Partner specificity in legume-rhizobia symbiosis involves perception of rhizobial signals by plant lysin motif receptor-like kinases (LysM-RLKs) leading to the formation of nitrogen-fixing root nodules. This work describes PUB1, a protein interactor of LYK3 LysM-RLK, which is involved in regulating the specificity of interaction between Medicago truncatula and Sinorhizobium meliloti.
Abstract
LYK3 is a lysin motif receptor-like kinase of Medicago truncatula, which is essential for the establishment of the nitrogen-fixing, root nodule symbiosis with Sinorhizobium meliloti. LYK3 is a putative receptor of S. meliloti Nod factor signals, but little is known of how it is regulated and how it transduces these symbiotic signals. In a screen for LYK3-interacting proteins, we identified M. truncatula Plant U-box protein 1 (PUB1) as an interactor of the kinase domain. In planta, both proteins are localized and interact in the plasma membrane. In M. truncatula, PUB1 is expressed specifically in symbiotic conditions, is induced by Nod factors, and shows an overlapping expression pattern with LYK3 during nodulation. Biochemical studies show that PUB1 has a U-box–dependent E3 ubiquitin ligase activity and is phosphorylated by the LYK3 kinase domain. Overexpression and RNA interference studies in M. truncatula show that PUB1 is a negative regulator of the LYK3 signaling pathway leading to infection and nodulation and is important for the discrimination of rhizobia strains producing variant Nod factors. The potential role of PUB E3 ubiquitin ligases in controlling plant–microbe interactions and development through interacting with receptor-like kinases is discussed.
INTRODUCTION
The ability of legumes to form a symbiosis with nitrogen-fixing bacteria termed rhizobia is of immense agronomic and ecological importance (Graham and Vance, 2003). The symbiosis is manifested by the production of nodules on the roots of the plant in which the bacteria fix nitrogen in exchange for plant carbohydrate and a safe niche. Genetic studies on both the bacterium and the plant have identified Nod factor signaling as an essential step in the establishment of the symbiosis in most, but not all studied legume–rhizobia interactions (Dénarié et al., 1996; Oldroyd and Downie, 2008; Masson-Boivin et al., 2009). Nod factors (NFs) are rhizobial lipochitooligosaccharides with a chitin fragment backbone of four or five N-acetyl glucosamines (tetrameric or pentameric factors) in which the terminal nonreducing sugar is de-N-acetylated and acylated with a fatty acid. The structure of the fatty acid and the type of chemical substitutions on the chitin fragment backbone are important elements for host–microbe recognition, infection, and nodulation. For example, the NFs produced by Sinorhizobium meliloti, which nodulates Medicago truncatula and related genera, are mainly tetrameric with a C16:2 fatty acid and a 6-O-acetyl substitution on the terminal nonreducing sugar and a 6-O-sulphuryl group on the reducing sugar (Roche et al., 1991a). The O-sulfate is important for all NF activities, whereas the substitutions on the terminal, nonreducing sugar are important for infection (Roche et al., 1991b; Ardourel et al., 1994).
Studies, particularly on model legumes such as M. truncatula and Lotus japonicus, have led to the identification of specific lysin motif receptor-like kinases (LysM-RLKs) as putative NF receptors (Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003). In both M. truncatula and L. japonicus, two probably orthologous pairs of genes are essential for nodulation. M. truncatula NFP and L. japonicus NFR5 are required for all NF responses and may play very similar roles (Radutoiu et al., 2003; Arrighi et al., 2006). However, whereas L. japonicus NFR1 is also required for early NF responses (Radutoiu et al., 2003), its putative ortholog in M. truncatula, LYK3, is not required for early responses but plays an essential role in regulating root hair curling and in promoting infection thread (IT) growth and nodulation (Catoira et al., 2001; Limpens et al., 2003; Smit et al., 2007).
It is important to understand how these receptors perceive NFs and transduce the signal to downstream components. Plant genetic studies have revealed that downstream of the LysM-RLKs, NF signal transduction requires a calcium signaling pathway, called the common symbiotic or DMI (for Does not Make Infections) pathway that is also required for establishment of the symbiosis with endomycorrhizal fungi (Oldroyd and Downie, 2008). In M. truncatula, this pathway is defined by three genes: DMI2 (encoding a leucine-rich repeat [LRR] RLK), DMI1 (encoding an ion channel protein), and DMI3 (encoding a calcium calmodulin protein kinase). Whereas DMI1 and DMI2 are required for calcium spiking, DMI3 is required for transducing the calcium signal. Further downstream, specific transcription factors are required for gene activation, including expression of early nodulin genes such as ENOD11 (Andriankaja et al., 2007; Hirsch et al., 2009). Another regulatory gene, NIN, is not necessary for early NF responses but is required for nodulation and infection (Marsh et al., 2007).
A key question remains on how these pathways are activated by the LysM-RLKs. In M. truncatula, studies on plant responses to rhizobial nodulation mutants led to the suggestion that there are two NF receptors: the signaling receptor, which has a nonstringent requirement for the structure of the nonreducing end of NFs and leads to initial NF responses, and the entry receptor, which requires NFs with precise modifications on the nonreducing end and leads to infection (Ardourel et al., 1994). This hypothesis was supported by the different phenotypes of nfp and lyk3 mutants, with NFP being proposed to be a major component of the signaling receptor as it is required for all early signaling responses, including calcium spiking and gene expression (Ben Amor et al., 2003; Arrighi et al., 2006). As the DMI pathway is required for the majority of the signaling responses, it is proposed that this pathway is activated via NFP. Mutants in LYK3 (initially named HCL) were identified due to their aberrant root hair curling and infection (Catoira et al., 2001), and later the gene was cloned through potential synteny with a gene from pea (Pisum sativum) that is involved in NF structure-dependent infection (Limpens et al., 2003; Smit et al., 2007). These mutants still show NF signaling responses, suggesting that LYK3 is not involved in the early NF recognition step. As rhizobial infection in a weak lyk3 (hcl-4) mutant and LYK3-RNA interference (RNAi) plants is dependent on the structure of the modifications on the terminal nonreducing sugar, it has been suggested that LYK3 is a component of the entry receptor with a stringent demand toward the NF structure (Smit et al., 2007). Transcriptomics using a lyk3 mutant (Mitra et al., 2004) have shown that LYK3 is required for the regulation of a subset of genes that are differentially expressed in the plant after 24 h exposure to S. meliloti, suggesting that its activity leads to specific gene expression. To date, how LYK3 is regulated and couples to this specific gene expression, root hair curling, infection, and nodulation is completely unknown.
Although forward genetics on model legumes has proved a powerful tool for identifying genes essential for establishment of the symbiosis, it is likely that other key players have been overlooked either due to redundancy or playing pleiotropic roles. In this work, we set out to identify such genes through the identification of protein interactors of LYK3 using a yeast two-hybrid screen.
RESULTS
PUB1, an UND-PUB-ARM Protein, Is an Interactor of the Intracellular Region of LYK3
To identify potential interacting proteins of LYK3, the intracellular region of the protein (LYK3-IR: amino acids 257 to 620), containing the juxta membrane, kinase domain, and C-tail, was used as bait to screen a GAL4-based yeast two-hybrid cDNA library prepared from the root hair cells of M. truncatula roots treated with NFs (Andriankaja et al., 2007). Thirty-two clones contained a coding region related to the same gene, annotated as Mt5g090510 in the M. truncatula genome sequencing project. From the cDNA sequences of our clones and those in the M. truncatula Expressed Sequence Tag (MtEST) database, it is clear that the C terminus of the protein has not been correctly deduced from the automatic gene discovery/annotation programs in the M. truncatula genome sequencing project; thus, our sequence has been submitted to the GenBank databases and can be found under accession number BK007068.
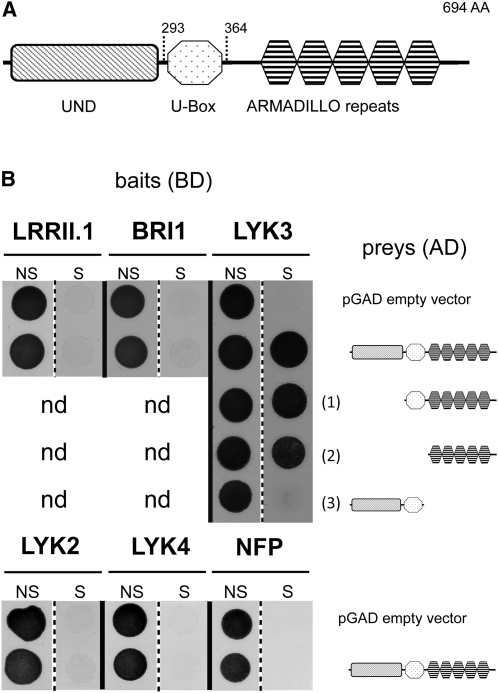
Using the BLAST algorithm, interrogation of the National Centre for Biotechnology Information nonredundant protein database revealed that the encoded protein is related to the Plant U-box (PUB) E3 ubiquitin ligase family (Azevedo et al., 2001; Yee and Goring, 2009). The gene was thus named PUB1. Domain structure analysis showed that the U-box in this protein is highly conserved and followed by a region containing at least five ARMADILLO (ARM) repeats, some of which are highly variant compared with the consensus sequence. PUB1 also contains the large U-box N-terminal Domain (UND), present in a subset of PUB-ARM proteins (Mudgil et al., 2004), including the well-characterized Brassica napus protein ARC1 (Gu et al., 1998; Stone et al., 1999). An alignment to homologous plant proteins, including ARC1 (see Supplemental Figure 1 online) shows that the UND domain is poorly conserved compared with the U-box and ARM repeat domains. In summary, PUB1 has the typical structure of an UND-PUB-ARM protein (Figure 1A).
Figure 1.
Characterization of PUB1 as a Protein Interactor of the LYK3 Intracellular Region.
(A) Domain organization of the PUB1 protein. The U-box domain (amino acids 293 to 364) is located between the UND domain at the N terminus and the ARMADILLO repeats near the C terminus.
(B) PUB1 interacts with LYK3 through the ARM repeats in yeast. Yeast cells expressing either LRRII.1, BRI1, LYK2, LYK4, NFP, or LYK3 intracellular regions cloned in the pBD bait vector were cotransformed with prey constructs cloned in the pGAD vector. The negative control prey consists of the pGAD vector. A full-size PUB1 prey fusion protein was tested for interaction with all bait fusion proteins. Three deletion variants, PUB1ΔUND (1), PUB1ΔUNDΔU-Box (2), and PUB1ΔARM (3), were tested for interaction with the LYK3 bait. Cotransformation of yeast cells with bait and prey vectors is reported by yeast growth on nonselective (NS) medium lacking Trp and Leu. Positive interactions are reported by yeast growth on selective (S) medium lacking Trp, Leu, and His and supplemented with 5 mM 3-amino-triazol. Yeast growth is in black. nd, not determined.
