This study examines the molecular differences between sexual reproduction and apomixis (i.e., asexual reproduction through seeds). Profiling of sexually reproducing maize and apomictic maize-Tripsacum hybrids revealed that four loci involved in gene silencing via DNA methylation are dowregulated in apomictic plants. Plants deficient for two of these genes mimic aspects of apomictic reproduction.
Abstract
Apomictic plants reproduce asexually through seeds by avoiding both meiosis and fertilization. Although apomixis is genetically regulated, its core genetic component(s) has not been determined yet. Using profiling experiments comparing sexual development in maize (Zea mays) to apomixis in maize-Tripsacum hybrids, we identified six loci that are specifically downregulated in ovules of apomictic plants. Four of them share strong homology with members of the RNA-directed DNA methylation pathway, which in Arabidopsis thaliana is involved in silencing via DNA methylation. Analyzing loss-of-function alleles for two maize DNA methyltransferase genes belonging to that subset, dmt102 and dmt103, which are downregulated in the ovules of apomictic plants and are homologous to the Arabidopsis CHROMOMETHYLASEs and DOMAINS REARRANGED METHYLTRANSFERASE families, revealed phenotypes reminiscent of apomictic development, including the production of unreduced gametes and formation of multiple embryo sacs in the ovule. Loss of DMT102 activity in ovules resulted in the establishment of a transcriptionally competent chromatin state in the archesporial tissue and in the egg cell that mimics the chromatin state found in apomicts. Interestingly, dmt102 and dmt103 expression in the ovule is found in a restricted domain in and around the germ cells, indicating that a DNA methylation pathway active during reproduction is essential for gametophyte development in maize and likely plays a critical role in the differentiation between apomictic and sexual reproduction.
INTRODUCTION
Sexual reproduction in angiosperms takes place within a multicellular ovule in which the formation of the female gametes entails two consecutive steps: megasporogenesis (spore formation) and megagametogenesis (gamete formation). Megasporogenesis initiates with the differentiation of the megaspore mother cell (MMC), which undergoes meiosis, producing four haploid spores. Three of them usually degenerate, leaving a single functional megaspore. In the Polygonum type of megagametogenesis, the most common type in angiosperms, the functional megaspore undergoes three rounds of mitotic divisions to form the embryo sac (ES) containing the female gametes (the egg cell and the central cell), two synergids at the micropylar pole, and three antipodal cells at the chalazal pole (Reiser and Fischer, 1993). In male reproductive organs (the anthers), all four meiotic products form male gametophytes (pollen grains), which consist of two reproductive sperm cells embedded in the vegetative cell. The fertilization of the egg cell and the central cell gives rise to the embryo and the endosperm, respectively.
Apomixis refers to a diverse group of reproductive behaviors that result in asexual reproduction through seeds (Nogler, 1984). Apomictic plants bypass both meiotic reduction (a process called apomeiosis) and egg cell fertilization (via parthenogenesis), thus producing offspring that are exact genetic replicas of the mother plant. In the diplosporous type of apomixis, which occurs, for example, in Tripsacum (Leblanc et al., 1995), a wild relative of maize (Zea mays), or Boechera species (Naumova et al., 2001), which are related to Arabidopsis thaliana, the MMC differentiates but fails to complete meiosis and finally produces an unreduced functional megaspore. Megagametogenesis then proceeds similarly to sexual plants, but the embryo develops from the unreduced egg cell without fertilization (i.e., by parthenogenesis). In the aposporous type of apomixis, megasporogenesis is omitted and one or more ESs can develop directly from somatic nucellar cells (aposporous initials), adjacent to the surviving sexually derived megaspore. The embryo then similarly develops parthenogenetically. Development of the endosperm of apomictic plants is either pseudogamous (i.e., it arises from the fertilization of the central cell by one male sperm) or autonomous when it occurs without fertilization of the central cell.
In many apomictic species, meiotic abnormalities are not restricted to the female reproductive cells. Unreduced pollen grain formation has been reported, for example, in Boechera (Schranz et al., 2006) and Tripsacum (Grimanelli et al., 2003), suggesting that male and female reproductive paths are affected in similar ways. Also, in most if not all documented cases, apomixis and sexual reproduction are not mutually exclusive, since they usually coexist in the same ecotypes with varying levels of relative expressivity (Nogler, 1984). Most diplosporous apomicts produce both apomeiotic (unreduced) and meiotic (reduced) spores and female gametes. Similarly, in the case of aposporous apomixis, the ovule usually contains both meiotically derived and apomeiotically derived ESs. Interestingly, while the formation of multiple ESs is a hallmark of aposporous apomixis, some diplosporous plants also develop extra ES-like structures (e.g., Paspalum minus; Bonilla and Quarin, 1997). Therefore, the frontier between both types of apomixis and between apomixis and sexual reproduction appears relatively blurry, fueling the perception that apomixis might be a deregulated form of sexual reproduction rather than a new function. Indeed, previous studies showed that apomixis and sexual reproduction share key regulatory mechanisms (Tucker et al., 2003). Thus, it has been proposed that apomixis could result from a temporal or spatial deregulation of the transcriptional programs that regulate sexual reproduction (Grimanelli et al., 2003; Koltunow and Grossniklaus, 2003; Bicknell and Koltunow, 2004; Bradley et al., 2007). The model postulates that sexual reproduction involves several transitions (from somatic cells to reproductive cells, from sporogenesis to gametogenesis, from gametogenesis to embryogenesis) whose orderly progression is altered in apomicts, resulting in ectopic or heterochronic expression of the core developmental program.
Recent studies have further demonstrated the crucial role of chromatin-based regulation in vegetative developmental transitions (Poethig, 2003; Bezhani et al., 2007). Similarly, several lines of evidence suggest that transitions during reproduction and early seed development are epigenetically regulated through dynamic changes in chromatin state (Huanca-Mamani et al., 2005; Xiao et al., 2006; Baroux et al., 2007; Curtis and Grossniklaus, 2008; Olmedo-Monfil et al., 2010). Whether alterations in the epigenetic regulations that orchestrate these transitions are involved in the differentiation between apomictic and sexual reproduction is currently unknown. However, recent analyses of Arabidopsis plants defective in the ARGONAUTE9 (AGO9) gene (Olmedo-Monfil et al., 2010) suggest this might be the case. AGO9 is part of a non-cell-autonomous small RNA pathway expressed in the somatic tissues of the ovule. Mutants in AGO9 affect the specification of the precursor cells of the gametes in the Arabidopsis ovule and display a multiple-spore, aposporous-like phenotype. While the resulting spores are sterile, this is evidence that epigenome-level regulations are important to direct female germ cell development toward sexual reproduction. Also, it was recently shown that parthenogenetic embryos can be generated at a relatively high frequency in Arabidopsis transgenic lines expressing a modified centromere-specific histone CENH3 protein (Ravi and Chan, 2010). However, whether these results are illustrative of apomictic mechanisms in the wild is unclear.
In this study, we compared the expression patterns of diverse chromatin-modifying enzymes (CMEs) during reproduction in sexual and apomictic plants to identify possible chromatin-level regulators of apomixis. Two key mechanisms affecting the transcriptional competence of chromatin are covalent modifications of histone tail residues and DNA (cytosine) methylation (Vaillant and Paszkowski, 2007). DNA methylation in plants takes place on CG, CNG, and CNN nucleotide groups. In Arabidopsis, where the process is best understood, at least three classes of DNA methyltransferases (DMTs) are active in methylation pathways. DOMAINS REARRANGED METHYL TRANSFERASE2 (DRM2) is the main de novo DMT, involved in all sequence contexts (Cao and Jacobsen, 2002). DRM2 also plays a role, together with CHROMOMETHYLTRANSFERASE3 (CMT3), in the maintenance of methylation at non-CG sites (Cao et al., 2003). Another methyltransferase, METHYLTRANSFERASE1 (MET1), is involved in maintaining DNA methylation at CG dinucleotides. Finally, DECREASE IN DNA METHYLATION1 (DDM1), a SWI2/SNF2-like chromatin-remodeling factor, also participates in the maintenance of methylation at both CG and non-GC sites (Jeddeloh et al., 1999). Along with DMTs, small interfering RNAs, histone-modifying enzymes, and RNA interference (RNAi) proteins are involved in mediating both DNA and histone methylation via the so-called RNA-directed DNA methylation (RdDM) pathway (Huettel et al., 2007). The silencing effect of the RdDM pathway is reinforced by the establishment of repressive histone marks, in particular methylation of Lys-9 on histone H3 (H3K9me), by the activity of KRYPTONITE, the main H3K9 methyltransferase in Arabidopsis (Jackson et al., 2002).
Here, we show that apomictic development and sexual development differ in the expression of a few CMEs that are expressed in the maize ovule and downregulated in ovules of apomictic ecotypes. We further demonstrate that dmt102 and dmt103, homologous to CMTs and DRMs in Arabidopsis, respectively, play key roles in ovule development in maize. Loss-of-function mutants result in phenotypes that are strikingly reminiscent of apomictic development, suggesting that, in addition to a crucial role in gametophyte development, DNA methylation in the maize ovule might regulate transcriptional expression of genes involved in the differentiation between apomixis and sexual reproduction.
RESULTS
Identification of CMEs Differentially Expressed in Sexual and Apomictic Plants
To address the possible role of chromatin structure in the regulation of apomixis, we first performed a comparative analysis of transcription activity of CMEs during sexual and apomictic reproduction. We used the B73 maize inbred line as a sexually reproducing control. The apomictic plant used in this analysis, referred to as 38C, is an apomictic hybrid obtained from an original F1 plant between maize and its apomictic relative, Tripsacum dactyloides 65-1234. Ecotype 38C expresses apomixis with high penetrance (>99%). It contains one diploid set of chromosomes from maize (2n=2x=20) and one haploid set of chromosomes from Tripsacum (1n=1x=18). 38C reproduces via diplosporous apomixis: an unreduced ES is derived from a MMC and premature divisions of the unreduced egg cell result in a precocious, parthenogenetic pro-embryo, formed before fertilization. This apomictic model plant has been described in previous publications (Leblanc et al., 1996, 2009; Grimanelli et al., 2003; Grimanelli et al., 2005). This is an imperfect model for expression studies because of its very complex genetic background. However, there are no apomictic isolines of B73 (or any other maize inbred line), and 38C is, to our knowledge, the closest ecotype to B73.
We reanalyzed a set of microarray data comparing nonpollinated mature ovules of maize and 38C to identify maize CMEs expressed during sexual and apomictic development (Grimanelli et al., 2005). In addition, we included in the analysis a set of CMEs reported in The Chromatin Database (www.chromdb.org) as being expressed in maize ears (see Supplemental Table 1 online). Collectively, the data sets contained 384 loci annotated in the B73 genome as CMEs. Based on data indicative of expression within reproductive tissues (see Methods for the selection criteria), we selected 56 loci, belonging to 15 different protein families (see Supplemental Table 2 online). We then used RT-PCR to monitor, at precise stages of female reproductive development, the expression of the corresponding alleles during sexual reproduction in maize and after their introduction in the 38C apomictic background. These stages included sporogenesis (ovules were dissected to contain female meiocytes, thus including meiosis and apomeiosis), mature ESs before fertilization (ovules containing either an egg cell or a parthenogenetic pro-embryo in sexual and apomictic plants, respectively), and early embryogenesis (seeds were collected 3 d after pollination [DAP]) (see Supplemental Figure 1 online). Each candidate locus was analyzed with at least three independent biological replicates (corresponding to sampling of sexually produced and apomictic ovules from different plants), and RT-PCR assays showing potential differences were repeated at least three times with independent reverse transcriptions. Differences in expression profiles indicated that the introduction of a given maize allele in the apomictic plants resulted in altered expression at the same developmental stages and served as markers to screen for loci that are subjected to differential regulation that is potentially linked, directly or indirectly, to divergent reproductive behavior.
