This work shows that PAM68 is a previously unknown PSII assembly factor that is required for efficient accumulation of photosystem II complexes in Arabidopsis thaliana and that is involved in early steps of PSII assembly in Synechocystis and Arabidopsis.
Abstract
Photosystem II (PSII) is a multiprotein complex that functions as a light-driven water:plastoquinone oxidoreductase in photosynthesis. Assembly of PSII proceeds through a number of distinct intermediate states and requires auxiliary proteins. The photosynthesis affected mutant 68 (pam68) of Arabidopsis thaliana displays drastically altered chlorophyll fluorescence and abnormally low levels of the PSII core subunits D1, D2, CP43, and CP47. We show that these phenotypes result from a specific decrease in the stability and maturation of D1. This is associated with a marked increase in the synthesis of RC (the PSII reaction center-like assembly complex) at the expense of PSII dimers and supercomplexes. PAM68 is a conserved integral membrane protein found in cyanobacterial and eukaryotic thylakoids and interacts in split-ubiquitin assays with several PSII core proteins and known PSII assembly factors. Biochemical analyses of thylakoids from Arabidopsis and Synechocystis sp PCC 6803 suggest that, during PSII assembly, PAM68 proteins associate with an early intermediate complex that might contain D1 and the assembly factor LPA1. Inactivation of cyanobacterial PAM68 destabilizes RC but does not affect larger PSII assembly complexes. Our data imply that PAM68 proteins promote early steps in PSII biogenesis in cyanobacteria and plants, but their inactivation is differently compensated for in the two classes of organisms.
INTRODUCTION
Photosystem II (PSII) is a multiprotein-pigment complex that functions as a light-driven water:plastoquinone oxidoreductase in the thylakoid membranes of cyanobacteria and chloroplasts (Wollman et al., 1999; Iwata and Barber, 2004; Nelson and Yocum, 2006). The reaction center of PSII is composed of the subunits D1 and D2, which bind the pigment cofactors chlorophyll, pheophytin, and plastoquinone, the α and β subunits of cytochrome (Cyt) b559 (hereafter designated as PsbE and PsbF, the E and F subunits of PSII), and PsbI. Peripherally attached are CP43 and CP47, which bind chlorophyll a and β-carotene; CP43 and D1 together provide ligands for the CaMn4 cluster that is involved in water oxidation (Ferreira et al., 2004; Guskov et al., 2009). Surrounding these subunits are several low molecular weight subunits of PSII (Shi and Schröder, 2004), and extrinsic subunits on the lumenal side of PSII stabilize the CaMn4 cluster (reviewed in Roose et al., 2007; Enami et al., 2008). Moreover, the extrinsic phycobilisomes (in cyanobacteria) and the intrinsic chlorophyll a/b binding light-harvesting complex of PSII (LHCII) (in chloroplasts) operate as distal, pigment-containing, light-harvesting complexes of PSII.
In cyanobacteria and chloroplasts, assembly of PSII occurs in a stepwise manner (Baena-Gonzalez and Aro, 2002; Rokka et al., 2005; Mulo et al., 2008; Nixon et al., 2010). In chloroplasts, this process is modulated by a combination of translational auto- and transregulation of PSII subunit synthesis (Choquet et al., 2001; Choquet and Wollman, 2002). Radioactive pulse-chase experiments, together with sucrose gradient and native gel analyses, provided initial support for the stepwise assembly of the chloroplast PSII complex, involving a number of discrete PSII subcomplexes and processing of the D1 protein (van Wijk et al., 1995, 1996, 1997). On the basis of two-dimensional analysis of radioactively labeled thylakoid proteins, Rokka et al. (2005) described the sequential appearance of distinct PSII assembly states in chloroplasts. First, the precursor of the D1 subunit (pD1) is assembled into the receptor complex (consisting of D2, PsbE, PsbF, and PsbI) to form RC, the PSII reaction center-like assembly complex. Next, CP47 attaches to RC to generate the so-called CP47-RC complex. In two further steps, the PsbH, PsbM, PsbTc, and PsbR subunits are added to form CP43-free PSII monomers (or CP43-PSII). Addition of CP43 and other subunits then generates PSII core monomers, which in turn form PSII core dimers and PSII supercomplexes, the native forms of functional PSII units that in vivo collect light energy, convert it into electro-chemical energy, and drive electron transfer from water to plastoquinone (Minagawa and Takahashi, 2004). In the PSII supercomplexes, LHCII trimers are attached to PSII core dimers (Boekema et al., 1995; Hankamer et al., 1997), probably via the proteins CP29, CP26, and CP24 that serve as linkers (Nelson and Yocum, 2006). During the biogenesis of PSII, subsequent to the insertion of pD1 in RC, the C-terminal extension of pD1 is cleaved by the C-terminal peptidase (CtpA) on the luminal side of the thylakoid membrane to form mature D1 (mD1) (Diner et al., 1988; Taylor et al., 1988; Anbudurai et al., 1994; Shestakov et al., 1994). The processing of assembled pD1 is a prerequisite for the ligation of the catalytic manganese cluster and, thus, for the formation of PSII complexes capable of splitting water (Diner et al., 1988; Nixon et al., 1992; Hatano-Iwasaki et al., 2000; Roose and Pakrasi, 2004).
In addition to the structural subunits of PSII, several auxiliary proteins are involved in PSII biogenesis (reviewed in Mulo et al., 2008; Nixon et al., 2010). Moreover, many of the auxiliary proteins involved in the de novo biogenesis of PSII also participate in the PSII repair cycle, which involves the replacement of damaged subunits, mainly the D1 subunit, by a newly synthesized copy and occurs much more frequently than does de novo biogenesis in mature chloroplasts (Adir et al., 2003; Aro et al., 2005; Nixon et al., 2005; Edelman and Mattoo, 2008; Mulo et al., 2008; Kato and Sakamoto, 2009). Some, but not all, PSII assembly factors are conserved in cyanobacteria and chloroplasts. These include HIGH CHLOROPHYLL FLUORESCENCE 136 (HCF136)/HYPOTHETICAL CHLOROPLAST OPEN READING FRAME 48 (YCF48) (Meurer et al., 1998; Komenda et al., 2008), ALBINO3 (ALB3)/Slr1471 (Sundberg et al., 1997; Spence et al., 2004), Psb27 (Kashino et al., 2002; Roose and Pakrasi, 2004; Chen et al., 2006; Nowaczyk et al., 2006; Wei et al., 2010), Psb28 (Kashino et al., 2002; Dobakova et al., 2009), Psb29 (Kashino et al., 2002; Keren et al., 2005), YCF39 (Ermakova-Gerdes and Vermaas, 1999), as well as immunophilins like CYCLOPHILIN38/THYLAKOID LUMEN PEPTIDYL-PROLYL ISOMERASE OF 40 kD (Sirpiö et al., 2008). Presumably, therefore, their roles in PSII biogenesis or maintenance have been retained during evolution. Other factors are specific to cyanobacteria (for instance, PROCESSING ASSOCIATED TPR PROTEIN [PratA] [Klinkert et al., 2004; Schottkowski et al., 2009a], Slr0286 [Kufryk and Vermaas, 2001], and Slr2013 [Kufryk and Vermaas, 2003]) or to chloroplasts (such as LOW PSII ACCUMULATION1 [LPA1]/PSII REPAIR 27 [REP27] [Peng et al., 2006; Park et al., 2007] and LPA2 [Ma et al., 2007]) and may represent evolutionary adaptations to specific environments.
HCF136, LPA1, LPA2, and ALB3 are the best studied PSII assembly factors in Arabidopsis thaliana. ALB3 performs auxiliary functions during the assembly of several thylakoid protein complexes, and ALB3 gene inactivation in Arabidopsis, Chlamydomonas reinhardtii, and Synechocystis sp. PCC 6803 results in severe phenotypes (Sundberg et al., 1997; Bellafiore et al., 2002; Spence et al., 2004). In chloroplasts, ALB3 proteins have been shown to interact with D1, D2, and CP47, as well as with subunits from photosystem I (PSI) and the ATP synthase (Ossenbühl et al., 2004; Pasch et al., 2005; Göhre et al., 2006), whereas the Synechocystis ALB3 ortholog has been proposed to play a role in the integration of pD1 into the membrane (Ossenbühl et al., 2006). The gene for HCF136 was identified in the Arabidopsis high chlorophyll fluorescence mutant hcf136, which contains only traces of PSII but synthesizes PSII subunits at normal rates (Meurer et al., 1998). HCF136 predominantly associates with a very early PSII assembly complex containing D2, PsbE, and PsbF (Plücken et al., 2002), and YCF48, the Synechocystis HCF136 homolog, has been detected in RC, where it interacts with unassembled pD1 (Komenda et al., 2008). In Arabidopsis, LPA1 binds to D1 during de novo biogenesis of PSII (Peng et al., 2006), whereas its Chlamydomonas ortholog, REP27, has also been suggested to play a role in D1 synthesis, albeit during PSII repair rather than during assembly (Park et al., 2007; Dewez et al., 2009). The Arabidopsis LPA2, which has no counterpart in cyanobacteria or Chlamydomonas, interacts specifically with CP43 and ALB3 (Ma et al., 2007).
In this study, we describe the identification and characterization of PAM68, a protein required for normal accumulation of PSII in Arabidopsis. The thylakoid protein PAM68, like HCF136 and ALB3, is conserved in cyanobacteria and photosynthetic eukaryotes. PAM68 also interacts with these two proteins, as well as with several structural subunits of PSII and further PSII assembly factors. Our data imply that PAM68 is a previously undiscovered PSII assembly factor that acts at the level of D1 maturation and stability, promoting the transition from the RC assembly state to larger PSII assembly complexes. In Synechocystis, inactivation of the homolog of PAM68, Sll0933, has less severe effects on photosynthesis and prevents accumulation of RC to detectable levels under steady state conditions, indicating that lack of PAM68 is differently compensated for in plants and cyanobacteria.
RESULTS
Mutations in PAM68 Severely Affect Growth Rate and PSII Function
Screening of the Arabidopsis GABI-KAT T-DNA insertion collection (Rosso et al., 2003) for lines that show alterations in ΦII, the effective quantum yield of PSII, resulted in the recovery of a set of mutants with defects in photosynthesis (Varotto et al., 2000a, 2000b). In the line GABI_152D07, renamed as the photosynthesis affected mutant 68-1 (pam68-1), ΦII was drastically reduced (Table 1), and this character segregated as a recessive trait. Moreover, homozygous pam68-1 mutant plants had pale-green cotyledons and leaves (Figure 1A). Analysis of chlorophyll a fluorescence showed a clear increase in the minimum fluorescence (F0; Figure 1A) and a dramatic decrease in maximum quantum yield of PSII (Fv/Fm; Figure 1A, Table 1). This combination is characteristic for mutants with abnormally low levels of PSII, such as hcf136 (Meurer et al., 1998), lpa1 (Peng et al., 2006; see Supplemental Figure 1 online), and lpa2 (Ma et al., 2007) mutants. Closer inspection of the chlorophyll a fluorescence induction curves employed to determine Fv/Fm and ΦII showed that, in pam68-1, chlorophyll a fluorescence transiently dropped below F0 after the initial rise induced by exposure to actinic light (Figure 1B). This too is typical of mutants in which the ratio PSII/PSI is reduced (Meurer et al., 1998; Peng et al., 2006; Ma et al., 2007; see Supplemental Figure 1 online for lpa1-2) and can be attributed to the higher initial activity of PSI relative to PSII.
Table 1.
Parameters of Chlorophyll a Fluorescence in Mutant (pam68-1, pam68-2, and lpa1-2) and Corresponding Wild-Type (Col-0 and Ler) Leaves
| Parameter | pam68-1 | pam68-2 | Col-0 | lpa1-2 | Ler |
| Fv/Fm | 0.44 ± 0.01 | 0.45 ± 0.02 | 0.83 ± 0.01 | 0.56 ± 0.01 | 0.83 ± 0.01 |
| ΦII | 0.28 ± 0.01 | 0.29 ± 0.02 | 0.72 ± 0.00 | 0.42 ± 0.02 | 0.72 ± 0.01 |
| 1-qP | 0.24 ± 0.03 | 0.23 ± 0.02 | 0.09 ± 0.01 | 0.13 ± 0.03 | 0.09 ± 0.01 |
| NPQ | 0.13 ± 0.02 | 0.14 ± 0.01 | 0.18 ± 0.03 | 0.11 ± 0.00 | 0.21 ± 0.01 |
At least five leaves from different plants were measured. Actinic light intensity was 80 μmol photons m−2 s−1. Fv/Fm, maximum quantum yield of PSII; ΦII, effective quantum yield of PSII; 1-qP, excitation pressure; NPQ, nonphotochemical quenching. Mean values ± sd are provided.
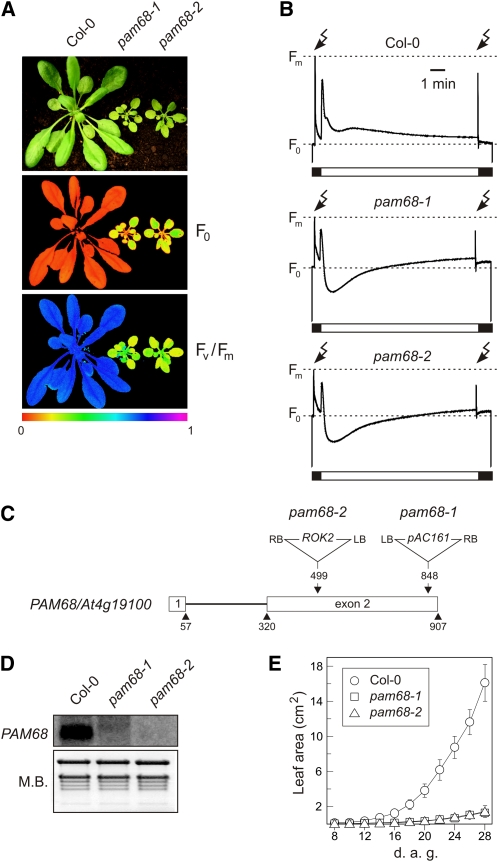
Figure 1.
Identification and Characterization of the Mutants pam68-1 and pam68-2.
(A) Five-week-old wild type (Col-0) and mutant (pam68-1 and pam68-2) plants were grown in the greenhouse (top panel), and the photosynthetic parameters F0 (minimum chlorophyll a fluorescence) and Fv/Fm (maximum quantum yield of PSII) were measured as described in Methods. Signal intensities for F0 and Fv/Fm are indicated in accordance with the color scale at the bottom of the figure.
