The processing of complex DNA intermediates in replication and repair is essential. This work defines the role of two ATPases, RAD5A and RECQ4A, and the endonuclease MUS81 in DNA repair and recombination in Arabidopsis. It shows that all three proteins are involved in different pathways of DNA repair and have specific roles in double-strand break–induced homologous recombination.
Abstract
Complex DNA structures, such as double Holliday junctions and stalled replication forks, arise during DNA replication and DNA repair. Factors processing these intermediates include the endonuclease MUS81, helicases of the RecQ family, and the yeast SNF2 ATPase RAD5 and its Arabidopsis thaliana homolog RAD5A. By testing sensitivity of mutant plants to DNA-damaging agents, we defined the roles of these factors in Arabidopsis. rad5A recq4A and rad5A mus81 double mutants are more sensitive to cross-linking and methylating agents, showing that RAD5A is required for damage-induced DNA repair, independent of MUS81 and RECQ4A. The lethality of the recq4A mus81 double mutant indicates that MUS81 and RECQ4A also define parallel DNA repair pathways. The recq4A/mus81 lethality is suppressed by blocking homologous recombination (HR) through disruption of RAD51C, showing that RECQ4A and MUS81 are required for processing recombination-induced aberrant intermediates during replication. Thus, plants possess at least three different pathways to process DNA repair intermediates. We also examined HR-mediated double-strand break (DSB) repair using recombination substrates with inducible site-specific DSBs: MUS81 and RECQ4A are required for efficient synthesis-dependent strand annealing (SDSA) but only to a small extent for single-strand annealing (SSA). Interestingly, RAD5A plays a significant role in SDSA but not in SSA.
INTRODUCTION
The DNA integrity of living organisms is constantly threatened. DNA damage can be induced by endogenous sources, such as oxidative stress, or by exogenous sources, such as UV radiation and toxic substances. Organisms have developed several DNA repair mechanisms to cope with DNA damage. While some types of DNA damage are primarily repaired by the action of a single, specific repair pathway, most types of damages are repaired by more than one pathway. One type of DNA damage, DNA cross-links, can be triggered by different agents, including mitomycin C (MMC) and cisplatin (CDDP). MMC mainly forms interstrand cross-links on DNA (Rink et al., 1996), whereas the structures preferentially formed by CDDP are intrastrand cross-links (Eastman, 1985). Both agents can also cause, to a minor degree, a wide variety of other kinds of DNA damage. In Saccharomyces cerevisiae, it has been proposed that three different repair mechanisms are involved in repairing intrastrand cross-links: recombination, postreplication repair (PRR), and nucleotide excision repair (Grossmann et al., 2001).
Sc RAD5, a member of the RAD6 epistasis group, is involved in error-free PRR (Johnson et al., 1992; Xiao et al., 2000). The rad5 (rev2) mutants of yeast are sensitive to UV light and other DNA-damaging agents (Johnson et al., 1992; Hishida et al., 2009). RAD5 is a DNA-dependent ATPase of the SNF2 family with a HIRAN and a RING-finger (C3HC4) domain (Bang et al., 1992; Johnson et al., 1992; Iyer et al., 2006).
DNA lesions in a template strand block replication fork progression. At such damaged sites in the genome, cells use a number of PRR mechanisms to complete DNA replication. Thus, PRR guarantees the completion of DNA replication in the presence of DNA damage that blocks replication fork progression. PRR can be divided into two main pathways: bypass repair that is either error prone or error free and another error-free gap-filling pathway that uses a strand invasion mechanism to complement gaps with DNA intermediates (Andersen et al., 2008; Unk et al., 2010). Both error-free repair pathways depend on different template-switching mechanisms. In yeast, it has been demonstrated that Sc RAD5 physically interacts with different proteins involved in PRR, such as RAD18, an ATPase with a RING finger, and the E2-conjugating enzyme UBC13 (Ulrich and Jentsch, 2000). When RAD5 recruits the protein complex of UBC13 and MMS2 to DNA (Brusky et al., 2000; Ulrich and Jentsch, 2000), the error-free pathway of PRR is initiated; this is followed by proliferating cell nuclear antigen (PCNA) polyubiquitination through a Lys-63–linked ubiquitin chain (Hoege et al., 2002; Parker and Ulrich, 2009). In this process, RAD5 acts as a ubiquitin ligase (E3) due to the C3HC4 motif characteristic for this class of proteins (Johnson et al., 1992). However, RAD5 also seems to be mechanistically involved in the error-free bypass pathway of PRR by its translocase activity: as a result of a so-called overshoot synthesis, the newly synthesized strand on the undamaged parental strand is further elongated. By regressing the replication fork, a special type of Holliday junction, the chicken foot, is formed, and the two newly synthesized strands anneal to each other. Such a conformation allows the longer strand to serve as a template for synthesizing the blocked strand (Figure 1A). Fork regression was recently demonstrated in vitro: RAD5 readily promoted a four-way junction containing moveable homologous arms into a Y-shaped fork (Blastyák et al., 2007). Recently, a report was published demonstrating that Sc rad5 mutant lines were defective in restarting stalled replication forks. The RAD5-dependent formation of Holliday junction containing intermediates indicates that RAD5 may coordinate template switch events between sister chromatids at stalled forks together with proteins involved in HR (Minca and Kowalski, 2010).
Figure 1.
Model for Different RAD5-Dependent Pathways in PRR.
(A) If replication stalls at an unrepaired DNA lesion, error-free repair proceeds with overshoot synthesis at the undamaged parental strand. By the regression of the replication fork, a so-called chicken-foot structure is formed, and through a template switch, the newly synthesized sister strands can anneal; therefore, the longer one can function as a template for synthesizing the shorter one.
(B) Alternatively, error-free repair proceeds through gap filling/strand invasion, refilling a single-strand gap opposite the damage behind the replication fork. In this case, the gap can be filled by a template-switching mechanism: the 3′-end of the newly synthesized strand can invade the undamaged sister chromatid and use the homologous region of the undamaged sister chromatid as a template.
Another way to repair DNA damage is the gap-filling/strand invasion mechanism (Figure 1B). It has been proposed that gap-filling repair requires homologous recombination (HR) as well as RAD5-mediated PCNA polyubiquitination (Branzei et al., 2008). It is thought that with RAD51 and other proteins, a gap can be filled by a strand invasion-dependent pathway: the 3′-end of the newly synthesized strand can invade and get elongated with the sequence information copied from the homologous region of the undamaged sister chromatid (Unk et al., 2010).
The general importance of RAD5 function is highlighted by the fact that there are two RAD5 homologs in humans, SHPRH and HLTF, both of which are tumor suppressor candidates (Motegi et al., 2006, 2008; Unk et al., 2006; MacKay et al., 2009; Unk et al., 2010). Recently, HLTF has been reported to function in the reversal of blocked replication forks (Blastyák et al., 2010). While many studies on RAD5 have centered on its role in PRR, the function of RAD5 in HR is rather elusive. In yeast rad5 mutants, HR has been found to be altered in different ways depending on the reporter system that is applied (Ahne et al., 1997; Liefshitz et al., 1998; Friedl et al., 2001). Additionally, yeast RAD5 seems to suppress nonhomologous end-joining (Ahne et al., 1997; Moertl et al., 2008). Previously, we identified two Arabidopsis thaliana homologs of RAD5, RAD5A (At5g22750) and RAD5B (At5g43530). Both genes are more closely related to each other than to Sc RAD5 and seem to have arisen by a duplication, but only RAD5A is required for DNA repair and HR (Chen et al., 2008). rad5B mutant lines show no effect compared with wild-type plants neither in sensitivity assays with different DNA-damaging agents nor in HR (Chen et al., 2008).
However, mutant plants of RAD5A showed high sensitivity to DNA-damaging agents that induce mainly cross-links (MMC and CDDP) or methylate bases (methyl methanesulfonate [MMS]).
