to 15 d after flowering (DAF) with a yeast two-hybrid (Y2H) system. We used bait plasmids producing FLO2 protein fragments [FLO2(N), FLO2(M), and FLO2(C)] that correspond to regions between the N terminus and amino acid residue 898, the region between amino acid 535 and 1189, and the region between amino acid 900 residue and the C terminus, respectively [FLO2(1–898), FLO2(535–1189), and FLO2(900–1720)].
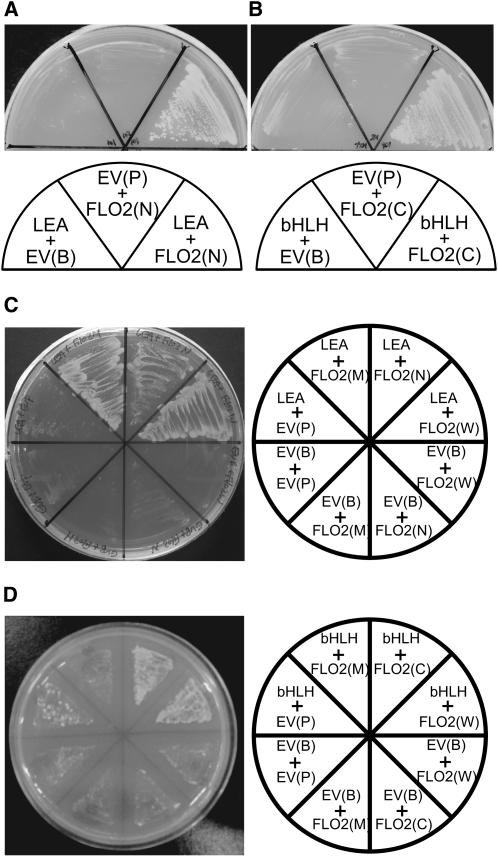
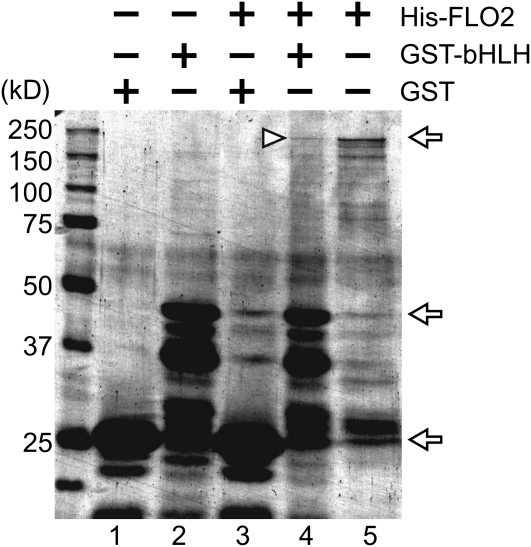
Positive clones obtained by the first screening were further screened for growth on selective media, resulting in 29 candidates showing strong affinity for FLO2 (Table 1). Eight out of 29 clones contained cDNA for late embryogenesis abundant (LEA) proteins, and seven corresponded to a putative basic helix-loop-helix (bHLH) protein (Table 1, Figures 7A and 7B). To confirm the interaction between these LEA and bHLH proteins and FLO2, we performed a reciprocal swapping Y2H experiment. The clones containing the cDNA for the full-length FLO2 on the prey plasmid, along with each of the cDNAs for the LEA protein and the bHLH as the bait plasmid, grew on the strict selection media plates, suggesting that these proteins may interact with the entire FLO2 protein. We found that the clones expressing the LEA protein in combination with FLO2(N) or FLO2(M), but not FLO2(C), were able to grow on the selection media (Figure 7C). This result suggested that the LEA protein interacted with the region containing amino acids 535 to 898 in the FLO2 protein. As for the bHLH protein, clones expressing the full-length FLO2 [FLO2(W)] or FLO2(900–1720) were able to grow in combination with the bHLH protein on the selection media (Figure 7D), suggesting that the bHLH protein interacted with FLO2 in the region lying between amino acids 1189 and the C terminus (Table 1). The interaction between the full-length FLO2 and the bHLH protein was also confirmed by an in vitro pull-down experiment (Figure 8).
Table 1.
Candidates for Interactors with FLO2
| Redundancyc |
|||||
| Accession No.a | Annotationb | Total | N | M | C |
| AK061818 | Late embryogenesis abundant protein | 8 | 5 | 3 | 0 |
| AK070651 | bHLH transcription factor | 7 | 0 | 0 | 7 |
| AK121079 | Complex 1 family protein/LVR family protein | 2 | 0 | 0 | 2 |
| AK067257 | Bowman-Birk–type bran trypsin inhibitor | 2 | 0 | 0 | 2 |
| AK068710 | Carboxyvinyl-carboxyphosphonate phosphorylmutase | 2 | 0 | 0 | 2 |
| AK121590 | Polyubiquitin | 1 | 1 | 0 | 0 |
| AK061085 | Phospho-2-dehydro-3-deoxyheptanate aldolase | 1 | 0 | 0 | 1 |
| AK069237 | β-Tubulin | 1 | 0 | 0 | 1 |
| AK072707 | K+ channel protein | 1 | 0 | 0 | 1 |
| AK106782 | rcd1-like cell differentiation protein | 1 | 0 | 0 | 1 |
| AK110542 | β-1,3-Glucanase | 1 | 0 | 0 | 1 |
| AK062356 | Unknown | 1 | 0 | 0 | 1 |
| AK101806 | β-Expansin | 1 | 0 | 1 | 0 |
| Total | 29 | 6 | 4 | 19 | |
The predicted gene products were listed as the candidate clones by the Y2H screening.
The corresponding full-length cDNA in the database, in which the isolated cDNA was contained.
Gene names identified from the database or by similarity.
Number of clones contained in the pool of candidates isolated from Y2H screening using each part of truncated FLO2 corresponding to FLO2(1–898), FLO2(535–1189), and FLO2(900–1720), which are shown by N, M, and C, respectively. The total number of these clones on each candidate is shown in the column “Total.”
Figure 7.
Interaction of LEA and bHLH with FLO2.
(A) and (B) Y2H analysis of FLO2 with a LEA protein (accession number AK061818) (A) and a bHLH protein (accession number AK070651) (B). LEA, bHLH, FLO2(N), and FLO2(C) indicate yeast cells containing plasmids for expression of the LEA, bHLH, FLO2(1–898), and FLO2(900–1720), respectively. EV(B) and EV(P) show those containing the empty bait and prey vectors instead of the corresponding genes, respectively.