To test the specificity of the LYK3-IR/PUB1 interaction, the intracellular region of two RLKs from the LRR subfamilies was tested: the root-expressed LRR-RLK (LRRII.1) from M. truncatula (Lefebvre et al., 2010) and the brassinosteroid RLK (BRI1) from Arabidopsis thaliana (Li and Chory, 1997). No interaction between these RLKs and PUB1 could be detected (Figure 1B). Specificity was then tested against members of the two subfamilies of LysM-RLKs: LYK2, LYK4, and NFP. No interaction with PUB1 could be detected. The shorter clones isolated from the library lacked the UND domain (PUB1ΔUND), indicating that this domain is not essential for interacting with LYK3-IR as shown by pairwise interactions in Figure 1B. Further interacting-domain mapping revealed that the region containing the ARM repeats is necessary and sufficient for the LYK3-IR/PUB1 interaction in yeast (Figure 1B).
PUB1 Associates with Membranes and Interacts with LYK3 in Planta
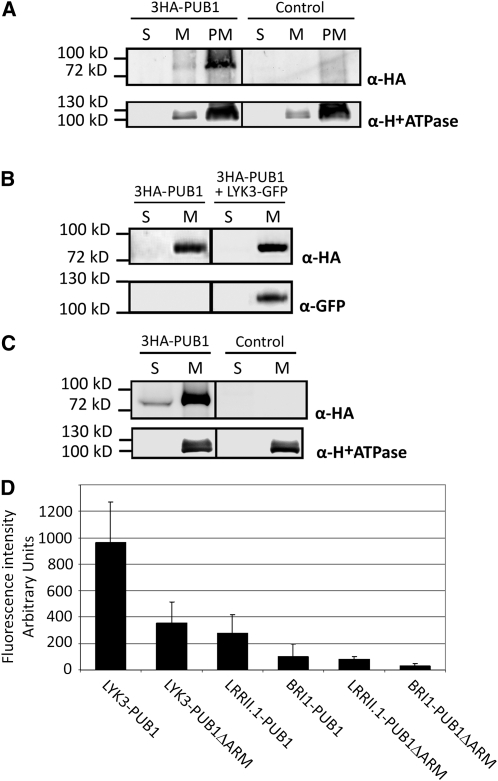
To investigate the subcellular localization of PUB1, we first attempted to express yellow fluorescent protein (YFP)-tagged PUB1 proteins in Nicotiana benthamiana leaves. However, the fusion proteins showed partial cleavage, thus making localization based on YFP fluorescence unreliable. The subcellular localization of PUB1 was therefore determined by cellular fractionation using an N-terminal 3HA-tagged fusion protein (3HA-PUB1). The construct was expressed in N. benthamiana leaves and in M. truncatula roots by Agrobacterium tumefaciens and Agrobacterium rhizogenes transformation, respectively. Immunoblot analyses with an anti-HA antibody specifically detected the expressed 3HA-PUB1 fusion protein in both plant systems at the expected size of 81 kD (Figures 2A to 2C).
Figure 2.
Subcellular Localization of PUB1 and Interaction with LYK3 in Planta.
For all biochemical experiments ([A] to [C]), plant material was fractionated into plasma membrane (PM), total membrane (M), or soluble (S) fractions. S and M fractions were concentrated and resuspended in the same volume and equal volumes were loaded. M and PM fractions contained equal protein amounts. The three fractions were used in immunoblots with indicated antibodies.
(A) 3HA-PUB1 is associated with the plasma membrane in N. benthamiana. N. benthamiana leaves were transformed with the 3HA-PUB1 construct or the vector alone (control).
(B) The membrane localization of PUB1 is not altered by coexpression with LYK3 in N. benthamiana. N. benthamiana leaves expressed 3HA-PUB1 alone or together with LYK3-GFP.
(C) 3HA-PUB1 is predominantly associated with membranes in M. truncatula and is also present in the soluble fraction. M. truncatula roots were transformed with either the 3HA-PUB1 construct or control vector expressing GFP.
(D) PUB1 interacts with LYK3 and not with BRI1 or LRRII.1 in N. benthamiana. BiFC experiments were performed by coexpression of the indicated split-YFP pair combinations: The C-terminal domain of YFP (Yc) was fused to the C terminus of the RLKs, whereas the N-terminal domain of YFP (Yn) was fused to the N terminus of PUB1 or a derivative lacking the ARM repeats (PUB1ΔARM). Leaves were imaged by epifluorescence using identical exposure settings and quantified. Fluorescence intensities (arbitrary units) and standard deviations are shown.
Soluble (S), total membrane (M), and plasma membrane (PM) fractions were prepared from leaves of N. benthamiana either expressing 3HA-PUB1 or transformed with a vector control. Immunoblot analysis showed strong enrichment of the H+ATPase PM marker proteins (Maudoux et al., 2000) in the PM fraction compared with the M fraction, whereas this protein was not detected in the S fraction (Figure 2A). A similar partitioning between the three fractions for 3HA-PUB1 and the H+ATPase marker (Figure 2A) suggests that full-length PUB1 is associated with the PM. To investigate whether the presence of the LYK3 receptor could cause dissociation of PUB1 from the membranes, both proteins were coexpressed in N. benthamiana leaves. The 3HA-PUB1 expressed alone or together with a green fluorescent protein (GFP)-tagged LYK3 fusion protein was detected only in the M fraction (Figure 2B). S and M fractions were also prepared from M. truncatula roots expressing 3HA-PUB1. Detection of the H+ATPase marker exclusively in the M fraction shows that the S fraction was not contaminated with membranes (Figure 2C). PUB1 was found to be preferentially associated with membranes in M. truncatula roots, although a small proportion of the protein appeared to be soluble (Figure 2C). Together, the data show that PUB1 is predominantly associated with membranes, independently of the presence of LYK3.
Bimolecular fluorescence complementation (BiFC) was next used to confirm the interaction between LYK3 and PUB1 in planta. This system has recently been used to demonstrate the specific interaction between LYK3 and a PM-associated protein, SYMREM1 (Lefebvre et al., 2010). YFP or its C-terminal part (Yc) was cloned as a translational fusion to the C terminus of the LYK3, LRRII.1, and BRI1 RLKs. The N-terminal part (Yn) was cloned as a translational fusion to the N terminus of PUB1. Fusion proteins were expressed in N. benthamiana leaves by A. tumefaciens–mediated transformation. All the RLKs were detected at the PM but LYK3 appeared to be relatively poorly expressed (see Supplemental Figure 2 online, left panels). However, coexpression of LYK3-Yc together with Yn-PUB1 led to a clear YFP signal at the PM of leaf epidermal cells whereas relatively weak fluorescence signals were detected when the BRI1-Yc/Yn-PUB1 or the LRRII.1-Yc/Yn-PUB1 pairs were used (Figure 2D; see Supplemental Figure 2 online). In all cases, PUB1 lacking the ARM repeats (Yn-PUB1ΔARM) gave weaker fluorescence signals than the full-length protein. Thus, although the ARM repeats are clearly important for the interaction with LYK3 in planta, it is possible that other domains also contribute. In summary, these results show that PUB1 is capable of interacting with the full-length LYK3 receptor at the PM in living plant cells.
PUB1 Is an E3 Ubiquitin Ligase and Is Phosphorylated by LYK3 in Vitro
The presence of the U-box domain suggests that PUB1 is an E3 ubiquitin ligase and thus functions in protein ubiquitination, a reaction that requires ubiquitin (Ub), an E1 (Ub-activating) protein, an E2 (Ub-conjugating) protein, and a protein substrate. Ubiquitination is a posttranslational modification specific to eukaryotes and in the past years has been recognized as a major means of protein regulation. In many cases, ubiquitination marks the protein for degradation, although in some cases, it modulates the activity or localization of the protein (Komander, 2009).
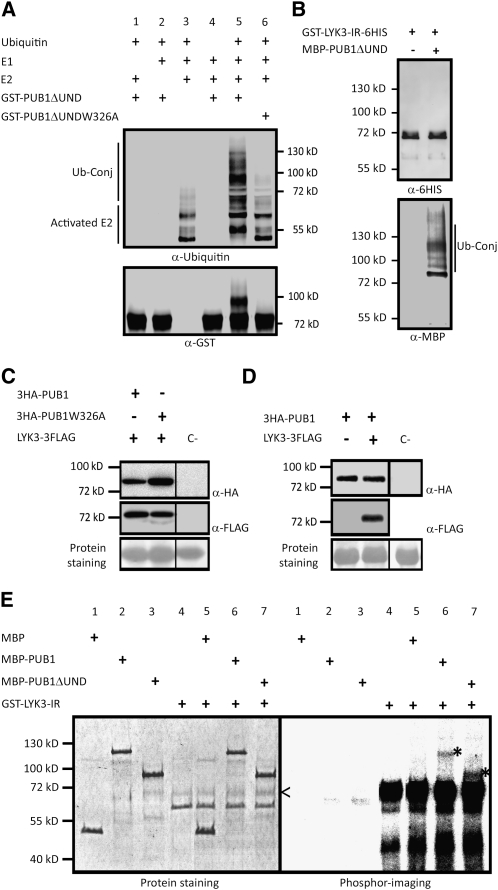
To examine whether PUB1 is a functional E3 ubiquitin ligase, we performed in vitro ubiquitination assays with PUB1 lacking the UND domain (PUB1ΔUND), purified from Escherichia coli as a glutathione S-transferase (GST) fusion protein. In addition, a construct was made in which a highly conserved Trp residue in the U-box domain was mutated to Ala (PUB1ΔUNDW326A), a modification that has previously been shown to abolish E3 ubiquitin ligase activity (Trujillo et al., 2008). A yeast E1 protein and the Arabidopsis E2 protein UBC8 were used in these assays. As expected, the E2 protein was activated by ubiquitination in the presence of the E1 and ubiquitin, independently of PUB1ΔUND (Figure 3A, lane 3). When PUB1ΔUND was added to the reaction, a protein ladder and a band higher than PUB1ΔUND could be detected using anti-ubiquitin and anti-GST antibodies, respectively (Figure 3A, lane 5). Together, these results show that PUB1ΔUND has E3 ubiquitin ligase activity and strongly suggest that the protein autoubiquitinates. The observed E3 ubiquitin ligase activity of PUB1 was dependent on the presence of Ub, E1, and E2 and was abolished by the W326A mutation in the U-box domain (Figure 3A, lane 6). Thus, PUB1 is a functional E3 ubiquitin ligase whose enzymatic activity depends on the structure of the U-box domain.
Figure 3.
In Vitro E3 Ubiquitin Ligase Activity of PUB1, Trans-Phosphorylation of PUB1 by LYK3, and Coexpression Studies of PUB1 and LYK3 in N. benthamiana.
(A) PUB1 possesses an E3 ubiquitin ligase activity in vitro. GST-PUB1ΔUND was tested for E3 ubiquitin ligase activity in presence or absence of yeast E1, Arabidopsis E2 (UBC8), and ubiquitin. Anti-Ub and anti-GST antibodies were used to detect ubiquitinated proteins and GST-PUB1ΔUND, respectively. A range of proteins conjugated with ubiquitin (Ub-conj) were formed when GST-PUB1ΔUND was added in the mix and not with GST-PUB1ΔUND-W326A, an inactive U-box variant.