RecQ helicases are also postulated to play a major role in replication fork regression. Mutations in three human RecQ homologs have been shown to cause severe autosomal recessive hereditary diseases, such as Bloom syndrome (BLM), Werner syndrome, and Rothmund-Thomson syndrome that result from biallelic loss-of-function mutations in the genes BLM, WRN, and RECQ4, respectively. All of these syndromes exhibit a set of common characteristics, such as genomic instability and a predisposition to malignant cancers. An elevated sister chromatid exchange rate is a hallmark characteristic in blm fibroblasts and is due to an increased HR frequency (Chaganti et al., 1974; Bohr, 2008; Chu and Hickson, 2009; Vindigni and Hickson, 2009). In Arabidopsis, there are seven different RecQ-like genes (Hartung et al., 2000; Hartung and Puchta, 2006). At the sequence level, two of them, RECQ4A and RECQ4B, can be considered as putative BLM homologs. Interestingly, knockout of RECQ4A and RECQ4B leads to antagonistic phenotypes. Similar to BLM mutant cells, recq4A mutant cells show elevated sensitivity to DNA-damaging agents and an increased HR frequency compared with the wild type. By contrast, the recq4B mutant is not mutagen sensitive but is strongly impaired in HR (Hartung et al., 2007).
MUS81 is a highly conserved protein that forms, together with EME1 (also known as MMS4 in S. cerevisiae) (Boddy et al., 2000; Interthal and Heyer, 2000; Chen et al., 2001), a highly efficient nuclease complex that can resolve intermediate DNA structures, such as 3′-flaps, replication fork structures, displacement loops (D-loops), and nicked Holliday junctions (Boddy et al., 2001; Chen et al., 2001; Kaliraman et al., 2001; Constantinou et al., 2002; Doe et al., 2002; Abraham et al., 2003; Ciccia et al., 2003; Gaillard et al., 2003; Osman et al., 2003; Fricke et al., 2005; Taylor and McGowan, 2008). Besides a MUS81 homolog, two EME1 homologs (EME1A and EME1B) are present in Arabidopsis. We could show that both MUS81/EME1A and MUS81/EME1B exhibit endonucleolytic activity on 3′-flap substrates as well as intact and nicked Holliday junctions (Geuting et al., 2009). Furthermore, we and others recently identified a strong sensitivity to the genotoxic agents MMC, MMS, and CDDP and to ionizing radiation in mus81 mutant lines (Hartung et al., 2006; Berchowitz et al., 2007). Additionally, we observed a reduction of HR frequency after inducing genotoxic stress in the mus81 mutant lines (Hartung et al., 2006). These results indicate a function of MUS81 in HR and probably in resolving intermediates during DNA repair. Furthermore, we showed that recq4A mus81 double mutants exhibited developmental defects and died within about 2 weeks. The growth defect of the recq4A mus81 double mutant is reminiscent of the lethality of mutants of the respective MUS81 and RECQ homologs of budding and fission yeast, SGS1 and RQH1 (Mullen et al., 2001; Doe et al., 2002; Fabre et al., 2002), indicating a high level of conservation of the somatic RECQ and MUS81 functions as well as the involvement of the two proteins in resolving stalled replication forks through two parallel pathways.
To further define the roles of RAD5A, RECQ4A, and MUS81 in DNA repair and HR, we examined the effects of the rad5A, recq4A, and mus81 mutations in different HR reporter lines and the viability of various double mutants prior to and after their exposure to different DNA-damaging agents.
RESULTS
Role of RAD5A in Cross-Link– and Methylation-Induced DNA Repair in Relation to RECQ4A
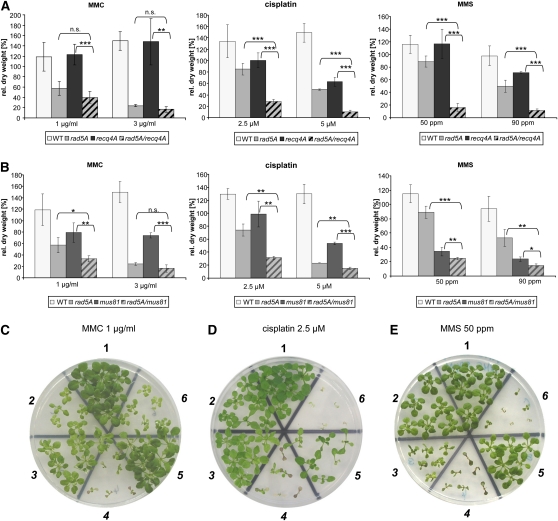
We previously demonstrated that rad5A mutants are sensitive to MMS, which methylates DNA bases, as well as to the DNA cross-linking agents MMC and CDDP (Chen et al., 2008). RecQ helicases have been postulated to be involved in the regression of replication forks in a similar way as RAD5. It was therefore interesting to test whether RECQ4A, which in many ways is the functional equivalent of BLM in humans, is epistatic to RAD5A. recq4A showed a strong sensitivity to both MMS and the intrastrand cross-linking agent CDDP. The mutant lines recq4A-4 and rad5A-2 (Hartung et al., 2007; Chen et al., 2008) were crossed, and the respective rad5A recq4A double mutants were further characterized. These plants were viable and showed no differences from the wild-type phenotype under standard growth conditions. Using two different concentrations for each genotoxin, we examined the sensitivity after 3 weeks of growth on solid GM medium and determined the dry weight of the plantlets in comparison to the wild-type plantlets and plantlets of both single mutants (Figure 2A). The results were statistically evaluated by t tests (see Supplemental Table 1 online). Interestingly, the rad5A recq4A double mutants showed hypersensitivity to CDDP: with the treatment of CDDP at a concentration of 5 μM, the plants died shortly after germination, and this effect was more than additive. Interestingly, with MMC treatment (which mainly causes interstrand cross-links in DNA; Iyer and Szybalski, 1963), we did not detect any effect on the recq4A single mutants. The rad5A recq4A double mutant line displayed no significant difference in dry weight change compared with the rad5A single mutant line. Thus, RAD5A and RECQ4A act through two independent pathways for intrastrand cross-link repair. In addition, only RAD5A is involved in the repair of interstrand cross-links. We also treated the double mutants with another mutagen (MMS) that methylates DNA bases (Lawley, 1989; Beranek, 1990). An effect of the single mutant lines could be detected if the MMS concentration was at least 90 ppm. When the rad5A recq4A double mutants were treated with 90 ppm MMS, a strong hypersensitive phenotype was observed, in contrast with the single mutants (Figure 2A). Treated with 50 ppm MMS, the double mutants displayed a >80% reduction in dry weight compared with the wild type, whereas the rad5A single mutant line showed a 25% reduction, and the recq4A single mutant showed no sensitivity compared with the wild type. Therefore, RAD5A and RECQ4A act through two independent pathways to repair methylated DNA.
Figure 2.
Sensitivity of Single and Double Mutants to DNA-Damaging Agents.
(A) and (B) Depicted are the rad5A-2 and recq4A-4 single mutant lines, as well as the rad5A recq4A double mutant line (A) and the mus81-1 single mutant line, as well as the rad5A mus81 double mutant line (B), treated with the cross-linking agents MMC (1 and 3 μg/mL), CDDP (2.5 and 5 μM) or the methylating agent MMS (50 and 90 ppm). The dry weight of treated seedlings was compared with that of untreated ones. The results were obtained from at least three independent experiments. Error bars indicate sd. Asterisks indicate P values from unpaired t tests. P values are given in Supplemental Table 1 online (***P < 0.001, extremely significant; **P = 0.001 to 0.01, very significant; *P = 0.01 to 0.05, significant; P > 0.05, not significant (n.s.). WT, wild type.
(C) to (E) Wild type (1), rad5A (2), mus81 (3), rad5A mus81 (4), recq4A (5), and rad5A recq4A (6) Arabidopsis sown on media containing 1 μg/mL MMC (C), 2.5 μM CDDP (D), and 50 ppm MMS (E) after 3 weeks of growth.