(C) and (D) Reciprocal swapping using LEA and FLO2 (C) and bHLH and FLO2 (D). FLO2(W) and FLO2(M) indicate yeast cells containing plasmids for expression of the full-length FLO2 and FLO2(535–1189), respectively. These cells were grown on agar plates lacking Trp, Leu, and His in the medium.
Figure 8.
Detection of the FLO2–bHLH Interaction by an in Vitro Pull-Down Experiment.
Proteins on SDS-PAGE were detected by Coomassie blue staining. Lanes 1 to 5 indicate GST, GST-fused bHLH, 6xHis tagged FLO2 pulled down with GST, 6xHis tagged FLO2 pulled down with GST-fused bHLH, and 6xHis-tagged FLO2 without GST-fused bHLH, respectively. Arrows indicate the size of 6xHis-tagged FLO2 (215 kD), GST-fused bHLH (48 kD), and GST (25 kD). The triangle indicates the predicted position of the 6xHis-tagged FLO2 bound with GST-fused bHLH. Size markers and their molecular sizes are shown on the left.
Spatial Expression Patterns of the FLO2 Gene
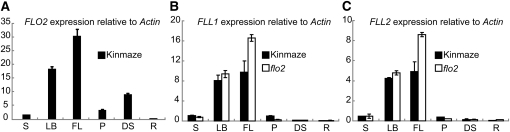
We analyzed the spatial expression of FLO2, FLL1, and FLL2 by real-time RT-PCR. The transcript of FLO2 was abundant in immature seeds and mature leaves, but not abundant in panicles before heading, stems, and roots (Figure 9A). Both FLL1 and FLL2 were highly expressed in leaves but poorly expressed in developing seeds (Figures 9B and 9C).
Figure 9.
Expression Pattern of FLO2, FLL1, and FLL2.
(A) Expression of FLO2 in the stem (S), leaf blade (LB), flag leaf (FL), panicles before heading (P), 10 DAF developing seed (DS), and root (R). Amounts of transcripts are shown as the relative values to those of Actin I. Error bar shows sd (n = 3).
(B) and (C) Expression level of FLL1 (B) and FLL2 (C) in the stem (S), leaf blade (LB), flag leaf (FL), panicles before heading (P), 10 DAF developing seed (DS), and root (R). Closed box, wild type (Kinmaze); open box, flo2 mutant (EM37). Amounts of transcripts are shown as the relative values to those of Actin I. Error bar shows sd (n = 3).
Transgenic Overexpression of the FLO2 Gene
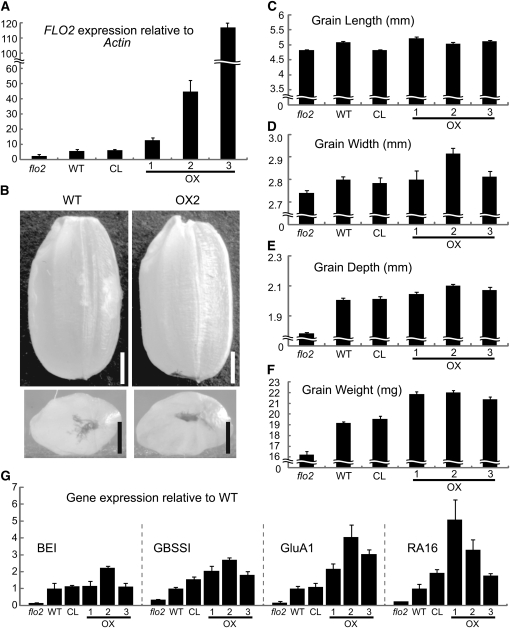
We created transgenic CP lines harboring the entire wild-type FLO2 gene with its own promoter in the flo2 mutant background. Among 82 CP lines, we defined FLO2-overexpression lines (designated OX) as those that have more than double the amount of FLO2 transcripts compared with the wild type. OX1, OX2, and OX3 had 2.0-, 4.3-, and 14.1-fold amounts of FLO2 transcripts compared with the wild type, respectively (Figure 10A). We measured grain length, width, depth, and weight of mature grains and found that the flo2 mutants produced shorter, narrower, thinner, and lighter grains, while the OX lines produced larger and heavier grains compared with the wild type (Figures 10B to 10F). Student’s t tests confirmed a statistically significant difference in grain weight of OX lines (P < 0.001) from the wild type. These facts suggest that increased FLO2 expression may enlarge grain size by expanding the width and depth and, hence, increasing grain weight. The expression levels of the representative genes involved in storage starch (BEI and GBSSI), storage proteins (GluA1), and 14- to 16-kD allergenic proteins (RA16) also increased in the OX lines (Figure 10G).
Figure 10.
Transgenic Lines Overexpressing FLO2.
(A) Expression level of FLO2 in the developing seed at 20 DAF of the flo2 mutant (EM37), the wild type (Kinmaze), a representative CL, and three overexpression lines (OX) in which the wild-type FLO2 gene including its own promoter was introduced in the flo2 mutant. The number of each OX line is indicated. The amount of the FLO2 transcript was determined by real-time RT-PCR and shown as relative values of the FLO2 transcript to those of rice Actin I. Error bar shows sd (n = 3).
(B) Shapes of the grains of the wild type and OX (line 2). The top panel indicates the pictures of grains. The bottom panel shows the grains of the wild type (Kinmaze) and the OX line from a style side. Bars = 0.1 cm.
(C) to (F) Averaged grain length (C), grain width (D), grain depth (E), and grain weight (F) of these lines. Error bar shows sd (n = 20).
(G) Expression levels of representative genes in the flo2 mutant, wild-type, CL, and OX lines. Total RNA extracted from 10 DAF developing seeds was used for real-time RT-PCR analysis. GluA1, glutelin A1; RA16, a 16-kD rice allergenic protein. Expression levels of the genes are shown normalized to the wild type, which is set at 1. Error bar shows sd (n = 3).
