The β-cyanoalanine synthase enzyme is responsible for detoxifying the cyanide generated during cellular metabolism, primarily in the synthesis of ethylene. This analysis of a null mutant of this enzyme indicates that cyanide can act in some developmental processes as a signaling molecule.
Abstract
Cyanide is stoichiometrically produced as a coproduct of the ethylene biosynthesis pathway and is detoxified by β-cyanoalanine synthase enzymes. The molecular and phenotypical analysis of T-DNA insertion mutants of the mitochondrial β-cyanoalanine synthase CYS-C1 suggests that discrete accumulation of cyanide is not toxic for the plant and does not alter mitochondrial respiration rates but does act as a strong inhibitor of root hair development. The cys-c1 null allele is defective in root hair formation and accumulates cyanide in root tissues. The root hair defect is phenocopied in wild-type plants by the exogenous addition of cyanide to the growth medium and is reversed by the addition of hydroxocobalamin or by genetic complementation with the CYS-C1 gene. Hydroxocobalamin not only recovers the root phenotype of the mutant but also the formation of reactive oxygen species at the initial step of root hair tip growth. Transcriptional profiling of the cys-c1 mutant reveals that cyanide accumulation acts as a repressive signal for several genes encoding enzymes involved in cell wall rebuilding and the formation of the root hair tip as well as genes involved in ethylene signaling and metabolism. Our results demonstrate that mitochondrial β-cyanoalanine synthase activity is essential to maintain a low level of cyanide for proper root hair development.
INTRODUCTION
Hydrogen cyanide (HCN) is a colorless and highly volatile liquid. The anion cyanide (CN−) is very toxic because it reacts with keto compounds and Schiff base intermediates to give cyanohydrins and stable nitrile derivatives, respectively, and because it chelates di- and trivalent metal ions in the prosthetic groups of several metalloenzymes. In mitochondria, cyanide binds to the heme iron of cytochrome c oxidase, thereby blocking the utilization of oxygen in cellular functions (Donato et al., 2007). Despite its toxicity, cyanide is naturally produced in microorganisms and plant cells from several biochemical processes. In cyanogenic plants, cyanide is produced during the degradation of cyanogenic lipids and from the catabolism of cyanogenic glycosides (Poulton, 1990). Cyanide and cyanogenic compounds play an important role in plant defense against herbivores (Zagrobelny et al., 2008).
In noncyanogenic species, such as Arabidopsis thaliana, cyanide is produced from other biochemical processes, including stoichiometric production as a by-product of the ethylene biosynthesis pathway (Peiser et al., 1984). The first committed step of ethylene biosynthesis is the conversion of S-AdoMet to 1-aminocyclopropane-1-carboxylic acid (ACC) by ACC synthase (S-adenosyl-l-methionine methylthioadenosine-lyase). ACC is finally oxidized by ACC oxidase (ACO) to form ethylene and cyanoformic acid, which is then spontaneously degraded to carbon dioxide and cyanide (Peiser et al., 1984; Wang et al., 2002). Both ACS and ACC oxidase are encoded by multigene families whose members are differentially expressed in response to different environmental and developmental factors (Babula et al., 2006).
Cyanide must be rapidly detoxified and metabolized to keep its concentration below toxic levels. The main cyanide detoxification processes described in plant cells are catalyzed by β-cyanoalanine synthase (CAS) and to a lesser extent by rhodanese and mercaptopyruvate sulfurtransferase.
β-Cyanoalanine synthase is a pyridoxal phosphate-dependent enzyme that converts Cys and cyanide to hydrogen sulfide and β-cyanoalanine. Phylogenetic analysis shows that the β-cyanoalanine synthase in Arabidopsis is encoded by a small gene family of three members, CYS-C1 (At3g61440), CYS-D1 (At3g04940), and CYS-D2 (At5g28020) (Jost et al., 2000). The most abundant CAS enzyme is CYS-C1, which is localized in the mitochondria and contributes most of the CAS activity in root and leaf tissue (Watanabe et al., 2008). The isoforms CYS-D1 and CYS-D2 are localized in the cytosol and are much less abundant than CYS-C1 based on the numbers of ESTs as well as DNA microarray data and real-time PCR data (Watanabe et al., 2008). However, β-cyanoalanine synthase activity has not been observed in vitro for these enzymes (Hatzfeld et al., 2000; Yamaguchi et al., 2000).
Although most of the cyanide produced in plants is detoxified, little attention has been given to the potential physiological and molecular roles of cyanide and CAS enzymes. Several authors have mentioned that cyanide may act as a signaling molecule in the control of some metabolic processes (Bogatek et al., 1991; Grossmann, 1996; Siegien and Bogatek, 2006). There are a number of references that have suggested cyanide as a regulator of seed germination and dormancy release in rice (Oryza sativa), apple (Malus domestica), Helianthus tuberosus, and Arabidopsis (Cohn and Hughes, 1986; Fol et al., 1989; Bogatek and Lewak, 1991; Bethke et al., 2006). Treatment with HCN or sodium nitroprusside breaks Arabidopsis seed dormancy, and the emission of HCN has been observed during the pregermination period of many seeds (Esashi et al., 1991; Bethke et al., 2006).
Similar observations have been reported for the effect of HCN on resistance to biotic and abiotic stresses after ethylene production. For example, cyanide enhances the resistance of tobacco (Nicotiana tabacum) and Arabidopsis leaves to tobacco mosaic virus and turnip vein clearing virus, respectively (Chivasa and Carr, 1998; Wong et al., 2002). It has also been suggested that cyanide, and not ethylene, is responsible for the resistance to blast fungus infection in young rice plants (Iwai et al., 2006).
The role of cyanide as a regulatory molecule is not restricted to plants, and it has been suggested that cyanide can act as a neuromodulator of the central nervous system (Gunasekar et al., 2004; Cipollone and Visca, 2007). Cyanide in the central nervous system and leukocytes is generated from Gly via a reaction catalyzed by peroxidases (Zgliczynski and Stelmaszynska, 1979; Gunasekar et al., 2000), which can be stimulated by opiate receptors (Gunasekar et al., 2004).
The aim of this work is to learn more about the specific function of the mitochondrially localized β-cyanoalanine synthase and its role in the plant. For this purpose, we investigated knockout mutants of the β-cyanoalanine synthase genes in Arabidopsis and observed the potential regulatory effect of cyanide accumulated in the mitochondria of these mutants.
RESULTS
Arabidopsis cys-c1 Knockout Mutant Has Altered Root Hair Development
To address the functional roles of β-cyanoalanine synthases in plants, we characterized T-DNA insertional mutants of the mitochondrial β-cyanoalanine synthase, CYS-C1, which contributes most of the CAS activity in leaves and roots (Watanabe et al., 2008). Several homozygous mutants from the SALK collection (SALK_022479, SALK_068045, and SALK_067713) were screened by RT-PCR analysis, but only the mutant SALK_022479 lacked a detectable CYS-C1 transcript (see Supplemental Figure 1A online). This null allele was named cys-c1. DNA gel blot analysis of this mutant clearly showed one insertion of the T-DNA into the genome (see Supplemental Figure 1B online). Biochemical characterization of the mutant showed a significant reduction in CAS activity to 23% of the level measured in wild-type plants (see Supplemental Figure 1C online). The other SALK lines showed wild-type levels of CYS-C1 transcript and CAS activity and thus were not considered for further characterization.
Phenotypic analysis of soil-grown homozygous cys-c1 plants revealed no apparent change in the development and growth of the aerial part of the plant, as previously described (Watanabe et al., 2008). However, mutant seedlings grown on vertical Murashige and Skoog (MS) plates clearly showed a defect in root hair formation (Figures 1A to 1F). The root hairs of the mutant roots correctly started to grow out and away from the root surface and developed a small bulge; however, they did not further elongate to form normal hairs.
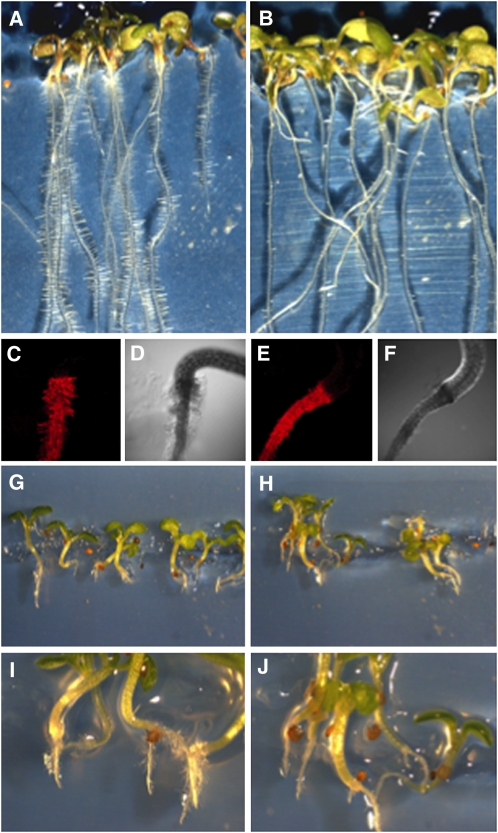
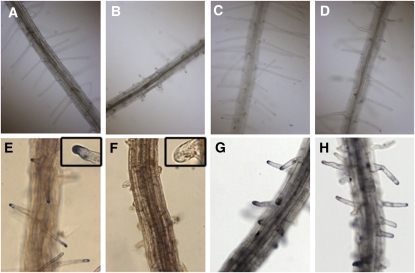
Figure 1.
