Norcoclaurine synthase (NCS) catalyzes the first step in the formation of benzylisoquinoline alkaloids, which include the painkillers morphine and codeine. Two different enzymes identified as norcoclaurine synthase have been reported. Characterization of NCS in opium poppy and meadow rue has shown that only the enzyme belonging to the PR10/Bet v1 protein family is involved in norcoclaurine biosynthesis.
Abstract
Norcoclaurine synthase (NCS) catalyzes the first committed step in the biosynthesis of benzylisoquinoline alkaloids (BIAs). NCS from Thalictrum flavum (Tf NCS), Papaver somniferum (Ps NCS1 and Ps NCS2), and Coptis japonica (Cj PR10A) share substantial identity with pathogen-related 10 (PR10) and Bet v1 proteins, whose functions are not well understood. A distinct enzyme (Cj NCS1) with similarity to 2-oxoglutarate–dependent dioxygenases was suggested as the bona fide NCS in C. japonica. Here, we validate the exclusive role of PR10/Bet v1-type NCS enzymes in BIA metabolism. Immunolocalization of Ps NCS2 revealed its cell type–specific occurrence in phloem sieve elements, which contain all other known BIA biosynthetic enzymes. In opium poppy, NCS transcripts and proteins were abundant in root and stem, but at low levels in leaf and carpel. Silencing of NCS in opium poppy profoundly reduced alkaloid levels compared with controls. Immunoprecipitation of NCS from total protein extracts of T. flavum cells resulted in a nearly complete attenuation of NCS activity. A Ps NCS2–green fluorescent protein fusion introduced by microprojectile bombardment into opium poppy cells initially localized to the endoplasmic reticulum but subsequently sorted to the vacuole. In our hands, Cj NCS1 did not catalyze the formation of (S)-norcoclaurine from dopamine and 4-hydroxyphenylacetaldehyde.
INTRODUCTION
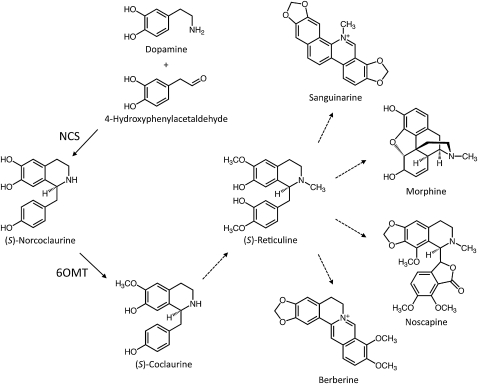
Norcoclaurine synthase (NCS; EC 4.2.1.78) catalyzes the stereoselective Pictet-Spengler condensation of the l-Tyr derivatives dopamine and 4-hydroxyphenylacetaldehyde (4-HPAA), yielding (S)-norcoclaurine, which serves as the central precursor to myriad benzylisoquinoline alkaloids (BIAs), a large and diverse group of natural products found primarily in a several related plant families (Stadler et al., 1989). Sequential 6-O-methylation of (S)-norcoclaurine followed by N-methylation, cytochrome P450–dependent 3′-hydroxylation, and 4’-O-methylation lead to the formation of the central branch point intermediate (S)-reticuline (Ziegler and Facchini, 2008). Subsequent internal carbon-carbon and carbon-oxygen phenol coupling of (S)-reticuline results in the formation of a wide variety of backbone structures, including the morphinan (morphine), phthalideisoquinoline (noscapine), benzophenanthridine (sanguinarine), and protoberberine (berberine) alkaloid subclasses (Figure 1), among others. Many of the estimated 2500 known and natural BIAs are pharmacologically active, such as the narcotic analgesics and cough suppressants codeine and morphine, the vasodilator papaverine, the potential anticancer drug noscapine, and the antimicrobial agents sanguinarine and berberine. Opium poppy (Papaver somniferum; Papaveraceae) produces all of these compounds (except for berberine) and remains the only commercial source of morphine, codeine, thebaine, and oripavine; the latter two morphinan alkaloids are used for the preparation of semisynthetic derivatives such as oxycodone, buprenorphine, naltrexone, and naloxone.
Figure 1.
NCS Catalyzes the First Step in BIA Metabolism.
Dopamine and 4-HPAA condense to form (S)-norcoclaurine, which undergoes O-methylation to yield (S)-coclaurine. Additional N-methylation, aromatic ring hydroxylation, and O-methylation yield the central branch point intermediate (S)-reticuline. Subsequent carbon-carbon and carbon-oxygen phenol coupling and functional group substitutions lead to an estimated 2500 known compounds belonging to a variety of structural categories, including sanguinarine, morphine, noscapine, and berberine. 6OMT, norcoclaurine 6-O-methyltransferase.
A gene encoding NCS was initially isolated based on empirical amino acid sequences of peptides obtained by tryptic digestion of the purified enzyme from meadow rue (Thalictrum flavum; Ranunculaceae), which primarily accumulates berberine (Samanani and Facchini, 2001, 2002; Samanani et al., 2004). The T. flavum NCS (Tf NCS) sequence was used to query an opium poppy EST database, which resulted in the isolation of two isoforms (Ps NCS1 and Ps NCS2) sharing 89% amino acid identity, each displaying 40% identity with Tf NCS and efficiently catalyzing the formation of (S)-norcoclaurine from dopamine and 4-HPAA (Liscombe et al., 2005). The amino acid sequence of Tf NCS showed ~30 to 40% identity (~50 to 60% similarity) to members of the pathogenesis-related 10 (PR10) protein/Bet v1 allergen family. Subsequently, the reaction mechanism leading to the formation of (S)-norcoclaurine was shown to proceed via a two-step cyclization of a putative iminium ion intermediate (Luk et al., 2007; Bonamore et al., 2010). The overall structure of Tf NCS was initially obtained by NMR spectroscopy coupled with homology modeling of Bet v1 proteins (Berkner et al., 2008), and the specific structural determinants responsible for the stereoselective Pictet-Spengler cyclization were established by x-ray crystallographic analysis (Ilari et al., 2009). NCS is the only known member of the PR10/Bet v1 family shown unequivocally to possess catalytic activity in plants. Another well-established fungal enzyme, TcmN aromatase/cyclase, which participates in the cyclization and structural diversification of aromatic polyketides, has also been shown to contain a PR10/Bet v1 fold (Ames et al., 2008). Generally, PR10 proteins are thought to participate in the defense of plants against microorganisms and fungi (Chadha and Das, 2006), although putative roles in plant metabolism have been proposed (Swoboda et al., 1996; Fujimoto et al., 1998; Bais et al., 2003; Michalska et al., 2010). A Bet v1 protein from white birch (Betula alba) was also reported to possess ribonuclease activity (Bufe et al., 1996), whereas others from plants such as Arabidopsis thaliana, pepper (Capsicum annuum), and tobacco (Nicotiana tabacum) have been characterized as homologs of opium poppy major latex proteins (MLPs; Osmark et al., 1998).
A protein designated Cj NCS1 and sharing substantial amino acid similarity with 2-oxoglutarate (2OG)/Fe(II)-dependent dioxygenases was subsequently reported as the bona fide enzyme responsible for the formation of (S)-norcoclaurine in Japanese goldthread (Coptis japonica), a member of the Ranunculaceae related to meadow rue, despite the concurrent isolation of a protein (designated Cj PR10A) displaying 62% amino acid identity to Tf NCS and possessing NCS activity (Minami et al., 2007). All 2OG/Fe(II)-dependent dioxygenases require Fe(II) ion as a cofactor and use O2 as a cosubstrate (Hausinger, 2004; Kovaleva and Lipscomb, 2008; Loenarz and Schofield, 2008). Most of these enzymes are 2OG dependent, including hyoscyamine 6β-hydroxylase (Matsuda et al., 1991) and desacetoxyvindoline-4-hydroxylase (Vazquez-Flota et al., 1997), which are involved in the biosynthesis of tropane and monoterpenoid indole alkaloids, respectively, and anthocyanidin synthase (Davies, 1993), flavonol synthase (Holton et al., 1993), and flavanone 3β-hydroxylase (Britsch et al., 1993), functioning in flavonoid metabolism. A few notable exceptions that do not require 2OG include 1-aminocyclopropane-1-carboxylate oxidase (Holdsworth et al., 1987) and fungal isopenicillin N-synthase (Ramón et al., 1987). Cj NCS1 was reported to require Fe(II) ion but was not dependent on 2OG or O2 and was proposed to lack a 2OG binding domain (Minami et al., 2007). Recently, we identified two 2OG/Fe(II)-dependent dioxygenases, thebaine 6-O-demthylase (T6ODM), and codeine O-demethylase (CODM), responsible for the O-demethylation steps involved in the conversion of thebaine to morphine in opium poppy (Hagel and Facchini, 2010a, 2010b). In opium poppy, T6ODM and CODM belong to a phylogenetic clade containing members that share ~40% identity with Cj NCS1.
The chemical reactivity of dopamine and 4-HPAA leads to the spontaneous formation of (R,S)-norcoclaurine especially under certain conditions (Samanani and Facchini, 2001; Minami et al., 2007); thus, a variety of enzymes might be associated with the ability to encourage the coupling of these substrates. However, in vitro activity is insufficient to implicate an enzyme in a specific metabolic pathway. Given the pivotal role of NCS as the entry point into BIA metabolism, it is important to establish identity of the enzyme(s) responsible for the formation of (S)-norcoclaurine in planta. In this article, we report on a series of biochemical, physiological, and gene-silencing experiments that unequivocally support the role of PR10/Bet v1-type NCS enzymes in BIA metabolism in opium poppy and related plants.
