Cassava is a very important staple crop, especially in the arid tropics where it is a chief source of carbohydrates, but the provitamin A content in the storage root is insufficient to sustain a healthy life. The work presented shows that a single amino acid exchange in a conserved region of the enzyme phytoene synthase leads to substantive accumulation of β-carotene (provitamin A) in the roots.
Abstract
Cassava (Manihot esculenta) is an important staple crop, especially in the arid tropics. Because roots of commercial cassava cultivars contain a limited amount of provitamin A carotenoids, both conventional breeding and genetic modification are being applied to increase their production and accumulation to fight vitamin A deficiency disorders. We show here that an allelic polymorphism in one of the two expressed phytoene synthase (PSY) genes is capable of enhancing the flux of carbon through carotenogenesis, thus leading to the accumulation of colored provitamin A carotenoids in storage roots. A single nucleotide polymorphism present only in yellow-rooted cultivars cosegregates with colored roots in a breeding pedigree. The resulting amino acid exchange in a highly conserved region of PSY provides increased catalytic activity in vitro and is able to increase carotenoid production in recombinant yeast and Escherichia coli cells. Consequently, cassava plants overexpressing a PSY transgene produce yellow-fleshed, high-carotenoid roots. This newly characterized PSY allele provides means to improve cassava provitamin A content in cassava roots through both breeding and genetic modification.
INTRODUCTION
In plants, carotenoids perform essential physiological functions in photosynthesis and as precursors of regulatory molecules, such as abscisic acid (Schwartz et al., 1997) and strigolactones (Bouwmeester et al., 2007; Gomez-Roldan et al., 2008; Umehara et al., 2008). Their formation is initiated by reactions catalyzed by carotenoid cleavage dioxygenases (CCDs; for review, see Bouvier et al., 2005; Moise et al., 2005; Auldridge et al., 2006). In animals, including humans, provitamin A carotenoids containing at least one unsubstituted β-ionone ring serve as precursors of retinoids, synthesized through cleavage at the central C15-C15′ double bond, which is catalyzed by the β-carotene cleavage monoxygenase 1 (reviewed in Moise et al., 2005).
Because of the widespread prevalence of vitamin A deficiency diseases in the tropics (WHO, 2009), efforts are underway to increase the provitamin A content of edible plant tissues, an approach commonly referred to as biofortification (for review, see Mayer et al., 2008). This can be achieved through breeding, if sufficient genetic variability for the trait is present, but requires genetic modification in its absence. Golden Rice, for example, was the logical consequence of the absence of carotenoid production in the endosperm of the rice (Oryza sativa) grain (Ye et al., 2000; Paine et al., 2005). A further barrier to breeding is sterility, such as in commercial banana (Musa spp) cultivars, or where breeding is complicated, as for some vegetatively propagated crops, such as potato (Solanum tuberosum). Cultivated potato suffers from serious inbreeding depression upon self-pollination and represents a highly heterozygous tetraploid. Consequently, genetic transformation was employed to increase provitamin A content in potato tubers (Diretto et al., 2007). Provitamin A carotenoids in cassava (Manihot esculenta) have been increased threefold through conventional breeding; however, the heterozygous nature of the crop renders varietal recovery difficult, and long breeding cycles slow down the progress of this endeavor (Rojas et al., 2009).
Cassava is quoted as the fifth most important staple crop worldwide but ranks first in many arid regions, such as in sub-Saharan Africa. Like with other staples, the cassava storage root is rich in starch but poor in protein and micronutrients like iron, zinc, and provitamin A. Yellow-rooted cultivars producing provitamin A carotenoids are rare, although some yellow landraces have been identified in Amazonia (Ferreira et al., 2008; Nassar et al., 2009); most breeding populations are white rooted. Because of the difficult and unwieldy breeding of cassava, genetic modification is being considered as a viable option but was hindered in the past by less-well-developed basic knowledge of the crop compared with other staples, such as maize (Zea mays), wheat (Triticum aestivum), and rice. It is only during recent years that research has been done to identify storage root-specific promoters (Zhang et al., 2003; Arango et al., 2010a; Beltrán et al., 2010) and that genomic sequence information has become available (http://www.phytozome.net/cassava).
It is desirable to gain insight into the carotenoid biosynthetic pathway of a given plant tissue to be able to design cDNA constructs capable of overcoming rate limitations of the biosynthetic pathway. This has become possible thanks to the almost complete molecular elucidation of the pathway (for review, see DellaPenna and Pogson, 2006).
In plant tissues exhibiting low-level carotenoid production, the limiting factor is commonly the expression level of phytoene synthase (PSY), which catalyzes the first carotenoid-specific reaction of prenyl lipid metabolism in the plastid (Figure 1). This has been shown to be the case in canola (Brassica napus; Shewmaker et al., 1999) and Arabidopsis thaliana seeds (Lindgren et al., 2003). PSY expression can be driven to levels at which β-carotene crystals are formed, as in Arabidopsis cells and in carrot (Daucus carota) roots (Maass et al., 2009). However, there are cases where additional transgenes are needed to raise carotenoid production levels, such as in Golden Rice, in which the carotene desaturase CrtI had to be introduced along with PSY (Burkhardt et al., 1997; Ye et al., 2000; Schaub et al., 2005) or potato, in which a mini pathway of three carotenoid genes was necessary to achieve high levels of production (Diretto et al., 2007). In the light of such variable requirements, a combinatorial transgenic approach has been proposed and shown to function with maize (Zhu et al., 2008).
Figure 1.
The Carotenoid Biosynthetic Pathway in Plants.
Note that PSY is central, catalyzing the first committed biosynthetic step in carotenogenesis. IPP, isopentenyl-diphosphate; DMAPP, dimethylallyl-diphosphate; GGPP, geranylgeranyl-diphosphate; GGPS, geranylgeranyl-diphosphate synthase; PDS, phytoene desaturase; ZISO, ζ-carotene isomerase; ZDS, ζ-carotene desaturase; CRTISO, carotene isomerase; LCY, lycopene β and ε-cyclase. The dashed arrow represents multiple enzymatic steps.
To understand the biosynthetic pathway in cassava, we undertook comparative research using carotenoid-accumulating and white-rooted cultivars. Previous research has shown the presence of three PSY genes (PSY1, PSY2, and PSY3) out of which only two (PSY1 and PSY2) were found to be expressed under standard growth conditions as well as under drought and salt stress. Quantitative expression analysis showed that PSY2 expression in roots is ~10 times higher than that of PSY1 (Arango et al., 2010b).