Temporal Expression of the FLO2 Gene and Its Response to the High-Temperature Stress during Seed Development
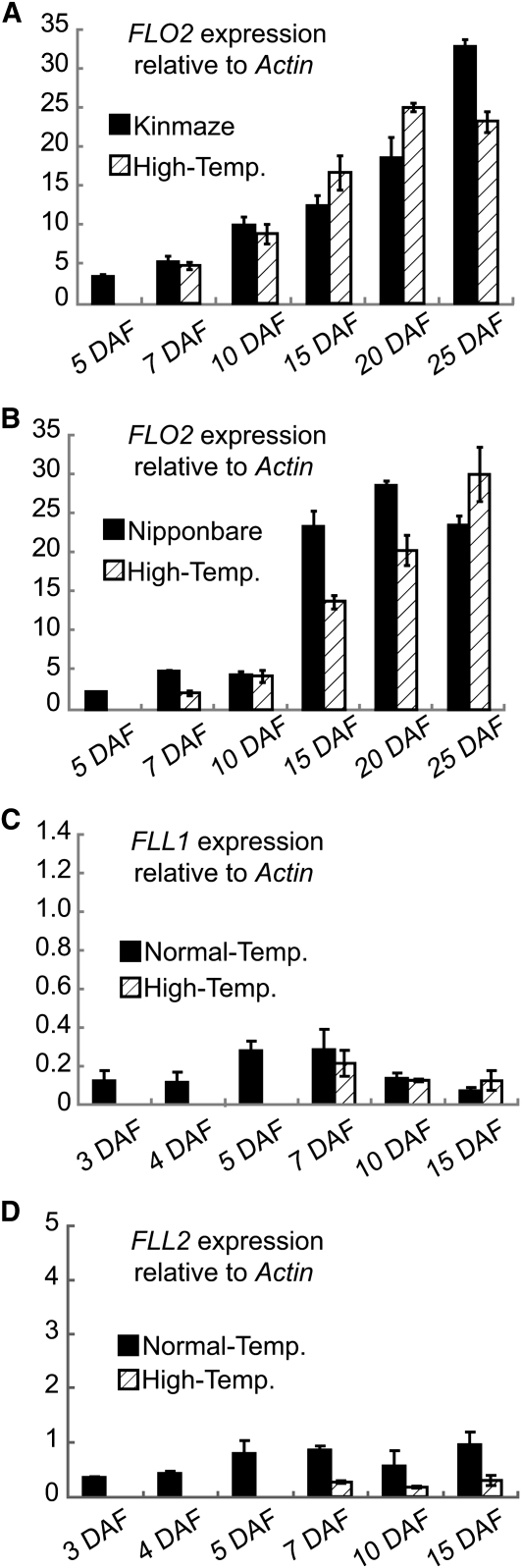
It has been shown that the Nipponbare rice cultivar is very sensitive to high-temperature stress, leading to reduced grain quality and productivity, while Kinmaze, the wild-type cultivar for the flo2 mutant, is relatively tolerant to high-temperature stress during seed development (She et al., 2010). We analyzed the temporal expression pattern of FLO2 in these cultivars during seed development under normal and high-temperature conditions. To determine the response of FLO2 expression to high-temperature stress, plants were transferred into the high-temperature environment at 5 DAF, and the seeds were ripened in the high-temperature conditions. Under the normal environmental conditions, both Nipponbare and Kinmaze showed FLO2 expression that gradually increased as seed development progressed (Figures 11A and 11B). Under high-temperature conditions, developing seeds of Nipponbare showed a significant reduction in FLO2 transcript levels at 15 and 20 DAF when high levels of FLO2 expression were shown in the normal condition (Figure 11B). By contrast, FLO2 expression in developing Kinmaze seeds increased at 15 and 20 DAF under high-temperature stress (Figure 11A). The amounts of FLL1 and FLL2 transcripts remained at a very low level in developing seeds through the seed development stages in high-temperature conditions (Figures 11C and 11D).
Figure 11.
Expression of FLO2, FLL1, and FLL2 during Seed Development in Normal and High-Temperature Environments.
(A) and (B) Expression of FLO2 in Kinmaze (A) and Nipponbare (B) during seed development at 5, 7, 10, 15, 20, and 25 DAF. Closed box, the wild type (Kinmaze and Nipponbare, respectively) under the normal temperature condition; hatched box, the wild type treated with high-temperature stress after 5 DAF. Amounts of transcripts are shown normalized to Actin I, which is set at 1. Error bars show sd (n = 3).
(C) and (D) Expression levels of FLL1 (C) and FLL2 (D) during seed development at 3, 4, 5, 7, 10, and 15 DAF. Closed box, the wild type (Kinmaze) under the normal temperature conditions; hatched box, the wild type treated with high-temperature stress after 5 DAF. Amounts of transcripts are shown normalized to Actin I, which is set at 1. Error bars show sd (n = 3).
DISCUSSION
Storage starch and storage proteins accumulate rapidly during seed development. It is predicted that there are some regulatory systems that control endosperm development (Sabelli and Larkins, 2009), but currently little is known about this regulation. We focused on FLO2, which may coregulate expression of the genes participating in starch synthesis in developing seeds (Kawasaki et al., 1996). Our results indicate that FLO2 is involved in regulation of production of the storage proteins as well as storage starch in rice endosperm.
Our map-based cloning method successfully identified the FLO2 locus on chromosome 4 of the rice genome (Figure 5). We found that seven independently isolated flo2 alleles had point mutations in the candidate gene, Os04g0645100. Furthermore, the phenotype of the flo2 mutation was efficiently complemented by introduction of the wild-type Os04g0645100 gene. These results indicate that the Os04g0645100 gene is indeed responsible for the flo2 mutation.
The FLO2 gene belongs to a gene family that consists of FLO2, FLL1, and FLL2 in the rice genome. Orthologs of these genes widely exist in plant species, including mosses, but none was found in yeast or animals. The phylogenetic tree indicates that there are three major clades for higher plants, each of which consists of the orthologs of FLO2, FLL1, and FLL2 (Figure 6). The interspecies similarity between members is larger than the similarities among FLO2, FLL1, and FLL2 within a species. These facts suggest that this gene family was acquired in an ancestor plant and diverged within the plant kingdom. The FLO2 homologs in moss (P. patens) form an independent branch in the phylogenetic tree, suggesting that the moss FLO2 family members may have evolved independently.