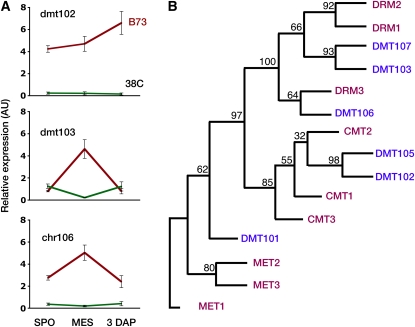
As previously described (Grimanelli et al., 2005), global expression profiles between apomictic and sexual plants differed only marginally; accordingly, we identified only six loci with clear qualitative differential expression between the two reproductive modes (see Supplemental Figures 2 and 3A online): chr106, dmt102, dmt103, dmt105, hdt104, and hon101. For all six loci, the PCR primer pairs amplified both the maize and Tripsacum alleles on genomic DNA. All primer pairs amplified mRNA in at least one combination of stage/reproductive mode, indicating that the primer pairs were able to amplify reverse-transcribed mRNAs. All six loci showed downregulation in the apomictic versus sexual materials. Four of the six loci showed downregulation at all three developmental stages, while the two remaining ones exhibited heterochronic expression. The absence of any RT-PCR product for dmt102, dmt103, and chr106 in 38C suggests that neither the maize nor the Tripsacum alleles were expressed in the apomictic ecotype. These results were corroborated using real-time PCR for dmt102, dmt103, and chr106 (Figure 1A). BLAST analyses against the Arabidopsis and maize databases together with the phylogenetic relationships among Arabidopsis and maize DMTs indicated probable functions for all six proteins (Table 1, Figure 1B; see Supplemental Data Set 1 online). DMT102, DMT103, DMT105, and CHR106 are homologous to well-characterized enzymes involved in DNA methylation in Arabidopsis. dmt102 and dmt105 encode DMT enzymes, closely related to Arabidopsis CHROMOMETHYLASEs. DMT102 is required for cytosine methylation at CNG sites, and it is likely involved in a maintenance function (Papa et al., 2001; Makarevitch et al., 2007). dmt103 is a close homolog of DRM1 and/or 2; it encodes a putative DMT that shows rearranged catalytic domains conserved in others eukaryotic proteins, but its function remains uncharacterized in maize. chr106 is homologous to Arabidopsis DDM1. All four genes were broadly expressed during sexual development in maize but totally absent during apomictic reproduction. Interestingly, dmt101, the closest homolog of the Arabidopsis MET1 gene, showed similar expression patterns during sexual and apomictic development (see Supplemental Figure 2A online). The set of deregulated genes also included hdt104, a member of the plant-specific histone deacetylase family (homologous to Arabidopsis HD2A), and hon101, a histone H1 linker protein gene.
Figure 1.
Identification of CMEs Deregulated during Apomictic Reproduction.
(A) Quantitative RT-PCR profiles of CMEs during apomictic development. SPO, sporogenesis; MES, mature embryo sac; B73, sexual maize; 38C, apomictic maize-Tripsacum hybrid. Target gene expression profiles were determined relative to gpm120 and nfe101 expression levels (see Methods). Error bars indicate se; four replicates for two different cDNA samples were used for each data point. AU, arbitrary units.
(B) Phylogenetic relationships among selected DMTs in maize (blue) and their homologs in Arabidopsis (red). Consensus tree following 1000 bootstraps. The numbers on the branches indicate the number of times the partition into the two sets that are separated by that branch occurred among the trees, out of 100 trees.
Table 1.
BLAST Analysis of Selected Maize CMEs with the Arabidopsis Protein Database
| Protein Family | Maize Locus | At Homolog | At Gene | Function | % Identity | E Values |
| SNF2 | chr106 | AT5G66750 | DDM1 | Maintenance of Cyt methylation | 63 | e-126 |
| DMT | dmt102 | AT1G69770 | CMT3 | Maintenance of CNG methylation | 50 | 0 |
| DMT | dmt103 | AT5G14620 | DRM2 | de novo methylation | 51 | e-151 |
| DMT | dmt103 | AT5G15380 | DRM1 | de novo methylation | 51 | e-149 |
| DMT | dmt105 | AT4G19020 | CMT3 | Maintenance of CNG methylation | 51 | 0 |
| HDA | hdt104 | AT3G44750 | HD2A | Histone deacetylase | 42 | 2e-51 |
| Histone | hon101 | AT2G18050 | HIS1-3 | Histone linker | 52 | e-24 |
The maize locus nomenclature is according to CHROMDB. Percentage of identity was calculated using the complete length of the predicted maize protein.
Previous studies have linked changes in DNA methylation with changes in polyploidy levels (Wang et al., 2004). It is also known that interspecific hybridizations can induce epigenetic variations (Madlung et al., 2002; Chen et al., 2008). Interestingly, both hybridization and polyploidy have been proposed as direct causes for the induction of apomixis, through unknown routes (Carman, 1997; Koltunow and Grossniklaus, 2003; Paun et al., 2006). Apomictic 38C was derived by interspecific hybridization between maize and Tripsacum, and it carries a polyploid set of chromosomes (Leblanc et al., 1996). To evaluate the possibility that interspecific hybridization, polyploidy, or both might have induced the expression patterns detected in 38C, we compared the expression of dmt102, dmt103, and chr106 in mature ESs of T. dactyloides 65-1234, the natural apomictic plant used to produce the 38C hybrid, with that observed in diploid and tetraploid sexual maize (diploid B73 inbred and tetraploid N108A stocks, respectively) and in the maize-Tripsacum 38C hybrid (see Supplemental Figure 3B online). Genes with conserved expression levels in sexual maize and the natural apomictic ecotype, but altered in the 38C interspecific ecotype, were classified as sensitive to interspecific hybridization. Similarly, genes with altered expression between diploid and tetraploid sexual maize lines were classified as ploidy dependent. We found that expression of chr106 was identical in diploid and tetraploid sexual maize lines. chr106 was also similarly downregulated in 65-1234 and 38C apomicts for both the maize and Tripsacum alleles. Thus, the deregulation of chr106 in apomicts was independent of either polyploidy or interspecific hybridization. By contrast, while dmt102 and dmt103 transcripts corresponding to maize and Tripsacum alleles were neither detected in Tripsacum nor in 38C apomictic contexts, both genes were strongly expressed in diploid maize but downregulated in the autotetraploid. Their expression might therefore be ploidy dependent. We cannot discard an additional negative transregulation by the Tripsacum genome background, but probably not as a consequence of interspecific hybridization.
Apomixis Correlates with the Downregulation of CMT3 and DRM1/2 Homologs in Both Tripsacum and Boechera
Apomixis in plants can occur by different developmental routes, and it is an open question whether the different varieties of apomictic development rely on similar or unrelated regulatory pathways. To address this, we analyzed the expression of DRM1, DRM2, CMT3, and DDM1 in growing ovules of Arabidopsis and Boechera holboellii, a diplosporous apomictic relative of Arabidopsis. We performed RT-PCR on two biological and two technical replicates of ovules dissected at the MMC and mature ES stages using primer pairs that amplified genomic fragments in both species (see Supplemental Figure 3C online). We found that both DRM genes were deregulated, though in a different manner, in apomictic Boechera when compared with sexual Arabidopsis. DRM2 was downregulated during sporogenesis in apomeiotic ovules of Boechera but not in mature ESs. DRM1, on the other hand, showed clear expression at both stages in the sexual plants but no expression in apomictic ovules. Finally, CMT3 and DDM1 had similar expression patterns in both species, suggesting differential regulation of the homologous genes in maize and Arabidopsis. However, we cannot discard a possible downregulation specifically within Boechera gametophytes that does not affect expression in the surrounding somatic tissues, and more detailed studies are required to assess the role of CMT3 and DDM1 in apomixis in Boechera. These results nonetheless show that at least DRM1 and DMR2, or their homologs, are deregulated in two unrelated apomictic species.
dmt102 and dmt103 Are Expressed in the Reproductive Cells of Maize Ovules
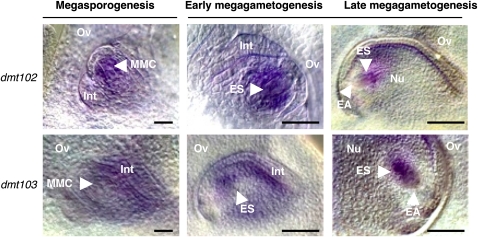
Real-time PCR and RT-PCR analyses (Figure 1; see Supplemental Figure 2 online) showed that in sexual maize, dmt102 and dmt103 are strongly expressed during early reproductive stages, including in male reproductive organs (see Supplemental Figure 3D online). To further characterize their expression patterns in reproductive tissues, we performed in situ RNA hybridization in ovules of wild-type maize plants (Figure 2). Both were expressed in a restricted portion of the nucellus during megasporogenesis. The dmt102 expression domain encompassed the reproductive cell and a few layers of nucellar cells around it (n > 25 whole-mount ovules), while dmt103 was detected only in the epidermal layer and integuments of the growing ovule (n > 25). dmt102 conserved the same spatial expression during early gametogenesis, while dmt103 expression expanded to include the ES (n > 25 each). At the end of gametogenesis, the expression of dmt102 became confined to the chalazal region of the ES area, with no detectable expression around the egg apparatus (n > 25). dmt103 signals were also restricted to the ES area, also with a strong signal at the chalazal pole, and weaker expression around the egg apparatus (n > 25).
Figure 2.
Expression Patterns of dmt102 and dmt103 during Ovule Development in Maize as Revealed by mRNA in Situ Hybridization on Whole-Mount Ovules.
EA, egg apparatus; ES, embryo sac; Int, ovule integuments; Nu, nucellus; Ov, ovary walls. Bars = 10 μm.
Downregulation of dmt102 and dmt103 in Sexual Maize Promotes the Formation of Unreduced Gametes
To further define the biological role of these CMEs in reproductive development, we wanted to analyze the phenotypes of loss-of-function lines for dmt102 and dmt103 in sexual maize. Mutant and wild-type lines were grown under identical experimental conditions (same time, same location) and analyzed for both morphological and reproductive phenotypes. For dmt102, we used a previously characterized mutant line that carries a Mutator element inserted within the DMT domain and produces an aberrant protein (Papa et al., 2001). The mutated allele originated from a highly active and genetically undefined population of Mu-active plants and was subsequently introgressed by five successive backcrossings into the B73 background. The mutant line (either homozygous or heterozygous for the dmt102::Mu alelle) had no apparent morphological or seed phenotype, and the dmt102::Mu alelle was normally transmitted via both male and female gametes in reciprocal crosses of a heterozygous line to wild-type (B73 line) plants. For dmt103, we analyzed two dmt103-specific RNAi lines, hereafter referred to as D103-2 and D103-4, produced by the Maize Chromatin Consortium (McGinnis et al., 2005) and one EMS allele (dmt103-6) recovered from the maize TILLING population that contains two nonsynonymous substitutions in the coding region of the gene (R49I and R272Q). The tilling allele was obtained in the B73 background. The RNAi lines, while produced in the A188 background, were systematically backcrossed to B73 at each subsequent generation, from T2 to T4. None of these dmt103 loss-of-function lines showed morphological phenotypes during vegetative development. The T3 progenies of both RNAi lines exhibited severe defects in seed morphology. When the RNAi line was used as female and pollinated with wild-type (B73) pollen, all seeds produced by heterozygous D103-4 T3 lines were at least partially defective (see Supplemental Figure 4 online), with miniature and aborted kernels, the latter representing 53% (±11) of the kernels (n > 500, five independent plants). Crossing heterozygous D103-2 with wild-type (B73) pollen also resulted in normal (21%, ±8), miniature (37%, ±8), and aborted (42%, ±11; five independent ears each) kernels (see Supplemental Figure 4 online). In controlled cross-pollinations between wild-type B73 plants grown in the same conditions, by contrast, 94% (±3) normal seeds (n = 500, five independent ears) were produced. This suggests at least a maternal sporophytic effect of Dmt103 inactivation when the RNAi-inducing sequence is driven by the 35S promoter. Additional gametophytic effects could not be excluded. Note that the 35S promoter is not expressed in the female gametophyte in maize. Therefore, gametophytic effects in the RNAi lines would be an indirect consequence of the sporophytic deregulation of the gene, rather than a gametophyte-specific requirement for DMT103 activity.