We describe here allelic variation in PSY leading to enhanced enzymatic activity. A single nucleotide polymorphism in PSY2, causing a nonconservative amino acid exchange, leads to markedly increased carotenoid formation and accumulation in cassava storage roots.
RESULTS
No Correlation between Root Color and Expression of Major Carotenoid Biosynthesis Genes
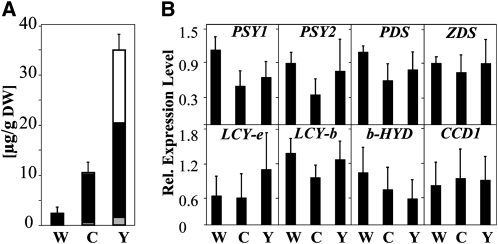
To determine the rate-limiting step in the carotenoid biosynthetic pathway in storage roots, the expression levels of major carotenoid biosynthesis genes was determined for cultivars differing in carotenoid content. The cultivars analyzed were CM 3306-4 with low, CM 2772-3 with intermediate, and MBRA 253 with high root carotenoid content (from here on referred to as white [W], cream [C], and yellow [Y], respectively). The real-time RT-PCR study included the two expressed cassava PSY genes (Arango et al., 2010b), phytoene desaturase (PDS), ζ-carotene desaturase (ZDS), lycopene β- and ε-cyclase (LCY-b and LCY-e), and β-carotene hydroxylase (b-HYD). CCD1 was included as a representative of an unspecific carotenoid degrading enzyme. As shown in Figure 2, the pronounced differences in carotenoid content in the roots of these cultivars (see Supplemental Table 1 online) were not reflected in the expression levels of these genes. Consequently, the rate limitation in W and C must stem from differential expression of pathway genes not considered in the analysis or be based on entirely different principles.
Figure 2.
Carotenoid Content of Three Different Cassava Landraces and Expression Levels of Carotenoid Biosynthetic Genes.
(A) Carotenoid content in 9-month-old field-grown roots of W (cv CM 3306-4), C (cv CM 2772-3), and Y (cv MBRA 253). White, xanthophylls; black, β-carotene; gray, phytoene.
(B) RNA expression levels of carotenoid biosynthetic genes in the roots analyzed in (A). Transcript levels were determined by real-time RT-PCR, normalized to 18S rRNA levels, and are expressed relative to the expression level of one sample from W.
Error bars in (A) and (B) represent the se of three biological replicates.
An Allelic Variant of One Cassava PSY Gene Defines Cassava Root Color
Sequencing of the two cassava full-length PSY cDNAs (PSY1 and PSY2) revealed the occurrence of single nucleotide polymorphisms (SNPs) in PSY2 (see Supplemental Figure 1 online). Two of these, SNP1 and SNP2, were present in the Y cultivar, defining the alleles PSY2-Y-1 and PSY2-Y-2 (Figure 3), while the C cultivar contained only SNP2. SNP3 was restricted to W and was homonymous and therefore not considered further. By contrast, PSY2-Y-1 carried a G521C nucleotide substitution equivalent to an R174T amino acid exchange, and PSY2-Y-2 carried a C572A nucleotide substitution leading to an A191D amino acid exchange. The authenticity of these sequence differences was verified by RT-PCR amplification and sequencing of the individual cDNA variants. Because of the positive color/SNP correlation, we hypothesized that the SNPs discovered were crucial for root color formation.
Figure 3.
Variants of PSY2 in Cassava Cultivars Differing in their Root Carotenoid Levels.
(A) Occurrence of SNPs identified in PSY2 in white- and yellow-rooted cultivars, defining the PSY alleles W-1, W-2, Y-1, and Y-2. SNP3 (asterisk) has no effect on the amino acid sequence.
(B) Sequencing chromatograms of the two nonsynonymous SNPs of PSY2 in cultivars W and Y (see Supplemental Figure 1 online for full sequence information). Root transverse sections of the corresponding cultivars are shown on the left side.
(C) Amino acid alignment of PSYs, including PSY2 from the white-colored (Me2-W) and the nonsynonymous allelic variants present in the yellow-colored cassava landrace (Me2-Y-1 and Me2-Y-2). The relevant A191D amino acid exchange is given in red. The R174T exchange (red) has no contribution to increased root carotenoid formation. The position of the A/D exchange introduced into three additional PSYs by site-directed mutagenesis is shown in blue. Me, Manihot esculenta; Os, Oryza sativa; Sl, Solanum lycopersicum; Np, Narcissus pseudonarcissus; At, Arabidopsis thaliana; Zm, Zea mays. Identical amino acids are shown with a gray background.
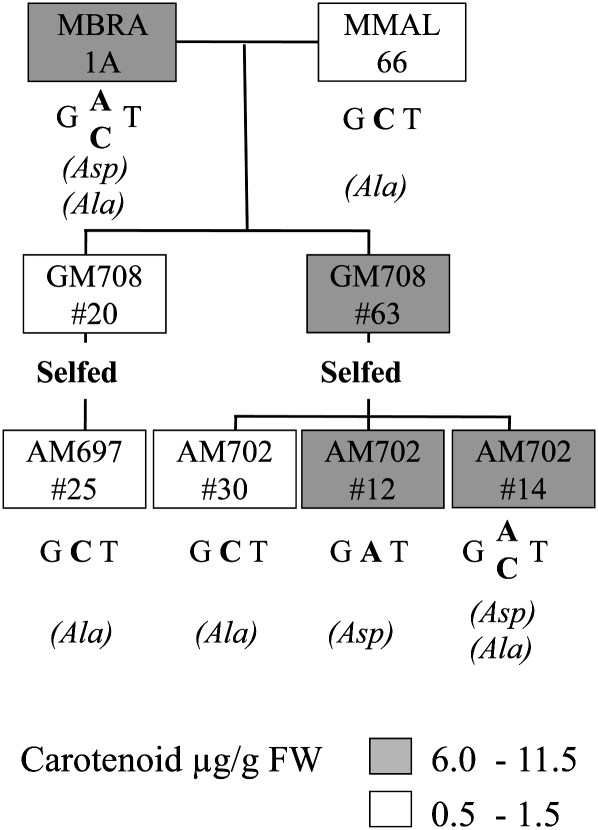
To investigate further, we took advantage of a breeding population selected for their wide variation in provitamin A root phenotypes (Figure 4). The population was based on an initial cross between cultivars other than those mentioned above, namely, the white-rooted cultivar MMAL 66 and the yellow-rooted MBRA 1A. SNP1 was not detected in either parent. The R174T exchange was therefore ruled out as a cause for the color phenotype. By contrast, SNP2 (resulting in A191D) was present in the yellow-rooted parent and absent from the white. Self-pollination of white F1 (GM 708-20) and yellow F1 (GM 708-63) genotypes resulted in contrasting S1 families. A total of 37 S1 individuals made up family AM 697 (from the W genotype GM 708-20) and all had white roots. Only two of these genotypes were therefore analyzed, showing carotenoid levels below 1 μg g−1 (see Supplemental Table 2 online). By contrast, there was a wide segregation among the S1 individuals of family AM 702, derived from the yellow genotype GM 708-63 (Figure 4). Carotenoid analysis and DNA sequencing revealed that plants producing white storage roots containing 0.6 to 1.5 μg carotenoids per g fresh weight (~15% phytoene) were homozygous for C572 (coding for Ala-191). Ala-572 (coding for Asp-191), on the other hand, segregated with yellow root color (6.0 to 11.5 μg g−1 fresh weight). Homozygosity for SNP2 was not required for the yellow storage root phenotype, hinting at dominance of this allele.
