Abstract
The molecular mechanisms driving the conserved metazoan developmental shift referred to as the mid-blastula transition (MBT) remain mysterious. Typically, cleavage divisions give way to longer asynchronous cell cycles with the acquisition of a gap phase. In Drosophila, rapid synchronous nuclear divisions must pause at the MBT to allow the formation of a cellular blastoderm through a special form of cytokinesis termed cellularization. Drosophila Fragile X mental retardation protein (dFMRP; FMR1), a transcript-specific translational regulator, is required for cellularization. The role of FMRP has been most extensively studied in the nervous system because the loss of FMRP activity in neurons causes the misexpression of specific mRNAs required for synaptic plasticity, resulting in mental retardation and autism in humans. Here, we show that in the early embryo dFMRP associates specifically with Caprin, another transcript-specific translational regulator implicated in synaptic plasticity, and with eIF4G, a key regulator of translational initiation. dFMRP and Caprin collaborate to control the cell cycle at the MBT by directly mediating the normal repression of maternal Cyclin B mRNA and the activation of zygotic frühstart mRNA. These findings identify two new targets of dFMRP regulation and implicate conserved translational regulatory mechanisms in processes as diverse as learning, memory and early embryonic development.
Keywords: Cyclin B, Fragile X syndrome, Frühstart (Z600), Drosophila
INTRODUCTION
The mid-blastula transition (MBT) is defined as the first developmental event that requires zygotic gene activity and represents a critical transition in animal development, but the molecular regulatory mechanisms that control the proper timing of the MBT are only partially understood (reviewed by Tadros and Lipshitz, 2009). Initially, embryos are subdivided through cleavage without cell growth. However, when a species-specific nucleo-cytoplasmic (N:C) ratio is achieved, the MBT is triggered, a developmental event typically characterized by a dramatic increase in the length and asynchrony of subsequent cleavage division cycles. Preceding the MBT, animal embryos must undergo a controlled degradation of maternal transcripts and activation of the zygotic genes in a precise hand-off of genetic control known as the maternal-to-zygotic transition (MZT).
In Drosophila, both the degradation of maternal transcripts and the wholesale activation of the zygotic genome are largely driven by a timing mechanism and are independent of the N:C ratio, although they can profoundly impact the morphological events of the MBT (Benoit et al., 2009; Lu et al., 2009). For example, loss of function of smaug (smg), a key regulator of maternal mRNA decay, results in the failure of many downstream processes, including activation of the DNA damage checkpoint, cell cycle slowing, cellularization and the transcription of many zygotic genes (Benoit et al., 2009). However, many specific aspects of the Drosophila MBT, including cell formation at nuclear cycle 14 (NC14) and the activation of specific zygotic genes, are believed to be triggered by unknown signals stemming from the N:C ratio (Edgar et al., 1986; Lu et al., 2009). This has been demonstrated in part through analysis of Drosophila maternal haploid (mh) mutants. The haploid embryos derived from mh mothers develop normally until NC14. However, they then undergo an additional nuclear division to achieve the necessary N:C ratio prior to extending interphase and undergoing cellularization (Edgar et al., 1986). The molecular nature of the N:C signal remains elusive, but ultimately impacts Cyclin-dependent kinase 1 (CDK1, also known as CDC2) through multiple mechanisms. These include modulation of Cyclin B (CYCB) levels through rounds of protein synthesis and degradation (Edgar et al., 1994; Huang and Raff, 1999; Raff et al., 2002), activation of the grp (Chk1) and mei-41 (atr) DNA damage checkpoint pathway (Fogarty et al., 1997; Sibon et al., 1997; Sibon et al., 1999; Royou et al., 2008), and precisely timed zygotic transcription of the mitotic cyclin-dependent kinase (M-CDK1) inhibitors frühstart (frs; Z600 – FlyBase) and tribbles (trbl) (Grosshans and Wieschaus, 2000; Mata et al., 2000; Grosshans et al., 2003; Gawlinski et al., 2007).
It is clear that degradation of maternal mRNA and initiation of zygotic transcription contribute to the timing and morphological events of the MBT (Arbeitman et al., 2002; Tadros and Lipshitz, 2005; Pilot et al., 2006; De Renzis et al., 2007; Lu et al., 2009; Tadros and Lipshitz, 2009); however, the translational regulatory mechanisms that modulate rates of protein synthesis during this transition are largely unexplored. We previously found that the transcript-specific translational regulator, dFMRP (FMR1 – FlyBase), is required for the major morphological event of the MBT, i.e. cellularization (Monzo et al., 2006). In this study, we identify proteins that are associated with dFMRP and demonstrate that dFMRP collaborates with one of these, Caprin, to ensure correct timing of the MBT. dFMRP and Caprin associate with both CycB and frs mRNAs, but function to activate translation of one target while repressing translation of the other to appropriately modulate the cell cycle at the MBT. The identification of Caprin as a partner for dFMRP in this novel context, the MBT, suggests that these proteins might respond together to diverse developmental signals.
MATERIALS AND METHODS
Genetics
Stocks were reared on standard cornmeal molasses media. Oregon-R (wild type), w1118, w1118; Df(3R)Exel6265, P{XP-U}Exel6265/TM6B Tb1, y1w67c23; P{EPgy2}CG18811EY06062, w; Df(3L)Cat ri sbd e/TM3 Ser, and w; CycB2/CyO, P{ftz/lacB}E3 were from the Bloomington Stock Center (Bloomington, IN, USA), w1118; fmr13/TM6C, Tb, Sb from T. Jongens (University of Pennsylvania, Philadelphia, PA, USA), and CyO, P{w+mC=GAL4-twi.G}2.2, P{UAS-2xEGFP}AH2.2 (referred to as Cyo-GFP) was from D. Stein (The University of Texas at Austin, TX, USA).
To generate Capr alleles, homozygous y1w67c23; P{EPgy2}CG18811EY06062 females were crossed to y, w/Y; ry Sb Δ2-3/TM6 males. Genomic DNA from single w/Y; Pexcision/Balancer males was screened by PCR (Gloor et al., 1993) to characterize deletions, using primers 2+ (5′-GACTATGTTAGGGTTTATGCGG-3′), 3– (5′-ACTGCGTCAACAACTTGC-3′) and 4– (5′-GCGATAGGACTCCAGTTTG-3′). The corresponding genomic region of four viable deletions (520, 648, 707 and 973 bp) and a precise excision event designated Caprrvt were sequenced from homozygous flies. Df(3L)Cat (http://flybase.org/reports/FBab0002301.html) removes ∼750 kb in the region 75C1-75C2;75F1. To generate the Df(3L)Cat, fmr13 recombinant chromosome, progeny of w; Df(3L)Cat ri sbd e/fmr13 females were recovered over TM3 ri e Sb. Stocks were generated for eleven individuals over either TM2 Ubx e or TM6B Hu e selected for the presence of ri and loss of sbd e. The desired recombinant was identified by immunoblotting recombinant/Df(3R)Exel6265, P{XP-U}Exel6265/TM6B Tb1 fly extracts with the 5A11 monoclonal antibody and recombinant/Capr1 fly extracts with anti-CAPR antibodies.