FLL1 and FLL2 have similar protein structures to FLO2, and presumably have similar functions, but have somewhat different expression patterns. FLO2 was expressed in organs such as leaves and developing seeds (Figure 9A) and is therefore assumed to function in these organs. However, the flo2 mutant showed no apparent phenotype other than in seeds. In both the wild type and the flo2 mutant, a normal level of expression of both FLL1 and FLL2 was observed in leaves, whereas they were poorly expressed in developing seeds. It is presumed that the function of FLO2 is specific to developing seeds.
Expression of the genes involved in storage starch production was greatly reduced in the flo2 mutant (Figure 3). The molecular structure of the storage starch accumulated in flo2 endosperm was also quite different from that of the wild type. Phenotypes similar to those of the flo2 mutant are found in several mutants of the enzymes involved in starch biosynthesis, but many differences are evident in their features. For example, the amylopectin properties of the flo2 grain largely resemble those of the mutant lacking BEI, although the endosperm of this mutant seed exhibits a normal phenotype and contains the same amount of starch as the wild type (Satoh et al., 2003). The flo2 mutant showed a decrease of BEI protein (Figure 4), suggesting a reduction of BEI activity. Therefore, it is reasonable to state that the reduced BEI activity in the flo2 mutant greatly contributed to the aberrant structure of storage starch. The mutant of BEIIb also produces a white-core endosperm, but its amylopectin structure differs greatly from that of the flo2 mutant where the long side chains contained in its amylopectin are highly enriched in the BEIIb mutant (Nishi et al., 2001). Amylose is mainly synthesized by the activity of GBSS (Wang et al., 1995), suggesting that the decrease of amylose content could be attributed to the reduced expression of GBSSI in the flo2 mutant. It has been shown that AGPase activity strongly affects the yield of grain starch (Kawagoe et al., 2005). ISA1 and PUL, both of which have activities as starch debranching enzymes, are essential for the formation of the highly organized clustered structure of amylopectin (Kubo et al., 1999). Therefore, aberrant features of storage starch in the flo2 endosperm are ascribed to the combined effect of reduced expression of these genes.
The flo2 mutant also showed significantly decreased expression of storage protein genes. In addition, decreased expression was detected for several enzymes involved in metabolism (Figure 3). In particular, Susy is considered a key enzyme for the production of storage starch as well as cellulose synthesis (Chourey et al., 1998). PPDK has been reported as the gene responsible for the rice flo4 mutant, which has a similar appearance to the flo2 mutant (Kang et al., 2005). PGI is important for both glycolysis and gluconeogenesis by catalyzing the reversible isomerization of glucose-6-phosphate to fructose-6-phosphate (Grauvogel et al., 2007). Alanine aminotransferase is involved in the efficiency of nitrogen uptake and grain weight increase (Shrawat et al., 2008). These facts suggest that FLO2 widely affects the processes involved in storage substance accumulation in rice endosperm.
There are several reports on the regulatory factors involved in the production of storage substances in endosperm (Sabelli and Larkins, 2009). It has been suggested that there are some regulatory factors that directly or indirectly control the genes involved in the individual enzymes or genes encoding the storage proteins. In the case of direct regulation of gene expression, specific regulatory factors, which interact with the specific nucleotide sequences in the promoter region of the target gene, are proposed (e.g., Onodera et al., 2001; Marzabel et al., 2008).
FLO2 has a TPR motif composed of tandem repeats of 34 amino acid residues. The TPR motif adopts a helix-turn-helix structure and may mediate protein–protein interactions (D’Andrea and Regan, 2003; Chadli et al., 2008). We detected two candidate proteins, an LEA protein and a bHLH protein, that interact with FLO2 (Table 1, Figures 7 and 8); however, our results indicate that neither protein interacts with the TPR motif region of FLO2, suggesting that additional interactors may remain to be identified. The LEA proteins, constituting a large gene family in rice (Hundertmark and Hincha, 2008), and bHLH proteins are involved in many functions, including a multiple stress response through transcriptional regulation (Ogo et al., 2007), although their individual functions remain unclear. The flo2 mutant showed reduced expression of multiple genes involved in storage starch and storage proteins (Figures 3 and 4). Therefore, it is suggested that FLO2 might interact with the bHLH protein in a transcriptional complex that could regulate storage starch and storage proteins directly or indirectly through influencing the development of endosperm in rice.
In the flo2 mutant, we detected little alteration in the expression of several genes participating in storage starch and storage protein biosynthesis, such as BEIIa, GBSSII, and GluB4 (Figure 3). This result indicates that several genes involved in production of the storage substance may not be regulated by FLO2. Expression profiles of genes involved in starch biosynthesis have been analyzed. Expression patterns of the genes participating in accumulation of storage starch differ depending on the individual genes; GBSSI, SSI, SSIIa, SSIIIa, SSIVb, BEI, BEIIb, AGPL2, and AGPS2b genes are specifically expressed in the endosperm of developing rice seeds at 10 to 20 DAF, whereas GBSSII, SSIIb, SSIIc, SSIVa, BEIIa, AGPL1, and AGPS1 genes are expressed in leaves as well as developing seeds (Hirose et al., 2006). We observed that expression of all genes in the former group was significantly reduced in the flo2 mutants, whereas little decrease in expression levels was shown in most genes of the latter group (Figure 3). It has been reported that the patterns of gene expression in seeds are tissue and developmental stage specific. Among the genes working in the endosperm, GBSSI, SSIIa, SSIIIa, BEI, BEIIb, AGPS2b, AGPL2, ISA1, and PUL genes, which have transcripts that are low at onset and then rise steeply at the start of starch synthesis in the endosperm, are thought to play essential roles in endosperm starch synthesis at the middle stage of seed development (Ohdan et al., 2005). These genes corresponded to the genes specifically expressed in the endosperm of developing seeds reported by Hirose et al. (2006). Our observation revealed that FLO2 expression was highly elevated at 15 to 25 DAF of seed development (Figure 11). Consequently, it is suggested that FLO2 selectively affects the expression of these genes, whose expression patterns were consistent with that of the FLO2 gene.
It has been reported that high-temperature stress during seed development apparently reduces the accumulation of storage starch in rice grains (Yamakawa et al., 2007). In the case of Nipponbare, which is a rice cultivar sensitive to high-temperature stress, high temperature during seed development resulted in significant reduction of FLO2 expression, while FLO2 was expressed most abundantly under normal ripening conditions. The timing of elevation of FLO2 expression seemed to be delayed for several days in response to high-temperature stress (Figure 11B). This result suggests that FLO2 expression was apparently repressed by high-temperature stress in this cultivar. On the other hand, in Kinmaze, which is relatively tolerant to high-temperature stress, it seemed that FLO2 expression was upregulated by high-temperature stress (Figure 11A). These facts support the idea that FLO2 plays an important role in the trait for tolerance to high-temperature stress (She et al., 2010).