(B) LYK3 in vitro ubiquitination assay. GST-LYK3-IR-6HIS with E1, E2, and ubiquitin was incubated in the presence or absence of MBP-PUB1ΔUND. Immunoblot analyses were performed with indicated antibodies.
(C) PUB1 does not affect LYK3 abundance in N. benthamiana. 3HA-PUB1 or the inactive 3HA-PUB1W326A mutated protein was coexpressed with LYK3-3FLAG. Protein combinations were expressed in different halves of the same N. benthamiana leaf to avoid leaf expression effects. Total extracts from 1-cm diameter leaf discs were analyzed by immunobloting with the corresponding antibodies. Nontransformed leaves were used as the immunoblotting control (C-). Ponceau red staining shows that equal amount of protein were loaded on the gel.
(D) LYK3 does not affect PUB1 abundance in N. benthamiana. 3HA-PUB1 was expressed with or without LYK3-3FLAG. Experiments were performed as in (C).
(E) PUB1 is phosphorylated by LYK3 in vitro. The auto- and trans-phosphorylation activities of GST-LYK3 (intracellular region) were assayed alone or by addition of MBP, MBP-PUB1, or MBP-PUB1ΔUND. On the left panel, protein staining shows the size and quantity of the purified proteins. On the right panel, phosphor imaging reveals LYK3 kinase-dependent phosphorylation. The ~70-kD recombinant LYK3 kinase (<) as detected by immunoblot using anti-GST antibodies (see Supplemental Figure 3 online) shows autophosphorylation. Trans-phosphorylated forms of MBP-PUB1 and MBP-PUB1ΔUND are visible at the expected size of 117 and 87 kD, respectively (*). No radiolabeling of MBP is observed.
We also tested whether the LYK3 intracellular region could be a substrate for the E3 ubiquitin ligase activity of PUB1. In vitro ubiquitination assays using a maltose binding protein (MBP) fusion of PUB1ΔUND and GST-LYK3-IR-6HIS did not succeed in detecting ubiquitinated forms of the LYK3-IR (Figure 3B) even after long exposure of the immunoblot.
A way to follow whether PUB1 ubiquitinates LYK3 in a plant system leading to its degradation is to follow LYK3 stability in the presence of PUB1. Despite many efforts (production of LYK3 specific antibodies and purifying plasma membrane), we were unable to detect LYK3 in M. truncatula, likely due to extremely low levels of the protein. Also, we were unable to produce stable transgenic lines of M. truncatula overexpressing LYK3, as expression was lost in all transgenic plants following propagation by seed. As an alternative approach, we coexpressed 3HA-PUB1 or 3HA-PUB1W326A with a LYK3-3FLAG fusion protein in N. benthamiana leaves. Similar levels of the LYK3 protein were detected using the active and inactive PUB1 proteins, suggesting that LYK3 is not preferentially degraded through the E3 ubiquitin ligase activity of PUB1 (Figure 3C). Therefore, we have no evidence to suggest that LYK3 is a substrate of the E3 ubiquitin ligase activity of PUB1, leading to its degradation. We then investigated whether PUB1 protein stability could be regulated by LYK3. For that purpose, PUB1 protein abundance was monitored after expression of 3HA-PUB1 with or without LYK3-3FLAG in N. benthamiana (Figure 3D). No difference in PUB1 protein abundance was detected in either case, suggesting that interaction with LYK3 does not lead to degradation of PUB1.
LYK3 possesses an active kinase domain as demonstrated previously by an in vitro autophosphorylation assay of a GST-LYK3-IR fusion protein (Arrighi et al., 2006). We therefore investigated whether the LYK3 kinase could trans-phosphorylate PUB1. PUB1 and PUB1ΔUND were expressed as MBP fusion proteins and purified from E. coli. In vitro protein phosphorylation was followed by incorporation of radiolabeled phosphate. Strong autophosphorylation of LYK3-IR was observed as expected (Figure 3E, lanes 4 to 7; see Supplemental Figure 3 online). In the presence of GST-LYK3, addition of MBP-PUB1 or MBP-PUB1ΔUND led to phosphorylation of both proteins (lanes 6 and 7) by the kinase, whereas MBP alone was not phosphorylated (Figure 3E, lane 5). These results suggest that LYK3 phosphorylates the PUB-ARM region of PUB1.
PUB1 Is a Divergent Member of the M. truncatula UND-PUB-ARM Family
Higher plants contain a family of UND-PUB-ARM proteins (Mudgil et al., 2004; Samuel et al., 2008); for example, there are 17 such genes in Arabidopsis. Only a few UND-PUB-ARM genes have been functionally characterized; these include Arabidopsis PUB17 and its potential tobacco ortholog, ACRE276 (Yang et al., 2006), B. napus ARC1 (Gu et al., 1998; Stone et al., 1999), tobacco PUB1 (Kim et al., 2003), and Oryza sativa SPL11 (Zeng et al., 2004).
To identify potential homologs of Medicago PUB1, the current M. truncatula genome release (version 3.0) was searched for genes closely related to Mt PUB1 and encoding UND-PUB-ARM proteins (see Supplemental Methods online). First, a low stringency similarity search for U-box domains using the BLAST-P algorithm and Mt PUB1 U-box domain as reference was performed against all M. truncatula predicted proteins. In a second step, we looked for the presence of ARM repeats in the previously retrieved protein subset using the most conserved ARM repeat of Mt PUB1. Finally, a subset was made of those proteins containing an UND domain. To ensure that no proteins closely related to Mt PUB1 were missed, further searches were made with the complete Mt PUB1 protein and its three component domain regions. Two sequences on chromosome 2 showing the highest similarity to Mt PUB1 were found by this analysis (Mt2g007770/80), but they do not appear to contain an UND domain. The predicted proteins have an incomplete U-box, and there is no evidence from the EST database that they are expressed. Seven genes encoding complete UND-PUB-ARM proteins, in addition to Mt PUB1, were identified. These eight M. truncatula proteins, 17 proteins from Arabidopsis, and the four functionally characterized proteins (Nt PUB4, Nt ACRE276, Bn ARC1, and Os SPL11) were aligned (see Supplemental Data Set 1 online) and used in phylogenetic analyses.
These analyses identified M. truncatula proteins that are potentially orthologous to several of the Arabidopsis UND-PUB-ARM proteins and of Nt ACRE276 and Bn ARC1 (see Supplemental Figure 4 online). Mt PUB1, however, does not appear to be closely related to any of the proteins used in the analysis. To look more closely at the evolution of this gene, similarity searching using the BLAST-P algorithm was used to identify UND-PUB-ARM homologs from other plant species whose genomes are highly sequenced, including legumes (Glycine max and L. japonicus), other dicots (Vitis vinifera and Populus trichocarpa), and a monocot (O. sativa). Alignment and phylogenetic analysis of these proteins show that Mt PUB1 falls into a discrete subclade (see Supplemental Data Set 2 and Supplemental Figure 5 online). This subclade comprises two L. japonicus, one G. max, one V. vinifera, and two P. trichocarpa proteins, which are thus the potential orthologs of Mt PUB1.
PUB1 Is Expressed during Symbiosis and Is Induced by NFs
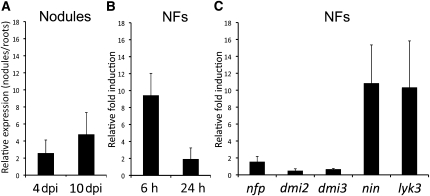
To identify the expression profiles of PUB1 and its closest homologs, we first used the M. truncatula Gene Expression Atlas, which is based on microarray analysis (http://bioinfo.noble.org/gene-atlas/v2/; Benedito et al., 2008; He et al., 2009). PUB1 and its previously described M. truncatula homologs, with the exception of Mt7g088030, are represented in this database. Most of these M. truncatula UND-PUB-ARM genes are expressed in both roots and aerial organs, in a variety of conditions (see Supplemental Data Set 3 online). By contrast, PUB1 has a very different and much narrower expression pattern. It shows very low expression in all organs during nonsymbiotic growth and is strongly induced in roots during nodulation and to a lesser extent following mycorrhization. These Gene Expression Atlas data for PUB1 induction during nodulation were confirmed by quantitative RT-PCR (qRT-PCR) (Figure 4A). Quantification of PUB1 mRNA abundance was performed in nitrogen-starved roots of M. truncatula and in bumps and developing nodules at 4 and 10 d after inoculation (DAI), respectively. Ratios of gene induction obtained by qRT-PCR in 4 and 10 d samples in relation to the roots (Figure 4A) were similar to the ratios calculated from the Gene Expression Atlas data (see Supplemental Data Set 3 online).
Figure 4.
Quantification of PUB1 Expression upon Rhizobia Inoculation and NF Treatment Using qRT-PCR.
Each bar represents the average of values from three independent biological replicates normalized using two reference genes. Standard deviation between biological replicates is represented.
(A) PUB1 is induced in nodules at 4 and 10 DAI (dpi) compared with noninoculated roots.
(B) PUB1 is induced after 6 and 24 h of NF versus water treatment.
(C) PUB1 is not induced after 6 h of NF versus water treatment in nfp, dmi2, and dmi3 mutants but is still induced in nin and lyk3 mutants.
NFs are the major elicitor of transcriptional changes occurring at early stages of the nitrogen fixing symbiosis (Mitra et al., 2004); therefore, we investigated whether they could elicit changes in PUB1 expression. PUB1 expression in roots of M. truncatula seedlings treated with NFs was compared with water-treated samples by qRT-PCR. PUB1 showed a very early and transient induction after NF application in M. truncatula (Figure 4B). To assess if this induction was dependent on the NF signaling pathway, the same experiment was performed after 6 h of NF application to roots of various symbiotic mutants (Figure 4C). No induction of PUB1 expression was detected in the nfp, dmi2, and dmi3 mutants. By contrast, induction of PUB1 expression was still observed in the nin and lyk3 mutants (Figure 4C). These results suggest that PUB1 induction by NFs is dependent on the signaling receptor (NFP) and the DMI pathway rather than on the entry receptor (LYK3).
