Abstract
Bile salts are the major end-metabolites of cholesterol and are important in lipid digestion and shaping of the gut microflora. There have been limited studies of bile-salt variation in birds. The purpose of our study was to determine bile-salt variation among birds and relate this variation to current avian phylogenies and hypotheses on the evolution of bile salt pathways. We determined the biliary bile-salt composition of 405 phylogenetically diverse bird species, including 7 paleognath species. Bile salt profiles were generally stable within bird families. Complex bile-salt profiles were more common in omnivores and herbivores than in carnivores. The structural variation of bile salts in birds is extensive and comparable to that seen in surveys of bile salts in reptiles and mammals. Birds produce many of the bile salts found throughout nonavian vertebrates and some previously uncharacterized bile salts. One difference between birds and other vertebrates is extensive hydroxylation of carbon-16 of bile salts in bird species. Comparison of our data set of bird bile salts with that of other vertebrates, especially reptiles, allowed us to infer evolutionary changes in the bile salt synthetic pathway.
Keywords: bile acids, cholesterol, enzymes, mass spectrometry, molecular evolution, steroids
Cholesterol is essential for vertebrate species, which take advantage of this versatile molecule to regulate cell membrane fluidity, insulate nerve fibers, and serve as a precursor for the synthesis of steroid hormones and other endogenous compounds. The synthesis of bile salts from the cholesterol molecule and subsequent excretion into bile is the major pathway for eliminating cholesterol from the body in vertebrates (Moschetta et al. 2005). Bile salts also have multiple other functions. They are amphipathic molecules (positively charged on one half, negatively charged on the other) that are essential for the absorption of dietary lipids and fat-soluble vitamins. Bile salts help with digestion of dietary proteins by enhancing the action of proteases from the pancreas (Gass et al. 2007) and also act as hormones that influence carbohydrate and fat metabolism (Hylemon et al. 2009). Bile salts are produced in the liver, excreted in the hepatic bile ducts, stored in the gallbladder (for animals possessing this organ), and then travel via the common bile duct to be released into the duodenum. In addition, bile salts have antibacterial properties and likely influence microbial colonization of the intestinal tract (Hofmann and Eckmann 2006, Hofmann and Hagey 2008).
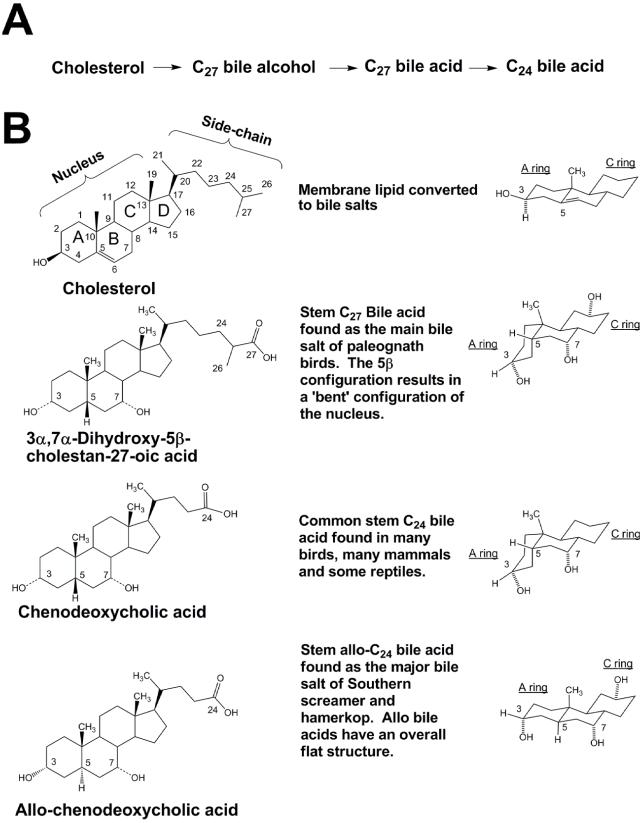
Bile salts are produced by every class of vertebrate animals and show substantial cross-species diversity (Haslewood 1967, Une and Hoshita 1994, Hofmann et al. 2010). Indeed, no other class of small molecules shows such striking variety across vertebrates. The diversity in bile salt chemical structures originates from differences in the two basic structural components of bile salt molecules. These are (1) the 19-carbon (C19) steroid nucleus and (2) a side-chain of variable length, most commonly containing five or eight carbon atoms (Fig. 1). The stereochemistry of the juncture between the A and B rings is variable and influences the overall shape of the bile salt, with 5β-bile salts having a “bent” orientation, and 5α(‘allo’)-bile salts having a flat (planar) structure (Hofmann and Hagey 2008). The middle two structures of Figure 1B are bent 5β-bile salts, and the bottom structure is a planar allo-bile salt. Unless otherwise specified, all bile salts referred to here are 5β in orientation.