FLO2 was abundantly expressed in immature seeds (Figures 9 and 11), and its expression pattern was similar to that of genes involved in the production of storage starch and storage proteins. This temporal expression may account for the appearance of the loss-of-function phenotype observed in the endosperm of the flo2 mutant seeds, such as reduced production of these storage substances. While the flo2 mutant produced smaller grains, the OX lines had larger seeds than those of the wild type (Figures 2 and 10), exhibiting the gain-of-function phenotype of the FLO2 gene. The increase in gene expression for enzymes involved in starch biosynthesis, however, is not directly linked to the level of FLO2 expression. As a matter of fact, the increases in some of the parameters of grain size measured are barely significant (Figure 10). This could be attributed to the fact that expression of the analyzed genes was measured at a single time point and that the impact of FLO2 may not coincide with its expression level at that time. This work suggests that FLO2 has a pivotal role in the accumulation of storage substances in the endosperm and sets the foundation for the understanding of grain size and quality regulation in rice.
METHODS
Plant Materials
flo2 mutants were isolated following chemical mutagenisis of the Japonica rice (Oryza sativa) cultivar, Kinmaze, by MNU (Satoh and Omura, 1981). Nipponbare is a Japonica rice cultivar that has been determined to be sensitive to the high-temperature stress during seed development, while Kinmaze is relatively tolerant (She et al., 2010). Seeds were germinated at 30°C in a dark chamber and grown in pots in the summer season at our growth area under the natural open-air condition until flowering. After flowering, the rice plants were cultivated in the growth chamber with 12-h-light (28°C) and 12-h-dark (25°C) condition. For the preparation of grains developed under high-temperature stress, plants were temporarily moved to a growth chamber and grown during 5 to 15 DAF with 70% humidity under a light cycle with 12 h light (33°C) and 12 h dark (28°C), as described previously (Yamakawa et al., 2007).
PCR and Real-Time Quantitative RT-PCR
Genomic DNA was extracted according to the method of Murray and Thompson (1980). PCR was performed using Blend-taq thermo-stable DNA polymerase (Toyobo). Total RNA was extracted from each tissue as described previously (Imamura et al., 2007). The first-strand cDNA was synthesized from 1 μg of total RNA using a ReverTra-Ace cDNA synthesis kit (Toyobo) with an oligo-dT(20) primer. Real-time quantitative PCR was performed using the ABI PRISM 7000 sequence detection system (Applied Biosystems) with an SYBR Green Real-time PCR mix (Toyobo). The value of Actin I mRNA (accession number AK100267) was used for data normalization as a positive control. Primer pairs used for PCR are listed in Supplemental Table 1 online.
Scanning Electron Microscopy Analysis
Scanning electron microscopy observation was performed as described previously (Imai et al., 2006). Rice samples were transversely cut by knife and put on the stage of a HITACHI S-3500N scanning electron microscope to take images. All procedures were according to the manufacturer’s protocol.
Analysis of Amylose Content and Determination of the Chain Length of α-1,4-Glucan in Amylopectin
The apparent amylose content of rice seeds was measured using an iodine colorimetric method (Juliano, 1971). The chain length distribution of amylopectin was determined using high-performance anion-exchange chromatography with pulsed amperometric detection, after being hydrolyzed by Pseudomonas amyloderamosa isoamylase. Side chain lengths in amylopectin and other grain properties were characterized as previously described (Yamakawa et al., 2007).
Protein Extraction, SDS-PAGE, and Protein Gel Blot Analysis
Proteins were extracted from the powdered seeds as described previously (Asano et al., 2002). SDS-PAGE and protein gel blot analysis were performed according to Ausubel et al. (1987). In this experiment, 10 grains of each line were combined and used for preparation of total proteins. The total rice glutelin fraction, including the glutelin precursor proteins, α-subunits of glutelin, and β-subunits of glutelin, was prepared from the total proteins according to Robert et al. (1985). For detection of BEI and RA16 by protein gel blot analysis, a rabbit antiserum raised against these proteins was used. RA16 antiserum was provided by K. Kadowaki. BEI antiserum was prepared using a fusion protein of rice BEI (accession number AK65121) with glutathione S-transferase (GST) that was expressed using an Escherichia coli expression system (GE Healthcare Life Sciences).
Genome Mapping of the flo2 Locus and DNA Sequence Analysis
Approximately 4000 F2 seeds were obtained from F1 seeds crossed between the flo2 mutant EM37 (a Japonica cultivar background) and Kasalath (an Indica cultivar). Among them, 639 recessive flo2 lines were used for genome mapping according to Shen et al. (2004). Fine mapping of the flo2 locus was performed by PCR on sequence tag sites using DNA markers, which were previously and newly established primers based on the nucleotide polymorphisms in the corresponding regions between the Japonica and Indica cultivars. Previously established public markers can be found on Rice Genome Research Program website (http://rgp.dna.affrc.go.jp/whoga/index.html). Newly established markers are listed in Supplemental Table 2 online.
Complementation of the flo2 Mutant
A 14,355-nucleotide genomic fragment of the wild-type plant corresponding to the rice Os04g0645100 gene that contained the region between the position of 1037 nucleotide upstream the position of the initiation codon and the position of 538 nucleotide downstream the stop codon was cloned into a binary vector, pGWB1 (provided by T. Nakagawa), in which the native promoter of the FLO2 gene as well as the region for the entire ORF encoding the predicted protein was expected to be included. It was then introduced into the flo2 mutant, EM37, using an Agrobacterium tumefaciens–mediated method (Hiei et al., 1994). Transformants were grown in a greenhouse. Expression of the FLO2 gene was determined by real-time RT-PCR using RNA prepared from immature seeds.
Phylogenetic Analysis
FLO2 protein amino acid sequences were aligned using ClustalX program from the Clustal website (http://www.clustal.org/). The phylogenetic tree was constructed using Phylip (Felsenstein, 2005) neighbor-joining method with bootstrap values from 1000 neighbor-joining bootstrap replicates. The tree was visualized using the program TreeView (Page, 1996).