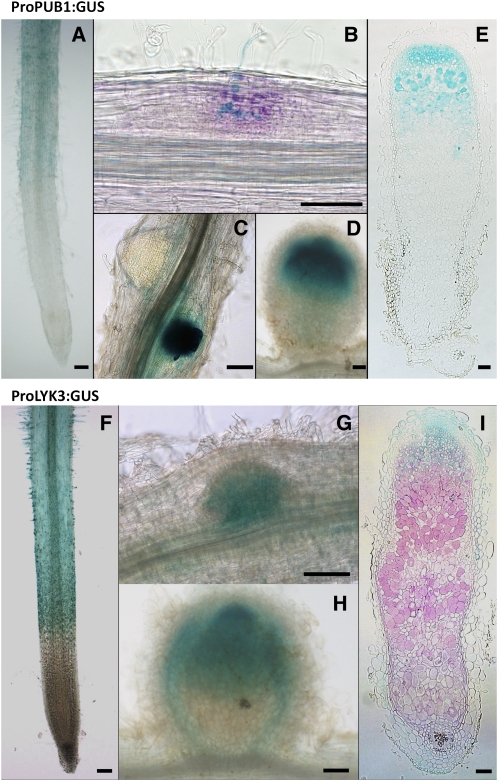
To examine the spatial expression pattern of PUB1 during nodulation, A. rhizogenes was used to produce transgenic roots containing fusions of a 3-kb promoter region of PUB1 to the β-glucuronidase (GUS) reporter gene (ProPUB1:GUS). In noninoculated roots, the GUS reporter was expressed weakly, including in the hairs of the developing root hair zone, which are most susceptible to rhizobial infection (Figure 5A). Following inoculation, expression of the reporter was noticeably stronger in cortical cells in close contact to rhizobial infections (Figure 5B). Strong GUS expression was seen in the nodule primordia compared with lateral root primordia (Figure 5C). In young nodules, the expression was confined to the apical regions (Figure 5D). In mature nodules GUS expression was evident in a broad apical region encompassing the preinfection, infection, and early nitrogen fixation zones (Figure 5E). Thus, PUB1 shows an expression pattern compatible with a specific role during early stages of nodulation and during infection. Because PUB1 is a protein interactor of LYK3, we also examined the spatial expression pattern of LYK3 to assess if the two patterns of expression overlap. Transgenic roots containing fusions of a 2.6-kb promoter region of LYK3 to the GUS reporter gene (ProLYK3:GUS) were generated by A. rhizogenes transformation. In noninoculated roots, the reporter was clearly expressed in the epidermal and root hair cells of the developing root hair zone (Figure 5F). Following inoculation, strong induction occurred in the nodule primordia (Figure 5G). In young nodules, GUS expression was evident in a broad apical region, like PUB1, and in the vasculature (Figure 5H). In mature nodules, expression was predominantly in the apical part of the nodule encompassing the preinfection and infection zones (Figure 5I). Thus, LYK3 and PUB1 patterns of expression show strong overlap before and during nodulation.
Figure 5.
Histochemical Localization of ProPUB1:GUS and ProLYK3:GUS Expression during Nodulation of M. truncatula.
PUB1 expression pattern was determined using a 3-kb promoter region of the gene cloned in front of the GUS coding sequence. LYK3 expression pattern was determined using the 2.6-kb promoter region of LYK3 cloned in front of the GUS-GFP fusion coding sequence. M. truncatula roots transformed by A. rhizogenes were used for ProPUB1:GUS or ProLYK3:GUS detection during nodulation using either X-Gluc (blue in [A] and [C] to [I]) or Magenta-Gluc (magenta in [B]). Rhizobia were detected by β-galactosidase (LacZ) activity using X-Gal (blue in [B]) or Magenta-Gal (magenta in [I]). Bars = 100 μm.
(A) ProPUB1:GUS is weakly expressed in noninoculated roots, including in the root hair development zone.
(B) After inoculation, ProPUB1:GUS is expressed predominantly in the nodule primordium and in cells associated with infection.
(C) ProPUB1:GUS is expressed strongly in nodule compared with root primordia.
(D) ProPUB1:GUS is expressed in the apical region of young, developing nodules.
(E) In mature nodules, ProPUB1:GUS is expressed in the apical region, including the preinfection and infection zones and the early nitrogen-fixing zone.
(F) ProLYK3:GUS is expressed in the developing root hair zone of noninoculated roots, including the epidermal cells.
(G) ProLYK3:GUS is expressed in the nodule primordium.
(H) In young nodules ProLYK3:GUS is expressed predominantly in the apical region of the nodule and to some extent in the vascular tissue.
(I) In mature nodules, ProLYK3:GUS expression is predominantly in the apical region, including the preinfection and infection zones.
PUB1 Is a Negative Regulator of Infection and Nodulation
To examine the physiological role of PUB1, we attempted to identify mutants in the gene. However, despite screening the major M. truncatula mutant populations obtained by fast-neutron (Rogers et al., 2009), Tnt1 transposon insertion (d’Erfurth et al., 2003; Tadege et al., 2008), and chemical (ethyl methanesulfonate) mutagenesis (Le Signor et al., 2009), we were unable to recover null mutants in PUB1. We thus altered expression of the gene by overexpression with the Pro35S and knockdown using RNAi in A. rhizogenes–transformed roots.
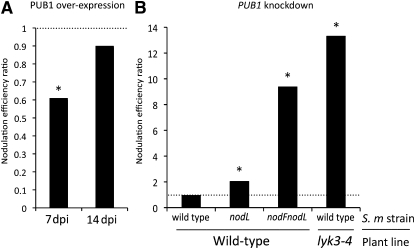
We previously showed (Figure 2C) that expression with the Pro35S of 3HA-PUB1 in wild-type M. truncatula leads to the production of the protein of the expected size. Following inoculation with the wild-type S. meliloti strain, the plants expressing 3HA-PUB1 showed a reduction of ~40% in the number of nodules per plant compared with the control at 7 DAI in two large-scale experiments (Table 1). At 14 DAI, the difference was no longer significant, showing that overexpression of PUB1 caused a delay in nodulation. These results are graphically represented in Figure 6A by showing the ratios of the mean number of nodules in the assay versus the control using data pooled from the two independent experiments; this ratio is referred to as the nodulation efficiency ratio and should be 1.0 if the genetic modification has no effect. The decreased nodulation efficiency ratio at 7 DAI (Figure 6A) suggests that PUB1 could play a negative role in the establishment of the nitrogen-fixing symbiosis.
Table 1.
Nodulation Phenotype of M. truncatula Roots Overexpressing PUB1
| Mean No. of Nodules per Plant |
||||
| DAI | Experiment | Control | 3HA-PUB1 | P Valuea |
| 7 DAI | Experiment 1 | 6.86 ± 5.81 (n = 96) | 4.18 ± 4.19 (n = 87) | 5.27 × 10−4*** |
| Experiment 2 | 6.95 ± 5.21 (n = 43) | 4.29 ± 3.44 (n = 42) | 7.43 × 10−3** | |
| 14 DAI | Experiment 1 | 9.09 ± 5.96 (n = 96) | 8.42 ± 5.32 (n = 87) | 0.42 |
| Experiment 2 | 8.8 ± 5.23 (n = 43) | 7.28 ± 3.99 (n = 42) | 0.15 | |
Transformed wild-type M. truncatula plants overexpressing 3HA-PUB1 or GFP (control) were inoculated with wild-type S. meliloti. Two independent experiments were performed, and the means of the number of nodules formed at 7 and 14 DAI is shown as well as the standard deviations. The number of scored plants is shown in parentheses.
Statistical analysis of nodulation of 3HA-PUB1 versus control plants. P values below 0.01 and 0.001 are marked with two and three asterisks, respectively.
Figure 6.
Analysis of the Role of PUB1 in Nodulation using Overexpression and Knockdown Strategies.
Graphs represent the nodulation efficiency ratios: average number of nodules in the assay versus average number of nodules in the control. Each ratio is calculated using data from two independent experiments shown in Tables 1 and 2. The dashed line represents no difference of the assay plants to the vector control plants. *, Data showing statistical differences (P values below 0.01) in replicate experiments (see Tables 1 and 2).
(A) Overexpression of 3HA-PUB1 induces a significant delay in nodulation. M. truncatula wild-type plants, overexpressing 3HA-PUB1 or GFP (control) under the control of Pro35S, were inoculated with wild-type S. meliloti, and nodules were scored at 7 and 14 DAI (dpi) in growth pouches.
(B) PUB1-RNAi plants show increased nodulation efficiency of M. truncatula wild-type plants with S. meliloti strains altered in NF structure and of M. truncatula lyk3-4 plants with the S. meliloti wild-type strain. M. truncatula wild-type or lyk3-4 plants, transformed with the PUB1-RNAi construct or vector control, were inoculated with wild-type, nodL, or nodFnodL S. meliloti strains. Nodules were scored at 21 DAI in pots.
To investigate this possible negative role by another approach, a double-stranded RNA construct (RNAi) was prepared to knock down PUB1 expression by targeting the 450-bp 5′ region of the gene transcript. None of the seven Mt UND-PUB-ARM homologs described above (see Supplemental Figure 4 online) show sufficient homology in this region (21 identical and consecutive base pairs) to be affected by the RNAi construct. Analysis of roots from PUB1-RNAi plants pretreated with NFs showed a 70% reduction in PUB1 transcripts on average compared with the control plants (see Supplemental Figure 6 online). The construct therefore appears to be efficient in downregulating expression of PUB1 and was used to investigate the effect of PUB1 knockdown on nodulation. M. truncatula plants transformed with this construct were inoculated with the wild-type S. meliloti strain. No difference was observed in the number of nodules (nodulation efficiency ratio of 0.97; Table 2, Figure 6B), a result also obtained in LYK3-RNAi plants (Limpens et al., 2003). However, LYK3-RNAi plants (Limpens et al., 2003) show reduced nodulation and a stronger phenotype with strains producing NFs modified at the nonreducing end. We thus tested nodulation of PUB1-RNAi plants using two strains that produce NFs harboring such alterations. The S. meliloti nodL mutant produces NFs that are not O-acetylated on the terminal nonreducing sugar and shows reduced nodulation on M. truncatula (Ardourel et al., 1994; Smit et al., 2007). The S. meliloti nodFL mutant produces non-O-acetylated NFs in which a C18:1 fatty acid generally replaces the specific C16:2 acyl chain. Such rhizobial mutants show very poor nodulation on M. truncatula (Ardourel et al., 1994). Nodulation experiments with such S. meliloti mutant strains show a very strong increase in the number of nodules produced on PUB1-RNAi plants compared with vector control plants, especially with the nodFnodL strain (Table 2, Figure 6B). Thus, the negative role in nodulation of PUB1, suggested by overexpressing PUB1, is confirmed using a knockdown strategy.
Table 2.
Nodulation Phenotype of PUB1 Knockdown Roots of M. truncatula
| Mean No. of Nodules per Plant |
|||||
| M. truncatula | S. meliloti | Experiment | Control | PUB1 RNAi | P Valuea |
| Wild type | Wild type | Experiment 1 | 10.18 ± 4.77 (n = 44) | 10.95 ± 5.71 (n = 60) | 0.47 |
| Experiment 2 | 18.16 ± 7.48 (n = 62) | 17.71 ± 7.89 (n = 63) | 0.76 | ||
| Wild type | nodL | Experiment 1 | 3.34 ± 3.43 (n = 44) | 7.30 ± 4.26 (n = 52) | 3.90 × 10−6*** |
| Experiment 2 | 5.82 ± 3.59 (n = 65) | 12.15 ± 5.92 (n = 61) | 3.85 × 10−11*** | ||
| Wild type | nodFnodL | Experiment 1 | 0.22 ± 0.64 (n = 67) | 1.53 ± 2.09 (n = 58) | 3.77 × 10−6*** |
| Experiment 2 | 0.17 ± 0.59 (n = 66) | 2.11 ± 2.14 (n = 65) | 1.04 × 10−10*** | ||
| lyk3-4 | Wild type | Experiment 1 | 0.11 ± 0.42 (n = 26) | 2.79 ± 3.34 (n = 23) | 1.34 × 10−4*** |
| Experiment 2 | 0.28 ± 0.56 (n = 86) | 3.39 ± 2.99 (n = 90) | 1.60 × 10−16*** | ||
Wild-type or lyk3-4 (hcl-4) mutant plants, transformed with the PUB1 RNAi construct or empty vector (control), were inoculated with wild-type or nod mutant bacteria. For each combination, two independent experiments were performed, and the mean number of nodules formed at 21 DAI is shown as well as the standard deviations. The number of scored plants is shown in parentheses.
