Abstract
The germline of multicellular animals is segregated from somatic tissues, which is an essential developmental process for the next generation. Although certain ecdysozoans and chordates segregate their germline during embryogenesis, animals from other taxa segregate their germline after embryogenesis from multipotent progenitor cells. An overlapping set of genes, including vasa, nanos and piwi, operate in both multipotent precursors and in the germline. As we propose here, this conservation implies the existence of an underlying germline multipotency program in these cell types that has a previously underappreciated and conserved function in maintaining multipotency.
Keywords: Germline, Stem cell, Vasa, Piwi, Nanos, Sea urchin
Introduction
Multicellular animals consist of specialized cells, most of which are differentiated and devoid of reproductive potential. However, some cells, such as stem cells, retain both the ability to self-renew and to create new differentiated cells. One such stem cell, the germline stem cell (GSC), gives rise to a continual supply of germ cells, which are specialized for the task of reproduction. Yet once egg and sperm fuse, the resulting zygote is totipotent, and thus can give rise to all cell types of the animal. Although GSCs normally give rise only to germ cells in vivo, and thus are unipotent, a significant body of evidence shows that these cells maintain the potential to acquire many cell fates. For example, mouse primordial germ cells (PGCs) acquire the morphological and cytological characteristics of embryonic stem (ES) cells when cultured in the presence of three growth factors: steel factor, leukemia inhibitory factor (LIF) and fibroblast growth factor (FGF) (Matsui et al., 1992). These cells create teratomas when injected into adult mice and chimeric pups when injected into blastocysts, thus demonstrating their pluripotency. Isolated human PGCs can also be converted to pluripotent stem cells without genetic manipulation (Shamblott et al., 1998). Furthermore, pluripotent cells spontaneously arise from isolated adult mouse spermatogonial stem cells under specific culture conditions (Guan et al., 2006). This phenomenon is not limited to vertebrates as mutations in the translational regulators mex-3 and gld-1 in Caenorhabditis elegans leads to transdifferentiation of germ cells into muscle, neurons and intestinal cells (Ciosk et al., 2006). Thus, although germ cells are highly specialized, they seemingly retain the capacity to give rise to all cell types. As such, it is possible that the developmental potential of GSCs in these organisms is not intrinsically limited but might instead be restricted by environment. What, then, is the underlying regulatory program that controls this highly potent cell type?
The molecular regulation of the germline is best understood in a handful of animals, including in C. elegans, Drosophila melanogaster and the mouse. Here, we explore the diversity of germline origins more broadly by examining germline segregation mechanisms across diverse taxa for which molecular data are available, including in cnidarians, lophotrochozoans and echinoderms. From the synthesis of such data collected from a wide variety of animals with diverse and often complex life histories, we propose the likely existence of a highly conserved germline multipotency program (GMP) that operates in both multipotent cells and germ cells. Furthermore, we highlight key taxa for which molecular data are lacking, thus helping to guide future work in uncovering the conserved and critical aspects of this GMP.
Germline segregation: an overview
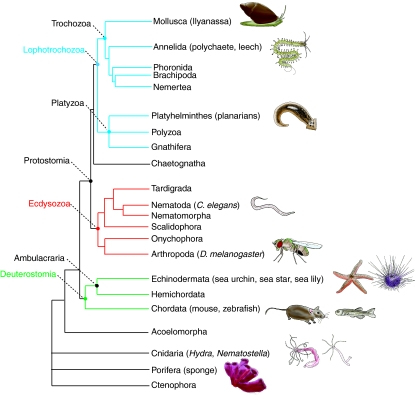
Sexually reproducing metazoans generally segregate their germline away from their somatic tissue before they make gametes; in Drosophila, C. elegans and mouse, this occurs during embryogenesis. In both Drosophila and C. elegans, germline specification is cell autonomous, whereas the mouse germline is specified by inductive signals (Extavour and Akam, 2003; Illmensee and Mahowald, 1974; Illmensee and Mahowald, 1976; Lawson and Hage, 1994; Strome and Wood, 1982; Tam and Zhou, 1996). As these differences have been previously well reviewed (e.g. Extavour and Akam, 2003), we instead focus here on an important similarity: that all three animals establish a population of PGCs during embryogenesis, which migrate into the developing somatic gonad and become exclusively GSCs in the adult (Richardson and Lehmann, 2010) (Fig. 1A). Embryonic segregation of the germline appears to be widely used by both ecdysozoans (such as flies and nematodes) and by chordates (which includes ascidians, fish, amphibians and mammals) (Brown et al., 2009; Extavour and Akam, 2003; Johnson et al., 2001; Johnson et al., 2003; Takamura et al., 2002; Whitington and Dixon, 1975; Yoon et al., 1997) (Fig. 2).
Fig. 1.
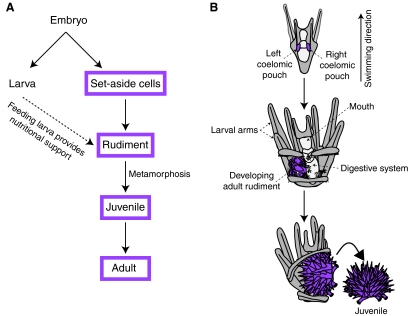
Germline segregation strategies. (A) In animals that segregate their germline during embryogenesis, primordial germ cells (PGCs) are specified in the embryo. PGCs give rise exclusively to either male or female germline stem cells (GSCs), which both self-renew (as indicated by the curved arrows) and give rise to a constant supply of gametes. Gametes are highly specialized cells, but when they fuse at fertilization they create a totipotent zygote. (B) In animals that segregate their germline after embryogenesis, a multipotent progenitor is established in the embryo from which the germline is segregated after embryogenesis is completed. We propose that the red cells in both panels operate a conserved germline multipotency program.
Fig. 2.
Metazoan phylogenetic tree. The evolutionary relationships among the major animal phyla. Animals mentioned in the text are noted in parentheses. Deuterostome phyla are shown in green, Ecdysozoa phyla in red and Lophotrochozoa phyla in blue. Drawings by S.Z.S. and Elizabeth Schroeder.
However, data collected from animal taxa that lie outside of the chordates and ecdysozoans demonstrate that such embryonic germline segregation is not universal. For example, in the lophotrochozoans Ilyanassa (marine snail) and P. dumerilii (polychaete annelid), and in echinoderms, such as the sea urchin, the germline is segregated after embryogenesis completes. In these animals, multipotent progenitor cells are established during embryogenesis that give rise to both germ and somatic cells in the developing juvenile (Findley et al., 2003; Juliano et al., 2010; Rabinowitz et al., 2008; Rebscher et al., 2007; Swartz et al., 2008; Voronina et al., 2008) (Fig. 1B). We broadly refer to these cells as multipotent progenitors because of their developmental potential, but refrain from using the term `stem cell' in cases where the capacity to self-renew remains undetermined. These multipotent cells are unique in that they are stably undifferentiated cells that are set aside and persist beyond the completion of embryogenesis. In some cases, they remain mitotically quiescent until they are used later in development, such as in the sea urchin (Tanaka and Dan, 1990); in others, they are self-renewing and can thus be called stem cells. For example, in the planarian flat worm, another lophotrochozoan, germ cells are continually segregated from a self-renewing population of totipotent adult stem cells called neoblasts (Newmark et al., 2008). A similar strategy is used in at least some cnidarians and sponges, which have populations of adult multipotent or totipotent stem cells that continually give rise to both germ and somatic cells (Bosch and David, 1987; Funayama, 2010; Muller et al., 2004).
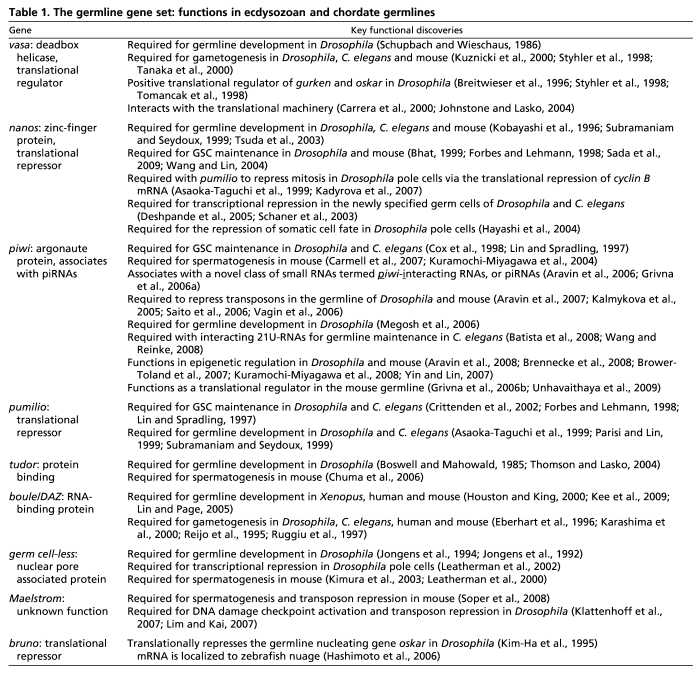
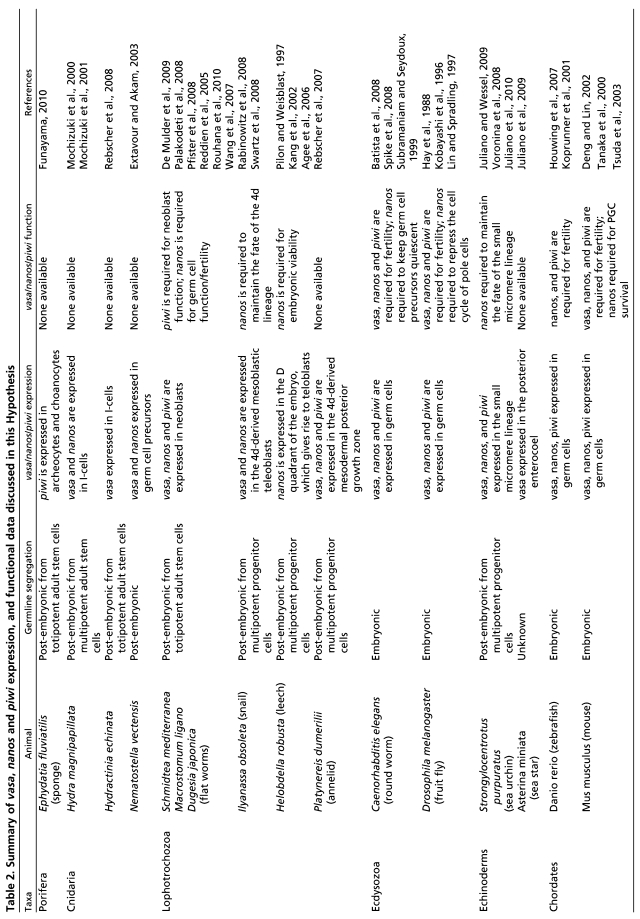
Regardless of the strategy used, the same set of genes appears both to specify and maintain PGCs during embryonic germline segregation and to maintain long-term multipotent progenitor cells in animals that segregate their germline after embryogenesis. Extensive functional studies in chordates and ecdysozoans have begun to identify members of this gene set, including vasa, nanos and piwi. Loss-of-function studies demonstrate that these genes are required initially to specify the germline and/or to maintain adult GSCs in ecdysozoans and chordates. The molecular functions of these genes are under intense investigation (see Table 1 for a summary of functional data on the GMP genes and for relevant references), and the recent identification of their expression patterns and functions outside of the chordates and ecdysozoans demonstrate the persistence of this conserved and overlapping gene set in PGCs, GSCs and multipotent progenitors.