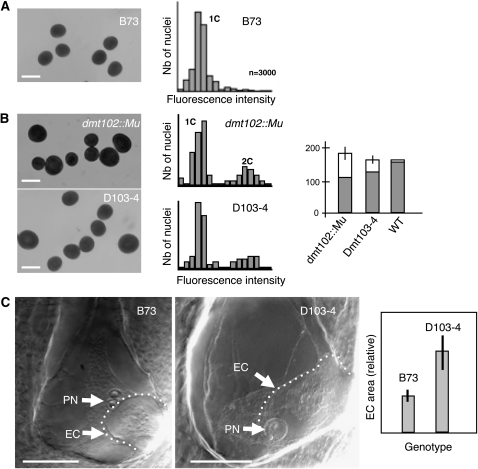
The dmt102 and dmt103 mutant lines produced abundant pollen grains following normal anther dehiscence. However, cytological analyses of male gametophytes in dmt102::Mu mutants and D103 lines revealed a high proportion of abnormally large pollen grains (i.e., ~40 and ~25% larger than the wild type, respectively) (Figures 3A and 3B). This was observed in 10 independent homozygous plants that were highly isogenic to B73 but not in any wild-type B73 plant grown under identical conditions (Figure 3A, n = 10 independent plants). To discard the possibly that additional mutations in the Mutator stock were responsible for the pollen phenotype in the dmt102::Mu line, we further analyzed wild-type and mutant siblings produced from a single heterozygous Dmt102/dmt102::Mu line (dmt102::Mu/dmt102::Mu × B73). Fifteen plants, determined as being homozygous dmt102::Mu/dmt102::Mu by genotyping, showed a clear mix of wild-type size and abnormally large pollen grains, as above. By contrast, 32 plants, either heterozygous or homozygous wild type, had pollen grain of wild-type size. The phenotype was thus only found in homozygous dmt102 mutants, suggesting that this is likely a sporophytic maternal effect rather than a gametophytic effect of dmt102 loss of function. By design, the DMT103 RNAi lines generate dominant mutations. However, similar to the female sterility phenotype, the 35S promoter is only expressed in the sporophytic tissues in male inflorescences, suggesting that sporophytic downregulation of dmt103 was sufficient to induce a pollen phenotype. To verify that large pollen size correlated with unreduced gamete formation, we quantified DNA contents in bulks of mature pollen grains by flow cytometry. Quantification of relative fluorescence intensity confirmed the production of reduced, but also unreduced and aneuploid, pollen grains. The frequency of unreduced/aneuploid pollen grains estimated by flow cytometry was similar to the frequency of large pollen grains estimated cytologically (Figure 3B; four independent flow cytometry estimates for each genotype). This indicates that downregulation of both dmt102 and dmt103 resulted in the production of unreduced male gametes.
Figure 3.
dmt102 and dmt103 Mutants Produce Unreduced Male Gametes.
(A) From left to right: morphology and 1C DNA contents in wild-type pollen grains. DNA content in pollen grains was determined by flow cytometry. The peak corresponding to haploid DNA content (1C) was determined in the B73 sample. x axis, relative fluorescence intensity; y axis, number of nuclei. Bar = 100 μm.
(B) From left to right: morphology, DNA content in mutant pollen grains from homozygous dmt102 mutant, and heterozygous dmt103 RNAi lines (the 2C peak represents unreduced and aneuploid pollen grains) and size distribution of pollen grains (gray bars, wild-type [WT] sized grains; white bars, larger grains with area more than twice that of the wild type; error bars indicate se; n > 500 for each sample). Bars = 100 μm.
(C) Left, whole-mount clearing of mature ovules of wild-type maize B73 line and D103-4. In the mutant, an abnormally large female gamete is visible at the micropylar pole. EC, egg cell; PN, polar nuclei; the dashed line delineates the egg cell. Right, quantification of size difference based on optical sections; n = 25, with 100% occurrence. Bars = 100 μm.
Whether unreduced female gametes were also produced was technically more difficult to assess. All the viable seeds produced in both mutants contained diploid embryos, as determined by flow cytometry on the resulting seedlings, when pollinated by wild-type pollen (dmt102::Mu/dmt102::Mu, n = 100; DMT103-2: n = 110; DMT103-4: n = 95; 100% diploid seedlings each). This shows that the viable embryos originated from reduced female gametes. dmt102::Mu homozygous lines produced nearly full seed sets, indicating that most, if not all, female gametes were meiotically derived. By contrast, the high proportion of abortive seeds in D103 lines might reflect endosperm defects resulting from altered genome parental dosage due to the occurrence of unreduced or aneuploid female gametes or poor viability of unreduced or aneuploid gametes (Birchler, 1993). Interestingly, cleared ovules from both D103 lines at the mature gametophyte stage exhibited surprisingly large female gametes (Figure 3C) (n = 25, with 100% occurrence). Whether they reflect nonreduction is unclear and requires further examination.
Downregulation of dmt102 and dmt103 in Sexual Maize Induces the Formation of Multiple ESs in the Ovule
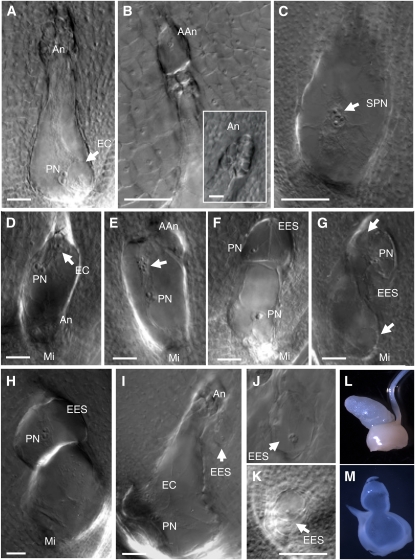
To better characterize the effect of dmt102 and dmt103 loss of function on ES development, we analyzed whole-mount ovule clearings of the mutant lines and of wild-type controls at different developmental stages. The dmt102 mutant allele had been backcrossed to B73 five time and was thus compared with B73 as the wild-type control. Both dmt103 RNAi lines (following three recurrent crosses to B73) were analyzed by comparing segregating heterozygous plants carrying the transgene to transgene-free plants originating from the same mother plants, all grown under similar conditions. Interestingly, we observed related phenotypes in the ESs of both mutants. We found that the ESs of dmt102::Mu plants exhibited normal development until cellularization (n = 50). The mature ES conserved the Polygonum-type organization occurring in maize. In particular, the three antipodal cells of wild-type maize proliferated before pollination (Figure 4A), thus producing an ephemeral structure of ~50 to 100 cells, which shares protoplasm content because of incomplete cell wall formation (Diboll and Larson, 1966). ESs in dmt102::Mu/dmt102::Mu ovules followed the wild-type behavior in terms of antipodal cell proliferation. However, at late stages (mature ES), we observed the development at the chalazal pole of large structures (in ~25% of ES, n = 86; Figure 4B), presumably originating from abnormal growth of antipodal cells and strongly resembling small uninucleated ESs, as typically seen before the first division of the functional megaspore. No further development was observed since they remained arrested as large chalazal cells until the multicellular antipodal structure degenerated. We never observed such structures in the B73 control (n = 50). This suggests that DMT102 might be involved in the maintenance of antipodal cell fate during late gametogenesis.
Figure 4.
Downregulation of dmt102 and dmt103 in Sexual Maize Induces the Formation of Multiple ESs in the Ovule.
(A) Wild-type MES. An, antipodal cells; EC, egg cell; PN, polar nuclei.
(B) MES in a dmt102::Mu line with abnormal development of antipodal cells (AAn). The caption shows wild-type antipodal development in B73.
(C) to (K) Abnormal gametophyte development in the dmt103 RNAi lines. (C) and (D) DMT103-4. (E) to (K) DMT103-2.
(C) MES with supernumerary, but normally positioned, polar nuclei (SPN).
(D) MES in inverted position showing antipodal cells at the micropylar side (Mi).
(E) MES with proliferating nuclei (arrow). Note also the absence of normal antipodal cells and the development of enlarged cells (AAn), as shown in (B).
(F) to (H) MESs showing development of two (F) or one ([G] and [H]) extra ES (EES) at the chalazal end containing clearly differentiated polar nuclei and egg cells (arrows).
(I) and (J) Development of an enlarged cell (arrow and magnified in [J]) in the nucellus.
(K) Development of multiple uninucleate gametophytes (arrows) in young ovules.
(L) to (M) Overproliferation of nucellar cells in dmt103-6 (observed in 46% of the ovaries) that extrude from the ovule; intact mature ovary prior to fertilization (L) and section at the same stage (M) showing continuity of nucellar tissues.
Bars = 25 μm.
[See online article for color version of this figure.]
In D103-2 and D103-4 lines, a similar but much more severe phenotype was found. Despite normal development of ovule primordia during sporogenesis, we observed remarkable sporophytic and gametophytic defects by the time of megagametogenesis in 29 and 43% of the ovules, respectively (Figures 4C to 4H; detailed counts for all biological replicates are provided in Tables 2 and 3). First, we noticed an incomplete ovule rotation that caused drastic changes in ovule polarity and in ES position, which frequently extruded from the nucellus or collapsed on the ovary wall (see Supplemental Figure 4B online). Furthermore, the total number of gametophytic nuclei was unpredictable. These were clustered in a central position within ESs, eventually acquiring the typical shape and large nucleoli associated with polar nuclei (Figure 4C). However, extra egg cells were not seen in individual ESs. In addition, loss of polarity of the ES was observed in morphologically normal ovules, resulting in an inverted orientation of the cells (Figure 4D). Strikingly, D103-2 individuals also showed frequent production of extra ESs, usually positioned at the chalazal pole (Figures 4E to 4K, Table 2). The number, position, and size of extra ESs varied, but the presence of differentiated egg and polar nuclei was a consistent feature of these structures. In particular, individual ESs with up to eight unfused polar nuclei were observed. Proliferating nuclei of unclear identity were also observed in atypical positions within central cells (Figure 4E), similar to what has been described in the ig1 (indeterminate gametophyte1) mutant of maize (Evans, 2007). Contrary to the ES positioned at the micropyle, the apical-basal polarity of extra ESs was usually inverted (Figure 4G). The multiple ES phenotype was clear in D103-2 but rare for D103-4 plants (Table 2). However, observation of cleared sections of dmt103-6 ovules also revealed abnormal gametophytic development, including multiple ES formation (Table 2). Collectively, this suggests that DMT103 is essential for several aspects of gametophytic development in maize, with a specific role in limiting the number of ESs in the ovule.
Table 2.