Biochemistry and molecular biology
Except as noted, procedures were performed according to Sisson et al. (Sisson et al., 2000). `Protein null' embryos were obtained from Capr2/Df(3L)Cat (Capr–) and dfmr13/Df(3R)Exel6265 (dfmr1–) females. CDC2 phospho-isoforms were resolved on a 20-cm long, 1-mm thick 10.5% PAGE gel at 300V for 8 hours at 4°C prior to transfer to PVDF membrane. Extracts for immunoprecipitations were as described (Monzo et al., 2006), except where noted. Briefly, for MudPIT analysis, gradients were loaded with 0.9 mg extract from cellularizing embryos. Immunoprecipitations were performed on pooled sucrose gradient fractions 1-3, pellets were washed with TKT100 buffer, rinsed in 10 mM Tris-HCl pH 8.0, 50 mM KCl, and eluted for 20 minutes at room temperature in fresh 8 M urea, 100 mM Tris-HCl pH 7.0. For Fig. 1D, the extract was prepared from NC13 to early NC14 embryos. Immunoprecipitate pellets were incubated in either TKT100 or 100 μg/ml RNase A in TKT100 for 15 minutes at 25°C and collected again by centrifugation. Supernatants were saved and pellets washed twice prior to analysis by immunoblot. Single adult flies for immunoblots were homogenized in 30 μl SDS sample buffer and 11 μl was loaded.
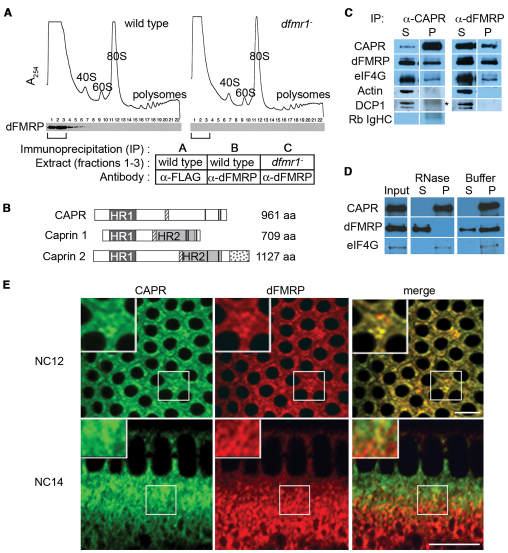
Fig. 1.
dFMRP associates with Caprin. (A) Extracts from Drosophila embryos laid by wild-type or dfmr– mutant [dfmr13/Df(3R)Exel6265] females were fractionated on sucrose gradients. UV traces (A254) show positions of ribosomal material (40S, 60S, 80S, polysomes). Fractions (1-22) were analyzed by immunoblotting for dFMRP. Fractions 1-3 (brackets) from each gradient were separately pooled for immunoprecipitations using anti-FLAG (sample A) or anti-dFMRP (samples B and C) antibodies. (B) Alignment of Drosophila Caprin (CAPR) (CG18811) with human CAPRIN1 and 2 showing Homology Region 1 and 2 (shaded boxes HR1 and HR2) (Grill et al., 2004), G3BP-binding motif (hatched boxes) (Solomon et al., 2007), Caprin 2-specific C1q-related domain (stippled box) (Aerbajinai et al., 2004; Grill et al., 2004), RNA-binding RGG motifs (thick black bars) (Shiina et al., 2005; Wang et al., 2005) and lengths (aa, amino acid). For Drosophila CAPR, the percentage identity/similarity to human CAPRIN1 HR1 is 32%/52% and to human CAPRIN2 HR1 is 51%/73%. (C) Immunoblot of supernatants (S) and pellets (P) from anti-CAPR and anti-dFMRP immunoprecipitations from wild-type nuclear cycle (NC) 13 to early NC14 embryo extracts probed with antibodies to the proteins listed on the left. DCP1, an mRNA decapping protein, and Actin served as controls and were not present in either pellet. Asterisk marks a non-specific signal (Rb IgHC) detected by goat anti-rabbit HRP secondary antibody alone (compare with bottom panel). (D) Immunoblot of anti-CAPR immunoprecipitate from wild-type embryo extract (Input) and equal percentages of supernatants (S) and pellets (P) from Input incubated with RNase A or buffer. Probing was with antibodies to the proteins listed on the left. (E) Immunofluorescence analysis of fixed wild-type embryos reveals partial colocalization between CAPR (green) and dFMRP (red). Optical sections are through the apical cytoplasm of an NC12 embryo and the sagittal plane of an NC14 embryo. The boxed regions are shown at higher magnification in the insets. Scale bars: 10 μm.
Polyclonal antisera to CAPR were generated by injection of two rabbits with KLH-conjugated C-terminal peptide (CRQNQSQRMPLGLENKN) (Covance Research Products, Denver, PA, USA). Antibodies against the following additional proteins were used: (1) rabbit polyclonals: DCP1 (58-2) (Barbee et al., 2006), eIF4G (Zapata et al., 1994), FRS (Grosshans et al., 2003), Cyclin B3 (Jacobs et al., 1998), α-Tubulin 67C (Matthews et al., 1993) and CDC20 (FZY) (Raff et al., 2002); (2) mouse monoclonals: dFMRP (5A11 for immunoblots only), Actin (JLA20) and Cyclin A (A12) (all from the Developmental Studies Hybridoma Bank at the University of Iowa), Lamin (ADL101) (Stuurman et al., 1995), phospho-Histone H3-Ser10 (6G3, Cell Signaling Technology, Danvers, MA, USA), and CDC2 (PSTAIR) and α-Tubulin (DM1A) (both from Sigma-Aldrich, St Louis, MO, USA); and (3) goat polyclonal Cyclin B (dN-17, Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Total RNA was extracted from 1.75- to 2.75-hour-old embryos (late NC13 to early NC14), immunoprecipitated material, and corresponding input S10B using TRIzol-LS reagent (Invitrogen, Carlsbad, CA, USA). Calculations performed are described in the legend to Fig. 5. For quantitative PCR, total embryonic RNA (1 μg), immunoprecipitated RNA (100 ng) or S10B RNA (100 ng) was digested with amplification-grade DNAaseI (Invitrogen) according to the manufacturer's protocol followed by reverse transcription using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) and RNase inhibitors (Promega, Madison, WI, USA). Quantitative PCR was performed with Power SYBR PCR Master Mix in a 7900HT sequence detector (Applied Biosystems). Levels of specific RNAs were quantified as described (Monzo et al., 2006). For primer sequences, see Table S1 in the supplementary material.