Figure 4.
SNP2 (D191) Cosegregates with Colored Roots in a Cassava Pedigree.
Cultivars MBRA1A (yellow-rooted) and MMAL66 (white-rooted) were crossed using MMAL66 as the male parent. Siblings of this cross were self-pollinated, and the resulting offspring grown in the field. Randomly selected white and colored roots were harvested after 9 months of growth and analyzed for carotenoid content and composition. Leaf DNA was used for SNP2 identification through sequencing of the respective PCR-amplified PSY region. The presence of Asp at position 191, replacing the conserved Ala, was decisive for carotenoid accumulation (see Supplemental Table 2 online).
The A191D Exchange in PSY Results in Increased Enzymatic Activity
The open reading frames (ORFs) coding for PSYA191 or PSYD191 were independently coexpressed in yeast cells supplemented with geranylgeranyl-diphosphate synthase (GGPS) from white mustard (Sinapis alba) to produce the PSY substrate geranylgeranyl-diphosphate (GGPP). HPLC quantification revealed that PSYD191 (present in yellow-rooted cassava) produced approximately twice as much phytoene as the PSYA191 version of white-rooted cultivars, indicating increased enzymatic activity (Figure 5A). To provide further evidence, an Escherichia coli–based system expressing GGPS and the bacterial carotene desaturase CrtI was used, and the conversion of phytoene into red-colored lycopene was measured photometrically. These experiments were complemented by coexpressing the PSY genes from daffodil (Narcissus pseudonarcissus; Np-PSY), Arabidopsis (At-PSY), and maize (Zm-PSY1), all containing the A-to-D exchange mutation in the conserved position (Figure 3C). As shown in Figure 5B, Np-PSY and At-PSY, known to be less active than Zm-PSY1 (Paine et al., 2005), responded to the amino acid exchange by an approximate doubling of lycopene production at comparable protein levels, similar to the equivalent cassava PSY versions tested in yeast. The coexpression of the mutagenized Zm-PSY1 resulted in ~20% higher lycopene production.
Figure 5.
Production of Phytoene by PSY Variants in Yeast and E. coli Cells.
(A) Cassava PSY2D191 from the Y cultivar and PSY2A191 from the W cultivar were individually expressed in yeast cells coexpressing the GGPS cDNA from S. alba. The phytoene formed was quantified by HPLC. Cell lysate (30 μg protein) was used to estimate PSY levels using an anti-At-PSY antibody. The levels of HA-tagged GGPS were determined using anti-HA antibodies (loading control).
(B) Production of lycopene in recombinant E. coli cells. Site-directed mutagenesis was performed to exchange the wild type A for D (as given in Figure 3C) with cDNAs from N. pseudonarcissus (Np-PSY), Arabidopsis (At-PSY), and Z. mays (Zm-PSY1). Wild-type and mutated PSY versions were expressed separately in an E. coli strain expressing GGPS and the bacterial phytoene desaturase (CrtI), thus capable of producing lycopene.
Error bars in (A) and (B) represent the se of three biological replicates. Immunoblots were conducted using anti-6*His-Tag antibodies and cell lysate corresponding to 1.5 μg protein per lane.
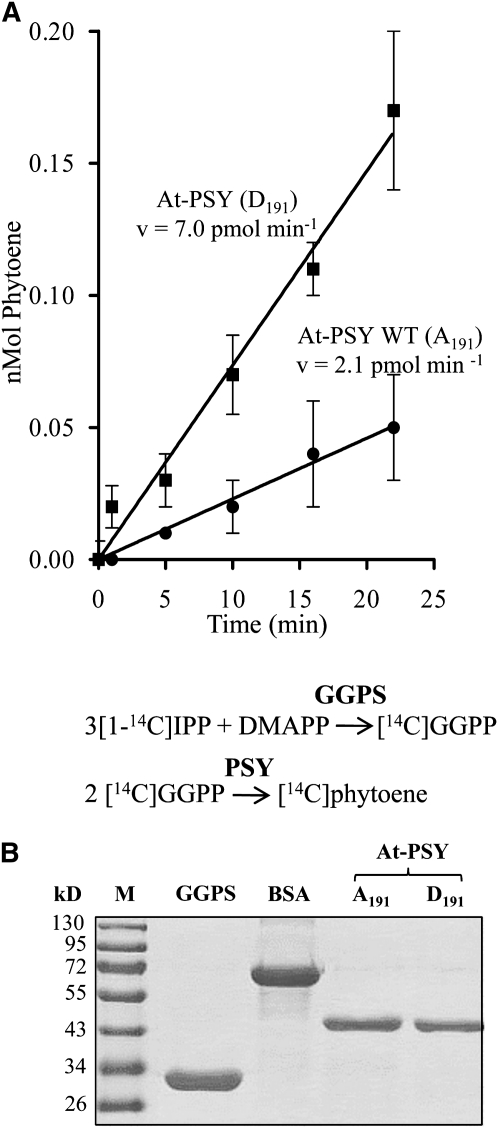
To confirm that increased enzymatic PSY activity is the underlying cause of the yellow phenotype, we used the wild type and the A-to-D mutagenized At-PSY in vitro (Figure 3C). The system consisted of purified N-His6-GGPS (Kloer et al., 2006) delivering the PSY substrate GGPP and the N-His6-fused PSY versions in a 2:1 GGPS/PSY molar ratio in a biphasic (liposome-containing) assay (Figure 6). The reaction was started with [1-14C]isopentenyl-diphosphate (IPP) and dimethylallyl-diphosphate. Phytoene was quantified by liquid scintillation counting. Corroborating the previous results, the mutagenized PSY version led to approximately threefold higher specific activities.
