This work examines the molecular lesions responsible for the low-nicotine phenotype of a classical tobacco mutant. Extensive duplications of genes encoding jasmonate-responsive transcription factors, found at the NIC2 locus, offer new insights into how regulatory genes for alkaloid biosynthesis are organized and evolved.
Abstract
Tobacco (Nicotiana tabacum) synthesizes nicotine and related pyridine alkaloids in the root, and their synthesis increases upon herbivory on the leaf via a jasmonate-mediated signaling cascade. Regulatory NIC loci that positively regulate nicotine biosynthesis have been genetically identified, and their mutant alleles have been used to breed low-nicotine tobacco varieties. Here, we report that the NIC2 locus, originally called locus B, comprises clustered transcription factor genes of an ethylene response factor (ERF) subfamily; in the nic2 mutant, at least seven ERF genes are deleted altogether. Overexpression, suppression, and dominant repression experiments using transgenic tobacco roots showed both functional redundancy and divergence among the NIC2-locus ERF genes. These transcription factors recognized a GCC-box element in the promoter of a nicotine pathway gene and specifically activated all known structural genes in the pathway. The NIC2-locus ERF genes are expressed in the root and upregulated by jasmonate with kinetics that are distinct among the members. Thus, gene duplication events generated a cluster of highly homologous transcription factor genes with transcriptional and functional diversity. The NIC2-locus ERFs are close homologs of ORCA3, a jasmonate-responsive transcriptional activator of indole alkaloid biosynthesis in Catharanthus roseus, indicating that the NIC2/ORCA3 ERF subfamily was recruited independently to regulate jasmonate-inducible secondary metabolism in distinct plant lineages.
INTRODUCTION
Plants have evolved the ability to synthesize a tremendous repertoire of structurally diverse secondary metabolites, which often confer physiological or ecological advantages in a changing environment occupied by heterogeneous competitors (Crozier et al., 2006; Bednarek and Osbourn, 2009). Many natural products of plant origin have long been used to enhance human health and social life. Among such bioactive natural products, nitrogen-containing low molecular weight compounds, collectively called alkaloids, are known to function in the chemical defense of plants against herbivores and pathogens, and their potent biological activities have variously been exploited as pharmaceuticals, stimulants, narcotics, and poisons (Roberts and Wink, 1998). Nicotine is arguably the most popular alkaloid due to the prevalence and long history of smoking. Smoking of nicotine-containing tobacco leaf products can be traced back to as early as 5000 BC, as a component of religious rituals (Gately, 2001).
Tobacco remains one of the most commercially important agricultural crops, although cigarette consumption in many developed countries is declining due to health concerns and government regulations. Most of the commercial tobaccos produced in the world are Nicotiana tabacum, hereafter referred to simply as tobacco. N. tabacum is a natural allotetraploid combining two ancestral diploid genomes that are closely related to modern Nicotiana sylvestris and Nicotiana tomentosiformis (Murad et al., 2002); the two coresident parental genomes do not appear to show extensive recombination with each other (Matassi et al., 1991).
Industrial tobacco breeding targets several commercially important traits, including leaf nicotine levels (Davis and Nielsen, 1999). Low-nicotine traits initially found in strains of Cuban cigar tobacco varieties were introduced into cigarette varieties through a series of backcrosses (Valleau, 1949). Low-nicotine tobacco germplasms were subsequently registered in the genetic backgrounds of cultivars Burley 21 (Legg et al., 1970) and NC 95 (Chaplin, 1975). Careful genetic studies using the low-nicotine Burley 21 lines indicated that two unlinked loci contribute to nicotine levels in the tobacco leaf (Legg and Collins, 1971). The two loci were originally described as A and B (Legg and Collins, 1971), which we call NIC1 and NIC2, respectively (Hibi et al., 1994). The nic1 and nic2 mutations are semidominant or show dose-dependent effects on nicotine levels, with the effects of nic1 ~2.4 times stronger than those of nic2. There are synergistic effects at both loci; phenotypes of the weak nic2 mutation are clearly measured only in the presence of the nic1 mutation (Legg and Collins, 1971). We have previously exploited tobacco nic mutants to discover new enzyme and transporter genes involved in nicotine biosynthesis and transport by screening structural genes that are downregulated in the mutant backgrounds (Hibi et al., 1994; Katoh et al., 2007; Shoji et al., 2009). Downregulation of nicotine biosynthesis genes in the nic mutants has been confirmed in other studies (Reed and Jelesko, 2004; Cane et al., 2005). These studies suggest collectively that NIC1 and NIC2 are the regulatory loci that specifically control the expression of nicotine-related structural genes.
Nicotine is the predominant alkaloid accumulating in the leaf of most commercial tobacco varieties, whereas other related pyridine alkaloids are also present at substantial levels in tobacco roots, in elicited cultured tobacco cells, or in wild Nicotiana species (Saito et al., 1985; Shoji and Hashimoto, 2010). Nicotine is composed of a pyridine ring and a pyrrolidine ring (see Supplemental Figure 1 online); ornithine decarboxylase (ODC; Imanishi et al., 1998), putrescine N-methyltransferase (PMT; Hibi et al., 1994), and N-methylputrescine oxidase (MPO; Heim et al., 2007; Katoh et al., 2007) are involved in the formation of the pyrrolidine ring, whereas the pyridine ring uses enzymes involved in the early steps of NAD biosynthesis, such as aspartate oxidase (AO), quinolinic acid synthase (QS), and quinolinic acid phosphoribosyl transferase (QPT) (Sinclair et al., 2000; Katoh et al., 2006). A PIP family oxidoreductase, A622, is also required for the biosynthesis of tobacco alkaloids, possibly to form a nicotinic acid-derived precursor (Deboer et al., 2009; Kajikawa et al., 2009). Biosynthesis of nicotine and nornicotine requires both pyrrolidine and pyridine pathways, whereas anatabine and anabasine are synthesized from the pyridine pathway alone (Shoji and Hashimoto, 2008). Tonoplast-localized MATE family transporters MATE1 and MATE2 (MATE1/2) contribute to the accumulation of tobacco alkaloids in the vacuoles of tobacco roots (Shoji et al., 2009). Collectively, the tobacco genes encoding these proteins are called the structural genes of the nicotine pathway. Arginine decarboxylase (ADC) catalyzes the first step in an alternative pathway to form putrescine, whereas spermidine synthase (SPDS), S-adenosylmethionine decarboxylase (SAMDC), and S-adenosylmethionine synthase (SAMS) are involved in polyamine metabolism (see Supplemental Figure 1 online). These enzymes are treated in this study as control metabolic enzymes since there is no evidence that they contribute significantly to nicotine biosynthesis in tobacco plants.
Damage to tobacco leaves by browsing herbivores causes a dramatic increase in de novo nicotine synthesis in roots (Baldwin, 1989). This wounding-inducible nicotine biosynthesis is mediated by the jasmonate signaling cascade conserved in plants (Staswick, 2008). When a tobacco ortholog of the jasmonate receptor CORONATINE INSENSITIVE1 was suppressed or when tobacco JASMONATE ZIM domain (JAZ) transcriptional repressors were expressed in dominant-negative forms, the jasmonate-inducible expression of PMT and increase in tobacco alkaloids were both effectively inhibited in the tobacco roots (Shoji et al., 2008). In Arabidopsis thaliana and tomato (Solanum lycopersicum), JAZ proteins physically associate with the basic helix-loop-helix leucine zipper-type transcription factor MYC2 and repress its transcriptional activity in the jasmonate-uninduced state (Chini et al., 2007). MYC2 homologs of N. benthamiana were recently shown to bind the PMT promoter and function as positive regulators of nicotine biosynthesis (Todd et al., 2010). Virus-induced gene silencing and transient overexpression assays were used to identify several other transcription factors that might regulate nicotine biosynthesis (De Sutter et al., 2005; Todd et al., 2010), but their potential functions have not been studied in stably transformed tobacco plants.
To identify and characterize tobacco regulatory genes that specifically activate structural genes involved in nicotine biosynthesis, we examined molecular lesions in the classical NIC2 regulatory locus. Extensive duplications of genes encoding jasmonate-responsive transcription factors, found at the NIC2 locus, offer new insights into how regulatory genes for alkaloid biosynthesis are organized and evolved and should stimulate future studies on how the general jasmonate signaling pathway effectively activates a downstream transcriptional network for particular alkaloids.
RESULTS
Several Ethylene Response Factor Genes Are Downregulated in nic Mutants
To analyze comprehensively root transcriptomes associated with the tobacco low-nicotine nic mutants, we used a tobacco oligonucleotide microarray that represented >20,000 independent transcripts. Total RNA from the roots of the wild type and a nic1 nic2 double mutant (cv Burley 21) was labeled with different fluorescent dyes and reciprocally hybridized to the array (see Methods for details). We thus obtained a list of tobacco genes that may be downregulated in the roots of the double mutant compared with the wild type (see Supplemental Table 1 online). Almost all known structural genes that encode enzymes for nicotine biosynthesis (PMT and A622), enzymes for the synthesis of nicotine precursors (ODC, AO, QS, and QPT), and a putative nicotine transporter (MATE1) were found in the top 200 genes, thereby supporting the validity of the experiment. The nicotine pathway-related gene MPO is known to be less tightly regulated by the NIC loci (Shoji and Hashimoto, 2008) and was ranked 391st on the list.
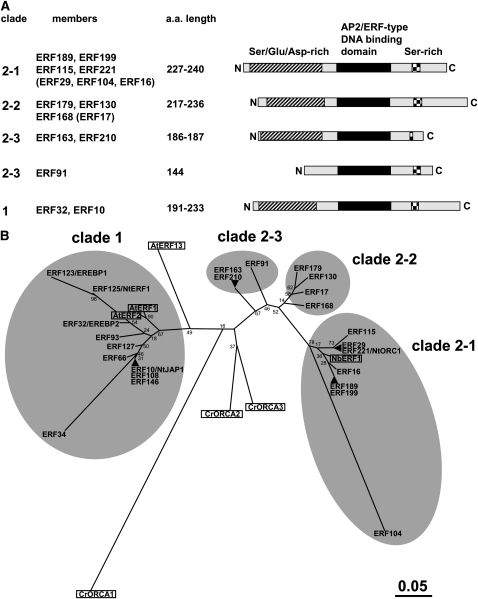
Among the most severely affected genes in the list is ethylene response factor 189 (ERF189), a member of the AP2/ERF transcription factor superfamily. The tobacco genome contains at least 239 AP2/ERF members (Rushton et al., 2008a), among which ERF189 belongs to the group IXa subfamily (Nakano et al., 2006). Multiple sequence alignment of the AP2/ERF DNA binding domain sequences generated a phylogenetic tree of tobacco IXa ERFs, in which we also included homologous members from Arabidopsis (At ERF1, 2, and 13) and Catharanthus roseus (Cr ORCA1-3) (Figure 1). ORCA3 has been shown to activate specifically several structural genes involved in terpenoid indole alkaloid biosynthesis upon the elicitation of cultured cells with methyl-jasmonate (MeJA) (van der Fits and Memelink, 2000). Here, we further grouped the tobacco IXa subfamily into clade 1 and clade 2, the latter further divided into three subclades (2-1 to 2-3). At ERF1 and At ERF2 belong to clade 1, whereas ORCA2 and ORCA3 are more closely related to clade 2. Tobacco ERF189 is a member of clade 2-1, which includes ERF221 (also known as ORC1; De Sutter et al., 2005) and N. benthamiana ERF1 (Todd et al., 2010). ORC1 significantly activated the PMT promoter in a transient overexpression assay using tobacco BY-2 protoplasts (De Sutter et al., 2005), whereas downregulation of ERF1 by virus-induced gene silencing considerably reduced nicotine levels in N. benthamiana plants (Todd et al., 2010). Tobacco ERF104 cDNA contains a frame-shift mutation within the AP2/ERF domain, thereby producing a truncated protein, but is included in clade 2-1. When entire coding regions of ERF proteins were used for an alignment, the group IXa ERFs were organized into the same clades as in the AP2/ERF domain–based phylogenetic tree, and clade 2-1 could be further grouped into the ERF189/199 cluster and the ERF115/29/221 cluster (see Supplemental Figure 2 and Supplemental Data Set 2 online).
Figure 1.
Group IXa ERF Members in Tobacco.