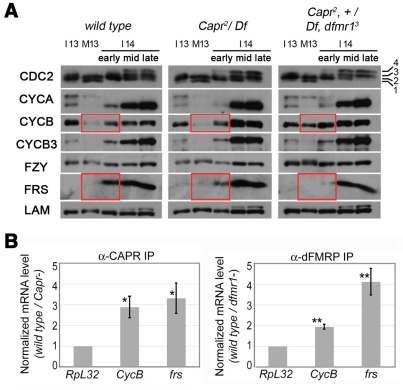
Fig. 5.
dFMRP and CAPR associate with CycB and frs mRNAs in wild-type embryos and specifically alter CYCB and FRS protein expression at the MBT. (A) An immunoblot of ten Drosophila embryos per lane of the indicated genotypes (top) was probed for the indicated proteins (left), which included Lamin (LAM) as a loading control. Hand-sorted embryos were of the indicated stages: NC13 interphase (I13) and mitosis (M13), or NC14 early, middle and late interphase (I14). Phospho-isoforms of CDC2 (Edgar et al., 1994) are labeled (1-4). Red boxes highlight differences in protein expression between genotypes. (B) mRNA specifically immunoprecipitating from NC13 to early NC14 embryo extracts with CAPR or dFMRP as determined by quantitative PCR. Levels are presented as ratios (wild type/mutant) of RpL32 (control), CycB and frs mRNAs in immunoprecipitates. Error bars indicate s.d. Significance was determined for normalized mRNA values using a two-tailed Student's t-test. *, P=0.022 for CycB and P=0.020 for frs; **, P=0.003 for CycB and P=0.00005 for frs.
Multidimensional protein identification technology (MudPIT)
Protein samples were digested with trypsin as previously described (Link et al., 1999). The protein digest was pressure-loaded onto a fused silica capillary biphasic column containing 3 cm of 5 μm Aqua C18 material (Phenomenex, Ventura, CA, USA) followed by 2 cm of 5 μm Partisphere strong cation exchanger (Whatman, Clifton, NJ, USA) packed into a 250 μm internal diameter capillary with a 2 μm filtered union (UpChurch Scientific, Oak Harbor, WA, USA). The biphasic column was washed with buffer A (94.9% water, 5% acetonitrile, 0.1% formic acid). After desalting, a 100 μm internal diameter capillary with a 5 μm pulled tip packed with 10 cm 3 μm Aqua C18 material and the entire split-column (biphasic column-filter union-analytical column) were placed inline with a 1100 quaternary HPLC (Agilent, Palo Alto, CA, USA) and analyzed using a modified four-step separation as described previously (Washburn et al., 2001). Step 1 consisted of a 100-minute gradient from 0-100% buffer B (80% acetonitrile/0.1% formic acid). Steps 2-4 were: 3 minutes of 100% buffer A, 2 minutes of X% buffer C (500 mM ammonium acetate/5% acetonitrile/0.1% formic acid), a 10-minute gradient from 0-15% buffer B, and a 97-minute gradient from 15-45% buffer B; the 2-minute buffer C percentages (X) were 20, 40 and 100%, respectively, for the four-step analysis.
Eluted peptides were electrosprayed directly into an LTQ two-dimensional ion-trap mass spectrometer (ThermoFinnigan, Palo Alto, CA, USA) with the application of a distal 2.4 kV spray voltage. A cycle of one full-scan mass spectrum (400-1700 m/z) followed by eight data-dependent MS/MS spectra at a 35% normalized collision energy was repeated continuously throughout each step of the multidimensional separation. Scan functions and solvent gradients were controlled by the Xcalibur data system. Poor quality MS/MS spectra were removed from the dataset using an automated algorithm (Bern et al., 2004). Remaining MS/MS spectra were searched with the SEQUEST algorithm (Edgar et al., 1994) against the EBI-IPI Drosophila 17, 05/18/06 concatenated to a decoy database of reversed sequences (Peng et al., 2003). All searches were parallelized and performed on a Beowulf computer cluster consisting of 100 1.2 GHz Athlon CPUs (Sadygov et al., 2002). No enzyme specificity was considered for any search. SEQUEST results were assembled and filtered using the DTASelect (version 2.0) program (Tabb et al., 2002; Cociorva et al., 2007). The false positive rates (5% in this analysis) are estimated by the program from the number and quality of spectral matches to the decoy database. The MS/MS spectra for the modified peptides were manually evaluated using criteria reported previously (Link et al., 1999).
Imaging
Live analysis of cellularization was performed as described (Monzo et al., 2006). Time-lapse differential interference contrast (DIC) imaging of cortical nuclear divisions in live embryos was performed using a Zeiss AxioVert 200 microscope with a 40× EC Plan Neofluar objective. Imaging began just prior to NC10, shortly after pole bud formation, and ended when the furrow front ingressed 35 μm. Durations of interphase and mitosis were determined by measuring the time between nuclear envelope breakdown and reassembly. Penetrance of precocious mitosis 14 was determined by counting the total number of embryos imaged that either partially or completely cellularized during interphase of NC15. Nuclear densities at the cortex were measured to confirm that mutant embryos begin cortical divisions relative to pole bud formation normally. To test the role of zygotic Cyclin B in the precocious mitosis 14 phenotype, embryos were imaged by time-lapse DIC microscopy and genotypes were subsequently determined by scoring the presence or absence of GFP expression by fluorescence microscopy. Fixed immunofluorescence analysis was performed as described (Papoulas et al., 2005). For visualization of spindle microtubules, Paclitaxel (Sigma-Aldrich) was added to a final concentration of 2.5 μM as described (Maldonado-Codina and Glover, 1992). DNA was visualized with TO-PRO-3 iodide (Invitrogen Molecular Probes).
RESULTS
dFMRP associates with Caprin and eIF4G in early Drosophila embryos
To define the mechanism of dFMRP function during the MBT we set out to identify protein binding partners of dFMRP and to characterize their function during embryogenesis. Extracts from wild-type and dfmr1– (see Materials and methods) embryos were subjected to sucrose gradient velocity centrifugation in parallel (Fig. 1A). dFMRP sedimented at the top of the sucrose gradient in wild-type extracts, away from ribosomal subunits and active polyribosomes, and was absent from dfmr1– extracts (Fig. 1A). The top fractions from each gradient were pooled and subjected to specific and control immunoprecipitations (Fig. 1A). Proteins within each immunoprecipitate were then identified by multidimensional protein identification technology (MudPIT). Proteins immunoprecipitated with anti-dFMRP antibody from wild-type extract, but not found in either control immunoprecipitate (non-specific antibody and wild-type extract, or anti-dFMRP antibody and dfmr1– protein extract) were considered specific dFMRP-associated proteins. These stringent criteria identified only two proteins: eukaryotic initiation factor (eIF) 4G and the previously uncharacterized Drosophila homolog of the vertebrate cell cycle-associated protein (Caprin). eIF4G mediates the binding of all translationally competent mRNAs to the 40S ribosome to form a pre-initiation complex and is a major target of translational regulation (Pestova et al., 2007). Vertebrate Caprins are transcript-specific RNA-binding proteins implicated in translational regulation (Shiina et al., 2005; Solomon et al., 2007). Drosophila Caprin (CAPR) and human Caprins share a highly conserved Homology Region 1 (HR1) (32% identical/52% similar to human CAPRIN1), G3BP/Rasputin-binding domain (7/7 consensus core residues) and three RGG RNA-binding domains (Fig. 1B). The HR1 domain is the most highly conserved domain among the Caprin family members and Capr (CG18811) is the only HR1-containing gene in the Drosophila genome (Grill et al., 2004). The identification of only two dFMRP-interacting proteins, both translational regulators, validates the stringency of our screen and suggests that the proteins identified are relevant to the mechanism of dFMRP function.