(B) Chlorophyll a fluorescence induction curves of wild-type (Col-0) and mutant (pam68-1 and pam68-2) leaves. The white bar indicates exposure to actinic light (80 μmol photons m−2 s−1) and the lightning symbols the application of saturation light pulses (0.8 s; 5000 μmol photons m−2 s−1 white light). F0 and Fm are indicated for each genotype.
(C) T-DNA tagging of the PAM68/At4g19100 locus. Exons are numbered and shown as white boxes and the intron as a black line. Locations and orientations of T-DNA insertions are indicated. The pam68-1 allele was found in the GABI-KAT line GABI_152D07; pam68-2 corresponds to the SALK_044323 line from the SALK collection. Note that the T-DNAs are not drawn to scale.
(D) Effect of the T-DNA insertions on steady state levels of PAM68 RNA. Aliquots (30 μg) of total leaf RNA were fractionated on a denaturing agarose gel, transferred to a positively charged nylon membrane, and hybridized with a PAM68 cDNA probe. rRNA was stained with methylene blue (M.B.) as a loading control.
(E) Growth kinetics of wild type (Col-0) and mutant (pam68-1 and pam68-2) plants (n ≥ 10). Leaf area was measured during the period from 8 to 28 d after germination (d.a.g.). Bars indicate sd.
DNA gel blot analysis of a population of 30 plants segregating for the pam68-1 mutation using the 5′-end of the AC106 T-DNA as a probe revealed the presence of only one T-DNA copy, which cosegregated with the pam68-1 phenotype. A DNA fragment flanking the left border of the T-DNA was isolated and was found to be identical to the second exon of the At4g19100 gene (Figure 1C). A second line with a T-DNA insertion in this gene, pam68-2, was obtained from the SALK T-DNA insertion collection (Alonso et al., 2003). The pam68-2 plants behaved like the pam68-1 line with respect to growth, leaf coloration, and chlorophyll a fluorescence parameters (Table 1) and induction curve characteristics (Figures 1A and 1B), confirming that disruption of At4g19100 is responsible for the observed pam68 phenotypes.
The effect of the T-DNA insertions on the expression of PAM68 was monitored by RNA gel blot analysis. In pam68-1, few PAM68/At4g19100 transcripts with increased size were detected, whereas transcripts were undetectable in the pam68-2 allele (Figure 1D). To quantify the effect of loss of PAM68 function on growth behavior, we compared mutant and wild-type plants using noninvasive image analysis (Leister et al., 1999). Even under optimal greenhouse conditions, the leaf area of the mutants was reduced by ~90% relative to the wild type at 28 d after germination (Figure 1E). Because the two pam68 alleles behaved very similarly in all analyses, the pam68-2 allele was used in the following experiments. To evaluate quantitatively the altered coloration of pam68-2 leaves, leaf pigments were analyzed by HPLC. As expected, the mutant contained only 65% of wild-type levels of total chlorophyll (chlorophyll a + b) (pam68-2, 1339 ± 123; Columbia-0 [Col-0], 2060 ± 180 nmol/g fresh weight). The chlorophyll a/b ratio was slightly decreased (pam68-2, 2.7; wild type, 3.1), indicating either a higher PSII/PSI ratio or, more probably in light of the chlorophyll a fluorescence analyses described above, an increase in the size of the peripheral antenna relative to the chlorophyll a binding reaction centers.
Taken together, the data suggest that disruption of PAM68/At4g19100 affects photosynthetic electron flow, particularly at the level of PSII, resulting in severely reduced growth rates and altered leaf pigment composition.
Relative Levels of PSII Subunits Are Perturbed in pam68-2 Plants
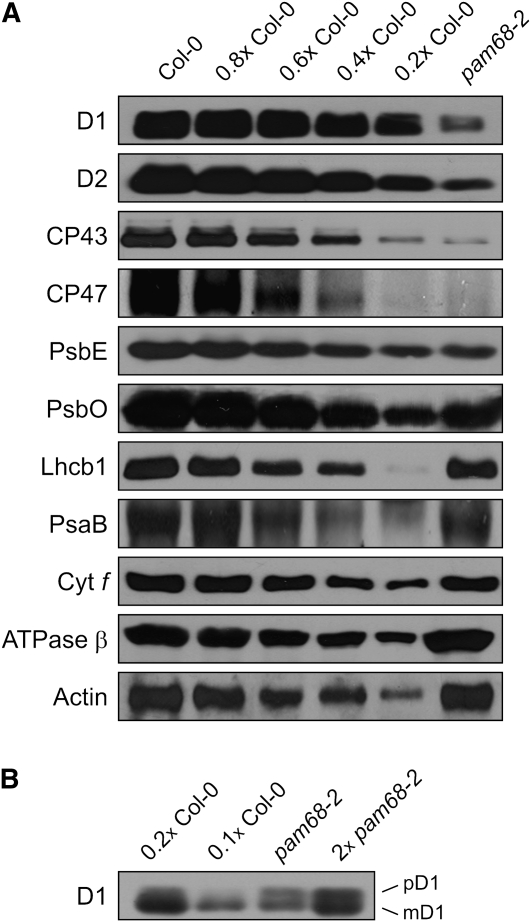
To determine whether the defect in PSII activity found in the pam68 mutants reflects a reduction in the abundance of PSII subunits, and to assess the levels of representative members of other thylakoid multiprotein complexes, immunoblot analyses were performed on total protein extracts from leaves. Marked reductions in the levels of the PSII core subunits D1, D2, CP43, CP47, and PsbE, to about ~10, ~10, ~15, ~15, and ~30% of those seen in the wild type, respectively, were indeed observed on the basis of estimating the intensity of mutant signals relative to the ones of different dilutions of wild-type samples (Figure 2A). PsbO (a subunit of the oxygen-evolving complex) was reduced to ~40% of wild-type levels, Lhcb1 (a subunit of LHCII) to 80%, and the B subunit of PSI (PsaB) and Cyt f (part of the Cyt b6/f complex) to ~70%. Only the chloroplast ATPase β-subunit appeared to show an increase relative to the wild type (Figure 2A). Interestingly, in pam68-2, two distinct D1 bands were obtained, representing the precursor (pD1) and mature (mD1) forms of the protein (Figure 2A). To clarify whether pD1 is indeed overrepresented in pam68-2 thylakoids, total protein extracts containing similar amounts of total D1 protein were compared (wild type, 8 and 4 μg; pam68-2, 40 and 80 μg of total leaf protein) and signal intensities quantified (see Methods) (Figure 2B). In the wild type, the pD1/mD1 ratio was ~0.15, but in pam68-2, it was ~0.85, indicating that processes which trigger conversion of pD1 into mD1 are affected in the mutant.
Figure 2.
Immunoblot Analysis of Representative Thylakoid Proteins.
(A) Total leaf proteins from pam68-2 and wild-type (Col-0) plants were fractionated by SDS-PAGE, and blots were probed with antibodies raised against individual subunits of PSII (D1, D2, CP43, CP47, PsbE, and PsbO), LHCII (Lhcb1), PSI (PsaB), the Cyt b6/f complex (Cyt f) and the chloroplast ATP synthase (β-subunit). Decreasing levels of wild-type proteins were loaded in the lanes marked 0.8x Col-0, 0.6x Col-0, 0.4 Col-0, and 0.2x Col-0. Actin served as loading control.
(B) Total leaf protein extracts containing similar amounts of total D1 protein were loaded (wild type, 4 and 8 μg; pam68-2, 40 and 80 μg of protein), fractionated and treated as in (A). pD1, precursor D1; mD1, mature D1.
Taken together, our results imply that the phenotype of pam68-2 plants is attributable primarily to changes in steady state concentrations of PSII proteins. In particular, levels of PSII core subunits are much lower than in the wild type, while the pD1/mD1 ratio is much higher.
Absence of PAM68 Affects the Maturation and Stability of Newly Synthesized D1
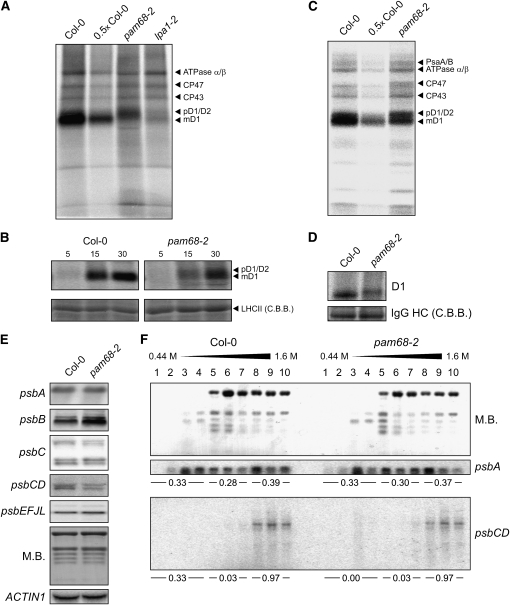
To investigate whether the fall in steady state levels of PSII proteins in pam68-2 is caused by a decrease in de novo synthesis of its plastid-encoded subunits, rates of synthesis of thylakoid membrane proteins were analyzed by in vivo pulse labeling experiments. The results for pam68-2 leaves were compared with those obtained for another PSII mutant, lpa1-2 (see Supplemental Figure 1 online). The labeling experiments with [35S]Met were performed under low light (20 μmol photons m−2 s−1) on leaves from greenhouse-grown plants to avoid activating the PSII repair cycle, which would lead to preferential labeling of the D1 protein (Figure 3A). In the presence of cycloheximide to block synthesis of nucleus-encoded proteins, newly translated chloroplast-encoded proteins were labeled for 20 min and thylakoid proteins were isolated. In both mutant genotypes, rates of synthesis of the PSII core proteins CP43 and CP47, as well as the α- and β-subunits of the cpATPase, were almost unchanged relative to the respective wild-type rates. However, incorporation of [35S]Met into D1 + D2 was reduced to ~60 and 35% of wild-type levels in pam68-2 and lpa1-2 mutant plants, respectively, on the basis of quantification of signal intensities (see Methods) (Figure 3A). Moreover, and in contrast with lpa1-2, in pam68-2, the D1 + D2 band was slightly shifted toward higher molecular weight products (Figure 3A). In fact, immunoblot analysis (see Supplemental Figure 2 online) allowed the conclusion that the position of the upper band in pam68-2 corresponds to the position of the signals for pD1 and D2, whereas the lower signal should exclusively derive from mD1. To clarify whether the drop in the accumulation of labeled D1 + D2 was caused by a decrease in synthesis or by rapid degradation of newly synthesized D1 proteins, shorter pulse times (5 and 15 min) were applied (Figure 3B). In all cases, markedly less radioactivity was incorporated into D1 + D2 in pam68-2 plants relative to the wild type. These results imply either that D1 + D2 synthesis is impaired or that newly synthesized D1 + D2 polypeptides are very rapidly degraded.
Figure 3.
Translation Profiling of Chloroplast-Encoded Proteins.
(A) Incorporation of [35S]Met into thylakoid membrane proteins of 4-week-old greenhouse-grown wild-type (Col-0) and mutant (pam68-2 and lpa1-2) plants at low light (20 μmol photons m−2 s−1). After pulse labeling of leaves with [35S]Met for 20 min in the presence of cycloheximide, thylakoid membranes were isolated, and proteins were fractionated by SDS-PAGE and detected by autoradiography. In pam68-2 and lpa1-2, incorporation of [35S]Met into D1 was reduced to 61% ± 3% and 35% ± 7% of wild-type levels, respectively, as determined by quantification of the intensity of signals shown here and of three further repetitions of the experiment. Note that Ler, the genetic background of lpa1-2, behaved like Col-0.
(B) Incorporation of [35S]Met into thylakoid membrane proteins of 4-week-old wild-type (Col-0) und mutant (pam68-2) plants as in (A), except that pulses of 5, 15, and 30 min were applied. As loading control, nonlabeled LHCII was visualized by staining with Coomassie blue (C.B.B.).
(C) Incorporation of [35S]Met into thylakoid membrane proteins of 4-week-old wild-type (Col-0) und mutant (pam68-2) plants grown heterotrophically (see Methods) under very low light levels (5 μmol photons m−2 s−1). Pulse labeling for 60 min and signal detection were performed as in (A).
(D) SDS-solubilized membrane proteins with the equivalent of 200,000 incorporated cpm were used for immunoprecipitation with a D1-specific antibody (see Methods). As loading control, IgG heavy chains (HC) were visualized by staining with Coomassie blue. Incorporation of [35S]Met into D1 was reduced to 53% ± 4% of wild-type levels, as determined by quantification of the intensity of signals shown here and of two further repetitions of the experiment.
(E) Transcript analysis in wild-type (Col-0) and mutant (pam68-2) plants. Ten-microgram aliquots of total leaf RNA from 4-week-old plants were fractionated by denaturing agarose gel electrophoresis, blotted onto nylon membrane, and hybridized with probes specific either for the first 500 bp of the coding region (psbA, psbB, psbC, and psbD) or the complete sequence (psbE) of the transcript. Note that the psbC probe recognizes both psbC (bottom bands) and psbCD (top bands) transcripts, the psbD probe detects the bicistronic psbCD transcripts (corresponding to the top two bands of the psbC probed membrane), and psbE labels psbEFJL transcripts. To control for loading, replicate blots were stained with methylene blue (M.B.) and hybridized with a probe specific for ACTIN1.
(F) Association of psbA and psbD mRNAs with polysomes. Whole-cell extracts from Col-0 and pam68-2 plants were fractionated in linear 0.44 to 1.6 M (15 to 55%) sucrose gradients by ultracentrifugation. Gradients were divided into 10 fractions, and RNA was isolated from equal volumes. RNA gel blots were stained with methylene blue (M.B.) to visualize the distribution of rRNAs and then hybridized with probes specific for psbA and psbD (recognizing psbCD polycistronic transcripts). Signals were quantified (see Methods), and relative values of fractions 1 to 4, 5 to 7, and 8 to 10 calculated and given below the psbA and psbCD panels.
To minimize further any secondary effects on chloroplast translation due to the reduction in photosynthesis in the pam68-2 mutants, labeling experiments were performed under very low light (5 μmol photons m−2 s−1) on leaves from heterotrophically grown Col-0 and pam68-2 plants (Figure 3C). Labeling was performed for 60 min to visualize better the synthesis of thylakoid proteins other than D1. Whereas rates of synthesis of the PSI reaction center PsaA/B proteins and PSII core proteins CP43 and CP47, as well as the α- and β-subunits of the cpATPase, were at most barely affected, incorporation of [35S]Met into the PSII subunit D1 + D2 was reduced to ~70% of wild-type levels in the pam68-2 mutant on the basis of quantification of signal intensities (Figure 3C). Once again, in pam68-2, but not in Col-0, a prominent signal for pD1 + D2 was detected. To quantify unambiguously the effect of the pam68-2 mutation on the rate of D1 synthesis, immunoprecipitation of the translation products employing a D1-specific antibody was performed. This experiment confirmed that D1 synthesis in pam68-2 was severely reduced and corresponded to ~50% of wild-type levels, based on quantification of signal intensities (Figure 3D). This also implies that the upper band in Figures 3A and 3C mostly derives from pD1.