Root Phenotype of the cys-c1 Mutant.
(A) and (B) Bright-field image of 2-week-old wild-type and cys-c1 mutant seedlings, respectively, growing on MS medium.
(C) and (D) Confocal fluorescent and transmitted light images of a wild-type root stained with propidium iodide for cell wall imaging, respectively.
(E) and (F) Confocal fluorescent and transmitted light micrographs of a cys-c1 root stained with propidium iodide, respectively.
(G) and (H) Bright-field image of 2-week-old wild-type and mutant seedlings, respectively, grown on MS medium containing 50 μM ACC.
(I) and (J) Magnified images of (G) and (H).
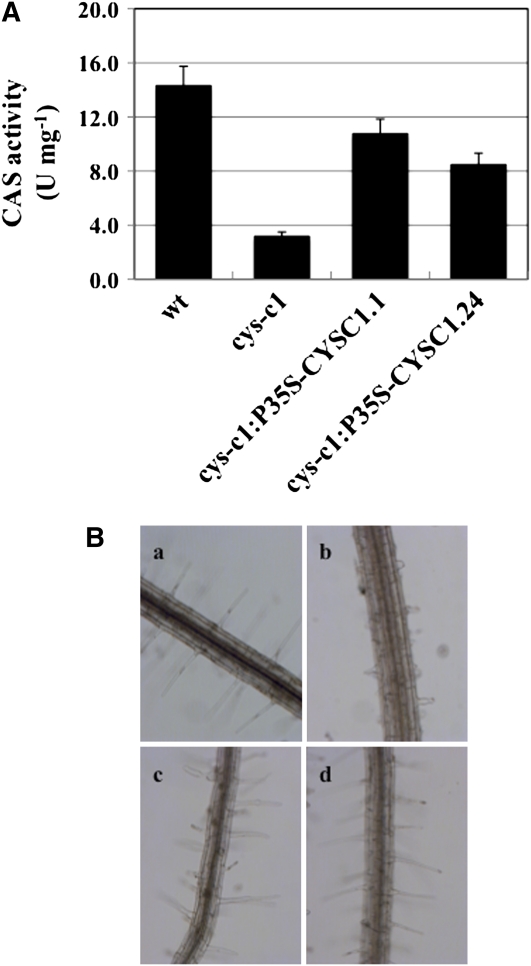
Phenotypic analysis of null mutants of the cytosolic isoforms CYS-D1 (mutant line SALK_ 092696) and CYS-D2 (mutant line SALK_ 097875) (Watanabe et al., 2008) grown on vertical MS plates showed normal root hair development. However, these isoforms are much less abundant and contribute less to the synthesis of β-cyanoalanine and to cyanide detoxification (Watanabe et al., 2008). To verify that the observed phenotype of the cys-c1 mutant plants was indeed due to disruption of the CYS-C1 gene, the mutant was complemented using the full-length cDNA fragment. The complemented cys-c1:P35S-CYSC1.1 and cys-c1:P35S-CYSC1.24 lines showed 75 and 59% of the CAS activity observed in the wild type, respectively, compared with only 23% CAS activity in cys-c1 (Figure 2A). These complemented lines rescued the root phenotype of the mutant, although the root hairs appeared to be slightly shorter than in the wild type (Figure 2B). Therefore, the observed root phenotype of the cys-c1 mutant is specific to the loss of the mitochondrial CAS enzyme.
Figure 2.
Characterization of the Complemented cys-c1Mutant Lines.
(A) β-Cyanoalananine synthase activity of the complemented cys-c1:P35S-CYSC1.1 and cys-c1:P35S-CYSC1.24 lines was quantified in root tissue of 4-week-old plants by measuring the formation of sulfide. One unit of activity corresponds to the formation of 1 nmol min−1 of sulfide. Values are means ± sd of three independent experiments.
(B) Root phenotype of the cys-c1 and complemented mutant lines. Bright-field image of 2-week-old seedlings growing on MS medium. a, The wild type; b, cys-c1 mutant line; c, complemented cys-c1:P35S-CYSC1.1 mutant line; d, complemented cys-c1:P35S-CYSC1.24 mutant line.
Cyanide Accumulates in the cys-c1 Mutant and Is Responsible for Its Phenotype
Since ethylene biosynthesis is the major source of cyanide in Arabidopsis, seedlings were grown on vertical MS plates containing 50 μM ACC, the precursor of ethylene via ACC oxidases (De Paepe and Van der Straeten, 2005). Both wild-type and mutant seedlings showed an inhibition of root and shoot elongation but mutant cys-c1 seedlings were not able to develop root hairs in the absence of ACC (Figures 1G to 1J).
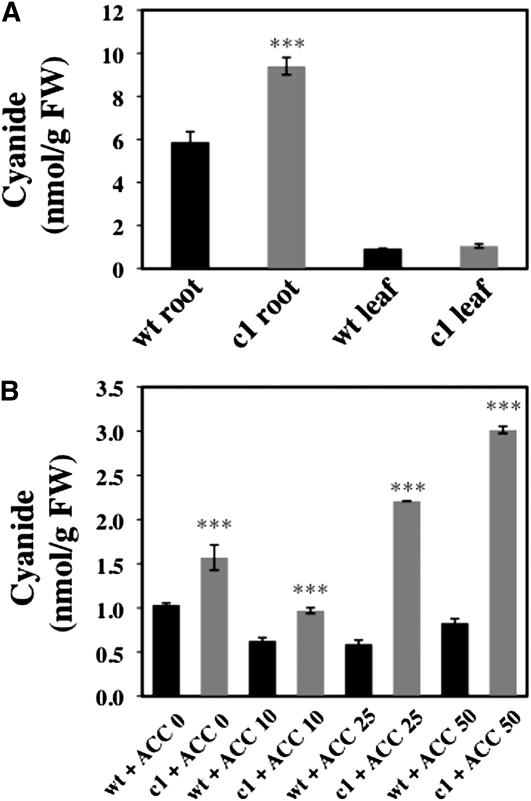
To address whether the observed phenotype of the mutant correlates with a defect in the detoxification of cyanide, we measured the accumulation of cyanide. In root tissue from 2-week-old seedlings grown on vertical MS medium, cys-c1 accumulated 62% more cyanide than did wild-type plants. By comparison with roots, leaf tissues accumulated less cyanide and showed no significant differences between the wild type and mutant (Figure 3A). In plant seedlings treated with ACC, we were unable to isolate enough root tissue because of the inhibition in root growth after ACC treatment. Therefore, we measured the accumulation of cyanide in whole seedlings treated with 0, 10, 25, and 50 μM ACC. We detected a 3.6-fold increase in the cyanide content in the null allele treated with 50 μM ACC, with levels ranging from 0.83 ± 0.05 in the wild type to 3.02 ± 0.04 nmol of CN− per g fresh weight (FW) in mutant plants. This effect is dependent of the concentration of ACC used (Figure 3B). Therefore, cyanide cannot be properly detoxified in the cys-c1 mutant, and differences are mainly observed in root tissues.
Figure 3.
Cyanide Determination in Root and Leaf Tissues.
(A) Two-week-old seedlings grown on vertical MS medium were collected and root and leaf tissues were assayed for cyanide content.
(B) Whole 2-week-old seedlings grown on vertical MS medium in the absence or presence of 10, 25, or 50 μM ACC were also collected and their cyanide content was determined. Values are means ± sd of three independent experiments. ***P < 0.001.