Fig. 1.
Bile salt pathways and representative bile salt structure. (A) Simplified version of the bile salt synthetic pathway, showing the three major classes of compounds that can serve as primary bile salts (C27 bile alcohols, C27 bile acids, and C24 bile acids). (B) All bile salts are derived from cholesterol (topmost structure), illustrated with the carbon atoms numbered and the steroid rings labeled A, B, C, and D. Cholesterol has a relatively planar structure (see representation of A, B, and C rings on the right side). Paleognath birds utilize C27 bile acids such as 3α,7α-dihydroxy-5β-cholestan-27-oic acid that have an overall “bent” configuration of the steroid rings (see representation of A, B, and C rings on the right side). This C27 bile acid is considered a “stem” bile salt because it likely possesses the minimum number of hydroxyl groups sufficient to support typical bile salt function. Many neognath birds secrete the stem C24 bile acid chenodeoxycholic acid (CDCA). A small number of birds, including the Southern Screamer and Hamerkop, utilize C24 bile acids such as allo-CDCA that have an overall planar and extended structure of the steroid rings (see representation of A, B, and C rings on the right side).
The most common modifications to the bile salt nucleus are addition of hydroxyl (−OH) groups, a function carried out by specialized enzymes known as hydroxylases (Russell 2003, Norlin and Wikvall 2007). A less common modification is the addition of an oxo (= O) group. Hydroxyl groups can theoretically be added at any carbon atom in the bile salt molecule and are labeled α or β, depending on whether they are axial (perpendicular) or equatorial (parallel) to the plane of the steroid nucleus. The biochemical pathway for synthesizing bile acids (known only in humans and rodents) is long and complex, involving up to 16 enzymes and multiple organelles within the liver cell (Supplemental Figure 1; see Acknowledgments for link to supplemental materials; Russell 2003, Norlin and Wikvall 2007).
Some bile-salt hydroxylations are very common. For example, all primary bile salts have a hydroxyl group at C-3 (as they are formed from cholesterol) and at C-7. In addition to hydroxyl groups at C-3 and C-7, bile salts may have a third hydroxyl group on the steroid nucleus. The most common sites for the third hydroxyl group in birds are at C-12 or C-16, but other sites of hydroxylation (some quite rare, e.g., C-4 and C-5) can occur, as we will describe here.
The second source of structural variation for bile salts is in the side-chain. For bile acids, the side-chain ends in a carboxyl (−COOH) group. For bile alcohols, the side-chain ends in a hydroxyl group. Bile salts that retain all the carbon atoms of cholesterol have a total of 27 carbon atoms (C27), possessing an eight-carbon (C8) side-chain in addition to the C19 nucleus. Such bile salts occur in jawless fish, cartilaginous fish, lobe-finned fish, amphibians, some reptiles (e.g., crocodilians, turtles, varanid lizards), and a small number of mammals (e.g., elephants, manatees, rhinoceroses, and rock hyrax), as well as in some birds, as we will describe here. In many species (including most birds and mammals), the side-chain is shortened by three carbon atoms, resulting in C24 bile acids with a C5 side-chain. Like the steroid nucleus, the bile salt side-chain can be modified by the addition of hydroxyl groups. We use the term “bile salts” to refer to the broad class of cholesterol end-metabolites (bile acids and bile alcohols). Most bile salts can be assigned to one of three broad classes (Hofmann et al. 2010), (1) C27 bile alcohols, (2) C27 bile acids, or (3) C24 bile acids, and we use this classification here.