(A) Structure of tobacco IXa ERFs. Diagrams show the positions of the conserved AP2/ERF DNA binding domain, a Ser/Asp/Glu-rich region, and a short Ser stretch in four clades (1, 2-1, 2-2, and 2-3). Available cDNA clones for ERF29, ERF104, ERF16, and ERF17 (shown in parentheses) do not contain full-length coding sequences. ERF91 is truncated at the N terminus. a.a., amino acid.
(B) Phylogenetic tree of tobacco IXa ERFs and other homologous proteins based on the alignment of their AP2/ERF domain sequences (available as Supplemental Data Set 1 online). The tree was generated using MEGA4 software (Tamura et al., 2007) with the neighbor-joining algorithm. Bootstrap values are indicated at branch nodes, and the scale bar indicates the number of amino acid substitutions per site. Two or three ERFs located at the same place (e.g., ERF189 and ERF199) indicate homologous tobacco ERF proteins whose AP2/ERF domains are identical in amino acid sequence. At ERF1 (At4g17500), At ERF2 (At5g47220), and At ERF13 (At2g44840) are from Arabidopsis, Cr ORCA1, Cr ORCA2, and Cr ORCA3 are from C. roseus, and Nb ERF1 is from N. benthamiana.
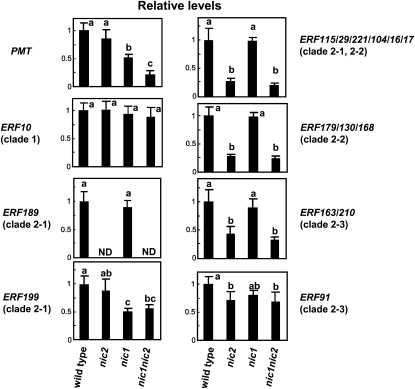
To validate the microarray expression data of ERF189 and to examine expression profiles of homologous tobacco genes, quantitative real-time PCR (qRT-PCR) was used to assay transcript abundance of IXa ERF genes in the roots of wild-type, nic2, nic1, and nic1 nic2 plants (Figure 2). Primers designed for qRT-PCR (see Supplemental Table 2 online) amplified one target gene specifically or highly homologous genes of the same or related clades, as revealed by sequencing the amplified fragments from the root RNA samples (see Supplemental Figure 3 online). Transcript levels of PMT, a NIC-regulated gene involved in nicotine biosynthesis, decreased in the order of the wild type, nic2, nic1, and nic1 nic2, as reported previously (Hibi et al., 1994). Expression of ERF10 (clade 1) and ERF91 (clade 2-3) were not affected or were only moderately affected by the nic mutations, whereas ERF199 (clade 2-1) expression was downregulated by 50% in the nic1-containing genetic backgrounds. Interestingly, transcript levels of ERF189 (clade 2-1), ERF115/29/221/104/16/17 (clades 2-1 and 2-2), ERF130/168/179 (clade 2-2), and ERF163/210 (clade 2-3) were reduced in nic2 and nic1 nic2. Notably, we could not detect any transcripts of ERF189 in the nic2-containing mutant roots.
Figure 2.
Expression Levels of Clade 2 ERF Genes Are Affected in nic Mutant Roots.
Transcript levels of clade 2 ERFs, ERF10 (clade 1), and PMT in the tobacco roots of different NIC genotypes were measured by qRT-PCR and are shown relative to the wild-type levels. ERF115/29/221/104/16/17, ERF179/130/168, and ERF163/210 were collectively amplified by the PCR primers used (see Supplemental Figure 3 online for breakdown of the amplified ERF proportions). Error bars represent sd of three replicates. Significant differences among the genotypes at P < 0.05 were determined by one-way analysis of variance (ANOVA) followed by the Tukey-Kramer test and are indicated by different letters. ND, not detected.
The NIC2 Regulatory Locus Encodes Clustered ERF Genes
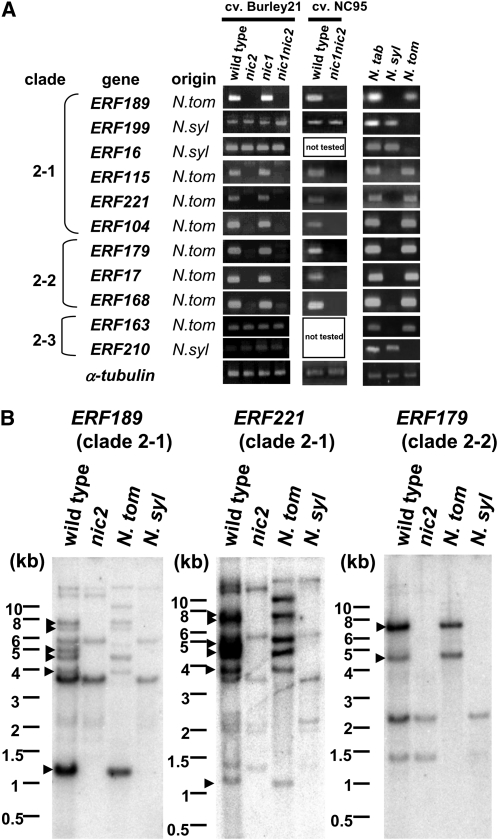
The absence of ERF189 transcripts in nic2 could be caused by an absolute dependence of ERF189 expression on the NIC2 activity. Alternatively, ERF189 may be deleted in nic2. To address the latter possibility, ERF genes in the IXa subfamily were amplified from genomic DNA of N. tabacum with different NIC genotypes (wild type, nic2, nic1, and nic1 nic2 of cv Burley 21). NC 95 cultivars in which the nic loci were independently introgressed (wild type and nic1 nic2) were also examined, together with two diploid Nicotiana species, N. sylvestris and N. tomentosiformis, which are both considered to be progenitors of the allotetraploid N. tabacum (Clarkson et al., 2005). Because requirements to design PCR primers were less stringent for genomic PCR, compared with qRT-PCR, we could specifically amplify many single ERF genes from the Nicotiana genomes (Figure 3A; see Supplemental Table 3 online).
Figure 3.
Seven ERF Genes Originating from N. tomentosiformis Are Absent in nic2 Mutants.
(A) Genomic PCR analysis of clade 2 ERF genes. Genomic DNA templates from tobacco (N. tabacum; cv Burley 21 of the wild type, nic2, nic1, and nic1 nic2 and cv NC 95 of the wild type and nic1 nic2), N. sylvestris (N. syl), and N. tomentosiformis (N. tom) were amplified with the primers specific to the indicated ERF genes. The tobacco α-tubulin gene was analyzed as a control.
(B) Genomic DNA gel bolt analysis of clade 2 ERF genes. Genomic DNA samples from tobacco (the wild type and nic2), N. sylvestris (N. syl), and N. tomentosiformis (N. tom) were digested by HindIII, blotted on a membrane, and probed with cDNA fragments from ERF189, ERF221, or ERF179, which showed high, but not exclusive, specificity to the target genes (see Supplemental Figure 4A online). DNA fragments missing in nic2 are marked by arrowheads. The numbers on the left sides of blots show the sizes of marker fragments in kilobases. The hybridization data for clade 2-3 ERFs are available in Supplemental Figure 4B online.
ERF189 was amplified only from the tobacco genomes containing the wild-type NIC2 locus in two independent cultivars, indicating that ERF189 is deleted in nic2. ERF189 probably originates from the N. tomentosiformis progenitor since its amplification was observed from the genome of N. tomentosiformis, but not N. sylvestris. Remarkably, the same results were obtained for ERF115, ERF221, ERF104, ERF179, ERF17, and ERF168; these ERF genes belonging to clades 2-1 and 2-2 were amplified only from the wild-type NIC2 genome and from the N. tomentosiformis genome. The amplification of ERF199 and ERF16 (both in clade 2-1) was independent of the nic genotypes, and these genes were derived from the N. sylvestris progenitor. Two ERF genes of the 2-3 clade, ERF163 and ERF210, were amplified from all the tobacco genotypes tested and originated from N. tomentosiformis and N. sylvestris, respectively. These results based on the genomic PCR experiments suggest that seven ERF genes of clades 2-1 and 2-2 are missing in the nic2 background and all originated from N. tomentosiformis.
Unsuccessful PCR amplification could result from diverged DNA sequences to which primers fail to anneal. To provide further evidence that these seven ERF genes are indeed deleted in the nic2 mutants, DNA gel blots were prepared from wild-type and nic2 tobacco genomes and from genomes of N. tomentosiformis and N. sylvestris and were then hybridized with radiolabeled probes of ERF189 (clade 2-1), ERF221 (clade 2-1), or ERF179 (clade 2-2) (Figure 3B). These probes were derived from relatively variable N-terminal portions of ERFs and recognized target ERF genes with fairly high specificity, although some cross hybridization to highly related ERF genes was observed (see Supplemental Figure 4A online). These ERF probes recognized one to five HindIII-digested DNA fragments strongly and a few DNA fragments weakly in N. tomentosiformis, whereas a few weakly hybridizing fragments were observed in N. sylvestris, consistent with these ERF genes being derived from N. tomentosiformis and with these probes also recognizing highly related ERF members. In wild-type tobacco, more hybridizing DNA fragments were observed and in most cases were the sum of DNA fragments found in the two progenitor species. Notably, several DNA fragments found in the wild type (arrowheads) were missing in the genomic DNA gel blots of nic2, and these missing fragments generally corresponded to those found in N. tomentosiformis. On the other hand, when the clade 2-3 genes (ERF163 and ERF91) were used as hybridization probes, there were no differences found in the DNA fragment patterns between the wild type and nic2 (see Supplemental Figure 4B online). These results show that at least seven ERF genes of clades 2-1 and 2-2 and of N. tomentosiformis origin were deleted in the nic2 mutant.
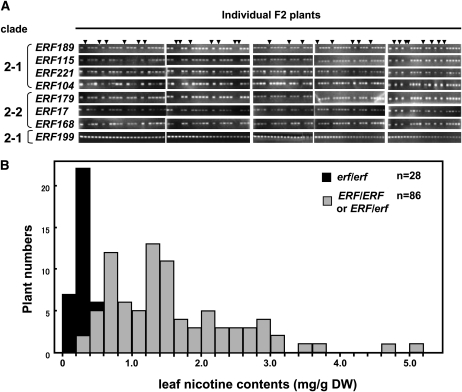
We next asked whether the deletion of these ERF genes is genetically associated with the low-nicotine nic2 phenotype. The impact of the NIC2 locus on leaf nicotine content is quite mild on its own but becomes considerably stronger in the presence of the nic1 mutation (Legg and Collins, 1971). Therefore, we crossed nic1 with nic1 nic2 to produce a F1 plant that was allowed to self-pollinate to generate a F2 population, in which NIC2/nic2 segregated in the nic1 mutant background. One hundred and fourteen F2 individuals were first analyzed for deletion of ERF genes by genomic PCR, which would detect homozygous deletions (Figure 4A). In ~25% of the F2 plants (28/114), ERF189, ERF115, ERF221, ERF104, ERF179, ERF17, and ERF168 (the seven ERF genes deleted in nic2) were found to be deleted. Remarkably, the deletion of these genes segregated together, indicating they are genetically linked within a chromosomal region of <0.9 centimorgans. Next, nicotine levels were measured in the leaves of mature F2 plants (Figure 4B). Here, we refer to the genotype of a homozygous ERF deletion as erf/erf, whereas heterozygous and wild-type genotypes are called ERF/erf and ERF/ERF, respectively. In the homozygous erf/erf individuals (n = 28), the nicotine content was very low, with a mean value of 0.26 mg/g dry weight. By contrast, the remaining plants with the ERF/erf or ERF/ERF genotype (n = 86) showed variable but generally high levels of nicotine in leaf, with a mean value of 1.6 mg/g dry weight. The difference in leaf nicotine content was statistically significant between erf/erf individuals and the individuals with the ERF/erf or ERF/ERF genotype (Student’s t test; P < 0.001).
Figure 4.
Seven ERF Genes Are Clustered at the NIC2 Locus.
(A) Cosegregation of ERF genes. A genetic cross between nic1 and nic1 nic2 produced a population of tobacco F2 plants (n = 114) in which deletions of ERF genes found in nic2 segregated in the nic1 background. The presence or absence of ERF genes was analyzed by genomic PCR. Arrowheads indicate individual F2 plants in which homozygous deletions of ERF genes (erf/erf) were observed. ERF199 was analyzed as a control for nondeleted ERF genes.