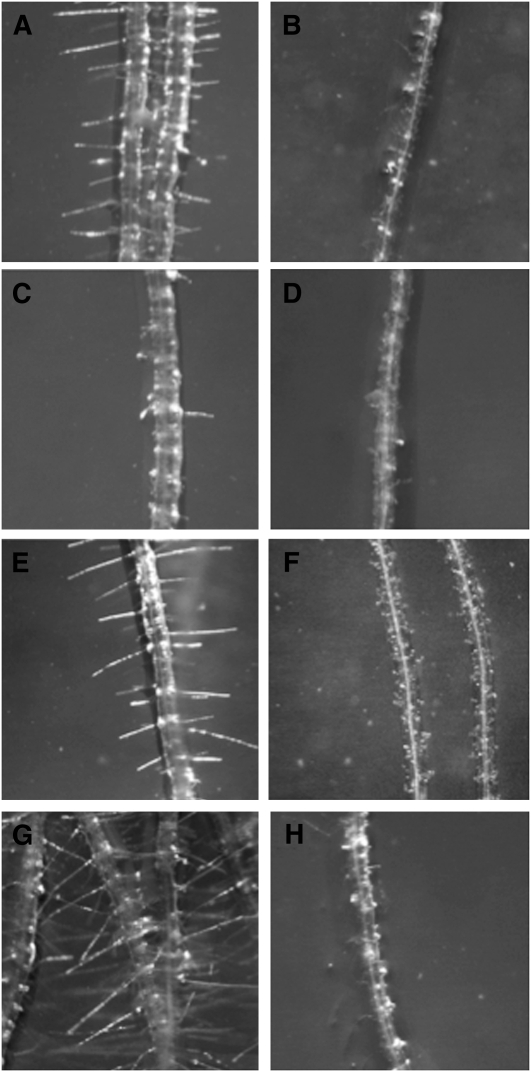
We explored the effect of exogenous added cyanide in the development of root hairs by transferring 5-d-old seedlings that had been growing in the absence of cyanide to MS medium containing KCN. The addition of up to 50 μM cyanide to the growth medium of wild-type plants phenocopied the defect in root hair formation observed in cys-c1 (Figures 4A and 4B), while the addition of cyanide to the growth medium of mutant plants did not have any effect on the mutant (Figures 4C and 4D). The effect on root hair formation was observed in those zones of the roots that penetrated slightly into the medium and were intimately in contact with cyanide. Lower concentrations of cyanide were less effective and only produced a reduction in the elongation of the root hair. Inhibition of root hair formation in the presence of cyanide was also observed in the cys-d1 and cys-d2 mutants. These mutants, despite having defects in the putative cytosolic CAS enzymes, showed normal root hair development in cyanide-free MS growth medium (Figures 4E to 4H).
Figure 4.
Cyanide Effect on Root Hair Formation.
Seedlings from wild-type and β-cyanoalanine mutant lines grown for 5 d on vertical MS medium were transferred to MS medium containing cyanide and grown for two additional days.
(A) and (B) Root hair phenotype of wild-type plants grown in the absence or the presence of 50 μM KCN, respectively.
(C) and (D) Root hair phenotype of cys-c1 mutant plants grown in the absence or the presence of 50 μM KCN, respectively.
(E) and (F) Root hair phenotype of cys-d1 mutant plants grown in the absence or the presence of 50 μM KCN, respectively.
(G) and (H) Root hair phenotype of cys-d2 mutant plants grown in the absence or the presence of 50 μM KCN, respectively.
Hydroxocobalamin, a natural form of vitamin B12, is a heme-like molecule with a complexed cobalt (Co) atom. Since cyanide reacts with high affinity with metals such as ferric iron and cobalt, hydroxocobalamin is the most commonly used antidote for severe acute cyanide poisoning in humans by ingestion or inhalation (Borron et al., 2007; Hall et al., 2007). Hydroxocobalamin can penetrate cells and bind cyanide intracellularly to form nontoxic cyanocobalamin, which is excreted in the urine (Astier and Baud, 1996). Although plants can synthesize several of the B complex vitamins, there is no evidence of B12 biosynthesis in vascular plants (Roje, 2007; Smith et al., 2007). Addition of up to 5 mM hydroxocobalamin to the MS plate medium had no effect on the phenotype of wild-type roots, but it partially restored the development of root hairs in the cys-c1 mutant seedlings (Figures 5A to 5D).
Figure 5.
Hydroxocobalamin Effect on Root Hair Formation and ROS Localization at the Root Hair Tips.
(A) and (B) Root hair phenotype of wild-type and cys-c1 mutant plants, respectively, germinated and grown for 5 d in the absence of hydroxocobalamin.
(C) and (D) Root hair of wild-type and cys-c1 mutant plants, respectively, germinated and grown for 5 d in the presence of 5 mM hydroxocobalamin.
(E) and (F) Five-day-old wild-type and cys-c1 roots stained with NBT for superoxide detection.
(G) and (H) Five-day-old wild-type and cys-c1 mutant roots grown on hydroxocobalamin medium stained with NBT.
The cys-c1 Mutant Does Not Generate Reactive Oxygen Species at the Root Tip
The function of reactive oxygen species (ROS) in root hair development is well documented (Carol and Dolan, 2006; Takeda et al., 2008). At the initial stages, ROS accumulate within the hair at the dome of the trichoblast. As tip growth begins, ROS become concentrated at the tip and continue to accumulate during hair growth. In this process, RHD2 NADPH oxidase is involved in the localized production of ROS that elevates the concentration of cytoplasmic Ca2+. Therefore, Arabidopsis rhd2 mutants do not form root hairs (Foreman et al., 2003; Carol and Dolan, 2006). To test whether cys-c1 has reduced ROS formation on root hair outgrowths, we examined ROS production by nitroblue tetrazolium (NBT) staining. NBT staining clearly showed blue formazan deposition in wild-type root hair tips, but staining was absent in cys-c1 root outgrowths (Figures 5E and 5F). However, cys-c1 seedlings grown in hydroxocobalamin-containing MS medium developed root hairs and NBT staining similar to the wild type during root hair outgrowth. This effect was mainly observed in the surface of the roots that was in contact with the growth medium containing hydroxocobalamin (Figures 5G and 5H).
Respiration Rates and Mitochondria Distribution in Root Tips
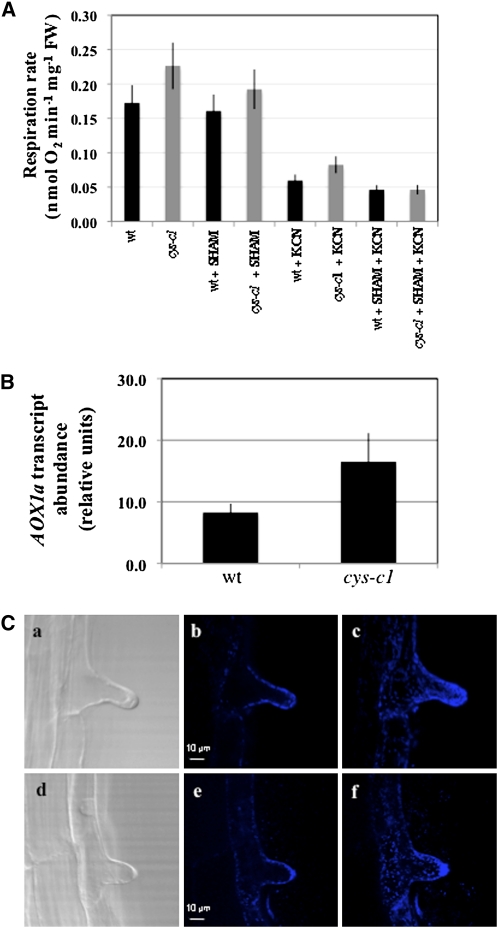
Since cyanide is a potent inhibitor of the cytochrome respiration pathway (COP), we measured the respiration rates of whole-root tissue by measuring oxygen uptake. The cys-c1 mutant shows a 30% increase in respiration rates compared with the wild type, and this increase is partially reduced by the addition of salicylhydroxamic acid (SHAM), an inhibitor of the alternative oxidase pathway. The addition of KCN or KCN plus SHAM reduces oxygen uptake to basal levels (Figure 6A). The increase of the alternative oxidase pathway in the cys-c1 mutant correlates with an increase in transcript abundance of the AOX1a gene, which is induced by alteration of the COP (Figure 6B) (Albury et al., 2009).
Figure 6.
Respiration Rates and Mitochondrial Localization in Root Hair.
(A) Respiration rate in the wild type and cys-c1 mutant line in 15-d-old root tissues. Cyanide-independent and alternative oxidase respiration was determined in the presence of 0.5 mM KCN or 4 mM SHAM, respectively. The results are expressed as the mean ± sd from at least three replica samples.
(B) Transcription level of the alternative oxidase gene AOX1a in 15-d-old wild-type and cys-c1 mutant root tissue was analyzed by quantitative real-time RT-PCR using the primers qAOX1a-F and qAOX1a-R (see Supplemental Table 4 online). The expression level was normalized to that of the constitutive UBQ10 gene by subtracting the CT value of UBQ10 from the CT value of the gene (ΔCT). Values are means ± sd from three replica samples.
(C) Visualization of stained mitochondria with MitoTracker Deep Red 633 dye. a and d, Transmitted light imaging of 5-d-old root tissues from wild-type or cys-c1 mutant plants, respectively. b and e, Single optical sections of stained roots from wild-type or cys-c1 mutant plants, respectively. c and d, Maximum projection of 18 optical sections of stained roots from wild-type or cys-c1 mutant plants, respectively.