Based on both parsimony and biochemical considerations, we have proposed that the “ancestral” (plesiomorphic) character state of bile salts is most likely a C27 bile salt, probably a C27 bile alcohol (Reschly et al. 2008, Hagey et al. 2010a). Here, we will refer to C27 bile alcohols and acids as the ancestral or plesiomorphic state. C27 bile alcohols are the dominant bile salts of jawless, cartilaginous, and lobe-finned fish but are otherwise uncommon in vertebrates (Anderson and Haslewood 1964, Haslewood 1966, Reschly et al. 2008). Bile alcohols are almost always present in bile as sulfate “conjugates.” Cholic acid (CA; 3α,7α,12α-trihydroxy-5β-cholan-24-oic acid) and chenodeoxycholic acid (CDCA; 3α,7α-dihydroxy-5β-cholan-24-oic acid) are examples of C24 bile acids that we believe are examples of the evolutionarily “derived” (apomorphic) character state of bile salts that require a more complex biochemical synthetic pathway (Fig. 1B; Supplemental Fig. 1). Bile acids are typically found “conjugated” with taurine or glycine. In prior surveys of bile salts, conjugation (technically N-acylamidation) of bile acids with taurine was much more common than with glycine (Haslewood 1967, Moschetta et al. 2005). Conjugation makes bile salts more soluble in water and helps maintain high concentrations of bile salts in bile and intestinal content.
The physiological importance of bile salt structural variation has not been determined. A logical hypothesis is that the variation is related to diet, but extensive surveys of bile salts in fish, reptiles, and mammals have shown no clear correlation with diet. For example, common bile salts have been found in animals with a variety of diets, and complexity of bile salt profiles is not correlated with complexity of diet in fish, reptiles, or mammals (Hagey et al. 2010a, b).
An alternative hypothesis to explain bile salt variation is that the variation is related to interactions between bile salts and intestinal bacteria. When bile acids are excreted into the intestine, they undergo efficient absorption by the small intestine, with only a fraction (usually <10%) excreted in the feces as the main elimination route of cholesterol. Following absorption from the intestine, bile acids are returned to the liver, efficiently transported through the hepatocyte (liver cell) into bile, and once again delivered to the small intestine by a cycle known as the enterohepatic circulation.
From studies in mammals, it is known that anaerobic bacteria in the distal intestine may alter (“damage”) bile acids, principally by deconjugation (e.g., removal of the taurine or glycine) and removal of hydroxyl groups. Such bile salts are called “secondary” to distinguish them from “primary” bile salts that are formed in the liver from cholesterol. The most common bacterial modification of bile acids is the removal of the hydroxyl group at C-7 (7-dehydroxylation) to form 7-deoxy bile salts that can be toxic. The most toxic secondary bile acid in mammals is lithocholic acid (3α-hydroxy-5β-cholan-24-oic acid, LCA), which is formed by deconjugation and 7-dehydroxylation of the common primary bile acid CDCA. LCA has poor water-solubility, having only a single hydrophilic (“water-loving”) hydroxyl group, and can cause a variety of problems, including damage to the intestinal mucosa and formation of gallstones (Hofmann 2004, Hofmann and Hagey 2008). We have hypothesized that bile salt diversity in mammals may be an evolutionary mechanism to either prevent the formation of LCA or mitigate its toxicity (Hofmann 2004, Hagey et al. 2010b, Hofmann et al. 2010).
Prior surveys of bile salts in vertebrates have included very few avian species (Haslewood 1967, Une and Hoshita 1994, Moschetta et al. 2005), so the present study greatly extends knowledge of bile salt diversity in birds. A recent review of bile salt composition in vertebrates, which included a smaller number of bird species, was recently published (Hofmann et al. 2010). One of our main purposes here is to relate changes in bird bile salt structures to current models of avian phylogeny. We overlay bile salt variation on current avian phylogenies and also discuss the implications for the evolution of the biochemical pathway for bile salt synthesis. We also provide a study of bile salts in the feces of three birds to determine whether there is microbial alteration of bile salts in the intestine of birds.