Ovule Phenotypes Observed in Individuals from dmt103 Single and Double Mutants
| Lines | No. | Phenotypes |
||||
| Wild Type | Abnormal | Aborted | Extra ESs | n | ||
| D103-2 | 4 | 28 | 6 | 0 | 4 | 38 |
| D103-2 | 7 | 10 | 0 | 4 | 0 | 14 |
| D103-2 | 11 | 22 | 12 | 2 | 7 | 43 |
| D103-2 | 16 | 25 | 2 | 0 | 2 | 29 |
| D103-2 | 22 | 22 | 3 | 0 | 1 | 26 |
| Total (%) | 107 (71) | 23 (15) | 6 (4) | 14 (9) | 150 | |
| D103-4 | 2 | 21 | 5 | 0 | 0 | 26 |
| D103-4 | 3 | 2 | 0 | 33 | 0 | 35 |
| D103-4 | 4 | 17 | 13 | 0 | 0 | 30 |
| D103-4 | 6 | 30 | 0 | 0 | 0 | 30 |
| D103-4 | 8 | 1 | 12 | 0 | 0 | 13 |
| D103-4 | 9 | 9 | 2 | 0 | 0 | 11 |
| D103-4 | 10 | 6 | 0 | 0 | 1 | 7 |
| Total (%) | 86 (57) | 32 (21) | 33 (22) | 1 (1) | 152 | |
| dmt103-6 | 1 | 3 | 15 | 0 | 3 | 21 |
| dmt103-6 | 4 | 7 | 14 | 0 | 1 | 22 |
| dmt103-6 | 6 | 5 | 12 | 0 | 4 | 21 |
| Total (%) | 15 (23) | 41 (64) | 0 (0) | 8 (13) | 64 | |
| Dmt102/dmt102::Mu D103.4 | 12 | 9 | 0 | 0 | 8 | 17 |
| % | 53 | 0 | 0 | 47 | ||
| F2#3 | 1 | 16 | 1 | 0 | 0 | 17 |
| F2#3 | 6 | 18 | 1 | 0 | 0 | 19 |
| F2#3 | 11 | 19 | 0 | 1 | 0 | 20 |
| F2#3 | 12 | 28 | 0 | 2 | 0 | 30 |
| Total (%) | 81 (94) | 2 (2) | 3 (3) | 0 (0) | 86 | |
| F2#12 | 1 | 10 | 6 | 0 | 0 | 16 |
| F2#12 | 2 | 9 | 4 | 1 | 0 | 14 |
| F2#12 | 3 | 4 | 6 | 0 | 1 | 11 |
| F2#12 | 4 | 6 | 1 | 1 | 2 | 10 |
| F2#12 | 5 | 12 | 1 | 0 | 11 | 24 |
| F2#12 | 6 | 8 | 2 | 0 | 0 | 10 |
| F2#12 | 7 | 28 | 0 | 0 | 0 | 28 |
| Total (%) | 77 (68) | 20 (18) | 2 (2) | 14 (12) | 113 | |
| B73 control | 1 | 93 | 2 | 5 | 0 | 100 |
F2#3, double mutant line (homozygous dmt102::Mu/dmt102::Mu) carrying a heterozygous RNAi transgene from DMT103-4.
Table 3.
ES Phenotypes Observed in Individuals from dmt103 Single and Double Mutants
| Lines | No. | Phenotype |
|||||
| Normal | Extra Nuclei | Inverted ESs | Abnormal Ansa | Multiple ES | n | ||
| D103-2 | 1 | 28 | 2 | 0 | 0 | 2 | 32 |
| D103-2 | 6 | 25 | 0 | 0 | 0 | 1 | 26 |
| D103-2 | 11 | 22 | 2 | 1 | 0 | 8 | 33 |
| D103-2 | 16 | 25 | 0 | 0 | 2 | 0 | 27 |
| D103-2 | 17 | 18 | 0 | 0 | 2 | 0 | 20 |
| D103-2 | 18 | 27 | 0 | 0 | 1 | 0 | 28 |
| D103-2 | 22 | 22 | 1 | 0 | 0 | 2 | 25 |
| D103-2 | 23 | 13 | 1 | 0 | 2 | 0 | 16 |
| D103-2 | 24 | 21 | 0 | 0 | 1 | 0 | 22 |
| Total (%) | 201 (88) | 6 (3) | 1 (0) | 8 (3) | 13 (6) | 229 | |
| DMT103-4 | 4 | 6 | 0 | 0 | 0 | 1 | 7 |
| DMT103-4 | 8 | 1 | 0 | 0 | 0 | 0 | 1 |
| DMT103-4 | 9 | 9 | 0 | 2 | 0 | 0 | 11 |
| Total (%) | 16 (84) | 0 (0) | 2 (11) | 0 (0) | 1 (5) | 19 | |
| Dmt102/dmt102-D103.4 | 12 | 10 (59) | 0 (0) | 0 (0) | 0 (0) | 7 (41) | 17 |
| B73 control | 1 | 100 | 0 | 0 | 0 | 0 | 100 |
Ans, antipodals.
An important question regarding the extra ESs is whether they originated from haploid gametophytic cells or from diploid somatic (nucellar) cells in the ovule. Our observations suggest that both are possible. At maturity, it was hard to differentiate ESs originating from either cell types because the supernumerary gametophytes occupied a large nucellar volume adjacent to the micropylar ES (Figures 4F and 4G), and collapsed antipodal and nucellar cells were difficult to characterize. However, a large proportion (65%) of the mature ESs seen in the most severely affected dmt103 RNAi line (D103-2) was deprived of antipodal cells (Figures 4F to 4H). Instead, and very similar to the dmt102::Mu mutant, large undifferentiated cells were localized at the chalazal pole, in the position normally occupied by the antipodal cells. This suggests that the extra ESs were derived from the cellularized gametophyte, either before or after the definition of the antipodal cells. However, we also observed (in 14% of all ovules in D103 lines) the development of large, undifferentiated cells in the nucellus, similar to uninucleate ESs in wild-type plants (Figures 4I and 4J). One or a small number of cell layers separates these undifferentiated cells from the ES proper, thus showing that these undifferentiated cells have a distinct origin from the antipodal cells. At even earlier stages, we also noticed (28%, n = 50) the formation of multiple uninucleate ESs (Figure 4K). Whether these uninucleate ESs arose from sister (haploid) megaspores or somatic nucellar cells cannot be determined due to a lack of appropriate markers. Collectively, however, the data indicate that extra ESs can be produced from both gametophytic cells and nucellar cells in the ovule. Additionally, we observed an overproliferation of nucellar tissue (Figures 4L and 4M), which suggests an important deregulation of cell activity in this tissue.
To further investigate the function of DMT102 and DMT103, we generated Dmt102/dmt102::Mu F1 individuals carrying the RNAi transgene from D103-4 (dmt102::Mu D103.4). While dmt103 inactivation in the D103-4 line resulted in a lower frequency of multiple ESs compared with that in D103-2, F1 double mutant plants exhibited an increased severity of the dmt103 RNAi phenotype, as the frequency of multiple ESs increased significantly (from 15 to 40%; Tables 2 and 3). Thus, combining loss of function for both genes significantly enhanced the phenotypic effect observed in D103-4. We next examined segregating F2 families derived from two F1 individuals (F2#3 and F2#12). We found that 78% of the plants produced unreduced male gametophytes, but without additive effects, since the proportion of unreduced gametophytes remained similar in the double or single mutants. Multiple ES development was found in 28% of the F2s and cosegregated with the RNAi transgene only (Figure 5). These data indicate that DMT102 and DMT103 likely act on a common process in the ovule, consistent with their mRNA colocalization in this tissue but that the formation of fully developed supernumerary ESs seems to be more specific of dmt103 loss of function.
Figure 5.
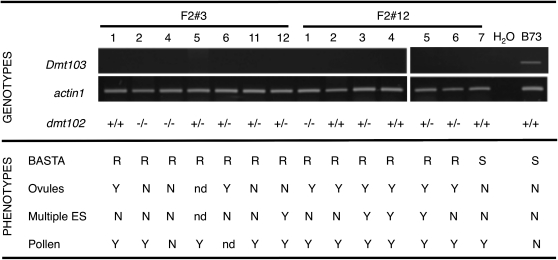
Genotypes and Phenotypes in Two F2 Progeny (F2#3 and F2#12) Derived from Dmt102/dmt102::Mu Plants Carrying the RNAi Transgene from D103-4.
RT-PCR was used to determine expression of Dmt103 in F2 plants; four technical replicates were performed for each F2 plants. Genotypes at the dmt102 locus were determined by PCR: wild type (+/+), heterozygous (+/−), or homozygous recessive (−/−). Herbicide resistance tests indicated resistant (R) and susceptible (S) plants. Ovule, ES, and pollen defects were scored as positive (Y) or negative (N), depending on whether some of the defects observed in single dmt- mutants were observed. nd, not determined.
Patterns of Chromatin Modification in dmt102 Mutant Lines Mimic the Effect of Apomixis during Sporogenesis and Gametogenesis
Both DMT102 and DMT103 are predicted to function as DMT proteins. We were therefore interested in comparing DNA methylation patterns in the reproductive cells of sexual, mutant and apomictic plants. The restricted domains of expression of both dmt102 and dmt103 within the ovule, however, rendered chromatin immunoprecipitation or related experiments technically challenging. Alternatively, we used immunolocalization experiments to determine chromatin states during sporo- and gametogenesis by determining the global distribution of two antagonist marks, dimethylation of H3K9 (a repressive mark) and acetylation of H3K9 (a permissive mark). Previous studies in Arabidopsis demonstrated the close interplay of DNA and histone K9 covalent modifications (Jackson et al., 2002; Soppe et al., 2002; Tariq et al., 2003; Lindroth et al., 2004; Mathieu et al., 2005; Johnson et al., 2007). For these experiments, we used the stable dmt102::Mu mutant line rather than the D103 RNAi lines, reasoning that the relative instability of RNAi induction might result in excessive variability when comparing individual ovules.
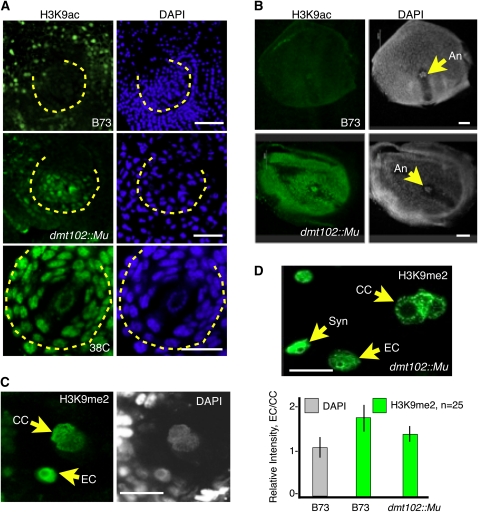
H3K9 acetylation (H3K9ac) patterns differed markedly between wild-type and dmt102::Mu maize ovules. During sporogenesis in wild-type ovules, the reproductive cells, as well as the surrounding somatic cells, were conspicuously deprived of H3K9ac signals (Figure 6A, n > 25). This pattern followed closely the spatial localization of dmt102 mRNA, as observed by in situ hybridization analysis at the same developmental stage (Figure 2). By contrast, H3K9ac signal in the dmt102::Mu line was visible in the reproductive cells and the surrounding somatic cells (Figure 6A, n > 25). Thus, DMT102 is necessary to maintain a deacetylated H3K9 state in a spatially limited domain of the ovule, which contains the reproductive cells and expresses dmt102 in sexually producing maize. Strikingly, the pattern seen in the dmt102::Mu mutant line was also observed during apomeiosis and gametogenesis in apomictic 38C plants (Figure 6A, n > 25), with the spatial domain encompassing the archespore/MMC similarly hyperacetylated.
Figure 6.
Patterns of H3K9 Acetylation and Dimethylation in dmt102::Mu Mutant Lines Mimic the Effect of Apomixis during Sporogenesis and Gametogenesis.