We also tested whether cyanide accumulation alters the localization of mitochondria within the root hair. Mitochondria are present at a high density in tip-growing cells, and during root hair formation they are spatially associated with the initiation bulge (Ciamporova et al., 2003). To visualize mitochondria in root hairs, we stained root tissues with MitoTracker Deep Red, which passively diffuses across the plasma membrane and accumulates in active mitochondria. We observed in single xy-planes an even distribution of mitochondrial staining throughout the cytoplasmic space, both in the wild type and in cys-c1 mutant root hair bulges (Figure 6C, b and e). A z-projection of several xy-single planes clearly shows a high density of mitochondrial staining at the root hair tip of both lines (Figure 6C, c and f).
Root Hair Development and Ethylene-Related Genes Are Repressed in cys-c1 Roots
Using Affymetrix ATH1 GeneChips, we performed a comparative transcriptomic analysis of roots of the cys-c1 and wild-type plants. Total RNA was extracted from roots of 14-d-old plants grown under identical conditions on MS medium (three biological replicates for each genotype), and these samples were used to prepare complementary RNA and hybridize the chips (Gene Expression Omnibus repository GSE 19242). The normalized data from the replicates showed differential expression of 160 genes in the cys-c1 mutant, with 97 genes downregulated (see Supplemental Data Set 1 online) and 63 genes upregulated (see Supplemental Data Set 2 online) more than twofold [−1 < M < 1, where M is the log-transformed ratio calculated as log2(cys-c1/wild type)] with an false discovery rate value <0.05. Among the repressed genes, several were assigned to functional groups previously shown to be involved in root hair morphogenesis, such as cell wall enzymes, arabinogalactan proteins, peroxidases, and glycosylphosphatidylinositol (GPI)-anchored proteins, and others were involved in ethylene signaling and metabolism.
Two of the most strongly repressed genes in the cys-c1 mutant (see Supplemental Data Set 1 online), MRH5 and MRH6, were identified from a root hair morphogenesis transcriptome analysis and from the screening of their corresponding T-DNA insertional lines (Jones et al., 2006). The similar root hair phenotypes and complementation test results strongly suggest that mrh5 is identical to the previously isolated root hair mutant shv3 (Parker et al., 2000; Jones et al., 2006). The gene MRH5 (At4g26690) encodes a glycerophosphoryl diester phosphodiesterase-like GPI-anchored protein, and the T-DNA knockout mutant of this gene shows short wide burst hairs resembling the cys-c1 root phenotype. The MRH6 (At2g03720) gene encodes a protein similar to the Escherichia coli universal stress protein A, which is essential for the survival of bacterial cells in the stationary phase.
Eleven genes that are functionally classified in the cell wall group are also repressed in the cys-c1 mutant, including two arabinogalactan proteins (At2g20520, FLA6; At4g40090, AGP3), two xyloglucan:xyloglucosyl transferases (At4g28850 and At5g57530), one xyloglucan endotransglycosylase (At4g25820, XTR9), as well as two pectinesterases and one pectate lyase protein (At5g04960, At4g22080, and At3g10710). Pharmacological and genetic evidence has correlated cell surface arabinogalactan with the organization of cortical microtubules that influence root epidermal expansion and morphogenesis (Ding and Zhu, 1997; Nguema-Ona et al., 2007). There is also evidence that root hair initiation is accompanied by a localized increase in xyloglucan endotransglycosylase at the site of future bulge formation, where the trichoblast locally loosens its cell wall (Vissenberg et al., 2001).
Ethylene and other hormones such as auxin and jasmonate are also involved in the regulation of root hair development. The ethylene response mutant ctr1 has ectopic root hairs on the atrichoblast and produces very few root hairs. In addition, treatment of wild-type plants with ACC induces the ectopic formation of root hairs in Arabidopsis (Tanimoto et al., 1995; Zhu et al., 2006). The transcript profile of the cys-c1 mutant showed reduced expression of several genes related to ethylene signaling and metabolism. In this group, we found repression of one member of the ACC synthase gene family (ACS6, At4g11280).
The transcript profile of the cys-c1 mutant also showed 63 upregulated genes belonging to eight functional groups (see Supplemental Data Set 2 online). The most highly induced gene was assigned to the RNA functional group and encodes RNS1 (At2g02990), a member of the ribonuclease T2 family (Taylor and Green, 1991). RNS1 responds to inorganic phosphate starvation and inhibits the production of anthocyanin. It is also involved in wound-induced signaling independent of jasmonic acid and oligosaccharide elicitors (Bariola et al., 1994; LeBrasseur et al., 2002). Two peroxidases, PER59 and PER10, were also significantly induced, but their functional roles are unknown.
The gene induction and repression that we observed by microarray analysis was confirmed by quantitative real-time PCR for selected genes such as MRH5, CYP81 (a cytochrome P450), ECS1, FLA6 (fasciclin-like arabinogalactan-protein), PER10, PER59, and RNS1. The changes in transcript abundance showed the same pattern of expression, thus validating the data obtained by the array (see Supplemental Figure 2 online).
The microarray data for the cys-c1 mutant were meta-analyzed using the Bio-Array Resource for Arabidopsis Functional Genomics (Toufighi et al., 2005). All databases available for the Expression Browser tools (tissue, stress, hormone and chemical, pathogen, and seed series) were compared separately with the cys-c1 transcript profiles. Although we could not find significant correlation with the upregulated genes, a significant correlation was found between the downregulated genes and the hormone-chemical series (Toufighi et al., 2005).
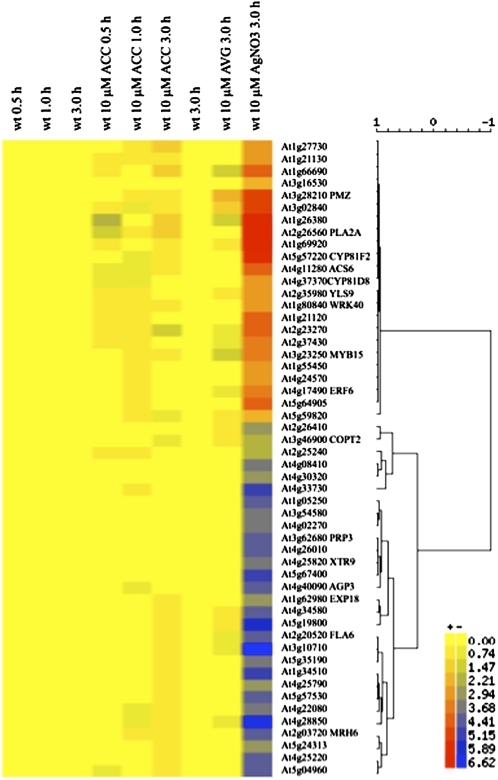
A higher correlation was observed in the hormone-chemical series with genes regulated by the ethylene response inhibitor silver nitrate (AgNO3). At least 52 genes repressed in the cys-c1 mutant are coregulated at the level of gene expression by >80% (as measured by the Pearson correlation coefficient) with those in the data set of 10 μM AgNO3-treated seedlings (Figure 7). However, no significant correlation was observed in the data set of hormone treatment (ACC, zeatin, methyl jasmonate, indolacetic acid, abscisic acid, gibberellic acid, brassinolide, and cytokinin) or by treatment with aminoethoxyvinylglycine (AVG), an inhibitor of ethylene biosynthesis. Most of the genes coregulated by cyanide and silver nitrate are related to root hair development, such as MRH6, FLA6, AGP3, and XTR9.
Figure 7.
Graphic Display of Hierarchical Cluster Analysis of cys-c1 Downregulated Genes in Seedlings Treated with Ethylene Inhibitors or Precursors.
Hierarchical clustering analysis was applied to a set of 52 downregulated genes in the cys-c1 mutant, as well as ACC-, AVG-, and AgNO3-treated seedlings. Each column represents the time course and treatment samples indicated at its top, and each row refers to a gene. A dendrogram representing hierarchical relationships among treatments is shown, and the scale at the top marks the correlation coefficient represented by the length of the branches that connect pairs of nodes. The color scale indicates the log2 level of expression above (red) or below (blue) the median. The data sets of the ACC time course and of the ethylene inhibitor treatments are from the AtGenExpress Consortium, NASCarray experiment reference numbers 172 and 188, respectively.
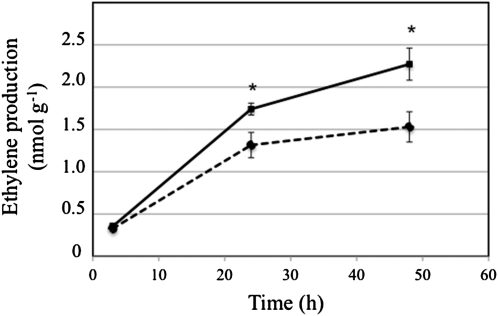
Ethylene Production Is Reduced in the cys-c1 Mutant
Since the transcription profile of the mutant showed a repression of the ACS6 gene and a coregulation of several genes by treatment with the inhibitor AgNO3, we investigated whether the biosynthesis of ethylene is altered in the cys-c1 mutant. We measured by gas chromatography the production of ethylene in wild-type and cys-c1 seedlings grown for 19 d on vertical MS plates. The production of ethylene after 3 h was not statistically different between wild-type and mutant seedlings, with values around 110 pmol of ethylene per h per g FW tissue (Figure 8). Nevertheless, a significant difference was apparent after 24 h, at which point the cys-c1 mutant line had 25% less ethylene production than the wild-type line. After 2 d, the rate of ethylene production declined significantly in both lines to 72 and 55 pmol of ethylene per h per g FW tissue in the wild type and mutant, respectively, falling to 60% of the 3-h rate in the wild-type seedlings. However, the inhibition of ethylene production after 2 d was greater in the mutant line.