(B) The low-nicotine phenotype of the nic2 mutation is genetically linked with the ERF deletion. The F2 plants analyzed in (A) (n = 114) were used to determine nicotine levels in the leaves. The plants in which the seven ERF genes were not detected (erf/erf; n = 28) are indicated with black bars, while the plants in which these ERF genes were present (ERF/ERF or ERF/erf; n = 86) are shown with gray bars. DW, dry weight.
The semidominant nature of the nic2 mutation (Legg and Collins, 1971) may underlie the wide variation in nicotine content in ERF/erf or ERF/ERF plants. To examine the exact genotypes, we selected 10 F2 plants with the lowest nicotine content (from 0.29 to 0.64 mg/g dry weight) and 10 F2 plants with the highest nicotine content (from 2.8 to 5.1 mg/g dry weight) from the ERF/erf or ERF/ERF plants and determined their genotypes by genomic PCR analysis of F3 progenies after self-pollination. The low-nicotine F2 plants (n = 10) were all found to be ERF/erf, whereas two individuals were ERF/erf and the other eight individuals were ERF/ERF in high-nicotine F2 plants (see Supplemental Figure 5 online). Therefore, the heterozygous deletion at this ERF locus contributes, at least partly, to decreased nicotine content in the leaf.
Together, these results suggest that the NIC2 locus contains seven clustered ERF genes in clades 2-1 and 2-2 of the IXa AP2/ERF subfamily, which we hereafter refer to as the NIC2-locus ERF genes, and that a relatively large deletion encompassing the NIC2 locus of N. tomentosiformis origin is responsible for the low-nicotine phenotype associated with the nic2 mutation.
Suppression of NIC2-Locus ERF Genes Decreases Nicotine Biosynthesis
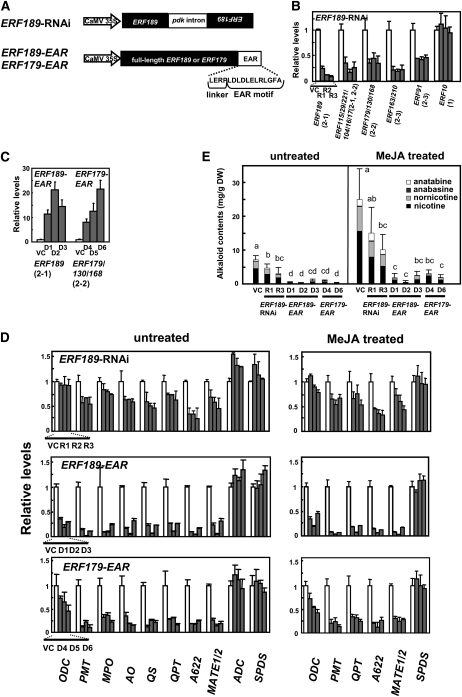
To provide further evidence that NIC2-locus ERF genes are required for nicotine biosynthesis, we adopted two different strategies to interfere with ERF functions (Figure 5). First, a 0.4-kb region of the ERF189 cDNA that showed high homology to the corresponding regions of the clade 2 ERFs was used to downregulate the expression of ERF189 and other related ERFs by an RNA interference (RNAi) mechanism. We also converted ERF 189 and ERF179 into dominant repressors by attaching to their C termini an ERF-associated amphiphilic repression (EAR) motif. When attached to a transcription factor, the EAR motif has been shown actively to repress expression of target genes of the transcription factor (Hiratsu et al., 2003). The ERF189-RNAi construct and the ERF189/ERF178-EAR constructs (Figure 5A) were placed under the control of the cauliflower mosaic virus (CaMV) 35S promoter and used to transform wild-type tobacco by Agrobacterium rhizogenes, thereby generating three transgenic hairy root lines for each construct, as well as a control line which was transformed with an empty vector (VC). In the ERF189-RNAi lines (R1 to R3), expression of the primary target ERF189 decreased by 75 to 91% compared with the vector control, while other ERF genes of clade 2 were also downregulated by 54 to 83%. Suppression appeared to be restricted to clade 2 ERFs since expression of ERF10, which belonged to clade 1, was not affected (Figure 5B). In the ERF189-EAR lines (D1 to D3) and the ERF179-EAR lines (D4 to D6), the transgenes were highly expressed; the transcripts of the transgenes were at least 8 to 22 times more abundant than the endogenous wild-type transcripts (Figure 6C).
Figure 5.
Downregulation or Dominant Repression of NIC2-Locus ERF Genes Inhibits Nicotine Biosynthesis in Transgenic Tobacco Roots.
Transcript levels are shown relative to empty vector control values, and error bars indicate the sd for three biological replicates. Clade numbers are indicated in parentheses.
(A) Schematic diagrams of the ERF189-RNAi, ERF189-EAR, and ERF179-EAR constructs. The ERF189-RNAi construct contains an intron of PDK (pyruvate orthophosphate dikinase from Flaveria trinerva) and two flanking ERF189 fragments in opposite orientations. ERF189-EAR and ERF179-EAR are fusion proteins with an EAR motif attached to the C termini of ERF189 and ERF179 via a short linker.
(B) Transcript levels of ERF genes in three ERF189 lines (R1 to R3; gray bars) and a vector control line (VC; white bars). Expression levels of clade 2 ERFs and clade 1 ERF10 were analyzed by qRT-PCR.
(C) Transcript levels of ERF genes in three ERF189-EAR lines (D1 to D3), three ERF179-EAR lines (D4 to D6), and a vector control line (VC). qRT-PCR was used to measure expression levels of both the endogenous ERF genes and the introduced transgenes.
(D) Transcript levels of tobacco metabolic enzyme genes. Tobacco hairy roots were cultured in the absence or presence of 100 μM MeJA for 24 h and analyzed for expression of the structural genes of nicotine biosynthesis (ODC, MPO, PMT, AO, QS, QPT, A622, and MATE1/2) and the control genes (ADC and SPDS) by qRT-PCR. In the VC line, MeJA increased the transcript levels as follows: ODC, 5.6-fold; PMT, 9.5-fold; QPT, 8.4-fold; A622, 9.2-fold; MATE1/2, 7.2-fold; and SPDS, 1.1-fold.
(E) Alkaloid levels in the transgenic root lines. In one experiment, tobacco hairy roots were treated with 100 μM MeJA for 3 d. Significant differences among the lines were determined at P < 0.05 by one-way analysis of variance, followed by the Tukey-Kramer test, and are indicated by different letters. DW, dry weight.
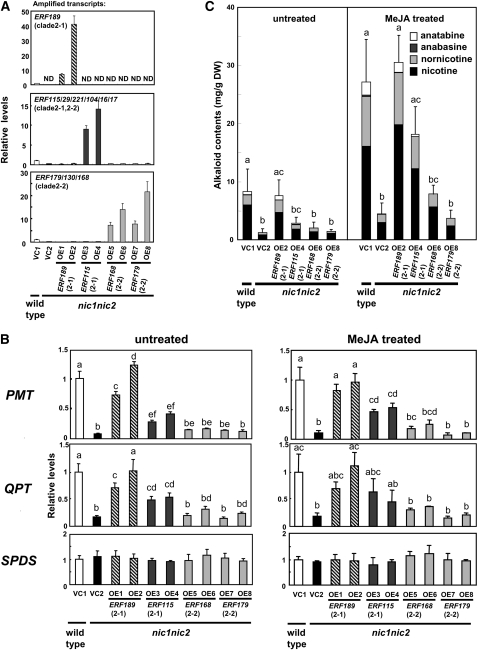
Figure 6.
Complementation of nic1 nic2 Roots by Overexpression of ERF Genes.
Transcript levels are shown relative to the values for the wild type transformed with the empty vector (VC1), and error bars indicate the sd for three biological replicates. Significant differences among the lines were determined at P < 0.05 by one-way ANOVA, followed by the Tukey-Kramer test, and are indicated by different letters. Clade numbers are indicated in parentheses.
(A) Overexpression of ERF189, ERF115, ERF168, or ERF179 in nic1 nic2 tobacco hairy roots. The vector control lines VC1 and VC2 were generated by transforming wild-type and nic1 nic2 plants, respectively, with an empty vector. Transcript levels include both the endogenous genes and the transgenes. ND, not detected.
(B) Relative expression levels of PMT, QPT, and SPDS in the transgenic root lines. For MeJA treatment, the hairy roots were elicited with 100 μM MeJA for 24 h. In the VC1 line, MeJA increased the transcript levels as follows: PMT, 8.7-fold; QPT, 7.9-fold; and SPDS, 1.2-fold.
(C) Alkaloid levels in the transgenic root lines. The right graph shows data from tobacco hairy roots treated with 100 μM MeJA for 3 d. DW, dry weight.
We first examined the expression of several metabolic and transporter genes by qRT-PCR in the transgenic hairy root lines (Figure 5D). In the ERF189-RNAi lines, moderate but significant reductions (by 17 to 74%) were observed for the transcripts encoding nicotine biosynthesis-related enzymes (PMT, MPO, AO, QS, QPT, and A622) and a transporter (MATE1/2). Expression levels of ODC, ADC, and SPDS were not reduced. In the ERF189-EAR lines and the ERF179-EAR lines, we found much higher reductions (by 64 to 98%) in levels of transcripts for the above-mentioned nicotine-related genes, as well as considerable decreases (by 27 to 81%) in levels of the ODC transcripts. The metabolic genes that are not involved in nicotine biosynthesis (ADC and SPDS) were not affected. Despite their similar overexpression levels (Figure 5C), the ERF189-EAR construct appeared to be slightly more effective than the ERF179-EAR construct, as clearly evident from their effects on ODC expression. The expression was also analyzed in the hairy roots that had been treated with 100 μM MeJA for 24 h. MeJA treatment increased transcript levels of the nicotine structural genes 4- to 10-fold (see the legend of Figure 5D). Compared with the expression in the vector control line, transcript levels of ODC, PMT, QPT, A622, and MATE1/2 were reduced, while those of SPDS were not affected in the transgenic hairy root lines ERF189-RNAi, ERF189-EAR, and ERF179-EAR. Thus, in both uninduced and MeJA-induced conditions, knockdown of ERF189 and closely related genes or dominant-negative inhibition of ERF189/179 transactivation functions inhibited the expression of tobacco genes involved in nicotine biosynthesis and transport but generally did not affect the expression of other metabolic genes.
Tobacco hairy roots accumulate nicotine as a major alkaloid but also contain other metabolically related alkaloids (nornicotine, anatabine, and anabasine, in order of decreasing abundance). The levels of these tobacco alkaloids increased 2- to 3-fold after elicitation with MeJA, but their relative abundance remained constant (Figure 5E). In the ERF189-RNAi lines, alkaloid levels decreased by 34 to 59%, in comparison with those in the control line, in both untreated and MeJA-elicited conditions. The reduction was much more severe (by 87 to 98%) in the ERF189-EAR and ERF179-EAR lines. The alkaloid profiles did not markedly change, indicating that the genes for the enzymes required for the synthesis of these tobacco alkaloids are similarly regulated by NIC2-locus ERF genes.
In accordance with a transcription factor function of NIC2-locus ERFs, green fluorescent protein–tagged ERF189 was located in the nucleus when transiently expressed in onion epidermal cells (see Supplemental Figure 6 online).
NIC2-Locus ERF Genes Are Not Equally Effective in Promoting Nicotine Biosynthesis
The above results show that NIC2-locus ERF genes are required for nicotine biosynthesis but do not address whether these ERF transcription factors are functionally equivalent or differ qualitatively or quantitatively in activating structural genes of nicotine biosynthesis. To assess in vivo functions of NIC2-locus ERF proteins, we overexpressed four related ERFs individually in the nic2 mutant background to test how effectively they complement the nic2 phenotypes. ERF189 and ERF115 belong to clade 2-1, whereas ERF168 and ERF179 are members of clade 2-2. Transgenic hairy root lines were established that expressed one of these ERFs under the control of the CaMV 35S promoter. The nic1 nic2 tobacco mutant was used for the transformation host since the dosage effects of NIC2-locus ERFs were much more pronounced in the presence of the nic1 mutation. Wild-type and nic1 nic2 hairy roots transformed with an empty vector (VC1 and VC2) served as the controls with high and low NIC2 activities, respectively. Transcript levels were measured by qRT-PCR and expressed as relative values compared with the high vector control line (VC1).