Figure 6.
In Vitro Assays Conducted with Purified GGPS and PSY.
(A) Purified recombinant GGPS from S. alba (5 μg mL−1) was combined separately with 3.3 μg mL−1 purified wild type (Ala-191) and mutagenized Arabidopsis PSY (At-PSYD191; see Figure 3C for positions) in a molar ratio of 2:1 GGPS/PSY. This showed the higher effectiveness of the system employing the PSYD191 version as demonstrated by a tripling of the reaction velocity at identical protein concentrations. Data represent the mean ± se of phytoene productions from each of three independent GGPS and PSY purifications.
(B) SDS-PAGE of GGPS and At-PSY versions used in the enzymatic assays. GGPS was purified to homogeneity as published (Kloer et al., 2006) and the A191D-mutagenized At-PSY version as given in Methods. Stain, Coomassie blue; M, molecular mass marker proteins.
PSY Is Rate Limiting in the Carotenoid Biosynthetic Pathway of Cassava Roots
Given that improved enzymatic activity of PSY can increase the flux through the carotenoid pathway in cassava roots, we hypothesized that the root-specific overexpression of any PSY should result in the formation of colored carotenoids by increasing phytoene synthesizing capacity. To test our hypothesis, we constructed the binary vector pCAS-Phyt harboring an RbcS transit peptide-bacterial PSY gene (TP-CrtB) under the control of the cassava CP1 promoter (Beltrán et al., 2010). Agrobacterium tumefaciens–mediated transformation was used with the white-rooted cassava cultivar TMS 60444. Transgenics were confirmed by DNA gel blot analysis (see Supplemental Figure 2 online). After 6 months of growth, carotenoid analysis was performed on roots using quantitative HPLC (Table 1). This showed that all transgenic events had a higher carotenoid content over the wild-type control. Among these, event 12 roots had the deepest orange color (Figure 7). However, the carotenoid pattern showed accumulation of a substantial fraction of the noncolored carotene intermediates phytoene and phytofluene (see Discussion). Thus, in cassava roots, PSY activity, replaceable by increased PSY expression levels, represents an important control point in the biosynthesis of downstream carotenoids.
Table 1.
Carotenoid Profile Determined by HPLC from Roots of 6-Month-Old Field-Grown Transgenic Plants Expressing the Bacterial PSY CrtB (CP1:TP-CrtB)
| Event | Total Car. | Phytoene | Phytofluene | β-Carotene | Xanthophylls |
| Wild type | 0.65 ± 0.06 | 0.00 ± 0.00 (nd) | 0.00 ± 0.00 (nd) | 0.41 ± 0.01 (63) | 0.24 ± 0.05 (37) |
| 84 | 1.18 ± 0.32 | 0.21 ± 0.10 (16) | 0.30 ± 0.00 (25) | 0.38 ± 0.03 (32) | 0.30 ± 0.09 (25) |
| 11 | 1.19 ± 0.03 | 0.09 ± 0.04 (8) | 0.10 ± 0.01 (8) | 0.41 ± 0.01 (35) | 0.59 ± 0.11 (49) |
| 220 | 1.20 ± 0.21 | 0.15 ± 0.06 (12) | 0.18 ± 0.00 (15) | 0.38 ± 0.07 (32) | 0.50 ± 0.10 (41) |
| 122 | 1.27 ± 0.40 | 0.24 ± 0.07 (19) | 0.13 ± 0.02 (10) | 0.29 ± 0.09 (23) | 0.62 ± 0.15 (48) |
| 139 | 1.58 ± 0.60 | 0.16 ± 0.05 (10) | 0.22 ± 0.02 (14) | 0.45 ± 0.10 (28) | 0.74 ± 0.14 (47) |
| 509 | 1.95 ± 0.08 | 0.26 ± 0.04 (13) | 0.36 ± 0.01 (19) | 1.09 ± 0.01 (56) | 0.23 ± 0.09 (12) |
| 12 | 21.84 ± 0.84 | 8.91 ± 0.37 (39) | 4.30 ± 0.05 (20) | 6.67 ± 0.93 (32) | 1.96 ± 0.25 (9) |
All transgenic events had increased carotenoid (Car.) levels over the wild type. nd, not detectable. Carotenoid levels are given in μg g−1 dry weight; the relative distribution of individual carotenoids in percentage is given in parentheses. Data represent the mean ± se of three technical replicates.
Figure 7.
Overexpression of PSY Is Sufficient to Enable the Accumulation of Colored Carotenoids in Cassava Roots.
Yellow roots represent transformation event no. 12, and white roots represent the wild-type control. A photograph of representative peeled roots is shown in the top panel; transverse roots sections are shown below.
DISCUSSION
The enzyme PSY has been shown to be rate determining in carotenogenesis for a number of different plant tissues. Paine et al. (2005) also showed that the source of the PSY transgene was critical for the increased carotenoid production in Golden Rice 2. Whereas the PSY paralogs from daffodil, tomato (Solanum lycopersicum), and pepper (Capsicum annuum), known to enable high carotenoid overproduction in chromoplasts, were fairly ineffective, those from rice (not developing chromoplasts) and maize were in this case the best to bolster carotenoid production in the grain endosperm. Because of the high similarity between plant PSYs, sequence variations affecting the catalytic efficiency of the enzyme can be as subtle as one single amino acid difference, as we are able to show here.
In the past, association mapping and linkage analysis have pointed to alleles favoring a desired carotenoid composition. In different maize genotypes, polymorphisms capable of increasing the proportion of provitamin A carotenoids have been identified in the genes for lycopene ε-cyclase (Harjes et al., 2008) and β-carotene hydroxylase 1 (Yan et al., 2010). A single SNP leading to an amino acid replacement in a conserved position of LCY-b, and probably leading to impaired catalytic function, was found to define red- and yellow-fleshed watermelon (Citrullis vulgaris) due to the accumulation of lycopene and cyclic carotenoids, respectively (Bang et al., 2007). A single amino acid substitution in the enzyme LCY-e is responsible for the introduction of one or two ionone rings (Cunningham and Gantt, 2001).