Figure 8.
Ethylene Production in the cys-c1 Mutant.
Ethylene accumulation was determined in wild-type (continuous line) and cys-c1 mutant (discontinuous line) seedlings grown for 19 d on vertical MS plates by gas chromatography. The accumulation of ethylene was determined at 3, 24, and 48 h after the vials were sealed. Values are means ± sd of at least 20 samples per plant line. *P < 0.05.
DISCUSSION
It is well established that ethylene plays an essential role in root development by stimulation and inhibition of cell growth. It acts as a strong inhibitor of root growth by regulating cell elongation both at the root tip zone and the elongation zone (Dugardeyn and Van Der Straeten, 2008). However ethylene can also act as an inducer of root hair formation or of division of root cells in the quiescent center (Tanimoto et al., 1995; Ortega-Martinez et al., 2007; Dugardeyn and Van Der Straeten, 2008). So, in the presence of ACC, ectopic root hair formation is induced, while in the presence of AVG, ectopic root hair formation is repressed. Cyanide is stoichiometrically produced as a coproduct of the ethylene biosynthesis pathway, and it is generally accepted that this cyanide is effectively detoxified by β-cyanoalanine synthases or other less abundant enzymes such as rhodanese and mercaptopyruvate sulfurtransferase.
Our results from the analysis of the cys-c1 null allele point out that the accumulation of cyanide within the mitochondrion can act as an inhibitor of root hair development. Since ACC oxidase is localized to the cytoplasm, the cyanide coproduced during ethylene biosynthesis must be handled by the cytosolic β-cyanoalanine synthases, CYS-D1 and CYS-D2, but their knockout mutants do not show changes in β-cyanoalanine synthase activity or in phenotype under laboratory-controlled conditions of growth (Watanabe et al., 2008). However, even under controlled growth conditions and low ethylene production, CYS-D1 and CYS-D2 are not able to handle all of the cyanide produced in the cytoplasm, since loss of the mitochondrial CYS-C1 results in cyanide accumulation. This effect is enhanced in plants treated with ACC, which is a stimulator of ethylene synthesis and concomitantly of cyanide synthesis. It is conceivable that cyanide could accumulate in the cytoplasm and reach the mitochondria where CYS-C1 maintains discrete levels of cyanide. The loss of CYS-C1 activity in the null mutant results in increased levels of cyanide, probably within the mitochondria, where it seems to act as an inhibitor or a repressor of the formation of root hair. However, the accumulation of cyanide does not alter the elongation of other root cell types where ethylene is also produced.
A similar phenotype of inhibition of root hair formation is also observed in wild-type plants as well as in the cys-d1 and cys-d2 mutants by exogenous application of cyanide to the growth medium. Moreover, the fact that the exogenous addition of hydroxocobalamin alleviates the effect and recovers the development of the hairs supports our conclusion that the phenotype observed in the cys-c1 mutant is due to the accumulation of cyanide. Besides, it is completely dependent on the loss of function of the mitochondrial CYS-C1 activity as the root hair defect phenotype is fully restored in the complemented lines.
The addition of hydroxocobalamin to the MS medium also recovers the ROS staining that is detectable in the wild-type hair tip and that has been described as being produced by the RHD2 NADPH oxidase (Foreman et al., 2003). This plasma membrane enzyme is necessary for the establishment of the tip-based Ca2+ gradient, which is, in turn, essential for root hair polar growth (Foreman et al., 2003). Furthermore, tip-localized mitochondria seem to be important for the clearance of the high cytoplasmic concentration of calcium in the growing tip (Levina and Lew, 2006).
Although the mechanism by which the cyanide molecule inhibits the elongation of root hairs is unknown, several hypotheses can be postulated. It is possible that cyanide acts locally by inhibiting RHD2 activity through the formation of a complex with its heminic iron. In fact, a similar effect has been observed in wild-type roots treated with diphenylene iodonium chloride, which covalently binds the reaction center of flavoproteins, thus inhibiting their activity and phenocopying the rhd2 mutant hairs. Resistance to inhibition by cyanide has been used as a criterion to identify the enzymatic source of superoxide generation as NADPH oxidase rather than NADPH peroxidase (Bolwell et al., 1998). However, there are no in vitro data about the enzymatic sensitivity to cyanide of RHD2 and of the RBOH enzyme family in general (Torres and Dangl, 2005).
A second hypothesis is that cyanide triggers a repressing signaling pathway generated in the mitochondria that regulates the plasma membrane NADPH oxidase activity that, in turn, regulates the initiation of root hairs. This theory is in agreement with the data of the transcriptomic profile of the cys-c1 mutant. This analysis shows that cyanide accumulation in the mitochondria triggers a developmental program that leads to the inhibition of several well-known genes related to root hair formation, such as the glycerophosphoryl diester phosphodiesterase-like GPI-anchored protein, MRH5, and the universal stress protein A, MRH6. Also, several genes coding for cell wall enzymes are repressed in the mutant, including one xyloglucan endotransglycosylase (XTR9), which is involved in loosening of the cell wall for root hair initiation (Vissenberg et al., 2001). The regulation of these genes is in agreement with a model proposed for the formation of root hairs. The model includes a relaxation of the cell wall in the zone of elongation of the hair, right at the tip, and a simultaneous reinforcement of the cell wall in the flank of the hair, behind the apex, to prevent its expansion (Knight, 2007; Monshausen et al., 2007). In addition, comparison of the transcript profile obtained in the mutant with microarray data publicly available reveals that many of the genes involved in root hair formation are also coregulated by silver nitrate, which is an artificial inhibitor of ethylene action (Beyer, 1976). Therefore, endogenous cyanide seems to act as a natural inhibitor of ethylene action.
Some of the genes repressed in cys-c1 root tissue are ACS6, encoding an ACC synthase, and three ERF/AP2 transcription factors. The Arabidopsis genome encodes 12 enzymatically active ACC synthase polypeptides, which are differentially regulated by a number of factors (De Paepe and Van der Straeten, 2005). We might expect that, when cyanide accumulates in the mitochondria, ethylene biosynthesis must be regulated to avoid further accumulation of cyanide to a toxic level. Nevertheless, it has been reported that the addition of cyanide at a low concentration transiently induces the transcript level of ACS6, although this induction was not observed in the ethylene-insensitive mutant, ein-2 (Smith and Arteca, 2000). The measurement of ethylene production in the cys-c1 mutant seedlings clearly shows a decrease in the rate of ethylene synthesis.
We excluded the possibility that the observed phenotype is due to a cytotoxic effect of cyanide within the mitochondria because the mutant is completely viable, neither programmed cell death nor necrotic tissue is observable, and the only appreciable phenotype is the inhibition of root hair development. In addition, it is well established that impairment of mitochondrial function leads to a severe defect in respiration and to cytoplasmic male sterility resulting in phenotypes that range from defective male flower organs to drastically altered flower architecture (Karpova et al., 2002; Linke and Borner, 2005). However, we have not detected any phenotypic effect on pollen development or viability in the cys-c1 mutant. The respiration rate measured in whole-root tissue of the mutant shows that oxygen consumption is slightly induced by an increase in the alterative oxidase pathway as demonstrated by the effect of SHAM on the respiration rate and by the elevation of the expression of the AOXa1 gene. However, the level of respiration due to the COP is similar in the wild type and the mutant line in spite of the presence of cyanide. In addition, we have not observed changes in the cytological localization and viability of mitochondria in the polarized root hair cell. At the molecular level, we do not observe the induction of genes associated with poisoning of the mitochondria in the transcriptomic profile of the cys-c1 mutant. Chemically induced dysfunction of mitochondria triggers significant changes in the transcript levels of genes related to mitochondrial function, such as those encoding the mitochondrial import components (translocases on the inner mitochondrial membrane), molecular chaperones (heat shock proteins), and proteins involved in respiratory chain assembly (Lister et al., 2004). However, none of these genes have altered transcripts levels in the cys-c1 mutant.