Role of RAD5A in Cross-Link– and Methylation-Induced DNA Repair in Relation to MUS81
The nuclease MUS81 is also involved in resolving aberrant replication intermediates (Hartung et al., 2006; Geuting et al., 2009). The mus81 mutant line was characterized as strongly sensitive to both the methylating agent MMS and the cross-linking agent MMC (Hartung et al., 2006). We generated a double mutant line by crossing mus81-1 and rad5A-2. The rad5A mus81 double mutants were viable and showed no differences from the wild-type controls under standard growth conditions. Treatment with 90 ppm MMS reduced the dry weight of the double mutants to nearly one-fifth of the respective wild-type values (Figure 2B). The mus81 mutants also displayed sensitivity to CDDP (Berchowitz et al., 2007). Treatment with 2.5 μM CDDP reduced the dry weight of the double mutants to one-third of the untreated control values. By contrast, both single mutant lines showed only minor dry weight reductions (Figure 2B). Treatment with 1 μg/mL MMC induced a statistically significant additive effect in the rad5A mus81 double mutants in comparison to the single mutants. However, using a higher concentration (3 μg/mL) of MMC, the rad5a single mutant line seemed to be so drastically damaged that we were not able to see an additive effect in the rad5A mus81 double mutant anymore (Figure 2B). Notably, the rad5A single mutant line was more sensitive to MMC and CDDP than the mus81 single mutant line, whereas the mus81 mutant line revealed a stronger sensitivity to MMS than the rad5A mutant line (Figure 2B). These results (t test performed, P values; see Supplemental Table 1 online) indicate that RAD5A and MUS81 are not epistatic in DNA repair and that they are involved in independent pathways for repairing MMS-, CDDP-, and MMC-induced damages in Arabidopsis.
Roles of RECQ4A and MUS81 in Processing HR-Induced Aberrant Replication Intermediates
Our results indicate that both RECQ4A and MUS81 define RAD5A-independent pathways of DNA repair. Therefore, the obvious question was whether the two proteins share a common pathway or act through different pathways. To address this question, we crossed these two mutants and found that the recq4A mus81 double mutants showed dramatic growth defects (Hartung et al., 2006). In comparison to the wild type, less than half of the seeds germinated. Most of the homozygous double mutants died early during development (within the first 2 to 3 weeks); very few plants survived longer, and those showed severe growth defects and died after 7 to 8 weeks (Figure 3B). These results demonstrate that RECQ4A and MUS81 act in parallel pathways presumably to resolve replication-associated DNA intermediates. If both pathways fail, further replication is massively hindered, and the seedlings will die early.
Figure 3.
Rescue of the Lethal recq4A/mus81 Phenotype by rad51C.
(A) All pictures display 5-week-old plants. The recq4A-4 single mutants show normal growth and fertility.
(B) The recq4A-4 mus81-1 double mutants exhibit severe growth reduction in soil.
(C) The recq4A mus81 rad51C-1 homozygous triple mutants show normal vegetative growth, together with sterility attributed to the phenotype of rad51C.
To analyze whether the intermediate formation responsible for the lethality depends on HR, we crossed the double mutants with rad51C-1 mutants (Bleuyard et al., 2005). Arabidopsis RAD51C has been shown to play a prominent role in HR (Abe et al., 2005; Bleuyard et al., 2005). As expected, in the rad51C background, the lethality of the recq4A-4 mus81-1 double mutants was rescued. The resulting triple mutants showed normal vegetative growth (Figure 3C), but they were sterile. The meiotic phenotype was attributed to the parental rad51C mutants that were sterile themselves (Bleuyard et al., 2005). Therefore, HR is responsible for producing replication DNA intermediates that must be removed by RECQ4A, MUS81, or both to allow complete replication.
Analysis of HR-Mediated Double-Strand Break Repair
Aside from their involvement in the resolution of DNA repair intermediates, mutations in RAD5A, MUS81, and RECQ4A all could affect HR in somatic cells, as we have previously demonstrated: the rad5A mutants, as well as the mus81 mutants, exhibited a reduced frequency of intrachromosomal HR after the application of the double-strand break (DSB)-inducing agent bleomycin (Hartung et al., 2006; Chen et al., 2008), whereas only the recq4A mutants exhibited an increase of somatic HR frequency when no genotoxin was applied (Hartung et al., 2007).
Unfortunately, the reporter line 651 (Swoboda et al., 1994), which was applied in the previous studies, did not allow us to conclusively discriminate between different HR pathways. Previously, we demonstrated that, in plants, a genomic DSB can be repaired by at least two HR pathways (Figures 4A and 4C): single-strand annealing (SSA) (Siebert and Puchta, 2002) and gene conversion through synthesis-dependent strand annealing (SDSA) (Puchta, 1998). In SDSA, DSBs are processed into 3′-single-strand overhangs by exonuclease-catalyzed digestion (Mimitou and Symington, 2009). By forming a D-loop, a free 3′-end invades the double-stranded donor; therefore, repair synthesis can occur. After elongation of the 3′-end of the acceptor molecule and the disruption of the D-loop, the single-stranded overhangs of the acceptor can anneal, and the sequence of the broken region is restored (Figure 4C). The reaction is conservative because no sequence information is lost. By contrast, in the SSA model, a DSB is induced between repetitive sequences. By exonuclease-catalyzed digestion, 3′-single-strand overhangs are produced. The complementary sequences of the overhangs anneal, and a chimeric DNA molecule is formed. The repair is completed by trimming the overhanging sequences or by filling the single-stranded gaps in DNA. The reaction is nonconservative because all information between the repetitive sequences is lost (Figure 4A).
Figure 4.
Recombination Frequency of rad5A, recq4A, or mus81 Mutant Plants.
Models of the SSA and the SDSA pathway are presented in (A) (SSA) and (C) (SDSA). The HR events are determined with the reporter lines DGU.US-1 (SSA) and IU.GUS-8 (SDSA), which are depicted in (B) and (D). In DGU.US-1, the I-SceI site is flanked by two halves of a GUS gene harboring an overlap of 557 bp. The 5′-GUS fragment was fused with a 35S promoter of the cauliflower mosaic virus and the 3′-GUS fragment to a NOS terminator. Furthermore, the DGU.US-1 construct harbors a resistance gene for phosphinothricin resistance flanked by a 35S promoter and a 35S terminator (B). In IU.GUS-8, a nonfunctional internal fragment of the GUS gene is arranged in inverted orientation to the GUS gene, in which the I-SceI site is embedded. The IU.GUS-8 construct harbors a hygromycin resistance gene fused with a NOS promoter and a NOS terminator (D). The relative recombination frequency is calculated as the mean value of three independent experiments. Error bars indicate the sd. The recombination frequencies of the rad5A (E), recq4A (F), or mus81 (G) mutant plants are presented in relation to the frequency of the corresponding wild-type (WT) control plants (100%). BAR, phosphinothricin acetyltransferase; hygromycin, hygromycin phosphotransferase; RB, right border; LB, left border; NOS, nopaline synthase.
Two reporter lines, DGU.US-1 and IU.GUS-8 (Figures 4B and 4D; Orel et al., 2003), enabling us to discriminate between the two different HR pathways, were crossed into the rad5A, recq4A, or mus81 mutant background (see Supplemental Figure 1 online). Restoration of a β-glucuronidase (GUS) gene in the DGU.US reporter line occurred through the SSA pathway (Figure 4B), whereas restoration in the IU.GUS line depended on gene conversion through the SDSA pathway (Figure 4D). DSBs were induced at a defined position in the respective reporter substrates by a highly specific meganuclease, I-SceI (Colleaux et al., 1986; Puchta et al., 1993).
Requirement of RAD5A for Efficient Homologous Recombination by the SDSA Pathway but Not by the SSA Pathway
For IU.GUS, the recombination frequencies were reduced by half in the rad5A mutant background compared with the wild-type control. By contrast, no significant difference was observed between the mutants and the wild-type controls with the DGU.US reporter system. Thus, RAD5A is involved in gene conversion but not in the SSA pathway of HR in somatic plant cells (Figure 4E).
MUS81 and RECQ4A Play Only Minor Roles in the SSA Pathway but Major Roles in the SDSA Pathway
In contrast with rad5A, a slight reduction of the HR efficiency in the SSA pathway was observed in both the recq4A and the mus81 mutants (Figures 4F and 4G). The reduction was less than a third, arguing that other factors might have more prominent roles or that they were able to substitute for the nuclease or for the helicase. Interestingly, a much stronger effect was observed with the SDSA substrate, in which the HR efficiency was reduced to less than half for both single mutants (Figures 4F and 4G). This observation clearly indicates that both MUS81 and RECQ4A play important roles in SDSA that cannot be fully complemented by other factors. The result is especially interesting for RECQ4A because this protein is involved in suppressing HR when no DSBs are induced (Bagherieh-Najjar et al., 2005; Hartung et al., 2007). Thus, RECQ4A seems to have antagonistic functions in HR depending on whether the reaction is initiated by a DSB. Taken together, we conclude that loss of RAD5A, MUS81, or RECQ4A affects HR frequency, demonstrating that each of the three proteins has a specific role in HR.