The SNP in PSY2 is associated with root color due to increased PSY catalytic activity, as we showed using yeast and E. coli as heterologous expression systems as well as through in vitro enzymology with the purified enzymes. At this point, we cannot distinguish whether the improved catalysis resides in PSY per se or in an improved interaction with GGPS. All of the PSYs from different organisms we tested in vitro did not accept GGPP as substrate, but required its de novo synthesis and delivery through GGPS. Therefore, it is not possible to determine the Km for PSYs. Moreover, all PSY-GGPS combinations we used so far showed a variable degree of leakiness (i.e., the release of GGPP as a side-product). However, there is no available evidence for the free in vivo diffusion of the extremely amphipathic intermediate GGPP, which is thought to be channeled into the different prenyl lipid biosynthetic routes (Figure 1). Evidence has been presented in the older literature for the existence of a PSY complex comprising at least GGPS and PSY (reviewed in Cunningham and Gantt, 1998). The observation that the increment of increased activity upon mutagenesis depends on the basal activity of the respective wild-type PSY (Figure 5B) together with the fact that increased pathway flux was also observed in cassava (where PSY interacts with a cassava GGPS; thus, a different GGPS than the one from S. alba that we used in vitro), and the in vitro data obtained are altogether in support of increased intrinsic PSY activity as the underlying cause for provitamin A accumulation.
Finally, a metabolite channel may exist between PSY and PDS. Although the presence of protein-free liposomes (as used here) are sufficient for effective phytoene production in vitro, the product pattern in the transgenic cassava events generated suggests that protein–protein interaction may occur between PSY and PDS. Large amounts of phytoene accumulated upon the expression of the bacterial PSY (CrtB, Table 1), which shares only 40% similarity with cassava PSYs. This low level of similarity might result in an impaired interaction between the two proteins, thus producing membrane-bound phytoene that is less accessible to PDS. Alternatively, PDS may simply represent the subsequent rate-limiting step in the pathway of the cultivar used, which is revealed upon relieving the first.
A threefold increase in the in vitro conversion rate contrasts with an up to 20-fold increase in the carotenoid content of roots carrying the PSYD191 allele. The other in vivo systems used, yeast and E. coli, showed a clear increase in lycopene and β-carotene production, although more in tune with PSY activity. The reason for the discrepancy between our in vitro and in vivo observations most probably lies in the long incubation time (i.e., the potential of the carotenoid biosynthesis pathway to be active over several months of secondary root development). In the presence of a comparatively lower β-carotene turnover, accumulation can take place at high levels. Kinetic considerations, such as substrate concentrations substantially below the Km, as generally present in cells, and other unknown factors determining the biochemical environment of amyloplasts, however, render any quantitative comparisons between the in vitro and the in vivo situations difficult.
The A191D amino acid exchange is in a highly conserved sequence region of PSY (Figure 3). The sequence diversity of PSY1 in grasses has been assessed (Fu et al., 2010), revealing a hot spot of nonsynonymous substitutions residing in a region located ~115 amino acids closer to the N terminus than the exchange reported here. In addition, allelic PSY1 variants of durum wheat were found to be associated with high endosperm carotenoid contents (Zhang and Dubcovsky, 2008; Singh et al., 2009), but none of these mutations were in a position equivalent to the one reported here. Whereas A191D appears to be a rare variant, its activity-increasing effect was similar in all the mutagenized PSY homologs investigated. Therefore, we conclude that the mutation has generally applicable structural and functional implications. Crystallization of PSY is currently underway to elucidate the catalytic mechanisms involved.
The PSY mutation described here provides a previously unknown means for the improvement of cassava provitamin A content. A transgenic approach employing root-specific promoters and targeting relevant cassava varieties can take advantage of this PSY variant. Alternatively, the finding presented here can provide breeders with the necessary tools to increase the levels of provitamin A carotenoids of cassava roots using marker-assisted selection.
METHODS
Cassava Plant Materials and Genetic Transformation
Cassava (Manihot esculenta) plants were grown in the field at the International Center for Tropical Agriculture (CIAT). Earlier studies identified that a hybrid family from the cross of two landraces (MMAL 66 × MBRA1A) segregated widely for carotenoid contents. Two contrasting individuals with high and low carotenoid content were selected from this full sib family (GM 708-20 and GM 708-63) and self-pollinated to produce S1 families AM 697 (from GM 708-20) and AM 702 (from GM708-63). In addition, four landraces were analyzed: CM 3306-4 and MPER 183 with low, CM 2772-3 with an intermediate, and MBRA 253 with a high root carotenoid content.
Cassava was transformed as described elsewhere (Beltrán et al., 2010) using embryogenic callus of the genotype TMS 60444 (coded as Nga 11 in the germplasm collection at CIAT).
DNA Gel Blot Analysis
Cassava genomic DNA was isolated according to Sambrook and Russell (2001). Fifteen micrograms of genomic DNA were digested with HindIII and fractioned electrophoretically through a 0.8% (w/v) agarose gel before capillary transfer and immobilization on nylon membrane (Hybond N+; GE Healthcare). A PCR fragment containing 470 bp of the CrtB/nos terminator region (see Supplemental Table 3 online for primer information) was amplified from the vector pCAS-Phyt and DIG-dUTP labeled using a PCR DIG probe synthesis kit (Roche Diagnostics) according to the manufacturer’s instructions. Hybridization was performed according to standard procedures.
cDNA Cloning, Vector Construction, and Expression Analysis
cDNA sequences from cassava PSY1, PSY2, and PSY3 (Arango et al., 2010b), PDS, ZDS, LCY-b, LCY-e, b-HYD, and CCD1 were amplified by RT-PCR using cultivar CM 3306-4 (for primers and accession numbers, see Supplemental Table 3 online). The use of primer pair SNP-sen and SNP-asen allowed amplifying PSY1 and PSY2 fragments encompassing SNP1 and SNP2; primer SNP-seq was used for sequencing.
For construction of the binary vector pCas-Phyt, the TP-CrtB gene (Diretto et al., 2007) encoding the PSY CrtB from Pantoea ananatis fused with the pea (Pisum sativum) RbcS transit peptide (TP; Misawa et al., 1993) was isolated from pTrio1, a pBluescript KS derivative, using SacI and NcoI. The fragment thus obtained was ligated into the accordingly digested pCas-CrtI, a pBluescipt derivative harboring a TP-CrtI expression cassette driven by the cassava CP1 promoter (Beltrán et al., 2010), to yield pCas-CrtB. The plasmid pCas-CrtB was digested with ClaI, blunt-ended with T4-DNA polymerase, and treated with XbaI to isolate CP1-TP-CrtB. The fragment thus obtained was then ligated into PmlI/XbaI-digested pCAMBIA1305.2 (Canberra) to yield pCas-Phyt.