Methods
Collection and preservation of bile samples
Bile samples were obtained from a number of sources (see Acknowledgments) over the course of two decades of analysis. Most samples were obtained during necropsy of animals that died in captivity at the San Diego Zoo. Bile samples were collected at necropsy by aspirating bile from the gallbladder, or from the common bile duct in species that do not have a gallbladder (pigeons and some psittacine species). Cages were checked for bird deaths daily, so the maximal interval between death and bile collection was 24 h. Bile was dispersed in at least 4 volumes of isopropanol saturated with nitrogen in brown bottles and stored at 4°C. Some of the samples analyzed were from an extensive collection of bile samples donated by the late G. A. D. Haslewood. These samples were dispersed in several volumes of ethanol, heated to denature proteins, and filtered. The filtrate was taken to dryness and the residual powder or gum was stored in a test tube and kept at 4°C. Fecal samples from birds were generously provided by the National Aviary (Pittsburgh, Pennsylvania) and were stored frozen at −20°C until analyzed. No birds were sacrificed specifically for the purposes of the present study.
Bile salts are stable molecules (Hofmann and Hagey 2008). If left wet and exposed to fungi and some bacteria, the amino acids (e.g., taurine and glycine) can be cleaved off and some of the side-chain hydroxyls oxidized to ketones. Because it is quite rare to have unconjugated bile acids in animal bile in any proportion and the first event for microbial modification is deconjugation, the presence of microbial effects in a sample is readily observable. In our experience with more than 10,000 samples, there has not been any signs of decomposition in bile salts preserved in isopropanol or dried for long-term storage, including samples from the Haslewood collection that are >60 years old. In a recent publication, we have recovered and analyzed intact conjugated bile acids from coprolites in dry desert caves from an extinct giant ground sloth (~11,000 years old) and ancient humans (~7,000 years old) (Hagey et al. 2010b), demonstrating the stability of bile salts in dry conditions.
Analysis of bile salts: Thin-layer chromatography
Whole bile was separated on silica gel G (E. Merck, Darmstadt, Germany) using two solvent systems: (1) isoamyl acetate:propionic acid:1-propanol:water 4:3:2:1 (v/v) (Hofmann 1962); and (2) a double development system for the resolution of conjugated bile acids, in which the plates were developed first in chloroform:methanol:water:acetone:propionic acid 10:2:1:4:1 (v/v) and then, after drying overnight, in 1-butanol:propionic acid:water 10:1:1 (v/v) (Oude Elferink et al. 1989). Bile acids were visualized by spray reagents for hydroxyl groups (phosphomolybdic acid, 10% w/v in ethanol), oxo groups (2,4-dinitrophenyl-hydrazine), sugars (naphtholresorcinol), or vicinal hydroxyl group (lead tetra-acetate).
Analysis of bile salts: High-performance liquid chromatography
Conjugated bile acids were analyzed by high-performance liquid chromatography (HPLC) using a modification of a previously reported technique (Rossi et al. 1987). An octadecylsilane column (RP C-18; Beckman Instruments, Fullerton, California) was used with isocratic elution at 0.75 mL min−1. The eluting solution was composed of a mixture of methanol and 0.01 M KH2PO4 (67.4% v/v), adjusted to an apparent pH of 5.35 with H3PO4. Conjugated bile acids were quantified by measuring the absorbance of their amide bond at 205 nm. Unconjugated bile acids and bile alcohol sulfates are not detected by this method. Bile acids were tentatively identified by matching their relative retention times with those of known standards.
Analysis of bile salts: Electrospray ionization mass spectrometry–mass spectrometry
Gallbladder contents were dissolved and diluted in methanol (Burdick and Jackson, Muskegon, Michigan) and analyzed using electrospray-ionization tandem mass spectrometry (ESI–MS–MS) on a Hewlett-Packard HP 1100 MSD operated in the negative mode. The HPLC column was removed and the injector output coupled directly to the ESI inlet. Samples (2 μL) were injected in methanol:water 90:10 (v/v) mobile phase running at a flow rate of 0.35 mL min−1. The fragmenter was set to 200 V and the capillary voltage was set to 5,000 V.