The drop in accumulation of newly synthesized D1 in the pam68-2 mutant could be due to a defect in translation, in posttranslational processing of pD1 and/or a decrease in the stability of D1. To distinguish between these possibilities, levels of total and polysome-associated chloroplast transcripts encoding PSII subunits were analyzed. RNA gel blot analysis revealed that the psbA transcript encoding the D1 protein was present at wild-type levels in pam68-2 leaves (Figure 3E). The levels of the polycistronic psbEFJL transcript and the psbC transcript resembled those in the wild type, whereas the amounts of psbB mRNA encoding CP47 were increased and the polycistronic psbCD transcript was slightly decreased (Figure 3E). The effects of the pam68-2 mutation on the translation of the psbA transcript were further examined by analyzing the association of psbA mRNA with ribosomes. To this end, whole-cell extracts from Col-0 and pam68-2 plants leaves were fractionated in sucrose gradients under conditions that keep polysomes intact. Efficiently translated RNAs are almost all associated with ribosomes and migrate deep into the gradient. Plastidic and cytosolic rRNAs in wild-type and pam68-2 polysome gradients showed an equal distribution, as determined by methylene blue staining and quantification of signals, indicating that there is no general difference in the distribution of ribosomes between the two genotypes (Figure 3F). Specific mRNAs were localized in the gradients by performing RNA gel blot hybridizations with RNA purified from gradient fractions. In both genotypes, the distribution of psbA mRNA within the gradient was very similar. For the psbCD polycistronic transcript also, no difference in degree of association with polysomes was detected between the two genotypes (Figure 3F).
Therefore, our data suggest that the reduction in synthesis of D1 does not result from a decrease in the availability of psbA transcripts or from altered translation initiation in terms of their loading onto polysomes. Instead, (1) stability of the D1 protein is specifically affected in the pam68-2 mutant; (2) the defect in D1 accumulation in pam68-2 is less pronounced than that found in the lpa1-2 mutant, which is impaired in the synthesis of D1 and D2 (Peng et al., 2006); and (3) the drastic accumulation of pD1 is characteristic for pam68-2 but not for lpa1.
Formation and Stability of PSII Assembly Complexes Are Perturbed in pam68-2 Plants
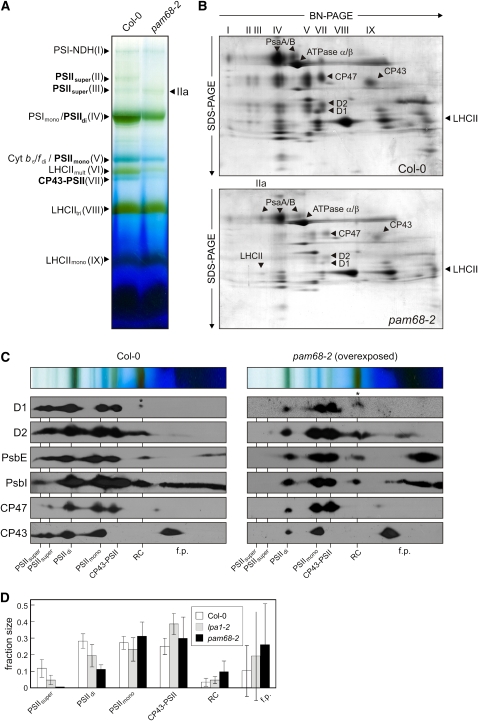
To investigate the effects of altered D1 levels on the biogenesis of PSII, Blue-Native PAGE (BN-PAGE) analysis was performed. Thylakoids were prepared from equal amounts of leaf material and solubilized with n-dodecyl β-d-maltoside (β-DM) (Schägger et al., 1988), and proteins were separated in the presence of Coomassie blue G 250. Nine prominent bands were resolved and assigned to thylakoid multiprotein complexes based on earlier reports (Granvogl et al., 2006; Schwenkert et al., 2006; Peng et al., 2008) as follows: PSI-NDH supercomplex (band I), PSII supercomplexes (bands II and III), PSI monomers and PSII core dimers (band IV), PSII core monomers and Cyt b6/f dimers (band V), multimeric LHCII (band VI), CP43-free PSII monomers (band VII), and trimeric (band VIII) and monomeric (band IX) LHCII (Figure 4A). Only the signals for LHCII monomers and trimers, and for the PSI-NDH supercomplex, showed similar intensities in wild-type and pam68-2 mutant plants. All other bands were markedly weaker in the mutant (Figure 4A).
Figure 4.
Accumulation of PSII Assembly Complexes under Steady State Conditions.
(A) BN-PAGE analysis of thylakoid multiprotein complexes. Thylakoids were isolated from equal amounts of fresh leaf material (100 mg) obtained from wild-type (Col-0) and mutant (pam68-2) plants and solubilized with 1.5% (w/v) β-DM. The extracts were then fractionated by BN-PAGE. The bands detected were identified with specific protein complexes in accordance with previously published profiles (Granvogl et al., 2006; Schwenkert et al., 2006; Peng et al., 2008): PSI-NDH supercomplex (PSI-NDH; band I), PSII supercomplexes (PSIIsuper; bands II and III), PSI-LHCII complex (band IIa), PSI monomers and PSII dimers (PSImono and PSIIdi; band IV), PSII monomers and dimeric Cyt b6/f (PSIImono and Cyt b6/fdi; band V), multimeric LHCII (LHCIImult; band VI), CP43-free PSII monomers (CP43-PSII; band VII), trimeric LHCII (LHCIItri; band VIII), and monomeric LHCII (LHCIImono; band IX). PSII assembly complexes are highlighted in bold.
(B) 2D BN/SDS-PAGE separation of thylakoid protein complexes. Individual lanes from BN-PA gels as in (A) were analyzed in the presence of SDS by electrophoresis on 10 to 16% PA gradient gels, which were then stained with colloidal Coomassie blue (G 250). The identity of relevant proteins is indicated by arrows.
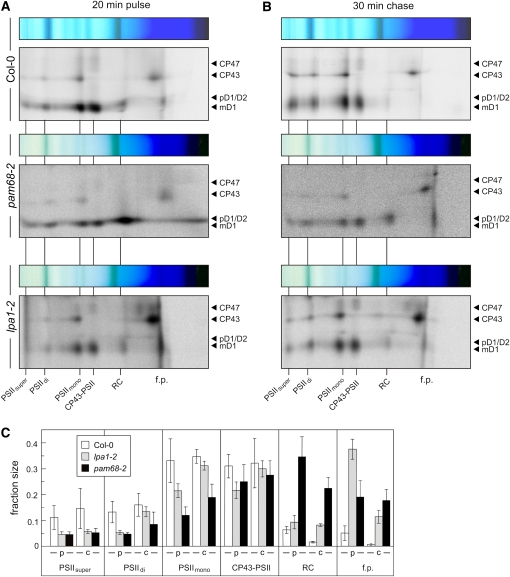
(C) Detection of PSII assembly complexes by immunoblot analyses of 2D BN/SDS gels as in (B) with antibodies against D1, D2, PsbE, PsbI, CP47, and CP43. The positions of PSII assembly complexes (PSIIsuper, PSII supercomplexes; PSIIdi, PSII dimers; PSIImono, PSII monomers; CP43-PSII, CP43-free PSII monomers; RC, reaction center-like complex) and free proteins (f.p.) are indicated. Signals were obtained by chemiluminescence. Exposure times were ~2 min (wild type) and ~10 min (mutant). Accumulation of pD1 is indicated by asterisks.
(D) Signals obtained for D1, D2, PsbE, PsbI, CP43, and CP47 in (C), and for the same proteins from the lpa1-2 mutant (see Supplemental Figure 1 online), were quantified for each PSII assembly complex and for free proteins. The relative amounts of all PSII assembly complexes were calculated for each protein (summing to 1 for each protein), and mean values and standard deviations for each PSII assembly complex were determined from three replicates. Note that Ler, the genetic background of lpa1-2, behaved similar to Col-0.
The complexes resolved by BN-PAGE were then separated into their subunits in the second dimension by electrophoresis on SDS-PAGE gels and stained with Coomassie blue G 250 (Figure 4B). In pam68-2 plants, levels of the chloroplast ATP synthase, Cyt b6/f, and LHCII were equivalent to those in the wild type; amounts of PSI-forming proteins were slightly lower (Figure 4B). As expected (Figure 2A), the amounts of the PSII core subunits D1, D2, CP43, and CP47 were drastically reduced in the mutant. CP43-free PSII monomers (band VII), as well as PSII monomers (band V) and dimers (band IV), were still detectable, but PSII supercomplexes (bands II and III) were not found in pam68-2 plants (Figures 4A and 4B). The two-dimensional (2D) BN/SDS-PAGE analysis also revealed that pam68-2 plants contain a stable PSI-LHCII supercomplex (designated as band IIa), which migrates in the same region as band III. This PSI-LHCII supercomplex can be formed when the plastoquinone pool becomes overreduced (Pesaresi et al., 2002, 2009; Ihnatowicz et al., 2008). Although 1-qP, a standard measure for the reduction state of plastoquinone, was indeed found to be increased in pam68-2 plants (Table 1), it is more likely that the plastoquinone pool is more oxidized because of the marked drop in PSII abundance relative to PSI in the mutant (Figure 2A). Therefore, it appears plausible that the relative abundance of LHCII with respect to PSII (Figures 2A, 4A, and 4B) should favor increased docking of LHCII to PSI.
To investigate further the changes in the PSII assembly process in the pam68-2 mutant, immunoblot analysis with antibodies directed against several PSII core subunits (D1, D2, PsbE, PsbI, CP47, and CP43) was performed on replicate 2D BN/SDS-PAGE gels (Figure 4C), and signals were quantified (Figure 4D). This analysis confirmed that PSII supercomplexes were virtually absent and PSII dimers were drastically reduced in amount, relative to PSII monomers and CP43-free PSII monomers. In addition, an increase in free PSII proteins was observed. Moreover, the PSII assembly intermediate RC, which comprises pD1, D2, PsbE, PsbF, and PsbI (Rokka et al., 2005), was detected, migrating just below the LHCII trimers (band VIII). Compared with the wild type, about twice as much pD1, D2, PsbE, and PsbI was present in the pam68-2 RC complex (Figures 4C and 4D). Interestingly, in the lpa1-2 mutant, PSII monomers and CP43-free PSII monomers are the predominant PSII assembly states, and RC does not accumulate to the same degree as in pam68-2 (see Supplemental Figure 1 online; Figure 4D). Moreover, PSII supercomplexes are detectable in the lpa1-2 line, but not in pam68-2 (see Supplemental Figure 1 online; Figure 4D).
To study the kinetics of PSII assembly, thylakoid membrane proteins from Col-0, pam68-2, and lpa1-2 plants were separated by 2D BN/SDS-PAGE after labeling with [35S]Met for 20 min (Figure 5A), as well as after a subsequent 30-min chase with unlabeled Met (Figure 5B). The various PSII assembly intermediates, RC, CP43-free PSII monomers (CP43-PSII), PSII monomers, dimers, and supercomplexes, as well as free PSII proteins, were visualized autoradiographically (Figures 5A and 5B) and quantified (Figure 5C). After pulse labeling, in Col-0 and lpa1-2, most of the assembled radiolabeled D1 was found in PSII monomers and CP43-PSII, and the two genotypes differed only with respect to free unassembled D1 protein, the level of which was about sevenfold higher in lpa1-2. Strikingly, the pam68-2 mutant exhibited a sixfold increase (relative to the wild type) in newly synthesized RC, which clearly contained pD1 instead of mD1 (Figures 5A and 5C). After the chase, the relative abundance of CP43-PSII and PSII monomers and dimers was increased in pam68-2 plants at the expense of RC, suggesting that the mutant RC complex was assembled into larger PSII intermediates (Figure 5C). When the relative accumulation of PSII intermediates under steady state conditions was also considered (Figure 4D), it became clear that the PSII assembly in pam68-2 plants is delayed. Thus, directly after pulse labeling, radiolabeled D1 was found predominantly in the RC complex in pam68-2 plants, whereas Col-0 and lpa1-2 showed labeling in PSII monomers and CP43-PSII. After the chase, pam68-2 plants showed labeled D1 mostly in CP43-less monomers, while Col-0 and lpa1-2 had mostly labeled PSII monomers and CP43-PSII. Under steady state conditions, the predominant PSII intermediates, as determined by immunoblot analyses (Figures 4C and 4D), were PSII monomers and CP43-PSII in pam68-2 plants, PSII monomers and dimers in Col-0, and PSII monomers and CP43-PSII in lpa1-2 (Figure 5C).
Figure 5.
Rates of Synthesis of PSII Assembly Complexes as Detected by Pulse-Chase Analysis.
(A) and (B) 2D BN/SDS-PAGE analysis of [35S]Met incorporation into thylakoid membrane protein complexes. After pulse labeling of 4-week-old leaves with [35S]Met for 20 min in the presence of cycloheximide (A), a chase of unlabeled Met for 30 min was applied (B). After thylakoid membrane isolation, proteins were fractionated by 2D BN/SDS-PAGE and complexes visualized by autoradiography. The positions of the different PSII assembly complexes and free proteins (f.p.) are indicated as in Figure 4C.
(C) Signals obtained for D1 + D2 in (A) and (B) were quantified for each PSII assembly complex as in Figure 4D. p, pulse; c, chase.
The discrepancy between the sixfold increase in the synthesis of RC and the only twofold increase under steady state conditions on one side (Figures 4D and 5C) and between the rate of D1 synthesis of 60% of wild-type levels versus only 10% of wild-type accumulation under steady state conditions on the other side (Figures 2A and 3A) clearly suggests a decrease in the stability of pD1 or D1 in pam68-2 plants. In fact, quantification of the signals of D1 + D2 obtained in pulse-chase experiments as shown in Figures 5A and 5B indicated that D1 degradation already occurs during 30 min of chase (see Supplemental Figure 3A online). Moreover, the half-life of D1 in pam68-2 plants is much shorter than that of wild-type D1, as determined by time-course experiments with plants treated with lincomycin (see Supplemental Figure 3B online).