Transcripts of target ERFs accumulated at high levels (~7- to 42-fold increase compared with those in VC1), with the caveat that our qRT-PCR system amplified multiple highly homologous genes, with the exception of ERF189, and accumulating transcripts could come from any of the potentially amplifiable ERF genes (Figure 6A). However, we did not observe induced expression of off-target ERF genes after overexpression of a target ERF gene. In the ERF189-overexpressing lines (OE1 and OE2), the expression levels of PMT and QPT increased dramatically from the low control levels of VC2 to levels comparable to the high control levels of VC1, in both untreated and MeJA-elicited conditions (Figure 6B). Increases of PMT and QPT transcript levels in the ERF115-overexpressing lines (OE3 and OE4) were significant but smaller than those in the ERF189-overexpressing lines. Overexpression of ERF168 or ERF179 (OE5-OE8), on the other hand, caused negligible or marginal increases on the transcript levels of PMT and QPT. The transcript levels of a nicotine-unrelated gene, SPDS, did not vary in any of the transgenic root lines. Transcript profiles of the most effective ERF189-overexpressing lines (OE1 and OE2) were examined in more detail and showed that all the known structural genes for nicotine biosynthesis and transport were considerably upregulated, but the unrelated genes were not (see Supplemental Figure 7 online).
Alkaloid profiles of the transgenic hairy roots in untreated and MeJA-elicited conditions (Figure 6C) reflected the transcript profiles; the amounts of tobacco alkaloids increased dramatically from low VC2 levels to reach high VC1 levels in a ERF189-overexpressing line (OE2), restored to 35 to 67% of VC1 in a ERF115-overexpressing line (OE4), and scarcely or marginally increased in the hairy root lines overexpressing ERF168 or ERF179 (OE6 and OE8). We conclude that the clade 2-1 ERFs (ERF189 and ERF115) promote nicotine biosynthesis via upregulation of the target biosynthetic genes, with ERF189 being more effective than ERF115. The clade 2-2 ERFs (ERF168 and ERF179) are far less effective or ineffective in promoting nicotine biosynthesis in our overexpression assay.
We then asked whether ERF189 can further stimulate alkaloid production when overexpressed in a wild-type genetic background. Transgenic hairy root tobacco lines that overexpressed ERF189 (in the wild-type NIC1 NIC2 background) had 2- to 3-fold more of the transcripts of PMT and QPT than did the vector control line (see Supplemental Figure 8A online). ERF189 overexpression caused 2- to 3-fold higher levels of tobacco alkaloids compared with the control (see Supplemental Figure 8B online). Therefore, the activity of ERF189 is not saturated in wild-type tobacco roots.
Direct in Vivo Activation of Genes for Nicotine Synthesis by ERF189
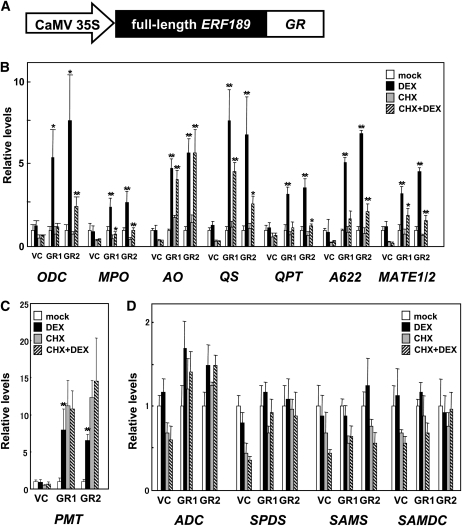
To examine whether ERF189 directly activates the structural genes of nicotine biosynthesis, we employed a steroid receptor-based inducible expression system (Picard, 1993). ERF189-fused glucocorticoid receptor (ERF189-GR) is kept inactive in the cytoplasm by heat shock proteins until exogenously supplied dexamethasone (DEX) binds to it. Then, it is released and translocated to the nucleus to activate the ERF189 target genes in a translation-independent manner. We generated two transgenic hairy root tobacco lines (GR1 and GR2) that constitutively expressed ERF189-GR (Figure 7A) and a control hairy root line (VC) that had been transformed with an empty vector and treated these lines with DEX. DEX-triggered activation of ERF189-GR resulted in a 2.5- to 7.8-fold increase in the expression of genes for nicotine biosynthesis (ODC, PMT, MPO, AO, QS, QPT, A622, ADC, SPDS, SAMS, SAMDC, and MATE; Figures 7B and 7C) but little (up to 1.7-fold) or no increase in the expression of the control genes (ADC, SAMS, and SAMDC; Figure 7D).
Figure 7.
Steroid-Induced Activation of ERF189-GR Increases the Expression of Genes for Nicotine Biosynthesis in Tobacco Roots.
Hairy root lines were treated with DEX, CHX, or both (DEX+CHX) at 10 μM for 4 h. Transcript levels are shown relative to the solvent-treated mock values, and error bars indicate the sd for three biological replicates. Significant differences were determined by Student’s t test for DEX treatment versus the mock control and for DEX+CHX treatment versus CHX treatment. *P < 0.05 and **P < 0.01.
(A) Schematic diagram of ERF189-GR construct. GR, glucocorticoid receptor.
(B) Expression levels of the genes for nicotine biosynthesis in two independent ERF189-GR–transformed lines (GR1 and GR2) and a vector-transformed line (VC).
(C) Expression levels of PMT. Transcript levels of PMT increased dramatically after the CHX treatment in the GR lines.
(D) Expression levels of the control genes.
Next, we treated the tobacco roots with a protein synthesis inhibitor, cycloheximide (CHX), or with both CHX and DEX. In the presence of CHX, only the immediate target genes of ERF189 are expected to be transcribed after the treatment with DEX. We found that PMT, but not other genes, was strongly induced by the treatment with CHX alone in the GR lines (Figure 7C). Although we cannot offer plausible explanations for this unusual finding, it precludes further analysis of PMT in the presence of CHX. Other genes involved in nicotine biosynthesis were induced to express by DEX even in the presence of CHX, although the change in levels (CHX plus DEX versus CHX alone) varied from a 3.9-fold increase in AO to a 1.6-fold increase in QPT (Figure 7B). The control genes were scarcely induced in the presence of CHX and DEX (Figure 7D). These results show that most (if not all) of the genes for nicotine biosynthesis are at least partly activated by ERF189 without nascent protein synthesis and therefore appear to be the direct targets of ERF189.
ERF189 Recognizes a GCC-Box Element in the PMT Promoter
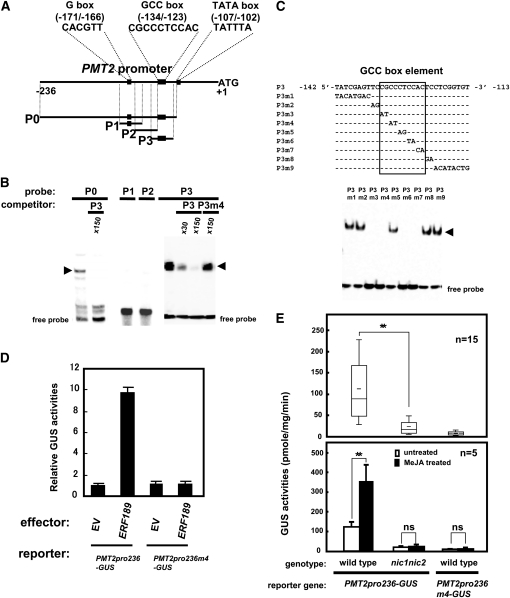
To test whether ERF189 directly binds to the promoter region of a gene for nicotine biosynthesis, a previously characterized promoter region of N. sylvestris PMT2 was incubated with a full-length recombinant ERF189 protein that had been expressed in bacteria and then affinity-purified using an N-terminal His-tag. The 236-bp promoter region of PMT2 faithfully reported the spatio-temporal expression patterns of PMT and contained two functionally important cis-elements, a G-box element and a GCC-box element (Shoji et al., 2000b; Oki and Hashimoto, 2004; Xu and Timko, 2004). We first tested a DNA probe that covered the entire promoter region upstream of the predicted TATA box (P0; −236 to –105, numbered from the first ATG) and three overlapping 30-bp probes within P0 (P1, −182 to −153; P2, −162 to −133; and P3, −142 to −113) (Figure 8A). An electrophoresis mobility shift assay (EMSA) was used to analyze the DNA–protein interaction. The P0 and P3 probes, but not the P1 and P2 probes, yielded retarded bands of the DNA-ERF189 complex, which could be competed out with excess amounts of the unlabeled P3 probe but not with its mutant version R3m4 (Figure 8B, also see Figure 8C for the P3m4 sequence). To determine specificity, the P3 sequence containing the core GCC-box element was mutated with a series of 2- or 8-bp substitutions to generate nine mutant probes (P3m1 to P3m9). No detectable binding was observed for P3m3, P3m4, P3m6, and P3m7 (Figure 8C), thus defining an in vitro ERF189 binding sequence as (5′-CGCCCTCCAC-3′) in which the central CT (underlined) is dispensable.
Figure 8.
In Vitro Binding of Recombinant ERF189 to the GCC-Box Element in the PMT2 Promoter.
(A) Schematic representation of a 236-bp N. sylvestris PMT2 proximal promoter region. Positions of the probes P0 (−236 to −105), P1 (−182 to −153), P2 (−162 to −133), and P3 (−142 to −113) are shown. Core sequences of G-box (−171 to 186), GCC-box (−134 to −123), and TATA-box (−107 to −102) are indicated in black boxes.
(B) EMSA of recombinant ERF189 in a complex with P0, P1, P2, or P3. Excess molar amounts (×30 or ×150) of unlabeled probes (P3 or P3m4) were used as competitors. The arrowheads indicate the ERF189-probe complexes.
(C) Binding of recombinant ERF189 to a series of mutant P3 probes (P3m1 to P3m9). Substituted nucleotide sequences are shown, while the unchanged sequences are indicated with dashes. A core GCC-box element is boxed.
(D) Transactivation of the PMT2pro236-GUS reporter and its mutant PMT2pro236m4-GUS. Cultured tobacco cells were bombarded with a combination of the reporter plasmid, a luciferase-expressing control plasmid, and either an ERF189-expressing effector plasmid or an empty plasmid (EV). GUS activity in the cell extracts is shown relative to the luciferase activity. The error bars indicate the sd for three biological replicates.
(E) Activity of the PMT2 promoter depends on the NIC loci and the GCC-box in transgenic tobacco roots. PMT2pro236-GUS and PMT2pro236m4-GUS reporter constructs were expressed in wild-type or nic1 nic2 tobacco hairy roots. Mean (horizontal line segment in box) and 95 (top of bar), 75 (top of box), 50 (horizontal line in box), 25 (bottom of box), and 5 (bottom of bar) percentile values of GUS activity from 15 independent transgenic lines are shown in the top panel. Five lines showing near-mean GUS activity were chosen from each group and then assayed under both untreated and MeJA-treated conditions (bottom panel). Tobacco hairy roots were treated with 100 μM MeJA for 24 h. Error bars represent sd. Significant differences were determined by Student’s t test between the indicated pairs. **P < 0.01; ns, not significant.
To assay the activation of the PMT promoter by ERF189 in vivo, a PMT2 promoter (−236 to −1) fused to the β-glucuronidase (GUS) reporter (PMT2pro236-GUS), a CaMV 35S promoter–driven luciferase reference reporter, and a CaMV 35S promoter–driven ERF189 effector (35S-ERF189) were together delivered into cultured tobacco BY-2 cells by particle bombardment and were expressed transiently. The GUS activity of the cell extracts was normalized to the reference luciferase activity and expressed as relative activity (Figure 8D). The PMT2 promoter was strongly activated (~10-fold) by the transient overexpression of ERF189 compared with the empty vector–transformed control. When the m4 mutation, which completely abolished the binding of the recombinant ERF189 to the probe in vitro (Figure 8C), was introduced into the GCC-box element of the PMT promoter, the overexpressed ERF189 could no longer activate the reporter, suggesting that the GCC-box element indeed mediated the transactivation in the assay.