RESULTS
Immunoprecipitation of Tf NCS Abolishes NCS Activity in Total Protein Extracts
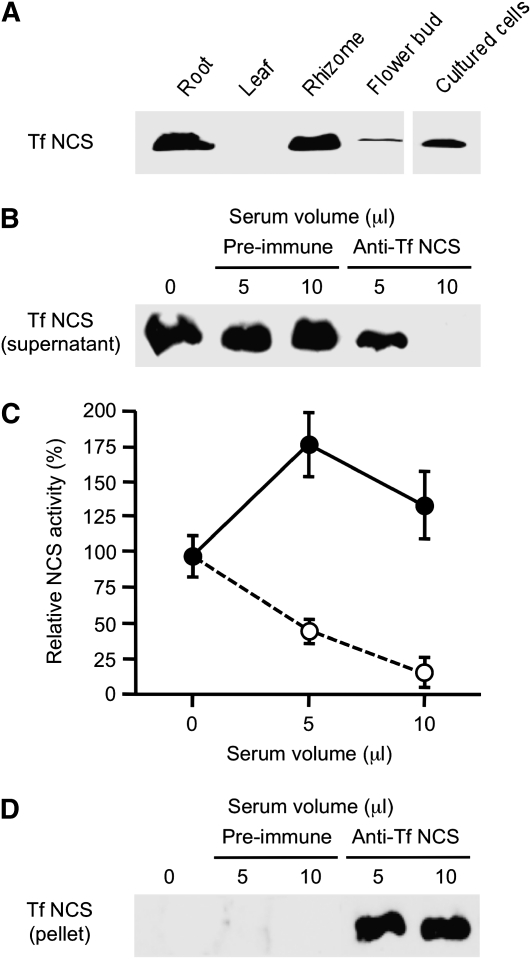
Tf NCS antiserum prepared using purified recombinant enzyme showed specificity toward a single ~20-kD polypeptide on immunoblots of total T. flavum protein extracted from various plant organs (Figure 2A). Tf NCS was detected in roots, rhizomes, flower buds, and cultured cells of T. flavum (Figure 2A). Preimmune or anti-Tf NCS serum was incubated with total soluble protein extracts from T. flavum cell cultures, and the IgG enzyme complex was precipitated using protein A-Sepharose. The supernatants were divided into two aliquots. Proteins from one aliquot were subjected to immunoblot analysis using Tf NCS antiserum. Preincubation with preimmune serum had no effect on the relative abundance of Tf NCS in total soluble protein extracts from T. flavum cell cultures compared with the control (Figure 2B). However, preincubation with 5 μL of Tf NCS antiserum reduced the relative abundance of Tf NCS compared with the use of a corresponding volume of preimmune serum. Tf NCS was not detected on the immunoblot when 10 μL of Tf NCS antiserum was used for immunoprecipitation. The remaining supernatant aliquot was used to measure residual NCS activity, which was reduced proportional to the dose of Tf NCS antiserum preincubated with the total soluble protein extracts (Figure 2C). By contrast, NSC activity increased in response to preincubation with corresponding volumes of preimmune serum. The immunoprecipitation of Tf NCS was confirmed by immunoblot analysis of the IgG-associated enzyme bound to protein A-Sepharose (Figure 2D). Serum IgGs were also visible on the immunoblot, but the relatively low molecular mass NCS protein was clearly separated.
Figure 2.
Immunoprecipitation of NCS Using Tf NCS Antiserum.
(A) Immunoblot analysis using Tf NCS antiserum. Soluble protein was prepared from various organs and cultured cells of T. flavum.
(B) Immunoprecipitation was performed by incubating total protein extracts of T. flavum cell cultures with either preimmune or Tf NCS antiserum conjugated protein A-Sepharose. Residual Tf NCS protein in the supernatant after immunoprecipitation was visualized by immunoblot analysis using Tf NCS antiserum.
(C) NCS activity remaining in supernatant fractions after immunoprecipitation using different volumes of preimmune (closed circles) or Tf NCS (open circles) antisera. Enzyme activity was determined by measuring the quantity of (S)-norcoclaurine produced after 2 h incubation with 4-HPAA and dopamine. Data represent the mean ± sd of four replicates.
(D) NCS protein in pellet fractions after immunoprecipitation was confirmed by immunoblot analysis using Tf NCS antiserum.
Occurrence of NCS in Opium Poppy Plant Organs and Elicitor-Treated Cell Cultures
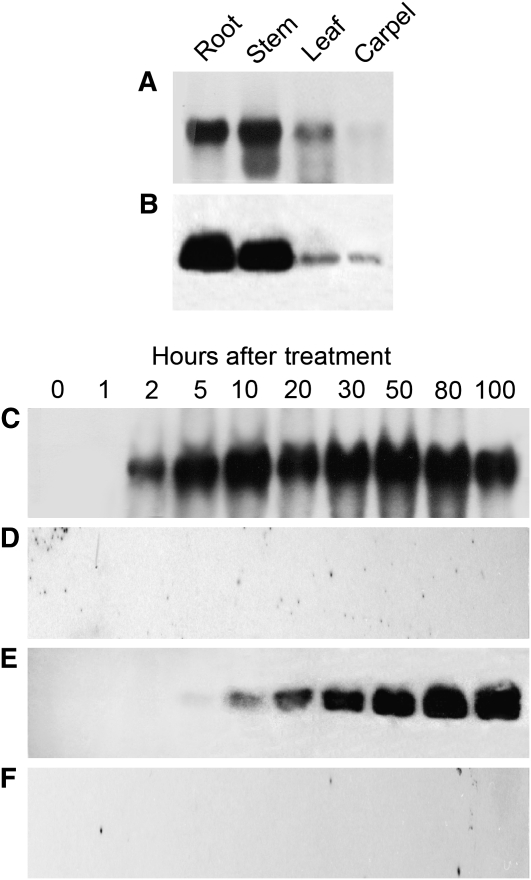
The occurrence of NCS transcripts and proteins in opium poppy plant organs and elicitor-induced cell cultures were analyzed by RNA gel blot hybridization analysis using Ps NCS2 cDNA as a probe and by immunoblot analysis using a polyclonal antibody raised against Ps NCS2 (Figure 3). NCS transcripts (Figure 3A) and proteins (Figure 3B) were substantially more abundant in root and stem compared with leaf and carpel. Treatment of opium poppy cell cultures with a fungal elicitor resulted in the coordinated induction of NCS transcripts (Figure 3C) and proteins (Figure 3E). NCS transcripts were initially absent but were detected 2 h after the addition of the elicitor, reached maximum levels between 5 and 10 h, and remained elevated for the duration of the 100-h time course (Figure 3C). Similarly, NCS proteins were initially not detected but increased continuously for the duration of the time course beginning 5 h after addition of the elicitor (Figure 3E). NCS transcripts and proteins were not detected in control cultures treated with an equal volume of water rather than the elicitor (Figures 3D and 3F). The Ps NSC2 antiserum showed specificity toward a single ~20-kD polypeptide in crude opium poppy protein extracts (Figures 3B and 3E).
Figure 3.
Relative Abundance of NCS Transcripts and Proteins in Opium Poppy Plants and Elicitor-Induced Cell Cultures.
Total RNA and protein extracts were used for RNA gel blot ([A], [C], and [D]) and immunoblot ([B], [E], and [F]) analyses of opium poppy plant organs ([A] and [B]) and cell cultures sampled at different times after treatment with a fungal-derived elicitor ([C] and [E]) or water ([D] and [F]).
NCS Is Localized to Sieve Elements of the Phloem in Opium Poppy
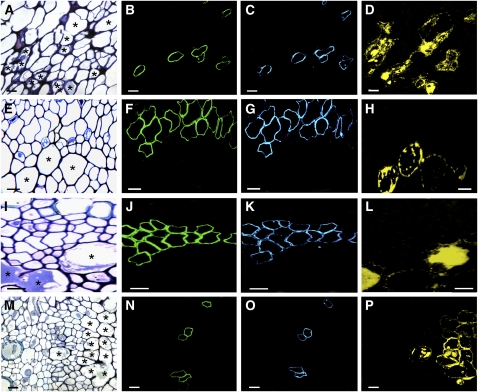
The cellular localization of NCS in opium poppy was investigated by immunocytochemical staining (Figure 4). The Ps NCS2 antiserum revealed the cell type–specific occurrence of NCS in the vascular bundles of all opium poppy organs. Previously, we showed that other known BIA biosynthetic enzymes are localized to sieve elements of the phloem (Bird et al., 2003; Samanani et al., 2006). To determine whether NCS was also associated with phloem sieve elements, the subsequent enzyme in the BIA pathway (6OMT) responsible for the conversion of (S)-norcoclaurine to (S)-coclaurine was colocalized in serial sections of each plant organ using the antiserum raised against 6OMT from opium poppy (Samanani et al., 2006). MLP antiserum was used in additional serial sections to show the relative location of laticifers, the cell type in which BIAs accumulate. NCS and 6OMT were detected in the same cells of root (Figures 4B and 4C), stem (Figures 4F and 4G), leaf (Figures 4J and 4K), and carpel (Figures 4N and 4O). As reported previously for 6OMT and other known BIA biosynthetic enzymes (Bird et al., 2003; Samanani et al., 2006), NCS is localized to sieve elements of the phloem, which are proximal or adjacent to laticifers in the vascular bundles of all plant organs.
Figure 4.
Immunolocalization of NCS in Opium Poppy.
Primary NCS (Ps NCS2), norcoclaurine-6-O-methyltransferase (6OMT), or MLP antisera were used to visualize the cellular location of NCS (green), 6OMT (blue), and MLP (yellow) in serial sections of each organ using fluorescent-labeled secondary antibodies. Bar = 10 μm for all micrographs. Asterisks indicate several laticifers in (A), (E), (I), and (M) stained with toludine blue O.
(A) to (D) Phloem of serial root cross sections.
(E) to (H) Phloem of serial stem cross sections.
(I) to (L) Phloem of serial leaf cross sections.
(M) to (P) Phloem of serial carpel cross sections.
Virus-Induced Gene Silencing of NCS in Opium Poppy
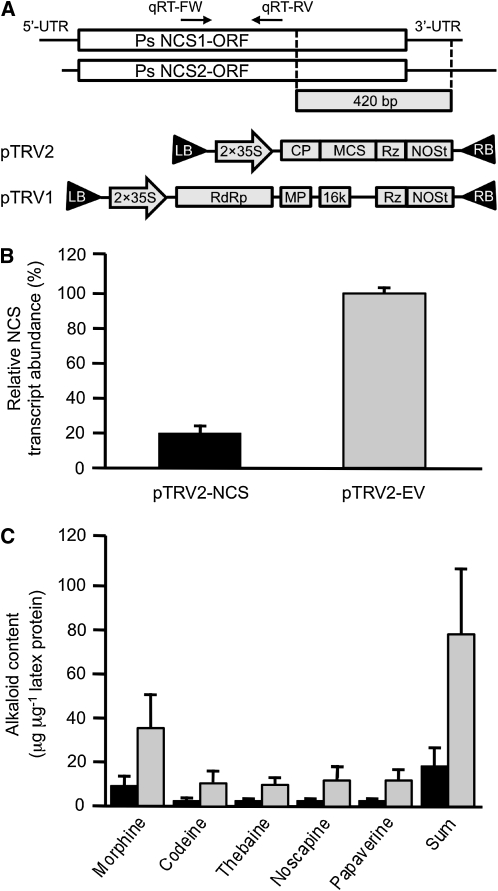
Virus-induced gene silencing (VIGS) was used to validate the role of NCS in BIA metabolism in opium poppy plants (Figure 5). An identical 420-bp fragment occurring in both Ps NCS1 and Ps NCS2 was cloned into pTRV2 (pTRV2-PsNCS; Figure 5A). A mixed culture of Agrobacterium tumefaciens harboring either pTRV2-PsNCS or pTRV1 was infiltrated into opium poppy seedlings ~2 weeks after germination. Control seedlings were infiltrated with a mixture of A. tumefaciens cultures harboring the empty pTRV2 vector (pTRV2-EV) or pTRV1 (Figure 5A). Plants were analyzed ~8 weeks after infiltration. No developmental differences were observed in plants exposed to either pTRV2-PsNCS or pTRV2-EV. To ensure that infiltrated plants contained the viral vector, the occurrence of tobacco rattle virus (TRV) coat protein transcripts was initially confirmed by RT-PCR (see Supplemental Figure 1 online). Relative NCS transcript levels were determined in infiltrated opium poppy plants containing TRV coat protein transcripts. Primers for quantitative RT-PCR (qRT-PCR) were specific to NCS transcript sequences outside the region used to create pTRV2-PsNCS (Figure 5A). The abundance of NCS transcripts was reduced by >80% (P value of <0.001) in stem tissue from plants infiltrated with A. tumefaciens harboring pTRV2-PsNCS compared with those exposed to bacteria containing pTRV2-EV (Figure 5B). NCS transcript levels were normalized relative to the abundance of transcripts of the constitutive opium poppy elongation factor 1a (EL1A). Levels of the five major alkaloids in opium poppy latex (morphine, codeine, thebaine, noscapine, and papaveine) were reduced between 75 and 82% (P value of <0.01) in plants infiltrated with A. tumefaciens harboring pTRV2-PsNCS and showing a reduction in NCS transcript levels compared with plants exposed to bacteria containing pTRV2-EV (Figure 5C).