Taken together, our results imply that in pam68-2 plants, the assembly of PSII is delayed. The assembly step leading from RC to larger PSII assembly intermediates seems to be impaired in pam68-2, as RC accumulates together with pD1 (see Figures 5A to 5C). However, the finding that CP43-less PSII monomers and PSII monomers, and not RC, are the predominant forms of PSII under steady state conditions (Figures 4C and 4D), along with the decreased stability of D1 in the mutant (see Supplemental Figure 3 online), leads to the conclusion that a fraction of the RC complex becomes disassembled and degraded in pam68-2. This interpretation is corroborated by the observation that the accumulation of free PSII proteins is increased. The marked drop in steady state levels of PSII dimers and PSII supercomplexes observed in pam68-2 (Figures 4C and 4D) follows their lowered synthesis rates, as determined by in vivo labeling (Figure 5). This effect on late steps in PSII assembly is most likely the consequence of the decreased availability of the early assembly states due to delayed transition of RC into later assembly intermediates. Furthermore, because the lpa1-2 mutant, which shows a more pronounced drop in levels of newly synthesized D1 than does pam68-2 (Figure 3A), does not display a pam68-like increase in RC accumulation (Figure 5C), it can be concluded that PAM68 and LPA1 exert different functions on the assembly of D1 into PSII.
PAM68 Is Conserved in Photosynthetic Eukaryotes and Cyanobacteria
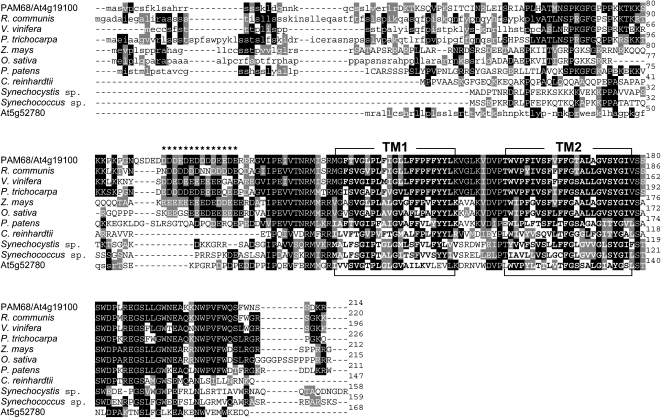
PAM68 encodes a protein of 214 amino acids with a predicted molecular mass of 24.3 kD. The ChloroP program (see Methods) predicts a chloroplast targeting signal of 35 amino acids, resulting in a mature protein of 20.3 kD (Figure 6). Orthologs of PAM68 exist in all sequenced photosynthetic eukaryotes and cyanobacteria. For instance, the mature Arabidopsis PAM68 protein and its cyanobacterial orthologs in Synechocystis and Synechococcus share 42/61% and 38/55% amino acid sequence identity/similarity, respectively. Even higher levels of homology were found between Arabidopsis PAM68 and its orthologs in photoautotrophic eukaryotes, ranging from 43/67% identity/similarity in C. reinhardtii to 76/89% identity/similarity in Ricinus communis. In addition, the product of the At5g52780 gene in Arabidopsis shares 38/58% sequence identity/similarity with PAM68.
Figure 6.
Sequence Alignment of Arabidopsis PAM68 and Its Homologs from Other Species.
The sequence of the PAM68 protein was compared with related sequences from R. communis, Vitis vinifera, Populus trichocarpa, Zea mays, Oryza sativa, Physcomitrella patens subsp patens, C. reinhardtii, Synechocystis sp PCC 6803 and Synechococcus sp PCC 7002, and the Arabidopsis homolog At5g52780. The sequences were aligned using ClustalW and BoxShade (see Methods). Transit peptide sequences predicted by ChloroP are shown in lowercase letters. The two TM domains (TM1 and TM2) of each protein predicted by TMHMM (see Methods) are highlighted in bold; residues comprising the acidic domains are indicated by asterisks. Identical and closely related amino acids that are conserved in at least 30% of the aligned sequences are highlighted by black and gray shading, respectively.
All PAM68 proteins contain two transmembrane domains (TMs), as predicted by the TMHMM program (see Methods) (Figure 6; amino acids 126 to 145 and 155 to 177 for PAM68). In addition, the PAM68 proteins in vascular plants contain an N-terminal stretch composed of acidic amino acids only, which is referred to in the following as the acidic domain. The PAM68 sequences of Physcomitrella, Synechocystis, Synechococcus, and the Arabidopsis At5g52780 homolog at least partially lack this acidic domain.
Two lines of evidence argue against the idea that PAM68 and At5g52780 have redundant functions in Arabidopsis. First, the acidic domain is not conserved in At5g52780 (see above) and, second, PAM68 and At5g52780 do not derive from a recent segmental duplication in the genome of the Arabidopsis lineage (Arabidopsis Paralogon database; http://wolfe.gen.tcd.ie/athal/dup; Blanc et al., 2003). To clarify unambiguously whether At5g52780 and PAM68 encode proteins with overlapping functions, the At5g52780 T-DNA insertion mutant SALK_143426, designated at5g52780-1, was identified in the SALK collection and characterized (see Supplemental Figure 4 online). The at5g52780-1 mutant plants totally lacked the At5g52780 protein, as revealed by immunoblot analysis with an antibody specific for the protein, and displayed wild-type-like growth rate and leaf coloration. Moreover, at5g52780-1 chloroplasts showed wild-type-like levels of the D1 protein. A pam68-2 at5g52780-1 double mutant was generated by crossing and found to behave like pam68-2 plants with respect to growth rate, leaf coloration, and accumulation of the D1 protein (see Supplemental Figure 4 online). This clearly indicates that PAM68 and At5g52780 do not exercise redundant functions.
Synechocystis PAM68 Also Promotes Early Steps in PSII Biogenesis
To test whether the function of PAM68 was conserved during evolution, Sll0933, the PAM68 ortholog from Synechocystis sp PCC 6803 (hereafter, Synechocystis), was disrupted by insertion of a kanamycin resistance cassette (see Supplemental Figure 5A online). The resulting mutant ins0933 showed complete segregation of the mutated gene (see Supplemental Figure 5B online), and the absence of the Sll0933 protein was verified by immunoblot analysis using an Sll0933-specific antibody (see Supplemental Figure 5C online). The ins0933 mutant grew like the wild type under both mixotrophic and photoautotrophic conditions (see Supplemental Figure 5D online). No differences between the wild type and ins0933 were observed in fluorescence emission spectra at low temperature (77K); immunoblot analyses likewise revealed no changes in levels of PSII and PSI subunits (see Supplemental Figures 5C and 5E online). Furthermore, light-dependent oxygen evolution was very similar in ins0933 and the wild type (ins0933, 1490 ± 54 μmol O2 mg Chl−1·h−1; wild type, 1440 ± 55 μmol O2 mg Chl−1·h−1). This indicates that Sll0933 is less important for efficient photosynthesis than is its plant counterpart PAM68. In ins0933, late PSII assembly complexes were present in normal amounts, but the PSII reaction center complexes RCa and RCb, which in wild-type cells contain pD1, mD1, D2, PsbE, PsbF, and PsbI and probably different associated assembly factors (reviewed in Nixon et al., 2010), were not detectable by 2D BN/SDS-PAGE analysis (Figure 7). Moreover, in wild-type cells, the Sll0933 protein was detected in complexes corresponding in molecular weight to early PSII assembly intermediates (Figure 7).
Figure 7.
2D BN/SDS-PAGE Analysis of PSII Complexes from Synechocystis Wild-Type and ins0933 Cells.
Membrane samples (each equivalent to 20 μg of chlorophyll) were solubilized with β-DM, subjected to 2D BN/SDS-PAGE, and blotted onto a nitrocellulose membrane. The PSII proteins D1, pD1, CP47, CP43, and Sll0933 (the Synechocystis PAM68 homolog) were detected using appropriate antibodies. Designation of complexes: RCC1, PSII core monomers; RC47, PSII core complex lacking CP43; RCa and RCb, reaction center complexes a and b, respectively. Positions of size marker bands are indicated at the top.
Taken together, the data indicate that PAM68 and its cyanobacterial counterpart are involved in early step(s) in PSII biogenesis. However, defects in this early step lead to very different effects in the two species: levels of RC and pD1 increase in pam68-2, whereas RCa and RCb, as well as pD1, become undetectable in ins0933. Furthermore, while absence of PAM68 has marked effects on the accumulation of PSII dimers and supercomplexes and, thus, on the photosynthesis rate, ins0933 mutants behave like the wild type with respect to photosynthesis and growth.
PAM68 Is an Integral Thylakoid Protein
Several attempts were made to generate an antibody that specifically recognized PAM68 (see Methods), but only an antibody raised against an epitope located in the acidic stretch of PAM68 was able to detect minimal traces of PAM68 in wild-type plants. However, this antibody also recognized multiple additional proteins in the pam68-2 mutant and was therefore unsuitable for conventional immunoblot analyses (see Supplemental Figure 6 online). Therefore, plants that overexpressed the PAM68 protein were constructed for subcellular localization experiments (see Methods). The PAM68 overexpressor (35S:PAM68 in the pam68-2 background, referred to as oePAM68) plants showed an ~30-fold increase in PAM68 levels, which reversed the effects of the pam68-2 mutation with respect to growth, pigmentation, and D1 accumulation (see Supplemental Figure 6 online). Chloroplasts from oePAM68 plants were fractionated into stroma and thylakoid fractions. Both fractions were subjected to immunoblot analysis, and the purity of fractions was tested by monitoring the stromal CSP41a and thylakoid D2 proteins (Figure 8A). The PAM68 protein was exclusively detected in the thylakoid fraction. To clarify whether PAM68 constitutes an integral or peripheral thylakoid protein, thylakoids from oePAM68 plants were treated with alkaline and chaotropic salts to release membrane-associated proteins (Figure 8B). In this assay, PAM68 behaved like the integral protein Lhcb1 and not like the peripheral PsaD1, indicating that PAM68 represents an integral membrane protein, as already suggested by the presence of two predicted TMs (Figure 6). As expected, fractionation and solubilization experiments similar to those presented in Figures 8A and 8B revealed that Synechocystis Sll0933 too is an integral thylakoid protein (see Supplemental Figures 5F and 5G online).
Figure 8.
Subcellular Localization and Topology of PAM68.
(A) Suborganellar localization of PAM68. Chloroplasts, stroma, and thylakoids were isolated from oePAM68 (35S:PAM68 pam68-2) plants, fractionated by SDS-PAGE, transferred to poly(vinylidene difluoride) membrane, and visualized using antibodies raised against the acidic domain of PAM68, CSP41a (as a control for stromal proteins), or D2 (as a control for thylakoid proteins).
(B) Extraction of thylakoid-associated proteins with chaotropic salt solutions or alkaline pH. Thylakoid membranes from oePAM68 plants were resuspended at 0.5 mg chlorophyll/mL in 10 mM HEPES/KOH, pH 7.5, containing either 2 M NaCl, 0.1 M Na2CO3, 2 M NaSCN, 0.1 M NaOH, or no additive. After incubation for 30 min on ice, supernatants containing the extracted proteins (s) and membrane fractions (p) were separated by SDS-PAGE and immunolabeled with antibodies raised against the acidic domain of PAM68, PsaD (as a control for peripheral membrane proteins), or Lhcb1 (as a control for integral membrane proteins).
(C) Schematic representation of the two possible topologies of PAM68, with the two TMs indicated by black boxes, the acidic domain as a gray box, and the trypsin cleavage sites depicted by asterisks. In the bottom half of the panel, relevant proteolytic fragments are indicated.
(D) Immunoblot analysis of thylakoid membrane preparations with antisera specific for the acidic domain of PAM68 or PsbO (as a control for luminal thylakoid proteins) before (−Trypsin) and after (+Trypsin) treatment with trypsin. In intact thylakoids, only stroma-exposed polypeptides are accessible to the enzyme.
To determine the topology of PAM68 in the thylakoid membrane, thylakoids were subjected to mild digestion with trypsin, such that only the stroma-exposed face was accessible to the protease. If the N and C termini of PAM68 faced the stroma (Topology 1 in Figure 8C), tryptic digestion should generate a 2-kD peptide detectable with the PAM68 antibody raised against the acidic domain. Tryptic digestion of an oppositely oriented PAM68 (with the N and C termini facing the lumen; Topology 2 in Figure 8C) at two sites within the inter-TM region is expected to result in a 13-kD fragment. As expected, PsbO on the lumen side of the thylakoid membrane was not affected by trypsination, whereas PAM68 was efficiently digested without leaving any detectable proteolytic fragment sized between 8 and 20 kD (Figure 8D). The missing detection of the 2-kD fragment can be explained by failed precipitation with acetone, which was used for trypsin inactivation, or inefficient membrane transfer of the small fragment during electroblotting. Nevertheless, the absence of the 13-kD fragment clearly implies that Topology 1 depicted in Figure 8C represents the actual topology of PAM68.
PAM68 Forms Part of a Small Multiprotein Complex That Requires LPA1 for Its Stability
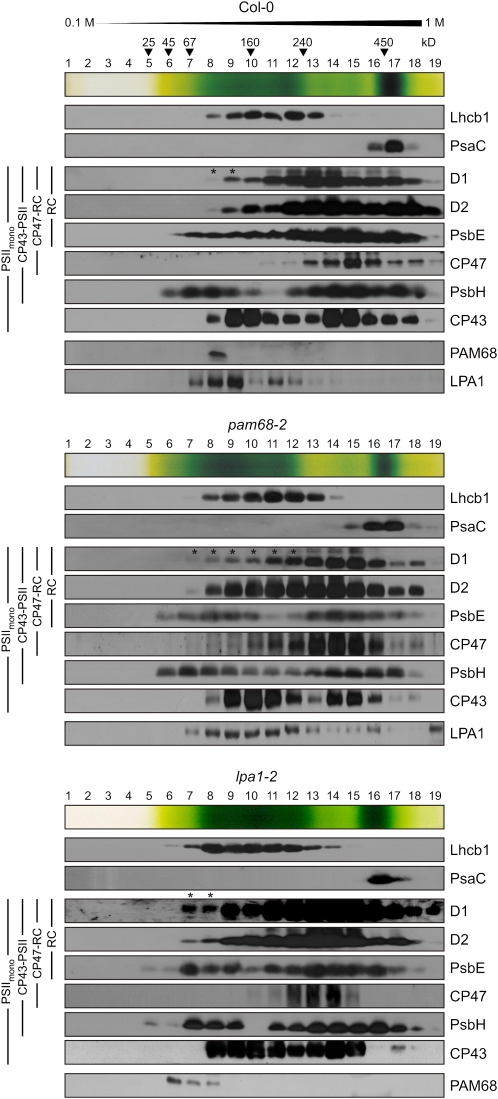
The above results suggest that Arabidopsis PAM68 promotes early steps in PSII biogenesis and that the Synechocystis PAM68 ortholog is present in low molecular weight complexes corresponding to early PSII assembly intermediates. Because our PAM68 antibodies were not suitable for detection of PAM68 on 2D BN/SDS gels in which only limited amounts of thylakoid complexes can be loaded and separated (see above), we used sucrose gradient centrifugation to fractionate increased amounts of assembly intermediates in Arabidopsis. To this end, thylakoids were isolated from Col-0 and pam68-2 plants and solubilized with β-DM. Thylakoid complexes were then separated by ultracentifugation on a linear 0.1 to 1 M sucrose gradient. After centrifugation, 19 fractions were collected (numbered from top to bottom), and proteins were precipitated and subjected to immunoblot analysis. Several additional unspecific bands of various sizes were detected with the PAM68 antibody, as expected from the PAM68 immunoblot experiments with total thylakoid fractions (see Supplemental Figure 6 online), but these signals were distributed throughout the gradient and much weaker than the distinct PAM68 signal present in Col-0 but not in pam68-2. Thus, in Col-0, but not in pam68-2, a distinct signal for PAM68 was identified in fraction 8 (Figure 9). Fraction 8 from both the wild type and pam68-2 also contained CP43, PsbE, PsbH, and LPA1, together with traces of pD1 and D2, but no CP47 (Figure 9). In pam68-2, traces of pD1 and D2, but no CP43, were also detected in fraction 7, possibly indicating that loss of PAM68 affects a complex that contains pD1 and D2, but not CP43. Interestingly, the distribution of LPA1 was shifted toward denser fractions in pam68-2 (fractions 7 to 19) compared with the wild type (fractions 7 to 14).