Transcript levels were determined by real-time RT-PCR using 6FAM-labeled TaqMan probes for detection of PSY, PDS, ZDS, LCY-b, and LCY-e; those of b-HYD and CCD1 were determined with SYBR green (for primers and probes, see Supplemental Table 3 online). The 18S rRNA levels were quantified with the eukaryotic 18S rRNA endogenous control kit (Applied Biosystems) and used for normalization. The relative quantity of the transcripts was calculated using the comparative threshold cycle method (Livak, 1997). Data were normalized first to the 18S rRNA level of the corresponding sample and then calculated relative to one sample from the white-rooted cassava cultivar.
Carotenoid Analysis
Carotenoids were extracted from 500 mg of lyophilized ground roots and analyzed by HPLC (Welsch et al., 2008). Phytoene from yeast cells was extracted accordingly, but 100 μL of β-carotene (Sigma-Aldrich; 60 μg mL−1 in acetone) were added as an internal standard. Extracts were saponified prior to HPLC analysis. Photometric quantification of cyclic carotenoids was done using ε452nm = 134,500 liters mol−1 cm−1. Escherichia coli cell pellets were extracted twice with chloroform/methanol (2/1, v/v). Lycopene concentration was determined photometrically in petroleum benzene using ε470nm = 185,230 liters mol−1 cm−1.
Heterologous Expression of PSY Variants in Yeast and E. coli Cells
For the investigation of cassava PSY2 versions in vivo, the split-ubiquitin system was used according to Obrdlik et al. (2004), including vector and yeast strains described herein. The ORF of Sinapis alba GGPS, with its putative transit peptide truncated as indicated by Welsch et al. (2000), was cloned in vivo in the vector pNXgate using the yeast strain THY.AP5. The resulting vector pNubGGPSdTP encoded GGPS as a translational fusion with an N-terminal HA tag. The ORFs of PSY2 allelic versions, with their putative transit peptides truncated (Arango et al., 2010b), were cloned (primers MePSY2B1 and MePSY2B2) in the vector pMetYCgate using the yeast strain THY.AP4. Cells carrying pNubGGPSdTP were mated with each of the THY.AP4, each transformed with either of the PSY2 variants. Diploid cells were grown and subcultured in synthetic complete medium containing adenine and histidine. The phytoene formed was quantified by HPLC.
For the coexpression of GGPS with PSYs of different origins and CrtI from P. ananatis in E. coli strain BL21-CodonPlus-RIL, the DUET vector set (Novagen-Merck) was used. pET-GGPS contained the S. alba GGPS ORF, with its putative transit peptide truncated. It was obtained by subcloning a BamHI/SalI fragment from pGGPS (Welsch et al., 2000) into the accordingly digested vector pETDuet-1. For pCDF-CrtI, the CrtI ORF was amplified from pBaal2 (Ye et al., 2000) with the primers CrtIEco and CrtIHind, the PCR fragment digested with EcoRI and HindIII and subcloned into the accordingly digested vector pCDFDuet-1. pRSF-PSY contained the ORF of PSY versions, with their putative transit peptide removed. Vectors coding for the A/D amino acid exchange equivalent to cassava PSY2 were generated by overlapping extension PCR using the corresponding pRSF vectors with wild-type sequences as templates. Primers are given in Supplemental Table 3 online; positions of the first amino acid and of the A/D exchange are as follows: At-PSY, Arabidopsis thaliana, start, #58, A187D; Np-PSY, Narcissus pseudonarcissus, start, #65, A184D; Zm-PSY1, Zea mays, start, #52, A174D.
Overnight grown cultures from E. coli cells transformed with pET-GGPS, pCDF-CrtI, and one PSY version in pRSF were subcultured in 50 mL of fresh medium and grown to OD600nm = 0.5 at 37°C. Expression was induced with 0.5 mM isopropyl-β-D-thiogalactopyranoside, and cultures were incubated at 28°C. Two, three, and four hours after induction, the OD600nm was recorded; 4.5 mL of culture spun down and used for lycopene extraction as described above. The mean value from these measurements was recorded.
Immunoblot Analysis, Protein Purification, and Enzymatic Assays
E. coli or yeast cells in PBS were disrupted with a French press. After centrifugation (3100g), the protein concentration was determined in the supernatant. Proteins (30 μg yeast lysate; 1.5 μg E. coli lysate) were separated by SDS-PAGE and blotted onto polyvinylidene fluoride membranes. Immunodetection of PSY2 in yeast protein was performed with anti-At-PSY antibodies (Maass et al., 2009). GGPS in yeast was detected using an anti-HA-tag antibody (Cell Signaling Technology). His6-GGPS and His6-PSY2 were detected with an anti-6*His-Tag antibody (Sigma-Aldrich).
GGPS from S. alba was purified as published (Kloer et al., 2006). The Arabidopsis PSY ORF with its transit peptide sequence removed at position #58 was subcloned into pCOLDI (Takara-Clontech) to yield an N-terminal His6 fusion as described (Maass et al., 2009). From this plasmid, a second vector coding for an Asp instead of Ala at position #187 was generated by overlapping extension PCR (for primers, see Supplemental Table 3 online). Cell pellets from 1 liter of induced culture were resuspended in 20 mL lysis buffer [50 mM phosphate buffer, pH 7.6, 1 mM MgCl2, and 1 mM TCEP (Tris [2-carboxyethyl]phosphine hydrochloride)] and kept for 30 min on ice after adding 10 units of benzonase and lysozyme (Sigma-Aldrich). After sonication and centrifugation (16,000g, 40 min), the resulting pellet was washed twice with water, once with 1% (v/v) Triton X-100, and once with 2 M urea. Inclusion bodies obtained were solubilized in 10 mL of 6 M GuHCl containing 1 mM TCEP. Refolding was achieved by adding 450 mL of a solution containing 50 mM HEPES-KOH, pH 7.6, 1 mM TCEP, 0.2% (w/v) N,N-dimethyldodecylamine N-oxide (LDAO), 1 mM MgCl2, and 150 mM NaCl. After centrifugation (10,000g, 20 min), 2 mL of Co2+ Fractogel (Merck) was added. The resin was washed with 50 mM Tris-HCl, pH 7.6, 300 mM NaCl, 1 mM TCEP, 1 mM MgCl2, and 0.035% (w/v) LDAO and treated with 3 mL of elution buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1 mM TCEP, 0.035% [w/v] LDAO, and 100 mM EDTA) for 20 min. After centrifugation, the supernatant was dialyzed against 500 mL of dialysis buffer (50 mM Tris-HCl, pH 7.6, 600 mM NaCl, 1 mM TCEP, 1 mM MgCl2, and 0.035% [w/v] LDAO) for 3 h.