We wished to characterize further the ERF189 binding GCC-box element using stable tobacco transformants. The PMT2pro236-GUS reporter, previously shown to confer transcriptional upregulation in response to the application of MeJA and root-specific expression in transgenic tobacco plants (Shoji et al., 2000b), was introduced into wild-type tobacco or the nic1 nic2 mutant to generate transgenic hairy root lines. Absolute GUS activity in the cell extracts varied among the transgenic lines, but the nic double mutant lines showed significantly lower levels of activity than the wild-type lines (P < 0.001 by Student’s t test) and had ~21% activity, on average, compared with the wild type (Figure 8E). The GCC-box mutant version of the reporter (PMT2pro236m4-GUS) was nearly inactive even in the wild-type tobacco roots. Upregulation of the GUS reporters by the application of MeJA was examined in five lines for each transgenic group, which showed GUS activity close to the average values of the group under the MeJA-untreated conditions. In contrast with the clear 3-fold increase in PMT2pro236-GUS expression in the wild type, no significant increases in GUS activities were observed for PMT2pro236-GUS in nic1 nic2 and for PMT2pro236m4-GUS in the wild type (Figure 8E). These results indicate that the NIC loci target the 0.2-kb promoter region of PMT2 for transcriptional activation, including MeJA-mediated upregulation, and that the GCC-box in the promoter region is required for the responses.
Functional Redundancy and Divergence among the IXa Subfamily ERFs
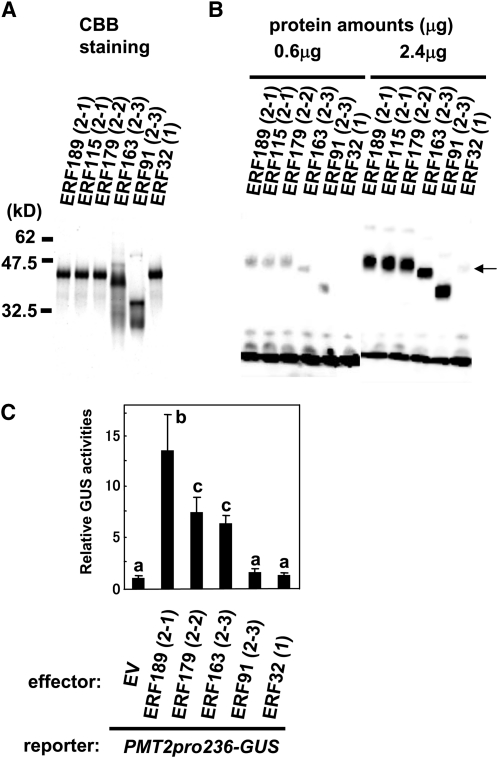
Efficient binding and activation of the GCC-box–containing PMT promoter by ERF189 raises the question of how much this transcriptional activation function is shared by other members of the IXa subfamily. Here, we selected representative members from clade 1 (ERF32), clade 2-1 (ERF189 and ERF115), clade 2-2 (ERF179), and clade 2-3 (ERF163 and ERF91) and prepared purified recombinant proteins for these ERFs (Figure 9A). EMSA showed clear binding of ERF189, ERF115, ERF179, ERF163, and ERF91 to the P3 probe derived from the PMT2 promoter (Figure 8), while ERF32 did not recognize this probe (Figure 9B). The relative binding efficiencies of ERF189, ERF115, ERF179, ERF163, and ERF91 appeared to be comparable, judging from similar band intensities with 0.6 μg of the recombinant proteins. Thus, binding to the PMT2 GCC-box element differentiates the clade 2 ERFs from the clade 1 ERF.
Figure 9.
Clade 2 ERFs Differentially Bind and Activate a PMT2 Promoter.
(A) The purity of recombinant ERF189, ERF115, ERF179, ERF163, ERF91, and ERF32 proteins was analyzed by separation on a 12% SDS-PAGE gel and subsequent staining with Coomassie Brilliant Blue (CBB). The molecular mass of marker proteins is shown on the left in kilodaltons. Clade numbers are indicated in parentheses.
(B) EMSA of recombinant ERF proteins with the probe P3 of the PMT2 promoter (see Figure 8A). Recombinant proteins were used at either 2.4 or 0.6 μg. The arrow on the right indicates a missing ERF32-P3 complex.
(C) Transactivation of the PMT2pro236-GUS reporter with various ERF effectors. Cultured tobacco cells were bombarded with a combination of the reporter plasmid, a luciferase-expressing control plasmid, and either an ERF-expressing effector plasmid or an empty plasmid (EV). GUS activities in the cell extracts were shown relative to the luciferase activities. Error bars indicate the sd for three biological replicates. Significant differences among the effectors were determined at P < 0.05 by one-way ANOVA, followed by the Tukey-Kramer test and are indicated by different letters.
Transactivation of the PMT2 promoter-driven GUS reporter (PMT2pro236-GUS) by these ERFs was examined in cultured tobacco cells as reported in Figure 8D. Among the ERFs tested, ERF189 was most effective, increasing the reporter activity 14-fold (Figure 9C). ERF179 and ERF163 also increased the GUS activity 8- and 6-fold, respectively. We did not observe significant upregulation of the reporter activities by the transient overexpression of either ERF91 or ERF32. ERF91 may be a defective activator despite its extensive in vitro binding to the PMT promoter (Figure 9B) because a conserved N-terminal region rich in Ser, Asp, and Glu, which is required for transcriptional activation in a closely related ERF, ORCA3 (van der Fits and Memelink, 2001), is absent in ERF91 (Figure 1A). These results suggest potential functional redundancy and divergence among the NIC2-locus ERF members in promoting nicotine biosynthesis.
NIC2-Locus ERF Genes Are Expressed in the Root and Induced by MeJA
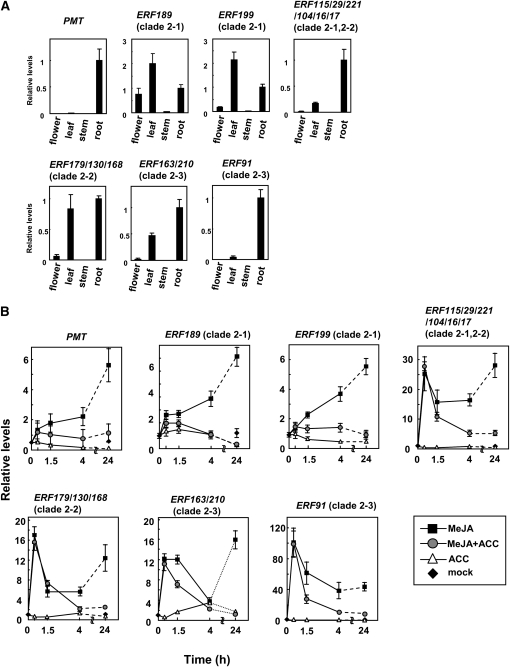
To analyze spatio-temporal expression patterns of NIC2-locus and homologous ERF genes and to see whether the expression patterns differ among them, qRT-PCR analyses were performed. First, we measured their relative expression levels in flowers, leaves, stems, and roots of 10-week-old wild-type tobacco plants. Most structural genes for nicotine biosynthesis, including PMT (Figure 10A), are expressed specifically in the root (Shoji and Hashimoto, 2010). All the ERF genes of clade 2 analyzed were substantially expressed in the root (Figure 10A; see Supplemental Figure 3 online) but were also expressed in other organs, especially in the leaf. The fact that expression of NIC2-locus ERFs is not restricted to the root indicates that these genes are necessary but not sufficient for the activation of nicotine biosynthesis-related genes. An unknown root-specific factor might work together with these ERFs to confer tissue specificity. Alternatively, an inhibitory factor might antagonize the function of NIC2-locus ERFs in the leaf.
Figure 10.
Expression Patterns of Clade 2 ERF Genes.
Transcript levels were analyzed by qRT-PCR. The error bars indicate the sd for three biological replicates. Expression patterns of PMT are also shown as references.
(A) Organ-specific expression. Expression levels in the flower, leaf, and stem of wild-type tobacco plants are shown relative to levels in the root.
(B) Responses to MeJA and ACC. Wild-type tobacco hairy roots were treated with 100 μM MeJA (black square), 100 μM ACC (white triangle), or both (gray circle) for 0.5, 1.5, 4, and 24 h. Transcript levels of mock-treated samples (black diamonds) are shown at 0 and 24 h. All values are relative to the transcript levels at 0 h.
MeJA coordinately induces the expression of most genes for nicotine biosynthesis (Imanishi et al., 1998; Shoji et al., 2000b, 2002; Goossens et al., 2003), whereas ethylene suppresses this jasmonate-mediated gene induction (Shoji et al., 2000a; Winz and Baldwin, 2001). Because many group IX ERF genes of tobacco were upregulated following jasmonate treatment (Rushton et al., 2008a), we carefully examined the time course of the induction of clade 2 ERF genes in response to jasmonate. Wild-type tobacco hairy roots were treated with MeJA, the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC), or both and cultured for up to 24 h (Figure 10B). Expression of PMT was induced at 30 min after the MeJA treatment and continued to increase for 24 h. While ACC alone did not affect PMT expression, it strongly suppressed the MeJA-triggered increase in the PMT transcript after 90 min. These response patterns of PMT resembled those of ERF189 and ERF199, which were upregulated gradually by MeJA and remained mostly uninduced when the roots were challenged with MeJA and ACC. The transcript levels of the other ERFs increased rapidly and strongly within 30 min of MeJA treatment, declined sharply thereafter, and began to increase gradually after 4 h. When amplified cDNA fragments were sequenced in the ERF115/29/221/104/16/17 qRT-PCR samples after 30 min of MeJA treatment, the cDNA population was composed of ERFs in a proportion similar to the 0-min control cDNA population (see Supplemental Figure 3 online), indicating that all ERF115/29/221/104/16/17 genes were rapidly upregulated soon after the MeJA treatment. The simultaneous treatment with ACC inhibited the second-phase gradual increase but not the initial sharp rise of the ERF transcripts. In conclusion, the MeJA-mediated induction profiles of ERF189 and ERF199 are distinct from those of the other ERFs in clade 2 and are similar to those of genes for nicotine biosynthesis.
DISCUSSION
The NIC2 Locus Comprises at Least Seven Clustered ERF Genes
In this study, we found that seven highly similar ERF genes are deleted at the NIC2 locus of tobacco in the nic2 mutant and showed that these ERF genes positively and specifically regulate the expression of structural genes involved in nicotine biosynthesis. Although ADC has been suggested to function in a nicotine biosynthesis pathway because it potentially supplies the pathway intermediate putrescine (e.g., Goossens et al., 2003), and although JAT1, another MATE family transporter, has been proposed to sequester nicotine into leaf vacuoles based on in vitro transport assays (Morita et al., 2009), neither gene is downregulated in the nic mutants or regulated by the NIC2-locus ERFs (Figures 5 and 7; see Supplemental Figures 7 and 9 online). To determine whether ADC and JAT1 are indeed involved in nicotine biosynthesis and accumulation in a NIC-independent manner, the effects of their suppression and overexpression on nicotine accumulation should be studied in tobacco roots or tobacco plants. Overexpression of ERF189 in wild-type tobacco roots increased the accumulation of tobacco alkaloids over the wild-type levels (see Supplemental Figure 8 online), indicating that NIC2 activity is not saturated in wild-type tobacco plants. Such a rate-limiting function explains the dose-dependent phenotype displayed by the semidominant nic2 mutation.
The NIC2-locus ERFs belong to clades 2-1 and 2-2 in group IXa and are derived from a diploid progenitor species, N. tomentosiformis. These and other ERF genes often form homologous pairs of almost identical sequences, in which one is derived from N. tomentosiformis and the other from N. sylvestris. Thus, there must be two corresponding NIC2 loci originating from two progenitor tobacco species in the allotetraploid tobacco, and in the nic2 mutant, the N. tomentosiformis–derived locus is mutated by a large chromosomal deletion while leaving the N. sylvestris–derived locus still functional. This interpretation is consistent with a weak phenotype of the nic2 mutation (Legg and Collins, 1971). Gene duplication at the NIC2 locus appeared to have occurred in the ancestral diploid Nicotiana species before allotetraploidization to form the cultivated tobacco. In the tobacco NIC2 locus, there exist two levels of redundancy: one caused by local gene duplications in a chromosomal region and the other originating from the allotetraploid history of the tobacco genome.