Figure 9.
Analysis of PAM68-Containing Complexes by Sucrose Gradient Sedimentation.
Thylakoids (1 mg chlorophyll/mL) from wild-type (Col-0) and mutant (pam68-2 and lpa1-2) plants were solubilized with 1% (w/v) β-DM and separated by centrifugation in a linear 0.1 to 1 M sucrose gradient. Note that because chlorophyll (a + b) levels in pam68-2 plants are equivalent to 65% of wild-type chlorophyll levels, the pam68-2 sample contained ~50% more protein. Nineteen fractions were collected (numbered from top to bottom) from two wild-type and four pam68-2 and lpa1-2 gradients, and fractions from the same genotype were pooled. Proteins were precipitated from each fraction, separated by SDS-PAGE, blotted onto poly(vinylidene difluoride) membranes, and detected with antibodies against D1, D2, PsbE, CP47, PsbH, CP43, PAM68, and LPA1, as well as PsaC and Lhcb1. At the top of each blot, an image of the sucrose gradient is shown, and the positions of molecular mass markers in the gradient are indicated. Note that Ler, which serves as wild-type control for lpa1-2, behaved very similarly to Col-0 (see Supplemental Figure 7 online) and that PAM68 and LPA1 were not detected in pam68-2 and lpa1-2 samples, respectively. Accumulation of pD1 is indicated by asterisks.
Because Col-0 LPA1 and PAM68 were found in the same fraction of the sucrose gradient, thylakoid complexes from lpa1-2 plants were also analyzed. In the corresponding wild type (Landsberg erecta [Ler]), as in Col-0 (Figure 9, top panel), the PAM68 protein was found in fraction 8, again together with traces of D1 and D2 (see Supplemental Figure 7 online). In lpa1-2 mutants, however, the distribution of PAM68 was shifted toward less dense fractions (fractions 6 to 8). In addition, pD1 and D2 also shifted to fraction 7 in the lpa1-2 mutant. The distribution of CP43, which was also present in fraction 8 from wild-type (Col-0 and Ler) plants, remained unchanged in lpa1-2.
Taken together, these data are consistent with the idea that Arabidopsis PAM68, like Synechocystis Sll0933 (Figure 7), is associated with a low molecular weight complex formed at an early step in the PSII assembly process. This complex might contain pD1 and D2, but it seems to lack CP47 or CP43. Because LPA1 is found in more dense fractions when PAM68 is absent, whereas PAM68 is found in less dense fractions when LPA1 is missing, one may speculate that PAM68 displaces LPA1 during PSII biogenesis. Thus, in the absence of LPA1, the PAM68 protein might bind to a smaller complex than usual. Conversely, in the absence of PAM68, the LPA1 protein remains associated with PSII or even larger complexes, such as ribosome-associated nascent chains at later assembly steps.
PAM68 Interacts with Several PSII Core Subunits and Assembly Factors
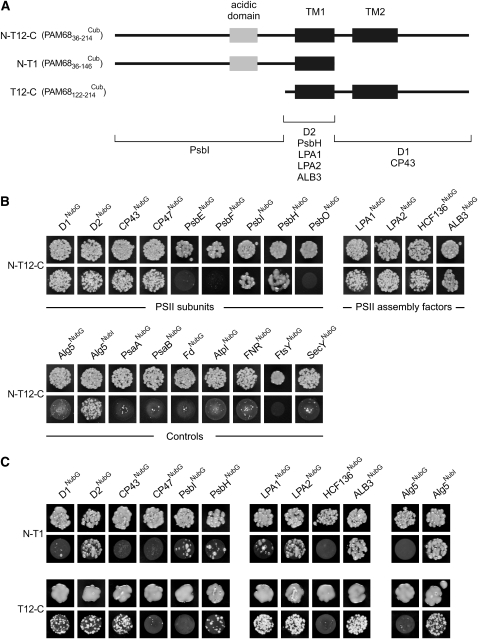
To test further for associations between PAM68 and structural or auxiliary PSII proteins, we examined the interaction of PAM68 with several thylakoid proteins using the split-ubiquitin system (Pasch et al., 2005). In this assay, the mature form of PAM68 (PAM6836-214; Figure 10A) failed to interact with representative components of PSI (PsaA, PsaB, ferredoxin [Fd], and the ferredoxin-NADP+ oxidoreductase [FNR]), subunit IV of the cpATPase (AtpI), or components of the signal recognition particle (FtsY; Kogata et al., 1999) or secretory (Sec) (SecY; Laidler et al., 1995; Roy and Barkan, 1998) thylakoid targeting pathways (Figure 10B). Strikingly, in the same assay, mature PAM68 interacted with most PSII core proteins tested (D1, D2, CP43, CP47, PsbH, and PsbI) but not with PsbE, PsbF, or PsbO. We next tested whether PAM68 interacts with known PSII assembly factors. Here, PAM68 was found to interact with HCF136, LPA1, LPA2, and ALB3 (Figure 10B).
Figure 10.
Interaction of PAM68 with Other Thylakoid Proteins.
(A) Schematic presentation of the fragments of PAM68 used to identify interaction domains. The acidic domain is shown as a gray box and the two TMs as black boxes. In the bottom half of the panel, the three interaction domains and the corresponding interacting proteins are shown.
(B) Split-ubiquitin assays for interactions between full-length PAM68 and selected thylakoid proteins. Assays were performed employing fusions to the C- (Cub) and N- (NubG) terminal halves of ubiquitin. Alg5NubI (the unrelated endoplasmic reticulum membrane protein Alg5 fused to the wild-type Nub) served as a positive control. Alg5 fused to NubG (Alg5NubG) was used as the negative control. To test for interactions involving the PAM68 protein, the mature PAM68 protein was fused to Cub (N-T12-C or PAM6836-214Cub), and the selected thylakoid proteins were fused to NubG. Yeast colonies were first plated on permissive (-LT, top panels) and then on selective medium (-LTH, bottom panels) (see Methods).
(C) Interaction mapping to distinct domains of PAM68. Two different fragments as shown in (A) were employed to detect interactions between domains of PAM68 and selected thylakoid proteins. Split-ubiquitin assays were performed as in (B).
To investigate whether the multiple interactions of PAM68 with PSII proteins and assembly factors depend on specific domains, deletion derivatives of PAM68 were analyzed for their interaction capacity. The PAM68 fragments tested were PAM6836-146 (representing the N-terminal region of the mature protein including TM1) and PAM68122-214, consisting of both TMs and the C terminus of PAM68 (Figures 10A and 10C). The deletion products interacted with most of the proteins that interacted with the full-length construct, and three different interaction domains could be identified, with the N terminus responsible for interaction with PsbI, the region including TM1 for interaction with D2, PsbH, LPA1, LPA2, and ALB3, and the C terminus required for interaction with D1 and CP43 (Figure 10A). The deletion derivatives failed to interact with CP47 and HCF136, indicating that multiple domains of PAM68 might be required for these interactions.
In summary, the interactions of PAM68 with D1, D2, HCF136, and LPA1 are compatible with the requirement for PAM68 for efficient D1 maturation and stability. Moreover, the multiple interactions of PAM68 with PSII subunits that are added later in PSII biogenesis might also provide an explanation for the decreased formation of PSII dimers and supercomplexes im pam68-2 plants (Figure 4), in the event that this is not simply a secondary effect of the delay in early PSII assembly steps.
DISCUSSION
Absence of Arabidopsis PAM68 Has Multiple Effects on PSII Assembly
Loss of PAM68 has drastic effects on PSII function and plant growth (Figure 1). These effects are similar to those seen in lpa1 (Peng et al., 2006; see Supplemental Figure 1 online) or lpa2 (Ma et al., 2007) mutants, but less severe than the seedling-lethal phenotype of the hcf136 mutant (Meurer et al., 1998). At the protein level, the abundance of PSII core subunits, particularly D1, D2, CP43, and CP47, was markedly decreased (Figures 2 and 4), and the maturation (Figure 2) and stability (Figure 3; see Supplemental Figure 3 online) of D1 are both impaired. Conversely, there is a clear increase in the accumulation of the RC assembly intermediate and free nonassembled proteins at the expense of PSII dimers and supercomplexes (Figure 4). In the lpa1 mutant, levels of newly synthesized D1 and D2 are even lower, but this has much less effect on PSII assembly (Figures 5A and 5B) and stability (Figures 4C and 4D). Hence, it can be concluded that the perturbation of PSII biogenesis in pam68 mutants cannot be attributed solely to the reduction in D1 abundance. The absence of PAM68, unlike that of LPA1, appears to have multiple effects on (1) maturation and stability of the D1 protein and (2) conversion of the RC complex into larger PSII intermediates. In addition, (3) the accumulation of PSII dimers and supercomplexes is highly reduced in pam68 plants under steady state conditions (Figure 4D), probably as a consequence of the delayed assembly of earlier PSII states, although it cannot be totally excluded that PAM68 might also play a role in the assembly of PSII dimers and supercomplexes (see below).
Cyanobacterial and Plant PAM68 Proteins Are Involved in Early PSII Biogenesis
Arabidopsis PAM68 (Figure 10) and LPA1 (Peng et al., 2006) interact with D1, and LPA2 binds CP43 (Ma et al., 2007). In all three cases, inactivation of the assembly factor by mutation is accompanied by a specific reduction in the synthesis of its binding partner. This implies that physical interaction of these assembly factors with specific PSII subunits is necessary for normal synthesis or stability of the latter. A dramatic increase in accumulation of pD1, like that seen in pam68-2 plants, has also been observed in hcf136 plants (Meurer et al., 1998), but not in lpa1 (Peng et al., 2006; Figure 3) or lpa2 (Ma et al., 2007) mutants. Comparison of the kinetics of the synthesis/processing of D1 and its incorporation into PSII showed that pD1 processing occurs early in PSII assembly (van Wijk et al., 1997), and in Synechocystis, an increase in pD1 has been noted when either PsbH or CP47 is absent, suggesting that the formation of the CP47-RC complex facilitates the maturation of D1 (Komenda et al., 2005). YCF48, the cyanobacterial HCF136, plays a role in the stabilization of newly synthesized pD1 and in its subsequent binding to D2-PsbE-PsbF; thus, absence of YCF48 dramatically decreases the level of pD1 (Komenda et al., 2008). The increase in pD1 observed in Arabidopsis hcf136 plants appears at first sight to be at variance with the ycf48 phenotype but can be explained by the fact that, in contrast with ycf48, the hcf136 mutant accumulates only trace amounts of D1 and PsbE/PsbF, while D2, CP43, and CP47 are undetectable (Meurer et al., 1998). Therefore, the formation of CP47-RC, which, by analogy to the situation in cyanobacteria, should be required for pD1 maturation, is prevented in the hcf136 genotype (Plücken et al., 2002). Why then does pam68 exhibit an increase in accumulation of pD1? This can be explained by assuming either a direct involvement of PAM68 in pD1 maturation or indirect effects caused by perturbations in the formation of CP47-RC in pam68-2, similar to the situation in hcf136. Although the accumulation of CP47-RC has not been determined here, the delayed assembly of RC into larger PSII intermediates in pam68-2 argues in favor of the latter scenario.
Inactivation of the Synechocystis gene for the Sll0933 protein prevents accumulation of both the RC complex and pD1 under steady state conditions, whereas the accumulation of later PSII assembly intermediates was unaltered (Figure 7). This clearly implies that, in contrast with the pam68-2 mutant, in the Synechocystis mutant the assembly of RC into larger intermediates is not delayed; instead, the transition from RC to CP47-RC might be even more efficient in the mutant so that both RC and pD1 are not detectable under steady state conditions. Assuming that PAM68 and its cyanobacterial homolog exert similar functions at the molecular level in both organisms, how can their absence lead to such contrasting effects? A general explanation for the more severe phenotype of the Arabidopsis pam68-2 mutation compared with the Synechocystis sll0933 mutation (and correspondingly for hcf136 in Arabidopsis and ycf48 in Synechocystis; see above) might be provided by the more effective elimination of nonassembled proteins and abnormal complexes by the proteolytic quality control system in chloroplasts (reviewed in Nixon et al., 2010). Alternatively, the delay in the assembly of RC into larger complexes evident in the Arabidopsis pam68-2 mutant might be bypassed in Synechocystis by pathways that have been lost during the evolution of the plant lineage. A third explanation could be that PAM68 contributes also to the PSII repair cycle, such that the contrasting mutant phenotypes in Synechocystis and Arabidopsis might result from the different consequences of perturbations in PSII repair in the two species.
At what stage in the assembly process do Arabidopsis and Synechocystis PAM68 proteins come into play? Synechocystis Sll0933 was detected in small complexes that might represent RC or even earlier assembly states (Figure 7). The sucrose gradient analysis of the Arabidopsis PAM68 protein (Figure 9) indicates that PAM68 may form a relatively small complex with LPA1, D1, and D2. This complex might again correspond to RC or earlier steps in PSII assembly. So whereas PAM68 proteins appear to associate with early assembly states, further analyses will be required for their unambiguous identification.
Do Assembly Factors Associate Only Transiently with PSII?