DISCUSSION
RAD5A, RECQ4A, and MUS81 Have Different Roles in DNA Repair
We previously demonstrated that in Arabidopsis, at least one homolog of yeast RAD5, RAD5A, is involved in DNA repair and HR. With the identification of two structural and functional RAD5 homologous proteins in humans (HLTF and SHPRH), it seems likely that basic RAD5 functions are conserved among yeast, plants, and humans, similar to RECQ and MUS81 functions.
Interestingly, the relationship between RecQ helicases and RAD5 homologs has not been characterized in detail. RECQ4A appears to be a functional homolog of the human BLM helicase (Hartung et al., 2007). By generating the recq4A rad5A double mutants, we demonstrated increased sensitivities of the double mutant plants to MMS and CDDP treatments that were more than additive. Therefore, we conclude that RAD5A and RECQ4A act through different DNA repair pathways. This finding was surprising because sensitivity data obtained with MMS treatment in yeast indicate that the RAD5 branch of PRR and the yeast RECQ homolog, SGS1, are involved in the same pathway of DNA repair (Karras and Jentsch, 2010). Thus, either Arabidopsis RECQ4A and yeast SGS1 or the yeast RAD5 homologs might not be functionally equivalent with respect to DNA repair. As in contrast with single homologs in yeast, there are gene families for both kinds of ATPases present in Arabidopsis. Therefore, the expectation of a one-to-one equivalence of proteins between Arabidopsis and yeast might be naive in this respect. In this context, it is especially interesting to note that RECQ4A and SGS1 also differ in their functions in HR (see the following discussion).
CDDP and MMS could be responsible for replication fork blockage in the S phase of the cell cycle, and the ATPases RAD5A and RECQ4A might have nonoverlapping functions in repairing a stalled replication fork. Homologs of both proteins have been shown to regress replication forks (Kanagaraj et al., 2006; Machwe et al., 2006; Ralf et al., 2006; Blastyák et al., 2007; Kobbe et al., 2009). Both single mutant lines showed a high sensitivity to DNA-damaging agents that can cause replication fork stalling. Nevertheless, our data indicate that the two ATPases cannot complement each other; therefore, they seem to target different types of substrates during DNA repair in vivo. Interestingly, recent in vitro analyses indicate that these two ATPases are indeed biochemically different: during the processing of homologous forks by RAD5, no single-stranded intermediates were formed, as revealed by kinetic analysis (Blastyák et al., 2007), whereas RecQ helicases displayed single-stranded DNA translocase activity, and, in contrast with RAD5 and HLTF, they could resolve heterologous model forks and catalyze the removal of single-stranded DNA from replication fork-like substrates (Kanagaraj et al., 2006; Unk et al., 2010).
The structure-specific endonuclease yeast MUS81 can process reversed but not unperturbed replication forks (Whitby et al., 2003). mus81 mutant lines are sensitive to DNA-damaging agents, such as MMS, camptothecin, hydroxyurea, and UV (Boddy et al., 2000; Interthal and Heyer, 2000; Doe et al., 2002). These agents cause different types of DNA damage, leading to the arrest or even a collapse of replication forks (Lehmann, 1972; Slater, 1973; Tsao et al., 1993; Tercero and Diffley, 2001). Because MMC and MMS have an additive effect on the rad5A mus81 double mutants in comparison to the single mutants, RAD5A and MUS81 seem to be involved in parallel pathways of DNA repair.
Because of the lethality of the recq4A mus81 double mutants, it is not possible to perform sensitivity analysis with these plants. Nevertheless, the lethality of the double mutants indicates that the two proteins are involved in parallel repair pathways of replicative DNA damage and cannot complement each other. RecQ homologs of various organisms have been reported to be part of the RTR (for RECQ-TOP3α-RMI1) complex. This complex is able to dissolve recombination intermediates, such as double Holliday junctions (dHJs) (Sharma et al., 2006; Mankouri and Hickson, 2007; Raynard et al., 2008). Because RECQ4A is part of the RTR complex in Arabidopsis (Hartung et al., 2008), it is tempting to speculate that as a component of the RTR complex it might act as a disolvase (Wu and Hickson, 2003; Hartung et al., 2008), whereas MUS81 might act as a resolvase on replicative intermediates that can be processed by both kinds of activities.
Indeed, we were able to suppress the phenotype of the recq4A mus81 double mutant by suppressing HR (by the disruption of RAD51C). It has been reported in yeast (Fabre et al., 2002; Bastin-Shanower et al., 2003) and Drosophila melanogaster (Trowbridge et al., 2007) that a mus81/sgs1 or mus81/blm phenotype can be suppressed by a rad51 mutation. Thus, it seems to be a common property for eukaryotes that replication intermediates are regularly processed by the HR machinery into structures receiving the best resolution or dissolution for a successful segregation. However, due to the experimental setup, we cannot discriminate whether the HR-dependent intermediates are processed by both RECQ4A and MUS81 or only by one or the other because either could be sufficient to restore viability.
In contrast with RECQ4A and MUS81, RAD5A seems to be most important for the repair of induced damage, especially cross-link repair induced by MMC and CDDP, because the rad5A single mutant line showed the highest sensitivity compared with either the recq4A or the mus81 single mutant line. RAD5A cannot complement the recq4A mus81 defect, indicating that it is no able to process certain classes of aberrant intermediates that arise during replication. The fact that recq4A mutant lines show a high sensitivity for CDDP but no sensitivity against MMC (Figure 2A) can be taken as indication that both chemicals induce at least partially different kinds of DNA damage, and RECQ4A is involved in the repair of a specific subset of damage induced by CDDP.
Thus, RAD5A, MUS81, and RECQ4A do not complement each other, and all of these proteins appear to be required for the processing of different types of aberrant DNA structures during DNA repair in vivo (Figure 5).
Figure 5.
Model for the Involvement of RAD5A, RECQ4A, and MUS81 in Different Pathways of DNA Repair in Somatic Cells of Arabidopsis.
(A) If replication stalls are due to endogenous DNA damage, RECQ4A and MUS81 are involved in processing such DNA intermediates through two independent pathways.
(B) CDDP- and MMS-induced damages can both be repaired by three different repair pathways defined by RAD5A, RECQ4A, and MUS81, respectively.
(C) Damages induced by MMC are repaired by an RAD5A-dependent or an MUS81-dependent pathway, and RECQ4A is not involved.
RAD5A, RECQ4A, and MUS81 Have Specific Roles in Homologous DSB Repair
Aside from their roles in DNA repair, RAD5A, RECQ4A, and MUS81 are also involved in HR in somatic cells. We previously showed that single mutant lines of each of the three genes exhibited a change of HR frequency in the assay line 651 (Swoboda et al., 1994; Hartung et al., 2006, 2007; Chen et al., 2008): in an untreated recq4A background, HR frequency increased, whereas it decreased after bleomycin treatment in the mus81 and the rad5A backgrounds. However, because different recombination reactions might lead to the restoration of the marker gene in line 651, from these experiments alone, it was impossible to draw conclusions regarding the mechanism by which the respective factors contribute to HR. To examine this question in more detail, we used two additional HR reporter lines and induced site-specific DSBs with I-SceI. We used the line IU.GUS to determine the efficiency of gene conversion and the line DGU.US for SSA (Orel et al., 2003) (Figures 4B and 4D). In principle, gene conversion can occur via the SDSA or the DSB repair pathway (Szostak et al., 1983). We previously demonstrated that in somatic plant cells, the main mechanism of gene conversion is SDSA (Puchta, 1998, 2005). Recent results in yeast indicate that, indeed, only a tiny fraction of gene conversions in somatic cells proceed through dHJs (best described by the DSB repair model) and that, in the majority of cases, the conversions proceed through SDSA (Bzymek et al., 2010; Mitchel et al., 2010). Therefore, the previous findings are in agreement with our conclusion that at least the vast majority of events detected with IU.GUS are indeed due to SDSA. Taken together, our data indicate that RAD5A, RECQ4A, and MUS81 play specific roles in HR.