Table 1.
The germline gene set: functions in ecdysozoan and chordate germlines
The sea urchin: a key to understanding the GMP
Uncovering the functionally conserved components of the GMP may reveal the most fundamental and absolutely required regions of this gene program. Echinoderms are key to understanding the GMP because, together with the hemichordates, they form the sister group to the chordates, and thus sit at a critical node at the base of the deuterostomes (Fig. 2). Embryogenesis in most echinoderms culminates in the creation of a free-swimming larva that feeds and supports the growth of the developing juvenile (Fig. 3) (Peterson et al., 1997; Raff, 2008). The cells that build the entire juvenile animal are insulated from the surrounding larval differentiation program during embryogenesis. These set-aside cells form the adult rudiment, which develops into the juvenile sea urchin that emerges at metamorphosis (Fig. 3). The germline of the sea urchin is clearly separate from the soma in the adult, but it is unknown when during the sea urchin life cycle the germline is initially segregated because direct observations are difficult within the developing juvenile (Houk and Hinegardner, 1980).
Fig. 3.
The juvenile sea urchin develops from embryonic multipotent cells. (A) Schematic of indirect development in sea urchin. During embryogenesis, cells are set aside for constructing the adult (the rudiment, shown in purple). The larva swims and feeds, providing protection and nutritional support to the developing adult structures. A S. purpuratus larva is competent to undergo metamorphosis after ∼6-8 weeks of feeding. (B) In the four-armed pluteus, the small micromere descendents are located in the left and right coelomic pouches (purple), where the adult rudiment will form. In the eight-armed pluteus, adult structures in the rudiment (purple) begin to form, such as the tube feet and spines. At metamorphosis, the juvenile emerges as an independent entity (purple) and larval tissues are lost.
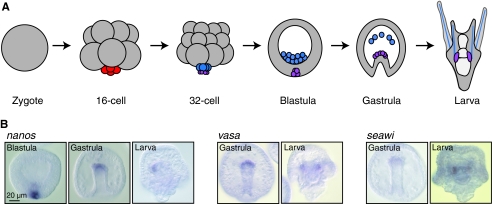
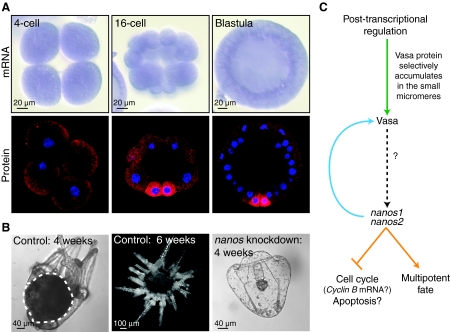
The small micromere lineage of the sea urchin embryo, which is hypothesized to give rise to the germline, is a conspicuous population of cells that contribute only to adult tissues (Pehrson and Cohen, 1986; Peterson et al., 1997). This lineage is a product of two unequal cleavage divisions: the fourth vegetal division, which produces four macromeres and four micromeres; and the subsequent unequal division of the micromeres, which produces four large micromeres and four small micromeres (Fig. 4A). Descendents of the small micromeres remain relatively quiescent through embryogenesis, dividing only once before gastrulation and then integrating into the larval coelomic pouches where the adult rudiment forms (Fig. 3). In support of the hypothesis that these cells give rise to the germline, the small micromeres of the sea urchin Strongylocentrotus purpuratus selectively accumulate vasa, nanos and piwi mRNA (Fig. 4B) (Juliano et al., 2006). However, once the small micromere descendents are incorporated into the coelomic pouches, they begin to proliferate, which is uncharacteristic of quiescent embryonic PGCs (Tanaka and Dan, 1990). Furthermore, removal of the small micromere precursors, the micromeres, results in cell fate transitions, which allow the embryo to develop successfully; the resulting larva gives rise to a gravid adult. Thus, it is clear that the micromeres do not contain any obligate germ cell factors (Ransick et al., 1996).
Fig. 4.
Multipotent progenitor cells in the sea urchin embryo. (A) A schematic of the early stages of sea urchin development during which the small micromere lineage (purple) is set aside. The vegetal fourth cleavage division is unequal, thus giving rise to a 16-cell embryo with four micromeres (red). The micromeres divide unequally to give rise to four large micromeres (blue) and four small micromeres (purple). The small micromeres reside at the vegetal plate of the blastula where they divide once more. The eight small micromere descendents are carried at the tip of the invaginating gut during gastrulation and upon larva formation, are incorporated into the larval coelomic pouches, the site of adult rudiment formation. The small micromere lineage gives rise exclusively to tissues of the adult. (B) nanos, vasa and seawi (a sea urchin piwi ortholog) whole-mount in situ RNA hybridizations of S. purpuratus embryos and larva at the indicated stages of development. nanos, vasa and seawi, members of the germline multipotency program (GMP) gene set, are expressed in the sea urchin small micromere lineage. Images reproduced, with permission, from Juliano et al. (Juliano et al., 2006).
An alternative hypothesis suggests that the small micromere descendents are multipotent progenitor cells that contribute to diverse adult tissues (Ransick et al., 1996; Tanaka and Dan, 1990; Voronina et al., 2008). The ultimate resolution of small micromere fate requires genetic lineage tracing because cell-labeling dyes do not last through the lengthy process of sea urchin larval and juvenile development. This can be achieved by creating chimeric embryos that contain small micromeres expressing a recombinant marker gene (such as GFP) from an incorporated BAC (Ettensohn et al., 2004b). In the meantime, a different approach has provided experimental support for small micromere multipotency: reduction of Nanos protein, a small micromere-specific gene and member of the GMP (Table 1), by the injection of a morpholino antisense oligonucleotide (MASO) in the S. purpuratus embryo completely disrupts formation of the adult rudiment (Fig. 5B) (Juliano et al., 2010). This finding strongly argues that the small micromeres are a multipotent lineage that contributes to diverse adult tissues in the developing juvenile. We therefore hypothesize that the small micromeres are embryonic multipotent progenitor cells within which the GMP operates. Furthermore, we propose that the germline is later segregated from these cells in the late larva or developing juvenile.
Fig. 5.
The multipotent small micromere program. (A) Images of S. purpuratus embryos at the four-cell, 16-cell and blastula stages of development, showing vasa whole-mount in situ RNA hybridizations (upper) (with vasa uniformly distributed throughout blastula formation) and Vasa protein (lower) enriched in the micromeres and small micromeres, respectively, on their formation [reproduced, with permission, from Juliano et al. (Juliano et al., 2006) and Voronina et al. (Voronina et al., 2008)]. (B) Control embryos produce larvae with well-formed adult rudiments around 4 weeks of age (broken white circle) that metamorphose around 6 weeks (middle panel). By contrast, when Nanos protein is knocked down in the embryo, the resultant larva fails to form an adult rudiment or undergo metamorphosis [reproduced, with permission, from Juliano et al. (Juliano et al., 2010)] (C). Vasa protein is post-transcriptionally enriched in the small micromere lineage (Voronina et al., 2008). nanos1 and nanos2 are expressed in this lineage at the 60-cell stage and are required for the continued enrichment of Vasa protein, for cell cycle repression and for the maintenance of the multipotent fate in this lineage (Juliano et al., 2010). Vasa protein enrichment precedes nanos transcription, but whether Vasa is required for nanos expression is unclear (question mark). Several similarities between the sea urchin small micromere germline multipotency program (GMP) and the Drosophila pole cell GMP are highlighted: Vasa protein (green) is selectively expressed in the pole cells as a result of regulated protein stability (Kugler et al., 2010; Styhler et al., 2002); nanos (blue) is required to maintain Vasa expression (Hayashi et al., 2004); and nanos (orange) is required to maintain cell fate by repression of the cell cycle and repression of apoptosis (Asaoka-Taguchi et al., 1999; Hayashi et al., 2004; Kadyrova et al., 2007).
Uncovering the regulatory program that operates in these multipotent cells requires that we understand both how these genes are selectively expressed (upstream network connections) and how they interact to affect the fate of the small micromeres (downstream network connections). The discovery of these molecular connections will allow the GMP across diverse taxa to be compared; conserved interactions are likely to reflect the key functions of the GMP. As described below, emerging molecular and functional data support the hypothesis that the small micromere GMP shares similarities with the germline program found in chordates and ecdysozoans.
Small micromere and PGC regulatory programs: conserved features
Vasa post-transcriptional regulation
vasa mRNA is ubiquitously present during early sea urchin embryogenesis, but Vasa protein is enriched only in the micromeres and the small micromeres, respectively, upon their formation (Fig. 5A) (Voronina et al., 2008). Thus, selective Vasa protein accumulation depends on post-transcriptional regulation, which appears to be a conserved feature of both embryonic (in PGCs) and post-embryonic (in multipotent progenitor cells) germline segregation across animal taxa. For example, by the cellular blastoderm stage in Drosophila embryos, vasa mRNA is uniformly distributed, whereas Vasa protein is restricted to the pole cells – the embryonic germline progenitors (Hay et al., 1990; Lasko and Ashburner, 1990). Vasa protein enrichment in the pole cells is driven by protein stability, and this requires both a deubiquitinating enzyme (encoded by fat facets) and the competing actions of two E3 ubiquitin ligases (encoded by gustavus and fsn) (Kugler et al., 2010; Styhler et al., 2002). In the zebrafish embryo, vasa mRNA is localized to the future germline by 6 hours post fertilization (hpf), but Vasa protein is uniformly distributed until 24 hpf. Microinjection of vasa-GFP chimeric RNA demonstrates that the vasa open reading frame (ORF) is sufficient to drive GFP enrichment in the germline, suggesting that regulation of protein stability directs eventual Vasa protein restriction to these cells (Wolke et al., 2002). The post-transcriptional regulation of vasa by protein stability might be a conserved feature of the GMP and, therefore, could be the mechanism by which Vasa protein is restricted to the sea urchin small micromeres. If this were the case, we predict that the sea urchin vasa ORF would be sufficient to drive the enrichment of Vasa protein in the small micromere lineage, which could be tested by injection of a GFP-Vasa chimera mRNA as was carried out in the zebrafish study (Wolke et al., 2002). Alternatively, if it is translational control that is required for Vasa protein enrichment, the vasa 3′UTRs would probably be required to drive GFP reporter enrichment in the small micromere lineage.