Our results indicate that PAM68 interacts with LPA1, LPA2, HCF136, and ALB3 in the split-ubiquitin assay (Figure 10), and the interaction with LPA1 is supported by sucrose gradient analysis (Figure 9). Because PSII assembly factors are generally thought to associate transiently with intermediates during PSII biogenesis, the action of PAM68 in early PSII biogenesis (see above) on one side, and its multiple interactions with other assembly factors thought to act on various PSII assembly complexes on the other side, is unexpected. However, although YCF48 has been found only in RC complexes (Komenda et al., 2008), its eukaryotic counterpart HCF136 has also been detected in larger PSII assembly complexes (Plücken et al., 2002). LPA2 is thought to promote CP43 assembly within PSII to form PSII core monomers, but it also interacts with ALB3 (Ma et al., 2007), which is thought to be involved in D1 biogenesis. Indeed, inspection of mRNA expression databases indicates that HCF136, ALB3, LPA1, and PAM68 are more or less constitutively expressed, whereas DEG1 (Kapri-Pardes et al., 2007; Sun et al., 2010) and VAR2 (Zaltsman et al., 2005), as examples for factors involved in PSII turnover and repair, vary markedly in their expression under different light conditions (see Supplemental Table 1 online). Therefore, PSII assembly factors might operate on more than one assembly state during PSII biogenesis. Alternatively, PAM68 might participate in complexes that contain other assembly factors but lack PSII subunits. Indeed, PAM68 (see Supplemental Figure 6 online) and HCF136 (P. Westhoff, personal communication) can accumulate many fold when overexpressed, implying that they do not require their PSII interaction partners for stability.
Conclusions
The PAM68 protein is involved in the assembly of PSII and appears to interact with early assembly intermediates. Intriguingly, its absence delays certain assembly steps in Arabidopsis, such that the RC complex accumulates, whereas in Synechocystis the transition from RC to later PSII intermediates seems to be accelerated. Moreover, PAM68 interacts in split-ubiquitin assays with some PSII proteins and assembly factors that appear at later stages of PSII assembly, and much lower levels of PSII dimers and supercomplexes accumulate under steady state conditions in pam68-2 than in the lpa1-2 mutant, which has a comparable synthesis rate of PSII dimers and supercomplexes (Figures 4D and 5C). Although the decrease in PSII dimer and supercomplex formation might be attributed, as a secondary effect, to the perturbation in early PSII assembly, it cannot be excluded that PAM68 might also support later assembly steps through transient interaction with the proteins identified by the split-ubiquitin assay, although PAM68 was detected only in smaller complexes (Figures 7 and 9). Future experiments have to clarify whether PAM68 might be sufficiently abundant for such an additional role in later PSII assembly steps and provide independent experimental evidence for its multiple interactions identified by the split-ubiquitin assay. Moreover, further experiments are needed to explain the different effects of the absence of PAM68 on PSII assembly in cyanobacteria and flowering plants.
METHODS
Plant Material, Propagation, and Growth Measurement
The pam68-1 mutant (GABI_152D07) is from the GABI-KAT collection (Rosso et al., 2003), and pam68-2 (SALK_044323) and at5g52780-1 (SALK_143426) are from the SALK T-DNA collection (http://signal.salk.edu/; Alonso et al., 2003). All three mutants have the Col-0 genetic background. The lpa1-2 mutant (CSHL_ET6851) originates from the Cold Spring Harbor Laboratory collection and carries a Dissociation element insertion in the Ler background (Martienssen, 1998).
Plants overexpressing PAM68 (oePAM68) were generated by introducing the PAM68 coding region (for primers, see Supplemental Table 2 online) into the Gateway plant expression vector pH2GW7 (Karimi et al., 2002) under the control of the 35S promoter of Cauliflower mosaic virus and then transforming flowers of pam68-2 mutant plants with the PAM68 overexpression construct as described (Clough and Bent, 1998). The plants were then transferred to the greenhouse, and seeds were collected after 3 weeks. Individual transgenic plants were selected on the basis of their resistance to hygromycin. The presence and expression of the transgene was confirmed by PCR, RNA gel blot, and protein immunoblot analyses.
Arabidopsis thaliana plants were grown on potting soil (Stender) under controlled greenhouse conditions (daylight supplemented with illumination from HQI Powerstar 400W/D, giving ~180 μmol photons m−2 s−1 on leaf surfaces, ~14/10 h light/dark cycle). Wuxal Super fertilizer (8% N, 8% P2O5, and 6% K2O; MANNA) was used according to the manufacturer's instructions. For in vivo translation assays, plants were grown on Murashige and Skoog medium (Duchefa) supplemented with 1% (w/v) sucrose. The screen for mutants exhibiting an altered photosynthetic performance was performed on 4-week-old, greenhouse-grown plants. Methods used for measurement of growth have been described previously (Leister et al., 1999).
Construction and Cultivation of Cyanobacterial Strains
Synechocystis sp PCC 6803 (referred to as the wild type) and mutant strains were grown under continuous irradiation (30 μmol of photons m−2 s−1) at 30°C on solid or in liquid BG 11 medium containing 5 mM glucose. To generate the sll0933 insertion mutant (ins0933), the Sll0933 coding sequence including its 5′- and 3′-flanking regions was amplified using the primers sll0933-5in (5′-TCTATCTGCTCCTCCTGAC-3′) and sll0933-3in (5′-TAAAGGTCTGACAGTAAATGC-3′) and subcloned into the pDrive vector (Qiagen). The fragment obtained after restriction with BamHI and XhoI was ligated into the Bluescript pKS vector, and the sll0933 gene was disrupted by insertion of a kanamycin resistance cassette of the plasmid pBSL15 into the single PstI site located 77 bp downstream of the ATG start codon. Wild-type cells were transformed with this construct as described (Eaton-Rye, 2004). Complete segregation of the ins0933 mutant was confirmed by PCR analysis using the primer pair sll0933-5kom (5′-AACATATGGCTGACCCCACCAATC-3′) and sll0933-3kom (5′-AAATCGATTCAACGATCGCCATTGTCC-3′).
Chlorophyll Fluorescence Analysis
Mutants that show alterations in ΦII, the effective quantum yield of PSII [ΦII= (Fm’ − F0’)/Fm’] were identified using an automatic pulse amplitude modulation fluorometer system (Varotto et al., 2000a, 2000b). In vivo chlorophyll a fluorescence of leaves was measured using the Pulse Amplitude Modulation 101/103 as described (Varotto et al., 2000b). Five plants of each genotype were analyzed, and average values and standard deviations were calculated. Plants were dark adapted for 30 min and minimal fluorescence (F0) was measured. Then, pulses (0.8 s) of white light (5000 μmol photons m−2 s−1) were used to determine the maximum fluorescence (Fm) and the ratio (Fm − F0)/Fm = Fv/Fm (maximum quantum yield of PSII) was calculated. A 10-min exposure to actinic light (80 μmol photons m−2 s−1) served to drive electron transport between PSII and PSI. Then, steady state fluorescence (Fs) was measured, and Fm’ was determined after exposure to further saturation pulses (0.8 s, 5000 μmol photons m−2 s−1). The effective quantum yield of PSII (ΦII) was calculated, and the photosynthetic parameters qP [photochemical quenching (Fm’ – Fs)/(Fm’ – F0)] and nonphotochemical quenching [(Fm − Fm’)/Fm’] were determined. In vivo chlorophyll a fluorescence of whole plants was recorded using an imaging chlorophyll fluorometer (Walz Imaging PAM) by exposing dark-adapted plants to a pulsed, blue measuring beam (1 Hz, intensity 4; F0) and a saturating light flash (intensity 4) to obtain Fv/Fm. The 77K fluorescence emission spectra of Synechocystis cells were measured after chlorophyll excitation at 435 nm as described previously (Klinkert et al., 2004).
Nucleic Acid Analysis
Arabidopsis DNA was isolated (Ihnatowicz et al., 2004), and T-DNA insertion-junction sites were recovered by amplification of insertion-mutagenized sites according to Frey et al. (1998). For RNA analysis, total leaf RNA was extracted from fresh tissue using the TRIzol reagent (Invitrogen). RT-PCR was performed by synthesizing first-strand cDNA using SuperScript reverse transcriptase (Invitrogen) and dT oligomers, followed by PCR with specific primers (see Supplemental Table 2 online). RNA gel blot analyses were performed under stringent conditions, according to standard protocols. Blots were stained with 0.04% methylene blue in 0.5 M sodium acetate, pH 5.2. Probes complementary to PAM68, ACTIN1, psbA, psbB, psbC, psbD, and psbE amplified from cDNA (see Supplemental Table 2 online), labeled with 32P, were used for the hybridizations. Signals were quantified using a phosphor imager (Typhoon; GE Healthcare) and the program IMAGE QUANT for Macintosh (version 1.2; Molecular Dynamics).
Polysomes were isolated as described (Barkan, 1988). Leaf tissue (200 mg) was frozen with liquid nitrogen in a mortar and ground with a pestle. Subsequently, the microsomal membranes were solubilized with 1% (v/v) Triton X-100 and 0.5% (w/v) sodium deoxycholate. The solubilized material was layered onto 15/55% sucrose step gradients (corresponding to 0.44/1.6 M) and centrifuged at 250,000g for 65 min at 4°C. The step gradient was fractionated and the mRNA associated with polysomes was then extracted with phenol/chloroform/isoamyl alcohol (25:24:1), followed by precipitation at room temperature with 95% ethanol. All samples were then subjected to RNA gel blot analysis.
Leaf Pigment Analysis
Pigments were analyzed by reverse-phase HPLC as described (Färber et al., 1997). For pigment extraction, leaf discs were frozen in liquid nitrogen and disrupted with beads in microcentrifuge tubes in the presence of acetone. After a short centrifugation, pigment extracts were filtered through a membrane filter (pore size 0.2 μm) and either used directly for HPLC analysis or stored for up to 2 d at −20°C.
BN- and 2D-PAGE
Leaves were harvested from plants at the 12-leaf rosette stage, and thylakoids were prepared as described (Bassi et al., 1985). For BN-PAGE, thylakoid samples equivalent to 100 mg of fresh leaf material were solubilized in 750 mM 6-aminocaproic acid, 5 mM EDTA, pH 7, and 50 mM NaCl in the presence of 1.0% (w/v) β-DM for 10 min at 4°C. Following centrifugation (15 min, 21,000g), the solubilized material was fractionated using nondenaturing BN-PAGE at 4°C as described (Schägger et al., 1988). For 2D-PAGE, samples were subsequently fractionated by electrophoresis on denaturing gradient Tricine-SDS gels (10 to 16% acrylamide) supplemented with 4 M urea (Schägger and von Jagow, 1987). Second-dimension gels were either stained with colloidal Coomassie Brilliant Blue (Candiano et al., 2004) or subjected to immunoblot analysis with antibodies against D1, D2, CP43, CP47, PsbE, and PsbO as described below.
For 2D BN/SDS-PAGE of Synechocystis membranes, Synechocystis cells were harvested at OD750 ~2.2, and membranes were isolated according to Dühring et al. (2006). 2D BN/SDS-PAGE was performed as described previously (Schottkowski et al., 2009a, 2009b).
Immunoblot Analyses
For 2D-PAGE analysis, proteins were prepared as described above. For SDS-PAGE analysis, total proteins were prepared from 4-week-old Arabidopsis leaves as reported (Martinez-Garcia et al., 1999), then fractionated on SDS-PAGE gradient gels (10 to 16% acrylamide) supplemented with 4 M urea (Schägger and von Jagow, 1987). Proteins were transferred to poly(vinylidene difluoride) membranes (Ihnatowicz et al., 2004), and replicate filters were incubated with antibodies specific for the PSII subunits D1 (obtained from Jürgen Soll, University of Munich), D2 (Agrisera), CP47 (obtained from Roberto Barbato, University of Alessandria), CP43 (Agrisera), PsbE (Agrisera), PsbI (Agrisera), PsbO (Agrisera), Cyt f (Agrisera), ATP synthase β-subunit and Lhcb1 (Agrisera), PsaB (PSI; Agrisera), LPA1 (obtained from Lixin Zhang, Chinese Academy of Sciences), CSP41a (obtained from David Stern, Boyce Thompson Institute for Plant Research, Cornell University), and actin (Dianova) as control. Signals were detected by enhanced chemiluminescence (GE Healthcare) and quantified using IMAGE QUANT for Macintosh (version 1.2; Molecular Dynamics).
In Vivo Translation Assay and Immunoprecipitation of D1
Radioactive labeling of thylakoid proteins was performed essentially as described (Pesaresi et al., 2006). In short, leaves of plants at the 12-leaf rosette stage, grown either in the greenhouse or in sterile culture, were vacuum-infiltrated in a syringe containing 20 μg/mL cycloheximide in 10 mL of 10 mM Tris, 5 mM MgCl2, 20 mM KCl, pH 6.8, and 0.1% (v/v) Tween 20 and incubated for 30 min to block cytosolic translation. Then leaves were again infiltrated with the same solution containing 1 mCi of [35S]Met, transferred into the light (5 to 20 μmol m–2 s–1), and collected after 5, 15, 20, 30, or 60 min. For a chase after pulse labeling, 10 mM of unlabeled Met was applied. Subsequently, thylakoid proteins were prepared. Immunoprecipitation of D1 was performed as described (Schult et al., 2007). Thylakoid proteins were either fractionated on denaturing gradient Tricine-SDS gels (10 to 16% acrylamide) supplemented with 4 M urea, or complexes were separated using 2D BN/SDS-PAGE as described. Signals were detected and quantified using a phosphor imager as described above.
Computational Analyses
Protein sequences were retrieved from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/), the Joint Genome Initiative (http://genome.jgi-psf.org), or the Cyanobase database (http://genome.kazusa.or.jp/cyanobase/). Putative chloroplast transit peptides were predicted by ChloroP (http://www.cbs.dtu.dk/services/ChloroP/; Emanuelsson et al., 1999) and transmembrane domains by the program TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/; Krogh et al., 2001). Amino acid sequences were aligned using the ClustalW program (www.ebi.ac.uk/clustalw/; Chenna et al., 2003), and alignments were shaded according to sequence similarity using the Boxshade server 3.21 (www.ch.embnet.org/software/BOX_form.html). Sequence identities and similarities were calculated using NCBI BLAST 2 sequences (Tatusova and Madden, 1999).
Expression analyses of target genes in response to different light qualities derive from the Genevestigator platform (www.genevestigator.com). These data originate from microarrays with The Arabidopsis Information Resource accession number 1007966126.
Generation of Antibodies
For immunolocalization of At5g52780, an antibody against the specific peptide sequence RTSEKPGRPD was generated in rabbits. For immunolocalization of Arabidopsis PAM68, several antibodies against specific peptide sequences (SKKPKPGNQSDE and DEDDDDEDEDD: acidic stretch), or the hydrophilic N terminus (amino acids 54 to 143) expressed in Escherichia coli, were generated in rabbits. Peptide synthesis, generation of peptide antibodies in rabbits, and purification of monospecific antibodies were all performed by Biogenes. All antibodies were used in a 1:100 dilution.