In conclusion, our results demonstrate that the mitochondrial β-cyanoalanine synthase is essential to maintain a low level of cyanide for proper root hair development. However, we cannot exclude that discrete and transient accumulation of cyanide, during an extreme ethylene burst, may act as an intracellular regulator of root hair elongation. It is possible that cyanide is an undesirable by-product of the ethylene biosynthetic pathway that is generally detoxified by CAS enzymes, but, at specific developmental stages or subcellular locations, it can also serve as a direct signal for the transduction pathway of ethylene and HCN to diverge and activate specific plant responses. Since the observed phenotype of the cys-c1 mutant seems to depend on an enzyme with mitochondrial localization, the beginning of the signaling chain must be generated in the mitochondria. In one model, cyanide could partially inhibit cytochrome c oxidase, which is located in the mitochondrial respiratory complex IV. Inhibition of cytochrome c oxidase generates an overreduction of the ubiquinone pool that can be alleviated by means of the induction of the alternative oxidase, AOX1a, preventing the formation of ROS (Millenaar and Lambers, 2003; Navrot et al., 2007). Alternative oxidase has been implicated in the regulation of several processes and signaling events, such as thermogenesis, programmed cell death, salicylic acid–induced resistance to viruses, and a response to phosphorus deficiency (Vanlerberghe et al., 2002; Millenaar and Lambers, 2003; Singh et al., 2004). Interestingly, root temperature and P deficiency are environmental factors affecting root hair formation that are regulated by ethylene production (Michael, 2001).
This model is compatible with a direct role of cyanide in signaling by means of the direct inhibition of enzymatic activities responsible for the formation or detoxification of ROS, such as NADPH oxidase, NADPH peroxidase, or superoxide dismutase.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana (accession Columbia-0) and the SALK_022479, SALK_068045, SALK_067713 (cys-c1 alleles), SALK_092696 (cys-d1), and SALK_097875 (cys-d2) mutants were used in this work (Sessions et al., 2002; Alonso et al., 2003; Yamada et al., 2003). The plants were grown either in soil supplemented with Hoagland medium or in solid MS medium in Petri dishes supplemented with 1% sucrose. Plants were grown under a photoperiod of 16 h of white light (120 mE m−2 s−1) at 20°C/8 h dark at 18°C.
Plant seedlings from wild-type and β-cyanoalanine mutant lines grown for 5 d on vertical MS medium were transferred to MS medium containing 50 μM KCN (Sigma-Aldrich) and grown for two additional days. For hydroxocobalamin or ACC treatments, the seedlings were germinated in MS containing 5 mM hydroxocobalamin (Sigma-Aldrich) or 50 μM ACC (Sigma-Aldrich) and grown for 5 or 14 d, respectively.
To generate the cys-c1 complementation lines, the 1107-bp cDNA fragment containing the full-length coding sequence of CYS-C1 was obtained by RT-PCR amplification that was extended from the CACC at the 5′-end using the proofreading Platinum Pfx enzyme (Invitrogen) and the primers GW-C1F and GW-C1R (see Supplemental Table 1 online). The fragment was first cloned into the pENTR/D-TOPO vector (Invitrogen) and then transferred into the vector pMDC32 (Curtis and Grossniklaus, 2003) using Gateway technology (Invitrogen) according to the manufacturer’s instructions. The final construct was introduced by transformation into Agrobacterium tumefaciens, which was then introduced into the cys-c1 null plants using the floral dipping method (Clough and Bent, 1998).
DNA Isolation and DNA Gel Blot Analysis
Total DNA was isolated from Arabidopsis leaves and digested with BglII, a restriction enzyme that does not cut inside the CYS-C1 gene. DNA gel blot analysis was performed as previously described (Lopez-Martin et al., 2008).
RNA Extraction and Microarray Hybridization
Total RNA was isolated with Trizol reagent (Invitrogen) followed by cleaning with the RNeasy plant mini kit (Qiagen); this was used to synthesize biotinylated cRNA for hybridization to Arabidopsis ATH1 arrays (Affymetrix) using the 3′ Amplification One-Cycle Target Labeling Kit. Briefly, 4 mg of RNA was reverse transcribed to produce first-strand cDNA using a (dT)24 primer with a T7 RNA polymerase promoter site added to the 3′ end. After second-strand synthesis, in vitro transcription was performed using T7 RNA polymerase and biotinyated nucleotides to produce biotin-labeled cRNA. The cRNA preparations (15 μg) were fragmented into fragments of 35 to 200 bp at 95°C for 35 min. The fragmented cRNAs were hybridized to the Arabidopsis ATH1 microarrays at 45°C for 16 h. Each microarray was washed and stained in the Affymetrix Fluidics Station 400 following standard protocols. Microarrays were scanned using an Affymetrix GeneChip Scanner 3000.
Microarray Data Analysis
Microarray analysis was performed using the affylmGUI R package (Wettenhall et al., 2006). The robust multiarray analysis algorithm was used for background correction, normalization, and for summarizing expression levels (Irizarry et al., 2003). Differential expression analysis was performed using Bayes t-statistics using the linear models for microarray data (Limma), which is included in the affylmGUI package. P values were corrected for multiple testing using Benjamini-Hochberg’s method (false discovery rate) (Benjamini and Hochberg, 1995; Reiner et al., 2003). A cutoff value of a twofold change and P value of <0.05 were adopted to discriminate expression of genes that were differentially altered. Gene classification into functional groups was obtained from The Arabidopsis Information Resource (http://www.Arabidopsis.org), MapMan (http://gabi.rzpd.de/projects/MapMan/), and GENEVESTIGATOR (https://www.genevestigator.ethz.ch/at/). Microarray data were meta-analyzed using the Bio-Array Resource for Arabidopsis Functional Genomics (http://www.bar.utoronto.ca).
Cyanide Determination by HPLC
Plant tissues were homogenized using a mortar and pestle with liquid nitrogen and resuspended in cold borate-phosphate extraction buffer (2 mL per g FW) containing 27 mM sodium borate and 47 mM potassium phosphate, pH 8.0. Homogenates were centrifuged at 15,000g for 15 min in the cold. Extracted cyanide was subsequently quantified by reverse-phase HPLC after derivatization with 2,3-naphthalene dicarboxyaldehyde to form a 1-cyano-2-alkyl-benz[f]isoindole derivative by described methods (Lin et al., 2005). A cyanide standard solution (CertiPUR; Merck) was used to make a six-point calibration curve over the concentration range of 0 to 3.8 mM. HPLC analyses were performed on a LaChrom Elite system (VWR International). The cyano-alkyl-benz[f]isoindole derivative was separated on an RP 18 (150 mm × 3.9 mm, 5 μm; Waters) column. The mobile phase consisted of a mixture of acetonitrile and 0.1% trifluoroacetic acid in water (28:72 v/v) and was delivered isocratically at a flow rate of 1 mL/min. The injection volume was 10 μL.
Control experiments were conducted to check the recovery of cyanide from the plant tissue. A known quantity of KCN was added to the tissue mixture and processed and assayed for cyanide concentration as described above. The percentage of recovery was estimated by comparing the assayed value to the known amount of cyanide added to the incubation. A 69% rate of recovery was calculated.
Respiration Measurements in Roots
Wild-type and mutant plants were grown for 15 d on vertical MS plates with 1% sucrose and 3.5 g L−1 phytagel. Approximately 50 mg of root tissues were cut, dried on paper towels, and transferred into air-tight cuvettes containing 20 mM HEPES, pH 7.2, and 0.2 mM CaCl2, and oxygen uptake was measured as a decrease of O2 concentration in the dark using a Clark-type electrode. Cyanide-resistant O2 uptake was measured in the presence of 0.5 mM KCN. The respiration component due to the alternative oxidase pathway was measured in the presence of 4 mM of the inhibitor SHAM. The results were expressed as the mean ± sd from at least three replica samples, and the experiment was repeated three times from independent samples.
Ethylene Determination by Gas Chromatography
Between 5 and 10 seedlings (~0.2 g) were collected from the surface of the plate, weighed, placed inside an 11-mL vial, and sealed. Ethylene produced and released to the gas phase was determined by gas chromatography at the indicated times by injecting 1 mL of the head space on a HP 5890A GC equipped with an activated alumina column and a flame ionization detector. The oven and the detector temperatures were isothermically maintained at 100 and 150°C, respectively (Castellano and Vioque, 2002). The results were expressed as the mean ± sd from at least eight replica samples, and the experiment was repeated three times from independent samples.
Assay of β-Cyanoalanine Synthase Activity
Cyanoalanine synthase activity was measured by the release of sulfide from l-Cys and cyanide following the method described by Meyer et al. (2003) and using the extinction coefficient of 15 × 106 cm2 mol−1 (Papenbrock and Schmidt, 2000).
Detection of Superoxide
For the detection of superoxide, plant material was stained with NBT as described (Carol et al., 2005). Briefly, 5-d-old seedlings were covered with a solution of 0.5 mg mL−1 NBT in 0.1 M Tris-HCl, 0.1 M NaCl, and 0.05 M MgCl2, pH 9.5, and incubated at room temperature in the dark for 2 h. Roots were imaged under bright-field illumination on an Olympus BX50 microscope. Images were taken using a Leica DFC300 FX digital camera.