Figure 5.
VIGS of Ps NCS2 in Opium Poppy.
(A) Assembly of the pTRV2-NCS vector. A 420-bp fragment of the Ps NCS2 cDNA was inserted into the multiple cloning site (MCS) between a tandem repeat of the CaMV 35S promoter (2×35S) and the NOS terminator (NOSt). Sequences encoding viral coat protein (CP) and self-cleaving ribozyme (Rz) flanked the MCS. LB, left border of T-DNA; RB, right border of T-DNA; RdRP, RNA-dependent RNA polymerase; MP, movement protein; 16K, 16-kD Cys-rich protein; UTR, untranslated region. qRT-FW and qRT-RV are PsNCS-specific primers used for RT-qPCR. Assembly of the pTRV1 and pTRV2 vectors has been described previously (Dinesh-Kumar et al., 2003).
(B) Ps NCS transcript levels, determined by qRT-PCR, in plants infiltrated with A. tumefaciens harboring pTRV2-NCS (black bar) or empty pTRV2 (gray bar) vector. Data represent the mean ± se of triplicate technical replicates performed on cDNA prepared for each of eight individual plants infiltrated with A. tumefaciens containing each construct.
(C) Effect of suppressing Ps NCS transcript levels by VIGS on the alkaloid content of opium poppy. Latex extracts from plants infiltrated with A. tumefaciens harboring pTRV2-NCS (black bar) or empty pTRV2 (gray bar) vector were analyzed by HPLC. Data represent the mean ± se of values obtained by the analysis of latex extracts from eight individual plants infiltrated with A. tumefaciens containing each construct. The differences in the transcript levels and alkaloid contents of pTRV2-NCS and pTRV2-EV plants were analyzed by paired or unpaired two-tailed Student’s t test. P values of <0.05 were considered statistically significant.
Members of the PR10/Bet v1 Protein Family Show NCS Activity
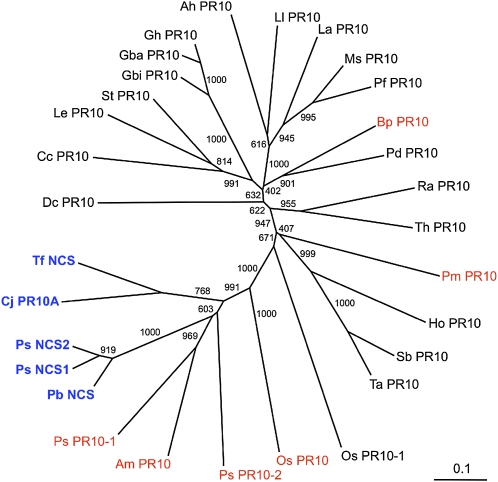
Two cDNAs encoding proteins with substantial amino acid identity to Ps NCS2 were identified in the EST databases of Persian poppy (Papaver bracteatum; 86% identity) and Mexican prickly poppy (Argemone mexicana; 43% identity). Phylogenetic relationships among several PR10/Bet v1 proteins from a variety of plants are shown in Figure 6. Tf NCS and Cj PR10A from T. flavum and C. japonica, respectively, which are members of the Ranunculaceae, share 62% amino acid identity with each other and ~40% identity with homologs from opium poppy and P. bracteatum. In the phylogeny of PR10/Bet v1 proteins, Pb NCS forms part of a clade that includes all known members of the family with NCS activity. Other members of the clade are PR10/Bet v1 proteins from opium poppy (Ps PR10-1 and Ps PR10-2), rice (Oryza sativa; Os PR10), and A. mexicana (Am PR10; Figure 6). Previously, we showed that the opium poppy and rice homologs, and other related proteins sharing less amino acid identity (Pm PR10 from Pinus monticola and Bp PR10 from Betula pendula), do not display NCS activity (Liscombe et al., 2005).
Figure 6.
Phlyogenetic Analysis of Known NCS and Related PR10/Bet v1-Type Proteins.
Amino acid sequences were aligned and analyzed for phylogenetic relationships using the neighbor-joining algorithm. Numbers refer to the bootstrap values for each node with 1000 iterations. Abbreviations are provided in Methods. Bar indicates 0.1 substitutions per position in the sequence. Blue labels indicate that the corresponding recombinant enzymes exhibited NCS activity, whereas red labels reflect recombinant proteins that did not catalyze the formation of (S)-norcoclaurine.
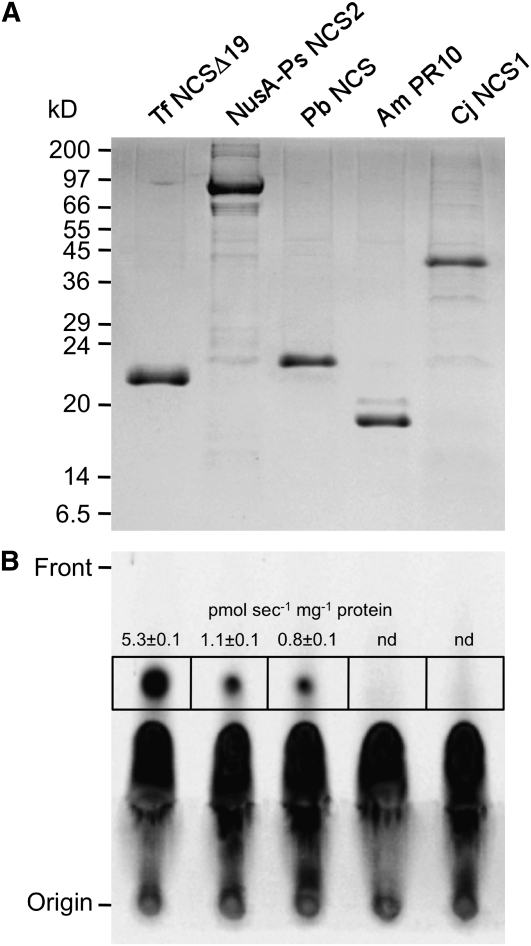
Codon-optimized genes encoding Pb NCS, Am PR10, and Cj NCS1 were expressed in Escherichia coli, and the His6-tagged proteins were purified by cobalt affinity chromatography (Figure 7). Recombinant Pb NCS, Am PR10, and Cj NCS1 exhibited the expected molecular masses of 22, 18, and 45 kD, respectively. In agreement with their relative positions in the PR10/Bet v1 protein phylogeny, Pb NCS showed NCS activity, but Am PR10 did not (Figure 7). It is notable that the NusA-Ps NCS2 fusion protein retained NCS activity as described for the native recombinant enzyme (Liscombe et al., 2005). Recombinant Cj NCS1 did not display NCS activity (Figure 7). Assays involving Cj NCS1 were performed as described previously, including the omission of exogenous Fe(II) in the reaction and the use of liquid chromatography–tandem mass spectrometry (LC-MS/MS) to detect the reaction product (Minami et al., 2007). However, substrate concentrations in the Cj NCS1 assay were reduced to 10 μM dopamine and 300 μM 4-HPAA, which were the conditions used to test PR10/Bet v1 proteins for NCS activity, owing to the spontaneous condensation of these compounds at higher levels (Samanani and Facchini, 2001).
Figure 7.
NCS Activity of Various Recombinant Proteins.
(A) His6-tagged recombinant Tf NCS, Ps NCS2, Pb NCS, Am PR10, and Cj NCS1 were purified from E. coli protein extracts by cobalt-affinity chromatography. Proteins were separated by SDS-PAGE and stained with Coomassie blue.
(B) Purified recombinant proteins (10 μg) were assayed for NCS activity using 4-HPAA and [8-14C]dopamine as substrates. [14C]Norcoclaurine was separated by thin layer chromatography and visualized by autoradiography. The radioactivity in the area on the thin layer chromatography plate represented by each black box, and corresponding to the migration distance of authentic (S)-norcoclaurine, was counted and used to calculate specific activity. Data represent the mean ± sd of three replicates. nd, not detectable.
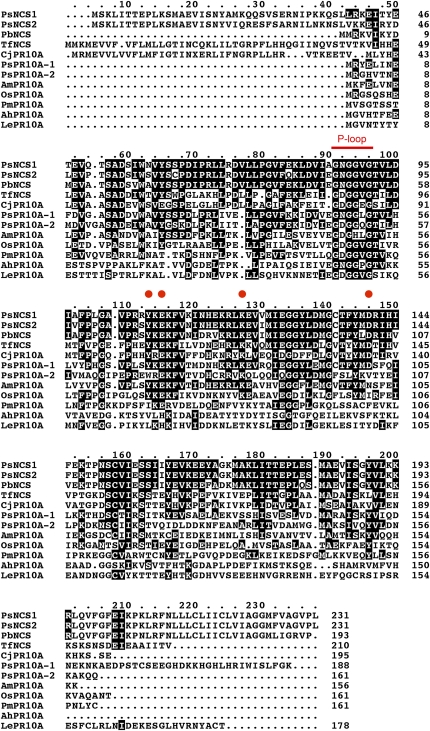
The amino acid sequences of all known members of the PR10/Bet v1 protein family exhibiting NCS activity (Ps NCS1, Ps NCS2, Pb NCS, Tf NCS, and Cj PR10A) were aligned with several inactive homologs (Os PR10, Ps PR10-1, Ps PR10-2, Am PR10, Arachis hypogaea Ah PR10, Pm PR10, and Lycopersicon esculentum PR10; Figure 8). As revealed by x-ray crystallographic analysis, the active site of Tf NCS is dominated by the geometry of the side chains of Tyr, Lys, Asp, and Glu residues that coordinate the binding of dopamine and 4-HPAA (Ilari et al., 2009) and are conserved in all proteins exhibiting NCS activity (Figure 8). Interestingly, these amino acids are conserved in Ps PR10A-1, which does not catalyze the formation of (S)-norcoclaurine, indicating that additional residues are important for NCS activity. The P-loop (GNGGPGT) sequence motif is found in all members of the PR10/Bet v1 family and was conserved in all aligned proteins (Figure 8). Finally, four out of the five known proteins with NCS activity (Ps NCS1, Ps NCS2, Tf NCS, and Cj PR10A) exhibit an extended N-terminal domain compared with other members of the PR10/Bet v1 family (Figure 8). Although the predicted translation product of Pb NCS did not possess this N-terminal extension, the cognate cDNA was only identified from a single EST; thus, we cannot rule out the possibility that the full-length amino acid sequence is not represented.