Vertebrate Caprin1 has been shown to localize to neuronal granules within dendrites and to repress the translation of specific mRNAs implicated in synaptic plasticity (Shiina et al., 2005; Solomon et al., 2007). Because Caprin and FMRP had individually been implicated in translational regulation and synaptic plasticity, we set out to characterize the functional significance of the CAPR-dFMRP interaction. Immune sera raised against a CAPR C-terminal peptide specifically recognized a single 140 kDa band in wild-type adult or embryo extracts (see Fig. S1 in the supplementary material; Fig. 2B). A significant proportion of dFMRP and CAPR, and a relatively small proportion of eIF4G, co-immunoprecipitated with both dFMRP and CAPR (Fig. 1C). Treatment of the immunoprecipitates with RNase A resulted in dissociation of the dFMRP-CAPR interaction but not that of eIF4G (Fig. 1D), indicating that dFMRP and CAPR co-immunoprecipitate through binding in a common ribonucleoprotein (RNP) complex and not through direct protein-protein interactions. Our immunofluorescence analysis of fixed wild-type cleavage stage embryos revealed that CAPR is cytoplasmic and appears to be enriched in previously described dFMRP-containing cytoplasmic RNP bodies (Monzo et al., 2006) (Fig. 1E). In gastrula stage embryos, CAPR and dFMRP are highly expressed in the central nervous system (see Fig. S2 in the supplementary material), consistent with the possibility that CAPR functions together with dFMRP in neurons as well as in the embryo.
Fig. 2.
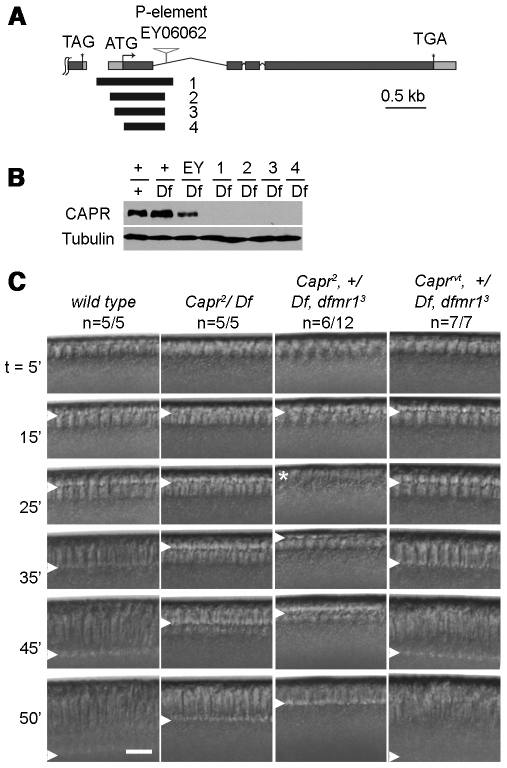
Maternal expression of Capr is required for the MBT. (A) Structure of Drosophila Capr showing start (ATG) and stop (TGA) codons, introns (thin lines), exons (gray boxes) and untranslated regions (light gray boxes). The P element transposon (EY06062) was excised to generate four deletion alleles. Black bars (1-4) indicate the deleted regions, none of which removes the 3′ end of the upstream gene (TAG stop codon). (B) Immunoblots with equal amounts of adult extracts were probed for CAPR or Tubulin. +, wild type; Df, Df(3L)Cat (which removes Capr); EY, transposon insertion EY06062; 1-4, Capr alleles. (C) Frames from representative DIC movies of single embryos from females of the genotypes indicated. Caprrvt is a perfect excision of EY06062, with no detectable phenotype. Frames show a sagittal portion of each embryo at times (in minutes) relative to NC14 onset; the number of embryos displaying the phenotype over the total period analyzed is indicated (n). Arrowheads indicate the furrow front. The asterisk indicates premature mitosis 14 with furrow disassembly.
dFMRP and Caprin collaborate to control timing of the MBT
To determine whether CAPR is required for embryogenesis, we generated mutations in Capr by imprecise P element transposon excision. Four `protein null' alleles were recovered (Fig. 2A,B), and the largest deletion that affected only the Capr transcript (Capr2) was characterized for phenotypes. Capr2/Df(3L)Cat flies were viable and showed no obvious morphological defects. Time-lapse DIC microscopy of embryos derived from these females (hereafter referred to as Capr– embryos) showed that they developed normally until the MBT (NC14); cellularization, however, then occurred at a significantly reduced rate (Fig. 2C). This appears to be identical to the previously reported phenotype observed in the majority of dfmr1– embryos (Monzo et al., 2006).
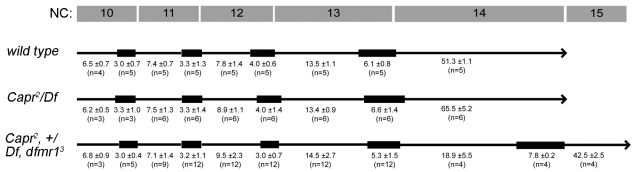
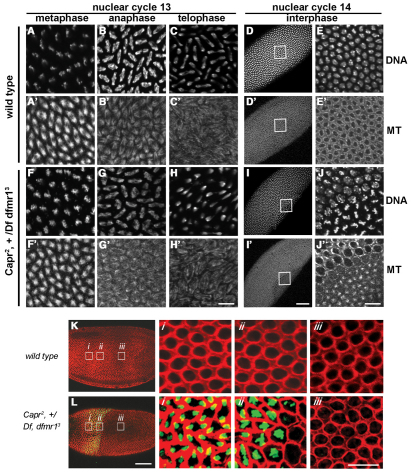
To test whether dFMRP and CAPR functionally interact, we examined the phenotype of Capr2, +/Df(3L)Cat, dfmr13 mutant flies (hereafter referred to as Capr–, fmr1– embryos), which lack both copies of Capr and one copy of dfmr1. These mutants are viable but embryos laid by Capr–, fmr1– females display a novel phenotype, as revealed by time-lapse DIC microscopy, that is not observed in embryos from females lacking dfmr1 or Capr alone. Remarkably, 50% of cleavage stage Capr–, fmr1– embryos displayed a dramatic disruption in MBT timing. They initiated cleavage furrow formation normally but, instead of undergoing a prolonged interphase, they entered mitosis 14 prematurely. A similar phenotype is observed in haploid embryos, which do not achieve the species-specific N:C ratio required to trigger the MBT until NC15 (Edgar et al., 1986). During the premature mitosis the nascent cleavage furrows regressed, only to reform during interphase of NC15, when the embryos attempted to complete cellularization (Fig. 2C; Fig. 3; compare Movies 1 and 2 in the supplementary material). In some embryos this occurred uniformly, whereas in others it occurred in large patches (Fig. 4I,I′,L; see Movie 2 in the supplementary material). In time-lapse DIC microscopy recordings of wild-type, Capr– and Capr–, fmr1– embryos, quantification of NC10-14 lengths showed no significant difference in nuclear cycle duration between wild-type and mutant embryos until NC14 (Fig. 3). Despite the dramatic disruption in the timing of the MBT, our analysis of live and fixed Capr–, fmr1– embryos revealed no signs of aberrant mitosis (Fig. 4). This is a significant departure from what has been observed in mutants of some other maternal-effect genes implicated in timing the Drosophila MBT (e.g. grp, mei-41 and smg), which display widespread mitotic spindle defects by NC13 (Fogarty et al., 1997; Sibon et al., 1997; Dahanukar et al., 1999; Sibon et al., 1999; Yu et al., 2000). Spindle morphology and nuclear spacing in Capr–, fmr1– embryos appeared normal during mitosis 13 (Fig. 4, compare A, B and C with F, G and H, and compare A′, B′ and C′ with F′, G′ and H′) as well as in those Capr–, fmr1– embryos undergoing premature mitosis 14 (Fig. 4I-J′,Li,Lii). Therefore, although Capr function, like that of dfmr1, is essential for efficient cellularization, the combined reduction of Capr and dfmr1 function produces a distinct, earlier phenotype resulting from disruption in timing of the two morphological aspects of the MBT: prolonged interphase and cellularization.