Tobacco cDNA sequence databases are fairly comprehensive but still incomplete (D’Agostino et al., 2009), and the DNA sequence information obtained by methylation filtering of the tobacco genome does not cover whole genomic regions (Rushton et al., 2008b). Therefore, we should keep in mind that several clade 2 ERF genes may not be included in the current tobacco databases. For example, ERF16 of clade 2-1 is derived from N. sylvestris (Figure 3A), but its counterpart of N. tomentosiformis origin has not been identified and might be deleted in nic2. The chromosomal deletion at the NIC2 locus thus may include more than seven ERF genes. Additionally, our present ability to map genetically the ERF genes to the NIC2 locus depended on their deletion in nic2; thus, this study does not exclude the possibility that more ERF genes (e.g., ERF163) may be present at the NIC2 locus adjacent to the deleted chromosomal region. The development of molecular markers for other potential NIC2-locus ERF genes, although the extent of genetic polymorphism is very limited among tobacco cultivars (Ren and Timko, 2001), or sequencing of the BAC clones containing the NIC2-locus ERF genes will reveal more completely the organization of the NIC2 locus.
Although we did not obtain a physical map of the clustered NIC2-locus ERF genes, it is informative to compare the corresponding loci in other Solanaceae species for which genome sequencing projects are underway. In the recently released version (2.30) of the tomato genome assembly, four tomato ERF genes that show the highest similarities to the tobacco NIC2-ERF genes (E-values below 5E-42) are tandemly arrayed within a 62-kb region of chromosome 1, and a fifth (E-value 2E-25) is located 42 kb from this cluster (International Tomato Genome Sequencing Project; http://solgenomics.net/genomes/Solanum_lycopersicum/index.pl). Apparently, tandem gene duplications of the ancestral ERF189-type gene have occurred in the Solanaceae family, and these tandem arrays may have expanded further in the Nicotiana genus.
Tandem arrays of two or more genes may frequently arise by unequal crossing over between homologous chromosomes (Gaut et al., 2007). Sequenced plant genomes contain substantial numbers of tandemly arrayed gene families, varying from a few percent to 15% or more of the identified genes (Jander and Barth, 2007; Huang et al., 2009). Most tandem arrays contain only two duplicated genes, and arrays with more than three members are rare (Rizzon et al., 2006). Although transcription factor families have much higher expansion rates in plants than in animals (Shiu et al., 2005), the Arabidopsis genome contains only one region with more than five duplicated transcription factor genes in tandem, whereas several such regions are present in soybean (Glycine max; Schmutz et al., 2010). Apparently, the clustering of more than seven duplicated transcription factor genes at the NIC2 locus is a rare event in the evolution of tobacco.
NIC2-Locus ERF Genes Function Differently in Nicotine Biosynthesis
Duplicated genes may follow three alternative outcomes at the level of expression or protein function: (1) nonfunctionalization through silencing or null mutations, (2) neofunctionalization through the gain of a novel function, and (3) subfunctionalization through the partitioning of different functional modules to each copy (Lynch and Conery, 2000). Since most duplicated Arabidopsis regulatory genes show evidence of divergent expression (Duarte et al., 2006), many plant regulatory genes may experience subfunctionalization and/or neofunctionalization. A clear example is found in a P1-rr locus of maize (Zea mays), containing genes encoding two tandemly duplicated Myb-like transcriptional activators (Zhang et al., 2000). One MYB-like gene is expressed in the kernel pericarp, cob, tassel glumes, and silk and is responsible for pigmentation in these tissues, whereas the other gene functions in the developing anther and silk.
The NIC2-locus ERF genes also experienced expression divergence. Whereas organ-specific expression patterns are not markedly different among them, with strong expression in the root and the leaf, their responses to jasmonate are divergent, with two distinct patterns (Figure 10). ERF189/199 genes are gradually upregulated over 24 h by jasmonate treatment, and their jasmonate-induced gene expression is antagonized by ethylene signaling, whereas the other ERF genes show an acute peak at 30 min after jasmonate treatment, which is insensitive to ethylene signaling. We also found functional divergence between ERFs of clade 2-1 and 2-2. Whereas the transcription factors belonging to either clade recognize the GCC-box element in the PMT2 promoter (Figure 9B), clade 2-2 ERFs were less efficient than clade 2-1 ERFs in transactivating the PMT2 promoter in a transient overexpression assay (Figure 9C) and nearly ineffective in inducing the expression of genes for nicotine biosynthesis and increasing tobacco alkaloids in transgenic roots (Figure 6). Our functional assays tentatively placed clade 2 ERFs in increasing order for promoting nicotine biosynthesis as follows: clade 2-3 (ERF210/163) and clade 2-2 (ERF168/130/170/17) << clade 2-1 (ERF115/29/221/16) ≤ clade 2-1 (ERF189/199). ERF104 (clade 2-1) and ERF91 (clade 2-3) are inactive, possibly due to their lack of a complete DNA binding domain and a potential activation domain, respectively. Subtle differences in amino acid sequences of the AP2/ERF DNA binding domain (i.e., ERF189/199 has a unique Glu-Gly insertion in the middle of the AP2/ERF domain) may partly contribute to such functional differences. Alternatively, more divergent regions outside of the conserved AP2/ERF domains may confer different efficiencies in the transcriptional activation of target genes among different clades. At any rate, when the marginal promoting effects on nicotine formation are considered, the metabolic significance of the rapid jasmonate-induced expression of clade 2-2 ERF genes is not apparent.
Gene duplications offer an opportunity for mutual regulation among the duplicated genes. Significantly decreased transcript levels of clade 2-3 ERF genes in the nic2 genotypes (Figure 2) might indicate that the expression of these genes is positively regulated by the NIC2-locus ERF genes. By contrast, we did not observe mutual upregulation of clade 2-1 and 2-2 ERF genes when one member was overexpressed in tobacco roots (Figure 6A). Further study is required to address whether there is crosstalk in the regulatory network of clade 2 ERF genes.
Tobacco NIC2-Locus ERFs Are Close Homologs of Periwinkle ORCA3
ORCA3 and another jasmonate-inducible homolog, ORCA2, of C. roseus are closely related to tobacco clade 2 ERFs in the group IXa subfamily (Figure 1B). ORCA3 was identified as a jasmonate-responsive transcription factor that, upon its overexpression, enhanced the gene expression of several, but not all, enzymes in the primary and secondary pathways for terpenoid indole alkaloids (van der Fits and Memelink, 2000). ORCA3 overexpression in C. roseus cell suspensions did not result in alkaloid synthesis but required feeding of a pathway intermediate for increased alkaloid production. A recombinant ORCA3 protein bound to the promoter fragments of three genes in the alkaloid pathway and was recruited to a GCC-box element in one of the promoters (van der Fits and Memelink, 2000, 2001). These and other results indicated that ORCA3 (and possibly ORCA2 as well) mediates the jasmonate-responsive expression of several structural genes for terpenoid indole alkaloid biosynthesis. It is remarkable that two distant plant species, tobacco (Solanaceae) and periwinkle (Apocynaceae), chose closely related transcription factor genes to control the expression of jasmonate-inducible structural genes for defensive secondary metabolites. The fact that many group IX ERF genes are upregulated by jasmonate may partly contribute to the molecular evolution of similar regulatory mechanisms in unrelated secondary metabolism.
It may be interesting to test whether the tobacco NIC2-locus ERFs and periwinkle ORCA3 are functionally interchangeable by overexpressing the tobacco proteins in periwinkle and vice versa. To suppress their functions in heterologous plant species, EAR-fused repressor versions of these ERFs should be useful in overcoming the genetic redundancy of endogenous genes, as we demonstrated in ERF189/179-EAR (Figure 5). An intriguing possibility is that homologs of NIC2/ORCA3 ERFs may be involved in the regulation of jasmonate-inducible secondary metabolism in commercially valuable medicinal plants. Biotechnological applications of these transcription factors are eagerly anticipated.
METHODS
Plant Materials
Sterilized seeds of Nicotiana species were germinated and grown to the seedling stage under continuous illumination on half-strength Gamborg B5 medium solidified with 0.3% (w/v) gellan gum and supplemented with 3% (w/v) sucrose. Two-week-old plantlets were transferred onto the soil in pots and grown to maturity in the greenhouse. The wild-type, nic1, nic2, and nic1 nic2 genotypes in the Burley 21 cultivar were obtained from the USDA, the wild-type and nic1 nic2 genotypes in the NC 95 cultivar from John Hamill (Monash University), and Nicotiana tabacum cv Petit Havana line SR1, N. sylvestris, and N. tomentosiformis from Japan Tobacco. For genetic crossing, the stigmas of nic1 nic2 (cv NC 95) were fertilized with the pollen of nic1 (cv Burley 21), and a resulting F1 plant was self-pollinated to provide an F2 segregating population. For chemical treatment of hairy roots cultured in liquid medium as described below, MeJA, ACC, DEX, or CHX solution was directly added to 3-d-old cultures to a final concentration of 100, 100, 10, or 10 μM, respectively.
Microarray Expression Analysis
Total RNA was isolated from 4-week-old wild-type and nic1 nic2 tobacco plants (cv Burley 21) using an RNeasy kit (Qiagen) and used to generate Cy3- or Cy5-labeled complementary RNA probes using a Quick-amp labeling kit (Agilent). Tobacco microarrays containing 44K 60-mer oligonucleotide probes (Agilent; product number G2519F) were hybridized in a dye-swap manner using two independent RNA samples from each genotype (four pairs of hybridization in total). Hybridized microarrays were simultaneously scanned for the Cy3- and Cy5-labeled probes with a microarray scanner G2505B (Agilent). Feature extraction software version 9.1 (Agilent) was run with the GE2-v5_91 protocol in the default setting and used to locate and delineate the hybridization signals on the array and to integrate the intensity, filtering, and normalization of individual signals.
Phylogenetic Analysis
Multiple alignments were created with ClustalW (Thompson et al., 1994) in the default setting. All positions containing gaps and missing data were eliminated from the data sets. Unrooted phylogenetic trees were constructed using the neighbor-joining algorithum with MEGA4 software (Tamura et al., 2007). Bootstrapping was performed with 1000 replications. Evolutionary distances were computed using the Poisson correction method.
qRT- PCR
Total RNA was isolated using an RNeasy kit (Qiagen) from plant samples that had been ground in liquid nitrogen and then converted to first-strand cDNA by SuperScriptII reverse transcriptase (Invitrogen) with an oligo(dT) primer. The cDNA templates were amplified using a LightCycler 480 (Roche) with SYBR Premix Ex Taq (Takara). The primer sequences are given in Supplemental Table 2 online. The thermal program was 5 min at 95°C followed with 60 cycles of 10 s at 95°C, 10 s at 55°C, and 10 s at 72°C. The specificity of the reactions was confirmed by the machine's standard melt curve method. EF1α was used as a reference gene. Each assay was repeated at least three times.
Genomic DNA Gel Blot and Genomic PCR Analyses
For the DNA gel blot analysis, 40 μg of total genomic DNA was isolated with a CTAB method (Murray and Thompson, 1980), digested with HindIII or EcoRI, and separated on 0.8% agarose gel at 20 V. After the gels were soaked in 0.25 n HCl and subsequently in 1.5 n NaOH and 0.5 M NaCl, DNA was transferred onto Hybond-N+ nylon membranes (Amersham Biosciences) in 0.4 n NaOH. To generate radiolabeled single-stranded DNA probes, the reaction mixtures contained 1.48 MBq [α-32P]-dCTP (Perkin-Elmer), 50 mM each of dATP, dGTP, and dTTP, 1× reaction buffer, 1.25 units of ExTaq DNA polymerase (Takara), 50 ng of the cDNA template, and 1 μM of a primer (ERF189, 5′-CGTCCAATCTCGCGGCGCG-3′; ERF221, 5′-TCTCCAATCTTGTGTCGCG-3′; ERF179, 5′-CCTCCAATATTGCTTCGTG-3′; ERF163, 5′-CTTCCAATCTTGTGGCGCG-3′; ERF91, 5′-GTTCCAATCCTGAGGTGCG-3′) in a total volume of 25 μL. The primers were complementary to the sense strands and synthesized single-stranded DNA probes encompassing the relatively variable N-terminal regions of the ERF genes. The reaction was performed with the following program: 32 cycles of 1.5 min at 94°C, 2 min at 50°C, and 4 min at 72°C. The blotted membranes were preincubated for 2 h and hybridized with the probes at a concentration of ~3 × 106 cpm/mL for 16 h in 0.25 M Na2HPO4, pH 7.4, 7% (w/v) SDS, and 1 mM EDTA at 65°C. The blots were washed at 65°C twice in 2× SSC, 0.1% (w/v) SDS for 10 min and three times in 0.1× SSC, 0.1% (w/v) SDS for 15 min. The hybridization signals were detected by BAS-2500 (Fujifilm). To test the specificity of the probes, the full-length coding regions of several ERFs amplified by PCR from cDNA clones (0.01 ng) were blotted on the membranes and analyzed in the same way.