Figure 8.
Alignment of the Amino Acid Sequences of NCS and Other PR10/Bet v1 Proteins from Various Plant Species.
Deduced amino acid sequences were aligned using the ClustalX algorithm. Identical amino acids are shaded in black boxes. Abbreviations are provided in the Methods. The red bar represents the P-loop domain conserved in PR10/Bet v1 family proteins. Red circles correspond to the Tyr, Lys, Asp, and Glu residues that form the catalytic site of Tf NCS (Ilari et al., 2009).
Opium Poppy NCS Has an N-Terminal Signal Peptide and Sorts to the Vacuole
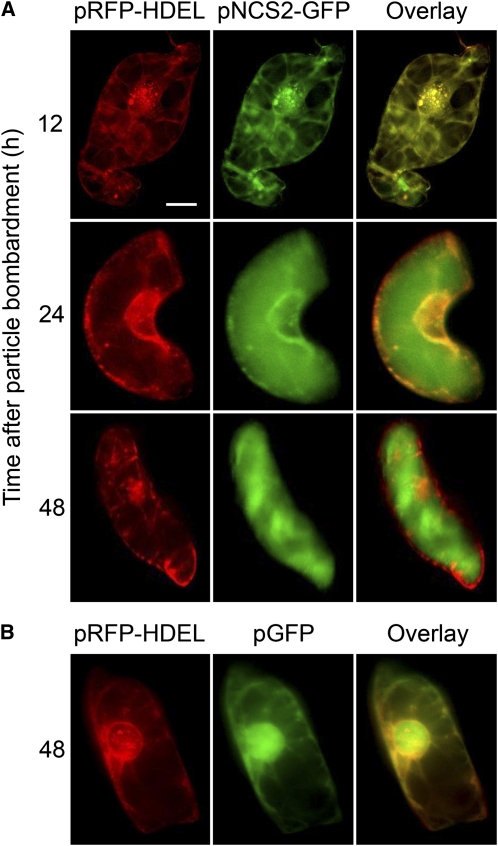
The first ~20 N-terminal amino acids of the predicted Tf NCS translation product were identified as a putative signal peptide using the SignalP Prediction Server (Samanani et al., 2004). To determine whether opium poppy NCS isoforms displays subcellular targeting resulting from the occurrence of a putative N-terminal signal peptide, a construct encoding a translational fusion between the C terminus of Ps NCS2 and the N terminus of the green fluorescent protein (GFP) and driven by the constitutive cauliflower mosaic virus (CaMV) 35S promoter was prepared. The vector (35S:PsNCS2-GFP) was introduced into cultured opium poppy cells by particle bombardment. The Ps NCS2-GFP fusion was initially observed in association with the endoplasmic reticulum (ER) 12 h after bombardment as revealed by the overlay of the GFP signal with the red fluorescent protein (RFP) marker resulting from cobombardment with the pRFP-HDEL vector (Figure 9A). The pRFP-HDEL construct encodes RFP fused to an N-terminal signal peptide and a C-terminal HDEL endomembrane retention signal and functions as a marker for the ER (Shockey et al., 2006). Transport of the Ps NCS2-GFP fusion from the ER to the central vacuole was observed between 12 and 24 h after bombardment (Figure 9A). By 48 h after bombardment, Ps NCS2-GFP was exclusively associated with the central vacuole as shown by the lack of overlay between the GFP and RFP signals. By contrast, native GFP was exclusively associated with the cytoplasm and nucleus for at least 48 h after bombardment with the pGFP vector (Figure 9B).
Figure 9.
Subcellular Localization of a Ps NCS2-GFP Fusion in Opium Poppy Cell Cultures at Various Times after Microprojectile Bombardment of the Corresponding Plasmid Vector.
(A) Opium poppy cells expressing either pNCS2-GFP and pRFP-HDEL were observed using a fluorescence microscope. Images were acquired 12, 24, and 48 h after bombardment. Signals from the GFP and RFP fusion proteins are shown as a red and green, respectively, and overlapping fluorescent regions are shown in yellow. Bar = 10 μm and applies to all micrographs.
(B) Opium poppy cells expressing pRFP-HDEL and pGFP. Images were acquired by fluorescence microscopy 48 h after microprojectile bombardment.
DISCUSSION
Pictet-Spengler reactions between amine and aldehyde substrates are responsible for establishing the core structures that define several different categories of compounds represented among the ~12,000 alkaloids produced by plants (Ziegler and Facchini, 2008). In addition to the formation of (S)-norcoclaurine as the first step in BIA metabolism, Pictet-Spengler condensations catalyzed by strictosidine synthase (Ma et al., 2006) and deacetylipecoside and deacetylisoipecoside synthases (De Eknamkul et al., 1997) yield central intermediates leading to many structurally diverse monoterpenoid indole and ipecac alkaloids, respectively. The Pictet-Spengler reaction involves the acid-catalyzed electrophilic addition of an iminium ion to a substituted benzyl or indole moiety via a two-step process whereby the iminium ion is first generated via condensation between the aldehyde carbonyl and the phenylethyl amine or tryptamine, followed by Mannich-based cyclization to yield the alkaloid product (Ma et al., 2006; Luk et al., 2007; Ilari et al., 2009; Bonamore et al., 2010). Despite a common reaction mechanism, at least two different enzyme families have been implicated in the catalysis of Pictet-Spengler condensations in plants. Strictosidine synthase, which yields strictosidine from tryptamine and secologanin, is a member of the amine lyase family and exhibits a novel six-bladed propeller fold unique among plant proteins (Ma et al., 2006). By contrast, Tf NCS shares no common features with the structure of strictosidine synthase but rather conforms to the overall fold of PR10/Bet v1 proteins (Ilari et al., 2009). It is notable that Pictet-Spengler condensations involving any aldehyde other than formaldehyde lead to the formation of a new chiral center and that strictosidine synthase and Tf NCS catalyze stereoselective reactions. Although the nonenzymatic coupling of dopamine and 4-HPAA leads to racemic (R,S)-norcoclaurine (Minami et al., 2007), only (S)-norcoclaurine has been identified in plants.
The substantial amino acid sequence identity among known biosynthetic enzymes suggests a monophyletic origin for BIA metabolism in plants belonging to the Papaveraceae, Ranunculaceae, Berberidaceae, and related families (Liscombe et al., 2005; Samanani et al., 2005). Proteins in the PR10/Bet v1 family that possess NCS activity have now been isolated from two members of the Papaveraceae (P. somniferum and P. bracteatum) and two members of the Ranunculaceae (T. flavum and C. japonica), all of which produce BIAs (Samanani et al., 2004; Liscombe et al., 2005; Minami et al., 2007; Figure 7). All of these NCS isoforms share at least 40% amino acid identity, belong to the same clade with respect to a large number of PR10/Bet v1 proteins reported from a wide variety of plant species (Figure 6), and exhibit common structural features including the strict conservation of catalytic residues identified in the active site of Tf NCS (Ilari et al., 2009; Figure 8) and the occurrence (with the exception of Pb NCS) of an N-terminal extension containing a signal peptide not found among PR10/Bet v1 proteins lacking NCS activity (Figures 8 and 9). It is notable that the Tf NCS cDNA was isolated via the empirical amino acid sequence of tryptic peptides derived from Tf NCS enzyme purified from T. flavum cell cultures (Samanani and Facchini, 2002; Samanani et al., 2004). The occurrence of a secondary activity not associated with column chromatography fractions containing Tf NCS was not detected, suggesting that no other enzyme contributed to the overall NCS activity in T. flavum cell cultures, which constitutively produce protoberberine alkaloids. The contribution of Tf NCS to the overall NCS activity in total protein extracts of T. flavum cell cultures was investigated by immunoprecipitation. The immunospecific precipitation of Tf NCS in crude protein to levels that were not detectable by immunoblot analysis resulted in a corresponding reduction in the formation of (R,S)-norcoclaurine to levels resulting from the spontaneous condensation of dopamine and 4-HPAA (Figure 2). Although we cannot rule out the lack of a contribution by another enzyme, the immunoprecipitation of the vast majority of the NCS activity in T. flavum cell cultures using a Tf NCS antiserum supports the role of the PR10/Bet v1–type enzyme in BIA metabolism.
In opium poppy, the abundance of NCS in root and stem is in agreement with the distribution of other BIA biosynthetic enzymes (Figure 3; Bird et al., 2003; Samanani et al., 2006), suggesting that although the mature carpel is a major site of alkaloid accumulation, BIA metabolism primarily occurs in stems and roots. The time course for the induction of NCS transcripts and protein in elicitor-treated opium poppy cell cultures is also in agreement with the transcriptional activation profiles of all other known enzymes involved in sanguinarine biosynthesis (Zulak et al., 2007). The localization of NCS to sieve elements of the phloem in all opium poppy organs was demonstrated by coimmunolocalization in serial sections of the sequential enzyme 6OMT, which was previously shown to reside in this cell type using a variety of cellular markers (Samanani et al., 2006). All successive enzymes in the pathway from (S)-norcoclaurine to (S)-reticuline, and several known enzymes operating in branch pathways leading to morphine and other end products, have also been localized to sieve elements of the phloem (Bird et al., 2003; Samanani et al., 2006); thus, NCS displays a consistent cellular localization with respect to other BIA biosynthetic enzymes. Similarly, transcripts encoding nine consecutive enzymes (including NCS) involved in berberine biosynthesis were localized by in situ hybridization to common cell types in the root and rhizome of T. flavum (Samanani et al., 2005). The affiliation of NCS with the same cell types containing other BIA biosynthetic enzymes in two different species further supports the role of PR10/Bet v1-type NCS in BIA metabolism.