Analysis of bile salts: Gas chromatography–mass spectrometry
Glycine and taurine conjugates of bile acids were deconjugated chemically using 1.0 N NaOH at 130°C for 4 h. Bile alcohol sulfates were deconjugated enzymatically. Unconjugated bile acids were isolated by acidification and extraction into ethyl acetate. They were then analyzed by capillary gas chromatography–mass spectrometry (GC–MS) as methyl ester acetates (prepared using acetic anhydride in acetic acid with perchloric acid catalyst) or as methyl ester trimethylsilyl derivatives (prepared using Tri-Sil; Pierce Chemicals, Rockford, Illinois). Gas chromatography was performed using a Hewlett-Packard 5890 Gas Chromatograph-5970 MSD, controlled by HP/UX CHEM STATION software. The column was a 30-m 0.25-mm ID intermediate polarity SPB-35 of 35% phenyl methyl silicone (Supelco, Bellefonte, Pennsylvania) operated at 277°C (isothermal). A splitless injection was used with an injection temperature of 290°C. Helium was used as the carrier gas with a 7 psi column head pressure. Relative retention times and fragmentation spectra of peaks obtained by GC–MS were compared with those of known standards for identification.
Comparison of bile salt profiles with avian phylogenies
We overlaid bile salt variation on six different avian phylogenies: (1) a nuclear DNA-based phylogeny (Hackett et al. 2008), (2, 3) two summary hypotheses (Cracraft et al. 2004, Harshman 2007), (4) a mitochondrial DNA-based phylogeny (Brown et al. 2008), (5) a morphological character-based phylogeny (Livezey and Zusi 2007), and (6) the “tapestry” based on DNA–DNA hybridization (Sibley and Ahlquist 1990). Seven bile salt phenotypes were classified as binary plesiomorphic (0) or apomorphic (1) characters. Details are presented in the respective figure legends. Homoplasy and tree length were determined in PAUP*, version 4 (Sinauer Associates, Sunderland, Massachusetts).
Classification of bile salt profiles
We followed the convention of grouping bile salts into three broad categories (C27 bile alcohols, C27 bile acids, and C24 bile acids). Species were further sorted into one of six categories based on which one or two bile salt categories are present at ≥5% of the biliary bile salts (Reschly et al. 2008): category I, C27 bile alcohols only; category II, C27 bile alcohols + C27 bile acids; category III, C27 bile alcohols + C24 bile acids; category IV, C27 bile acids only; category V, C27 bile acids + C24 bile acids; and category VI, C24 bile acids only (see summary information for Supplemental Table 1). This classification, modified from that proposed by Moschetta et al. (2005), makes no assumptions about the physiological functions or effectiveness of bile salts. Only 2 of the 405 species analyzed, the Spotted Nutcracker (Nucifraga caryocatactes) and Western Crowned Pigeon (Goura cristata), had all three categories of bile salts present in their bile pools at >5% of total bile salts. These two species were classified on the basis of two bile salt categories that comprise the majority of total bile salts.
Classification of bird diet
To examine how diet might be related to bile salt profile, we classified the birds in our study into two diet classifications. The first classification divided birds into the broad categories of carnivores, omnivores, and herbivores. The second classification used six categories: (1) mostly vertebrates; (2) vertebrates + invertebrates; (3) mostly invertebrates; (4) vertebrates, invertebrates, and plants; (5) invertebrates + plants; and (6) mostly plants. Information about bird diet was obtained from online (Avian Web; Biodiversity Explorer; Cornell Lab of Ornithology; IUCN Red List of Threatened Species; see Acknowledgments) and print sources (Doornbos 1979, Fuentes et al. 2004, Tella et al. 2004, Mwangomo et al. 2007, Hall et al. 2009).
Results
Overview of bile salt variation across birds
We analyzed the bile salts in biliary bile from 405 bird species, including 7 paleognath species. The species analyzed included birds from 29 orders and 85 families, with at least 2 species examined in each of 22 orders and 58 families. The bile salt profiles of all animals analyzed are provided in Supplemental Table 1. Representative ESI–MS–MS spectra of analysis of bird bile salts are given in Supplemental Figure 2. Five of the six main bile salt profile categories previously found in vertebrates (see above) were found in birds, with category VI (>95% C24 bile acids) being the most common in the birds analyzed (Supplemental Fig. 2H–M). Only the category I bile salt profile (>95% C27 bile alcohols) was not seen in any bird analyzed.