RAD5A Plays a Significant Role in Gene Conversion
Interestingly, while there was no difference between the rad5A mutant and the wild type when the DGU.US line was used, the recombination frequency in the rad5A mutants was only half of that in the wild-type controls in the IU.GUS line. Because gene conversion is the only perturbed pathway, it would seem improbable that RAD5A affected HR through an indirect mechanism, such as the suppression of nonhomologous end-joining, as previously suggested for yeast (Ahne et al., 1997). Otherwise, we would also expect a reduced efficiency in the SSA pathway. Thus, RAD5A seems to be involved in a step occurring in SDSA or a similar pathway that results in gene conversion. It is noteworthy that the classical SDSA model and the models used to explain RAD5 action in strand invasion to bypass replication damage by switching template have a row of striking similarities (Branzei et al., 2008; Blastyák et al., 2010; Unk et al., 2010). One is tempted to speculate that a break, similar to damaged bases in the parental strand, can be bridged by a gap-filling mechanism. RAD5A might be involved in any of the steps, such as D-loop formation, bridging, and D-loop disruption. Recently, it was demonstrated for yeast that RAD5 is able to act in a replication-independent manner outside of the S phase (Karras and Jentsch, 2010). Our result showing that RAD5A is involved in repairing nuclease-induced DSBs that are not linked to replication is also in agreement with a role of this protein in HR outside of the S phase. Biochemical studies with different DNA structures as templates might help to further define the role of RAD5A in DSB repair. Additionally, studies on the genetic interaction of RAD5A and other HR factors will shed light on the function of RAD5A in HR.
RECQ4A Plays Multiple Roles in HR
We and others have previously demonstrated that RECQ4A is involved in suppressing recombination in the absence of induced DSBs (Bagherieh-Najjar et al., 2005; Hartung et al., 2007). It was surprising to find that the SSA and the SDSA-like DSB repair pathway have a minor versus a major reduction of efficiency in the recq4A background compared with the wild-type background. This finding strongly indicates that depending on the recombination pathway, RECQ4A seems to have a row of different and partially antagonistic functions. The recombination suppression by RECQ4A might be related to the suppression of crossovers when recombination intermediates are dissolved by the RTR complex (Wu and Hickson, 2003). We have previously indicated that in Arabidopsis, RECQ4A, together with TOP3α and RMI1, is part of this complex (Hartung et al., 2008). Alternatively, suppression could also occur due to disruption of the RAD51-ssDNA filament, an active DNA intermediate structure that promotes HR (Bugreev et al., 2007).
It has been shown that BLM, as well as SGS1, is involved in double-stranded DNA resection after DSB induction (Gravel et al., 2008; Mimitou and Symington, 2008; Nimonkar et al., 2008; Zhu et al., 2008). In yeast, it has been suspected that DNA ends of a DSB may initially be engaged by the protein complex harboring the MRE11, RAD50, and XRS2 (NBS1 in humans) proteins. In conjunction with SAE2 (CtIP in humans), the MRX(N) complex initiates limited resection of the broken DNA ends (Lengsfeld et al., 2007; Sartori et al., 2007). The SGS1 helicase (BLM in humans) and the DNA2 helicase/nuclease or EXO1 are then required for further resection. The minor reduction of HR efficiency detected with the SSA substrate might well be due to lack of the RECQ4A function during resection in our experimental system. Alternatively, RECQ4A might also be involved in reannealing the single-strand ends because some RecQ helicases have been shown to possess annealing activity (Garcia et al., 2004; Cheok et al., 2005; Sharma et al., 2005; Kobbe et al., 2009; Chen and Brill, 2010).
Indeed, lack of RECQ4A resulted in a stronger decrease in HR efficiency in SDSA than in SSA. In yeast, it has been shown that the SGS1 helicase, as well as TOP3α and RMI1, is probably involved in the resection of 5′-ends (Zhu et al., 2008). Thus, RECQ4A might as well be involved in the resection in Arabidopsis. Although we cannot exclude that specificities of the particular recombination substrates used for the measurements might have an effect, it is conceivable that in Arabidopsis, less resection might be required to proceed the recombination reactions in SSA compared with that in SDSA; however, the evidence in yeast indicates otherwise (Mimitou and Symington, 2008, 2009). Therefore, we propose that RECQ4A might be required for a later step during SDSA. This hypothesis is especially suggestive because a similar finding has been reported for the BLM homolog in D. melanogaster. Studies examining DNA repair following P-element–induced DNA DSBs in blm mutant flies indicate a defective repair in the SDSA pathway (Adams et al., 2003; McVey et al., 2004). In these blm mutant flies, P-element excision was accompanied by an increase of deletions flanking the P-element donor site. These flanking deletions were suppressed in blm rad51 double mutant flies (McVey et al., 2004), indicating that BLM functions after RAD51-dependent strand invasion. Thus, the RecQ helicases seem to play a role in resolving recombination intermediates. Taking our data into account, it seems obvious that Arabidopsis RECQ4A may play a similar role as previously postulated for BLM. During SDSA, single-strand invasion results in a D-loop (Figure 4C). The 3′-end of the broken DNA strand is then used to initiate copying the template DNA. The D-loop structure is subsequently resolved, and the newly synthesized strand reanneals with the adjacent broken DNA end to yield a repair product (Figure 4C). For the resolution step, a helicase is required, which has been shown for BLM in vitro (Bachrati et al., 2006). RECQ4A might be able to resolve this SDSA-specific recombination intermediate.
It is interesting to compare our results to previous experiments performed with HO endonuclease in yeast. In the case of direct repeats, SSA is not altered when SGS1 is deleted, whereas in the case of gene conversion, a significant enhancement is observed in the sgs1 mutant background (Lo et al., 2006). Thus, SGS1 and RECQ4A deletions seem to have opposite effects on gene conversion, demonstrating (also seen in the previous discussion on MMS sensitivity) that SGS1 and RECQ4A are indeed not bona fide functional equivalents. Studies in chicken DT40 and murine cells indicate that I-SceI–induced gene targeting is reduced in blm knockout cells (Kikuchi et al., 2009; Larocque and Jasin, 2010). Thus, RECQ4A seems to be more similar to the BLM homolog of animals in this respect.
It is noteworthy that in case of Arabidopsis, the situation might be even more complicated than that in other organisms because the closely related RECQ4B, a second putative BLM homolog, is present in this organism. Because recq4B mutants are not sensitive to DNA-damaging agents, a role of this protein in damage-induced DNA repair can be excluded (Hartung et al., 2007). It has also been demonstrated that in contrast with the recq4A mutants, the recq4B mutants show no lethal phenotype in the mus81 background (Hartung et al., 2006). However, because the recq4B mutants show a severe defect in HR (Hartung et al., 2007), it will be interesting to test the role of this protein in SSA and SDSA.
Arabidopsis MUS81 Has a Significant Role in Processing Intermediates during SDSA
The MUS81 protein belongs to the XPF/MUS81 family of nucleases and forms a functional endonuclease complex with its interaction partner, EME1 (Boddy et al., 2000; Interthal and Heyer, 2000; Chen et al., 2001). Because two EME1 homologs are present in Arabidopsis, there are two functional complexes that are both able to cleave not only 3′-flap structures and nicked Holliday junctions but also intact Holliday junctions, although with reduced efficiency (Geuting et al., 2009). It has been shown that 3′-flap structures formed in SSA are efficiently processed by the RAD1/RAD10 heterodimer (Fishman-Lobell and Haber, 1992), a role also conserved in Arabidopsis (Dubest et al., 2002, 2004). The slight efficiency reduction in SSA in the Arabidopsis mus81 mutants might suggest that MUS81 could, to a minor extent, be involved in processing 3′-flap structures, a property of MUS81 that has been biochemically characterized previously (Geuting et al., 2009).
The function of MUS81 in resolving dHJs that result in crossovers is well documented in meiotic recombination (Berchowitz et al., 2007; Higgins et al., 2008). Because, in contrast with meiotic recombination, dHJs are not major products of DSB repair in somatic cells (Bzymek et al., 2010), the most likely function of MUS81 seems to be an involvement in disrupting the arising D-loop structure after repair synthesis. Because these structures might resemble nicked Holliday junctions, MUS81 might be especially active in processing such structures. Therefore, RAD5A, RECQ4A, and MUS81 might all be required for processing intermediates related to D-loop formation and dissolution during SDSA.