The occurrence of a putative N-terminal signal peptide was suggested as evidence that Cj PR10A could not serve as the enzyme producing (S)-norcoclaurine for BIA biosynthesis (Minami et al., 2007). We confirmed that the 40–amino acid N-terminal extension of Ps NCS2 included an ~25–amino acid signal peptide targeting the ER in opium poppy cells (Figure 9). The conservation of the 40–amino acid N-terminal extension in PR10/Bet v1–type NCS enzymes from different BIA-producing plant species in two different families and the lack of this domain in any other member of the PR10/Bet v1 protein family suggest that the formation of (S)-norcoclaurine occurs in the lumen of the ER and/or in association with ER-derived vesicles. The localization of NCS in the ER lumen rather than the cytosol, as suggested for Cj NCS1 (Minami et al., 2007), is consistent with the established targeting of the berberine bridge enzyme (BBE) as mediated by an N-terminal signal peptide and vacuolar-sorting determinant (Bird and Facchini, 2001; Alcantara et al., 2005). BBE is a branch point enzyme catalyzing the conversion of (S)-reticuline to (S)-scoulerine in the pathway leading to sanguinarine (Figure 1). BBE and (S)-tetrahydroprotoberberine oxidase, the final enzyme in the biosynthesis of berberine (Figure 1), have been associated with the ER in Berberis stolonifera (Berberidaceae) cell cultures (Amann et al., 1986). The interspersal of known cytosolic enzymes [various O- and N-methyltransferases, NADPH-dependent reductases, 2OG/Fe(II)-dependent dioxygenases, and an acetyltransferase], membrane-bound enzymes (several P450-dependent monooxygenases), and membrane-associated enzymes [NCS, BBE, and (S)-tetrahydroprotoberberine oxidase] in BIA metabolism suggests extensive transmembrane trafficking of pathway intermediates.
A major advancement in the establishment of opium poppy as a model system to investigate BIA metabolism has been achieved by demonstrating that VIGS is an effective method to correlate the posttranscriptional silencing of specific biosynthetic genes with targeted effects on alkaloid profile (Hileman et al., 2005). The specific silencing of T6ODM and CODM resulted in the profound accumulation of thebaine and codeine, respectively, which confirmed the in vitro catalytic activity of these enzymes in the plant (Hagel and Facchini, 2010a). The pTRV2-Ps NCS construct was expected to result in the silencing of Ps NCS1, Ps NCS2, and any other related members of the gene family (Figure 5A). Indeed, NCS transcripts were significantly reduced in plants infiltrated with A. tumefaciens harboring pTRV2-Ps NCS compared with plants infiltrated with the pTRV2-EV empty vector (Figure 5B). All plants selected for analysis contained TRV coat protein transcripts. The significant reduction in the levels of the five most abundant alkaloids in the latex of opium poppy (variety Bea’s Choice) in plants infiltrated with A. tumefaciens harboring pTRV2-Ps NCS compared with plants infiltrated with the pTRV2-EV empty vector strongly supports the role of PR10/Bet v1-type NCS in BIA metabolism (Figure 5C). The relative reduction by ~75% in the overall alkaloid content is in agreement with the decrease in NCS transcript levels by ~80% in plants exposed to pTRV2-Ps NCS compared with pTRV2-EV controls. The use of transient RNA interference to silence Cj NCS1 was reported to reduce berberine accumulation in C. japonica protoplasts (Minami et al., 2007).
Recombinant Cj NCS1 failed to catalyze the formation of (S)-norcoclaurine using assay conditions described previously (Minami et al., 2007) and dopamine and 4-HPAA at concentrations of 10 μM and 300 μM, respectively, which minimizes the nonenzymatic condensation these substrates. The inability of recombinant Cj NCS1 to catalyze the formation of (S)-norcoclaurine, in our hands, is underscored by the intriguing selection of Cj PR10A as the enzyme used for the reconstitution in E. coli of the early steps in BIA metabolism (Minami et al., 2008). Recombinant Cj NCS1 was purported to exhibit considerably lower activity than the native enzyme and was suggested as unsuitable for use in an efficient microbial production system (Minami et al., 2008). The basis for this conclusion cannot be substantiated because native Cj NCS1 has not been purified or characterized. The conclusion that Cj NCS1 is primarily responsible for NCS activity in C. japonica is difficult to reconcile given the occurrence of the much more efficient Cj PR10A.
We have established an EST database based on more than 3.5 million 454 GS-FLX Titanium (454 Life Sciences) pyrosequencing reads generated from opium poppy stem and cell culture cDNA libraries (Genebank Sequence Read Archive SRX012994 to SRX013002). The nearest homologs to Cj NCS1 in the opium poppy stem and cell culture transcriptomes were recently identified as 2OG/Fe(II)-dependent, regiospecific O-demethylases of morphinan and protoberberine alkaloids (Hagel and Facchini, 2010a, 2010b). T6ODM and CODM catalyze 6-O- and 3-O-demethylation, respectively, of morphinan alkaloids, whereas CODM and DIOX2 catalyze the 3-O- and 10-O-demethylation, respectively, of protoberberine alkaloids. Moreover, the knockdown of T6ODM and CODM using VIGS resulted in specific metabolic blocks only in late intermediates of morphine biosynthesis but did not affect overall alkaloid content (Hagel and Facchini, 2010a). We tested each enzyme for NCS activity under 2OG/Fe(II)-dependent dioxygenase (Hagel and Facchini, 2010a) and Cj NCS1 (Minami et al., 2007) assay conditions, but none was detected. Unlike the occurrence of PR10/Bet v1–type NCS enzymes in several BIA plant species (Figure 7), Cj NCS1 orthologs do not occur in opium poppy. To test the corollary possibility that Cj NCS1 plays a role in BIA metabolism other than the formation of (S)-norcoclaurine, we also performed assays using one of 10 available BIA substrates (see Supplemental Figure 2 online) under 2OG/Fe(II)-dependent dioxygenase reaction conditions (Hagel and Facchini, 2010a). No evidence of O-demethylation or hydroxylation was detected by LC-MS/MS, indicating that Cj NCS1 does not accept these particular substrates.
The Pictet-Spengler condensation of dopamine and 4-HPAA is central to the biosynthesis of myriad BIAs in plants, including several pharmacologically important compounds such as morphine and codeine. We unequivocally demonstrated that the enzyme responsible for this reaction is a member of the PR10/Bet v1 family of proteins.
METHODS
Plants and Cell Cultures
Opium poppy (Papaver somniferum) plants were cultivated in growth chambers (Conviron) at a day/night temperature of 20/18°C and under a photoperiod of 14 h. Plant organs were harvested 1 to 2 d after anthesis. Opium poppy cell suspension cultures were maintained on a medium consisting of B5 salts and vitamins (Gamborg et al., 1968), 100 mg L−1 myo-inositol, 1 g L−1 hydrolyzed casein, 20 g L−1 sucrose, and 1 mg L−1 2,4-D. Fungal elicitor was prepared from Botrytis spp as described previously (Facchini et al., 1996). Elicitor treatment was initiated by adding 1 mL of fungal homogenate to 50 mL of cell culture in rapid growth phase 2 d after subculture. Cultured cells were collected by vacuum filtration and stored at −80°C.
Chemicals and Substrates
[8-14C]Dopamine hydrochloride (50 mCi mmol−1) was purchased from American Radiolabeled Chemicals. Dopamine hydrochloride was purchased from Sigma-Aldrich. 4-HPAA was synthesized according to the method of Hirose et al. (2000). Codeine, thebaine, and oripavine were obtained as described previously (Hagel and Facchini, 2010a). (S)-Corytuberine was obtained from Sequoia Research Products, and (S)-reticuline was a gift from Tasmanian Alkaloids. Noscapine, papaverine, and (±)-pavine hydrochloride were purchased from Sigma-Aldrich. (S)-Scoulerine was purchased from Indofine Chemical. (R,S)-Tetrahydropalmatine from Corydalis yahsusuo was obtained from Ethnogarden Botanicals. All other chemicals were purchased from Sigma-Aldrich.
RNA Gel Blot Analysis
For RNA gel blot analysis, 15 μg of total RNA was isolated as described previously (Logemann et al., 1987), fractionated on a formaldehyde agarose gel, and blotted on a nylon membrane. Blots were hybridized with a full-length, random primer 32P-labeled NCS probe. Hybridizations were performed at 65°C in 250 mM sodium phosphate buffer, pH 8.0, 7% (w/v) SDS, 1% (w/v) BSA, and 1 mM EDTA. After hybridization, the membranes were washed at 65°C twice with 2× SSC containing 0.1% (w/v) SDS and twice with 0.2× SSC containing 0.1% (w/v) SDS and autoradiographed at −80°C.
Protein Extraction and Immunoblot Analysis
Plant tissues were ground under liquid nitrogen to a fine powder with a mortar and pestle. Ground samples were suspended in extraction buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.05% [v/v] Tween 20, 1 mM EDTA, and 250 μM phenylmethanesulphonyl fluoride) and incubated on ice for 15 min, and the supernatant was collected by centrifugation at 15,000g for 20 min. Protein concentrations were determined (Bradford, 1976) using BSA as the standard. Extracted proteins were resolved on 12% (w/v) SDS-PAGE gels, which were transferred to nitrocellulose membranes. Protein blots were blocked with 5% (w/v) skim milk and then incubated overnight with a 1:1000 (by volume) dilution of Ps NCS2 antiserum. After washing three times with PBST (137 mM NaCl, 8.1 mM Na2HPO4, 2.7 mM KCl, 1.5 mM KH2PO4, 0.1% [v/v] Tween 20), the blots were incubated for 1 h with a 1:10,000 (by volume) dilution of anti-mouse IgG antibodies conjugated with horseradish peroxidize (Bio-Rad). Immunoblots were washed several times with PBST, developed using SuperSignal West Pico chemiluminescent substrate (Pierce Biotechnology), and visualized by exposure to x-ray film.
Ps NCS2 Expression Construct
Full-length clones encoding two NCS isoforms (Ps NCS1 and Ps NCS2) were isolated from an opium poppy cDNA library (Liscombe et al., 2005). The Ps NCS2 open reading frame (ORF) was cloned into the SacI and HindIII sites of the expression vector pET43a(+) (EMD Chemicals). The pET43a(+) vector encodes an N-terminal NusA-tag/His6-tag/S-tag fusion upstream of thrombin and enterokinase cleavage sites, a multiple cloning site region, and a C-terminal His6-tag sequence. Escherichia coli ER2566pLysS (New England Biolabs) harboring the pET43a-NCS2 construct was cultured with shaking at 37°C in Luria-Bertani (LB) medium containing 50 μg mL−1 ampicillin and 34 μg mL−1 chloramphenicol to an OD600 of 0.5. Production of recombinant NCS2 fusion protein was induced by the addition of 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 20 h at room temperature. Cells were harvested by centrifugation at 8000g for 20 min and then sonicated in lysis buffer (50 mM Tris-HCl, pH 8.0, containing 50 mM NaCl, 1 mM EDTA, 1 mM DTT, and 1% [v/v] Triton X-100). After removal of cell debris by centrifugation, recombinant protein in the supernatant was purified using Talon His-Tag Purification Resin (Clontech) according to the manufacturer’s instructions.