(A) H3K9 acetylation in the ovule during sporogenesis. The dashed lines indicate the nucellar domain enclosed within the inner integuments. Bars = 10 μm.
(B) H3K9 acetylation in mature ovules. An, antipodal cell nuclei.
(C) and (D) H3K9 dimethylation in ESs of wild-type (C) and mutant (D) plants. EC, egg cell nucleus; CC, central cell nuclei; Syn, synergid nucleus. Quantification of signals intensity measures the ratio of egg cell to central cell signal per unit of DNA to take into account the dihaploid nature of the central cell. The same measure performed for DAPI signals produced the expected 1:1 ratio. The quantification shows a significant reduction in relative H3K9me2 in the egg cell in the mutant plant (n = 25 for each; error bars indicate se). No difference in signal intensity was detected in the synergids between the wild type and mutants.
Patterns of H3K9me2 in wild-type B73 and dmt102::Mu ovules were similar during gametophyte development (see Supplemental Figure 5 online). However, H3K9me2 signal in dmt102::Mu mature ESs differed from that observed in the wild type (Figures 6C and 6D, n = 25). In wild-type mature ESs, H3K9me2 signal was enhanced in the egg cell compared with the central cell. By contrast, in dmt102::Mu, H3K9me2 in the egg cell was significantly reduced relative to the central cell, suggesting that DMT102 activity is important for maintaining a repressive state of egg cell chromatin.
Differential Expression of CMEs in Apomictic and Sexual Plants Correlates with Different Transcription Patterns in the Mature ES and the Early Seed
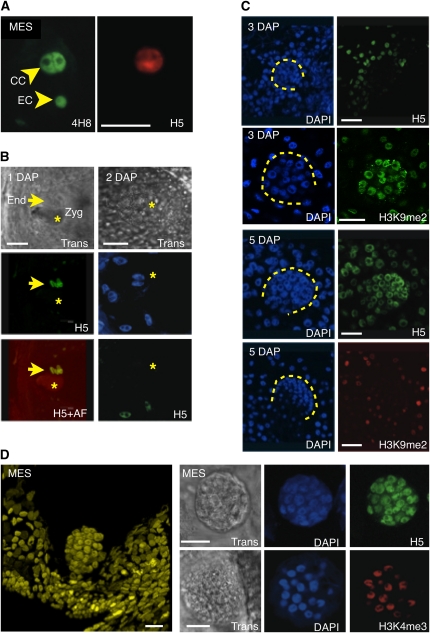
The differences detected at the level of histone marks suggested that apomictic and sexual plants likely differ in transcriptional activity in and around the germ cells in the ovule. To test this prediction, we analyzed global transcriptional patterns in ovules and early seeds of sexual and apomictic plants (see Methods and Supplemental Figure 6 online). Staining of maize ovules containing a mature ES with 4H8, an antibody against the heptamer repeats in the C-terminal domain of the main subunit of RNA POLYMERASE II (POLII), showed that most cells within the ovule contained some degree of POLII (n > 25), including the egg cell and the central cell (Figure 7A). However, a clear difference could be observed between the two cell types when immunostained with H5, which recognizes the same heptamers as 4H8 when phosphorylated on Ser-2. Phosphorylation of Ser-2 occurs concomitantly with transcript elongation; therefore, H5 is a mark of POLII molecules engaged in transcription (Palancade and Bensaude, 2003). While central cells showed strong staining for both 4H8 and H5, egg cells failed to produce a signal detectable above background level with H5 (Figure 7A, n > 25). Similar results were obtained with antibodies against H3K9ac and H3K4me3, two marks also broadly associated with a transcriptionally competent chromatin state. This is consistent with the pattern of the repressive mark, H3K9me2, indicating a much higher level of repressed chromatin in the egg cell than in the central cell (Figures 6C and 6D). Thus, complementary information suggests that the two gametes in the ES have distinct transcriptional activity, with an active central cell and a more quiescent egg cell coexisting in the ES.
Figure 7.
Transcription Patterns in ESs and Early Seeds of Maize and Apomictic 38C.
(A) Whole-mount immunostaining of 4H8 and H5 in mature maize ES prior to fertilization shows a sharp contrast between the egg cell (EC) and central cell (CC); both contain similar amounts of POLII, as revealed by the 4H8 antibody, but different amounts of the phosphorylated isoform, which is labeled with H5.
(B) Immunolocalization of H5 at 1 (left) or 2 (right) DAP in maize results in strong signals in the rapidly dividing endosperm nuclei (arrow) but undetectable signal in the zygote (Zyg; star), visible with transmitted light (Trans) or by autofluorescence (AF). Note that at 2 DAP, no clear signal is yet visible in the two nuclei of the zygote prior to the fist division.
(C) Immunolocalization experiments with H5 and H3K9me2 antibodies in developing maize seeds at 3 and 5 DAP, respectively, suggests a relative quiescence in the embryo (dashed lines) at 3 DAP and a dramatic increase in POLII activity at 5 DAP. Concomitantly, a sharp decrease in H3K9me2 is observed between 3 and 5 DAP.
(D) Whole-mount confocal imaging of a DAPI-stained parthenogenetic proembryo in the unfertilized ES of a 38C apomictic hybrid (left). Immunolocalization of H5 and H3K4me3 in proembryos indicates active transcription and a transcriptionally competent chromatin state. Trans, transmitted light observed with differential interference contrast optics.
Bars = 10 μm.
To determine whether these two patterns were maintained following fertilization, we probed 1 and 2 DAP seeds from wild-type B73 plants. Patterns of H5 (Figure 7B) were similar at 1 and 2 DAP, consistent with the pattern observed prior to fertilization: the dividing endosperm nuclei produced a strong signal, which was not detected in the zygote. This indicates that the egg cell-to-zygote transition in sexual maize occurs without massive transcriptional activation of the embryo genome. To determine more precisely the timing of zygotic genome activation, we probed growing seeds at various stages, from 3 to 6 DAP, with the same antibodies. The results showed that most embryos at 3 DAP (>90%, n > 50) had staining patterns similar to those of the zygote (Figure 7C), suggesting that zygotic transcriptional quiescence lasted at least until 3 DAP. However, a striking contrast was visible with most (>90%, n > 50) embryos at 5 DAP, where most if not all cells in the embryo showed strong H5 (Figure 7C) and H3K4me3 staining. Consistent with these observations, H3K9me2 staining showed a marked reduction at 5 DAP compared with 3 DAP (Figure 7C). These data collectively suggest that between 3 and 5 DAP, the embryo genome in sexual maize undergoes massive chromatin changes involving the release of repressive marks and global transcriptional activation.
To further compare apomictic and sexual plants, we looked at transcriptional activity in parthenogenetic embryos of the apomictic 38C ecotype. Pro-embryos in 38C develop precociously as part of the maturation of the ES. Thus, the mature ES contains a pro-embryo, arrested at the 16- to 64-cell stage, and an unfertilized central cell (Figure 7D). These precocious, arrested proembryos resume development after fertilization of the central cell. Immunostaining of H5 and H3K4me3 (Figure 7D) showed a strong signal in all proembryos (n > 50) regardless of their size (from 16 to ~60 cells). These results demonstrate that the precocious parthenogenetic embryos in mature ESs of apomictic 38C plants are transcriptionally active, by contrast to the gametes in mature ESs of sexual maize plants.
Parthenogenetic Pro-Embryos Have Lost Gametic Identity but Have Not Established Embryonic Patterning
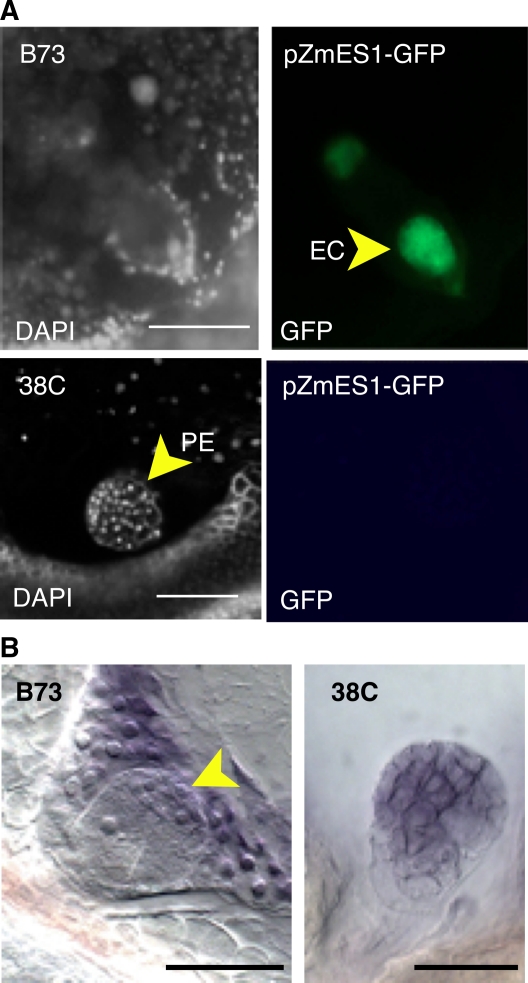
During embryo development, specific patterns of gene expression are tightly regulated in a spatial and temporal manner to either maintain or initiate changes in cell fate and development. We were thus interested in testing whether the differences that we observed between apomictic and sexual development correlated with changes in the expression patterns of gametic or embryonic cell identity markers. In particular, it remained unclear based on the above results whether the precocious embryo is an embryo or an overproliferating gamete. We analyzed the expression of a reporter gene consisting of the promoter region of the maize embryo sac1 (Zm ES1) gene fused to the green fluorescent protein (GFP) gene (Cordts et al., 2001; Figure 8A). In sexual maize, ES1 is specific to the egg apparatus, and its expression decreases strongly immediately after fertilization (Cordts et al., 2001). In the apomictic ecotype, expression was visible in the ES at the stage when the apomictic putative gamete consisted of a single cell but was absent in apomictic pro-embryos (n = 20; Figure 8A). This indicates that pro-embryos lost egg cell identity as they transitioned from unicellular to multicellular structures. To verify that these multicellular structures acquired proper embryonic identity, we further used in situ hybridization to localize transcripts of maize WUSCHEL-related homeobox2 (Zm Wox2) in parthenogenetic and sexual pro-embryos (Figure 8B). Zm Wox2 has been previously characterized in maize (Nardmann et al., 2007) and shows a pattern similar to its Arabidopsis homolog, WOX2: it is expressed specifically in the apical cells of the pro-embryo and marks the acquisition of clear apical-basal polarity in the embryo proper. Here, we found that Zm Wox2 expression was not specific to the apical cells in the apomictic pro-embryo but rather encompassed the entire pro-embryo (Figure 8B). This suggests that the parthenogenetic pro-embryos have not acquired the organization typically found in the sexual embryo.
Figure 8.
Cell Identity in Parthenogenetic Pro-Embryos.
(A) Expression of pZmES1-GFP in sexual maize and in apomictic 38C. EC, egg cell. pES1-GFP is specific to the egg apparatus in maize but absent from the apomictic pro-embryo (PE).
(B) Whole-mount in situ mRNA hybridization of Zm Wox2 probes in sexual maize early embryos and 38C pro-embryos. The arrowhead indicates the apical signal in the maize embryo. A signal is visible in the background of the sexual embryo and corresponds to Zm Wox2 expression in the endosperm. No such signal is visible in the apomictic ovule, which does not contain endosperm at that stage.
Bars = 10 μm.