Our analysis of seven paleognath species showed that all had very similar category II profiles (C27 bile alcohols and C27 bile acids), a plesiomorphic bile salt profile (Supplemental Fig. 2A–C). The two tinamou species analyzed, the Elegant Crested Tinamou (Eudromia elegans) and Red-Winged Tinamou (Rhynchotus rufescens), differed from the other five paleognath species, the Emu (Dromaius novaehollandiae), Greater Rhea (Rhea americana), Southern Cassowary (Casuarius casuarius), Southern Brown Kiwi (Apteryx australis), and Ostrich (Strutio camelus), in having 1β-hydroxyation of C27 bile acids, a bile salt so far unique to tinamous (Hagey et al. 2009), although 1β-hydroxylation of C24 bile acids was seen in some other bird species (Supplemental Table 1).
Overall, the bile salts of Neognathae are quite diverse, with extensive variation seen among orders of bird. Bile salts were generally stable within genera. For 61 genera, multiple species were analyzed; of these, only seven genera—Chlamydera (bowerbirds), Coracias (rollers), Crax (curassows), Dendrocygna (whistling ducks), Ducula (pigeons), Pteroglossus (aracaris), and Ptilinopus (fruit doves)—showed variation of bile salt structures. In these seven genera, this was mostly due to the presence of minor bile salts found in some species but not others within a genus. Bile salt profiles were also usually stable within bird families, with only 13 bird families (Accipitridae, Columbidae, Coraciidae, Corviidae, Cotungidae, Eurylaimidae, Muscicapidae, Passeridae, Pipridae, Psittacidae, Ptilonorhynchidae, Ramphastidae, and Turdidae) showing different bile salts among species. However, five of these families (Columbidae, Coraciidae, Cotingidae, Eurylaimidae, and Ramphastidae) had species with non-overlapping bile salt profiles. For example, some ramphastid species had C27 bile alcohols and acids and others had only C24 bile acids.
Bile salt variation in relation to avian phylogeny
We overlaid the bile salt variation on six avian phylogenies that represent either summary hypotheses (Cracraft et al. 2004, Harshman 2007) or studies of variation of nuclear DNA (Hackett et al. 2008), mitochondrial DNA (Brown et al. 2008), DNA–DNA hybridization (Sibley and Ahlquist 1990), or morphological characters (Livezey and Zusi 2007; Figs. 2 and 3 and Supplemental Figs. 3–6). Analysis of the trees in a parsimony framework showed high degrees of homoplasy for the bile salt character states (homoplasy indices ranged from 0.7500 to 0.8704 and retention indices ranged from 0.4524 to 0.6111; Supplemental Table 2).
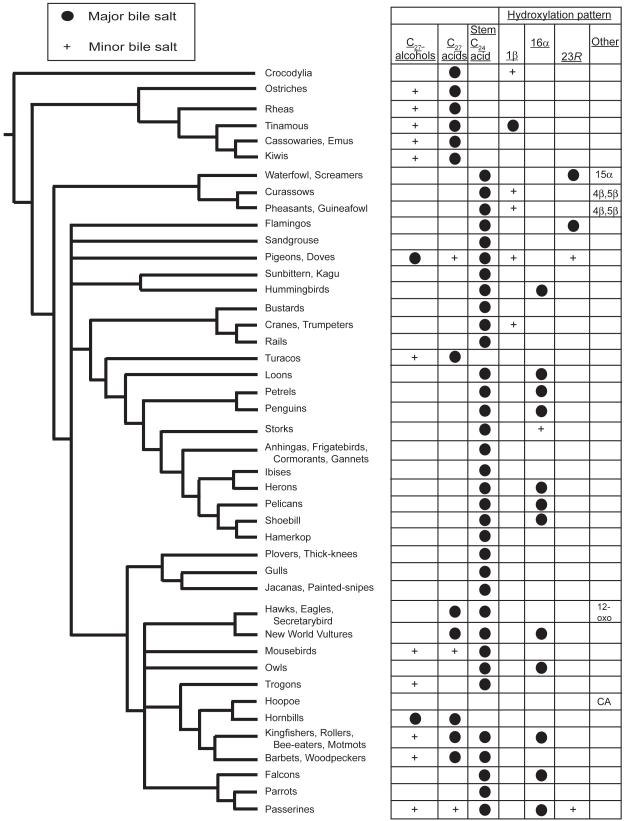
Fig. 2.