Mitochondria Staining
Wild-type and mutant plants were grown for 5 d on vertical MS plates with 1% sucrose and 3.5 g L−1 phytagel. Root tissues were cut and incubated with a 20 nM solution of MitoTracker Deep Red (Molecular Probes) for 10 min at room temperature. Samples were mounted onto a slide using a spacer between the slide and cover slip to avoid crushing the tissue and observed using a Leica HCX PLAN-APO ×63 1.4 NA oil immersion objectives attached to a Leica TCS SP2 spectral confocal microscope (Leica Microsystems). The dye was excited using the 644-nm line of a helium-neon laser, either in single confocal optical sections or serial optical sections of roots. Emitted light was collected through a triple dichroic beamsplitter (TD 488/543/633) and detected after spectral separation in the 650- to 700-nm range (pseudocolored blue).
Accession Numbers
Sequence and microarray data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CYS-C1 (At3g61440), CYS-D1 (At3g04940), CYS-D2 (At5g28020); CYS-C1 T-DNA mutants SALK_022479, SALK_068045, SALK_067713; CYS-D1 T-DNA mutant SALK_092696; CYS-D2 T-DNA mutant SALK_097875. The microarray Gene Expression Omnibus accession number is GSE19242
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Molecular Characterization of the Arabidopsis cys-c1 Mutant.
Supplemental Figure 2. Relative Expression Levels of Selected Genes in the cys-c1 Mutant Plant.
Supplemental Table 1. Oligonucleotides Used in This Work.
Supplemental Data Set 1. Groups of Functionally Related Genes Downregulated in Roots of the cysc1 Mutant When Compared with the Wild Type.
Supplemental Data Set 2. Groups of Functionally Related Genes Upregulated in Roots of the cys-c1 Mutant When Compared with the Wild Type.
Supplementary Material
Acknowledgments
This work was funded in part by the European Regional Development Fund through the Ministerio de Ciencia e Innovación (Grants BIO2007-62770 and BIO2010-15201) and the Junta de Andalucía (Grant BIO273). This work was also funded by the CONSOLIDER CSD2007-00057, Spain. I.G. is supported by a research contract from Junta de Andalucía (Programa de Retorno deInvestigadores).
References
- Albury M.S., Elliott C., Moore A.L. (2009). Towards a structural elucidation of the alternative oxidase in plants. Physiol. Plant. 137: 316–327 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Astier A., Baud F.J. (1996). Complexation of intracellular cyanide by hydroxocobalamin using a human cellular model. Hum. Exp. Toxicol. 15: 19–25 [DOI] [PubMed] [Google Scholar]
- Babula D., Misztal L.H., Jakubowicz M., Kaczmarek M., Nowak W., Sadowski J. (2006). Genes involved in biosynthesis and signalisation of ethylene in Brassica oleracea and Arabidopsis thaliana: Identification and genome comparative mapping of specific gene homologues. Theor. Appl. Genet. 112: 410–420 [DOI] [PubMed] [Google Scholar]
- Bariola P.A., Howard C.J., Taylor C.B., Verburg M.T., Jaglan V.D., Green P.J. (1994). The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J. 6: 673–685 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate - A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300 [Google Scholar]
- Bethke P.C., Libourel I.G., Reinöhl V., Jones R.L. (2006). Sodium nitroprusside, cyanide, nitrite, and nitrate break Arabidopsis seed dormancy in a nitric oxide-dependent manner. Planta 223: 805–812 [DOI] [PubMed] [Google Scholar]
- Beyer E.M., Jr (1976). A potent inhibitor of ethylene action in plants. Plant Physiol. 58: 268–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogatek R., Dziewanowska K., Lewak S. (1991). Hydrogen-cyanide and embryonal dormancy in apple seeds. Physiol. Plant. 83: 417–421 [Google Scholar]
- Bogatek R., Lewak S. (1991). Cyanide controls enzymes involved in lipid and sugar catabolism in dormant apple embryos during culture. Physiol. Plant. 83: 422–426 [Google Scholar]
- Bolwell G.P., Davies D.R., Gerrish C., Auh C.K., Murphy T.M. (1998). Comparative biochemistry of the oxidative burst produced by rose and french bean cells reveals two distinct mechanisms. Plant Physiol. 116: 1379–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borron S.W., Baud F.J., Mégarbane B., Bismuth C. (2007). Hydroxocobalamin for severe acute cyanide poisoning by ingestion or inhalation. Am. J. Emerg. Med. 25: 551–558 [DOI] [PubMed] [Google Scholar]
- Carol R.J., Dolan L. (2006). The role of reactive oxygen species in cell growth: Lessons from root hairs. J. Exp. Bot. 57: 1829–1834 [DOI] [PubMed] [Google Scholar]
- Carol R.J., Takeda S., Linstead P., Durrant M.C., Kakesova H., Derbyshire P., Drea S., Zarsky V., Dolan L. (2005). A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438: 1013–1016 [DOI] [PubMed] [Google Scholar]
- Castellano J.M., Vioque B. (2002). Characterisation of the ACC oxidase activity in transgenic auxin overproducing tomato during ripening. Plant Growth Regul. 38: 203–208 [Google Scholar]
- Chivasa S., Carr J.P. (1998). Cyanide restores N gene-mediated resistance to tobacco mosaic virus in transgenic tobacco expressing salicylic acid hydroxylase. Plant Cell 10: 1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciamporova M., Dekankova K., Hanackova Z., Peters P., Ovecka M., Baluska F. (2003). Structural aspects of bulge formation during root hair initiation. Plant Soil 255: 1–7 [Google Scholar]
- Cipollone R., Visca P. (2007). Is there evidence that cyanide can act as a neuromodulator? IUBMB Life 59: 187–189 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cohn M.A., Hughes J.A. (1986). Seed dormancy in red rice : v. Response to azide, hydroxylamine, and cyanide. Plant Physiol. 80: 531–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe A., Van der Straeten D. (2005). Ethylene biosynthesis and signaling: An overview. Vitam. Horm. 72: 399–430 [DOI] [PubMed] [Google Scholar]
- Ding L., Zhu J.K. (1997). A role for arabinogalactan-proteins in root epidermal cell expansion. Planta 203: 289–294 [DOI] [PubMed] [Google Scholar]
- Donato D.B., Nichols O., Possingham H., Moore M., Ricci P.F., Noller B.N. (2007). A critical review of the effects of gold cyanide-bearing tailings solutions on wildlife. Environ. Int. 33: 974–984 [DOI] [PubMed] [Google Scholar]
- Dugardeyn J., Van Der Straeten D. (2008). Ethylene: Fine-tuning plant growth and development by stimulation and inhibition of elongation. Plant Sci. 175: 59–70 [Google Scholar]
- Esashi Y., Isuzugawa K., Matsuyama S., Ashino H., Hasegawa R. (1991). Endogenous evolution of HCN during pre-germination periods in many seed species. Physiol. Plant. 83: 27–33 [Google Scholar]
- Fol M., Frachisse J.M., Petel G., Gendraud M. (1989). Effect of cyanide on the membrane-potential of Jerusalem artichoke (Helianthus tuberosus L) tuber-parenchyma - Different responses in relation to dormancy. C. R. Acad. Sci. III, Sci. Vie 309: 551–556 [Google Scholar]
- Foreman J., Demidchik V., Bothwell J.H., Mylona P., Miedema H., Torres M.A., Linstead P., Costa S., Brownlee C., Jones J.D., Davies J.M., Dolan L. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446 [DOI] [PubMed] [Google Scholar]
- Grossmann K. (1996). A role for cyanide, derived from ethylene biosynthesis, in the development of stress symptoms. Physiol. Plant. 97: 772–775 [Google Scholar]
- Gunasekar P.G., Borowitz J.L., Turek J.J., Van Horn D.A., Isom G.E. (2000). Endogenous generation of cyanide in neuronal tissue: involvement of a peroxidase system. J. Neurosci. Res. 61: 570–575 [DOI] [PubMed] [Google Scholar]
- Gunasekar P.G., Prabhakaran K., Li L., Zhang L., Isom G.E., Borowitz J.L. (2004). Receptor mechanisms mediating cyanide generation in PC 12 cells and rat brain. Neurol. Res. 49: 13–18 [DOI] [PubMed] [Google Scholar]
- Hall A.H., Dart R., Bogdan G. (2007). Sodium thiosulfate or hydroxocobalamin for the empiric treatment of cyanide poisoning? Ann. Emerg. Med. 49: 806–813 [DOI] [PubMed] [Google Scholar]
- Hatzfeld Y., Maruyama A., Schmidt A., Noji M., Ishizawa K., Saito K. (2000). beta-Cyanoalanine synthase is a mitochondrial cysteine synthase-like protein in spinach and Arabidopsis. Plant Physiol. 123: 1163–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Iwai T., Miyasaka A., Seo S., Ohashi Y. (2006). Contribution of ethylene biosynthesis for resistance to blast fungus infection in young rice plants. Plant Physiol. 142: 1202–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.A., Raymond M.J., Smirnoff N. (2006). Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J. 45: 83–100 [DOI] [PubMed] [Google Scholar]