Fig. 3.
Reduction of CAPR and dFMRP function specifically disrupts timing of the MBT. The length of interphase (black line) and mitosis (black box) for nuclear cycles (NC) 10-14 for Drosophila embryos derived from females of the indicated genotypes. Elapsed times (minutes ± s.d.) and number of embryos analyzed (n) are indicated beneath the line that represents each genotype. Note the precocious mitosis of embryos from Capr2, +/Df, dfmr13 females at 18.9 minutes into interphase of NC14. Minor differences noted in NC12 and NC13 interphase and mitosis lengths produced no net change in overall cycle length.
Fig. 4.
Mitosis appears normal in embryos with a disrupted MBT. (A-J′) Immunofluorescence analysis of Drosophila embryos laid by females of the indicated genotypes (left) during the indicated cell cycle phase (top), stained with TO-PRO-3 iodide (DNA, A-E,F-J) and with antibodies to α-Tubulin 67C to visualize microtubules (MT, A′-E′,F′-J′). The boxed regions of D, D′, I and I′ are enlarged as E, E′, J and J′, respectively. (K,L) Embryos of the indicated genotypes were imaged for F-actin (red) and phospho-Histone H3-Ser10, a marker for mitotic chromosomes (green). The boxed regions are enlarged as labeled (i-iii). The wild-type embryo (K) is in interphase of NC14, whereas a portion of the mutant embryo is in interphase of NC14 (iii) and another portion is undergoing precocious mitosis (i,ii). Scale bars: 50 μm in D,D′,I,I′,K,L; 10 μm in all other panels.
Loss of dFMRP and Caprin function specifically alters Cyclin B and Frühstart protein levels at the MBT
Intriguingly, vertebrate Caprin1 was originally identified as a mitotic phosphoprotein (Stukenberg et al., 1997) and is highly expressed in proliferating tissues (Grill et al., 2004; Shiina et al., 2005), including activated lymphocytes, in which it is required for normal cell cycle progression (Wang et al., 2005). To determine whether any of the cell cycle control proteins might be targets of dFMRP/CAPR-dependent regulation, we assessed protein expression. Lysates of stage-matched wild-type, Capr– and Capr–, fmr1– embryos were probed for proteins known to control the cell cycle during the MBT: CDC2, Cyclins A, B and B3, CDC20 (FZY) and Frühstart (FRS). Five developmental stages that span the MBT were analyzed for expression of candidate proteins (a single blot was probed for Fig. 5A; see Fig. S3 in the supplementary material). We looked for altered levels during the MBT in Capr–, fmr1– mutants relative to two controls (wild-type and Capr– embryos) that never undergo premature mitosis 14. In Capr–, fmr1– mutants the steady-state levels of CYCB were significantly elevated during mitosis 13 and levels of CYCA also appeared to be somewhat elevated (Fig. 5A). In addition to prematurely elevated CYCB levels, Capr–, fmr1– embryos displayed delayed accumulation of the known, zygotically transcribed CDK1 inhibitor, FRS (Grosshans et al., 2003; Gawlinski et al., 2007) (Fig. 5A). The other proteins examined were unaffected.
The phosphorylation state of CDK1 (CDC2) at specific residues is known to contribute to the activation and inhibition of Cyclin-CDK1 activity (Nurse, 1990; Edgar et al., 1994). Although antibodies are not available to the phosphatases String/CDC25 and Twine/CDC25, the normal stage-specific CDK1 phosphorylation profile in all three genotypes during the MBT suggested that the balance of phosphatase and kinase activities that regulates CDK1 was unaltered. mRNA levels were also unaffected: changes in CYCB and FRS accumulation occurred despite normal steady-state levels of CycB mRNA and normal zygotic transcriptional activation of frs (see Fig. S4 in the supplementary material). Together, these data suggest that CAPR and dFMRP act to promote the shift in cell cycle at the MBT through suppression of CycB expression and activation of frs expression.
dFMRP and Caprin specifically associate in vivo with mRNAs encoding Cyclin B and Frühstart
To determine whether CycB and frs are themselves direct targets of dFMRP/CAPR translational regulation we tested whether the mRNAs specifically associate with CAPR and/or dFMRP at this time in development. CAPR and dFMRP immunoprecipitations were conducted from extracts of wild-type embryos, and mock immunoprecipitations were conducted from extracts of stage-matched protein null mutant embryos. Levels of specific mRNAs in the starting extract (steady-state levels) and in the immunoprecipitates were determined by quantitative RT-PCR. Both CycB and frs mRNAs were enriched in immunoprecipitations performed from wild-type extracts (Fig. 5B) suggesting that they are part of a dFMRP- and CAPR-containing mRNA-protein complex in vivo and are likely to be direct targets of translational regulation.
Elevation of CYCB levels in Capr–, fmr1– embryos contributes to disrupted timing of the MBT
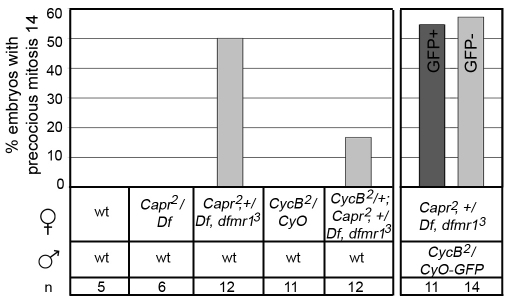
If the elevated CYCB expression specifically observed in Capr–, fmr1– mutants is relevant to the MBT phenotype, then reducing the expression of maternal CycB would be predicted to suppress the phenotype. The premature mitosis 14 phenotype was typically observed in 50% of Capr–, fmr1– embryos. Reduction of maternal CycB by one half in this background produced partial rescue, such that only 17% of embryos underwent premature mitosis (Fig. 6), indicating that protein derived from maternal CycB contributes to the phenotype. Normally, expression of zygotic CycB is believed to occur only after the completion of cellularization (during gastrulation) (Dalby and Glover, 1993). To determine whether premature expression of zygotic CycB also contributes to the premature mitosis 14 phenotype we used a GFP-marked balancer to follow the zygotic genotypes of live embryos. Capr–, fmr1– females were crossed to CycB2/Cyo-GFP males and their embryos were imaged by DIC and fluorescence microscopy. Reduction of the zygotic contribution of CycB had no effect on the premature mitosis phenotype (Fig. 6). Together, these results suggest that the elevated level of CYCB observed during mitosis 13 is derived exclusively from the premature translation of maternal CycB mRNA in Capr–, fmr1– mutants and contributes significantly to the observed MBT phenotype.