Preparation of Ps NCS2 Antibodies
The purified NusA-Ps NCS2 fusion protein was dialyzed against 146 mM NaCl, resuspended at a concentration of 200 μg mL−1, and emulsified 1:1 (by volume) with Freund’s complete adjuvant, and 100 μL was injected subcutaneously into mice. Booster injections were performed every 3 weeks until a sufficient titer was achieved. Preimmune serum was collected from each mouse before injecting the antigen.
Immunocytochemical Localization
Plant organs were fixed overnight at 4°C in 0.1 M phosphate buffer, pH 7.3, containing 4% (v/v) paraformaldehyde, cut into 1- to 3-mm sections, and further fixed for 4 to 6 h. Tissues were rinsed in 50 mM PIPES, pH 7.0, containing 5 μM phenylmethylsulfonyl fluoride, dehydrated in a graded 30 to 100% (v/v) ethanol series under vacuum, and infiltrated using LR White resin (London Resin Company) diluted 1:4 (by volume) with ethanol. The ratio of LR White to ethanol was increased to 1:3, 1:2, 1:1, 2:1, and 3:1 (by volume). Tissues were subsequently incubated at 4°C for 16 to 24 h in undiluted LR White resin, which was polymerized at 60°C for 16 h in gelatin capsules. Sections were cut 2-μm thick using an EM UC6 Ultramicrotome (Leica Microsystems).
Mouse NCS and 6OMT antisera (Samanani et al., 2006) and rabbit opium poppy MLP antiserum (Nessler et al., 1985) were used for immunocytochemical localization. Tissue sections were blocked for 1 h in Tris-buffered saline (TBS; 10 mM Tris-HCl, pH 7.2, 500 mM NaCl, 0.3% [v/v] Tween 20 containing 1% [w/v] BSA [Fraction V; Roche Diagnostics]), incubated with the primary antibody for 3 h in a humid chamber, and rinsed five times for 15 min each in TBS containing 1% (w/v) BSA. The sections were incubated for another 1 h with either Alexa 488–conjugated goat anti-mouse IgG or Alexa 594–conjugated goat anti-rabbit IgG (Molecular Probes), rinsed three times in TBS containing 1% (w/v) BSA, three times in TBS, and finally in filtered water. Slides were sealed with Fluoro-Gel (Electron Microscopy Sciences).
Microprojectile Bombardment of Cultured Opium Poppy Cells
The pNCS2-GFP construct was assembled by cloning the Ps NCS2 ORF in frame and upstream of the GFP coding region. Expression was achieved by inserting the fusion protein cassette between the constitutive CaMV 35S promoter and the nopaline synthase terminator in pBluescript. Construction of pRFP-HDEL encoding the RFP fused to an N-terminal signal sequence from Arabidopsis thaliana chitinase and a C-terminal HDEL retrieval signal was described previously (Shockey et al., 2006). Cultured opium poppy cells in rapid growth phase were collected on Whatman GF/D microfiber filters by a gentle vacuum filtration to form a thin layer. Microprojectile bombardment was performed using DNA-coated gold particles and a biolistic particle acceleration device (PDS 1000/He; Bio-Rad) as described previously (Bird and Facchini, 2001). After bombardment, 600 μL of sterile culture medium was added, and the cells were incubated at 23°C.
Fluorescence Microscopy and Imaging
Fluorescent images were captured using a Leica DM RA2 microscope (Leica Microsystems), a Retiga EX digital camera (QImaging), and Open Lab version 2.09 (Improvision). Light microscopy images were captured using the Leica microscope and the Retiga camera mounted with an RGB color liquid crystal filter (QImaging).
VIGS
A 420-bp fragment consisting of the 3′ end of the ORF and the 3′ untranslated region common to both Ps NCS1 and Ps NCS2 was amplified using a sense primer containing an EcoRI site (5′-CGCAATTCAGAAAACCCCAAACTCATG-3′) and an antisense primer containing a BglII site (5′-GAAGATCTTCAAACATCCGCCAGAAAC-3′). The fragment was cloned into the pTRV2 vector to generate pTRV2-NCS (Figure 5). Agrobacterium tumefaciens strain GV3101 harboring pTRV1, pTRV2-NCS, or the pTRV2-EV empty vector was cultured at 28°C in 300 mL of LB medium containing 10 mM MES, 20 μM acetosyringone, and 50 μg mL−1 kanamycin. Bacteria were pelleted at 3000g for 15 min and resuspended in infiltration buffer (10 mM MES, 200 μM acetosyringone, and 10 mM MgCl2) to an OD600 of 2.5. Two-week-old opium poppy seedlings were infiltrated using a 1-cc syringe with a 1:1 (by volume) mixture of A. tumefaciens cultures harboring pTRV1 and either pTRV2-NCS or pTRV2-EV. Infiltrated plants were analyzed at maturity (i.e., the emergence of flower buds). Stems were cut immediately below the flower bud, and 10 μL of exuding latex was collected. At the same time, young stem tissue was frozen in liquid nitrogen for qRT-PCR analysis.
qRT-PCR
Plant tissue was ground under liquid nitrogen and extracted in 0.8 M guanidinium thiocyanate, 0.4 M ammonium thiocyanate, 0.1 M sodium acetate, pH 5.0, 5% (v/v) glycerol, and 38% (v/v) Tris-buffered phenol. Subsequently, 200 μL CHCl3 was added and the mixture was emulsified. Samples were centrifuged, and 400 μL of the aqueous phase was precipitated with 500 μL isopropanol. After centrifugation, the supernatant was discarded and the pellet was washed with 70% (v/v) ethanol. The RNA was reduced to dryness and resuspended in 30 μL sterile water. First-strand cDNA was synthesized from 100 to 400 ng of total RNA using RevertAid M-MuLV reverse transcriptase (Fermentas) and an oligo(dT)20VN primer. Real-time PCR using SYBR Green detection was performed on an Applied Biosystems 7300 real-time PCR system. The opium poppy elongation factor 1a gene (Ps EL1A) was used as an endogenous reference. Forward and reverse primers for Ps NCS were 5′-GCAATGGTGGAGTTGGTAC-3′ and 5′-AAAGATATGGATCCTGTCCATGTAAAATG-3′, respectively. The forward and reverse primers for EL1A were 5′-TGCTCCTGTTCTGGATTGTC-3′ and 5′-GTCTCAACCACCATTGGCTTG-3′, respectively. TRV coat protein primers were designed as described previously (Rotenberg et al., 2006).
HPLC
HPLC was performed using a System Gold HPLC apparatus (Beckman-Coulter) equipped with a LiChrospher 60 RP Select B column (146 × 4.1 mm, 5 μm; Merck) and a mobile phase consisting of solvent A (2% [v/v] acetonitrile and 98% [v/v] water) and solvent B (98% [v/v] acetonitrile and 2% [v/v] water), each containing 0.01% (v/v) phosphoric acid. The column was equilibrated in solvent A, and alkaloids were eluted at a flow rate of 1.5 mL min−1 using the following gradient: 0 to 1 min to 10% (v/v) solvent B, 1 to 50 min to 100% (v/v) solvent B, 50 to 53 min to 2% (v/v) solvent B, 53 to 60 min hold at 2% (v/v) solvent B. Dextromethorphan was used as an internal standard. Peaks corresponding to morphine, codeine, thebaine, noscapine, papaverine, and dextromethorphan were monitored at 210 nm and identified on the basis of retention times and UV spectra compared with those of authentic standards.
Immunoprecipitation of NCS Activity
Crude protein extract was isolated from Thalitrum flavum cell cultures in extraction buffer (100 mM Tris-HCl, pH 7.0, and 12 mM 2-mercaptoethanol). Extracts were filtered and centrifuged at 15,000g for 20 min at 4°C, the supernatants were desalted using a PD-10 column (GE Healthcare), and the samples were pooled and concentrated. For immunoprecipitation, protein extracts were incubated overnight at 4°C with the different volumes of polyclonal Tf NCSΔ19 antiserum or preimmune serum. Assembly of the Tf NCSΔ19 expression construct in pET29b was described previously (Samanani et al., 2004). A 500-mL culture of E. coli ER2556 pLysS cells harboring pET29b-Tf NCSΔ19 was grown at 37°C in LB medium to an A600 of 0.6 and induced at room temperature for 4 h with 0.3 mM IPTG. Purification of the recombinant protein and the raising of antibodies against Tf NCSΔ19 were performed as described for the NusA-Ps NCS2 fusion protein. Fifty microliters of protein A-Sepharose 4B fast flow (Sigma-Aldrich) was washed twice with HNTG buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 0.1% [w/v] Triton X-100, and 10% glycerol). The enzyme antiserum mixture was incubated at 4°C for 7 h with the protein A-Sepharose. The supernatant was recovered by centrifugation at 6000g for 5 min and used for immunoblot analysis or enzyme assays. The pellet was washed three times with HNTG buffer, and proteins were eluted twice with 20 μL of 100 mM glycine, pH 2.5. The solution was neutralized with 1 M Tris-HCl, pH 9.5, eluted proteins were separated by SDS-PAGE, and immunoblot analysis was performed using Tf NCSΔ19 antiserum.
Expression and Purification of Recombinant Pb NCS, Am PR10, and Cj NCS1
Codon-optimized synthetic Pb NCS, Am PR10, and Cj NCS1 genes were purchased from GenScript. Pb NCS and Am PR10 were cloned into pQE30 (Qiagen), whereas Cj NCS1 was cloned into pRSETA (Invitrogen) using BamHI and PstI sites incorporated into the synthetic genes and flanking the ORFs. Cultures (500 mL) of E. coli BL21 harboring pQE30-PbNCS, pQE30-Am PR10, or pRSETA-Cj NCS1 were grown at 37°C in LB medium to an A600 of 0.3 and induced at room temperature for 4 h with 0.3 mM IPTG. Cells were harvested by centrifugation at 8000g for 20 min and sonicated in lysis buffer (50 mM Tris-HCl, pH 8.0, containing 50 mM NaCl, 1 mM EDTA, 1 mM DTT, and 1% [v/v] Triton X-100). After removal of cell debris by centrifugation, soluble His6-tagged proteins were purified using Talon His-Tag purification resin. Recombinant proteins were eluted using a 50 to 500 mM imidazole gradient, and peak fractions were identified by SDS-PAGE. Recombinant proteins were confirmed by immunoblot analysis using a His6-tag antibody. Purified proteins were desalted on a PD-10 column (GE Healthcare).