DISCUSSION
By comparing transcription profiles of a diverse set of maize CMEs in sexual and apomictic ovules, we identified a limited set of enzymes that showed qualitative expression differences between the two reproductive strategies. Four of these CMEs are either predicted (CHR106, DMT103, and DMT105) or have been shown (DMT102; Papa et al., 2001) to regulate DNA methylation. CHR106 shares close homology with the Arabidopsis DDM1. DMT102 and DMT105 are both closely related to CMT3, while DMT103 is highly homologous to DRM2 proteins. DDM1 is involved in the maintenance of methylation at both CG and non-CG sites, while CMT3 and DRM2 act redundantly to maintain non-CG methylation. Finally, de novo methylation is also a function of DRM2 in all sequence contexts (Cao and Jacobsen, 2002; Cao et al., 2003). Among the many different families of CMEs tested, the differentially expressed genes were not selected based on predicted functions; therefore, it is remarkable that most of them function, at least theoretically, in DNA methylation.
It has been suggested that apomixis might be a consequence of epigenomic shocks, such as interspecific hybridization and polyploidization, resulting in a broad deregulation of reproductive development. Because our experimental model consists of an apomictic hybrid of polyploid nature (as virtually all known apomictic plants) and interspecific origin, some or all the variation detected in gene expression patterns might be only indirectly related to apomixis. However, three arguments suggest otherwise. First, a similar deregulation pattern of DRM genes occurred in the two models we tested, T. dactyloides and B. holboellii, indicating that this deregulation might denote a conserved feature among apomictic plants. Second, deregulation affected genes with clear reproductive expression, as dmt102 and dmt103 transcription domains in maize ovules were confined to the germ cells and a few surrounding somatic nucellar cells. Finally, we also found that individual or combined downregulation of both genes resulted in phenotypes highly reminiscent of apomictic development. DMT102 loss of function resulted in the production of unreduced male gametes and in abnormal patterns of antipodal cell differentiation. It further resulted, as in apomictic 38C, in a hyperacetylated state of H3K9 in the archesporial domain of the ovule. Similar but more severe phenotypes were observed for dmt103 mutant lines, which additionally resulted in strong defects in ovule development, including ESs with supernumerary gametes, formation of extra ESs, possibly of both somatic and gametophytic origin, unreduced male gametes, and possibly unreduced female gametes. The presence of multiple ESs suggests that DMT103, and possibly DMT102, function in nucellar cells to ensure that a single gametophyte develops within each ovule. In addition, the occurrence of either differentiated or undifferentiated extra nuclei indicates that they might also regulate both cell identity and cell proliferation patterns during megagametogenesis. In addition to this novel role in gametophytic development, observation of unreduced gametes and supernumerary ESs is strikingly reminiscent of apomictic reproduction. Therefore, we propose that the inactivation of this specific set of genes might represent a crucial difference between apomixis and sexual reproduction. Further work will be required to demonstrate that restoring the activity of this set of genes in an apomictic background can reinstall sexual reproduction.
These phenotypes are somehow reminiscent of what is observed in loss-of-function alleles of AGO9 in Arabidopsis. ago9 mutations affect the specification of the precursor cells of the gametes in the Arabidopsis ovule in a dominant way, resulting in the production of multiple (probably diploid) spores that express ES identity markers (Olmedo-Monfil et al., 2010). These results suggest that a small RNA pathway dependent on AGO9 activity is essential to maintain a unique germ cell in the ovule, similar to our observations for dmt103. Whether these results point to convergent pathways in Arabidopsis and maize is unclear, and no association between DMT and the AGO9-dependent pathway has been reported. However, it has been shown that CMT3, which is homologous to dmt102, has a specific role in mediating both euchromatic and heterochromatic silencing in Arabidopsis germ cells (Pillot et al., 2010a, 2010b). Analyzing the phenotype of a loss of function in the maize homolog of AGO9 would therefore be a logical follow up.
Interestingly, the extra ES phenotype observed in dmt103 mutants is suggestive of aposporous development, which is not found in diplosporous Tripsacum. It has been an open question whether the different modes of apomictic development are genetically related. The strongest argument for a close relationship between apospory and diplospory is the coexistence of both forms in some species, for example in P. minus (Bonilla and Quarin, 1997). While it is possible that mutations allowing both developments might have accumulated independently in the same ecotypes, our data support the more parsimonious hypothesis that deregulating DNA methylation in reproductive cells participates in both phenotypes.
The expression domains of dmt102 and dmt103 together with the reproductive phenotypes of the mutants suggest the specialization, at least in maize, of a DNA methylation pathway acting in the germ cells. This is again reminiscent of the AGO9-dependent pathway in Arabidopsis, which is germ cell specific (Olmedo-Monfil et al., 2010). DMT102 in maize and the Arabidopsis homologs of DMT103 are involved in non-CG methylation (Papa et al., 2001; Vaillant and Paszkowski, 2007). Interestingly, DMT101, the maize homolog of MET1 (the key enzyme for the maintenance of methylation at CG sites), showed no difference in expression pattern in sexual and apomictic ovules. Although a direct effect on non-CG methylation of DMT103 still requires confirmation, this points to a specific role for non-CG methylation, and our results suggest that, during apomictic development in Tripsacum, a small ovule domain containing the reproductive cells possibly endures alterations in DNA methylation patterns at non-CG sites. Additionally, DRM2 is the main de novo methyltransferase in all sequence contexts, and we cannot exclude the possibility that de novo methylation activity might also be affected in apomictic progeny. Indeed, previous data showed that apomictic reproduction faithfully reproduces the genome through generations but often fails to properly replicate DNA methylation patterns (Leblanc et al., 2009).
Comparative cell-specific analysis of chromatin states within ovules revealed differences between sexual maize on the one hand and dmt mutants and apomictic 38C hybrids on the other. First, we observed that, in contrast with sexual maize, apomictic 38C plants and dmt102::Mu lines suffered ectopic H3K9 hyperacetylation in a nucellar domain overlapping that of dmt102 transcription. While changes in H3K9ac are likely an indirect consequence of DMT102 loss of function or apomixis, it represents a well-established mark of transcriptionally competent chromatin. Thus, this indicates that transcriptional activity in and around the germ cells in the ovule likely differs between apomictic and sexual plants. Second, transcriptional activity in the early sexually produced embryo and the parthenogenetic embryo at a similar stage of development strongly differed. Consistently with the aforementioned H3K9 hyperacetylation, methylation states at H3K9 and H3K4 indicated that the mature female gamete and the early seed in sexual maize are relatively quiescent until at least 3 DAP, when a major burst of transcriptional activity takes place. By contrast, parthenogenetic embryos showed active transcription at early stages. We previously demonstrated using microarray analysis (Grimanelli et al., 2005) that the mRNA populations present in ovules containing a parthenogenetic embryo and in sexual maize ovules are essentially similar. This showed that transcriptional activity in precocious pro-embryos was not the result of an early zygotic genome activation. Furthermore, this heterochronic activation of transcription does not correspond to the prolongation of a gametic program, as shown by the loss of egg cell-specific marker expression, and likely interferes with the correct expression of embryo patterning genes, such as Zm Wox2. That the same difference in transcriptional activity occurs at earlier stages of development was indirectly suggested by the differences observed for H3K9ac during sporogenesis. Although technically difficult, directly determining the extent of transcriptional repression during reproductive development remains critical to the understanding of both sexual and apomictic reproduction.
What could be the role of a relative quiescence in the reproductive cells? In animals, POLII-dependent transcription is repressed in the germ cells, a phenomenon that involves both inhibition of phosphorylation in the CTD of POLII and large-scale chromatin remodeling. This repression is thought to be important for the establishment and maintenance of germ-line fate, by preventing somatic differentiation (Seydoux and Braun, 2006). When transcription is not repressed, germ cells do not form, due to the transformation of their precursors into somatic cells. Transcriptional repression may also be a key factor in establishing transcriptional profiles compatible with totipotency (Seydoux and Braun, 2006). Plants do not have germ lines as such. Rather, somatic cells switch programs late during development to produce reproductive cells. Furthermore, the products of meiosis, which directly develop into gametes in animals, undergo additional cell division and differentiation steps to form multicellular gametophytes that contain the gametes. Despite these differences, recent data indicate that animal and plant germ cell development share mechanistic similarities, as illustrated by the essential silencing function of DDM1 in plant male gametes, which is reminiscent of PIWI-interacting RNA pathway function in animals (Slotkin et al., 2009) and the importance of ARGONAUTE protein members in both systems (Nonomura et al., 2007; Yin and Lin, 2007). Thus, is a relative transcriptional quiescence required for (sexual) plant female gamete definition? Here, we observed that, in contrast with sexually produced embryos, parthenogenetic embryos, which develop without fertilization, are transcriptionally active. Whether continuous transcription is sufficient to induce parthenogenesis is unclear; such a gain-of-function phenotype does not offer clear functional evidence. Yet, the data on both dmt102 and dmt103 mutants suggest that both gametophyte and egg cell development require transcriptional quiescence.
The genetic mechanism(s) underlying apomictic development in plants remains undetermined. Specific genes or alleles that regulate apomixis expression in any plant species have yet to be discovered. Although important advances have been made recently with the engineering of apomeiotic genotypes in Arabidopsis (Ravi et al., 2008; d’Erfurth et al., 2009), it is still unclear whether these phenotypes are related to apomixis in wild species. Furthermore, the genotypes generated so far in Arabidopsis alter sporogenesis without affecting gametogenesis or embryogenesis. Here, we took the reverse approach and analyzed apomixis regulation in an artificial interspecific hybrid as well as in natural ecotypes. Our data suggest that the downregulation of a reproductive RdDM-like pathway can result in alterations in both sporogenesis (which gives rise to unreduced gametes) and gametogenesis (which gives rise to extra ESs). This is in line with the fact that apomeiosis and parthenogenesis in Tripsacum are genetically linked (Leblanc et al., 2009). On the other hand, neither dmt102 nor dmt103 inactivation resulted in the production of fully apomictic maize progeny. To validate our hypothesis that downregulation of an ovule-specific chromatin-based silencing pathway in maize would result in apomictic reproduction, it will be necessary to identify the other members involved in this pathway and to explore the effects of their inactivation either individually or simultaneously.
METHODS
Plant Materials
The 38C apomictic maize-Tripsacum hybrid has been previously described in detail (Leblanc et al., 1996, 2009; Grimanelli et al., 2003). Transgenic version of 38C expressing pES1-GFP were produced as described by Leblanc et al. (1996) using a pZmES1-GFP maize (Zea mays) line provided by Thomas Dresselhaus (University of Regensburg, Germany). Plants carrying a dmt102::Mu defective allele (zmet2-m1::Mu; Papa et al., 2001) were provided by S.M. Kaeppler. They were recurrently backcrossed to B73 for five generations and selfed. In all experiments, we used seeds derived from a single dmt102::Mu/dmt102::Mu plant. The dmt103 RNAi materials (D103-2 and D103-4) were generated by the Maize Chromatin Consortium (for vectors and procedures, see McGinnis et al., 2005). The lines were backcrossed three times to B73 prior to analysis. These lines, the dmt103-6 tilling allele, stocks for the B73 maize inbred line, and N108A (autotetraploid line derived from B73) were provided by the Maize Genetic Cooperation Stock Center. The Tripsacum dactyloides 65-1234, which was used to derive 38C, is maintained at Centro Internacional de Mejoramiento de Maiz y Trigo, Mexico. Experiments in Arabidopsis thaliana were performed with the Columbia-0 ecotype. The apomictic Boechera holboellii accession was obtained from Enrico Perotti (Australian National University, Canberra).