Fig. 6.
Elevated levels of maternally derived CYCB promote premature mitosis at the MBT. The percentage of embryos (n, number of embryos analyzed) derived from the indicated crosses (beneath) that display a premature mitosis 14 phenotype. Progeny derived from Capr2, +/Df, dfmr13 females receiving either a CycB2 (light-gray bar, GFP–) or GFP-marked (dark-gray bar, GFP+) paternal chromosome displayed the premature mitosis 14 phenotype at similar frequencies. The reduction of maternal CycB mRNA partially rescues the precocious mitosis 14 phenotype (left panel, far right-hand bar), whereas the reduction of zygotic CycB mRNA does not (right panel, GFP–).
DISCUSSION
In summary, we propose that maternal dFMRP and CAPR associate to regulate translation of specific mRNAs and that this regulation is essential to the embryo at a critical and sensitive juncture – the MBT. Specifically, following normal degradation of cyclins during NC13 metaphase/anaphase, Capr–, fmr1– embryos synthesize CYCB prematurely from maternal mRNA and delay synthesis of FRS from zygotic mRNA. The resultant imbalance at this particular time disrupts the cell cycle. FRS expression normally first occurs at the MBT and, not surprisingly, premature ectopic expression can block earlier mitoses (Grosshans et al., 2003). The exquisite sensitivity of M-CDK1 activity to levels of CYCB at the MBT is consistent with evidence that exogenous CYCB protein can induce premature mitosis within the first 5 minutes of cellularization (NC14 interphase), whereas similar elevation of CYCB levels 15 minutes into cellularization cannot (Royou et al., 2008). It is also possible that the stochastic patches of precocious mitosis that we observe in our mutants result from small local differences in the level of CAPR- and dFMRP-containing mRNPs formed, as the responding machinery is particularly sensitive to levels of CYCB at this time. Levels of specific mRNAs are unaffected in our mutants at the initiation of NC14, so altered CYCB and FRS levels could arise in principle through altered protein synthesis or protein stability. However, it is unlikely that the elevated CYCB we observe reflects faulty degradation because chromosome segregation and mitotic exit were unaffected (Fig. 3, Fig. 4). Furthermore, all cyclins examined depend on anaphase-promoting complex/cyclosome (APC/C)-FZY/CDC20 for their degradation (Sigrist et al., 1995; Raff et al., 2002; Huang et al., 2007), and both FZY expression and APC/C activity, as evidenced by appropriate CYCB3 degradation, appeared to be normal (Fig. 5A). Therefore, we believe precise translational regulation by dFMRP and CAPR is necessary to successfully negotiate the MBT.
In order to understand how translational regulators can so accurately modulate complex and diverse programs, it will be necessary to determine the mechanism by which proteins such as dFMRP and CAPR are recruited to specific target mRNAs and modulate translation. Activation or repression could arise from distinct regulatory mRNPs assembled on each specific mRNA region (reviewed by Darnell et al., 2005a). For example, RNA structure could induce conformational changes as a protein binds, or vice versa. Several motifs have been reported to mediate RNA binding by FMRP in vitro or in vivo: G-quartets (Darnell et al., 2001), kissing complex (Darnell et al., 2005b) and SoSlip (Bechara et al., 2009). Our analysis using RNABOB (http://selab.janelia.org/software.html), BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) or Mfold (http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1.cgi) did not identify any of these in the frs and CycB mRNAs, suggesting the association of these mRNAs with dFMRP and CAPR might be mediated by novel sequence motifs or might rely on the binding of additional proteins or on RNA structures not identified by these algorithms. Once assembled, dFMRP- and CAPR-containing mRNPs might modulate translation rates directly or by affecting the localization or stability of specific mRNAs. It is intriguing in this regard that the absence of CAPR leads to partial stabilization of CycB mRNA subsequent to the observed precocious mitosis phenotype, and the stabilization is greater upon reduction of dFMRP (see Fig. S4 in the supplementary material). The binding of CAPR and dFMRP might therefore also be required to ensure appropriate degradation of CycB mRNA after the onset of the MBT.
How might dFMRP and CAPR directly modulate translation? In the case of FMRP there is general agreement that it is a transcript-specific translational regulator; however, whether it functions as a repressor or activator, and whether it modulates initiation, elongation and/or termination, remain controversial. The overall distribution of polyribosomes between wild-type and dfmr1– extracts was indistinguishable under our conditions, suggesting no gross change in the levels of translation (Fig. 1A). Since the majority of dFMRP does not co-sediment with active polyribosomes but can associate with eIF4G, a key scaffold for pre-initiation complex assembly (Fig. 1C,D), our data suggest that dFMRP in early embryos regulates the translational initiation of specific transcripts. Caprin has also been implicated in the control of local translation required for synaptic plasticity (Richter and Klann, 2009; Wang et al., 2010); however, as with FMRP, the mechanism remains unresolved. The presence of CAPR in cap-binding complexes from Drosophila ovaries (Pisa et al., 2009) suggests that CAPR, like dFMRP, might regulate initiation complex assembly through interactions with eIF4G, consistent with the RNA-independent association of eIF4G in our CAPR immunoprecipitates. Current data from vertebrate studies indicate mitotic phosphorylation and binding to G3BP, a Ras effector, as two likely modes for regulating CAPR function in response to developmental signals. In Drosophila, conservation of the G3BP-binding domain of CAPR, the high levels of expression of the G3BP ortholog Rasputin in early embryos (Pazman et al., 2000), and the presence of Rasputin in cap-binding complexes (Pisa et al., 2009), suggest that these regulatory mechanisms are likely to be conserved. Biochemical studies will be required to assess how eIF4G associates with dFMRP- and CAPR-containing complexes, and whether dFMRP or CAPR indeed affects the assembly of functional pre-initiation complexes or modulates translation through a distinct mechanism(s).
Although CycB and frs are targets of translational control during the MBT, it is likely that dFMRP and CAPR regulate additional targets, together or individually, to help ensure reliable completion of all aspects of this developmental transition and potentially other similarly dynamic transitions. Currently, FMRP is believed to regulate hundreds of mRNAs in neuronal dendrites (O'Donnell and Warren, 2002), and although CAPR is reported to function similarly, their interaction in this tissue has yet to be addressed. Similarly, CAPR has been implicated in the activation of lymphocytes (Grill et al., 2004; Wang et al., 2005), a role yet to be investigated for dFMRP. Intriguingly CAPR, dFMRP and Rasputin co-immunoprecipitate from Drosophila ovary extracts (Costa et al., 2005), suggesting that the germ line might be another tissue that requires their combined action. In addition to roles in development, FMRP and CAPR are implicated in rapid translational responses to stress. When tissues are exposed to particular forms of stress they respond with the formation of stress granules, which are RNA-sorting and -processing foci that contain 40S ribosomal subunits along with FMRP, CAPR and G3BP (reviewed by Anderson and Kedersha, 2009; Buchan and Parker, 2009). Our data indicate that FMRP and CAPR cooperate to modulate the timing of the MBT in early embryos and lead us to postulate that the ability to respond to developmental signals rapidly through transcript-specific translational regulation might underlie a number of developmentally significant transitions.