For the genomic PCR analysis, genomic DNA was prepared as described previously (Edwards et al., 1991) and used for PCR with Ex Taq DNA polymerase (Takara). The thermal program was 3 min at 94°C, followed by 35 cycles of 20 s at 95°C, 20 s at 55°C, and 1 min at 72°C.
Plant Transformation
To generate the ERF189-RNAi vector, two cDNA fragments of ERF189 (corresponding to amino acid residues from 35 to 169) were placed in front of a PDK intron at BamHI and EcoRI sites and just after it at HindIII and SacI sites in a reverse orientation in the pHANNIBAL vector (Wesley et al., 2001), and the resulting RNAi expression cassette was inserted into pBI121 (Clontech) at the BamHI and SacI sites. To construct the ERF189-EAR and ERF179-EAR vectors, full-length ERF189 or ERF179 cDNA and a synthetic DNA sequence (5′-TCTAGAGCGGCCGCTTGATTTGGATCTTGAACTCAGACTTGGATTTGCTTAGAGCTC-3′) encoding an EAR motif (LDLDLELRLGFA; Hiratsu et al., 2003) were inserted into BamHI and XbaI sites and XbaI and SacI sites of pBluescript II (Stratagene), respectively, and then the constructs were inserted into the BamHI and SacI sites in pBI121. To facilitate construction, a SacI site (GAGCTC) in ERF189 cDNA was converted to GAGCAC by PCR-based site-directed mutagenesis (Hemsley et al., 1989) with primers (5′-AGAGCACGGCTCAATTTTCCCCAC-3′ and 5′-CGAGCCGCGGATTTTGAAAGCG-3′) using high-fidelity Pyrobest DNA polymerase (Takara). For the overexpression vectors, full-length ERF189, ERF199, ERF115, ERF168, and ERF179 cDNAs were cloned into pGWB2 using Gateway cloning technology (Clontech). To generate the 35S-ERF189-GR vector, the full-length ERF189 cDNA and a rat glucocorticoid receptor (GR) sequence (Miesfeld et al., 1986) were inserted at BamHI and XbaI sites, and XbaI and SacI sites of pBluescriptII, respectively, and then the fusion constructs were placed into pBI121 at the BamHI and SacI sites. To generate the PMT2pro236-GUS binary vector for stable transformation, a promoter region of PMT2 (−236 to −1, numbered from the first ATG) was amplified from Nicotiana sylvestris genomic DNA by PCR and cloned upstream of the GUS sequence in pBI101 at SalI and BamHI sites. A mutation was introduced in the GCC-box element with primers (5′-TATCGAGTTCCGATCTCCACTCC-3′ and 5′-TAATAGAGTTAAAAAATCATTACAAC-3′) as described above to construct PMT2pro236m4-GUS.
To generate transgenic hairy roots, tobacco leaf discs were infected with Agrobacterium rhizogenes strain ATCC15834 harboring a binary vector (Horsch et al., 1985). Disinfection and drug resistance selection were done on solidified Murashige and Skoog medium containing 100 mg/L kanamycin sulfate and 250 mg/L cefotaxime sodium. The selected root lines were maintained by subculturing every 2 weeks in a liquid Gamborg B5 medium with constant shaking at 80 rpm in the dark. GUS activities were fluorometrically quantified as described (Shoji et al., 2008). Cultivar Petit Havana line SR1 was used for transformation in the experiments shown in Figures 5, 7, and 10, whereas wild-type and nic1 nic2 tobacco lines of cv NC 95 were transformed in Figures 6 and 8.
Alkaloid Analysis
Alkaloids were extracted from dry samples and measured by gas chromatography as described (Shoji et al., 2009), except in Figure 4B, where the thermal program of the GC analysis was shortened as follows to analyze a large number of samples: 100°C for 1 min, 36°C/min to 150°C, 5°C/min to 170°C, and 59°C/min to 300°C.
EMSA
Full-length protein-coding regions of ERF189, ERF221, ERF179, ERF163, ERF 91, and ERF32 were cloned in pET32b (Novagen) at BamHI and SalI sites for the first three and at NcoI and XbaI sites for the last two to express recombinant ERFs fused with thioredoxin, S-tag, and His-tag in tandem at their N-terminal ends. An Escherichia coli BL21 Star (DE3) strain (Novagen) was transformed with the vectors and grown at 37°C. When the optical density at 600 nm of the culture reached 1.0, isopropylthio-β-d-galactoside was added at a final concentration of 1 mM, and the bacteria were further cultured at 16°C overnight. The recombinant proteins were purified using Ni-NTA Magnetic Agarose Beads (Qiagen), quantified with Coomassie protein assay reagent (Thermo Scientific), and stained with Coomassie Brilliant Blue R 250 after separation by 12% SDS-PAGE.
Sense (P3s for P3, 5′-TATCGAGTTCCGCCCTCCACTCCTCGGTGT-3′, its mutant variants for P3m1 to P3m8, of which mutations were shown in Figure 8C; and P3m9s for P3m9, 5′-CAGTATGTGAGTGGAGGGCGGAACTCGATA-3′) and antisense (P3as for P3 and P3m1 to P3m8, 5′-ACACCGAG-3′; and P3m9as for P3m9, 5′-TATCGAGTT-3′) oligonucleotides were annealed during gradual cooling after boiling in 50 mM Tris-Cl, pH 7.5, 10 mM MgCl2, and 1 mM DTT, and the double-stranded probes were synthesized using the Klenow fragment (New England Biolabs). To label probes, biotin was attached to the antisense oligonucleotides at their 5′-ends. The biotin-labeled DNA probes (20 fmol) and the purified recombinant proteins (2 μg) were incubated in 10 mM Tris-Cl, 5 mM KCl, 5 mM MgCl2, 1 mM DTT, 2.5% (v/v) glycerol, 0.05% (v/v) Nonidet P-40, and 0.05 μg/μL poly(dI-dC) for 20 min at 25°C using the Light Shift Chemiluminescent EMSA kit (Pierce). For competition assay, excess molar amounts (×30 or ×150) of unlabeled DNA probes were included in the above mixtures. The reaction mixtures were separated on a 5% polyacrylamide gel and blotted onto Hybond-N+ nylon membrane using Trans-Blot SD (Bio-Rad). The membranes were probed with a streptavidin-horseradish peroxidase conjugate using a Chemiluminescent Nucleic Acid Detection Module (Pierce), after which the signals were imaged with a LAS-4000 (Fujifilm).
Transient Expression Analysis
The promoter region of PMT2 (−236 to −1) was amplified from N. sylvestris genomic DNA by PCR and cloned upstream of the GUS sequence in pBI221 at PstI and BamHI sites. Appropriate mutations were introduced as described above. ERF163, ERF189, ERF179, ERF91, and ERF32 cDNAs were cloned downstream of the CaMV 35S promoter in pBI221 at XbaI and SacI sites for ERF163 and at BamHI and SacI sites for the others, after a non-sense mutation was introduced into a SacI site in ERF179 with primers (5′-AGAGCACGGCTTAATTTTCCTC-3′ and 5′-CGAGCCGCGGATTTTAAAAGCG-3′) as described above The pBI221-LUC vector harboring firefly luciferase (LUC) under the control of the CaMV 35S promoter was cotransformed as an internal standard.
Gold particles (1.6 μm Gold Microcarriers; Bio-Rad) were coated with plasmid DNA (1.5 μg for each construct) that had been purified using a Plasmid Midi Kit (Qiagen) and were suspended in ethanol. Tobacco BY-2 cells cultured as described (Nagata et al., 1992) were spread evenly on filter paper that had been placed on the culture medium solidified with 0.3% (w/v) gellan gum. The tobacco cells were bombarded with the gold particles using the PDS-1000/He Biolistic Particle Delivery System (Bio-Rad) and incubated for 16 h in the dark. Cell extracts were prepared and used to measure GUS activity (Shoji et al., 2008). The LUC activity in the extracts was measured using the Luciferase Assay System (Promega).
Accession Numbers
Sequence data of tobacco ERF genes from this article can be found in the database of tobacco transcription factors (TOBFAC) under the names cited in this article, while the others are in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At4g17500 (At ERF1), At5g47220 (At ERF2), At2g44840 (At ERF13), AJ238739 (Cr ORCA1), AJ238740 (Cr ORCA2), EU072424 (Cr ORCA3), GQ859157 (Nb ERF1), AF127242 (ODC), D28506 (PMT), AB289456 (MPO), DW001381 (AO), AF154657 (QS), AB038494 (QPT), D28505 (A622), AB286961 (MATE1), AB286962 (MATE2), AF321137 (ADC), AF321139 (SPDS), AY445582 (SAMS), U91924 (SAMDC), D63396 (EF1α), AM991692 (JAT1), and AB004323 (PMT2). Additional accession numbers can be found in Supplemental Table 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Biosynthetic Pathways for Nicotine and Related Metabolites in Tobacco.
Supplemental Figure 2. Multiple Sequence Alignment and Phylogenetic Analysis of Amino Acid Sequences of Tobacco Clade 2 ERFs, Tobacco Clade 1 ERF32, and C. roseus ORCA3.
Supplemental Figure 3. Breakdown of cDNA Species Amplified by qRT-PCR.
Supplemental Figure 4. Genomic DNA Gel Blot Analysis of Clade 2 ERF Genes.
Supplemental Figure 5. Heterozygosity of the nic2 Mutation Contributes to Nicotine Contents in the Tobacco Leaf.
Supplemental Figure 6. Nuclear Localization of ERF189-GFP Fusion Protein in Onion Epidermal Cells.
Supplemental Figure 7. Structural Genes of Nicotine Biosynthesis Are Upregulated in the ERF189-Overexpressing Lines.
Supplemental Figure 8. Overexpression of ERF189 in Transgenic Hairy Roots of Wild-Type Tobacco Increases Nicotine Biosynthesis.
Supplemental Figure 9. Transcript Levels of JAT1 Measured by qRT-PCR.
Supplemental Table 1. Tobacco Genes That Showed Lower Expression in the nic1 nic2 Roots, Compared with the Wild-Type Roots, in Our Tobacco Microarray Analysis.
Supplemental Table 2. Primer Sequences for qRT-PCR Analysis.
Supplemental Table 3. Primer Sequences for Genomic PCR Analysis.
Supplemental Data Set 1. Text File of the Alignment Used to Generate the Phylogenetic Tree Shown in Figure 1B.
Supplemental Data Set 2. Text File of the Alignment Used to Generate the Phylogenetic Tree Shown in Supplemental Figure 2B.