Nanos maintains small micromere fate
Cell cycle lengthening is a conserved feature of newly specified PGCs and is exhibited by the sea urchin small micromeres (Tanaka and Dan, 1990). For example, the nuclei of the Drosophila pole cells exhibit a prolonged cell cycle immediately upon arrival at the posterior pole (Su et al., 1998). This requires nanos-dependent translational repression of the mitotic cyclin, cyclin B, which is present in the germ plasm but remains untranslated until just before mitotic divisions resume in the pole cells (Asaoka-Taguchi et al., 1999; Dalby and Glover, 1993; Kadyrova et al., 2007). Consistent with this mechanism, nanos-null pole cells divide precociously and then apoptose during mid embryogenesis. If the apoptotic pathway is suppressed in these nanos-null pole cells, they adopt a somatic cell fate (Asaoka-Taguchi et al., 1999; Hayashi et al., 2004). This result suggests that nanos is possibly doing more than repressing the cell cycle; it may repress other transcripts that inhibit PGC fate and that promote a somatic cell fate. Newly specified PGCs in both C. elegans and the mouse also enter a period of cell cycle quiescence. In C. elegans, the P4 lineage cell, which gives rise solely to the germline, is formed at the 24-cell stage and then divides only once to create the Z2 and Z3 cells. These cells remain arrested in G2 throughout embryogenesis until the larvae feed (Fukuyama et al., 2006; Subramaniam and Seydoux, 1999). Mouse PGCs also arrest at G2 (Seki et al., 2007), and in both animals, nanos expression is coincident with the interruption of the PGC cell cycle (Subramaniam and Seydoux, 1999; Suzuki et al., 2008). Loss of nanos function in C. elegans leads to the premature proliferation of the Z2 and Z3 cells in the larva (Subramaniam and Seydoux, 1999), and loss of nanos3 function in the mouse leads to PGC apoptosis (Suzuki et al., 2008). Thus, nanos has a conserved function in maintaining PGC fate, in part by repressing the cell cycle and apoptosis.
The function of nanos in the small micromeres of the sea urchin, S. purpuratus, appears to be analogous to its conserved germline functions in other animals. Small micromeres in nanos 1 and nanos2 knockdown sea urchin embryos appear to precociously divide, but then these descendents are not incorporated into the coelomic pouches; consequently, the adult rudiment does not form (Fig. 5B) (Juliano et al., 2010). Small micromere descendents in nanos1/2 knockdown embryos may undergo apoptosis, as do the nanos null pole cells of Drosophila and the nanos3-null PGCs of mice (Hayashi et al., 2004; Sato et al., 2007; Suzuki et al., 2008; Tsuda et al., 2003). Support for this idea has been demonstrated in the sea urchin Hemicentrotus pulcherrimus, in which a pan-caspase inhibitor appears to prolong the viability of the nanos-depleted small micromere descendents (Fujii et al., 2009). Thus, it is clear that nanos plays a conserved role in maintaining the fate of both multipotent small micromeres in the sea urchin and PGCs in chordates and flies. Furthermore, nanos achieves this in both cases via the repression of the cell cycle and of apoptosis.
In summary, several aspects of the GMP that function to preserve potency in PGCs and small micromeres are conserved, including: (1) the selective expression of nanos, vasa and piwi; (2) the post-transcriptional regulation of vasa potentially by protein stability; and (3) nanos-dependent cell fate via repression of the cell cycle and apoptosis (Fig. 5C).
Extensive work in model organisms, such as Drosophila, C. elegans, and the mouse has provided considerable insights into the molecular nature of the GMP (Table 1). A high percentage of germline-expressed genes appear to be RNA-binding proteins, indicating the crucial role of post-transcriptional control in this cell type. In support of this conclusion, germ cells in Drosophila, C. elegans and mouse undergo a period of global transcriptional quiescence after their specification (Nakamura and Seydoux, 2008). During this developmental period, the translational control of either maternally loaded mRNAs or of previously transcribed zygotic mRNAs might be crucial for controlling their fate. For example, vasa acts as a translational activator of two Drosophila mRNAs that localize to the oocyte: gurken (grk), which directs both anterior/posterior and dorsal/ventral polarity and oskar (osk), which directs germ plasm assembly at the posterior of the oocyte (Markussen et al., 1995; Styhler et al., 1998; Tomancak et al., 1998). However, as osk and grk are not conserved outside of flies, it is likely that Vasa regulates a broader suite of transcripts.
nanos, which contains two CCHC zinc fingers, is a translational repressor that acts with pumilio by binding the NRE (nanos response element) located in the 3′UTRs of nanos-regulated mRNAs. During Drosophila germ cell development, nanos acts as a translational repressor of cyclin b, to prevent premature divisions (Asaoka-Taguchi et al., 1999). In Xenopus laevis, nanos and pumilio regulate cyclin B1 translation during oocyte maturation, suggesting that the role of nanos as a repressor of cyclin b mRNA is conserved (Nakahata et al., 2001; Nakahata et al., 2003). Drosophila germ cells that lack nanos undergo apoptosis, or if apoptosis is repressed, the germ cells are incorporated into somatic tissues (Hayashi et al., 2004). This highlights the importance of nanos and of mitotic quiescence in retaining the germ cell fate. These cells appear to actively repress differentiation programs and maintain developmental potency at least in part through transcriptional and mitotic quiescence. This quiescence in both germ cells and multipotent germ cell precursors may serve to protect their DNA. In addition, piwi, another gene found in these cell types, contributes to protecting germ cell DNA, in conjunction with a class of small RNAs called piRNAs, by repressing transposons in these immortal cell types (O'Donnell and Boeke, 2007). Recent evidence in mice also indicates a role for vasa in transposon suppression by participating in piRNA biogenesis (Kuramochi-Miyagawa et al., 2010). Thus, the role of the GMP in part may be to protect the DNA, which will contribute to future generations.
The GMP in cnidarians, sponges and lophotrochozoans
Species from cnidarians, sponges and lophotrochozoans segregate their germline from the soma after embryogenesis, similar to the sea urchin. Furthermore, in these cases the GMP appears to operate in the multipotent progenitor cells, thus allowing for functional comparisons across these diverse animal taxa (Fig. 2; Table 2).
Table 2.
Summary of vasa, nanos and piwi expression, and functional data discussed in this Hypothesis
Multipotent germline progenitors in cnidarians
In adult Hydra, a well-studied cnidarian, multipotent interstitial stem cells, or I-cells, are interspersed among the epithelial cells. I-cells continually populate the interstitial cell lineage, which consists of both germ and somatic cell types (Bosch and David, 1987; Bosch and David, 1986; David and Murphy, 1977; Galliot et al., 2006). The adult I-cells selectively express vasa and nanos, implicating GMP control of these cells, but the functions of these genes in the adult stem cells of Hydra are not known (Mochizuki et al., 2001; Mochizuki et al., 2000). Morphological studies indicate that I-cells may be widespread in the Medusozoa branch of cnidarians, which includes all hydroids and jellyfish, but in most cases the molecular mechanisms of multipotency are unexplored (Ball et al., 2004; Boelsterli, 1977; Chapman, 1974; Ralph, 1960). A recent study in Hydractinia echinata, a colonial hydroid, has demonstrated that vasa mRNA accumulates specifically in I-cells of the larva, which is a totipotent lineage in this animal (Muller et al., 2004; Rebscher et al., 2008). Thus, the GMP may be involved in the initial specification of I-cells in Hydractinia.
I-cells have not been found in any Anthozoans, the branch of cnidarians that includes corals and sea anemones, even though germ cells in sea anemones segregate continuously in the adult (Extavour and Akam, 2003). In the sea anemone Nematostella vectensis, vasa and nanos transcripts are present in many somatic domains early in the embryo. Then in the larva, these transcripts specifically accumulate in two patches of cells that give rise to the adult mesenteries – in-foldings of epithelium that extend from the body wall into the body cavity (Extavour et al., 2005). Extavour and co-workers suggest that these cells are PGCs because gametes are found in the mesenteries of the adult (Extavour et al., 2005). However, the possibility remains that these cells are precursors to multipotent stem cells that give rise to both somatic and germ cells in the adult. This is an important issue to resolve in the future, as reconstructing the ancestral mode of germline segregation in cnidarians is key to understanding the evolution of this process in their sister group, the bilaterians (Peterson and Eernisse, 2001).
Sponge totipotent stem cells express piwi
Members of the phylum Porifera, or sponges, long ago diverged from the bilaterian lineage. The exact evolutionary relationships among the non-bilaterian phyla, such as among sponges, ctenophores and cnidarians, have been difficult to ascertain because, in order to resolve such relationships, significantly divergent sequences need to be compared, which requires deep sequencing of numerous taxa (DeSalle and Schierwater, 2008; Dunn et al., 2008; Holder and Lewis, 2003; Rokas et al., 2005). Regardless, these groups are important for discerning the evolutionary origins of the GMP. Sponges contain an adult totipotent stem cell, called an archeocyte, which gives rise to both differentiated somatic cells and oocytes. Archeocytes also give rise to the choanocytes, which function in food collection and can give rise to sperm. Choanocytes can transdifferentiate back into archeocytes, and thus probably maintain a totipotent capacity (Funayama, 2010). In the sponge Ephydatia fluviatilis, two piwi orthologs are specifically expressed in both the archeocytes and choancytes, potentially indicating the presence of the GMP in these totipotent germline precursors (Funayama et al., 2010).