Statistical analysis of nodulation of PUB1 RNAi versus control plants. P values below 0.001 are marked with asterisks.
To link this role to LYK3, nodulation experiments were performed using the weak lyk3-4 (hcl-4) allele, in which only ~8% of the LYK3 transcript appears to be correctly spliced. This mutant is poorly nodulated by wild-type rhizobia, resulting in <10% nodulation efficiency when compared with wild-type plants (Smit et al., 2007). In this mutant background, RNAi against PUB1 led to a dramatic increase in nodulation with the wild-type S. meliloti strain compared with the control (Table 2, Figure 6B). Nodules were formed in only 19% of the control plants; by contrast, 79% of the PUB1-RNAi plants produced nodules.
LYK3 is primarily involved in infection, and in the lyk3-4 allele, either the ITs are abnormally large, sac-like structures or the bacteria remain as microcolonies in the root hair curl (Smit et al., 2007). We thus used this allele to study the effect of PUB1 on infection. The number of ITs was dramatically increased in PUB1-RNAi lyk3-4 plants compared with the control (Table 3). In addition, numerous normal, long, and thin ITs were observed in PUB1 knockdown plants (Figure 7, left panel), while infections were restricted to large microcolonies and abnormal, short, and thick ITs at 5 DAI in control plants (Figure 7, right panel).
Table 3.
Infection Phenotype of PUB1 Knockdown Roots of M. truncatula lyk3-4
| Mean No. of ITs per Plant |
|||
| Experiment | Control | PUB1 RNAi | P Valuea |
| Experiment 1 | 1.92 ± 3.22 (n = 36) | 8.88 ± 10.48 (n = 33) | 3.18 × 10−4*** |
| Experiment 2 | 0.48 ± 0.87 (n = 25) | 6.41 ± 9.54 (n = 22) | 3.34 × 10−3** |
lyk3-4 (hcl-4) mutant plants, transformed with the PUB1 RNAi construct or empty vector (control), were inoculated with wild-type bacteria. For each combination, two independent experiments were performed, and the mean number of ITs formed at 5 DAI is shown as well as the standard deviations. The number of scored plants is shown in parentheses.
Statistical analysis of infection of PUB1 RNAi versus control plants. P values below 0.01 and 0.001 are marked with two and three asterisks, respectively.
Figure 7.
Analysis of ITs in PUB1 Knockdown Roots of lyk3-4 Plants.
lyk3-4 (hcl-4) mutant plants, transformed with the PUB1 RNAi construct (left panel) or empty vector (right panel), were inoculated with wild-type S. meliloti. Rhizobia were detected by β-galactosidase (LacZ) activity using X-Gal (blue). ITs were imaged at 5 DAI. Bars = 20 μm.
Thus, the negative role of PUB1 in nodulation is not confined to mutant strains producing altered NFs but is linked to the activity of LYK3 and the initiation and progression of the ITs.
DISCUSSION
LYK3 is essential for nodulation of M. truncatula but occupies an enigmatic position in the NF signaling pathways as little is known of how it links to downstream responses. In this article we describe an E3 ubiquitin ligase, PUB1, which interacts with LYK3 and provide evidence that this protein plays an important role as a negative regulator of the LYK3 signaling pathway controlling infection, nodulation and partner specificity.
The PUB1 gene was identified in a yeast two-hybrid screen of a root hair cDNA library using the intracellular region of LYK3 as bait. To validate its role as a LYK3 interactor, we have shown that the two proteins interact in planta and that LYK3 interacts and phosphorylates PUB1 in vitro (Figures 2 and 3). In M. truncatula, the two proteins locate to the same subcellular compartment, the plasma membrane (Figure 2), and their corresponding genes show a remarkable overlap in cellular expression patterns during nodulation (Figure 5), suggesting that the two proteins may encounter each other in vivo. Altogether, our data strongly suggest an interaction between LYK3 and PUB1 in M. truncatula, raising the question of the function and the role of PUB1 in LYK3-dependent nodulation. To address this question, we performed a detailed functional characterization of PUB1, including determining its enzymatic activity, its cellular function, and its biological role.
Our in vitro studies have shown that PUB1 has a functional E3 ubiquitin ligase activity, as shown for other UND-PUB-ARM proteins (Stone et al., 2003; Zeng et al., 2004; Yang et al., 2006). As for these other proteins, this activity is dependent on the U-box domain as demonstrated here (Figure 3) by mutation of a key Trp residue known to be involved in interaction with E2 proteins (Andersen et al., 2004). Ubiquitination is a common, eukaryote-specific posttranslational modification that occurs in very different proteins and regulates many cellular processes. Often it leads to polyubiquitination and labeling of the proteins for degradation. However, in other cases, ubiquitination is important for regulating the activity or trafficking of the target protein (Komander, 2009). The E3 ubiquitin ligases, which confer the specificity of substrate ubiquitination, are encoded by a large number of genes. For example, in Arabidopsis, there are >1200 E3 ubiquitin ligase genes (Smalle and Vierstra, 2004), including 64 encoding U-box proteins, many of which belong to the PUB-ARM subfamily (Yee and Goring, 2009).
Medicago PUB1 is a member of the plant PUB-ARM protein family and particularly of the subset of these proteins that contains an UND domain (Azevedo et al., 2001; Mudgil et al., 2004). This UND-PUB-ARM group of proteins includes several with important roles in recognition responses, including self-incompatibility (B. napus ARC1; Stone et al., 1999) and biotic plant interactions (rice SPL11 [Zeng et al., 2004], tobacco ACRE276, and its potential ortholog in Arabidopsis PUB17 [Yang et al., 2006]). In M. truncatula, other members of the UND-PUB-ARM family are more highly related to these proteins than PUB1 (see Supplemental Figure 4 online), suggesting that PUB1 is not orthologous to the previously characterized UND-PUB-ARM genes. Indeed, PUB1 is not closely related to any Arabidopsis protein but is the single M. truncatula member of a small clade of uncharacterized UND-PUB-ARM proteins, which includes two proteins from L. japonicus and one from G. max (see Supplemental Figure 5 online). Potential orthologs in nonleguminous dicotyledonous plants such as V. vinifera and P. trichocarpa suggest that this gene was not originally specialized for a role in nodulation, but such a function may have arisen during evolution of the legumes. However, this conclusion needs to await the completion of the M. truncatula genome sequencing project and the functional characterization of other legume proteins in the PUB1 subclade.
Several E3 ubiquitin ligases have been shown to be regulated and/or play a role in nodulation (Nishimura et al., 2002; Karlowski and Hirsch, 2003; Shimomura et al., 2006; Den Herder et al., 2008; Kiss et al., 2009; Yano et al., 2009). These E3 ubiquitin ligases belong to other subfamilies than PUB1 and have rather different protein structures. Their protein targets and modes of action are not yet known. The involvement of multiple E3 ubiquitin ligases in the control of a complex biological process, such as nodulation, is expected due to the key roles of these proteins in regulating protein level, localization, and activity. The targets and modes of action of PUB1 and these various E3 ubiquitin ligases are thus expected to be different.
Our results on over- and knockdown expression clearly show that PUB1 plays a physiological role in the negative control of nodulation. Overexpression leads to a delay in nodulation with wild-type S. meliloti (Table 1). Knockdown of expression demonstrates that the level of PUB1 is important in determining the specificity of nodulation to rhizobial species in a NF-dependent manner (Table 2). For example, RNAi against PUB1 leads to better nodulation of M. truncatula by rhizobial strains that produce nonoptimal NF structures (alteration in the acyl chain structure and non-O-acetylation of the terminal nonreducing sugar). This result is opposite to similar work on LYK3 where knockdown plants and a weak LYK3 mutant (lyk3-4) show increased specificity for rhizobial strains such that those producing nonoptimal NFs can no longer nodulate (Limpens et al., 2003; Smit et al., 2007). In addition, our demonstration that the weak LYK3 mutant can be partially complemented for nodulation and infection by knockdown of PUB1 (Figures 6 and 7, Tables 2 and 3) provides a genetic link between the two genes.
Thus, the use of rhizobial NF mutants has clearly shown a role of PUB1 in a NF-dependent selection of the rhizobial symbiont. PUB1 overexpression and the use of the weak LYK3 mutant have confirmed the role of PUB1 during nodulation with wild-type rhizobia and provide evidence that the role of PUB1 is LYK3 dependent. Although, knockdown of PUB1 did not lead to increased nodule number when inoculated with wild-type rhizobia, this result could be explained by the RNAi construct not being totally effective in downregulating PUB1 activity, that another mechanism of negative regulation still controls the nodule number in PUB1-RNAi plants, or that another UND-PUB-ARM protein has partial functional redundancy with PUB1. Indeed, several of the M. truncatula UND-PUB-ARM genes are expressed in roots and nodules, although only PUB1 shows an expression pattern specific to symbiosis (see Supplemental Data Set 3 online). Together, the data show that PUB1 controls nodulation as a negative regulator in the signaling pathway driven by LYK3.
There are two main modes of action by which PUB1 could exert an effect at the cellular level through interaction with LYK3. First, it could ubiquitinate LYK3, leading to its degradation. In animals, many RLKs are regulated by various E3 ubiquitin ligases leading to their degradation. For example, regulation of the Toll-like receptor family and their interacting proteins is important for regulating the innate immune response to invading microbes (Carpenter and O’Neill, 2009). In plants, direct evidence for regulation of RLKs by ubiquitination is starting to appear (Geldner and Robatzek, 2008; Göhre et al., 2008), but although several plant E3 ubiquitin ligases have been shown to interact with RLKs (Gu et al., 1998; Kim et al., 2003; Wang et al., 2006; Samuel et al., 2008), their direct role in RLK ubiquitination and degradation has not been shown. However, evidence for such a mechanism has been provided recently by the observation that a bacterial effector with E3 ubiquitin ligase activity (AvrPtoB), which is translocated to the plant cell cytoplasm, leads to ubiquitination and degradation of microbe-associated molecular pattern receptors and increases virulence of the pathogen (Göhre et al., 2008; Xiang et al., 2008; Gimenez-Ibanez et al., 2009). In our experiments, PUB1 did not ubiquitinate the LYK3 intracellular region in vitro, and coexpression of PUB1 with LYK3 in N. benthamiana did not lead to any changes in the level of LYK3 (Figure 3). By contrast, similar work using the same in vitro and heterologous systems showed ubiquitination and degradation of CERK1 by AvrPtoB (Gimenez-Ibanez et al., 2009). Together, our results have failed to provide evidence for a role of PUB1 in regulating the activity or degradation of LYK3 through ubiquitination. However, we cannot exclude the possibility that in M. truncatula such a mechanism may exist.