Conclusions
We were able to show that in Arabidopsis at least three different pathways are available to process DNA intermediates that arise during DNA replication and DNA repair defined by the two ATPases, RAD5A and RECQ4A, and the endonuclease MUS81. Interestingly, both RECQ4A and MUS81 are also involved in DSB-induced HR in somatic cells via single-strand annealing and gene conversion, whereas RAD5A is required for gene conversion only.
METHODS
Strains
The rad5A-2 (SALK_047150), recq4A-4 (GABI_203C07), and mus81-1 (GABI_113F11) mutant lines have been previously described (Hartung et al., 2006, 2007; Chen et al., 2008). All T-DNA mutant lines are in Columbia-0 background. To generate the rad5A-2 mus81-1 and the rad5A-2 recq4A-4 double mutants, respective homozygous plants were crossed, and the homozygous double mutants were identified in the F2 progeny by PCR (Hartung et al., 2007; Chen et al., 2008). The rad51C-1 mutant line (Salk_021960) has been described previously (Abe et al., 2005; Bleuyard et al., 2005). Because the homozygous rad51C mutant line is sterile, we used heterozygous plants for crossing. The seeds from the F2 generation of the recq4A mus81 double mutants (described in Hartung et al., 2006) and the recq4A mus81 rad51C triple mutants were sown on agar plates containing GM to visualize early lethal phenotypes and on substrate containing 1:1 Floraton 3 (Floragard) and Vermiculit (Deutsche Vermiculite; Dämmstoffe) for cultivation in the greenhouse.
The rad5A-2, recq4A-4, mus81-1, and the rad51C-1 T-DNA insertion lines were obtained in the public T-DNA Express database established by the Salk Institute Genomic Analysis Laboratory accessible from the SIGnAL website at http://signal.salk.edu (Alonso et al., 2003). The rad5A-2 (Salk_047150) and the rad51C (Salk_021960) mutant lines derived from the SALK-T-DNA collection, and the recq4A-4 (GABI_203C07) and the mus81-1 (GABI_113F11) mutant lines were from the GABI collection (Rosso et al., 2003).
Analysis of Recombination in Arabidopsis thaliana Using the DGU.US and IU.GUS Reporter Lines
The Arabidopsis plants used for the SSA and SDSA recombination assay have to be heterozygous for both the reporter construct and the I-SceI–expressing construct in a homozygous mutant or a respective wild-type background. To obtain such plants, we first crossed the reporter lines (Orel et al., 2003) and an I-SceI expressing line with the respective mutant lines independently. The I-SceI expression line was transformed with the plasmid pPZP221 (Hajdukiewicz et al., 1994) containing a gentamycin resistance cassette (with a double 35S promoter and a 35S terminator) and the artificial I-SceI open reading frame optimized for plant expression (Puchta et al., 1993) fused to a 35S promoter and an octopin terminator. In the F2 generation, plants homozygous for the I-SceI–expressing or the reporter construct in the respective mutant or the corresponding wild-type background were identified by PCR. Plants homozygous for either the I-SceI–expressing construct or the respective reporter construct in either the respective homozygous mutant background or the corresponding wild-type background were propagated. As a final step, these reporter substrates were crossed with the I-SceI–expressing line, either in the mutant or the corresponding wild-type background. In the next generation, all seeds were heterozygous for both the I-SceI–expressing line and the reporter line (see Supplemental Figure 1 online). These seeds were then sown out on Petri dishes containing solid GM medium supplemented with the according antibiotics to exclude plantlets from self-fertilization from the mother line (which was always the I-SceI–expressing line). The DGU.US reporter line contains a PPT resistance marker; the IU.GUS line contains a hygromycin resistance marker. After 2 weeks, plantlets were histochemically stained as described by Swoboda et al. (1994). Destaining of leaf pigments with 70% ethanol facilitated the following analysis of recombination events by counting blue sectors under a binocular.
Genotoxicity Assays
Seeds of Arabidopsis were sterilized in 70% ethanol followed by 6% sodium hypochlorite for 5 min and rinsed several times with sterile water. Plants were grown in chambers at 22°C under white light (16 h light/8 h dark). For genotoxicity tests, sterilized seeds were spread onto fresh solid germination medium (4.9 g/L Murashige and Skoog medium [micro and macro elements including vitamins and MES buffer; Duchefa], 10 g/L saccharose, adjusted to pH 5.7, and 7.5 g/L plant agar) containing different genotoxins (MMS and CDDP, Sigma-Aldrich; MMC and bleomycin, Duchefa). After 21 d, the effects of the individual genotoxins on plant growth were evaluated. The plants were dried overnight, and the dry weight of treated seedlings was compared with that of untreated controls.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: NM_122181 (At5g22750, RAD5A), AJ404473 (At1g10930, RECQ4A), and AB177892 (At4g30870, MUS81).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Crossing Scheme.
Supplemental Table 1. P Values of Unpaired t Tests.
Supplementary Material
Acknowledgments
We thank Manfred Focke and Alexander Knoll for thoroughly reading the manuscript and Katharina Wahala, Mandy Katzer, and Maren Nitze for technical assistance. This work was supported by the German-Israeli Foundation for Scientific Research and Development (Grant I-820-70.12/2004) and by the Deutsche Forschungsgemeinschaft (Grant Pu 137/8).
References
- Abe K., Osakabe K., Nakayama S., Endo M., Tagiri A., Todoriki S., Ichikawa H., Toki S. (2005). Arabidopsis RAD51C gene is important for homologous recombination in meiosis and mitosis. Plant Physiol. 139: 896–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham J., et al. (2003). Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. EMBO J. 22: 6137–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams M.D., McVey M., Sekelsky J.J. (2003). Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299: 265–267 [DOI] [PubMed] [Google Scholar]
- Ahne F., Jha B., Eckardt-Schupp F. (1997). The RAD5 gene product is involved in the avoidance of non-homologous end-joining of DNA double strand breaks in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 25: 743–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Andersen P.L., Xu F., Xiao W. (2008). Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 18: 162–173 [DOI] [PubMed] [Google Scholar]
- Bachrati C.Z., Borts R.H., Hickson I.D. (2006). Mobile D-loops are a preferred substrate for the Bloom’s syndrome helicase. Nucleic Acids Res. 34: 2269–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagherieh-Najjar M.B., de Vries O.M., Hille J., Dijkwel P.P. (2005). Arabidopsis RecQI4A suppresses homologous recombination and modulates DNA damage responses. Plant J. 43: 789–798 [DOI] [PubMed] [Google Scholar]
- Bang D.D., Verhage R., Goosen N., Brouwer J., van de Putte P. (1992). Molecular cloning of RAD16, a gene involved in differential repair in Saccharomyces cerevisiae. Nucleic Acids Res. 20: 3925–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin-Shanower S.A., Fricke W.M., Mullen J.R., Brill S.J. (2003). The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol. Cell. Biol. 23: 3487–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beranek D.T. (1990). Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat. Res. 231: 11–30 [DOI] [PubMed] [Google Scholar]
- Berchowitz L.E., Francis K.E., Bey A.L., Copenhaver G.P. (2007). The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 3: e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyák A., Hajdú I., Unk I., Haracska L. (2010). Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol. Cell. Biol. 30: 684–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyák A., Pintér L., Unk I., Prakash L., Prakash S., Haracska L. (2007). Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell 28: 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleuyard J.Y., Gallego M.E., Savigny F., White C.I. (2005). Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J. 41: 533–545 [DOI] [PubMed] [Google Scholar]
- Boddy M.N., Gaillard P.H., McDonald W.H., Shanahan P., Yates J.R., III, Russell P. (2001). Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107: 537–548 [DOI] [PubMed] [Google Scholar]
- Boddy M.N., Lopez-Girona A., Shanahan P., Interthal H., Heyer W.D., Russell P. (2000). Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 20: 8758–8766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr V.A. (2008). Rising from the RecQ-age: The role of human RecQ helicases in genome maintenance. Trends Biochem. Sci. 33: 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Vanoli F., Foiani M. (2008). SUMOylation regulates Rad18-mediated template switch. Nature 456: 915–920 [DOI] [PubMed] [Google Scholar]