Flatworms: totipotent stem cell maintenance and the GMP
Flatworms have an extraordinary ability to regenerate owing to a population of totipotent adult stem cells called neoblasts that gives rise to somatic cells and germ cells (Baguna et al., 1989; Newmark and Sanchez-Alvarado, 2000). The neoblasts of adult flatworms express tudor, pumilio, piwi, bruno and vasa, all of which were first identified in the germline of flies and mice and have conserved functions there (Table 1) (De Mulder et al., 2009; Guo et al., 2006; Palakodeti et al., 2008; Pfister et al., 2008; Reddien et al., 2005; Rouhana et al., 2010; Salvetti et al., 2005; Shibata et al., 1999; Solana et al., 2009). tudor, pumilio and bruno are required for neoblast maintenance, as neoblasts are lost when these genes are knocked down (Guo et al., 2006; Rouhana et al., 2010; Salvetti et al., 2005; Solana et al., 2009). The two piwi homologs in the planarian flatworm Schmidtea mediterranea, Smedwi-2 and Smedwi-3, are not required for maintenance, but are essential for neoblast differentiation (Guo et al., 2006; Palakodeti et al., 2008; Reddien et al., 2005; Salvetti et al., 2005; Solana et al., 2009). However, in the marine flatworm Macrostomum lignano, piwi is required for the maintenance of neoblasts, similar to its function in maintaining GSCs in the Drosophila ovary (Cox et al., 1998; De Mulder et al., 2009). In contrast to the other members of the conserved GMP, the S. mediterranea ortholog of nanos is not expressed in neoblasts, although it is required for gonad regeneration (Wang et al., 2007). In general, genes required for flatworm neoblast maintenance and differentiation include those required for germ cell specification and development in other organisms, implying that the GMP operates within these neoblasts.
GMP genes in lophotrochozoan multipotent progenitor cells
Many lophotrochozoans develop indirectly through a feeding larva, similar to the sea urchin, and some cells that arise during embryogenesis will be used later in building adult structures. One such population of cells is the conserved 4d lineage, which gives rise to the adult mesoderm and endoderm in lophotrochozoans (Boyer et al., 1996; Lambert, 2008). In the snail Ilyanassa obsoleta, the 4d cell divides to create the mesoblastic teloblasts – self-renewing stem cells that contribute to larval mesoderm and endoderm. These 4d-derived lineages contribute to the adult snail, but it is unclear whether the mesoblastic teloblasts themselves remain into adulthood. However, some evidence in another mollusc suggests that, after populating the mesodermal lineages, the mesoblastic teloblasts retain their uncommitted morphology until after metamorphosis and arrival into the adult gonad, whereupon they become definitive germ cells (Woods, 1931). Both vasa (mRNA) and nanos (mRNA and protein) are uniformly distributed during early cleavage in this snail and then become selectively expressed in the 4d lineage (Rabinowitz et al., 2008; Swartz et al., 2008). In a second sea snail, Haliotis asinina, both vasa and nanos mRNA are detected in the mesodermal region of the larva, thus supporting the conclusion that these genes are expressed in multipotent cells of snails (Kranz et al., 2010). Furthermore, nanos is required to maintain the fate of the 4d lineage in Ilyanassa; loss of nanos protein by morpholino injection into the embryo results in a loss of all 4d-derived structures (Rabinowitz et al., 2008). In the leech Helobdella robusta, a direct developing lophotrochozoan annelid, nanos is also expressed in the D quadrant early in embryogenesis, which gives rise to the mesodermal and ectodermal teloblasts (Kang et al., 2002; Pilon and Weisblat, 1997). A reduction in Nanos protein by morpholino injection severely disrupts development, leading to shorter germinal bands that cannot undergo epibolic movements and to eventual death (Agee et al., 2006). This phenotype may be due to subtle changes in division rates and patterns of the teloblast cells that give rise to the germinal band (Agee et al., 2006). Therefore, the GMP-related function of nanos in sea urchin, snail and leech embryos in the maintenance of cell fate via cell cycle control may be conserved.
In the polychaete P. dumerilii, another lophotrochozoan annelid, the 4d lineage gives rise to a group of proliferating cells, the mesodermal posterior growth zone (MPGZ), which contributes to the mesodermal tissues of the developing adult segments. Vasa protein is restricted to the 4d lineage by post-transcriptional mechanisms, which may again reflect a conserved feature of the GMP (Rebscher et al., 2007). Subsequently, vasa, nanos and piwi are expressed selectively in the MPGZ of the larva. The proliferating cells of the MPGZ give rise to all of the mesodermal tissues of the developing adult segments, including the germ cells. vasa, nanos and piwi expression is retained in the newly specified germ cells, and lost in other differentiated tissues (Rebscher et al., 2007). Thus, similar to the sea urchin, the GMP of lophotrochozoans is associated with the establishment of multipotent progenitor cells from which the germline is segregated after embryogenesis. Comparisons across these two clades will be useful in uncovering the conserved features of the GMP in multipotent precursors.
Models for the origin of post-embryonic germline segregation
What is the evolutionary relationship between multipotent progenitor cells (as found in sea urchins, snails and Hydra) and embryonic PGCs (as found in Drosophila and mice)? More specifically, how did the small micromere lineage of the sea urchin, the 4d lineage of lophotrochozoans, and the cnidarian stem cells obtain their GMPs? Below, we propose two mutually exclusive hypotheses.
Post-embryonic germline segregation is ancestral
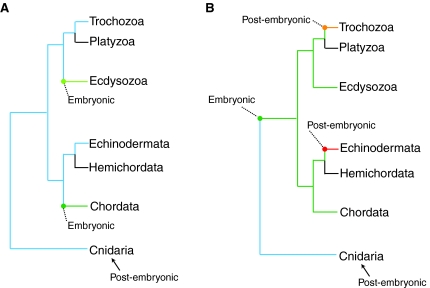
This model proposes that post-embryonic germline segregation from a multipotent precursor is ancestral, an idea that has been supported by several investigators (Fig. 6A) (Agata et al., 2006; Ewen-Campen et al., 2010; Extavour, 2007; Funayama, 2010; Juliano et al., 2010; Rebscher et al., 2007). In particular, this model argues that the protostome/deuterostome ancestor (the so-called `urbilaterian') segregated its germline through a pluripotent or multipotent stem cell (Agata et al., 2006; Extavour, 2007; Funayama, 2010). We offer a similar model here with the additional proposal that the urbilaterian germ cell multipotent progenitor was controlled by the GMP and that dedicated germ cells were segregated post-embryonically. This model also predicts that the acquisition of embryonic PGC specification in chordates and ecdysozoans must have occurred independently, perhaps by precocious specification of germ cells from the multipotent progenitor cells. A broader sampling of other taxa may reveal that the acquisition of embryonic PGC specification is common. For example, in the oyster C. gigas, a lophotrochozoan, vasa mRNA is localized to the vegetal pole of the unfertilized egg and is inherited by a small number of cells, indicative of embryonic PGC specification (Fabioux et al., 2004). This implies an advantage to this mode of germ cell specification that provides a selective pressure. Potentially, early segregation of the germline, which is then kept in a quiescent state, allows increased protection from mutations associated with DNA replication and/or additional morphogenetic freedom in subsequent development.
Fig. 6.
Models of the origin of post-embryonic germline segregation. (A) Model 1: post-embryonic germline segregation is ancestral. The echinoderms and lophotrochozoans use the same post-embryonic mechanism (blue) and obtained their multipotency program from a common origin. In this scenario, the acquisition of embryonic germline segregation in some chordates (dark green) and ecdysozoans (light green) must have occurred independently. (B) Model 2: post-embryonic germline segregation is derived. In this model, the last common bilaterian ancestor used embryonic germline specification (green), which is still used by at least some chordates and ecdysozoans. Thus, the acquisition of post-embryonic germline segregation in echinoderms (red) and lophotrochozoans (orange) must have occurred independently. In this scenario, the last common bilaterian ancestor probably acquired embryonic germline segregation after splitting from the cnidarians. Germline segregation in hemichordates and platyzoans remains largely unexplored (black).
Post-embryonic germline segregation is derived
In this model, the urbilaterian segregated its germline by specifying PGCs during embryogenesis using the GMP, as is currently exhibited by chordates and ecdysozoans. This model necessitates that the evolution of multipotent progenitor cells in lophotrochozoans and sea urchins occurred relatively recently and were independent acquisitions (Fig. 6B). For example, during the transition from direct to indirect development, the ancestor of the sea urchin small micromere lineage could have co-opted the GMP from the PGC specification pathway of the juvenile. The `intercalation hypothesis' suggests that plankton-feeding larvae evolved slowly by co-opting adult gene regulatory modules to build new larval structures (Sly et al., 2003). In support of this hypothesis, an almost identical set of regulatory genes appears to direct the formation of both the sea urchin larval and adult skeletons, thus suggesting co-option of the adult module by the larva (Gao and Davidson, 2008). Perhaps in a similar manner, the small micromere lineage co-opted the GMP from the germline cells of the adult, enabling it to remain multipotent. If this were the case, a similar scenario must also have occurred in the 4d lineage of lophotrochozoans.
Investigating the origins of the GMP
These models are based on data from a very limited number of animals and, in order to discern the evolutionary origins and conserved portions of the GMP, it is crucially important to further extend the phylogenetic tree with data from a wide variety of organisms. Given their important phylogenetic position as part of the chordate sister group, a better understanding of germline segregation in other echinoderm classes is essential to this endeavor. Is post-embryonic germline segregation through a multipotent progenitor ancestral for this phylum as is assumed by the first model? Or instead, is it specific to sea urchins, which diverged relatively recently within echinoderms? A better understanding of how the germline is segregated in the five echinoderm classes is necessary to discern between these two scenarios. Currently, in addition to the sea urchin, the sea star, sea cucumbers and brittle stars could be ideal model systems for future functional studies of the GMP as they are easily cultured in the laboratory and are amenable to morpholino-based gene knock down techniques (Ettensohn et al., 2004a). Some experimental evidence from the sea star, for example, indicates that a larval structure called the posterior enterocoel, which expresses Vasa, contributes to the adult germline; thus, the GMP may operate in these cells, although their developmental potential remains to be determined (Inoue et al., 1992; Juliano and Wessel, 2009).
A key class in which to further investigate the origins of the GMP is the crinoids (sea lilies and feather stars), which are the echinoderm out-group (Janies, 2001; Nakano et al., 2003). Currently crinoid studies are limited to expression analysis owing to the difficulty of culturing these animals in the laboratory (Nakano et al., 2003). However, expression analyses could reveal the mode of germline segregation used in this class, especially if these data are compared with those collected in the other echinoderm classes. If crinoids set aside multipotent progenitor cells from which the germline is segregated later, this would suggest that post-embryonic germline segregation is ancestral amongst echinoderms, thus supporting model one. However, if crinoids set aside a population of lineage-restricted PGCs during embryogenesis, this would call model one into question; model two, in which embryonic germline segregation is ancestral among echinoderms would be equally likely in this case. A confident reconstruction of the ancestral echinoderm mechanism is essential for inferring whether the urbilaterian segregated its germline embryonically through PGCs, or later through multipotent progenitor cells. Equally important to this effort is a more detailed analysis of other taxa by MASO- or RNAi-based approaches, including the lophotrochozoans, in which entire clades are unexplored, and the cnidarians, for which very little functional data have been collected. Expression data from diverse animals can also be used to extend the GMP tree.