The second possible mechanism is that LYK3 modulates the activity of PUB1, which then regulates downstream components, perhaps through their ubiquitination and degradation. In the interaction between the S-locus receptor kinase (SRK1) and ARC1, which regulates self-incompatibility in B. napus, SRK1 has been shown to phosphorylate ARC1 (Gu et al., 1998). Recently, a protein interacting with the UND domain of ARC1 has been identified and demonstrated to be ubiquitinated by ARC1. Functional analysis of this protein, EXO70A1, has shown that it is a downstream compatibility factor, which is inhibited in the incompatibility response leading to pollen rejection (Stone et al., 2003; Samuel et al., 2009). We have shown that PUB1 is phosphorylated by LYK3. PUB1 could then modulate the activity of downstream components in the LYK3 signal transduction pathway by ubiquitination, which could either lead to the degradation of positive regulators or the activation of negative regulators of nodulation. Isolation of PUB1 interactors could be a means to determine the mode of action of PUB1.
LYK3 is essential for nodulation and appears to be the key checkpoint for bacterial entry in M. truncatula (Limpens et al., 2003). Thus, its activity may need to be finely controlled to allow infection only by bacteria with the right passports (NFs) and to close the door to potential freeloaders and pathogens. The PUB1 E3 ubiquitin ligase may be a key component of this fine regulation playing a role as a negative regulator in signaling by LYK3. The transient induction of PUB1 by NFs via NFP and the common symbiotic pathway (Figure 4) could be a mechanism whereby a local increase in PUB1 could regulate LYK3-dependent activities. In this way, the signaling receptor (NFP) would regulate the activities of the entry receptor (LYK3) through PUB1 induction, leading to a fine control of infection and nodulation.
In summary, this work identified a key regulator of infection, nodulation, and partner specificity in the establishment of the legume-rhizobia symbiosis and provides further evidence for the role of UND-PUB-ARM E3 ubiquitin ligases in regulating plant development through modulating RLK signaling pathways. This work extends the pioneering work on SRK1/ARC1 in the Brassica self-incompatibility response to show that a RLK/UND-PUB-ARM protein interaction can affect specificity in a plant–microbe interaction. There are a large number of PUB-ARM genes in higher plants, some of which have been shown to play important physiological roles in plant development and plant–pathogen interactions (Yee and Goring, 2009). Our work, together with that on Bn ARC1, further suggests that RLKs are good candidates to be tested as interactors of this important class of E3 ubiquitin ligases.
METHODS
Plant Material
Medicago truncatula Jemalong A17 (wild type) and NF signaling mutants nfp-2, lyk3-1 (hcl-1), dmi2-1 (tr25), dmi3-1 (trv25), nin1.1, and lyk3-4 (hcl-4) were used in this study (Arrighi, et al., 2006; Catoira et al., 2000, 2001; Marsh et al., 2007; Smit et al., 2007). Seeds were surface-sterilized and germinated according to Boisson-Dernier et al. (2001) and either used for Agrobacterium rhizogenes transformation or transferred to nitrogen-free medium for 4 d before NF treatment or Sinorhizobium meliloti inoculation.
M. truncatula Transgenic Roots Generated by A. rhizogenes Transformation
Overexpression of a 3HA-Tagged PUB1
The 3HA-PUB1 construct consists of the coding sequence (3HA tag-AttB1 Gateway border-PUB1) under the control of Pro35S and cloned in a pGREEN vector: the control construct contained a Pro35S:GFP construct in the same vector (Navarro-Gochicoa et al., 2003). All primers used in this study are reported in Supplemental Table 1 online. For M. truncatula transformation, 7-d-old A17 seedlings grown on Farhaeus agar plates were wounded with a needle at the hypocotyl-root junction and cocultured with A. rhizogenes carrying the 3HA-PUB1 or GFP constructs by application of Luria-Bertani agar grown bacteria on the wounds. Plants were grown for 5 d at 25°C before cutting and discarding the primary untransformed root. A concentrated solution of A. rhizogenes (OD600 = 1) was again applied on the cut edge of the hypocotyl. At 2 weeks after the second inoculation, plants were transferred to growth pouches (Mega International; four plants/pouch) prior to inoculation with S. meliloti. Nodule number was scored at 7 and 14 DAI. Two independent biological replicates were performed. Student’s t test was used to compare the means between samples. For the localization studies, pieces of root expressing 3HA-PUB1 or GFP were cut and propagated at 25°C on M medium (for medium composition, see http://www.noble.org/MedicagoHandbook/) supplemented with 25 mg·L−1 kanamycin and 200 mg·L−1 augmentin.
Knockdown of PUB1 by RNAi
The first 450 bp of PUB1 coding sequence (1 to 450 bp starting from the ATG) was amplified by PCR using cDNA as template and cloned using the Gateway technology in the pK7GWiWG(II) binary vector, modified to express DsRED1 (Limpens et al., 2005). A version of the vector cured of the Gateway recombination sites was constructed by deletion of the sequence contained between the promoter and terminator 35S by ZraI/HindIII digestions and used as negative control. Germinated seedlings were transformed with A. rhizogenes as described by Boisson-Dernier et al. (2001). At 3 weeks after inoculation, kanamycin-resistant plants showing fluorescence of DsRed were transferred to 150-mL pots containing perlite (three plants/pot) and acclimatized for 5 d in a greenhouse prior to inoculation with S. meliloti strains. Nodule number was scored at 21 DAI and ITs at 5 DAI. For IT analysis, roots were fixed and bacterial LacZ activity was performed as described by Bersoult et al. (2005). Two independent biological replicates were performed. Student’s t test was used to compare the means between samples.
Rhizobium Inoculations
S. meliloti strains 2011 (GMI6526), nodL (GMI6563), and nodFnodL (GMI6630) mutants (Ardourel et al., 1994) harboring the hemA-lacZ transgene plasmid (pXLGD4) were grown for 3 d at 25°C on TY-agar medium supplemented with 6 mM CaCl2 and containing the appropriate antibiotic combinations. Suspension of bacteria diluted in water were used to inoculate plants in pouches (0.5 mL of an OD600 = 0.1 per plant) and perlite-containing pots (10 mL of an OD600 = 0.025 per pot).
Yeast Two-Hybrid Screen and Pairwise Interactions
The sequence coding the LYK3 intracellular region, containing the juxta membrane, the kinase, and the C-terminal regions (amino acids 257 to 620) was amplified by PCR and cloned by restriction digestion into the pBD vector (Clontech) and transformed into the yeast strain AH109 using the LiAc-mediated yeast transformation protocol as described in the Yeast Protocols Handbook (Clontech). The resulting yeast strain was then transformed with plasmid DNAs derived from a M. truncatula NF-elicited, root hair cell cDNA library (Andriankaja et al., 2007). Approximately 1.5 × 106 yeast transformants were screened for the activation of the HIS3 reporter gene on minimal medium lacking His, Trp, and Leu and supplemented with 10 mM 3-amino-triazol. The HIS3+ yeast colonies were recovered after 3 or 4 d of growth at 28°C and retested for growth on minimal medium lacking His and adenine. The cDNA inserts of positive clones were amplified by PCR from yeast cell extracts and sequenced after a PCR product purification step. Plasmid rescue of nonredundant candidates was then performed. The intracellular regions of the Arabidopsis thaliana BRI1 RLK (amino acids 815 to 1196), the M. truncatula LRRII.1 RLK (amino acids 230 to 597), the M. truncatula LysM-RLK LYK2 (amino acids 255 to 612), LYK4 (amino acids 257 to 624), and NFP (amino acids 274 to 595) were amplified by PCR and cloned using the Gateway technology into the pBD vector prior to specificity tests with the obtained candidates. For homogeneity reasons, the LYK3 intracellular region (amino acids 253 to 620) was cloned into the pBD vector using the Gateway technology when interactions were retested. In addition to the three PUB1 variants obtained in the screen (full-size PUB1, PUB1ΔUND [amino acids 271 to 694], and PUB1ΔUNDΔU-Box [amino acids 397 to 694]), the PUB1ΔARM (amino acids 1 to 387) construct was obtained by PCR and cloned into the pGAD vector (Andriankaja et al., 2007) using Gateway technology.
Transient Expression in Nicotiana benthamiana Leaves and BiFC Experiments
The pGREEN 3HA-PUB1, pCAMBIA LYK3-3FLAG, and pGREEN LYK3-GFP constructs, all under the control of the Pro35S, were introduced into the Agrobacterium tumefaciens LBA4404 strain for expression in N. benthamiana leaves. The pAMPAT destination vectors and the constructs LYK3-YFP, LYK3-Yc, LRRII.1-YFP, and LRRII.1-Yc are described by Lefebvre et al. (2010). Yn-PUB1, Yn-PUB1ΔARM, BRI1-YFP, and BRI1-Yc constructs were made using Gateway technology and corresponding pAMPAT destination vectors and introduced in A. tumefaciens GV3103 pmp90RK. A. tumefaciens–containing expression vectors were grown overnight at 28°C in liquid YEB medium supplemented with appropriate antibiotics. Bacteria were washed and diluted in 10 mM MES, pH 5.6, 10 mM MgCl2, and 100 μM acetosyringone prior to N. benthamiana leaf inoculation. Bacteria containing pGREEN, pCAMBIA, or pAMPAT vectors were diluted to OD600 = 0.5, OD600 = 0.05, and OD600 = 0.25, respectively. The p19 protein, which inhibits gene silencing (Voinnet et al., 2003), was coexpressed with the pAMPAT constructs using bacteria diluted to OD600 = 0.1. Leaves were imaged using a Zeiss epifluorescence Axiophot microscope with a ×5 objective. Two 8-bit TIFF images for each transformed leaf were quantified using ImageJ. Background was removed using the same threshold for all images. All pairs were tested in three different leaves in each experiment. At least three independent experiments were performed. Results from one experiment are shown.
Protein Extraction and Membrane Fractionation
For total extract protein analysis, N. benthamiana leaf discs of 1-cm diameter were frozen in liquid nitrogen and ground for 1 min in 2-mL eppendorf tubes with 4-mm metal beads using a Retsch miller. Extracts were solubilized directly by adding 2% SDS gel loading buffer and heating at 95°C for 5 min.
For fractionation, N. benthamiana leaves or M. truncatula roots were homogenized at 4°C in a blender with 250 mM Sorbitol, 50 mM Tris-HCl, pH 8.0, 2 mM EDTA, 0.6% polyvinylpolypyrrolidone, 5 mM DTT, protease inhibitors (1 mM phenylmethylsulfonyl fluoride and 1 μg/mL each of leupeptin, aprotinin, antipain, chymostatin, and pepstatin from Sigma-Aldrich). After centrifugation for 10 min at 3000g, the resulting supernatant was centrifuged for 30 min at 100,000g, and the pellet resuspended in 330 mM sucrose, 3 mM KCl, 5 mM KH2PO4, pH 7.8, and protease inhibitors to give the total membrane fraction. The supernatant was precipitated with 10% trichloroacetic acid, washed twice with 90% acetone, and resuspended in the same volume as the total membrane fraction. Plasma membrane fraction was purified from the total membrane fraction by phase partitioning using 6.5% PEG3350 and 6.5% Dextran T500 as described by Larsson et al. (1987) and resuspended in the same buffer as the total membrane fraction. Fractions were solubilized by adding 2% SDS gel loading buffer and heating at 95°C for 5 min.