Supplementary Material
Acknowledgments
We thank J. Sierra (Universidad Autónoma de Madrid), J. Großhans (ZMBH, Heidelberg), J. Wilhelm (U.C. San Diego), T. Kaufman (Indiana University), J. Raff (The Gurdon Institute, Cambridge, UK), P. Fisher (SUNY, Stonybrook) and C. Lehner (University of Zurich) for the generous gift of reagents, and D. Bilder (U.C. Berkeley) and the Patterson 2nd Floor Writing Workshop participants for helpful comments on the manuscript. This work was funded by NIH P41 RR011823 grant support to J.R.Y. and NIH R01 GM097562 to J.C.S. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.055046/-/DC1
References
- Aerbajinai W., Lee Y. T., Wojda U., Barr V. A., Miller J. L. (2004). Cloning and characterization of a gene expressed during terminal differentiation that encodes a novel inhibitor of growth. J. Biol. Chem. 279, 1916-1921 [DOI] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. (2009). RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10, 430-436 [DOI] [PubMed] [Google Scholar]
- Arbeitman M. N., Furlong E. E., Imam F., Johnson E., Null B. H., Baker B. S., Krasnow M. A., Scott M. P., Davis R. W., White K. P. (2002). Gene expression during the life cycle of Drosophila melanogaster. Science 297, 2270-2275 [DOI] [PubMed] [Google Scholar]
- Barbee S. A., Estes P. S., Cziko A. M., Hillebrand J., Luedeman R. A., Coller J. M., Johnson N., Howlett I. C., Geng C., Ueda R., et al. (2006). Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 52, 997-1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechara E. G., Didiot M. C., Melko M., Davidovic L., Bensaid M., Martin P., Castets M., Pognonec P., Khandjian E. W., Moine H., et al. (2009). A novel function for fragile X mental retardation protein in translational activation. PLoS Biol. 7, e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit B., He C. H., Zhang F., Votruba S. M., Tadros W., Westwood J. T., Smibert C. A., Lipshitz H. D., Theurkauf W. E. (2009). An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development 136, 923-932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bern M., Goldberg D., McDonald W. H., Yates J. R., 3rd (2004). Automatic quality assessment of peptide tandem mass spectra. Bioinformatics 20Suppl 1, i49-i54 [DOI] [PubMed] [Google Scholar]
- Buchan J. R., Parker R. (2009). Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932-941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cociorva D., L Tabb D., Yates J. R. (2007). Validation of tandem mass spectrometry database search results using DTASelect. Curr. Protoc. Bioinformatics Chapter 13, Unit 13.4 [DOI] [PubMed] [Google Scholar]
- Costa A., Wang Y., Dockendorff T. C., Erdjument-Bromage H., Tempst P., Schedl P., Jongens T. A. (2005). The Drosophila fragile X protein functions as a negative regulator in the orb autoregulatory pathway. Dev. Cell 8, 331-342 [DOI] [PubMed] [Google Scholar]
- Dahanukar A., Walker J. A., Wharton R. P. (1999). Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol. Cell 4, 209-218 [DOI] [PubMed] [Google Scholar]
- Dalby B., Glover D. M. (1993). Discrete sequence elements control posterior pole accumulation and translational repression of maternal cyclin B RNA in Drosophila. EMBO J. 12, 1219-1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J. C., Jensen K. B., Jin P., Brown V., Warren S. T., Darnell R. B. (2001). Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 107, 489-499 [DOI] [PubMed] [Google Scholar]
- Darnell J. C., Mostovetsky O., Darnell R. B. (2005a). FMRP RNA targets: identification and validation. Genes Brain Behav. 4, 341-349 [DOI] [PubMed] [Google Scholar]
- Darnell J. C., Fraser C. E., Mostovetsky O., Stefani G., Jones T. A., Eddy S. R., Darnell R. B. (2005b). Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 19, 903-918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Renzis S., Elemento O., Tavazoie S., Wieschaus E. F. (2007). Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 5, e117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B. A., Kiehle C. P., Schubiger G. (1986). Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell 44, 365-372 [DOI] [PubMed] [Google Scholar]
- Edgar B. A., Sprenger F., Duronio R. J., Leopold P., O'Farrell P. H. (1994). Distinct molecular mechanism regulate cell cycle timing at successive stages of Drosophila embryogenesis. Genes Dev. 8, 440-452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty P., Campbell S. D., Abu-Shumays R., Phalle B. S., Yu K. R., Uy G. L., Goldberg M. L., Sullivan W. (1997). The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity. Curr. Biol. 7, 418-426 [DOI] [PubMed] [Google Scholar]
- Gawlinski P., Nikolay R., Goursot C., Lawo S., Chaurasia B., Herz H. M., Kussler-Schneider Y., Ruppert T., Mayer M., Grosshans J. (2007). The Drosophila mitotic inhibitor Fruhstart specifically binds to the hydrophobic patch of cyclins. EMBO Rep. 8, 490-496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor G. B., Preston C. R., Johnson-Schlitz D. M., Nassif N. A., Phillis R. W., Benz W. K., Robertson H. M., Engels W. R. (1993). Type I repressors of P element mobility. Genetics 135, 81-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill B., Wilson G. M., Zhang K. X., Wang B., Doyonnas R., Quadroni M., Schrader J. W. (2004). Activation/division of lymphocytes results in increased levels of cytoplasmic activation/proliferation-associated protein-1: prototype of a new family of proteins. J. Immunol. 172, 2389-2400 [DOI] [PubMed] [Google Scholar]
- Grosshans J., Wieschaus E. (2000). A genetic link between morphogenesis and cell division during formation of the ventral furrow in Drosophila. Cell 101, 523-531 [DOI] [PubMed] [Google Scholar]
- Grosshans J., Muller H. A., Wieschaus E. (2003). Control of cleavage cycles in Drosophila embryos by fruhstart. Dev. Cell 5, 285-294 [DOI] [PubMed] [Google Scholar]
- Huang J., Raff J. W. (1999). The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J. 18, 2184-2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. Y., Morley G., Li D., Whitaker M. (2007). Cdk1 phosphorylation sites on Cdc27 are required for correct chromosomal localisation and APC/C function in syncytial Drosophila embryos. J. Cell Sci. 120, 1990-1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs H. W., Knoblich J. A., Lehner C. F. (1998). Drosophila Cyclin B3 is required for female fertility and is dispensable for mitosis like Cyclin B. Genes Dev. 12, 3741-3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link A. J., Eng J., Schieltz D. M., Carmack E., Mize G. J., Morris D. R., Garvik B. M., Yates J. R., 3rd (1999). Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17, 676-682 [DOI] [PubMed] [Google Scholar]