The possibility remains that the coincident expression of GMP genes in similar cell types is not indicative of an interdependent program of multipotency, but rather that each gene works independently. Several potential GMP members have distinct functions outside of multipotent and germline cells. For example, Drosophila piwi is expressed in the somatic cells of the ovary, where it is required in a non-cell autonomous manner for GSC maintenance (Cox et al., 1998). The jellyfish Podocoryne carnea exhibits a low level of piwi expression in all somatic cells, and this expression increases upon transdifferentiation, thus implicating piwi function in reprogramming (Seipel et al., 2004). Furthermore, human CD34+ hematopoetic stem cells are piwi positive, and lose piwi as they differentiate (Sharma et al., 2001). nanos also has known functions outside of multipotent or germline cells. For example, it was initially discovered as an early patterning gene in Drosophila, and Nanos1 is expressed in the mouse brain (Haraguchi et al., 2003; Irish et al., 1989). However, independent and/or alternate functions for these genes do not preclude them from acting together in a conserved program in multipotent and germline cells. Rather, they may have been independently co-opted by other cells types with similar requirements as multipotent progenitors and germ cells, such as mitotic quiescence and protection from transposons. Another alternative possibility is that the GMP described here is not required for multipotency itself within multipotent progenitors, but rather for specifically conferring germline competency to those progenitors. However, given the dramatic loss of many adult tissues when nanos is knocked down in the sea urchin and the snail, and the loss of regenerative ability when piwi is knocked down in planarians, we favor the interpretation that the GMP is integrally entwined in maintaining multipotency itself. Further elucidation of the conserved functions of GMP genes in multipotent and germline cells will help to discern whether these genes are actually acting in a conserved and interrelated network.
Conclusion
The data collected in sea urchins, sponges, cnidarians and several lophotrochozoans, demonstrate that genes traditionally classified as `germline genes' have a broad role in establishing and maintaining multipotency. The specification and maintenance of potency in all of the cell types that express genes, such as vasa, nanos and piwi, probably shares a common underlying mechanism of regulatory control. PGCs and multipotent progenitor cells appear to be sister cell types, each realizing their developmental potential differently, but still closely linked by a common regulatory program. A comparison of mechanisms of germline segregation across animal taxa and in animals that use varied developmental strategies will allow us to uncover the crucial and ancient parts of this regulatory program. Our understanding of the germline will be limited if we focus only on organisms that represent extremes in what is likely to be a continuum between embryonic PGC and adult germline segregation from multipotent stem cells. This does not necessarily require that several new model systems be established, but rather that more emphasis be placed on less well-studied organisms, such as the snail, polychaete, sea urchin, sea star, planarian, Hydra and sea anemone, which are amenable to experimental investigation (Chera et al., 2006; Ettensohn et al., 2004b; Fischer and Dorresteijn, 2004; Genikhovich and Technau, 2009; Lohmann et al., 1999; Rabinowitz et al., 2008; Sanchez Alvarado and Newmark, 1999). Expression data from diverse animals that are less amenable to such experimental manipulation can also be used to bolster our understanding of the evolution of germline development and of the GMP. Lineage-restricted germ cells can reacquire pluripotency, either experimentally or in natural human disease states when ovarian-derived teratomas form. Furthermore, piwi and vasa overexpression is found in somatic and human germline cancers (Hashimoto et al., 2008; Lee et al., 2006; Liu et al., 2006; Qiao et al., 2002). Given these observations, the molecular pathways that are uncovered in sea urchin, lophotrochozoan and cnidarian multipotent cells as a result of such studies may be of broader relevance.
Note added in proof
Recent results reveal that Vasa accumulation selectively in the small micromeres of the sea urchin, when its mRNA is uniformly present in the embryo, uses a mechanism of selective Vasa protein degradation in non-small micromeres. Injection of mRNA into eggs encoding full length Vasa linked to GFP results in selective accumulation of GFP into the small micromeres. Gustavus, the ubiquitylation E3 ligase, is involved in this regulation.
Acknowledgments
We thank Casey Dunn and John Coleman for critical discussions and feedback on this manuscript. The authors are funded by the NSF and the NIH. Deposited in PMC for release after 12 months.
References
- Agata K., Nakajima E., Funayama N., Shibata N., Saito Y., Umesono Y. (2006). Two different evolutionary origins of stem cell systems and their molecular basis. Semin. Cell Dev. Biol. 17, 503-509 [DOI] [PubMed] [Google Scholar]
- Agee S. J., Lyons D. C., Weisblat D. A. (2006). Maternal expression of a NANOS homolog is required for early development of the leech Helobdella robusta. Dev. Biol. 298, 1-11 [DOI] [PubMed] [Google Scholar]
- Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M. J., Kuramochi-Miyagawa S., Nakano T., et al. (2006). A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203-207 [DOI] [PubMed] [Google Scholar]
- Aravin A. A., Hannon G. J., Brennecke J. (2007). The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318, 761-764 [DOI] [PubMed] [Google Scholar]
- Aravin A. A., Sachidanandam R., Bourc'his D., Schaefer C., Pezic D., Toth K. F., Bestor T., Hannon G. J. (2008). A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785-799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaoka-Taguchi M., Yamada M., Nakamura A., Hanyu K., Kobayashi S. (1999). Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat. Cell Biol. 1, 431-437 [DOI] [PubMed] [Google Scholar]
- Baguna J., Salo E., Auladell C. (1989). Regeneration and pattern formation in planarians III. Evidence that neoblasts are totipotent stem cells and the source of blastema cells. Development 107, 77-86 [Google Scholar]
- Ball E. E., Hayward D. C., Saint R., Miller D. J. (2004). A simple plan-cnidarians and the origins of developmental mechanisms. Nat. Rev. Genet. 5, 567-577 [DOI] [PubMed] [Google Scholar]
- Batista P. J., Ruby J. G., Claycomb J. M., Chiang R., Fahlgren N., Kasschau K. D., Chaves D. A., Gu W., Vasale J. J., Duan S., et al. (2008). PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell 31, 67-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat K. M. (1999). The posterior determinant gene nanos is required for the maintenance of the adult germline stem cells during Drosophila oogenesis. Genetics 151, 1479-1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelsterli U. (1977). An electron microscopic study of early developmental stages, myogenesis, oogenesis and cnidogenesis in the anthomedusa, Podocoryne carnea M. Sars. J. Morphol. 154, 259-289 [DOI] [PubMed] [Google Scholar]
- Bosch T. C., David C. N. (1986). Male and female stem cells and sex reversal in Hydra polyps. Proc. Natl. Acad. Sci. USA 83, 9478-9482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch T. C., David C. N. (1987). Stem Cells of Hydra magnipapillata can differentiate into somatic cells and germ line cells. Dev. Biol. 121, 182-191 [Google Scholar]
- Boswell R. E., Mahowald A. P. (1985). tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell 43, 97-104 [DOI] [PubMed] [Google Scholar]
- Boyer B. C., Henry J. Q., Martindale M. Q. (1996). Dual origins of mesoderm in a basal spiralian: cell lineage analyses in the polyclad turbellarian Hoploplana inquilina. Dev. Biol. 179, 329-338 [DOI] [PubMed] [Google Scholar]
- Breitwieser W., Markussen F. H., Horstmann H., Ephrussi A. (1996). Oskar protein interaction with Vasa represents an essential step in polar granule assembly. Genes Dev. 10, 2179-2188 [DOI] [PubMed] [Google Scholar]
- Brennecke J., Malone C. D., Aravin A. A., Sachidanandam R., Stark A., Hannon G. J. (2008). An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322, 1387-1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland B., Findley S. D., Jiang L., Liu L., Yin H., Dus M., Zhou P., Elgin S. C., Lin H. (2007). Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 21, 2300-2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown F. D., Tiozzo S., Roux M. M., Ishizuka K., Swalla B. J., De Tomaso A. W. (2009). Early lineage specification of long-lived germline precursors in the colonial ascidian Botryllus schlosseri. Development 136, 3485-3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell M. A., Girard A., van de Kant H. J., Bourc'his D., Bestor T. H., de Rooij D. G., Hannon G. J. (2007). MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12, 503-514 [DOI] [PubMed] [Google Scholar]
- Carrera P., Johnstone O., Nakamura A., Casanova J., Jackle H., Lasko P. (2000). VASA mediates translation through interaction with a Drosophila yIF2 homolog. Mol. Cell 5, 181-187 [DOI] [PubMed] [Google Scholar]
- Chapman D. (1974). Cnidarian histology. In Coelenterate Biology (ed. Muscatine L., Lenhoff H. M.), pp. 2-92 New York: Academic Press; [Google Scholar]
- Chera S., de Rosa R., Miljkovic-Licina M., Dobretz K., Ghila L., Kaloulis K., Galliot B. (2006). Silencing of the hydra serine protease inhibitor Kazal1 gene mimics the human SPINK1 pancreatic phenotype. J. Cell Sci. 119, 846-857 [DOI] [PubMed] [Google Scholar]
- Chuma S., Hosokawa M., Kitamura K., Kasai S., Fujioka M., Hiyoshi M., Takamune K., Noce T., Nakatsuji N. (2006). Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc. Natl. Acad. Sci. USA 103, 15894-15899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R., DePalma M., Priess J. R. (2006). Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science 311, 851-853 [DOI] [PubMed] [Google Scholar]
- Cox D. N., Chao A., Baker J., Chang L., Qiao D., Lin H. (1998). A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12, 3715-3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden S. L., Bernstein D. S., Bachorik J. L., Thompson B. E., Gallegos M., Petcherski A. G., Moulder G., Barstead R., Wickens M., Kimble J. (2002). A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417, 660-663 [DOI] [PubMed] [Google Scholar]