For production of antibodies specific for Sll0933, the sequence encoding the soluble N-terminal part of Sll0933 (amino acid positions 1 to 63) was overexpressed as a glutathione S-transferase fusion protein in E. coli cells and purified. Polyclonal αSll0933 antiserum was raised in rabbit (Biogenes) and used in a 1:625 dilution.
Determination of PAM68 Topology
Intact Arabidopsis chloroplasts were isolated and purified from leaves of 4- to 5-week-old plants as described (Aronsson and Jarvis, 2002). Intact chloroplasts were ruptured by mixing with 10 volumes of lysis buffer (20 mM HEPES/KOH, pH 7.5, and 10 mM EDTA) and incubated on ice for 30 min. To separate thylakoid and stroma phases, ruptured chloroplasts were centrifuged (42,000g, 30 min; 4°C).
For salt washes of thylakoids, according to Karnauchov et al. (1997), isolated thylakoids were resuspended in 50 mM HEPES/KOH, pH 7.5, at a chlorophyll concentration of 0.5 mg/mL. Extraction with 2 M NaCl, 0.1 M Na2CO3, 2 M NaSCN, or 0.1 M NaOH was performed for 30 min on ice, soluble and membrane proteins were separated by centrifugation for 10 min at 10,000g and 4°C, and immunoblot analysis was performed on both fractions using antibodies specific for PAM68, Lhcb1 (Agrisera), and PsaD (Agrisera).
For tryptic proteolysis experiments, thylakoid membranes were isolated as described above (omitting PMSF) and then resuspended in 50 mM HEPES/KOH, pH 8.0, and 300 mM sorbitol at a chlorophyll concentration of 1 mg/mL. Trypsin was added to a concentration of 10 μg/mL. Samples were taken 10 min later, and proteins were precipitated with 10 volumes of acetone and resuspended in SDS loading dye containing 5 mM of the Ser endopeptidase inhibitor PMSF.
Split-Ubiquitin Assay
In the split-ubiquitin assay, NubG and Cub are able to reconstitute ubiquitin only when brought into close proximity by two interacting test proteins that are expressed as fusion proteins with NubG and Cub. To test for interactions between PAM68 and other proteins, corresponding proteins or their fragments were fused to the C and N terminus of NubG and Cub, respectively. The coding sequences for the mature PAM68 protein and its respective fragments were cloned in the multiple cloning site of pAMBV4 (Dualsystems Biotech) and used as bait in interaction studies with prey proteins generated by cloning the coding sequences of mature thylakoid proteins (D1, D2, CP47, CP43, PsbI, PsbH, PsbE, PsbF, PsbO, LPA1, and LPA2; HCF136, Alb3, FtsY, SecY, PsaA, PsaB, Fd, AtpI, and FNR) into the multiple cloning site of pADSL (Dualsystems Biotech). Interaction studies were performed using the Dual-Membrane kit (Dualsystems Biotech) as described (Pasch et al., 2005). As negative controls, the plasmid pAlg5-NubG, which encodes the endoplasmic reticulum membrane protein Alg5 fused to NubG (Alg5NubG), and pADSL-Nx expressing soluble NubG were used for cotransformations. Because NubI (the wild-type Nub) and Cub spontaneously reassemble to reconstitute ubiquitin, Alg5 fused to NubI (Alg5NubI) was used as positive control. The specificity of the pADSL-Nx constructs was confirmed by cotransformation with a control vector encoding Alg5 fused to Cub (Alg5Cub). Yeast colonies were first plated on permissive medium (synthetic medium lacking Leu and Trp; -LT); the same colonies were later tested for their ability to grow on selective medium also lacking His (-LTH).
Sucrose Gradient Fractionation of Thylakoid Complexes
Thylakoids were washed twice with 5 mM EDTA, pH 7.8, and diluted in 20 mM Tricine/KOH, pH 7.5, to a chlorophyll concentration of 2 mg/mL. Solubilization of membrane complexes was performed by addition of an equal volume of 2% β-DM and incubation on ice for 10 min. Centrifugation at 16,000g for 5 min at 4°C removed nonsolubilized membranes. The supernatant was loaded onto a linear 0.1 to 1.0 M sucrose gradient in 20 mM Tricine/KOH, pH 7.5, and 0.06% (w/v) β-DM and centrifuged at 191,000g for 21 h at 4°C. The gradient was divided into 19 fractions (numbered from the top). Proteins were precipitated by extraction with methanol-chloroform (Wessel and Flügge, 1984) and separated on denaturing gradient Tricine-SDS gels (10 to 16% acrylamide) supplemented with 4 M urea. Immunoblot analyses were performed as described previously. The protein molecular mass standard contained proteins from 25 to 450 kD (Serva).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At4g19100 (Arabidopsis PAM68), At5g52780 (Arabidopsis homolog of PAM68), and the PAM68 homologs in Ricinus communis (GI:255557684), Vitis vinifera (GI:225466098), Populus trichocarpa (GI:224138363), Zea mays (GI:226491093), Oryza sativa (Os06g21530.1), Physcomitrella patens subsp patens (jgi|Phypa1_1|172321|estExt_fgenesh1_pg.C_3420021), Chlamydomonas reinhardtii (GI:159464826), Synechocystis sp PCC 6803 (Sll0933), and Synechococcus sp PCC 7002 (SYNPCC7002_A1824).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Characteristics of the lpa1-2 Mutant.
Supplemental Figure 2. Discrimination of Signals for D1 and D2 Proteins.
Supplemental Figure 3. Stability of the D1 Protein.
Supplemental Figure 4. Characterization of at5g52780-1 Mutants and the Double Mutant pam68-2 at5g52780-1.
Supplemental Figure 5. Synechocystis Sll0933: Generation and Characterization of Knockout Lines (ins0933) and Subcellular Localization and Topology.
Supplemental Figure 6. Overexpression of PAM68 Complements the pam68-2 Phenotype.
Supplemental Figure 7. Analysis of PAM68 Complex Formation.
Supplemental Table 1. Relative Transcript Levels of PSII Assembly Factors, as well as DEG1 and VAR2, under Different Light Conditions.
Supplemental Table 2. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Norio Murata and Yoshitaka Nishiyama for providing the pD1 antibody. Further antisera were obtained from Jürgen Soll, Elisabeth Ankele, and Monique Benz (against D1), Roberto Barbato (CP47), Lixin Zhang (LPA1), and David Stern (CSP41a). We thank Peter Nixon and Josef Komenda for discussions and suggestions, Paul Hardy for critical reading of the manuscript, and Christine Hümmer and Angela Dietzmann for technical support.
References
- Adir N., Zer H., Shochat S., Ohad I. (2003). Photoinhibition - A historical perspective. Photosynth. Res. 76: 343–370 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anbudurai P.R., Mor T.S., Ohad I., Shestakov S.V., Pakrasi H.B. (1994). The ctpA gene encodes the C-terminal processing protease for the D1 protein of the photosystem II reaction center complex. Proc. Natl. Acad. Sci. USA 91: 8082–8086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro E.M., Suorsa M., Rokka A., Allahverdiyeva Y., Paakkarinen V., Saleem A., Battchikova N., Rintamäki E. (2005). Dynamics of photosystem II: A proteomic approach to thylakoid protein complexes. J. Exp. Bot. 56: 347–356 [DOI] [PubMed] [Google Scholar]
- Aronsson H., Jarvis P. (2002). A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 529: 215–220 [DOI] [PubMed] [Google Scholar]
- Baena-González E., Aro E.M. (2002). Biogenesis, assembly and turnover of photosystem II units. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357: 1451–1459, discussion 1459–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1988). Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 7: 2637–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi R., dal Belin Peruffo A., Barbato R., Ghisi R. (1985). Differences in chlorophyll-protein complexes and composition of polypeptides between thylakoids from bundle sheaths and mesophyll cells in maize. Eur. J. Biochem. 146: 589–595 [DOI] [PubMed] [Google Scholar]
- Bellafiore S., Ferris P., Naver H., Göhre V., Rochaix J.D. (2002). Loss of Albino3 leads to the specific depletion of the light-harvesting system. Plant Cell 14: 2303–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Hokamp K., Wolfe K.H. (2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekema E.J., Hankamer B., Bald D., Kruip J., Nield J., Boonstra A.F., Barber J., Rögner M. (1995). Supramolecular structure of the photosystem II complex from green plants and cyanobacteria. Proc. Natl. Acad. Sci. USA 92: 175–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candiano G., Bruschi M., Musante L., Santucci L., Ghiggeri G.M., Carnemolla B., Orecchia P., Zardi L., Righetti P.G. (2004). Blue silver: A very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25: 1327–1333 [DOI] [PubMed] [Google Scholar]
- Chen H., Zhang D., Guo J., Wu H., Jin M., Lu Q., Lu C., Zhang L. (2006). A Psb27 homologue in Arabidopsis thaliana is required for efficient repair of photodamaged photosystem II. Plant Mol. Biol. 61: 567–575 [DOI] [PubMed] [Google Scholar]
- Chenna R., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D. (2003). Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y., Wollman F.A. (2002). Translational regulations as specific traits of chloroplast gene expression. FEBS Lett. 529: 39–42 [DOI] [PubMed] [Google Scholar]
- Choquet Y., Wostrikoff K., Rimbault B., Zito F., Girard-Bascou J., Drapier D., Wollman F.A. (2001). Assembly-controlled regulation of chloroplast gene translation. Biochem. Soc. Trans. 29: 421–426 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Diner B.A., Ries D.F., Cohen B.N., Metz J.G. (1988). COOH-terminal processing of polypeptide D1 of the photosystem II reaction center of Scenedesmus obliquus is necessary for the assembly of the oxygen-evolving complex. J. Biol. Chem. 263: 8972–8980 [PubMed] [Google Scholar]
- Dewez D., Park S., García-Cerdán J.G., Lindberg P., Melis A. (2009). Mechanism of REP27 protein action in the D1 protein turnover and photosystem II repair from photodamage. Plant Physiol. 151: 88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobáková M., Sobotka R., Tichý M., Komenda J. (2009). Psb28 protein is involved in the biogenesis of the photosystem II inner antenna CP47 (PsbB) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 149: 1076–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dühring U., Irrgang K.D., Lünser K., Kehr J., Wilde A. (2006). Analysis of photosynthetic complexes from a cyanobacterial ycf37 mutant. Biochim. Biophys. Acta 1757: 3–11 [DOI] [PubMed] [Google Scholar]
- Eaton-Rye J.J. (2004). The construction of gene knockouts in the cyanobacterium Synechocystis sp. PCC 6803. Methods Mol. Biol. 274: 309–324 [DOI] [PubMed] [Google Scholar]
- Edelman M., Mattoo A.K. (2008). D1-protein dynamics in photosystem II: The lingering enigma. Photosynth. Res. 98: 609–620 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., von Heijne G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enami I., Okumura A., Nagao R., Suzuki T., Iwai M., Shen J.R. (2008). Structures and functions of the extrinsic proteins of photosystem II from different species. Photosynth. Res. 98: 349–363 [DOI] [PubMed] [Google Scholar]
- Ermakova-Gerdes S., Vermaas W. (1999). Inactivation of the open reading frame slr0399 in Synechocystis sp. PCC 6803 functionally complements mutations near the Q(A) niche of photosystem II. A possible role of Slr0399 as a chaperone for quinone binding. J. Biol. Chem. 274: 30540–30549 [DOI] [PubMed] [Google Scholar]
- Färber A., Young A.J., Ruban A.V., Horton P., Jahns P. (1997). Dynamics of xanthophyll-cycle activity in different antenna subcomplexes in the photosynthetic membranes of higher plants. The relationship between zeaxanthin conversion and nonphotochemical fluorescence quenching. Plant Physiol. 115: 1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira K.N., Iverson T.M., Maghlaoui K., Barber J., Iwata S. (2004). Architecture of the photosynthetic oxygen-evolving center. Science 303: 1831–1838 [DOI] [PubMed] [Google Scholar]
- Frey M., Stettner C., Gierl A. (1998). A general method for gene isolation in tagging approaches: amplification of insertion mutagenised sites (AIMS). Plant J. 13: 717–721 [Google Scholar]
- Göhre V., Ossenbühl F., Crèvecoeur M., Eichacker L.A., Rochaix J.D. (2006). One of two alb3 proteins is essential for the assembly of the photosystems and for cell survival in Chlamydomonas. Plant Cell 18: 1454–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granvogl B., Reisinger V., Eichacker L.A. (2006). Mapping the proteome of thylakoid membranes by de novo sequencing of intermembrane peptide domains. Proteomics 6: 3681–3695 [DOI] [PubMed] [Google Scholar]
- Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. (2009). Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 16: 334–342 [DOI] [PubMed] [Google Scholar]
- Hankamer B., Nield J., Zheleva D., Boekema E., Jansson S., Barber J. (1997). Isolation and biochemical characterisation of monomeric and dimeric photosystem II complexes from spinach and their relevance to the organisation of photosystem II in vivo. Eur. J. Biochem. 243: 422–429 [DOI] [PubMed] [Google Scholar]
- Hatano-Iwasaki A., Minagawa J., Inoue Y., Takahashi Y. (2000). Characterization of chloroplast psbA transformants of Chlamydomonas reinhardtii with impaired processing of a precursor of a photosystem II reaction center protein, D1. Plant Mol. Biol. 42: 353–363 [DOI] [PubMed] [Google Scholar]
- Ihnatowicz A., Pesaresi P., Lohrig K., Wolters D., Müller B., Leister D. (2008). Impaired photosystem I oxidation induces STN7-dependent phosphorylation of the light-harvesting complex I protein Lhca4 in Arabidopsis thaliana. Planta 227: 717–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihnatowicz A., Pesaresi P., Varotto C., Richly E., Schneider A., Jahns P., Salamini F., Leister D. (2004). Mutants for photosystem I subunit D of Arabidopsis thaliana: Effects on photosynthesis, photosystem I stability and expression of nuclear genes for chloroplast functions. Plant J. 37: 839–852 [DOI] [PubMed] [Google Scholar]
- Iwata S., Barber J. (2004). Structure of photosystem II and molecular architecture of the oxygen-evolving centre. Curr. Opin. Struct. Biol. 14: 447–453 [DOI] [PubMed] [Google Scholar]
- Kapri-Pardes E., Naveh L., Adam Z. (2007). The thylakoid lumen protease Deg1 is involved in the repair of photosystem II from photoinhibition in Arabidopsis. Plant Cell 19: 1039–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., de Oliveira Manes C.L., Van Montagu M., Gheysen G. (2002). Activation of a pollenin promoter upon nematode infection. J. Nematol. 34: 75–79 [PMC free article] [PubMed] [Google Scholar]