Production of Recombinant Proteins and in Vitro Biochemical Assays
Recombinant Protein Purification
The LYK3-IR-6HIS, PUB1ΔUND, and At UBC8 GST-tagged proteins were expressed in Escherichia coli strain DH5α, and purifications were performed using Glutathione Sepharose 4B according to the manufacturer’s instructions (GE Healthcare). The PUB1 and PUB1ΔUND MBP-tagged proteins and MBP were expressed in E. coli strain DH5α, and purifications were performed using amylose resin according to the manufacturer’s instructions (New England Biolabs). For protein production, a 2.5-mL E. coli overnight culture was transferred to 250 mL of Luria-Bertani medium and grown at 37°C to a density of OD600 = 0.6. Isopropylthio-β-galactoside was added to 0.5 mM and cells were grown for 5 h at 20°C and 28°C for GST-tagged and MBP-tagged proteins, respectively. The cells were collected by centrifugation and resuspended in 20 mL of ice-cold lysis buffers supplemented with protease inhibitors and 1 mM benzamidine prior to cell disruption using a French press. Insoluble material was eliminated by centrifugation at 18,000g for 20 min at 4°C. The soluble fraction was run twice through columns prepared with 200 μL of prewashed glutathione sepharose 4B matrix or amylose resin. Bound proteins were washed with 10 mL of ice-cold lysis buffers prior to elution in 4 mL of GST or MBP elution buffers. Eluted proteins were concentrated to a final volume of 200 μL using Amicon Ultra Ultracel 10-K (Millipore). The purity of eluted proteins was checked visually by SDS-PAGE and staining with SimplyBlue SafeStain (Invitrogen).
Phosphorylation Assay
For kinase phosphorylation assays, proteins were incubated in 25 μL of 50 mM HEPES, pH 7.4, 10 mM MgCl2, 10 mM MnCl2, 1 mM DTT, and 100 nM (10 μCi) ϒ-P32ATP at 25°C for 1.5 h. Approximately equal quantities of the purified substrate proteins were used. Reactions were stopped by adding 2% SDS gel loading buffer and heating at 95°C for 5 min. Reactions were analyzed by SDS-PAGE, and after staining with SimplyBlue SafeStain, gels were dried and analyzed by phosphor imaging.
Ubiquitination Assay
In vitro ubiquitination assays were performed as follows: the reaction mixtures (20 μL) contained 0.5 μg of yeast (Saccharomyces cerevisiae) E1 enzyme (Sigma-Aldrich), 0.5 μg of purified Arabidopsis E2 enzyme UBC8, 0.5 μg of purified GST-PUB1ΔUND protein as the E3 ubiquitin ligase, and 1 μg of ubiquitin (Sigma-Aldrich) in 25 mM Tris-HCl, pH 7.5, 1 mM MgCl2, 1 mM ATP, and 0.5 mM DTT. Reactions were incubated at 30°C for 2 h and terminated by adding 2% SDS gel loading buffer and heating at 95°C for 5 min before SDS-PAGE and protein gel blot analysis.
Immunoblotting
Proteins were separated on 10% SDS-PAGE gels, transferred onto nitrocellulose or polyvinylidene difluoride membranes using the Mighty Small Transphor liquid transfer apparatus (GE healthcare), and detected with antibodies as indicated in each figure. The antibodies were diluted as follows: anti HA-HRP (Roche; 1:1000 to 1:4000), anti FLAG-HRP (Sigma; 1:4000), anti HIS-HRP (Sigma; 1:5000), anti GST-HRP (GE Healthcare; 1:10000), alone and mouse anti-ubiquitin (Santa Cruz Biotech; 1:5000), rabbit anti-GFP (Invitrogen; 1:5000) and rabbit anti-MBP (New England Biolabs; 1:7500) in combination with anti-rabbit-HRP (Millipore; 1:25000) or anti-mouse HRP (Santa Cruz Biotech; 1:20000). Horseradish peroxidase (HRP) was detected using the Immobilon protein gel blotting detection system (Millipore) and a digital camera (G-Box; Syngene).
NF Treatment and Gene Expression Quantification by qRT-PCR
Seedlings of aeroponically grown wild type and mutants, with the exception of dmi3-1 (treated on pouch paper), were transferred into tubes with the root system immersed in either Farhaeus medium alone as control or with NFs (10−9 M) purified from S. meliloti, containing predominantly NodSmIV (Ac, S, C16:2), prepared as described by Roche et al. (1991a). The plant material was incubated for 6 or 24 h before harvesting the roots. NF treatment of seedling roots grown on pouch paper support (Mega International) was performed as described by Sauviac et al. (2005) by covering the roots for 6 h in either Farhaeus medium alone or a 10−8 M concentration of purified NFs. To test RNAi efficiency, roots from 17 transgenic control plants and roots from 18 transgenic PUB1-RNAi plants [obtained using the lyk3-4 (hcl-4) mutant background and growing on Farhaeus agar plates] were transferred into tubes containing Farhaeus medium with purified NFs (10−9 M) and incubated for 8 h before harvesting of the roots.
Total RNA was extracted from roots of M. truncatula using the NucleoSpin RNA II total RNA isolation kit (Macherey-Nagel) according to the manufacturer’s instructions. Genomic DNA was removed via DNase treatment (TURBO DNase; Applied Biosystems) after RNA elution, following the manufacturer’s instructions. The integrity and quantity of the RNA samples were simultaneously checked using Agilent chips and the Agilent RNA 6000 Nano reagents kit (Agilent Technologies). First-strand cDNA synthesis was performed using 1 μg (or 200 ng for the RNAi efficiency experiment) of total RNA with an anchored oligo(dT) and Transcriptor reverse transcriptase (Roche) following the manufacturer’s protocol. Real-time PCR reactions were performed on Light Cycler 480 (Roche) according to the manufacturer’s instructions, using SYBR Green I Master and the following primer pairs: HLC (5′-GTACGAGGTCGGTGCTCTTGA-3′, 5′-GCAACCGAAAATTGCACCATA-3′), UBQ (5′-TTGTGTGTTGAATCCTAAGCA-3′, 5′-CAAGACCCATGCAACAAGTTC-3′), PUB1 (5′-AAAGCCGGTATAACCGACGATT-3′, 5′-TCCAACCCCTCAGAACAACCT-3′). Transcript levels of PUB1 were normalized to the endogenous HLC (Helicase MtD35391_GC) and UBQ (ubiquitin MtC00117.1_GC) gene transcript levels. Three independent biological replicates were performed for each condition tested.
Spatiotemporal Expression Analysis of the PUB1 and LYK3 Genes
For promoter GUS fusions, a 3-kb region upstream of PUB1 (−2968 to −1 bp from ATG) was cloned into the pBI101 binary vector. The ProLYK3:GUS-GFP expression construct used a 2.6-kb region upstream of LYK3 (−2600 to −4 bp from ATG). Transformation of M. truncatula and selection of kanamycin-resistant plants was performed as described previously. Plants were transferred to pouches or perlite-containing pots and inoculated with S. meliloti (GMI6526). Double detection of ProPUB1:GUS or ProLYK3:GUS expression and bacterial LacZ activity was performed as described by Bersoult et al. (2005) using Magenta-Gluc and X-Gal or X-Gluc and Magenta-Gal, respectively. Mature nodules were embedded in Technovit 7100 resin (Hareus Kulzer) before making 8-μm-thick sections with a Richert-Jung microtome. Observations were made with a bright-field Axiophot microscope. Digital images were taken with a Leica camera and software.
Accession Numbers
The PUB1 nucleotide sequence data reported are available in the Third Party Annotation Section of the GenBank/EMBL/DDBJ databases under the accession number TPA: BK007068. Other sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: BRI1 (AF017056), LRRII.1 (EU849167), LYK2 (BN001116), LYK4 (AY372403), and NFP (DQ496250). Accession numbers for proteins used in the alignments and phylogenetic analyses can be found in Supplemental Methods and Supplemental Figure 5 online.
Author Contributions
M.M. participated in all aspects of the work. C.H. and B.L. conceived and performed some of the experiments. S.C., L.D., D.K.-H., S.M., F.D.C.-N., S.F., S.R., and T.T. provided expert technical support and discussions on specific parts of the work. J.C., B.L., and C.H. conceived the project. The article was written by J.C., M.M., B.L., and C.H.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of Mt PUB1 with Closely Related Proteins.
Supplemental Figure 2. Interaction of PUB1 with LYK3 in N. benthamiana.
Supplemental Figure 3. Detection of GST LYK3-IR in the in Vitro Trans-Phosphorylation of PUB1 Experiment.
Supplemental Figure 4. Phylogenetic Relationships between the UND-PUB-ARM Proteins of M. truncatula and Selected UND-PUB-ARM Proteins from Other Higher Plants.
Supplemental Figure 5. Phylogenetic Analysis of Higher-Plant UND-PUB-ARM Proteins.
Supplemental 6. PUB1-RNAi Construct Efficiency.
Supplemental Table 1. Primers Used in This Study.
Supplemental Data Set 1. Text file of Alignment of UND-PUB-ARM Proteins of M. truncatula and Selected UND-PUB-ARM Proteins from Other Higher Plants.
Supplemental Data Set 2. Text file of Alignment of Higher-Plant UND-PUB-ARM Proteins.
Supplemental Data Set 3. Expression Pattern of the M. truncatula UND-PUB-ARM Family Based on the M. truncatula Gene Expression Atlas.
Supplemental Methods. Phylogenetic and Protein Sequence Analyses.
Acknowledgments
We gratefully acknowledge the contribution of the following persons at Wageningen University: René Geurts and Arend Streng for the gift of the ProLYK3:GUS construct and Eric Limpens for the modified RNAi plasmid. From our laboratory, we thank Fabienne Maillet for providing purified NFs and Clare Gough for critically reading the manuscript. This work was supported by the European Community funded Research Training Network “NODPERCEPTION” and by the French National Research Agency (ANR) contract “NodBindsLysM.” M.M. acknowledges a doctoral grant from the French Ministry for Higher Education and Research.