- Dalby B., Glover D. M. (1993). Discrete sequence elements control posterior pole accumulation and translational repression of maternal cyclin B RNA in Drosophila. EMBO J. 12, 1219-1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C. N., Murphy S. (1977). Characterization of interstitial stem cells in hydra by cloning. Dev. Biol. 58, 372-383 [DOI] [PubMed] [Google Scholar]
- De Mulder K., Pfister D., Kuales G., Egger B., Salvenmoser W., Willems M., Steger J., Fauster K., Micura R., Borgonie G., et al. (2009). Stem cells are differentially regulated during development, regeneration and homeostasis in flatworms. Dev. Biol. 334, 198-212 [DOI] [PubMed] [Google Scholar]
- Deng W., Lin H. (2002). miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell 2, 819-830 [DOI] [PubMed] [Google Scholar]
- DeSalle R., Schierwater B. (2008). An even ``newer'' animal phylogeny. BioEssays 30, 1043-1047 [DOI] [PubMed] [Google Scholar]
- Deshpande G., Calhoun G., Jinks T. M., Polydorides A. D., Schedl P. (2005). Nanos downregulates transcription and modulates CTD phosphorylation in the soma of early Drosophila embryos. Mech. Dev. 122, 645-657 [DOI] [PubMed] [Google Scholar]
- Dunn C. W., Hejnol A., Matus D. Q., Pang K., Browne W. E., Smith S. A., Seaver E., Rouse G. W., Obst M., Edgecombe G. D., et al. (2008). Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452, 745-749 [DOI] [PubMed] [Google Scholar]
- Eberhart C. G., Maines J. Z., Wasserman S. A. (1996). Meiotic cell cycle requirement for a fly homologue of human Deleted in Azoospermia. Nature 381, 783-785 [DOI] [PubMed] [Google Scholar]
- Ettensohn C. A., Wessel G. M., Wray G. (2004a). Development of Sea Urchins, Ascidians and Other Non-Vertebrate Deuterostomes: an Experimental Analysis. San Diego, CA: Elsevier Academic Press; [Google Scholar]
- Ettensohn C. A., Wessel G. M., Wray G. A. (2004b). The invertebrate deuterostomes: an introduction to their phylogeny, reproduction, development, and genomics. Methods Cell Biol. 74, 1-13 [DOI] [PubMed] [Google Scholar]
- Ewen-Campen B., Schwager E. E., Extavour C. G. (2010). The molecular machinery of germ line specification. Mol. Reprod. Dev. 77, 3-18 [DOI] [PubMed] [Google Scholar]
- Extavour C. (2007). Evolution of the bilaterian germ line: lineage origin and modulation of specification mechanisms. Integr. Comp. Biol. 47, 770-785 [DOI] [PubMed] [Google Scholar]
- Extavour C. G., Akam M. (2003). Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development 130, 5869-5884 [DOI] [PubMed] [Google Scholar]
- Extavour C. G., Pang K., Matus D. Q., Martindale M. Q. (2005). vasa and nanos expression patterns in a sea anemone and the evolution of bilaterian germ cell specification mechanisms. Evol. Dev. 7, 201-215 [DOI] [PubMed] [Google Scholar]
- Fabioux C., Huvet A., Lelong C., Robert R., Pouvreau S., Daniel J. Y., Minguant C., Le Pennec M. (2004). Oyster vasa-like gene as a marker of the germline cell development in Crassostrea gigas. Biochem. Biophys. Res. Commun. 320, 592-598 [DOI] [PubMed] [Google Scholar]
- Findley S. D., Tamanaha M., Clegg N. J., Ruohola-Baker H. (2003). Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development 130, 859-871 [DOI] [PubMed] [Google Scholar]
- Fischer A., Dorresteijn A. (2004). The polychaete Platynereis dumerilii (Annelida): a laboratory animal with spiralian cleavage, lifelong segment proliferation and a mixed benthic/pelagic life cycle. BioEssays 26, 314-325 [DOI] [PubMed] [Google Scholar]
- Forbes A., Lehmann R. (1998). Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 125, 679-690 [DOI] [PubMed] [Google Scholar]
- Fujii T., Sakamoto N., Ochiai H., Fujita K., Okamitsu Y., Sumiyoshi N., Minokawa T., Yamamoto T. (2009). Role of the nanos homolog during sea urchin development. Dev. Dyn. 238, 2511-2521 [DOI] [PubMed] [Google Scholar]
- Fukuyama M., Rougvie A. E., Rothman J. H. (2006). C. elegans DAF-18/PTEN mediates nutrient-dependent arrest of cell cycle and growth in the germline. Curr. Biol. 16, 773-779 [DOI] [PubMed] [Google Scholar]
- Funayama N. (2010). The stem cell system in demosponges: insights into the origin of somatic stem cells. Dev. Growth Differ. 52, 1-14 [DOI] [PubMed] [Google Scholar]
- Funayama N., Nakatsukasa M., Mohri K., Masuda Y., Agata K. (2010). Piwi expression in archeocytes and choanocytes in demosponges: insights into the stem cell system in demosponges. Evol. Dev. 12, 275-287 [DOI] [PubMed] [Google Scholar]
- Galliot B., Miljkovic-Licina M., de Rosa R., Chera S. (2006). Hydra, a niche for cell and developmental plasticity. Semin. Cell Dev. Biol. 17, 492-502 [DOI] [PubMed] [Google Scholar]
- Gao F., Davidson E. H. (2008). Transfer of a large gene regulatory apparatus to a new developmental address in echinoid evolution. Proc. Natl. Acad. Sci. USA 105, 6091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genikhovich G., Technau U. (2009). The starlet sea anemone Nematostella vectensis: an anthozoan model organism for studies in comparative genomics and functional evolutionary developmental biology. Cold Spring Harbor Protoc. 2009, pdb emo129 [DOI] [PubMed] [Google Scholar]
- Grivna S. T., Beyret E., Wang Z., Lin H. (2006a). A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 20, 1709-1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna S. T., Pyhtila B., Lin H. (2006b). MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc. Natl. Acad. Sci. USA 103, 13415-13420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K., Nayernia K., Maier L. S., Wagner S., Dressel R., Lee J. H., Nolte J., Wolf F., Li M., Engel W., et al. (2006). Pluripotency of spermatogonial stem cells from adult mouse testis. Nature 440, 1199-1203 [DOI] [PubMed] [Google Scholar]
- Guo T., Peters A. H., Newmark P. A. (2006). A Bruno-like gene is required for stem cell maintenance in planarians. Dev. Cell 11, 159-169 [DOI] [PubMed] [Google Scholar]
- Gustafson E. A., Yajima M., Juliano C. E., Wessel G. M. (2010). Post-translational regulation by gustavus contributes to selective Vasa protein accumulation in multipotent cells during embryogenesis. Dev. Biol. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi S., Tsuda M., Kitajima S., Sasaoka Y., Nomura-Kitabayashid A., Kurokawa K., Saga Y. (2003). nanos1: a mouse nanos gene expressed in the central nervous system is dispensable for normal development. Mech. Dev. 120, 721-731 [DOI] [PubMed] [Google Scholar]
- Hashimoto H., Sudo T., Mikami Y., Otani M., Takano M., Tsuda H., Itamochi H., Katabuchi H., Ito M., Nishimura R. (2008). Germ cell specific protein VASA is over-expressed in epithelial ovarian cancer and disrupts DNA damage-induced G2 checkpoint. Gynecol. Oncol. 111, 312-319 [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Suzuki H., Kageyama Y., Yasuda K., Inoue K. (2006). Bruno-like protein is localized to zebrafish germ plasm during the early cleavage stages. Gene Expr. Patterns 6, 201-205 [DOI] [PubMed] [Google Scholar]
- Hay B., Jan L. Y., Jan Y. N. (1988). A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell 55, 577-587 [DOI] [PubMed] [Google Scholar]
- Hay B., Jan L. Y., Jan Y. N. (1990). Localization of vasa, a component of Drosophila polar granules, in maternal-effect mutants that alter embryonic anteroposterior polarity. Development 109, 425-433 [DOI] [PubMed] [Google Scholar]
- Hayashi Y., Hayashi M., Kobayashi S. (2004). Nanos suppresses somatic cell fate in Drosophila germ line. Proc. Natl. Acad. Sci. USA 101, 10338-10342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holder M., Lewis P. O. (2003). Phylogeny estimation: traditional and Bayesian approaches. Nat. Rev. Genet. 4, 275-284 [DOI] [PubMed] [Google Scholar]
- Houk M. S., Hinegardner R. T. (1980). The formation and early differentiation of sea urchin gonads. Biol. Bull. 159, 280-294 [Google Scholar]
- Houston D. W., King M. L. (2000). A critical role for Xdazl, a germ plasm-localized RNA, in the differentiation of primordial germ cells in Xenopus. Development 127, 447-456 [DOI] [PubMed] [Google Scholar]
- Houwing S., Kamminga L. M., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D. V., Blaser H., Raz E., Moens C. B., et al. (2007). A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129, 69-82 [DOI] [PubMed] [Google Scholar]
- Illmensee K., Mahowald A. P. (1974). Transplantation of posterior polar plasm in Drosophila. Induction of germ cells at the anterior pole of the egg. Proc. Natl. Acad. Sci. USA 71, 1016-1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illmensee K., Mahowald A. P. (1976). The autonomous function of germ plasm in a somatic region of the Drosophila egg. Exp. Cell Res. 97, 127-140 [DOI] [PubMed] [Google Scholar]
- Inoue K., Kiyomoto M., Shirai H. (1992). Germ cell differentiation in starfish: the posterior enterocoel as the origin of germ cells in Asterina pectinifera. Dev. Growth Differ. 34, 413-418 [DOI] [PubMed] [Google Scholar]
- Irish V., Lehmann R., Akam M. (1989). The Drosophila posterior-group gene nanos functions by repressing hunchback activity. Nature 338, 646-648 [DOI] [PubMed] [Google Scholar]
- Janies D. (2001). Phylogenetic relationships of extant echinoderm classes. Can. J. Zool. 79, 1232-1250 [Google Scholar]
- Johnson A. D., Bachvarova R. F., Drum M., Masi T. (2001). Expression of axolotl DAZL RNA, a marker of germ plasm: widespread maternal RNA and onset of expression in germ cells approaching the gonad. Dev. Biol. 234, 402-415 [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Crother B., White M. E., Patient R., Bachvarova R. F., Drum M., Masi T. (2003). Regulative germ cell specification in axolotl embryos: a primitive trait conserved in the mammalian lineage. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 358, 1371-1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone O., Lasko P. (2004). Interaction with eIF5B is essential for Vasa function during development. Development 131, 4167-4178 [DOI] [PubMed] [Google Scholar]
- Jongens T. A., Hay B., Jan L. Y., Jan Y. N. (1992). The germ cell-less gene product: a posteriorly localized component necessary for germ cell development in Drosophila. Cell 70, 569-584 [DOI] [PubMed] [Google Scholar]
- Jongens T. A., Ackerman L. D., Swedlow J. R., Jan L. Y., Jan Y. N. (1994). Germ cell-less encodes a cell type-specific nuclear pore-associated protein and functions early in the germ-cell specification pathway of Drosophila. Genes Dev. 8, 2123-2136 [DOI] [PubMed] [Google Scholar]