Fifteen micrograms of recombinant purified His6-At-PSY and 22 μg of purified His6-GGPS were incubated in 4.5-mL reaction buffer (100 mM Tris-HCl, pH 7.6, 2 mM MnCl2, 1 mM TCEP, 0.08% [v/v] Tween 80, 10 mM MgCl2, and 20% [v/v] glycerol) and phosphatidylcholine liposomes in a final concentration of 62.5 ng μL−1. The reaction was initiated by adding 20 μM dimethylallyl-diphosphate (Isoprenoids) and 20 μM [1-14C] IPP (50 mCi mmol−1; ARC), fivefold isotope-diluted with unlabeled IPP (Isoprenoids) prior to use. Incubation took place at 20°C under continuous stirring. At given time intervals, 200-μL aliquots were withdrawn and the reaction stopped by adding EDTA to a final concentration of 40 mM. Samples were extracted by partitioning twice against 200 and 100 μL of butanol. The combined butanol phases containing geranygeranyl-diphosphate were mixed with 300 μL of a 1 M methanolic MgCl2 solution and partitioned against 300 μL heptane to recover the phytoene that was quantified by liquid scintillation counting.
Accession Numbers
Sequence data from this article can be found in the Genbank/EMBL/DDBJ databases under the following accession numbers: At-PSY, P37271; Np-PSY, P53797; Zm-PSY1, P49085; CrtB, M87280; Os-PSY1, NP_001058647; Os-PSY2, ABA99494; Os-PSY3, ACI62767; Sl-PSY1, EF534740; Sl-PSY2, EF534738; and S. alba GGPS, X98795. Cassava sequences were submitted to GenBank and received the following accession numbers: PDS, GU120072; ZDS, GU120073; LCY-b, GU120074; LCY-e, GU120075; b-HYD, GU120076; CCD1, GU120077; CCD4, GU120078; PSY2-W-1, GU111720; PSY2-W-2, GU111721; PSY2-Y-1, GU111722; and PSY2-Y-2, GU111723.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Allelic Variants of Cassava PSY2.
Supplemental Figure 2. DNA Gel Blot Analysis of Transgenic Cassava Events Transformed with pCAS-Phyt.
Supplemental Table 1. Carotenoid Content and Pattern of the Three Investigated Cassava Cultivars Differing in the Carotenoid Levels of Their Storage Roots.
Supplemental Table 2. Carotenoid Levels and SNP2 Presence in a Cassava Breeding Population.
Supplemental Table 3. Primers and Probes.
Supplementary Material
Acknowledgments
This research was supported by the HarvestPlus research consortium, which received a grant from the Bill & Melinda Gates Foundation. The valuable contribution by Yaneth Ladinob and Danilo López (CIAT) is gratefully acknowledged. We thank Petr Obrdlik for providing the vectors and yeast strains for the split-ubiquitin system. We thank Jorge Mayer for his critical reading of the manuscript.
References
- Arango J., Salazar B., Welsch R., Sarmiento F., Beyer P., Al-Babili S. (2010a). Putative storage root specific promoters from cassava and yam: Cloning and evaluation in transgenic carrots as a model system. Plant Cell Rep. 29: 651–659 [DOI] [PubMed] [Google Scholar]
- Arango J., Wüst F., Beyer P., Welsch R. (2010b). Characterization of phytoene synthases from cassava and their involvement in abiotic stress-mediated responses. Planta 232: 1251–1262 [DOI] [PubMed] [Google Scholar]
- Auldridge M.E., McCarty D.R., Klee H.J. (2006). Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr. Opin. Plant Biol. 9: 315–321 [DOI] [PubMed] [Google Scholar]
- Bang H., Kim S., Leskovar D., King S. (2007). Development of a codominant CAPS marker for allelic selection between canary yellow and red watermelon based on SNP in lycopene β-cyclase (LCYB) gene. Mol. Breed. 20: 63–72 [Google Scholar]
- Beltrán J., Prías M., Al-Babili S., Ladino Y., López D., Beyer P., Chavarriaga P., Tohme J. (2010). Expression pattern conferred by a glutamic acid-rich protein gene promoter in field-grown transgenic cassava (Manihot esculenta Crantz). Planta 231: 1413–1424 [DOI] [PubMed] [Google Scholar]
- Bouvier F., Isner J.C., Dogbo O., Camara B. (2005). Oxidative tailoring of carotenoids: A prospect towards novel functions in plants. Trends Plant Sci. 10: 187–194 [DOI] [PubMed] [Google Scholar]
- Bouwmeester H.J., Roux C., Lopez-Raez J.A., Bécard G. (2007). Rhizosphere communication of plants, parasitic plants and AM fungi. Trends Plant Sci. 12: 224–230 [DOI] [PubMed] [Google Scholar]
- Burkhardt P.K., Beyer P., Wünn J., Klöti A., Armstrong G.A., Schledz M., von Lintig J., Potrykus I. (1997). Transgenic rice (Oryza sativa) endosperm expressing daffodil (Narcissus pseudonarcissus) phytoene synthase accumulates phytoene, a key intermediate of provitamin A biosynthesis. Plant J. 11: 1071–1078 [DOI] [PubMed] [Google Scholar]
- Cunningham F.X., Jr, Gantt E. (1998). Genes and enzymes of carotenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 557–583 [DOI] [PubMed] [Google Scholar]
- Cunningham F.X., Jr, Gantt E. (2001). One ring or two? Determination of ring number in carotenoids by lycopene epsilon-cyclases. Proc. Natl. Acad. Sci. USA 98: 2905–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D., Pogson B.J. (2006). Vitamin synthesis in plants: Tocopherols and carotenoids. Annu. Rev. Plant Biol. 57: 711–738 [DOI] [PubMed] [Google Scholar]
- Diretto G., Al-Babili S., Tavazza R., Papacchioli V., Beyer P., Giuliano G. (2007). Metabolic engineering of potato carotenoid content through tuber-specific overexpression of a bacterial mini-pathway. PLoS ONE 2: e350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira C.F., Alves E., Pestana K.N., Junghans D.T., Kobayashi A.K., de Jesus Santos V., Silva R.P., Silva P.H., Soares E., Fukuda W. (2008). Molecular characterization of cassava (Manihot esculenta Crantz) with yellow-orange roots for beta-carotene improvement. Crop Breed. Appl. Biotechnol. 8: 23–29 [Google Scholar]