Genotyping
The genotype of the dmt102 locus was determined by PCR using a Mu-specific primer and a dmt102 internal primer as described by Papa et al. (2001). To genotype dmt103 RNAi lines, the herbicide resistance gene present in the transgenic construct (McGinnis et al., 2005) was used. First, a pool of T2 plants was screened for herbicide resistance and then verified by PCR for the presence of the transgene. Hemizygous plants were then maintained by crossing transgenic T2 plants as male or female progenitors with B73 plants. All subsequent analyses were performed in the resulting T3 to T5 segregating progeny obtained by recurrent backcrosses to B73, following the same procedure (herbicide resistance and transgene genotyping). Genotyping of the TILLed dmt103-6 allele is described elsewhere (http://genome.purdue.edu/maizetilling/). Primer sequences for genotype analysis are listed in Supplemental Table 3 online.
Selection of CMEs for RT-PCR Analysis
To identify CMEs expressed during sexual and/or apomictic ovule development, a set of microarrays from which global transcription profiles were previously reported (Grimanelli et al., 2005) was reanalyzed. The original experiment compared differential expression between ovules at the mature ES stage in apomictic and sexual plants. The results of BLAST searches were used to identify probes in the arrays corresponding to CMEs expressed either in sexual plants, apomictic plants, or both. Out of these, all loci (11) showing putative differential expression between the two reproductive modes were selected. Next, The Chromatin Database (www.chromdb.org) was used to identify an additional set of 45 genes with available RNA expression patterns that suggested higher expression in reproductive tissues than in vegetative parts. ID, CHROMDB accession numbers, and the corresponding protein classification are shown in Supplemental Tables 1 and 2 online.
RT-PCR
Immature ovaries were collected and carefully dissected to isolate ovule tissues. Bulks of isolated ovules were directly frozen in liquid nitrogen for further nucleic acid extraction. To determine the developmental stage within bulks (see Supplemental Figure 1 online for maize), small samples were set apart prior to freezing and cleared using benzyl:benzoate solutions (for maize and Tripsacum; Leblanc et al., 1995) or Herrs’s solution (for Arabidopsis and Boechera) to precisely determine the developmental stage of each sample. RNA extractions were performed with Trizol reagent (Invitrogen) followed by DNase treatment (DNase I; Invitrogen). RNA was reverse transcribed using the Superscript III RT-PCR kit (Invitrogen), following the provider’s instructions. For PCR, we used 10-μL reactions containing 1 μL of cDNA, 2 μL of ReadyMix Taq PCR reaction mix (Sigma-Aldrich), 1 μL of 10 μM each forward and reverse primers, and 5 μL Millipore water. PCR cycling program included 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, followed by an additional cycle of 2 min at 72°C. Amplification products were visualized using ethydium bromide and 1.5% agarose gels. For all experiments, maize actin1 or Arabidopsis ACTIN11 was used as a cDNA amplification control. Primers were designed to span an intron when possible (see Supplemental Table 3 online) to identify genomic DNA contaminations. Primer sequences are provided in Supplemental Table 3 online. To prevent cross-amplification of highly homologous sequences during PCR, the identity of the chr106, dmt102, dmt103, dmt105, hdt104, and hon101 amplification products was verified by direct sequencing.
Real-Time PCR
RNA was isolated from ovules at three developmental stages as described in the text (see Supplemental Figure 1 online), quantified by NanoDrop ND-1000 (NanoDrop Technologies), and verified for integrity by electrophoresis on a 1% (w/v) agarose gel. For each gene, real-time PCR was performed in four replicates for two different cDNA samples, each synthesized from 1 μg of total RNA using SuperScript III (Invitrogen). Because of the extremely low amounts of material that can be sampled during the early stages of ovule development, no biological replicate could be performed. cDNA was diluted 100-fold, and 5 μL was used in 20-μL PCR reactions using the Brilliant II SYBR Green QPCR (Stratagene) master mix and following the manufacturer’s instructions. Primers were designed using the Beacon Designer software (Stratagene) to target an amplicon size of 80 to 150 bp within the same region used in the initial screening for all target genes except dmt102 (see Supplemental Table 3 online). The specificity of primer combinations was examined by analyzing the dissociation curve of each reaction. PCR efficiency and data analyses were performed using the qbasePLUS software (www.biogazelle.com). To normalize the expression of target genes, we evaluated two commonly used housekeeping genes (tubulin4 and actin1), one housekeeping gene (gpm120) previously determined to be a control gene in reproductive tissues (ovaries and seeds 1 to 5 DAP; O. Leblanc, unpublished data), and three genes determined to be stable in transcriptional analyses (brat101, chr110, and nfe101; see Supplemental Table 2 online). Under our experimental conditions, gpm120 and nfe101 were the best choice for normalization in all samples (M and CV values < 0.2 and <0.05, respectively). Therefore, target gene expression profiles were determined relative to both gpm120 and nfe101 expression levels.
Imaging of Histone Modifications and Transcriptional Activity in Ovules and Seed Tissues
An approach first described by Laurie et al. (1999), referred to as semi-isolated ESs, was used. Fresh ovules or 1 DAP seeds were sliced with a vibratome (LeicaVT1000E) to produce sections containing intact ESs or developing embryos and endosperms. As described in the original article, these sections contain living materials that can be either fixed for further processing or grown on a basic growth medium. In our hands, most in vitro–cultured sections harvested after fertilization (>60%, n > 100) mimicked normal seed development during at least 6 DAP, therefore covering the time frame of our experiments. This allowed precise sampling and three-dimensional imaging of intact, whole-mount ESs, developing embryos, and syncitial endosperm tissue (see Supplemental Figure 6 online). This procedure was thus combined with immunostaining to monitor transcriptional activity and histone modifications during reproductive cell development. To achieve this, commercial antibodies (Abcam) that detected transcriptional activity (4H8 and H5) and several histone marks (i.e., H3K9me2, H3K9ac, and H3K4me3) were used. All these antibodies are routinely used in other organisms and produced (see below) results consistent with the literature (reviewed in Fuchs et al., 2006; see Supplemental Figure 6 online). ESs and early seeds were prepared following Laurie et al. (1999). Vibratome sections (~300 μm) were fixed for 3 h in 4% paraformaldehyde:1× PBS:2% Triton fixative and washed twice in 1× PBS. Samples were digested in an enzymatic solution (1% driselase, 0.5% cellulase, 1% pectolyase, and 1% BSA; all from Sigma-Aldrich) for 15 to 45 min at room temperature, depending on the developmental stage, subsequently rinsed three times in 1× PBS, and permeabilized for 1 to 2 h in 1× PBS, 2% Triton, and 1% BSA. They were then incubated overnight at 4°C with the primary antibodies (all from Abcam), used at the following concentrations: 1:400 for H3K9ac and 1:200 for 4H8, H5, H3K9me2, and H3K4me3. The sections were soaked for 8 to 12 h in 1× PBS and 0.2% Triton and incubated with secondary antibodies (either fluorescein isothiocyanate [FITC] conjugate or Cy3 conjugate [both from Sigma-Aldrich] or Alexa Fluor 488-conjugate [Molecular Probes]) used at 1:400 dilution. After washing in 1× PBS and 0.2% Triton for a minimum of 6 h, the sections were incubated with 4',6-diamidino-2-phenylindole (DAPI) (1 μg/mL in 1× PBS) for 1 h, washed for 1 h in 1× PBS, and mounted in PROLONG medium (Molecular Probes). A minimum of 50 ovules or seeds with interpretable staining patterns was scored for each developmental stage reported in this article. Images were captured on a Leica epifluorescence microscope equipped with a color CCD Leica camera and appropriate DAPI, FITC, or Cy3 filters. Three-dimensional ovule images were captured on a confocal laser scanning microscope (Leica SP2) equipped for 405-nm (DAPI), 488-nm (FITC/Alexa488), and 525-nm (Cy3/Alexa568) excitation and either ×20, ×40, or ×63 objectives. Maximum-intensity projections of selected optical sections were generated for this report and then edited using Graphic Converter (lemkeSOFT).
Whole-Mount Clearing of Ovule Samples
Ovaries were fixed in FAA (50% absolute ethanol, 5% glacial acetic acid, 10% formaldehyde, and 35% water) for 24 h at room temperature and stored in ethanol 70%. Fixed ovaries were sectioned by vibratome as described above. Then, sections were dehydrated throughout successive ethanol dilutions of 70, 85, and 100% and cleared according to Leblanc et al. (1995). Cleared sections were mounted in the clearing solution between two blocks of cover slips with similar thickness to that of samples and covered by a third cover slip. Samples were observed with differential interference contrast optics using a Zeiss Axio Imager.A1 microscope.
Whole-Mount in Situ mRNA Hybridizations
Vibratome sections of fresh ovaries were recovered and fixed as for immunolocalization assays. A standard in situ protocol, as described (García-Aguilar et al., 2005), was then followed using locked nucleic acid (LNA) probes with the following substitutions: dmt102, TAGGAALTGTCZCGTCCLAC; dmt103, TCGATCTZGTGATZGGTGGEAGTC; and Zm Wox2, CGCCGLGCCCPGCTCTCCEGC (E = A-LNA, L = C-LNA, Z = T-LNA, and P = G-LNA).
Phylogenetic Analysis
Sequence information for the maize and Arabidopsis loci were obtained from the CHROMDB database (www.chromdb.org) and aligned using a BLOSUM30 matrix and ClustalW, with an open gap penalty of 10 and an extend gap penalty of 0.1 in pairwise alignments, and an extend gap penalty of 0.05 and delay divergent setting of 40% in the multiple alignment (alignment provided as Supplemental Data Set 1 online). The resulting alignment was used for phylogenetic reconstruction using PHYLIP (Felsenstein, 1989). One thousand bootstrap replicates were performed to obtain an unrooted consensus tree.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At5g15380 (DRM1), At5g14620 (DRM2), At3g17310 (DRM3), At1g80740 (CMT1), At4g19020 (CMT2), At1g69770 (CMT3), At5g49160 (MET1), At4g14140 (MET2), At4g13610 (MET3), CA830161.1 (dmt102), CB329598.1 (dmt103), BT017721.1 (dmt105), CF052771, (dmt106), AW066779.1 (dmt101), BM336594.1 (chr106), AM234767 (Zm WOX2), AY733074 (ES1), DY532002.1 (hdt104), AI065477.2 (hon101), or in CHROMDB with their respective locus name.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Definition of Developmental Stages in the Ovule.
Supplemental Figure 2. Expression Patterns of Selected CMEs during Sexual and Apomictic Reproduction as Determined by RT-PCR.
Supplemental Figure 3. RT-PCR of CMEs Deregulated during Apomictic Reproduction.
Supplemental Figure 4. Phenotypes of D103 Mutant Lines.
Supplemental Figure 5. H3K9me2 Detection during Megagametogenesis in Sexual Maize (B73) and dmt102 Mutant Lines and in 38C Apomicts.
Supplemental Figure 6. Analysis of Chromatin States in Whole-Mount Seed Tissues.
Supplemental Table 1. List of CMEs Evaluated for Reproductive Tissue Expression as Determined by RNA Gel Blot Analyses (www.chromdb.org).
Supplemental Table 2. CMEs Selected for Transcriptional Analysis.
Supplemental Table 3. List of Primers.
Supplemental Data Set 1. Alignment of Maize and Arabidopsis Sequences Used for Phylogenetic Reconstructions.
Supplementary Material
Acknowledgments
We thank Bill Gordon-Kamm and Sheila Maddock at Pioneer Hi Bred for their training on manipulation of early seed development in maize and Martha Hernandez and Pablo Alva at the Centro Internacional de Mejoramiento de Maiz y Trigo for collecting the Tripsacum samples. We also thank T. Dresselhaus, S. Kaeppler, M. Sachs, the Maize Chromatin Consortium, and the Maize TILLING project members for sharing materials. This research was funded by an Apomixis Consortium grant from Pionner Hi Bred, Syngenta Seeds, Group Limagrain, and Institut de Recherche pour le Développement and by the Agence National de la Recherche (07-BLAN- 012001). M.G.-A. is the recipient of a Consejo Nacional de Ciencia y Tecnologia graduate fellowship from the Mexican government.