- Juliano C. E., Wessel G. M. (2009). An evolutionary transition of Vasa regulation in echinoderms. Evol. Dev. 11, 560-573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano C. E., Voronina E., Stack C., Aldrich M., Cameron A. R., Wessel G. M. (2006). Germ line determinants are not localized early in sea urchin development, but do accumulate in the small micromere lineage. Dev. Biol. 300, 406-415 [DOI] [PubMed] [Google Scholar]
- Juliano C. E., Yajima M., Wessel G. M. (2010). Nanos functions to maintain the fate of the small micromere lineage in the sea urchin embryo. Dev. Biol. 337, 220-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrova L. Y., Habara Y., Lee T. H., Wharton R. P. (2007). Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development 134, 1519-1527 [DOI] [PubMed] [Google Scholar]
- Kalmykova A. I., Klenov M. S., Gvozdev V. A. (2005). Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res. 33, 2052-2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D., Pilon M., Weisblat D. A. (2002). Maternal and zygotic expression of a nanos-class gene in the leech Helobdella robusta: primordial germ cells arise from segmental mesoderm. Dev. Biol. 245, 28-41 [DOI] [PubMed] [Google Scholar]
- Karashima T., Sugimoto A., Yamamoto M. (2000). Caenorhabditis elegans homologue of the human azoospermia factor DAZ is required for oogenesis but not for spermatogenesis. Development 127, 1069-1079 [DOI] [PubMed] [Google Scholar]
- Kee K., Angeles V. T., Flores M., Nguyen H. N., Reijo Pera R. A. (2009). Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature 462, 222-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J., Kerr K., Macdonald P. M. (1995). Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell 81, 403-412 [DOI] [PubMed] [Google Scholar]
- Kimura T., Ito C., Watanabe S., Takahashi T., Ikawa M., Yomogida K., Fujita Y., Ikeuchi M., Asada N., Matsumiya K., et al. (2003). Mouse germ cell-less as an essential component for nuclear integrity. Mol. Cell. Biol. 23, 1304-1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C., Bratu D. P., McGinnis-Schultz N., Koppetsch B. S., Cook H. A., Theurkauf W. E. (2007). Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell 12, 45-55 [DOI] [PubMed] [Google Scholar]
- Kobayashi S., Yamada M., Asaoka M., Kitamura T. (1996). Essential role of the posterior morphogen nanos for germline development in Drosophila. Nature 380, 708-711 [DOI] [PubMed] [Google Scholar]
- Koprunner M., Thisse C., Thisse B., Raz E. (2001). A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 15, 2877-2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz A. M., Tollenaere A., Norris B. J., Degnan B. M., Degnan S. M. (2010). Identifying the germline in an equally cleaving mollusc: Vasa and Nanos expression during embryonic and larval development of the vetigastropod Haliotis asinina. J. Exp. Zool. B. Mol. Dev. Evol. 314, 267-279 [DOI] [PubMed] [Google Scholar]
- Kugler J. M., Woo J. S., Oh B. H., Lasko P. (2010). Regulation of Drosophila vasa in vivo through paralogous cullin-RING E3 ligase specificity receptors. Mol. Cell. Biol. 30, 1769-1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S., Kimura T., Ijiri T. W., Isobe T., Asada N., Fujita Y., Ikawa M., Iwai N., Okabe M., Deng W., et al. (2004). Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131, 839-849 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S., Watanabe T., Gotoh K., Totoki Y., Toyoda A., Ikawa M., Asada N., Kojima K., Yamaguchi Y., Ijiri T. W., et al. (2008). DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 22, 908-917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S., Watanabe T., Gotoh K., Takamatsu K., Chuma S., Kojima-Kita K., Shiromoto Y., Asada N., Toyoda A., Fujiyama A., et al. (2010). MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 24, 887-892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznicki K. A., Smith P. A., Leung-Chiu W. M., Estevez A. O., Scott H. C., Bennett K. L. (2000). Combinatorial RNA interference indicates GLH-4 can compensate for GLH-1; these two P granule components are critical for fertility in C. elegans. Development 127, 2907-2916 [DOI] [PubMed] [Google Scholar]
- Lambert J. D. (2008). Mesoderm in spiralians: the organizer and the 4d cell. J. Exp. Zool. B. Mol. Dev. Evol. 310, 15-23 [DOI] [PubMed] [Google Scholar]
- Lasko P. F., Ashburner M. (1990). Posterior localization of vasa protein correlates with, but is not sufficient for, pole cell development. Genes Dev. 4, 905-921 [DOI] [PubMed] [Google Scholar]
- Lawson K. A., Hage W. J. (1994). Clonal analysis of the origin of primordial germ cells in the mouse. Ciba Found. Symp. 182, 68-84; discussion 84-91 [DOI] [PubMed] [Google Scholar]
- Leatherman J. L., Kaestner K. H., Jongens T. A. (2000). Identification of a mouse germ cell-less homologue with conserved activity in Drosophila. Mech. Dev. 92, 145-153 [DOI] [PubMed] [Google Scholar]
- Leatherman J. L., Levin L., Boero J., Jongens T. A. (2002). germ cell-less acts to repress transcription during the establishment of the Drosophila germ cell lineage. Curr. Biol. 12, 1681-1685 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Schutte D., Wulf G., Fuzesi L., Radzun H. J., Schweyer S., Engel W., Nayernia K. (2006). Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Hum. Mol. Genet. 15, 201-211 [DOI] [PubMed] [Google Scholar]
- Lim A. K., Kai T. (2007). Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 104, 6714-6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Spradling A. C. (1997). A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124, 2463-2476 [DOI] [PubMed] [Google Scholar]
- Lin Y., Page D. C. (2005). Dazl deficiency leads to embryonic arrest of germ cell development in XY C57BL/6 mice. Dev. Biol. 288, 309-316 [DOI] [PubMed] [Google Scholar]
- Liu X., Sun Y., Guo J., Ma H., Li J., Dong B., Jin G., Zhang J., Wu J., Meng L., et al. (2006). Expression of hiwi gene in human gastric cancer was associated with proliferation of cancer cells. Int. J. Cancer 118, 1922-1929 [DOI] [PubMed] [Google Scholar]
- Lohmann J. U., Endl I., Bosch T. C. (1999). Silencing of developmental genes in Hydra. Dev. Biol. 214, 211-214 [DOI] [PubMed] [Google Scholar]
- Markussen F. H., Michon A. M., Breitwieser W., Ephrussi A. (1995). Translational control of oskar generates short OSK, the isoform that induces pole plasma assembly. Development 121, 3723-3732 [DOI] [PubMed] [Google Scholar]
- Matsui Y., Zsebo K., Hogan B. L. (1992). Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell 70, 841-847 [DOI] [PubMed] [Google Scholar]
- Megosh H. B., Cox D. N., Campbell C., Lin H. (2006). The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr. Biol. 16, 1884-1894 [DOI] [PubMed] [Google Scholar]
- Mochizuki K., Sano H., Kobayashi S., Nishimiya-Fujisawa C., Fujisawa T. (2000). Expression and evolutionary conservation of nanos-related genes in Hydra. Dev. Genes Evol. 210, 591-602 [DOI] [PubMed] [Google Scholar]
- Mochizuki K., Nishimiya-Fujisawa C., Fujisawa T. (2001). Universal occurrence of the vasa-related genes among metazoans and their germline expression in Hydra. Dev. Genes Evol. 211, 299-308 [DOI] [PubMed] [Google Scholar]
- Muller W. A., Teo R., Frank U. (2004). Totipotent migratory stem cells in a hydroid. Dev. Biol. 275, 215-224 [DOI] [PubMed] [Google Scholar]
- Nakahata S., Katsu Y., Mita K., Inoue K., Nagahama Y., Yamashita M. (2001). Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J. Biol. Chem. 276, 20945-20953 [DOI] [PubMed] [Google Scholar]
- Nakahata S., Kotani T., Mita K., Kawasaki T., Katsu Y., Nagahama Y., Yamashita M. (2003). Involvement of Xenopus Pumilio in the translational regulation that is specific to cyclin B1 mRNA during oocyte maturation. Mech. Dev. 120, 865-880 [DOI] [PubMed] [Google Scholar]
- Nakamura A., Seydoux G. (2008). Less is more: specification of the germline by transcriptional repression. Development 135, 3817-3827 [DOI] [PubMed] [Google Scholar]
- Nakano H., Hibino T., Oji T., Hara Y., Amemiya S. (2003). Larval stages of a living sea lily (stalked crinoid echinoderm). Nature 421, 158-160 [DOI] [PubMed] [Google Scholar]
- Newmark P. A., Sanchez Alvarado A. (2000). Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev. Biol. 220, 142-153 [DOI] [PubMed] [Google Scholar]
- Newmark P. A., Wang Y., Chong T. (2008). Germ cell specification and regeneration in planarians. Cold Spring Harbor Symp. Quant. Biol. 73, 573-581 [DOI] [PubMed] [Google Scholar]
- O'Donnell K. A., Boeke J. D. (2007). Mighty Piwis defend the germline against genome intruders. Cell 129, 37-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palakodeti D., Smielewska M., Lu Y. C., Yeo G. W., Graveley B. R. (2008). The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA 14, 1174-1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M., Lin H. (1999). The Drosophila pumilio gene encodes two functional protein isoforms that play multiple roles in germline development, gonadogenesis, oogenesis and embryogenesis. Genetics 153, 235-250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson J. R., Cohen L. H. (1986). The fate of the small micromeres in sea urchin development. Dev. Biol. 113, 522-526 [DOI] [PubMed] [Google Scholar]
- Peterson K. J., Eernisse D. J. (2001). Animal phylogeny and the ancestry of bilaterians: inferences from morphology and 18S rDNA gene sequences. Evol. Dev. 3, 170-205 [DOI] [PubMed] [Google Scholar]
- Peterson K. J., Cameron R. A., Davidson E. H. (1997). Set-aside cells in maximal indirect development: evolutionary and developmental significance. BioEssays 19, 623-631 [DOI] [PubMed] [Google Scholar]
- Pfister D., De Mulder K., Hartenstein V., Kuales G., Borgonie G., Marx F., Morris J., Ladurner P. (2008). Flatworm stem cells and the germ line: developmental and evolutionary implications of macvasa expression in Macrostomum lignano. Dev. Biol. 319, 146-159 [DOI] [PubMed] [Google Scholar]
- Pilon M., Weisblat D. A. (1997). A nanos homolog in leech. Development 124, 1771-1780 [DOI] [PubMed] [Google Scholar]
- Qiao D., Zeeman A. M., Deng W., Looijenga L. H., Lin H. (2002). Molecular characterization of hiwi, a human member of the piwi gene family whose overexpression is correlated to seminomas. Oncogene 21, 3988-3999 [DOI] [PubMed] [Google Scholar]
- Rabinowitz J. S., Chan X. Y., Kingsley E. P., Duan Y., Lambert J. D. (2008). Nanos is required in somatic blast cell lineages in the posterior of a mollusk embryo. Curr. Biol. 18, 331-336 [DOI] [PubMed] [Google Scholar]