- Lu X., Li J. M., Elemento O., Tavazoie S., Wieschaus E. F. (2009). Coupling of zygotic transcription to mitotic control at the Drosophila mid-blastula transition. Development 136, 2101-2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Codina G., Glover D. M. (1992). Cyclins A and B associate with chromatin and the polar regions of spindles, respectively, and do not undergo complete degradation at anaphase in syncytial Drosophila embryos. J. Cell Biol. 116, 967-976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J., Curado S., Ephrussi A., Rorth P. (2000). Tribbles coordinates mitosis and morphogenesis in Drosophila by regulating string/CDC25 proteolysis. Cell 101, 511-522 [DOI] [PubMed] [Google Scholar]
- Matthews K. A., Rees D., Kaufman T. C. (1993). A functionally specialized alpha-tubulin is required for oocyte meiosis and cleavage mitoses in Drosophila. Development 117, 977-991 [DOI] [PubMed] [Google Scholar]
- Monzo K., Papoulas O., Cantin G. T., Wang Y., Yates J. R., 3rd, Sisson J. C. (2006). Fragile X mental retardation protein controls trailer hitch expression and cleavage furrow formation in Drosophila embryos. Proc. Natl. Acad. Sci. USA 103, 18160-18165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P. (1990). Universal control mechanism regulating onset of M-phase. Nature 344, 503-508 [DOI] [PubMed] [Google Scholar]
- O'Donnell W. T., Warren S. T. (2002). A decade of molecular studies of fragile X syndrome. Annu. Rev. Neurosci. 25, 315-338 [DOI] [PubMed] [Google Scholar]
- Papoulas O., Hays T. S., Sisson J. C. (2005). The golgin Lava lamp mediates dynein-based Golgi movements during Drosophila cellularization. Nat. Cell Biol. 7, 612-618 [DOI] [PubMed] [Google Scholar]
- Pazman C., Mayes C. A., Fanto M., Haynes S. R., Mlodzik M. (2000). Rasputin, the Drosophila homologue of the RasGAP SH3 binding protein, functions in ras- and Rho-mediated signaling. Development 127, 1715-1725 [DOI] [PubMed] [Google Scholar]
- Peng J., Elias J. E., Thoreen C. C., Licklider L. J., Gygi S. P. (2003). Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2, 43-50 [DOI] [PubMed] [Google Scholar]
- Pestova T. V., Lorsch J. R., Hellen C. U. T. (2007). The mechanism of translation initiation in eukaryotes. In Translational Control in Biology and Medicine (ed. Mathews M. B., Sonenberg N., Hershey J. W. B.), pp. 87-128 New York: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Pilot F., Philippe J. M., Lemmers C., Chauvin J. P., Lecuit T. (2006). Developmental control of nuclear morphogenesis and anchoring by charleston, identified in a functional genomic screen of Drosophila cellularisation. Development 133, 711-723 [DOI] [PubMed] [Google Scholar]
- Pisa V., Cozzolino M., Gargiulo S., Ottone C., Piccioni F., Monti M., Gigliotti S., Talamo F., Graziani F., Pucci P., et al. (2009). The molecular chaperone Hsp90 is a component of the cap-binding complex and interacts with the translational repressor Cup during Drosophila oogenesis. Gene 432, 67-74 [DOI] [PubMed] [Google Scholar]
- Raff J. W., Jeffers K., Huang J. Y. (2002). The roles of Fzy/Cdc20 and Fzr/Cdh1 in regulating the destruction of cyclin B in space and time. J. Cell Biol. 157, 1139-1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J. D., Klann E. (2009). Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev. 23, 1-11 [DOI] [PubMed] [Google Scholar]
- Royou A., McCusker D., Kellogg D. R., Sullivan W. (2008). Grapes(Chk1) prevents nuclear CDK1 activation by delaying cyclin B nuclear accumulation. J. Cell Biol. 183, 63-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadygov R. G., Eng J., Durr E., Saraf A., McDonald H., MacCoss M. J., Yates J. R., 3rd (2002). Code developments to improve the efficiency of automated MS/MS spectra interpretation. J. Proteome Res. 1, 211-215 [DOI] [PubMed] [Google Scholar]
- Shiina N., Shinkura K., Tokunaga M. (2005). A novel RNA-binding protein in neuronal RNA granules: regulatory machinery for local translation. J. Neurosci. 25, 4420-4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibon O. C., Stevenson V. A., Theurkauf W. E. (1997). DNA-replication checkpoint control at the Drosophila midblastula transition. Nature 388, 93-97 [DOI] [PubMed] [Google Scholar]
- Sibon O. C., Laurencon A., Hawley R., Theurkauf W. E. (1999). The Drosophila ATM homologue Mei-41 has an essential checkpoint function at the midblastula transition. Curr. Biol. 9, 302-312 [DOI] [PubMed] [Google Scholar]
- Sigrist S., Jacobs H., Stratmann R., Lehner C. F. (1995). Exit from mitosis is regulated by Drosophila fizzy and the sequential destruction of cyclins A, B and B3. EMBO J. 14, 4827-4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisson J. C., Field C., Ventura R., Royou A., Sullivan W. (2000). Lava lamp, a novel peripheral golgi protein, is required for Drosophila melanogaster cellularization. J. Cell Biol. 151, 905-918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon S., Xu Y., Wang B., David M. D., Schubert P., Kennedy D., Schrader J. W. (2007). Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol. Cell. Biol. 27, 2324-2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenberg P. T., Lustig K. D., McGarry T. J., King R. W., Kuang J., Kirschner M. W. (1997). Systematic identification of mitotic phosphoproteins. Curr. Biol. 7, 338-348 [DOI] [PubMed] [Google Scholar]
- Stuurman N., Maus N., Fisher P. A. (1995). Interphase phosphorylation of the Drosophila nuclear lamin: site-mapping using a monoclonal antibody. J. Cell Sci. 108, 3137-3144 [DOI] [PubMed] [Google Scholar]
- Tabb D. L., McDonald W. H., Yates J. R., 3rd (2002). DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W., Lipshitz H. D. (2005). Setting the stage for development: mRNA translation and stability during oocyte maturation and egg activation in Drosophila. Dev. Dyn. 232, 593-608 [DOI] [PubMed] [Google Scholar]
- Tadros W., Lipshitz H. D. (2009). The maternal-to-zygotic transition: a play in two acts. Development 136, 3033-3042 [DOI] [PubMed] [Google Scholar]
- Wang B., David M. D., Schrader J. W. (2005). Absence of caprin-1 results in defects in cellular proliferation. J. Immunol. 175, 4274-4282 [DOI] [PubMed] [Google Scholar]
- Wang D. O., Martin K. C., Zukin R. S. (2010). Spatially restricting gene expression by local translation at synapses. Trends Neurosci. 33, 173-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn M. P., Wolters D., Yates J. R., 3rd (2001). Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242-247 [DOI] [PubMed] [Google Scholar]
- Yu K. R., Saint R. B., Sullivan W. (2000). The Grapes checkpoint coordinates nuclear envelope breakdown and chromosome condensation. Nat. Cell Biol. 2, 609-615 [DOI] [PubMed] [Google Scholar]
- Zapata J. M., Martinez M. A., Sierra J. M. (1994). Purification and characterization of eukaryotic polypeptide chain initiation factor 4F from Drosophila melanogaster embryos. J. Biol. Chem. 269, 18047-18052 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.