- Brusky J., Zhu Y., Xiao W. (2000). UBC13, a DNA-damage-inducible gene, is a member of the error-free postreplication repair pathway in Saccharomyces cerevisiae. Curr. Genet. 37: 168–174 [DOI] [PubMed] [Google Scholar]
- Bugreev D.V., Yu X., Egelman E.H., Mazin A.V. (2007). Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 21: 3085–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzymek M., Thayer N.H., Oh S.D., Kleckner N., Hunter N. (2010). Double Holliday junctions are intermediates of DNA break repair. Nature 464: 937–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaganti R.S., Schonberg S., German J. (1974). A manyfold increase in sister chromatid exchanges in Bloom’s syndrome lymphocytes. Proc. Natl. Acad. Sci. USA 71: 4508–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.F., Brill S.J. (2010). An essential DNA strand-exchange activity is conserved in the divergent N-termini of BLM orthologs. EMBO J. 29: 1713–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.P., Mannuss A., Orel N., Heitzeberg F., Puchta H. (2008). A homolog of ScRAD5 is involved in DNA repair and homologous recombination in Arabidopsis. Plant Physiol. 146: 1786–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.B., Melchionna R., Denis C.M., Gaillard P.H., Blasina A., Van de Weyer I., Boddy M.N., Russell P., Vialard J., McGowan C.H. (2001). Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell 8: 1117–1127 [DOI] [PubMed] [Google Scholar]
- Cheok C.F., Wu L., Garcia P.L., Janscak P., Hickson I.D. (2005). The Bloom’s syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 33: 3932–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu W.K., Hickson I.D. (2009). RecQ helicases: Multifunctional genome caretakers. Nat. Rev. Cancer 9: 644–654 [DOI] [PubMed] [Google Scholar]
- Ciccia A., Constantinou A., West S.C. (2003). Identification and characterization of the human mus81-eme1 endonuclease. J. Biol. Chem. 278: 25172–25178 [DOI] [PubMed] [Google Scholar]
- Colleaux L., d’Auriol L., Betermier M., Cottarel G., Jacquier A., Galibert F., Dujon B. (1986). Universal code equivalent of a yeast mitochondrial intron reading frame is expressed into E. coli as a specific double strand endonuclease. Cell 44: 521–533 [DOI] [PubMed] [Google Scholar]
- Constantinou A., Chen X.B., McGowan C.H., West S.C. (2002). Holliday junction resolution in human cells: Two junction endonucleases with distinct substrate specificities. EMBO J. 21: 5577–5585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C.L., Ahn J.S., Dixon J., Whitby M.C. (2002). Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 277: 32753–32759 [DOI] [PubMed] [Google Scholar]
- Dubest S., Gallego M.E., White C.I. (2002). Role of the AtRad1p endonuclease in homologous recombination in plants. EMBO Rep. 3: 1049–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubest S., Gallego M.E., White C.I. (2004). Roles of the AtErcc1 protein in recombination. Plant J. 39: 334–342 [DOI] [PubMed] [Google Scholar]
- Eastman A. (1985). Interstrand cross-links and sequence specificity in the reaction of cis-dichloro(ethylenediamine)platinum(II) with DNA. Biochemistry 24: 5027–5032 [DOI] [PubMed] [Google Scholar]
- Fabre F., Chan A., Heyer W.D., Gangloff S. (2002). Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA 99: 16887–16892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman-Lobell J., Haber J.E. (1992). Removal of nonhomologous DNA ends in double-strand break recombination: The role of the yeast ultraviolet repair gene RAD1. Science 258: 480–484 [DOI] [PubMed] [Google Scholar]
- Fricke W.M., Bastin-Shanower S.A., Brill S.J. (2005). Substrate specificity of the Saccharomyces cerevisiae Mus81-Mms4 endonuclease. DNA Repair (Amst.) 4: 243–251 [DOI] [PubMed] [Google Scholar]
- Friedl A.A., Liefshitz B., Steinlauf R., Kupiec M. (2001). Deletion of the SRS2 gene suppresses elevated recombination and DNA damage sensitivity in rad5 and rad18 mutants of Saccharomyces cerevisiae. Mutat. Res. 486: 137–146 [DOI] [PubMed] [Google Scholar]
- Gaillard P.H., Noguchi E., Shanahan P., Russell P. (2003). The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol. Cell 12: 747–759 [DOI] [PubMed] [Google Scholar]
- Garcia P.L., Liu Y., Jiricny J., West S.C., Janscak P. (2004). Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 23: 2882–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuting V., Kobbe D., Hartung F., Dürr J., Focke M., Puchta H. (2009). Two distinct MUS81-EME1 complexes from Arabidopsis process Holliday junctions. Plant Physiol. 150: 1062–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S., Chapman J.R., Magill C., Jackson S.P. (2008). DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 22: 2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann K.F., Ward A.M., Matkovic M.E., Folias A.E., Moses R.E. (2001). S. cerevisiae has three pathways for DNA interstrand crosslink repair. Mutat. Res. 487: 73–83 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Hartung F., Plchová H., Puchta H. (2000). Molecular characterisation of RecQ homologues in Arabidopsis thaliana. Nucleic Acids Res. 28: 4275–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Puchta H. (2006). The RecQ gene family in plants. J. Plant Physiol. 163: 287–296 [DOI] [PubMed] [Google Scholar]
- Hartung F., Suer S., Bergmann T., Puchta H. (2006). The role of AtMUS81 in DNA repair and its genetic interaction with the helicase AtRecQ4A. Nucleic Acids Res. 34: 4438–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Suer S., Knoll A., Wurz-Wildersinn R., Puchta H. (2008). Topoisomerase 3alpha and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana. PLoS Genet. 4: e1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Suer S., Puchta H. (2007). Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 18836–18841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.D., Buckling E.F., Franklin F.C., Jones G.H. (2008). Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 54: 152–162 [DOI] [PubMed] [Google Scholar]
- Hishida T., Kubota Y., Carr A.M., Iwasaki H. (2009). RAD6-RAD18-RAD5-pathway-dependent tolerance to chronic low-dose ultraviolet light. Nature 457: 612–615 [DOI] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S. (2002). RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Interthal H., Heyer W.D. (2000). MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet. 263: 812–827 [DOI] [PubMed] [Google Scholar]
- Iyer L.M., Babu M.M., Aravind L. (2006). The HIRAN domain and recruitment of chromatin remodeling and repair activities to damaged DNA. Cell Cycle 5: 775–782 [DOI] [PubMed] [Google Scholar]
- Iyer V.N., Szybalski W. (1963). A molecular mechanism of mitomycin action: Linking of complementary DNA strands. Proc. Natl. Acad. Sci. USA 50: 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.E., Henderson S.T., Petes T.D., Prakash S., Bankmann M., Prakash L. (1992). Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol. Cell. Biol. 12: 3807–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliraman V., Mullen J.R., Fricke W.M., Bastin-Shanower S.A., Brill S.J. (2001). Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15: 2730–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagaraj R., Saydam N., Garcia P.L., Zheng L., Janscak P. (2006). Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 34: 5217–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras G.I., Jentsch S. (2010). The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell 141: 255–267 [DOI] [PubMed] [Google Scholar]
- Kikuchi K., Abdel-Aziz H.I., Taniguchi Y., Yamazoe M., Takeda S., Hirota K. (2009). Bloom DNA helicase facilitates homologous recombination between diverged homologous sequences. J. Biol. Chem. 284: 26360–26367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobbe D., Blanck S., Focke M., Puchta H. (2009). Biochemical characterization of AtRECQ3 reveals significant differences relative to other RecQ helicases. Plant Physiol. 151: 1658–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocque J.R., Jasin M. (2010). Mechanisms of recombination between diverged sequences in wild-type and BLM-deficient mouse and human cells. Mol. Cell. Biol. 30: 1887–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley P.D. (1989). Mutagens as carcinogens: Development of current concepts. Mutat. Res. 213: 3–25 [DOI] [PubMed] [Google Scholar]
- Lehmann A.R. (1972). Postreplication repair of DNA in ultraviolet-irradiated mammalian cells. J. Mol. Biol. 66: 319–337 [DOI] [PubMed] [Google Scholar]