NCS Assays
NCS activity was determined by monitoring (1) the formation of [14C]norcoclaurine using autoradiography, or (2) the production of (S)-norcoclaurine by LC-MS/MS. For the radioactive NCS assay, 30 μL reactions included 311 pmol (187 Bq) [8-14C]dopamime, 300 μM 4-HPAA, and 10 μg of purified recombinant protein in 100 mM Tris-HCl, pH 7.0, containing 12 mM 2-mercaptoethanol. After incubation for 2 h at 37°C, the reaction was applied to a silica gel 60 F254 TLC plate (EMD Chemicals), which was subsequently developed in n-butanol:acetic acid:water (4:1:5 by volume). Phosphor image screens were scanned in Molecular Imager FX (Bio-Rad), and [14C]norcoclaurine was quantified using Quantity One 4.5.1 (Bio-Rad) software.
For the nonradioactive NCS assays, 30-μL reactions contained 10 μM dopamine hydrochloride, 300 μM 4-HPAA, and soluble protein (i.e., remaining after immunoprecipitation) in 100 mM Tris-HCl, pH 7.0. After incubation for 2 h at 37°C, protein was precipitated with a 1:10 volume of 20% (v/v) trichloroacetic acid. For LC-MS/MS, the assays were diluted 1:10 (by volume) with solvent A (98% [v/v] water, 2% [v/v] acetonitrile, and 0.2% [v/v] phosphoric acid) and analyzed using a 6400 Triple Quadrupole LS-MS/MS system (Agilent Technologies). Liquid chromatography was performed at a flow rate of 0.4 mL min−1 using a Zorbax Eclipse Plus C18 column (2.1 × 50 mm, 1.8-μm particle size; Agilent Technologies). The column was equilibrated in solvent A, and (S)-norcoclaurine was eluted using the following gradient: 0 to 1 min to 5% (v/v) solvent B, 1 to 10 min to 25% (v/v) solvent B, and 10 to 12 min to 100% (v/v) solvent B (solvent B: 2% [v/v] water, 98% [v/v] acetonitrile, and 0.2% [v/v] phosphoric acid). Injection into the mass analyzer was performed using an electrospray ionization (ESI) probe inlet. Ions were generated and focused using an ESI voltage of 4000 kV, 10 liters min−1 gas flow, 30 p.s.i .nebulizing pressure, and gas temperature of 350°C. The signal was achieved in positive ion mode with ESI.
The MS/MS fragmentation patterns representing (S)-norcoclaurine were initially monitored in reactions containing 4-HPAA, dopamine, and purified recombinant Tf NCSΔ19 protein or total soluble proteins from T. flavum cell cultures. (S)-Norcoclaurine eluted from the C18 columns with a stable retention time and showed five significant [M+H]+ ions (i.e., mass-to-charge ratio [m/z] 272.1, 255.1, 161.0, 143.0, and 107.0) consistent with published ESI-MS/MS spectra (Schmidt et al., 2005). Analysis was performed by multireaction monitoring using the precursor ion at m/z 272.2 (collision energy of –5.0 eV) and the fragment ions at m/z 161.1 (collision energy of –15 eV) and 107 (collision energy of −28 eV) for (S)-norcoclaurine, and the precursor ion at m/z 272.0 (collision energy of −24 eV) and fragment ion at m/z 215.1 (collision energy of −24 eV), 171.1 (collision energy of −44 eV), and 147.1 (collision energy of −32 eV) for the internal standard dextromethorphan.
Standard curves for LC-MS/MS analysis were produced by linear regression of the peak area ratios for the analyte at m/z 107.0 [corresponding to (S)-norcoclaurine] versus the activity (in pmol s−1 mg−1) of NCS determined via radioactive assays using the same protein samples (i.e., different concentrations of purified Tf NCS protein and substrates). For more accurate quantification, an absolute amount of the internal standard was added into each sample for LC-MS/MS analysis and the peak intensity of the analyte at m/z 171.1 (corresponding to dextromethorphan) was used to normalize the actual intensity.
Phylogenetic Analysis
Multiple sequence alignment and molecular phylogenetic analysis of PR10/Bet v1-type proteins were performed with ClustalX and PHYLIP, respectively (see Supplemental Data Set 1 online). The graphical output of the bootstrapped figure was produced using Treeview (Page, 1996). The amino acid sequences used were Ah PR10 (Arachis hypogaea PR10 protein), Bp PR10 (Betula pendula Ypr10a protein), Am PR10 (Argemone mexicans PR10 protein), Cc PR10 (Capsicum chinense PR10 protein), Cj PR10A (Coptis japonica PR10 protein A), Dc PR10 (Daucus carota PR10 protein), Gba PR10 (Gossypium barbadense PR10-12-like protein), Gbi PR10 (Gossypium bicki PR10 protein), Gh PR10 (Gossypium hirsutum PR protein class 10), Ho PR10 (Hyacinthus orientalis PR10 protein), La PR10 (Lupinus albus PR10 protein), Le PR10 (Lycopersicon esculentum PR10 protein), Ll PR10 (Lupinus luteus PR10 protein), Ms PR10 (Medicago sativa PR10 protein), Os PR10 (Oryza sativa PR10 protein homolog), Os PR10-1 (Oryza sativa Japonica group root-specific PR10 protein), Pb NCS (Papaver bracteatum NCS), Pd PR10 (Prunus domestica PR10 protein), Pf PR10 (Pisum fulvum PR10 protein), Pm PR10 (Pinus monticola PR10 protein homolog 1.1), Ps NCS1 (Papaver somniferum NCS1), Ps NCS2 (P. somniferum NCS2), Ps PR10-1 (P. somniferum PR10 protein homolog-1), Ps PR10-2 (P. somniferum PR10 protein homolog-2), Ra PR10 (Rheum australe PR10 protein), Sb PR10 (Sorghum bicolar PR10 protein 10b), St PR10 (Solanum tuberosum PR10 protein), Ta PR10 (Triticum aestivum PR10 protein), Th PR10 (Tamarix hispida PR10 protein), and Tf NCS (Thalictrum flavum NCS).
2OG/Fe(II)-Dependent Dioxygenase Assay
Cj NCS1 was assayed in a 100-μL reaction containing 5 μg of purified recombinant protein in 100 mM Tris-HCl, pH 7.4, 100 μM alkaloid substrate, 100 μM 2OG, 10% (v/v) glycerol, 14 mM 2-mercaptoethanol, 10 mM sodium ascorbate, and 0.5 mM FeSO4. Assays were incubated at 30°C for 90 min and subjected to LC-MS/MS analysis. The release of aromatic methyl ethers or the introduction of a hydroxyl group was monitored by comparison of (1) the total ion current and (2) extracted ion chromatograms with respect to potential substrates and reaction products.
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession numbers: Ah PR10 (AY726607), Bp PR10 (AJ289771), Am PR10 (HM363760), Cc PR10 (AJ879115), Cj PR10A (AB267399), Dc PR10 (AB082377), Gba PR10 (AY560552), Gbi PR10 (AY704441), Gh PR10 (AF305067), Ho PR10 (AY389712), La PR10 (AJ000108), Le PR10 (Y15846), Ll PR10 (AF180941), Ms PR10 (AJ311050), Os PR10 (AK110687), Os PR10-1 (AB127580), Pb NCS (EU882990), Pd PR10 (EU117126), Pf PR10 (U65425), Pm PR10 (AY064202), Ps NCS1 (AY860500), Ps NCS2 (AY860501), Ps PR10-1 (AY861682), Ps PR10-2 (AY861683), Ra PR10 (EU931221), Sb PR10 (AY751554), St PR10 (M25156), Ta PR10 (EU908212), Th PR10 (FJ463256), Tf NCS (AY376412), synthetic Am PR10 (HM363761), synthetic Pb NCS (HM363762), and synthetic Cj NCS1 (HM363763).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Detection of tobacco rattle virus (TRV).
Supplemental Figure 2. Benzylisoquinoline alkaloids tested as possible substrates for recombinant Cj NCS1.
Supplemental Data Set 1. Alignment corresponding to phylogenetic analysis in Figure 6.
Supplementary Material
Acknowledgments
We thank Craig Nessler for the MLP antibodies, Robert Mullen for the pRFP-HDEL construct, and Savithramma Dinesh-Kumar for the TRV vector system. We thank Nailish Samanani for performing with the microprojectile bombardment, David Liscombe for constructing the pTRV2-NCS and pET43a-NCS2 vectors, Jörg Ziegler and Andrew Stopford for assistance with the LC-MS/MS analysis, Edward Yeung for helpful discussions, and Jillian Hagel for critical review of the manuscript. P.F. holds the Canada Research Chair in Plant Metabolic Processes Biotechnology. This work was funded by a Natural Sciences and Engineering Research Council of Canada Discovery Grant to P.F.