Yeast Two-Hybrid Analysis
Construction of a cDNA library from developing rice seeds (10 DAF) and detection of interacted proteins were performed using the Matchmaker Library Construction and Screening Kit (Takara Bio) and Matchmaker GAL4 Two-Hybrid System 3 (Takara Bio) following the manufacturer's instruction. Yeast cells were grown on three kinds of media plates prepared using Yeast Nitrogen Base without amino acids (BD Bioscience) supplemented with appropriate amino acids lacking Leu or Trp for the clone harboring a prey plasmid or a bait plasmid (−1 plate); His and Leu (−2 plate); His, Leu, and Trp (−3 plate) for 1st to 3rd screening, respectively. After the 3rd screening, the candidate clones were inoculated on the −3 plate to form colonies. To isolate the truly positive clones, this operation was performed five times, and the resultant clones were used for further analysis. In the case of the reciprocal swapping experiment, a prey plasmid and a bait plasmid were cointroduced into yeast cells and inoculated on the −3 plate to form the transformant colonies. pGADT7 and pGBKT7 (Takara Bio) were the vector plasmids for the prey plasmid and the bait plasmid, respectively, and also used as negative controls for the experiment.
Following cDNA fragments encoding three types of truncated FLO2 [FLO2(1–898), FLO2(535–1189), and FLO2(900–1720)] were prepared by PCR amplification using the full-length cDNA clone J09B0024C03, which was kindly provided by the Rice Genome Resource Center (Tsukuba, Japan) using the primer pairs listed in Supplemental Table 3 online. FLO2(1–898) was encoded by the fragment corresponding to the nucleotide position between the initiation codon of the predicted ORF and the 2694-nucleotide position downstream the initiation codon, FLO2(535–1189) was corresponding to the middle part of FLO2 between the positions of 1603 nucleotides and the 3567 nucleotides downstream the initiation codon in which the TPR motif is contained, and FLO2(900–1720) was corresponding to the last 2466 nucleotides upstream the stop codon. These fragments were used for construction of the bait plasmids. Clones expressing the full-length FLO2 [FLO2(W)], FLO2(1–898) [FLO2(N)], FLO2(535–1189) [FLO2(M)], and FLO2(900–1720) [FLO2(C)] were used for the analysis.
To confirm the results of the screening, prey plasmids containing the predicted full-length bHLH protein and the LEA protein were constructed, whose fragments were prepared by amplification from Nipponbare genomic DNA and the cDNA (accession number AK061818), respectively, by PCR using primer pairs listed in Supplemental Table 3 online, and cloned into pGADT7.
For the reciprocal swapping Y2H experiment, the prey plasmids containing the full-length FLO2, which was amplified using the primers FLO2-Head-BamHI and FLO2-Tail-XhoI (see Supplemental Table 3 online), as well as those for the truncated FLO2 fragments. The bait plasmids were constructed using the fragments for the bHLH protein and the LEA protein, which were amplified from the cDNAs (accession numbers AK070651 and AK061818) using primer pairs listed in Supplemental Table 3 online, and cloned into pGBKT7.
In Vitro Pull-Down Experiment
The 6xHistidine-tagged (6xHis-tagged) FLO2 protein and the GST-fused bHLH protein were used for the in vitro pull-down experiment. The gene for the 6xHis-tagged FLO2 protein was constructed using the DNA fragment encoding the full-length FLO2, which was amplified using primer set 5'-CGGATCCGTTGGGTTTGGAGCTTCA-3′ and 5'-CTTGTCGACAGCTGACAGAAGTGCAAA-3′ from the cDNA clone J09B0024C03 and inserted between BamHI and SalI sites in pET28a (Merck). The 6xHis-tagged FLO2 protein was purified using pET28a expression system (Merck). The gene for the GST-fused bHLH protein was constructed using the DNA fragment encoding the full-length bHLH protein, which was amplified from rice genomic DNA using the primers 5'-GGTGAGGGTGGAATTCATGGCGAAGGTGC-3′ and 5'-CCCGTCGACTCACCTCTCCATCTCGGTCTC-3′ and inserted between EcoRI and SalI sites of pGEX6p1 (GE Healthcare Life Sciences). The GST-fused bHLH protein was purified using the pGEX6p1 expression system (GE Healthcare Life Sciences). The 6xHis-tagged FLO2 protein was applied to the Glutathione Sepharose resin (GE Healthcare Life Sciences), and then the GST-fused bHLH protein was added to the resin. As the control experiment, GST was added instead of the GST-fused bHLH protein. The resin was washed with the buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM EDTA, 10 mM MgCl2, 1% Triton X-100, and 2% glycerol), and subsequently proteins were eluted by boiling the resin in the buffer (60 mM Tris-HCl, pH. 6.8, 2% SDS, 10% glycerol, and 1.5% DTT). The eluted fraction was analyzed by SDS-PAGE.
Accession Numbers
Sequence data from this article can be found in the Rice pipeline (http://cdna01.dna.affrc.go.jp/PIPE/index_ja.html) or GenBank/EMBL databases under the following accession numbers: FLO2 (Os04g0645100, CAE03171), FLL1 (Os02g0255700, EAZ22457), and FLL2 (Os07g0422000, EEE67056). Accession numbers for sequences used for RT-PCR and phylogenetic analysis can be found in Supplemental Table 2 online and Figure 6, respectively.
Author Contributions
H. Satoh created the flo2 mutant; K-C.S., H.K., and T. Aoyama determined characteristics of the mutant; and K-C.S., M.F., T. Ando, and M. Yano mapped and cloned the gene responsible. K-C.S., H.K., K.K., S.K., N.K., J.Y., and M. Yano performed expression analysis and database arrangement; K-C.S., H.K., T.I., N.N., Y.T., M. Yaeshima, K.M., E.I., and H. Shimada characterized FLO2; and H.Y. and M.H. analyzed grain starch. K-C.S., H.K., M.K., M. Yano, and T.S. performed complementation of the gene, and K-C.S., H.K., T.T., and H. Shimada designed the study, analyzed data, and wrote the article. K-C.S. and H.K. contributed equally to the study. All authors discussed the results and commented on the manuscript.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Primer Sequences Used for Real-Time RT-PCR.
Supplemental Table 2. Markers Used for FLO2 Mapping.
Supplemental Table 3. Primer Sequences Used for Yeast Two-Hybrid Experiments.
Supplemental Data Set 1. Text File of Alignment Corresponding to Phylogenetic Tree in Figure 6.