- Karnauchov I., Herrmann R.G., Klösgen R.B. (1997). Transmembrane topology of the Rieske Fe/S protein of the cytochrome b6/f complex from spinach chloroplasts. FEBS Lett. 408: 206–210 [DOI] [PubMed] [Google Scholar]
- Kashino Y., Lauber W.M., Carroll J.A., Wang Q., Whitmarsh J., Satoh K., Pakrasi H.B. (2002). Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 41: 8004–8012 [DOI] [PubMed] [Google Scholar]
- Kato Y., Sakamoto W. (2009). Protein quality control in chloroplasts: A current model of D1 protein degradation in the photosystem II repair cycle. J. Biochem. 146: 463–469 [DOI] [PubMed] [Google Scholar]
- Keren N., Ohkawa H., Welsh E.A., Liberton M., Pakrasi H.B. (2005). Psb29, a conserved 22-kD protein, functions in the biogenesis of photosystem II complexes in Synechocystis and Arabidopsis. Plant Cell 17: 2768–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkert B., Ossenbühl F., Sikorski M., Berry S., Eichacker L., Nickelsen J. (2004). PratA, a periplasmic tetratricopeptide repeat protein involved in biogenesis of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 279: 44639–44644 [DOI] [PubMed] [Google Scholar]
- Kogata N., Nishio K., Hirohashi T., Kikuchi S., Nakai M. (1999). Involvement of a chloroplast homologue of the signal recognition particle receptor protein, FtsY, in protein targeting to thylakoids. FEBS Lett. 447: 329–333 [DOI] [PubMed] [Google Scholar]
- Komenda J., Nickelsen J., Tichý M., Prásil O., Eichacker L.A., Nixon P.J. (2008). The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 283: 22390–22399 [DOI] [PubMed] [Google Scholar]
- Komenda J., Tichý M., Eichacker L.A. (2005). The PsbH protein is associated with the inner antenna CP47 and facilitates D1 processing and incorporation into PSII in the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol. 46: 1477–1483 [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Kufryk G.I., Vermaas W.F. (2001). A novel protein involved in the functional assembly of the oxygen-evolving complex of photosystem II in Synechocystis sp. PCC 6803. Biochemistry 40: 9247–9255 [DOI] [PubMed] [Google Scholar]
- Kufryk G.I., Vermaas W.F. (2003). Slr2013 is a novel protein regulating functional assembly of photosystem II in Synechocystis sp. strain PCC 6803. J. Bacteriol. 185: 6615–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidler V., Chaddock A.M., Knott T.G., Walker D., Robinson C. (1995). A SecY homolog in Arabidopsis thaliana. Sequence of a full-length cDNA clone and import of the precursor protein into chloroplasts. J. Biol. Chem. 270: 17664–17667 [DOI] [PubMed] [Google Scholar]
- Leister D., Varotto C., Pesaresi P., Niwergall A., Salamini F. (1999). Large-scale evaluation of plant growth in Arabidopsis thaliana by non-invasive image analysis. Plant Physiol. Biochem. 37: 671–678 [Google Scholar]
- Ma J., Peng L., Guo J., Lu Q., Lu C., Zhang L. (2007). LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 19: 1980–1993 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Martienssen R.A. (1998). Functional genomics: Probing plant gene function and expression with transposons. Proc. Natl. Acad. Sci. USA 95: 2021–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García J.F., Monte E., Quail P.H. (1999). A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 20: 251–257 [DOI] [PubMed] [Google Scholar]
- Meurer J., Plücken H., Kowallik K.V., Westhoff P. (1998). A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 17: 5286–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa J., Takahashi Y. (2004). Structure, function and assembly of photosystem II and its light-harvesting proteins. Photosynth. Res. 82: 241–263 [DOI] [PubMed] [Google Scholar]
- Mulo P., Sirpiö S., Suorsa M., Aro E.M. (2008). Auxiliary proteins involved in the assembly and sustenance of photosystem II. Photosynth. Res. 98: 489–501 [DOI] [PubMed] [Google Scholar]
- Nelson N., Yocum C.F. (2006). Structure and function of photosystems I and II. Annu. Rev. Plant Biol. 57: 521–565 [DOI] [PubMed] [Google Scholar]
- Nixon P.J., Barker M., Boehm M., de Vries R., Komenda J. (2005). FtsH-mediated repair of the photosystem II complex in response to light stress. J. Exp. Bot. 56: 357–363 [DOI] [PubMed] [Google Scholar]
- Nixon P.J., Michoux F., Yu J., Boehm M., Komenda J. (2010). Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. (Lond.) 106: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon P.J., Trost J.T., Diner B.A. (1992). Role of the carboxy terminus of polypeptide D1 in the assembly of a functional water-oxidizing manganese cluster in photosystem II of the cyanobacterium Synechocystis sp. PCC 6803: Assembly requires a free carboxyl group at C-terminal position 344. Biochemistry 31: 10859–10871 [DOI] [PubMed] [Google Scholar]
- Nowaczyk M.M., Hebeler R., Schlodder E., Meyer H.E., Warscheid B., Rögner M. (2006). Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of photosystem II. Plant Cell 18: 3121–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossenbühl F., Göhre V., Meurer J., Krieger-Liszkay A., Rochaix J.D., Eichacker L.A. (2004). Efficient assembly of photosystem II in Chlamydomonas reinhardtii requires Alb3.1p, a homolog of Arabidopsis ALBINO3. Plant Cell 16: 1790–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossenbühl F., Inaba-Sulpice M., Meurer J., Soll J., Eichacker L.A. (2006). The synechocystis sp PCC 6803 oxa1 homolog is essential for membrane integration of reaction center precursor protein pD1. Plant Cell 18: 2236–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Khamai P., Garcia-Cerdan J.G., Melis A. (2007). REP27, a tetratricopeptide repeat nuclear-encoded and chloroplast-localized protein, functions in D1/32-kD reaction center protein turnover and photosystem II repair from photodamage. Plant Physiol. 143: 1547–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasch J.C., Nickelsen J., Schünemann D. (2005). The yeast split-ubiquitin system to study chloroplast membrane protein interactions. Appl. Microbiol. Biotechnol. 69: 440–447 [DOI] [PubMed] [Google Scholar]
- Peng L., Ma J., Chi W., Guo J., Zhu S., Lu Q., Lu C., Zhang L. (2006). LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Shimizu H., Shikanai T. (2008). The chloroplast NAD(P)H dehydrogenase complex interacts with photosystem I in Arabidopsis. J. Biol. Chem. 283: 34873–34879 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pesaresi P., Hertle A., Pribil M., Kleine T., Wagner R., Strissel H., Ihnatowicz A., Bonardi V., Scharfenberg M., Schneider A., Pfannschmidt T., Leister D. (2009). Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. Plant Cell 21: 2402–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P., Lunde C., Jahns P., Tarantino D., Meurer J., Varotto C., Hirtz R.D., Soave C., Scheller H.V., Salamini F., Leister D. (2002). A stable LHCII-PSI aggregate and suppression of photosynthetic state transitions in the psae1-1 mutant of Arabidopsis thaliana. Planta 215: 940–948 [DOI] [PubMed] [Google Scholar]
- Pesaresi P., Masiero S., Eubel H., Braun H.P., Bhushan S., Glaser E., Salamini F., Leister D. (2006). Nuclear photosynthetic gene expression is synergistically modulated by rates of protein synthesis in chloroplasts and mitochondria. Plant Cell 18: 970–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plücken H., Müller B., Grohmann D., Westhoff P., Eichacker L.A. (2002). The HCF136 protein is essential for assembly of the photosystem II reaction center in Arabidopsis thaliana. FEBS Lett. 532: 85–90 [DOI] [PubMed] [Google Scholar]
- Rokka A., Suorsa M., Saleem A., Battchikova N., Aro E.M. (2005). Synthesis and assembly of thylakoid protein complexes: multiple assembly steps of photosystem II. Biochem. J. 388: 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose J.L., Pakrasi H.B. (2004). Evidence that D1 processing is required for manganese binding and extrinsic protein assembly into photosystem II. J. Biol. Chem. 279: 45417–45422 [DOI] [PubMed] [Google Scholar]
- Roose J.L., Wegener K.M., Pakrasi H.B. (2007). The extrinsic proteins of photosystem II. Photosynth. Res. 92: 369–387 [DOI] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Roy L.M., Barkan A. (1998). A SecY homologue is required for the elaboration of the chloroplast thylakoid membrane and for normal chloroplast gene expression. J. Cell Biol. 141: 385–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., Aquila H., Von Jagow G. (1988). Coomassie blue-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for direct visualization of polypeptides during electrophoresis. Anal. Biochem. 173: 201–205 [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166: 368–379 [DOI] [PubMed] [Google Scholar]
- Schottkowski M., Gkalympoudis S., Tzekova N., Stelljes C., Schünemann D., Ankele E., Nickelsen J. (2009a). Interaction of the periplasmic PratA factor and the PsbA (D1) protein during biogenesis of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 284: 1813–1819 [DOI] [PubMed] [Google Scholar]
- Schottkowski M., Ratke J., Oster U., Nowaczyk M., Nickelsen J. (2009b). Pitt, a novel tetratricopeptide repeat protein involved in light-dependent chlorophyll biosynthesis and thylakoid membrane biogenesis in Synechocystis sp. PCC 6803. Mol. Plant 2: 1289–1297 [DOI] [PubMed] [Google Scholar]
- Schult K., Meierhoff K., Paradies S., Töller T., Wolff P., Westhoff P. (2007). The nuclear-encoded factor HCF173 is involved in the initiation of translation of the psbA mRNA in Arabidopsis thaliana. Plant Cell 19: 1329–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenkert S., Umate P., Dal Bosco C., Volz S., Mlçochová L., Zoryan M., Eichacker L.A., Ohad I., Herrmann R.G., Meurer J. (2006). PsbI affects the stability, function, and phosphorylation patterns of photosystem II assemblies in tobacco. J. Biol. Chem. 281: 34227–34238 [DOI] [PubMed] [Google Scholar]
- Shestakov S.V., Anbudurai P.R., Stanbekova G.E., Gadzhiev A., Lind L.K., Pakrasi H.B. (1994). Molecular cloning and characterization of the ctpA gene encoding a carboxyl-terminal processing protease. Analysis of a spontaneous photosystem II-deficient mutant strain of the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 269: 19354–19359 [PubMed] [Google Scholar]
- Shi L.X., Schröder W.P. (2004). The low molecular mass subunits of the photosynthetic supracomplex, photosystem II. Biochim. Biophys. Acta 1608: 75–96 [DOI] [PubMed] [Google Scholar]
- Sirpiö S., Khrouchtchova A., Allahverdiyeva Y., Hansson M., Fristedt R., Vener A.V., Scheller H.V., Jensen P.E., Haldrup A., Aro E.M. (2008). AtCYP38 ensures early biogenesis, correct assembly and sustenance of photosystem II. Plant J. 55: 639–651 [DOI] [PubMed] [Google Scholar]
- Spence E., Bailey S., Nenninger A., Møller S.G., Robinson C. (2004). A homolog of Albino3/OxaI is essential for thylakoid biogenesis in the cyanobacterium Synechocystis sp. PCC6803. J. Biol. Chem. 279: 55792–55800 [DOI] [PubMed] [Google Scholar]
- Sun X., Ouyang M., Guo J., Ma J., Lu C., Adam Z., Zhang L. (2010). The thylakoid protease Deg1 is involved in photosystem-II assembly in Arabidopsis thaliana. Plant J. 62: 240–249 [DOI] [PubMed] [Google Scholar]
- Sundberg E., Slagter J.G., Fridborg I., Cleary S.P., Robinson C., Coupland G. (1997). ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell 9: 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova T.A., Madden T.L. (1999). BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174: 247–250 [DOI] [PubMed] [Google Scholar]
- Taylor M.A., Packer J.C.L., Bowyer J.R. (1988). Processing of the D1 polypeptide of the photosystem II reaction centre and photoactivation of a low fluorescence mutant (LF-1) of Scenedesmus obliquus. FEBS Lett. 237: 229–233 [Google Scholar]
- van Wijk K.J., Andersson B., Aro E.M. (1996). Kinetic resolution of the incorporation of the D1 protein into photosystem II and localization of assembly intermediates in thylakoid membranes of spinach chloroplasts. J. Biol. Chem. 271: 9627–9636 [DOI] [PubMed] [Google Scholar]
- van Wijk K.J., Bingsmark S., Aro E.M., Andersson B. (1995). In vitro synthesis and assembly of photosystem II core proteins. The D1 protein can be incorporated into photosystem II in isolated chloroplasts and thylakoids. J. Biol. Chem. 270: 25685–25695 [DOI] [PubMed] [Google Scholar]
- van Wijk K.J., Roobol-Boza M., Kettunen R., Andersson B., Aro E.M. (1997). Synthesis and assembly of the D1 protein into photosystem II: Processing of the C-terminus and identification of the initial assembly partners and complexes during photosystem II repair. Biochemistry 36: 6178–6186 [DOI] [PubMed] [Google Scholar]
- Varotto C., Pesaresi P., Maiwald D., Kurth J., Salamini F., Leister D. (2000a). Identification of photosynthetic mutants of Arabidopsis by automatic screening for altered effective quantum yield of photosystem 2. Photosynthetica 38: 497–504 [Google Scholar]
- Varotto C., Pesaresi P., Meurer J., Oelmüller R., Steiner-Lange S., Salamini F., Leister D. (2000b). Disruption of the Arabidopsis photosystem I gene psaE1 affects photosynthesis and impairs growth. Plant J. 22: 115–124 [DOI] [PubMed] [Google Scholar]
- Wei L., Guo J., Ouyang M., Sun X., Ma J., Chi W., Lu C., Zhang L. (2010). LPA19, a Psb27 homolog in Arabidopsis thaliana, facilitates D1 protein precursor processing during PSII biogenesis. J. Biol. Chem. 285: 21391–21398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D., Flügge U.I. (1984). A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138: 141–143 [DOI] [PubMed] [Google Scholar]
- Wollman F.A., Minai L., Nechushtai R. (1999). The biogenesis and assembly of photosynthetic proteins in thylakoid membranes1. Biochim. Biophys. Acta 1411: 21–85 [DOI] [PubMed] [Google Scholar]
- Zaltsman A., Feder A., Adam Z. (2005). Developmental and light effects on the accumulation of FtsH protease in Arabidopsis chloroplasts—Implications for thylakoid formation and photosystem II maintenance. Plant J. 42: 609–617 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.