References
- Alcantara J., Bird D.A., Franceschi V.R., Facchini P.J. (2005). Sanguinarine biosynthesis is associated with the endoplasmic reticulum in cultured opium poppy cells after elicitor treatment. Plant Physiol. 138: 173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann M., Wanner G., Zenk M.H. (1986). Intracellular compartmentation of two enzymes of berberine biosynthesis in plant cell cultures. Planta 167: 310–320 [DOI] [PubMed] [Google Scholar]
- Ames B.D., Korman T.P., Zhang W., Smith P., Vu T., Tang Y., Tsai S.-C. (2008). Crystal structure and functional analysis of tetracenomycin ARO/CYC: Implications for cyclization specificity of aromatic polyketides. Proc. Natl. Acad. Sci. USA 105: 5349–5354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais H.P., Vepachedu R., Lawrence C.B., Stermitz F.R., Vivanco J.M. (2003). Molecular and biochemical characterization of an enzyme responsible for the formation of hypericin in St. John’s wort (Hypericum perforatum L.). J. Biol. Chem. 278: 32413–32422 [DOI] [PubMed] [Google Scholar]
- Berkner H., Schweimer K., Matecko I., Rösch P. (2008). Conformation, catalytic site, and enzymatic mechanism of the PR10 allergen-related enzyme norcoclaurine synthase. Biochem. J. 413: 281–290 [DOI] [PubMed] [Google Scholar]
- Bonamore A., Barba M., Botta B., Boffi A., Macone A. (2010). Norcoclaurine synthase: Mechanism of an enantioselective pictet-spengler catalyzing enzyme. Molecules 15: 2070–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bird D.A., Facchini P.J. (2001). Berberine bridge enzyme, a key branch-point enzyme in benzylisoquinoline alkaloid biosynthesis, contains a vacuolar sorting determinant. Planta 213: 888–897 [DOI] [PubMed] [Google Scholar]
- Bird D.A., Franceschi V.R., Facchini P.J. (2003). A tale of three cell types: alkaloid biosynthesis is localized to sieve elements in opium poppy. Plant Cell 15: 2626–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bufe A., Spangfort M.D., Kahlert H., Schlaak M., Becker W.M. (1996). The major birch pollen allergen, Bet v 1, shows ribonuclease activity. Planta 199: 413–415 [DOI] [PubMed] [Google Scholar]
- Britsch L., Dedio J., Saedler H., Forkmann G. (1993). Molecular characterization of flavanone 3 β-hydroxylases. Consensus sequence, comparison with related enzymes and the role of conserved histidine residues. Eur. J. Biochem. 217: 745–754 [DOI] [PubMed] [Google Scholar]
- Chadha P., Das R.H. (2006). A pathogenesis related protein, AhPR10 from peanut: An insight of its mode of antifungal activity. Planta 225: 213–222 [DOI] [PubMed] [Google Scholar]
- Davies K.M. (1993). A Malus cDNA with homology to Anthirrhinum candida and Zea A2 genes. Plant Physiol. 103: 1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-Eknamkul W., Ounaroon A., Tanahashi T., Kutchan T.M., Zenk M.H. (1997). Enzymatic condensation of dopamine and secologanin by cell-free extracts of Alangium lamarckii. Phytochemistry 45: 477–484 [Google Scholar]
- Dinesh-Kumar S.P., Anandalakshmi R., Marathe R., Schiff M., Liu Y. (2003). Virus-induced gene silencing. Methods Mol. Biol. 236: 287–294 [DOI] [PubMed] [Google Scholar]
- Facchini P.J., Johnson A.G., Poupart J., de Luca V. (1996). Uncoupled defense gene expression and antimicrobial alkaloid accumulation in elicited opium poppy cell cultures. Plant Physiol. 111: 687–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto Y., Nagata R., Fukasawa H., Yano K., Azuma M., Iida A., Sugimoto S., Shudo K., Hashimoto Y. (1998). Purification and cDNA cloning of cytokinin-specific binding protein from mung bean (Vigna radiata). Eur. J. Biochem. 258: 794–802 [DOI] [PubMed] [Google Scholar]
- Gamborg O.L., Miller R.A., Ojima K. (1968). Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50: 151–158 [DOI] [PubMed] [Google Scholar]
- Hagel J.M., Facchini P.J. (2010a). Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy. Nat. Chem. Biol. 6: 273–275 [DOI] [PubMed] [Google Scholar]
- Hagel J.M., Facchini P.J. (July 15, 2010b.) Biochemistry and occurrence of O-demethylation in plant metabolism. Front. Physio. 1 (online), doi/10.3389/fphys.2010.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausinger R.P. (2004). Fe(II)/α-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem. Mol. Biol. 39: 21–68 [DOI] [PubMed] [Google Scholar]
- Hileman L.C., Drea S., Martino G., Litt A., Irish V.F. (2005). Virus-induced gene silencing is an effective tool for assaying gene function in the basal eudicot species Papaver somniferum (opium poppy). Plant J. 44: 334–341 [DOI] [PubMed] [Google Scholar]
- Hirose T., Sunazuka T., Zhi-Ming T., Handa M., Uchida R., Shiomi K., Harigaya Y., Omura S. (2000). Total syntheses of kurasoins a and b, novel protein farnesyltransferase inhibitors, and absolute structures of kurasoins a and b. Heterocycles 53: 777–784 [Google Scholar]
- Holdsworth M.J., Bird C.R., Ray J., Schuch W., Grierson D. (1987). Structure and expression of an ethylene-related mRNA from tomato. Nucleic Acids Res. 15: 731–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton T.A., Brugliera F., Tanaka Y. (1993). Cloning and expression of flavonol synthase from Petunia hybrida. Plant J. 4: 1003–1010 [DOI] [PubMed] [Google Scholar]
- Ilari A., Franceschini S., Bonamore A., Arenghi F., Botta B., Macone A., Pasquo A., Bellucci L., Boffi A. (2009). Structural basis of enzymatic (S)-norcoclaurine biosynthesis. J. Biol. Chem. 284: 897–904 [DOI] [PubMed] [Google Scholar]
- Kovaleva E.G., Lipscomb J.D. (2008). Versatility of biological non-heme Fe(II) centers in oxygen activation reactions. Nat. Chem. Biol. 4: 186–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscombe D.K., MacLeod B.P., Loukanina N., Nandi O.I., Facchini P.J. (2005). Evidence for the monophyletic evolution of benzylisoquinoline alkaloid biosynthesis in angiosperms. Phytochemistry 66: 2501–2520 [DOI] [PubMed] [Google Scholar]
- Logemann J., Schell J., Willmitzer L. (1987). Improved method for the isolation of RNA from plant tissues. Anal. Biochem. 163: 16–20 [DOI] [PubMed] [Google Scholar]
- Loenarz C., Schofield C.J. (2008). Expanding chemical biology of 2-oxoglutarate oxygenases. Nat. Chem. Biol. 4: 152–156 [DOI] [PubMed] [Google Scholar]
- Luk L.Y., Bunn S., Liscombe D.K., Facchini P.J., Tanner M.E. (2007). Mechanistic studies on norcoclaurine synthase of benzylisoquinoline alkaloid biosynthesis: An enzymatic Pictet-Spengler reaction. Biochemistry 46: 10153–10161 [DOI] [PubMed] [Google Scholar]
- Ma X., Panjikar S., Koepke J., Loris E., Stöckigt J. (2006). The structure of Rauvolfia serpentina strictosidine synthase is a novel six-bladed β-propeller fold in plant proteins. Plant Cell 18: 907–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda J., Okabe S., Hashimoto T., Yamada Y. (1991). Molecular cloning of hyoscyamine 6 β-hydroxylase, a 2-oxoglutarate-dependent dioxygenase, from cultured roots of Hyoscyamus niger. J. Biol. Chem. 266: 9460–9464 [PubMed] [Google Scholar]
- Michalska K., Fernandes H., Sikorski M., Jaskolski M. (2010). Crystal structure of Hyp-1, a St. John’s wort protein implicated in the biosynthesis of hypericin. J. Struct. Biol. 169: 161–171 [DOI] [PubMed] [Google Scholar]
- Minami H., Dubouzet E., Iwasa K., Sato F. (2007). Functional analysis of norcoclaurine synthase in Coptis japonica. J. Biol. Chem. 282: 6274–6282 [DOI] [PubMed] [Google Scholar]
- Minami H., Kim J.-S., Ikezawa N., Takemura T., Katayama T., Kumagai H., Sato F. (2008). Microbial production of plant benzylisoquinoline alkaloids. Proc. Natl. Acad. Sci. USA 105: 7393–7398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nessler C.L., Allen R.D., Galewsky S. (1985). Identification and characterization of latex-specific proteins in opium poppy. Plant Physiol. 79: 499–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmark P., Boyle B., Brisson N. (1998). Sequential and structural homology between intracellular pathogenesis-related proteins and a group of latex proteins. Plant Mol. Biol. 38: 1243–1246 [DOI] [PubMed] [Google Scholar]
- Page R.D. (1996). TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Ramón D., Carramolino L., Patiño C., Sánchez F., Peñalva M.A. (1987). Cloning and characterization of the isopenicillin N synthetase gene mediating the formation of the β-lactam ring in Aspergillus nidulans. Gene 57: 171–181 [DOI] [PubMed] [Google Scholar]
- Rotenberg D., Thompson T.S., German T.L., Willis D.K. (2006). Methods for effective real-time RT-PCR analysis of virus-induced gene silencing. J. Virol. Methods 138: 49–59 [DOI] [PubMed] [Google Scholar]
- Samanani N., Alcantara J., Bourgault R., Zulak K.G., Facchini P.J. (2006). The role of phloem sieve elements and laticifers in the biosynthesis and accumulation of alkaloids in opium poppy. Plant J. 47: 547–563 [DOI] [PubMed] [Google Scholar]
- Samanani N., Facchini P.J. (2001). Isolation and partial characterization of norcoclaurine synthase, the first committed step in benzylisoquinoline alkaloid biosynthesis, from opium poppy. Planta 213: 898–906 [DOI] [PubMed] [Google Scholar]
- Samanani N., Facchini P.J. (2002). Purification and characterization of norcoclaurine synthase. The first committed enzyme in benzylisoquinoline alkaloid biosynthesis in plants. J. Biol. Chem. 277: 33878–33883 [DOI] [PubMed] [Google Scholar]
- Samanani N., Liscombe D.K., Facchini P.J. (2004). Molecular cloning and characterization of norcoclaurine synthase, an enzyme catalyzing the first committed step in benzylisoquinoline alkaloid biosynthesis. Plant J. 40: 302–313 [DOI] [PubMed] [Google Scholar]
- Samanani N., Park S.U., Facchini P.J. (2005). Cell type-specific localization of transcripts encoding nine consecutive enzymes involved in protoberberine alkaloid biosynthesis. Plant Cell 17: 915–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J., Raith K., Boettcher C., Zenk M.H. (2005). Analysis of benzylisoquinoline-type alkaloids by electrospray tandem mass spectrometry and atmospheric pressure photoionization. Eur. J. Mass Spectrom. (Chichester, Eng.) 11: 325–333 [DOI] [PubMed] [Google Scholar]
- Shockey J.M., Gidda S.K., Chapital D.C., Kuan J.-C., Dhanoa P.K., Bland J.M., Rothstein S.J., Mullen R.T., Dyer J.M. (2006). Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18: 2294–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R., Kutchan T.M., Zenk M.H. (1989). (S)-Norcoclaurine is the central intermediate in benzylisoquinoline alkaloid biosynthesis. Phytochemistry 28: 1083–1086 [Google Scholar]
- Swoboda I., Hoffmann-Sommergruber K., O’Ríordáin G., Scheiner O., Heberle-Bors E., Vicente O. (1996). Bet v1 proteins, the major birch pollen allergens and members of a family of conserved pathogenesis-related proteins, show ribonuclease activity in vitro. Physiol. Plant. 96: 433–438 [Google Scholar]
- Vazquez-Flota F., De Carolis E., Alarco A.M., De Luca V. (1997). Molecular cloning and characterization of desacetoxyvindoline-4-hydroxylase, a 2-oxoglutarate dependent-dioxygenase involved in the biosynthesis of vindoline in Catharanthus roseus (L.) G. Don. Plant Mol. Biol. 34: 935–948 [DOI] [PubMed] [Google Scholar]
- Ziegler J., Facchini P.J. (2008). Alkaloid biosynthesis: Metabolism and trafficking. Annu. Rev. Plant Biol. 59: 735–769 [DOI] [PubMed] [Google Scholar]
- Zulak K.G., Cornish A., Daskalchuk T.E., Deyholos M.K., Goodenowe D.B., Gordon P.M., Klassen D., Pelcher L.E., Sensen C.W., Facchini P.J. (2007). Gene transcript and metabolite profiling of elicitor-induced opium poppy cell cultures reveals the coordinate regulation of primary and secondary metabolism. Planta 225: 1085–1106 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.