References
- Baroux C., Pien S., Grossniklaus U. (2007). Chromatin modification and remodeling during early seed development. Curr. Opin. Genet. Dev. 17: 473–479 [DOI] [PubMed] [Google Scholar]
- Bezhani S., Winter C., Hershman S., Wagner J.D., Kennedy J.F., Kwon C.S., Pfluger J., Su Y., Wagner D. (2007). Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell 19: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell R.A., Koltunow A.M. (2004). Understanding apomixis: Recent advances and remaining conundrums. Plant Cell 16 (Suppl): S228–S245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A. (1993). Dosage analysis of maize endosperm development. Annu. Rev. Genet. 27: 181–204 [DOI] [PubMed] [Google Scholar]
- Bonilla J.R., Quarin C.L. (1997). Diplosporous and aposporous apomixis in a pentaploid race of Paspalum minus. Plant Sci. 127: 97–104 [Google Scholar]
- Bradley J.E., Carman G.C., Jamison M.S., Naumova T.N. (2007). Heterochonic feactures of the female germline among several sexual diploid Tripsacum L. (Andropogoneae, Poaceae). Sex. Plant Reprod. 20: 9–17 [Google Scholar]
- Cao X., Aufsatz W., Zilberman D., Mette M.F., Huang M.S., Matzke M., Jacobsen S.E. (2003). Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 13: 2212–2217 [DOI] [PubMed] [Google Scholar]
- Cao X., Jacobsen S.E. (2002). Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 12: 1138–1144 [DOI] [PubMed] [Google Scholar]
- Carman J.G. (1997). Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol. J. Linnean Soc. 61: 51–94 [Google Scholar]
- Chen M., Ha M., Lackey E., Wang J., Chen Z.J. (2008). RNAi of met1 reduces DNA methylation and induces genome-specific changes in gene expression and centromeric small RNA accumulation in Arabidopsis allopolyploids. Genetics 178: 1845–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordts S., Bantin J., Wittich P.E., Kranz E., Lörz H., Dresselhaus T. (2001). ZmES genes encode peptides with structural homology to defensins and are specifically expressed in the female gametophyte of maize. Plant J. 25: 103–114 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2008). Molecular control of autonomous embryo and endosperm development. Sex. Plant Reprod. 21: 79–88 [Google Scholar]
- d’Erfurth I., Jolivet S., Froger N., Catrice O., Novatchkova M., Mercier R. (2009). Turning meiosis into mitosis. PLoS Biol. 7: e1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diboll A.G., Larson D.A. (1966). An electron microscopic study of the mature megagametophyte in Zea mays. Am. J. Bot. 53: 391–402 [PubMed] [Google Scholar]
- Evans M.M. (2007). The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. Plant Cell 19: 46–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1989). PHYLIP–Phylogeny Inference Package (Version 3.2). Cladistics 5: 164–166 [Google Scholar]
- Fuchs J., Demidov D., Houben A., Schubert I. (2006). Chromosomal histone modification patterns—From conservation to diversity. Trends Plant Sci. 11: 199–208 [DOI] [PubMed] [Google Scholar]
- García-Aguilar M., Dorantes-Acosta A., Pérez-España V., Vielle-Calzada J.P. (2005). Whole-mount in situ mRNA localization in developing ovules and seeds of Arabidopsis. Plant Mol. Biol. Rep. 23: 279–289 [Google Scholar]
- Grimanelli D., García M., Kaszas E., Perotti E., Leblanc O. (2003). Heterochronic expression of sexual reproductive programs during apomictic development in Tripsacum. Genetics 165: 1521–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimanelli D., Perotti E., Ramirez J., Leblanc O. (2005). Timing of the maternal-to-zygotic transition during early seed development in maize. Plant Cell 17: 1061–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huanca-Mamani W., Garcia-Aguilar M., León-Martínez G., Grossniklaus U., Vielle-Calzada J.-P. (2005). CHR11, a chromatin-remodeling factor essential for nuclear proliferation during female gametogenesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102: 17231–17236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel B., Kanno T., Daxinger L., Bucher E., van der Winden J., Matzke A.J.M., Matzke M. (2007). RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants. Biochim. Biophys. Acta 1769: 358–374 [DOI] [PubMed] [Google Scholar]
- Jackson J.P., Lindroth A.M., Cao X., Jacobsen S.E. (2002). Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560 [DOI] [PubMed] [Google Scholar]
- Jeddeloh J.A., Stokes T.L., Richards E.J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22: 94–97 [DOI] [PubMed] [Google Scholar]
- Johnson L.M., Bostick M., Zhang X., Kraft E., Henderson I., Callis J., Jacobsen S.E. (2007). The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr. Biol. 17: 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow A.M., Grossniklaus U. (2003). Apomixis: A developmental perspective. Annu. Rev. Plant Biol. 54: 547–574 [DOI] [PubMed] [Google Scholar]
- Laurie J.D., Zhang D., Mcgann L.E., Gordon-kamm W.J., Cass D.D. (1999). A novel technique for the partial isolation of maize embryo sacs and subsequent regeneration of plants. In Vitro Cell. Dev. Biol. Plant 3: 320–325 [Google Scholar]
- Leblanc O., Grimanelli D., Hernandez-Rodriguez M., Galindo P.A., Soriano-Martinez A.M., Perotti E. (2009). Seed development and inheritance studies in apomictic maize-Tripsacum hybrids reveal barriers for the transfer of apomixis into sexual crops. Int. J. Dev. Biol. 53: 585–596 [DOI] [PubMed] [Google Scholar]
- Leblanc O., Grimanelli D., Islam-Faridi N., Berthaud J., Savidan Y. (1996). Reproductive behavior in maize-Tripsacum polyhaploid plants: Implications for the transfer of apomixis into maize. J. Hered. 87: 108–111 [Google Scholar]
- Leblanc O., Peel M.D., Carman J.G., Savidan Y. (1995). Megasporogenesis and megagametogenesis in several Tripsacum species (Poaceae). Am. J. Bot. 82: 57–63 [Google Scholar]
- Lindroth A.M., et al. (2004). Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 23: 4286–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung A., Masuelli R.W., Watson B., Reynolds S.H., Davison J., Comai L. (2002). Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol. 129: 733–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch I., Stupar R.M., Iniguez A.L., Haun W.J., Barbazuk W.B., Kaeppler S.M., Springer N.M. (2007). Natural variation for alleles under epigenetic control by the maize chromomethylase zmet2. Genetics 177: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O., Probst A.V., Paszkowski J. (2005). Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J. 24: 2783–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis K., Chandler V., Cone K., Kaeppler H., Kaeppler S., Kerschen A., Pikaard C., Richards E., Sidorenko L., Smith T., Springer N., Wulan T. (2005). Transgene-induced RNA interference as a tool for plant functional genomics. Methods Enzymol. 392: 1–24 [DOI] [PubMed] [Google Scholar]
- Nardmann J., Zimmermann R., Durantini D., Kranz E., Werr W. (2007). WOX gene phylogeny in Poaceae: A comparative approach addressing leaf and embryo development. Mol. Biol. Evol. 24: 2474–2484 [DOI] [PubMed] [Google Scholar]
- Naumova T.N., Van der laak J., Osadtchiy J., Matzk F., Kravtchenko A., Bergervoet J., Ramulu K.S., Boutilier K. (2001). Reproductive development in apomictic populations of Arabis holboellii (Brassicaceae). Sex. Plant Reprod. 14: 195–200 [DOI] [PubMed] [Google Scholar]
- Nogler G.A. (1984). Gametophytic apomixis. Embryology of Angiosperms, Johri B.M., (Berlin: Springer-Verlag; ), pp. 475–518 [Google Scholar]
- Nonomura K.I., Morohoshi A., Nakano M., Eiguchi M., Miyao A., Hirochika H., Kurata N. (2007). A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19: 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo-Monfil V., Durán-Figueroa N., Arteaga-Vázquez M., Demesa-Arévalo E., Autran D., Grimanelli D., Slotkin R.K., Martienssen R.A., Vielle-Calzada J.P. (2010). Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464: 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B., Bensaude O. (2003). Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur. J. Biochem. 270: 3859–3870 [DOI] [PubMed] [Google Scholar]
- Papa C.M., Springer N.M., Muszynski M.G., Meeley R., Kaeppler S.M. (2001). Maize chromomethylase Zea methyltransferase2 is required for CpNpG methylation. Plant Cell 13: 1919–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paun O., Stuessy T.F., Hörandl E. (2006). The role of hybridization, polyploidization and glaciation in the origin and evolution of the apomictic Ranunculus cassubicus complex. New Phytol. 171: 223–236 [DOI] [PubMed] [Google Scholar]
- Pillot M., Autran D., Leblanc O., Grimanelli D. (2010a). A role for CHROMOMETHYLASE3 in mediating transposon and euchromatin silencing during egg cell reprogramming in Arabidopsis. Plant Signal. Behav. 5: 1167–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillot M., Baroux C., Vazquez M.A., Autran D., Leblanc O., Vielle-Calzada J.P., Grossniklaus U., Grimanelli D. (2010b). Embryo and endosperm inherit distinct chromatin and transcriptional states from the female gametes in Arabidopsis. Plant Cell 22: 307–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R.S. (2003). Phase change and the regulation of developmental timing in plants. Science 301: 334–336 [DOI] [PubMed] [Google Scholar]
- Ravi M., Chan S.W. (2010). Haploid plants produced by centromere-mediated genome elimination. Nature 464: 615–618 [DOI] [PubMed] [Google Scholar]
- Ravi M., Marimuthu M.P., Siddiqi I. (2008). Gamete formation without meiosis in Arabidopsis. Nature 451: 1121–1124 [DOI] [PubMed] [Google Scholar]
- Reiser L., Fischer R.L. (1993). The ovule and the embryo sac. Plant Cell 5: 1291–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz M.E., Kantama L., de Jong H., Mitchell-Olds T. (2006). Asexual reproduction in a close relative of Arabidopsis: A genetic investigation of apomixis in Boechera (Brassicaceae). New Phytol. 171: 425–438 [DOI] [PubMed] [Google Scholar]
- Seydoux G., Braun R.E. (2006). Pathway to totipotency: Lessons from germ cells. Cell 127: 891–904 [DOI] [PubMed] [Google Scholar]
- Slotkin R.K., Vaughn M., Borges F., Tanurdzić M., Becker J.D., Feijó J.A., Martienssen R.A. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe W.J.J., Jasencakova Z., Houben A., Kakutani T., Meister A., Huang M.S., Jacobsen S.E., Schubert I., Fransz P.F. (2002). DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 21: 6549–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq M., Saze H., Probst A.V., Lichota J., Habu Y., Paszkowski J. (2003). Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc. Natl. Acad. Sci. USA 100: 8823–8827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M.R., Araujo A.C., Paech N.A., Hecht V., Schmidt E.D., Rossell J.B., De Vries S.C., Koltunow A.M. (2003). Sexual and apomictic reproduction in Hieracium subgenus pilosella are closely interrelated developmental pathways. Plant Cell 15: 1524–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant I., Paszkowski J. (2007). Role of histone and DNA methylation in gene regulation. Curr. Opin. Plant Biol. 10: 528–533 [DOI] [PubMed] [Google Scholar]
- Wang J., Tian L., Madlung A., Lee H.S., Chen M., Lee J.J., Watson B., Kagochi T., Comai L., Chen Z.J. (2004). Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics 167: 1961–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Custard K.D., Brown R.C., Lemmon B.E., Harada J.J., Goldberg R.B., Fischer R.L. (2006). DNA methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell 18: 805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Lin H. (2007). An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature 450: 304–308 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.