Supplementary Material
Acknowledgments
We thank K. Kadowaki for antibodies and K. Ono for technical assistance in generating transgenic plants. This work was supported by grants from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genome for Agricultural Innovation, MA2226, QTL-2006, and IPG-0022), the Program for Development of Strategic Research Center in Private University supported by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT), and a Grant-in-Aid for Scientific Research from MEXT (Grant 21570050) to H. Shimada.
References
- Albani D., Hammond-Kosack M.C., Smith C., Conlan S., Colot V., Holdsworth M., Bevan M.W. (1997). The wheat transcriptional activator SPA: A seed-specific bZIP protein that recognizes the GCN4-like motif in the bifactorial endosperm box of prolamin genes. Plant Cell 9: 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T., et al. (2002). Rice SPK, a calmodulin-like domain protein kinase, is required for storage product accumulation during seed development: phosphorylation of sucrose synthase is a possible factor. Plant Cell 14: 619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent R., Kingston R.E., Moore D.M., Seidman J.G., Smith J.A., Struhl K.E. (1987). Current Protocols in Molecular Biology. (New York: John Wiley & Sons; ). [Google Scholar]
- Chadli A., Bruinsma E.S., Stensgard B., Toft D. (2008). Analysis of Hsp90 cochaperone interactions reveals a novel mechanism for TPR protein recognition. Biochemistry 47: 2850–2857 [DOI] [PubMed] [Google Scholar]
- Chourey P.S., Taliercio E.W., Carlson S.J., Ruan Y.L. (1998). Genetic evidence that the two isozymes of sucrose synthase present in developing maize endosperm are critical, one for cell wall integrity and the other for starch biosynthesis. Mol. Gen. Genet. 259: 88–96 [DOI] [PubMed] [Google Scholar]
- Coleman C.E., Clore A.M., Ranch J.P., Higgins R., Lopes M.A., Larkins B.A. (1997). Expression of a mutant α-zein creates the floury2 phenotype in transgenic maize. Proc. Natl. Acad. Sci. USA 94: 7094–7097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan R.S., Hammond-Kosack M., Bevan M. (1999). Transcription activation mediated by the bZIP factor SPA on the endosperm box is modulated by ESBF-1 in vitro. Plant J. 19: 173–181 [DOI] [PubMed] [Google Scholar]
- D’Andrea L.D., Regan L. (2003). TPR proteins: The versatile helix. Trends Biochem. Sci. 28: 655–662 [DOI] [PubMed] [Google Scholar]
- Diaz I., Vicente-Carbajosa J., Abraham Z., Martínez M., Isabel-La Moneda I., Carbonero P. (2002). The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J. 29: 453–464 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (2005). PHYLIP (Phylogeny Inference Package) Version 3.69. http://evolution.genetics.washington.edu/phylip.html
- Fujita N., et al. (2007). Characterization of SSIIIa-deficient mutants of rice: The function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol. 144: 2009–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux M.J., Boyer C., Feix G., Hannah L.C. (1994). Coordinated transcriptional regulation of storage product genes in the maize endosperm. Plant Physiol. 106: 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grauvogel C., Brinkmann H., Petersen J. (2007). Evolution of the glucose-6-phosphate isomerase: The plasticity of primary metabolism in photosynthetic eukaryotes. Mol. Biol. Evol. 24: 1611–1621 [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hirose T., Ohdan T., Nakamura Y., Terao T. (2006). Expression profiling of genes related to starch synthesis in rice leaf sheaths during the heading period. Physiol. Plant. 128: 425–435 [Google Scholar]
- Hirose T., Terao T. (2004). A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.). Planta 220: 9–16 [DOI] [PubMed] [Google Scholar]
- Holding D.R., Otegui M.S., Li B., Meeley R.B., Dam T., Hunter B.G., Jung R., Larkins B.A. (2007). The maize floury1 gene encodes a novel endoplasmic reticulum protein involved in zein protein body formation. Plant Cell 19: 2569–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.Z., Qian Q., Liu Z.B., Sun H.Y., He S.Y., Luo D., Xia G.M., Chu C.C., Li J.Y., Fu X.D. (2009). Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41: 494–497 [DOI] [PubMed] [Google Scholar]
- Hundertmark M., Hincha D.K. (2008). LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K.K., Ohashi Y., Tsuge T., Yoshizumi T., Matsui M., Oka A., Aoyama T. (2006). The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell 18: 382–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura T., Kusano H., Kajigaya Y., Ichikawa M., Shimada H. (2007). A rice dihydrosphingosine C4 hydroxylase (DSH1) gene, which is abundantly expressed in the stigmas, vascular cells and apical meristem, may be involved in fertility. Plant Cell Physiol. 48: 1108–1120 [DOI] [PubMed] [Google Scholar]
- Juliano B.O. (1971). A simplified assay for milled-rice amylose. Cereal Sci. Today 16: 334–340, 360 [Google Scholar]
- Kang H.G., Park S., Matsuoka M., An G. (2005). White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J. 42: 901–911 [DOI] [PubMed] [Google Scholar]
- Kaushik R.P., Khush G.S. (1991). Genetic analysis of endosperm mutants in rice Oryza sativa L. Theor. Appl. Genet. 83: 146–152 [DOI] [PubMed] [Google Scholar]
- Kawagoe Y., Kubo A., Satoh H., Takaiwa F., Nakamura Y. (2005). Roles of isoamylase and ADP-glucose pyrophosphorylase in starch granule synthesis in rice endosperm. Plant J. 42: 164–174 [DOI] [PubMed] [Google Scholar]
- Kawakatsu T., Yamamoto M.P., Touno S.M., Yasuda H., Takaiwa F. (2009). Compensation and interaction between RISBZ1 and RPBF during grain filling in rice. Plant J. 59: 908–920 [DOI] [PubMed] [Google Scholar]
- Kawasaki T., Mizuno K., Baba T., Shimada H. (1993). Molecular analysis of the gene encoding a rice starch branching enzyme. Mol. Gen. Genet. 237: 10–16 [DOI] [PubMed] [Google Scholar]
- Kawasaki T., Mizuno K., Shimada H., Satoh H., Kishimoto N., Okumura S., Ichikawa N., Baba T. (1996). Coordinated regulation of the genes participating in starch biosynthesis by the rice floury-2 locus. Plant Physiol. 110: 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.S., Gibbon B.C., Gillikin J.W., Larkins B.A., Boston R.S., Jung R. (2006). The maize Mucronate mutation is a deletion in the 16-kDa γ-zein gene that induces the unfolded protein response. Plant J. 48: 440–451 [DOI] [PubMed] [Google Scholar]