- Jost R., Berkowitz O., Wirtz M., Hopkins L., Hawkesford M.J., Hell R. (2000). Genomic and functional characterization of the oas gene family encoding O-acetylserine (thiol) lyases, enzymes catalyzing the final step in cysteine biosynthesis in Arabidopsis thaliana. Gene 253: 237–247 [DOI] [PubMed] [Google Scholar]
- Karpova O.V., Kuzmin E.V., Elthon T.E., Newton K.J. (2002). Differential expression of alternative oxidase genes in maize mitochondrial mutants. Plant Cell 14: 3271–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight M.R. (2007). New ideas on root hair growth appear from the flanks. Proc. Natl. Acad. Sci. USA 104: 20649–20650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBrasseur N.D., MacIntosh G.C., Pérez-Amador M.A., Saitoh M., Green P.J. (2002). Local and systemic wound-induction of RNase and nuclease activities in Arabidopsis: RNS1 as a marker for a JA-independent systemic signaling pathway. Plant J. 29: 393–403 [DOI] [PubMed] [Google Scholar]
- Levina N.N., Lew R.R. (2006). The role of tip-localized mitochondria in hyphal growth. Fungal Genet. Biol. 43: 65–74 [DOI] [PubMed] [Google Scholar]
- Lin C.C., Wong B.K., Burgey C.S., Gibson C.R., Singh R. (2005). In vitro metabolism of a thrombin inhibitor and quantitation of metabolically generated cyanide. J. Pharm. Biomed. Anal. 39: 1014–1020 [DOI] [PubMed] [Google Scholar]
- Linke B., Börner T. (2005). Mitochondrial effects on flower and pollen development. Mitochondrion 5: 389–402 [DOI] [PubMed] [Google Scholar]
- Lister R., Chew O., Lee M.N., Heazlewood J.L., Clifton R., Parker K.L., Millar A.H., Whelan J. (2004). A transcriptomic and proteomic characterization of the Arabidopsis mitochondrial protein import apparatus and its response to mitochondrial dysfunction. Plant Physiol. 134: 777–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Martín M.C., Becana M., Romero L.C., Gotor C. (2008). Knocking out cytosolic cysteine synthesis compromises the antioxidant capacity of the cytosol to maintain discrete concentrations of hydrogen peroxide in Arabidopsis. Plant Physiol. 147: 562–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T., Burow M., Bauer M., Papenbrock J. (2003). Arabidopsis sulfurtransferases: investigation of their function during senescence and in cyanide detoxification. Planta 217: 1–10 [DOI] [PubMed] [Google Scholar]
- Michael G. (2001). The control of root hair formation: Suggested mechanisms. J. Plant Nutr. Soil Sci. 164: 111–119 [Google Scholar]
- Millenaar F.F., Lambers H. (2003). The alternative oxidase: in vivo regulation and function. Plant Biol. 5: 2–15 [Google Scholar]
- Monshausen G.B., Bibikova T.N., Messerli M.A., Shi C., Gilroy S. (2007). Oscillations in extracellular pH and reactive oxygen species modulate tip growth of Arabidopsis root hairs. Proc. Natl. Acad. Sci. USA 104: 20996–21001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navrot N., Rouhier N., Gelhaye E., Jacquot J.P. (2007). Reactive oxygen species generation and antioxidant systems in plant mitochondria. Physiol. Plant. 129: 185–195 [Google Scholar]
- Nguema-Ona E., Bannigan A., Chevalier L., Baskin T.I., Driouich A. (2007). Disruption of arabinogalactan proteins disorganizes cortical microtubules in the root of Arabidopsis thaliana. Plant J. 52: 240–251 [DOI] [PubMed] [Google Scholar]
- Ortega-Martínez O., Pernas M., Carol R.J., Dolan L. (2007). Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science 317: 507–510 [DOI] [PubMed] [Google Scholar]
- Papenbrock J., Schmidt A. (2000). Characterization of two sulfurtransferase isozymes from Arabidopsis thaliana. Eur. J. Biochem. 267: 5571–5579 [DOI] [PubMed] [Google Scholar]
- Parker J.S., Cavell A.C., Dolan L., Roberts K., Grierson C.S. (2000). Genetic interactions during root hair morphogenesis in Arabidopsis. Plant Cell 12: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiser G.D., Wang T.T., Hoffman N.E., Yang S.F., Liu H.W., Walsh C.T. (1984). Formation of cyanide from carbon 1 of 1-aminocyclopropane-1-carboxylic acid during its conversion to ethylene. Proc. Natl. Acad. Sci. USA 81: 3059–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton J.E. (1990). Cyanogenesis in plants. Plant Physiol. 94: 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A., Yekutieli D., Benjamini Y. (2003). Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19: 368–375 [DOI] [PubMed] [Google Scholar]
- Roje S. (2007). Vitamin B biosynthesis in plants. Phytochemistry 68: 1904–1921 [DOI] [PubMed] [Google Scholar]
- Sessions A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegien I., Bogatek R. (2006). Cyanide action in plants: From toxic to regulatory. Acta Physiol. Plant. 28: 483–497 [Google Scholar]
- Singh D.P., Moore C.A., Gilliland A., Carr J.P. (2004). Activation of multiple antiviral defence mechanisms by salicylic acid. Mol. Plant Pathol. 5: 57–63 [DOI] [PubMed] [Google Scholar]
- Smith A.G., Croft M.T., Moulin M., Webb M.E. (2007). Plants need their vitamins too. Curr. Opin. Plant Biol. 10: 266–275 [DOI] [PubMed] [Google Scholar]
- Smith J.M., Arteca R.N. (2000). Molecular control of ethylene production by cyanide in Arabidopsis thaliana. Physiol. Plant. 109: 180–187 [Google Scholar]
- Takeda S., Gapper C., Kaya H., Bell E., Kuchitsu K., Dolan L. (2008). Local positive feedback regulation determines cell shape in root hair cells. Science 319: 1241–1244 [DOI] [PubMed] [Google Scholar]
- Tanimoto M., Roberts K., Dolan L. (1995). Ethylene is a positive regulator of root hair development in Arabidopsis thaliana. Plant J. 8: 943–948 [DOI] [PubMed] [Google Scholar]
- Taylor C.B., Green P.J. (1991). Genes with homology to fungal and S-gene RNases are expressed in Arabidopsis thaliana. Plant Physiol. 96: 980–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M.A., Dangl J.L. (2005). Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 8: 397–403 [DOI] [PubMed] [Google Scholar]
- Toufighi K., Brady S.M., Austin R., Ly E., Provart N.J. (2005). The Botany Array Resource: e-Northerns, expression angling, and promoter analyses. Plant J. 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe G.C., Robson C.A., Yip J.Y.H. (2002). Induction of mitochondrial alternative oxidase in response to a cell signal pathway down-regulating the cytochrome pathway prevents programmed cell death. Plant Physiol. 129: 1829–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissenberg K., Fry S.C., Verbelen J.P. (2001). Root hair initiation is coupled to a highly localized increase of xyloglucan endotransglycosylase action in Arabidopsis roots. Plant Physiol. 127: 1125–1135 [PMC free article] [PubMed] [Google Scholar]
- Wang K.L.C., Li H., Ecker J.R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14(Suppl): S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Kusano M., Oikawa A., Fukushima A., Noji M., Saito K. (2008). Physiological roles of the beta-substituted alanine synthase gene family in Arabidopsis. Plant Physiol. 146: 310–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettenhall J.M., Simpson K.M., Satterley K., Smyth G.K. (2006). affylmGUI: A graphical user interface for linear modeling of single channel microarray data. Bioinformatics 22: 897–899 [DOI] [PubMed] [Google Scholar]
- Wong C.E., Carson R.A.J., Carr J.P. (2002). Chemically induced virus resistance in Arabidopsis thaliana is independent of pathogenesis-related protein expression and the NPR1 gene. Mol. Plant Microbe Interact. 15: 75–81 [DOI] [PubMed] [Google Scholar]
- Yamada K., et al. (2003). Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Nakamura T., Kusano T., Sano H. (2000). Three Arabidopsis genes encoding proteins with differential activities for cysteine synthase and beta-cyanoalanine synthase. Plant Cell Physiol. 41: 465–476 [DOI] [PubMed] [Google Scholar]
- Zagrobelny M., Bak S., Møller B.L. (2008). Cyanogenesis in plants and arthropods. Phytochemistry 69: 1457–1468 [DOI] [PubMed] [Google Scholar]
- Zgiczyński J.M., Stelmaszyńska T. (1979). Hydrogen cyanide and cyanogen chloride formation by the myeloperoxidase-H2O2-Cl- system. Biochim. Biophys. Acta 567: 309–314 [DOI] [PubMed] [Google Scholar]
- Zhu C., Gan L., Shen Z., Xia K. (2006). Interactions between jasmonates and ethylene in the regulation of root hair development in Arabidopsis. J. Exp. Bot. 57: 1299–1308 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.