References
- Amor B.B., Shaw S.L., Oldroyd G.E., Maillet F., Penmetsa R.V., Cook D., Long S.R., Dénarié J., Gough C. (2003). The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 34: 495–506 [DOI] [PubMed] [Google Scholar]
- Andersen P., Kragelund B.B., Olsen A.N., Larsen F.H., Chua N.H., Poulsen F.M., Skriver K. (2004). Structure and biochemical function of a prototypical Arabidopsis U-box domain. J. Biol. Chem. 279: 40053–40061 [DOI] [PubMed] [Google Scholar]
- Andriankaja A., Boisson-Dernier A., Frances L., Sauviac L., Jauneau A., Barker D.G., de Carvalho-Niebel F. (2007). AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19: 2866–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardourel M., Demont N., Debellé F., Maillet F., de Billy F., Promé J.C., Dénarié J., Truchet G. (1994). Rhizobium meliloti lipooligosaccharide nodulation factors: Different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6: 1357–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi J.F., et al. (2006). The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142: 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C., Santos-Rosa M.J., Shirasu K. (2001). The U-box protein family in plants. Trends Plant Sci. 6: 354–358 [DOI] [PubMed] [Google Scholar]
- Benedito V.A., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Bersoult A., Camut S., Perhald A., Kereszt A., Kiss G.B., Cullimore J.V. (2005). Expression of the Medicago truncatula DM12 gene suggests roles of the symbiotic nodulation receptor kinase in nodules and during early nodule development. Mol. Plant Microbe Interact. 18: 869–876 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A., Chabaud M., Garcia F., Bécard G., Rosenberg C., Barker D.G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14: 695–700 [DOI] [PubMed] [Google Scholar]
- Carpenter S., O’Neill L.A. (2009). Recent insights into the structure of Toll-like receptors and post-translational modifications of their associated signalling proteins. Biochem. J. 422: 1–10 [DOI] [PubMed] [Google Scholar]
- Catoira R., Galera C., de Billy F., Penmetsa R.V., Journet E.P., Maillet F., Rosenberg C., Cook D., Gough C., Dénarié J. (2000). Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12: 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R., Timmers A.C., Maillet F., Galera C., Penmetsa R.V., Cook D., Dénarié J., Gough C. (2001). The HCL gene of Medicago truncatula controls Rhizobium-induced root hair curling. Development 128: 1507–1518 [DOI] [PubMed] [Google Scholar]
- Dénarié J., Debellé F., Promé J.C. (1996). Rhizobium lipo-chitooligosaccharide nodulation factors: Signaling molecules mediating recognition and morphogenesis. Annu. Rev. Biochem. 65: 503–535 [DOI] [PubMed] [Google Scholar]
- Den Herder G., De Keyser A., De Rycke R., Rombauts S., Van de Velde W., Clemente M.R., Verplancke C., Mergaert P., Kondorosi E., Holsters M., Goormachtig S. (2008). Seven in absentia proteins affect plant growth and nodulation in Medicago truncatula. Plant Physiol. 148: 369–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Erfurth I., Cosson V., Eschstruth A., Lucas H., Kondorosi A., Ratet P. (2003). Efficient transposition of the Tnt1 tobacco retrotransposon in the model legume Medicago truncatula. Plant J. 34: 95–106 [DOI] [PubMed] [Google Scholar]
- Geldner N., Robatzek S. (2008). Plant receptors go endosomal: A moving view on signal transduction. Plant Physiol. 147: 1565–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S., Hann D.R., Ntoukakis V., Petutschnig E., Lipka V., Rathjen J.P. (2009). AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 19: 423–429 [DOI] [PubMed] [Google Scholar]
- Göhre V., Spallek T., Häweker H., Mersmann S., Mentzel T., Boller T., de Torres M., Mansfield J.W., Robatzek S. (2008). Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 18: 1824–1832 [DOI] [PubMed] [Google Scholar]
- Graham P.H., Vance C.P. (2003). Legumes: Importance and constraints to greater use. Plant Physiol. 131: 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T., Mazzurco M., Sulaman W., Matias D.D., Goring D.R. (1998). Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc. Natl. Acad. Sci. USA 95: 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Benedito V.A., Wang M., Murray J.D., Zhao P.X., Tang Y., Udvardi M.K. (2009). The Medicago truncatula gene expression atlas web server. BMC Bioinformatics 10: 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S., Kim J., Muñoz A., Heckmann A.B., Downie J.A., Oldroyd G.E. (2009). GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlowski W.M., Hirsch A.M. (2003). The over-expression of an alfalfa RING-H2 gene induces pleiotropic effects on plant growth and development. Plant Mol. Biol. 52: 121–133 [DOI] [PubMed] [Google Scholar]
- Kim M., Cho H.S., Kim D.M., Lee J.H., Pai H.S. (2003). CHRK1, a chitinase-related receptor-like kinase, interacts with NtPUB4, an armadillo repeat protein, in tobacco. Biochim. Biophys. Acta 1651: 50–59 [DOI] [PubMed] [Google Scholar]
- Kiss E., Oláh B., Kaló P., Morales M., Heckmann A.B., Borbola A., Lózsa A., Kontár K., Middleton P., Downie J.A., Oldroyd G.E., Endre G. (2009). LIN, a novel type of U-box/WD40 protein, controls early infection by rhizobia in legumes. Plant Physiol. 151: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D. (2009). The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 37: 937–953 [DOI] [PubMed] [Google Scholar]
- Larsson C., Widell S., Kjellbom B. (1987). Preparation of high-purity plasma membranes. Methods Enzymol. 148: 558–568 [Google Scholar]
- Lefebvre B., et al. (2010). A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc. Natl. Acad. Sci. USA 107: 2343–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Signor C., Savois V., Aubert G., Verdier J., Nicolas M., Pagny G., Moussy F., Sanchez M., Baker D., Clarke J., Thompson R. (2009). Optimizing TILLING populations for reverse genetics in Medicago truncatula. Plant Biotechnol. J. 7: 430–441 [DOI] [PubMed] [Google Scholar]
- Li J., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Limpens E., Franken C., Smit P., Willemse J., Bisseling T., Geurts R. (2003). LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630–633 [DOI] [PubMed] [Google Scholar]
- Limpens E., Mirabella R., Fedorova E., Franken C., Franssen H., Bisseling T., Geurts R. (2005). Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc. Natl. Acad. Sci. USA 102: 10375–10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen E.B., Madsen L.H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Marsh J.F., Rakocevic A., Mitra R.M., Brocard L., Sun J., Eschstruth A., Long S.R., Schultze M., Ratet P., Oldroyd G.E. (2007). Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 144: 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson-Boivin C., Giraud E., Perret X., Batut J. (2009). Establishing nitrogen-fixing symbiosis with legumes: How many rhizobium recipes? Trends Microbiol. 17: 458–466 [DOI] [PubMed] [Google Scholar]
- Maudoux O., Batoko H., Oecking C., Gevaert K., Vandekerckhove J., Boutry M., Morsomme P. (2000). A plant plasma membrane H+-ATPase expressed in yeast is activated by phosphorylation at its penultimate residue and binding of 14-3-3 regulatory proteins in the absence of fusicoccin. J. Biol. Chem. 275: 17762–17770 [DOI] [PubMed] [Google Scholar]
- Mitra R.M., Shaw S.L., Long S.R. (2004). Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. USA 101: 10217–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgil Y., Shiu S.H., Stone S.L., Salt J.N., Goring D.R. (2004). A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 134: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Gochicoa M.T., Camut S., Timmers A.C., Niebel A., Herve C., Boutet E., Bono J.J., Imberty A., Cullimore J.V. (2003). Characterization of four lectin-like receptor kinases expressed in roots of Medicago truncatula. Structure, location, regulation of expression, and potential role in the symbiosis with Sinorhizobium meliloti. Plant Physiol. 133: 1893–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura R., Ohmori M., Fujita H., Kawaguchi M. (2002). A Lotus basic leucine zipper protein with a RING-finger motif negatively regulates the developmental program of nodulation. Proc. Natl. Acad. Sci. USA 99: 15206–15210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd G.E., Downie J.A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Roche P., Debellé F., Maillet F., Lerouge P., Faucher C., Truchet G., Dénarié J., Promé J.C. (1991b). Molecular basis of symbiotic host specificity in Rhizobium meliloti: nodH and nodPQ genes encode the sulfation of lipo-oligosaccharide signals. Cell 67: 1131–1143 [DOI] [PubMed] [Google Scholar]
- Roche P., Lerouge P., Ponthus C., Promé J.C. (1991a). Structural determination of bacterial nodulation factors involved in the Rhizobium meliloti-alfalfa symbiosis. J. Biol. Chem. 266: 10933–10940 [PubMed] [Google Scholar]
- Rogers C., Wen J., Chen R., Oldroyd G. (2009). Deletion-based reverse genetics in Medicago truncatula. Plant Physiol. 151: 1077–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel M.A., Chong Y.T., Haasen K.E., Aldea-Brydges M.G., Stone S.L., Goring D.R. (2009). Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell 21: 2655–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel M.A., Mudgil Y., Salt J.N., Delmas F., Ramachandran S., Chilelli A., Goring D.R. (2008). Interactions between the S-domain receptor kinases and AtPUB-ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiol. 147: 2084–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauviac L., Niebel A., Boisson-Dernier A., Barker D.G., de Carvalho-Niebel F. (2005). Transcript enrichment of Nod factor-elicited early nodulin genes in purified root hair fractions of the model legume Medicago truncatula. J. Exp. Bot. 56: 2507–2513 [DOI] [PubMed] [Google Scholar]
- Shimomura K., Nomura M., Tajima S., Kouchi H. (2006). LjnsRING, a novel RING finger protein, is required for symbiotic interactions between Mesorhizobium loti and Lotus japonicus. Plant Cell Physiol. 47: 1572–1581 [DOI] [PubMed] [Google Scholar]
- Smalle J., Vierstra R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Smit P., Limpens E., Geurts R., Fedorova E., Dolgikh E., Gough C., Bisseling T. (2007). Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 145: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Anderson E.M., Mullen R.T., Goring D.R. (2003). ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15: 885–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Arnoldo M., Goring D.R. (1999). A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 286: 1729–1731 [DOI] [PubMed] [Google Scholar]
- Tadege M., Wen J., He J., Tu H., Kwak Y., Eschstruth A., Cayrel A., Endre G., Zhao P.X., Chabaud M., Ratet P., Mysore K.S. (2008). Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54: 335–347 [DOI] [PubMed] [Google Scholar]
- Trujillo M., Ichimura K., Casais C., Shirasu K. (2008). Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. Biol. 18: 1396–1401 [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wang Y.S., Pi L.Y., Chen X., Chakrabarty P.K., Jiang J., De Leon A.L., Liu G.Z., Li L., Benny U., Oard J., Ronald P.C., Song W.Y. (2006). Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell 18: 3635–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T., Zong N., Zou Y., Wu Y., Zhang J., Xing W., Li Y., Tang X., Zhu L., Chai J., Zhou J.M. (2008). Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 18: 74–80 [DOI] [PubMed] [Google Scholar]
- Yang C.W., González-Lamothe R., Ewan R.A., Rowland O., Yoshioka H., Shenton M., Ye H., O’Donnell E., Jones J.D., Sadanandom A. (2006). The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18: 1084–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K., et al. (2009). CERBERUS, a novel U-box protein containing WD-40 repeats, is required for formation of the infection thread and nodule development in the legume-Rhizobium symbiosis. Plant J. 60: 168–180 [DOI] [PubMed] [Google Scholar]
- Yee D., Goring D.R. (2009). The diversity of plant U-box E3 ubiquitin ligases: From upstream activators to downstream target substrates. J. Exp. Bot. 60: 1109–1121 [DOI] [PubMed] [Google Scholar]
- Zeng L.R., Qu S., Bordeos A., Yang C., Baraoidan M., Yan H., Xie Q., Nahm B.H., Leung H., Wang G.L. (2004). Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16: 2795–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]