- Fu Z., Yan J., Zheng Y., Warburton M.L., Crouch J.H., Li J.S. (2010). Nucleotide diversity and molecular evolution of the PSY1 gene in Zea mays compared to some other grass species. Theor. Appl. Genet. 120: 709–720 [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- Harjes C.E., Rocheford T.R., Bai L., Brutnell T.P., Kandianis C.B., Sowinski S.G., Stapleton A.E., Vallabhaneni R., Williams M., Wurtzel E.T., Yan J., Buckler E.S. (2008). Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 319: 330–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloer D.P., Welsch R., Beyer P., Schulz G.E. (2006). Structure and reaction geometry of geranylgeranyl diphosphate synthase from Sinapis alba. Biochemistry 45: 15197–15204 [DOI] [PubMed] [Google Scholar]
- Lindgren L.O., Stålberg K.G., Höglund A.S. (2003). Seed-specific overexpression of an endogenous Arabidopsis phytoene synthase gene results in delayed germination and increased levels of carotenoids, chlorophyll, and abscisic acid. Plant Physiol. 132: 779–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J. (1997). User Bulletin No. 2: ABI PRISM 7700 Sequence Detection System. (Foster City, CA: PE Applied Biosystems; ). [Google Scholar]
- Maass D., Arango J., Wüst F., Beyer P., Welsch R. (2009). Carotenoid crystal formation in Arabidopsis and carrot roots caused by increased phytoene synthase protein levels. PLoS ONE 4: e6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer J.E., Pfeiffer W.H., Beyer P. (2008). Biofortified crops to alleviate micronutrient malnutrition. Curr. Opin. Plant Biol. 11: 166–170 [DOI] [PubMed] [Google Scholar]
- Misawa N., Yamano S., Linden H., de Felipe M.R., Lucas M., Ikenaga H., Sandmann G. (1993). Functional expression of the Erwinia uredovora carotenoid biosynthesis gene crtl in transgenic plants showing an increase of beta-carotene biosynthesis activity and resistance to the bleaching herbicide norflurazon. Plant J. 4: 833–840 [DOI] [PubMed] [Google Scholar]
- Moise A.R., von Lintig J., Palczewski K. (2005). Related enzymes solve evolutionarily recurrent problems in the metabolism of carotenoids. Trends Plant Sci. 10: 178–186 [DOI] [PubMed] [Google Scholar]
- Nassar N.M.R., Fernandes P.C., Melani R.D., Pires O.R., Jr (2009). Amarelinha do Amapá: a carotenoid-rich cassava cultivar. Genet. Mol. Res. 8: 1051–1055 [DOI] [PubMed] [Google Scholar]
- Obrdlik P., et al. (2004). K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc. Natl. Acad. Sci. USA 101: 12242–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine J.A., Shipton C.A., Chaggar S., Howells R.M., Kennedy M.J., Vernon G., Wright S.Y., Hinchliffe E., Adams J.L., Silverstone A.L., Drake R. (2005). Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat. Biotechnol. 23: 429–430 [DOI] [PubMed] [Google Scholar]
- Rojas M.C., Pérez J.C., Ceballos H., Baena D., Morante N., Calle F. (2009). Analysis of inbreeding depression in eight S1 cassava families. Crop Sci. 49: 543–548 [Google Scholar]
- Sambrook J., Russell W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Schaub P., Al-Babili S., Drake R., Beyer P. (2005). Why is golden rice golden (yellow) instead of red? Plant Physiol. 138: 441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S.H., Tan B.C., Gage D.A., Zeevaart J.A., McCarty D.R. (1997). Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276: 1872–1874 [DOI] [PubMed] [Google Scholar]
- Shewmaker C.K., Sheehy J.A., Daley M., Colburn S., Ke D.Y. (1999). Seed-specific overexpression of phytoene synthase: Increase in carotenoids and other metabolic effects. Plant J. 20: 401–412X [DOI] [PubMed] [Google Scholar]
- Singh A., Reimer S., Pozniak C.J., Clarke F.R., Clarke J.M., Knox R.E., Singh A.K. (2009). Allelic variation at Psy1-A1 and association with yellow pigment in durum wheat grain. Theor. Appl. Genet. 118: 1539–1548 [DOI] [PubMed] [Google Scholar]
- Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., Magome H., Kamiya Y., Shirasu K., Yoneyama K., Kyozuka J., Yamaguchi S. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- Welsch R., Beyer P., Hugueney P., Kleinig H., von Lintig J. (2000). Regulation and activation of phytoene synthase, a key enzyme in carotenoid biosynthesis, during photomorphogenesis. Planta 211: 846–854 [DOI] [PubMed] [Google Scholar]
- Welsch R., Wüst F., Bär C., Al-Babili S., Beyer P. (2008). A third phytoene synthase is devoted to abiotic stress-induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes. Plant Physiol. 147: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2009). Global prevalence of vitamin A deficiency in populations at risk 1995–2005. WHO Global Database on Vitamin A Deficiency. (Geneva, Switzerland: World Health Organization; ). [Google Scholar]
- Yan J., et al. (2010). Rare genetic variation at Zea mays crtRB1 increases β-carotene in maize grain. Nat. Genet. 42: 322–327 [DOI] [PubMed] [Google Scholar]
- Ye X., Al-Babili S., Klöti A., Zhang J., Lucca P., Beyer P., Potrykus I. (2000). Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287: 303–305 [DOI] [PubMed] [Google Scholar]
- Zhang P., Bohl-Zenger S., Puonti-Kaerlas J., Potrykus I., Gruissem W. (2003). Two cassava promoters related to vascular expression and storage root formation. Planta 218: 192–203 [DOI] [PubMed] [Google Scholar]
- Zhang W., Dubcovsky J. (2008). Association between allelic variation at the Phytoene synthase 1 gene and yellow pigment content in the wheat grain. Theor. Appl. Genet. 116: 635–645 [DOI] [PubMed] [Google Scholar]
- Zhu C., Naqvi S., Breitenbach J., Sandmann G., Christou P., Capell T. (2008). Combinatorial genetic transformation generates a library of metabolic phenotypes for the carotenoid pathway in maize. Proc. Natl. Acad. Sci. USA 105: 18232–18237 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.