- Lengsfeld B.M., Rattray A.J., Bhaskara V., Ghirlando R., Paull T.T. (2007). Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell 28: 638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefshitz B., Steinlauf R., Friedl A., Eckardt-Schupp F., Kupiec M. (1998). Genetic interactions between mutants of the ‘error-prone’ repair group of Saccharomyces cerevisiae and their effect on recombination and mutagenesis. Mutat. Res. 407: 135–145 [DOI] [PubMed] [Google Scholar]
- Lo Y.C., Paffett K.S., Amit O., Clikeman J.A., Sterk R., Brenneman M.A., Nickoloff J.A. (2006). Sgs1 regulates gene conversion tract lengths and crossovers independently of its helicase activity. Mol. Cell. Biol. 26: 4086–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machwe A., Xiao L., Groden J., Orren D.K. (2006). The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry 45: 13939–13946 [DOI] [PubMed] [Google Scholar]
- MacKay C., Toth R., Rouse J. (2009). Biochemical characterisation of the SWI/SNF family member HLTF. Biochem. Biophys. Res. Commun. 390: 187–191 [DOI] [PubMed] [Google Scholar]
- Mankouri H.W., Hickson I.D. (2007). The RecQ helicase-topoisomerase III-Rmi1 complex: A DNA structure-specific ‘dissolvasome’? Trends Biochem. Sci. 32: 538–546 [DOI] [PubMed] [Google Scholar]
- McVey M., Larocque J.R., Adams M.D., Sekelsky J.J. (2004). Formation of deletions during double-strand break repair in Drosophila DmBlm mutants occurs after strand invasion. Proc. Natl. Acad. Sci. USA 101: 15694–15699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E.P., Symington L.S. (2008). Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E.P., Symington L.S. (2009). DNA end resection: Many nucleases make light work. DNA Repair (Amst.) 8: 983–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minca E.C., Kowalski D. (2010). Multiple Rad5 activities mediate sister chromatid recombination to bypass DNA damage at stalled replication forks. Mol. Cell 38: 649–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchel K., Zhang H., Welz-Voegele C., Jinks-Robertson S. (2010). Molecular structures of crossover and noncrossover intermediates during gap repair in yeast: implications for recombination. Mol. Cell 38: 211–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moertl S., Karras G.I., Wismüller T., Ahne F., Eckardt-Schupp F. (2008). Regulation of double-stranded DNA gap repair by the RAD6 pathway. DNA Repair (Amst.) 7: 1893–1906 [DOI] [PubMed] [Google Scholar]
- Motegi A., Liaw H.J., Lee K.Y., Roest H.P., Maas A., Wu X., Moinova H., Markowitz S.D., Ding H., Hoeijmakers J.H., Myung K. (2008). Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc. Natl. Acad. Sci. USA 105: 12411–12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi A., Sood R., Moinova H., Markowitz S.D., Liu P.P., Myung K. (2006). Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J. Cell Biol. 175: 703–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J.R., Kaliraman V., Ibrahim S.S., Brill S.J. (2001). Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157: 103–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar A.V., Ozsoy A.Z., Genschel J., Modrich P., Kowalczykowski S.C. (2008). Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl. Acad. Sci. USA 105: 16906–16911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orel N., Kyryk A., Puchta H. (2003). Different pathways of homologous recombination are used for the repair of double-strand breaks within tandemly arranged sequences in the plant genome. Plant J. 35: 604–612 [DOI] [PubMed] [Google Scholar]
- Osman F., Dixon J., Doe C.L., Whitby M.C. (2003). Generating crossovers by resolution of nicked Holliday junctions: A role for Mus81-Eme1 in meiosis. Mol. Cell 12: 761–774 [DOI] [PubMed] [Google Scholar]
- Parker J.L., Ulrich H.D. (2009). Mechanistic analysis of PCNA poly-ubiquitylation by the ubiquitin protein ligases Rad18 and Rad5. EMBO J. 28: 3657–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H. (1998). Repair of genomic double-strand breaks in somatic plant cells by one-sided invasion of homologous sequences. Plant J. 13: 331–339 [Google Scholar]
- Puchta H. (2005). The repair of double-strand breaks in plants: Mechanisms and consequences for genome evolution. J. Exp. Bot. 56: 1–14 [DOI] [PubMed] [Google Scholar]
- Puchta H., Dujon B., Hohn B. (1993). Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 21: 5034–5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralf C., Hickson I.D., Wu L. (2006). The Bloom’s syndrome helicase can promote the regression of a model replication fork. J. Biol. Chem. 281: 22839–22846 [DOI] [PubMed] [Google Scholar]
- Raynard S., Zhao W., Bussen W., Lu L., Ding Y.Y., Busygina V., Meetei A.R., Sung P. (2008). Functional role of BLAP75 in BLM-topoisomerase IIIalpha-dependent holliday junction processing. J. Biol. Chem. 283: 15701–15708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink S.M., Lipman R., Alley S.C., Hopkins P.B., Tomasz M. (1996). Bending of DNA by the mitomycin C-induced, GpG intrastrand cross-link. Chem. Res. Toxicol. 9: 382–389 [DOI] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P. (2007). Human CtIP promotes DNA end resection. Nature 450: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Doherty K.M., Brosh R.M., Jr (2006). Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem. J. 398: 319–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Sommers J.A., Choudhary S., Faulkner J.K., Cui S., Andreoli L., Muzzolini L., Vindigni A., Brosh R.M., Jr (2005). Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J. Biol. Chem. 280: 28072–28084 [DOI] [PubMed] [Google Scholar]
- Siebert R., Puchta H. (2002). Efficient repair of genomic double-strand breaks by homologous recombination between directly repeated sequences in the plant genome. Plant Cell 14: 1121–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater M.L. (1973). Effect of reversible inhibition of deoxyribonucleic acid synthesis on the yeast cell cycle. J. Bacteriol. 113: 263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda P., Gal S., Hohn B., Puchta H. (1994). Intrachromosomal homologous recombination in whole plants. EMBO J. 13: 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J.W., Orr-Weaver T.L., Rothstein R.J., Stahl F.W. (1983). The double-strand break repair model of recombination. Cell 33: 25–35 [DOI] [PubMed] [Google Scholar]
- Taylor E.R., McGowan C.H. (2008). Cleavage mechanism of human Mus81-Eme1 acting on Holliday-junction structures. Proc. Natl. Acad. Sci. USA 105: 3757–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero J.A., Diffley J.F. (2001). Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557 [DOI] [PubMed] [Google Scholar]
- Trowbridge K., McKim K., Brill S.J., Sekelsky J. (2007). Synthetic lethality of Drosophila in the absence of the MUS81 endonuclease and the DmBlm helicase is associated with elevated apoptosis. Genetics 176: 1993–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao Y.P., Russo A., Nyamuswa G., Silber R., Liu L.F. (1993). Interaction between replication forks and topoisomerase I-DNA cleavable complexes: Studies in a cell-free SV40 DNA replication system. Cancer Res. 53: 5908–5914 [PubMed] [Google Scholar]
- Ulrich H.D., Jentsch S. (2000). Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 19: 3388–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unk I., Hajdú I., Blastyák A., Haracska L. (2010). Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair (Amst.) 9: 257–267 [DOI] [PubMed] [Google Scholar]
- Unk I., Hajdú I., Fátyol K., Szakál B., Blastyák A., Bermudez V., Hurwitz J., Prakash L., Prakash S., Haracska L. (2006). Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc. Natl. Acad. Sci. USA 103: 18107–18112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vindigni A., Hickson I.D. (2009). RecQ helicases: Multiple structures for multiple functions? HFSP J 3: 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby M.C., Osman F., Dixon J. (2003). Cleavage of model replication forks by fission yeast Mus81-Eme1 and budding yeast Mus81-Mms4. J. Biol. Chem. 278: 6928–6935 [DOI] [PubMed] [Google Scholar]
- Wu L., Hickson I.D. (2003). The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874 [DOI] [PubMed] [Google Scholar]
- Xiao W., Chow B.L., Broomfield S., Hanna M. (2000). The Saccharomyces cerevisiae RAD6 group is composed of an error-prone and two error-free postreplication repair pathways. Genetics 155: 1633–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Chung W.H., Shim E.Y., Lee S.E., Ira G. (2008). Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.