Acknowledgments
We thank Albrecht von Arnim (University of Tennessee), Tsuyoshi Nakagawa (Shimane University), and Kazuyuki Hiratsuka (Yokohama National University) for providing the respective plasmids pAVA393, pGWB2, and pBI221-LUC, and the USDA, John Hamill (Monash University), and Japan Tobacco for providing the Nicotiana seeds. Mitsue Fukazawa and Makoto Hayashi (National Institute for Basic Biology) are acknowledged for technical assistance in the microarray analysis. We also thank The International Tomato Genome Sequencing Consortium for the tomato genome information, and Ian Smith for English editing. This study was supported in part by a grant [Grant-in-Aid for Young Scientists (B), No. 21770046] from the Japan Society for the Promotion of Science to T.S. and Global COE program of Nara Institute of Science and Technology (Frontier Biosciences: strategies for survival and adaptation in a changing global environment) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
References
- Baldwin I.T. (1989). Mechanism of damage-induced alkaloid production in wild tobacco. J. Chem. Ecol. 15: 1661–1680 [DOI] [PubMed] [Google Scholar]
- Bednarek P., Osbourn A. (2009). Plant-microbe interactions: Chemical diversity in plant defense. Science 324: 746–748 [DOI] [PubMed] [Google Scholar]
- Cane K.A., Mayer M., Lidgett A.J., Michael A., Hamill J.D. (2005). Molecular analysis of alkaloid metabolism in AABB v. aabb genotype Nicotiana tabacum in response to wounding of aerial tissues and methyl jasmonate treatment of cultured roots. Funct. Plant Biol. 32: 305–320 [DOI] [PubMed] [Google Scholar]
- Chaplin J.F. (1975). Registration of LAFC 53 tobacco germplasm. Crop Sci. 15: 282 [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Clarkson J.J., Lim K.Y., Kovarik A., Chase M.W., Knapp S., Leitch A.R. (2005). Long-term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae). New Phytol. 168: 241–252 [DOI] [PubMed] [Google Scholar]
- Crozier A., Clifford M.N., Ashihara H. (2006). Plant Secondary Metabolites: Occurrence, Structure and Role in the Human Diet. (Oxford, UK: Blackwell Publishing; ). [Google Scholar]
- D’Agostino N., Traini A., Frusciante L., Chiusano M.L. (2009). SolEST database: A “one-stop shop” approach to the study of Solanaceae transcriptomes. BMC Plant Biol. 9: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D.L., Nielsen M.T. (1999). Tobacco: Production, Chemistry and Technology. (Oxford, UK: Blackwell Science; ). [Google Scholar]
- Deboer K.D., Lye J.C., Aitken C.D., Su A.K., Hamill J.D. (2009). The A622 gene in Nicotiana glauca (tree tobacco): Evidence for a functional role in pyridine alkaloid synthesis. Plant Mol. Biol. 69: 299–312 [DOI] [PubMed] [Google Scholar]
- De Sutter V., Vanderhaeghen R., Tilleman S., Lammertyn F., Vanhoutte I., Karimi M., Inzé D., Goossens A., Hilson P. (2005). Exploration of jasmonate signalling via automated and standardized transient expression assays in tobacco cells. Plant J. 44: 1065–1076 [DOI] [PubMed] [Google Scholar]
- Duarte J.M., Cui L., Wall P.K., Zhang Q., Zhang X., Leebens-Mack J., Ma H., Altman N., dePamphilis C.W. (2006). Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol. Biol. Evol. 23: 469–478 [DOI] [PubMed] [Google Scholar]
- Edwards K., Johnstone C., Thompson C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gately I. (2001). Tobacco: A Cultural History of How an Exotic Plant Seduced Civilization. (New York: Grove Press; ). [PubMed] [Google Scholar]
- Gaut B.S., Wright S.I., Rizzon C., Dvorak J., Anderson L.K. (2007). Recombination: an underappreciated factor in the evolution of plant genomes. Nat. Rev. Genet. 8: 77–84 [DOI] [PubMed] [Google Scholar]
- Goossens A., Häkkinen S.T., Laakso I., Seppänen-Laakso T., Biondi S., De Sutter V., Lammertyn F., Nuutila A.M., Söderlund H., Zabeau M., Inzé D., Oksman-Caldentey K.M. (2003). A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc. Natl. Acad. Sci. USA 100: 8595–8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim W.G., Sykes K.A., Hildreth S.B., Sun J., Lu R.H., Jelesko J.G. (2007). Cloning and characterization of a Nicotiana tabacum methylputrescine oxidase transcript. Phytochemistry 68: 454–463 [DOI] [PubMed] [Google Scholar]
- Hemsley A., Arnheim N., Toney M.D., Cortopassi G., Galas D.J. (1989). A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res. 17: 6545–6551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibi N., Higashiguchi S., Hashimoto T., Yamada Y. (1994). Gene expression in tobacco low-nicotine mutants. Plant Cell 6: 723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Horsch R.B., Fry J.E., Hoffmann N.L., Eichholtz D., Rogers S.G., Fraley R.T. (1985). A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Huang S., et al. (2009). The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41: 1275–1281 [DOI] [PubMed] [Google Scholar]
- Imanishi S., Hashizume K., Nakakita M., Kojima H., Matsubayashi Y., Hashimoto T., Sakagami Y., Yamada Y., Nakamura K. (1998). Differential induction by methyl jasmonate of genes encoding ornithine decarboxylase and other enzymes involved in nicotine biosynthesis in tobacco cell cultures. Plant Mol. Biol. 38: 1101–1111 [DOI] [PubMed] [Google Scholar]
- Jander G., Barth C. (2007). Tandem gene arrays: A challenge for functional genomics. Trends Plant Sci. 12: 203–210 [DOI] [PubMed] [Google Scholar]
- Kajikawa M., Hirai N., Hashimoto T. (2009). A PIP-family protein is required for biosynthesis of tobacco alkaloids. Plant Mol. Biol. 69: 287–298 [DOI] [PubMed] [Google Scholar]
- Katoh A., Shoji T., Hashimoto T. (2007). Molecular cloning of N-methylputrescine oxidase from tobacco. Plant Cell Physiol. 48: 550–554 [DOI] [PubMed] [Google Scholar]
- Katoh A., Uenohara K., Akita M., Hashimoto T. (2006). Early steps in the biosynthesis of NAD in Arabidopsis start with aspartate and occur in the plastid. Plant Physiol. 141: 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legg P.D., Collins G.B. (1971). Inheritance of percent total alkaloids in Nicotiana tabacum L. II. genetic effects of two loci in Burley21 X LA Burley 21 populations. Can. J. Genet. Cytol. 13: 287–291 [Google Scholar]
- Legg P.D., Collins G.B., Litton C.C. (1970). Registration of LA Burley 21 tobacco germplasm. Crop Sci. 10: 212 [Google Scholar]
- Lynch M., Conery J.S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Matassi G., Melis R., Macaya G., Bernardi G. (1991). Compositional bimodality of the nuclear genome of tobacco. Nucleic Acids Res. 19: 5561–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesfeld R., Rusconi S., Godowski P.J., Maler B.A., Okret S., Wikström A.C., Gustafsson J.A., Yamamoto K.R. (1986). Genetic complementation of a glucocorticoid receptor deficiency by expression of cloned receptor cDNA. Cell 46: 389–399 [DOI] [PubMed] [Google Scholar]
- Morita M., Shitan N., Sawada K., Van Montagu M.C.E., Inzé D., Rischer H., Goossens A., Oksman-Caldentey K.-M., Moriyama Y., Yazaki K. (2009). Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (MATE) transporter in Nicotiana tabacum. Proc. Natl. Acad. Sci. USA 106: 2447–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad L., Lim K.Y., Christopodulou V., Matyasek R., Lichtenstein C., Kovarik A., Leitch A. (2002). The origin of tobacco’s T genome is traced to a particular lineage within Nicotiana tomentosiformis (Solanaceae). Am. J. Bot. 89: 921–928 [DOI] [PubMed] [Google Scholar]
- Murray M.G., Thompson W.F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T., Nemoto Y., Hasezawa S. (1992). Tobacco BY-2 cells as the HeLa cells in the cell biology of higher plants. Int. Rev. Cytol. 132: 1–30 [Google Scholar]
- Nakano T., Suzuki K., Fujimura T., Shinshi H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki H., Hashimoto T. (2004). Jasmonate-responsive regions in a Nicotiana sylvestris PMT gene involved in nicotine biosynthesis. Plant Biotechnol. 21: 269–274 [Google Scholar]
- Picard D. (1993). Steroid-binding domains for regulating the functions of heterologous proteins in cis. Trends Cell Biol. 3: 278–280 [DOI] [PubMed] [Google Scholar]
- Reed D.G., Jelesko J.G. (2004). The A and B loci of Nicotiana tabacum have non-equivalent effects on the mRNA levels of four alkaloid biosynthesis genes. Plant Sci. 167: 1123–1130 [Google Scholar]
- Ren N., Timko M.P. (2001). AFLP analysis of genetic polymorphism and evolutionary relationships among cultivated and wild Nicotiana species. Genome 44: 559–571 [PubMed] [Google Scholar]
- Rizzon C., Ponger L., Gaut B.S. (2006). Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLOS Comput. Biol. 2: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M.F., Wink M. (1998). Alkaloids: Bbiochemistry, Ecology, and Medicinal Applications. (New York: Plenum Press; ). [Google Scholar]
- Rushton P.J., Bokowiec M.T., Han S., Zhang H., Brannock J.F., Chen X., Laudeman T.W., Timko M.P. (2008a). Tobacco transcription factors: Novel insights into transcriptional regulation in the Solanaceae. Plant Physiol. 147: 280–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton P.J., Bokowiec M.T., Laudeman T.W., Brannock J.F., Chen X., Timko M.P. (2008b). TOBFAC: the database of tobacco transcription factors. BMC Bioinformatics 9: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Noma M., Kawashima N. (1985). The alkaloid contents of sixty Nicotiana species. Phytochemistry 24: 477–480 [Google Scholar]
- Schmutz J., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Shiu S.H., Shih M.C., Li W.H. (2005). Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiol. 139: 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T., Hashimoto T. (2008). Why does anatabine, but not nicotine, accumulate in jasmonate-elicited cultured tobacco BY-2 cells? Plant Cell Physiol. 49: 1209–1216 [DOI] [PubMed] [Google Scholar]
- Shoji T., Hashimoto T. (2010). Nicotine biosynthesis. Plant Metabolism and Biotechnology, Ashihara H., (New York: John Wiley and Sons; ), in press [Google Scholar]
- Shoji T., Inai K., Yazaki Y., Sato Y., Takase H., Shitan N., Yazaki K., Goto Y., Toyooka K., Matsuoka K., Hashimoto T. (2009). Multidrug and toxic compound extrusion-type transporters implicated in vacuolar sequestration of nicotine in tobacco roots. Plant Physiol. 149: 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T., Nakajima K., Hashimoto T. (2000a). Ethylene suppresses jasmonate-induced gene expression in nicotine biosynthesis. Plant Cell Physiol. 41: 1072–1076 [DOI] [PubMed] [Google Scholar]
- Shoji T., Ogawa T., Hashimoto T. (2008). Jasmonate-induced nicotine formation in tobacco is mediated by tobacco COI1 and JAZ genes. Plant Cell Physiol. 49: 1003–1012 [DOI] [PubMed] [Google Scholar]
- Shoji T., Winz R., Iwase T., Nakajima K., Yamada Y., Hashimoto T. (2002). Expression patterns of two tobacco isoflavone reductase-like genes and their possible roles in secondary metabolism in tobacco. Plant Mol. Biol. 50: 427–440 [DOI] [PubMed] [Google Scholar]
- Shoji T., Yamada Y., Hashimoto T. (2000b). Jasmonate induction of putrescine N-methyltransferase genes in the root of Nicotiana sylvestris. Plant Cell Physiol. 41: 831–839 [DOI] [PubMed] [Google Scholar]
- Sinclair S.J., Murphy K.J., Birch C.D., Hamill J.D. (2000). Molecular characterization of quinolinate phosphoribosyltransferase (QPRtase) in Nicotiana. Plant Mol. Biol. 44: 603–617 [DOI] [PubMed] [Google Scholar]
- Staswick P.E. (2008). JAZing up jasmonate signaling. Trends Plant Sci. 13: 66–71 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd A.T., Liu E., Polvi S.L., Pammett R.T., Page J.E. (2010). A functional genomics screen identifies diverse transcription factors that regulate alkaloid biosynthesis in Nicotiana benthamiana. Plant J. 62: 589–600 [DOI] [PubMed] [Google Scholar]
- Valleau W.D. (1949). Breeding low-nicotine tobacco. J. Agric. Res. 78: 171–181 [Google Scholar]
- van der Fits L., Memelink J. (2000). ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289: 295–297 [DOI] [PubMed] [Google Scholar]
- van der Fits L., Memelink J. (2001). The jasmonate-inducible AP2/ERF-domain transcriptional factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant J. 26: 43–53 [DOI] [PubMed] [Google Scholar]
- Wesley S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Winz R.A., Baldwin I.T. (2001). Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. IV. Insect-induced ethylene reduces jasmonate-induced nicotine accumulation by regulating putrescine N-methyltransferase transcripts. Plant Physiol. 125: 2189–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Timko M.P. (2004). Methyl jasmonate induced expression of the tobacco putrescine N -methyltransferase genes requires both G-box and GCC-motif elements. Plant Mol. Biol. 55: 743–761 [DOI] [PubMed] [Google Scholar]
- Zhang P., Chopra S., Peterson T. (2000). A segmental gene duplication generated differentially expressed myb-homologous genes in maize. Plant Cell 12: 2311–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]