- Kim C.S., Hunter B.G., Kraft J., Boston R.S., Yans S., Jung R., Larkins B.A. (2004). A defective signal peptide in a 19-kD α-zein protein causes the unfolded protein response and an opaque endosperm phenotype in the maize De*-B30 mutant. Plant Physiol. 134: 380–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A., Fujita N., Harada K., Matsuda T., Satoh H., Nakamura Y. (1999). The starch-debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiol. 121: 399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Smith A.M. (1995). Starch biosynthesis. Plant Cell 7: 971–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzábal P., Gas E., Fontanet P., Vicente-Carbajosa J., Torrent M., Ludevid M.D. (2008). The maize Dof protein PBF activates transcription of γ-zein during maize seed development. Plant Mol. Biol. 67: 441–454 [DOI] [PubMed] [Google Scholar]
- Mizuno K., Kawasaki T., Shimada H., Satoh H., Kobayashi E., Okumura S., Arai Y., Baba T. (1993). Alteration of the structural properties of starch components by the lack of an isoform of starch branching enzyme in rice seeds. J. Biol. Chem. 268: 19084–19091 [PubMed] [Google Scholar]
- Murray M.G., Thompson W.F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y. (2002). Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: Rice endosperm as a model tissue. Plant Cell Physiol. 43: 718–725 [DOI] [PubMed] [Google Scholar]
- Nishi A., Nakamura Y., Tanaka N., Satoh H. (2001). Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol. 127: 459–472 [PMC free article] [PubMed] [Google Scholar]
- Ogo Y., Itai R.N., Nakanishi H., Kobayashi T., Takahashi M., Mori S., Nishizawa N.K. (2007). The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant J. 51: 366–377 [DOI] [PubMed] [Google Scholar]
- Onodera Y., Suzuki A., Wu C.Y., Washida H., Takaiwa F. (2001). A rice functional transcriptional activator, RISBZ1, responsible for endosperm-specific expression of storage protein genes through GCN4 motif. J. Biol. Chem. 276: 14139–14152 [DOI] [PubMed] [Google Scholar]
- Ohdan T., Francisco P.B., Jr., Sawada T., Hirose T., Terao T., Satoh H., Nakamura Y. (2005). Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 56: 3229–3244 [DOI] [PubMed] [Google Scholar]
- Page R.D.M. (1996). TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Pysh L.D., Aukerman M.J., Schmidt R.J. (1993). OHP1: A maize basic domain/leucine zipper protein that interacts with opaque2. Plant Cell 5: 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert L.S., Nozzolillo C., Altosaar I. (1985). Homology between rice glutelin and oat 12 S globlin. Biochim. Biophys. Acta 829: 19–26 [Google Scholar]
- Ryoo N., Yu C., Park C.S., Baik M.Y., Park I.M., Cho M.H., Bhoo S.H., An G., Hahn T.R., Jeon J.S. (2007). Knockout of a starch synthase gene OsSSIIIa/Flo5 causes white-core floury endosperm in rice (Oryza sativa L.). Plant Cell Rep. 26: 1083–1095 [DOI] [PubMed] [Google Scholar]
- Sabelli P.A., Larkins B.A. (2009). The development of endosperm in grasses. Plant Physiol. 149: 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh H., Nishi A., Yamashita K., Takemoto Y., Tanaka Y., Hosaka Y., Sakurai A., Fujita N., Nakamura Y. (2003). Starch-branching enzyme I-deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiol. 133: 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh H., Omura T. (1981). New endosperm mutations induced by chemical mutagens in rice, Oryza sativa L. Jpn. J. Breed. 31: 316–326 [Google Scholar]
- Satoh H., et al. (2008). Mutation of the plastidial α-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm. Plant Cell 20: 1833–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R.J., Burr F.A., Aukerman M.J., Burr B. (1990). Maize regulatory gene opaque-2 encodes a protein with a “leucine-zipper” motif that binds to zein DNA. Proc. Natl. Acad. Sci. USA 87: 46–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R.J., Ketudat M., Aukerman M.J., Hoschek G. (1992). Opaque-2 is a transcriptional activator that recognizes a specific target site in 22-kD zein genes. Plant Cell 4: 689–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She K.-C., Kusano H., Yaeshima M., Sasaki T., Satoh H., Shimada H. (2010). Reduced rice grain production under high-temperature stress closely correlates with ATP shortage during seed development. Plant Biotechnol. 27: 67–73 [Google Scholar]
- Shen Y.J., et al. (2004). Development of genome-wide DNA polymorphism database for map-based cloning of rice genes. Plant Physiol. 135: 1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry P.R., Napier J.A., Tatham A.S. (1995). Seed storage proteins: structures and biosynthesis. Plant Cell 7: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrawat A.K., Carroll R.T., DePauw M., Taylor G.J., Good A.G. (2008). Genetic engineering of improved nitrogen use efficiency in rice by the tissue-specific expression of alanine aminotransferase. Plant Biotechnol. J. 6: 722–732 [DOI] [PubMed] [Google Scholar]
- Uauy C., Distelfeld A., Fahima T., Blechl A., Dubcovsky J. (2006). A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314: 1298–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa J., Moose S.P., Parsons R.L., Schmidt R.J. (1997). A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc. Natl. Acad. Sci. USA 94: 7685–7690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa J., Oñate L., Lara P., Diaz I., Carbonero P. (1998). Barley BLZ1: A bZIP transcriptional activator that interacts with endosperm-specific gene promoters. Plant J. 13: 629–640 [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Zheng F.Q., Shen G.Z., Gao J.P., Snustad D.P., Li M.G., Zhang J.L., Hong M.M. (1995). The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J. 7: 613–622 [DOI] [PubMed] [Google Scholar]
- Yamakawa H., Hirose T., Kuroda M., Yamaguchi T. (2007). Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray. Plant Physiol. 144: 258–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M.P., Onodera Y., Touno S.M., Takaiwa F. (2006). Synergism between RPBF Dof and RISBZ1 bZIP activators in the regulation of rice seed expression genes. Plant Physiol. 141: 1694–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.