- Raff R. A. (2008). Origins of the other metazoan body plans: the evolution of larval forms. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 363, 1473-1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph P. M. (1960). Tetraplatia, a coronate scyphomedusan. Proc. R. Soc. Lond. B. Biol. Sci. 152, 263-281 [DOI] [PubMed] [Google Scholar]
- Ransick A., Cameron R. A., Davidson E. H. (1996). Postembryonic segregation of the germ line in sea urchins in relation to indirect development. Proc. Natl. Acad. Sci. USA 93, 6759-6763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebscher N., Zelada-Gonzalez F., Banisch T. U., Raible F., Arendt D. (2007). Vasa unveils a common origin of germ cells and of somatic stem cells from the posterior growth zone in the polychaete Platynereis dumerilii. Dev. Biol. 306, 599-611 [DOI] [PubMed] [Google Scholar]
- Rebscher N., Volk C., Teo R., Plickert G. (2008). The germ plasm component vasa allows tracing of the interstitial stem cells in the cnidarian Hydractinia echinata. Dev. Dyn. 237, 1736-1745 [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Oviedo N. J., Jennings J. R., Jenkin J. C., Sanchez Alvarado A. (2005). SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310, 1327-1330 [DOI] [PubMed] [Google Scholar]
- Reijo R., Lee T. Y., Salo P., Alagappan R., Brown L. G., Rosenberg M., Rozen S., Jaffe T., Straus D., Hovatta O., et al. (1995). Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat. Genet. 10, 383-393 [DOI] [PubMed] [Google Scholar]
- Richardson B. E., Lehmann R. (2010). Mechanisms guiding primordial germ cell migration: strategies from different organisms. Nat. Rev. Mol. Cell Biol. 11, 37-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A., Kruger D., Carroll S. B. (2005). Animal evolution and the molecular signature of radiations compressed in time. Science 310, 1933-1938 [DOI] [PubMed] [Google Scholar]
- Rouhana L., Shibata N., Nishimura O., Agata K. (2010). Different requirements for conserved post-transcriptional regulators in planarian regeneration and stem cell maintenance. Dev. Biol. 341, 429-443 [DOI] [PubMed] [Google Scholar]
- Ruggiu M., Speed R., Taggart M., McKay S. J., Kilanowski F., Saunders P., Dorin J., Cooke H. J. (1997). The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature 389, 73-77 [DOI] [PubMed] [Google Scholar]
- Sada A., Suzuki A., Suzuki H., Saga Y. (2009). The RNA-binding protein NANOS2 is required to maintain murine spermatogonial stem cells. Science 325, 1394-1398 [DOI] [PubMed] [Google Scholar]
- Saito K., Nishida K. M., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M. C. (2006). Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 20, 2214-2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvetti A., Rossi L., Lena A., Batistoni R., Deri P., Rainaldi G., Locci M. T., Evangelista M., Gremigni V. (2005). DjPum, a homologue of Drosophila Pumilio, is essential to planarian stem cell maintenance. Development 132, 1863-1874 [DOI] [PubMed] [Google Scholar]
- Sanchez Alvarado A., Newmark P. A. (1999). Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc. Natl. Acad. Sci. USA 96, 5049-5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Hayashi Y., Ninomiya Y., Shigenobu S., Arita K., Mukai M., Kobayashi S. (2007). Maternal Nanos represses hid/skl-dependent apoptosis to maintain the germ line in Drosophila embryos. Proc. Natl. Acad. Sci. USA 104, 7455-7460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaner C. E., Deshpande G., Schedl P. D., Kelly W. G. (2003). A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Dev. Cell 5, 747-757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T., Wieschaus E. (1986). Germline autonomy of maternal-effect mutations altering the embryonic body pattern of Drosophila. Dev. Biol. 113, 443-448 [DOI] [PubMed] [Google Scholar]
- Seipel K., Yanze N., Schmid V. (2004). The germ line and somatic stem cell gene Cniwi in the jellyfish Podocoryne carnea. Int. J. Dev. Biol. 48, 1-7 [DOI] [PubMed] [Google Scholar]
- Seki Y., Yamaji M., Yabuta Y., Sano M., Shigeta M., Matsui Y., Saga Y., Tachibana M., Shinkai Y., Saitou M. (2007). Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development 134, 2627-2638 [DOI] [PubMed] [Google Scholar]
- Shamblott M. J., Axelman J., Wang S., Bugg E. M., Littlefield J. W., Donovan P. J., Blumenthal P. D., Huggins G. R., Gearhart J. D. (1998). Derivation of pluripotent stem cells from cultured human primordial germ cells. Proc. Natl. Acad. Sci. USA 95, 13726-13731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A. K., Nelson M. C., Brandt J. E., Wessman M., Mahmud N., Weller K. P., Hoffman R. (2001). Human CD34(+) stem cells express the hiwi gene, a human homologue of the Drosophila gene piwi. Blood 97, 426-434 [DOI] [PubMed] [Google Scholar]
- Shibata N., Umesono Y., Orii H., Sakurai T., Watanabe K., Agata K. (1999). Expression of vasa(vas)-related genes in germline cells and totipotent somatic stem cells of planarians. Dev. Biol. 206, 73-87 [DOI] [PubMed] [Google Scholar]
- Sly B. J., Snoke M. S., Raff R. A. (2003). Who came first-larvae or adults? Origins of bilaterian metazoan larvae. Int. J. Dev. Biol. 47, 623-632 [PubMed] [Google Scholar]
- Solana J., Lasko P., Romero R. (2009). Spoltud-1 is a chromatoid body component required for planarian long-term stem cell self-renewal. Dev. Biol. 328, 410-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper S. F. C., van der Heijden G. W., Hardiman T. C., Goodheart M., Martin S. L., de Boer P., Bortvin A. (2008). Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev. Cell 15, 285-297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike C., Meyer N., Racen E., Orsborn A., Kirchner J., Kuznicki K., Yee C., Bennett K., Strome S. (2008). Genetic analysis of the Caenorhabditis elegans GLH family of P-granule proteins. Genetics 178, 1973-1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S., Wood W. B. (1982). Immunofluorescence visualization of germ-line-specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 79, 1558-1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styhler S., Nakamura A., Swan A., Suter B., Lasko P. (1998). vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development 125, 1569-1578 [DOI] [PubMed] [Google Scholar]
- Styhler S., Nakamura A., Lasko P. (2002). VASA localization requires the SPRY-domain and SOCS-box containing protein, GUSTAVUS. Dev. Cell 3, 865-876 [DOI] [PubMed] [Google Scholar]
- Su T. T., Campbell S. D., O'Farrell P. H. (1998). The cell cycle program in germ cells of the Drosophila embryo. Dev. Biol. 196, 160-170 [DOI] [PubMed] [Google Scholar]
- Subramaniam K., Seydoux G. (1999). nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development 126, 4861-4871 [DOI] [PubMed] [Google Scholar]
- Suzuki H., Tsuda M., Kiso M., Saga Y. (2008). Nanos3 maintains the germ cell lineage in the mouse by suppressing both Bax-dependent and -independent apoptotic pathways. Dev. Biol. 318, 133-142 [DOI] [PubMed] [Google Scholar]
- Swartz S. Z., Chan X. Y., Lambert J. D. (2008). Localization of Vasa mRNA during early cleavage of the snail Ilyanassa. Dev. Genes Evol. 218, 107-113 [DOI] [PubMed] [Google Scholar]
- Takamura K., Fujimura M., Yamaguchi Y. (2002). Primordial germ cells originate from the endodermal strand cells in the ascidian Ciona intestinalis. Dev. Genes Evol. 212, 11-18 [DOI] [PubMed] [Google Scholar]
- Tam P. P., Zhou S. X. (1996). The allocation of epiblast cells to ectodermal and germ-line lineages is influenced by the position of the cells in the gastrulating mouse embryo. Dev. Biol. 178, 124-132 [DOI] [PubMed] [Google Scholar]
- Tanaka S., Dan K. (1990). Study of the lineage and cell cycle of small micromeres in embryos of the sea urchin, Hemicentrotus pulcherrimus. Dev. Growth Differ. 32, 145-156 [DOI] [PubMed] [Google Scholar]
- Tanaka S. S., Toyooka Y., Akasu R., Katoh-Fukui Y., Nakahara Y., Suzuki R., Yokoyama M., Noce T. (2000). The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 14, 841-853 [PMC free article] [PubMed] [Google Scholar]
- Thomson T., Lasko P. (2004). Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis 40, 164-170 [DOI] [PubMed] [Google Scholar]
- Tomancak P., Guichet A., Zavorszky P., Ephrussi A. (1998). Oocyte polarity depends on regulation of gurken by Vasa. Development 125, 1723-1732 [DOI] [PubMed] [Google Scholar]
- Tsuda M., Sasaoka Y., Kiso M., Abe K., Haraguchi S., Kobayashi S., Saga Y. (2003). Conserved role of nanos proteins in germ cell development. Science 301, 1239-1241 [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y., Hao Y., Beyret E., Yin H., Kuramochi-Miyagawa S., Nakano T., Lin H. (2009). MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J. Biol. Chem. 284, 6507-6519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin V. V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P. D. (2006). A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320-324 [DOI] [PubMed] [Google Scholar]
- Voronina E., Lopez M., Juliano C. E., Gustafson E., Song J. L., Extavour C., George S., Oliveri P., McClay D., Wessel G. (2008). Vasa protein expression is restricted to the small micromeres of the sea urchin, but is inducible in other lineages early in development. Dev. Biol. 314, 276-286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Reinke V. (2008). A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr. Biol. 18, 861-867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zayas R. M., Guo T., Newmark P. A. (2007). nanos function is essential for development and regeneration of planarian germ cells. Proc. Natl. Acad. Sci. USA 104, 5901-5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Lin H. (2004). Nanos maintains germline stem cell self-renewal by preventing differentiation. Science 303, 2016-2019 [DOI] [PubMed] [Google Scholar]
- Whitington P. M., Dixon K. E. (1975). Quantitative studies of germ plasm and germ cells during early embryogenesis of Xenopus laevis. J. Embryol. Exp. Morphol. 33, 57-74 [PubMed] [Google Scholar]
- Wolke U., Weidinger G., Koprunner M., Raz E. (2002). Multiple levels of posttranscriptional control lead to germ line-specific gene expression in the zebrafish. Curr. Biol. 12, 289-294 [DOI] [PubMed] [Google Scholar]
- Woods F. (1931). History of the germ cells in Sphaerium striatinum. J. Morphol. 51, 545-595 [Google Scholar]
- Yin H., Lin H. (2007). An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature 450, 304-308 [DOI] [PubMed] [Google Scholar]
- Yoon C., Kawakami K., Hopkins N. (1997). Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development 124, 3157-3165 [DOI] [PubMed] [Google Scholar]