Bile salt variation in birds in relation to a nuclear DNA-based phylogeny. Bile salt variation is overlaid on a tree from a large-scale phylogenomic analysis of birds (Hackett et al. 2008). Major bile salts are those that constitute >50% of total biliary bile salts. Minor bile salts account for <50% but >10% of total bile salts. The categories of bile salts are C27 bile alcohols, C27 bile acids, stem C24 bile acid (chenodeoxycholic acid), 1β-hydroxylated bile acids, 15α-hydroxylated bile acids, 16α-hydroxylated bile acids, and 23R-hydroxylated bile acids.
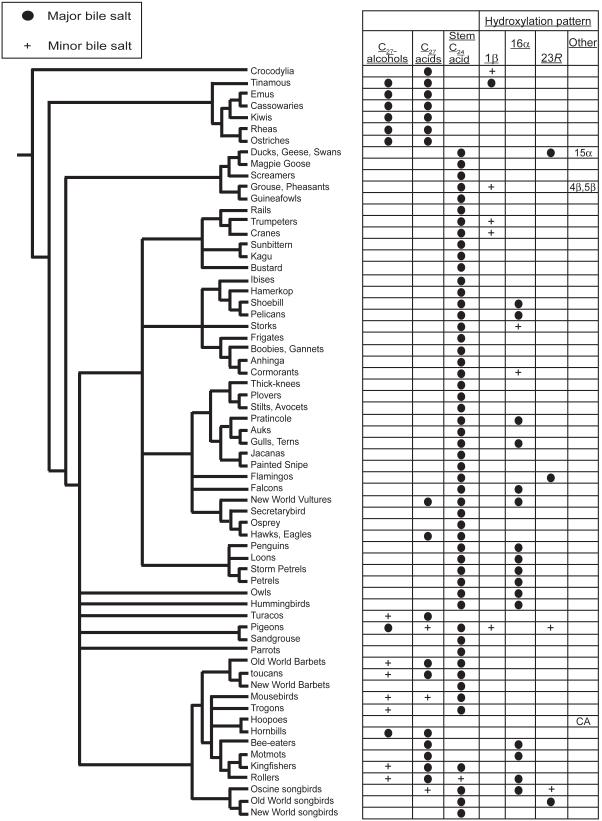
Fig. 3.
Bile salt variation in birds in relation to a summary hypothesis of avian phylogeny (Cracraft et al. 2004: fig. 27.10). Major bile salts are those that constitute >50% of total biliary bile salts. Minor bile salts account for <50% but >10% of total bile salts. The categories of bile salts are C27 bile alcohols, C27 bile acids, stem C24 bile acid (chenodeoxycholic acid), 1β-hydroxylated bile acids, 15α-hydroxylated bile acids, 16α-hydroxylated bile acids, and 23R-hydroxylated bile acids.
Table 1 summarizes bile salt variation within a subset of bird orders. Three of the orders analyzed (Apodiformes, Procellariiformes, and Sphenisciformes) showed no variation in bile salt structures among different species within the order. Within the broad group of Ciconiiformes–Pelecaniformes, a subset of birds that have been called the “core pelecanifoms”—Anhingidae (darters), Fregatidae (frigatebirds), Phalacrocoracidae (cormorants), and Sulidae (boobies)—all had a generic bile salt profile with CA and CDCA, whereas species analyzed in the remaining groups—Ardeidae (egrets and herons), Pelecanidae (pelicans), Balaenicipitridae (Shoebill [Balaeniceps rex])—all had 16α-hydroxylated bile acids. Within Coraciiformes, the species analyzed within Meropidae (bee-eaters), Momotidae (motmots), and Coraciidae (rollers) shared the phenotype of having C27 bile acids (plesiomorphic character state) with 16α-hydroxylation (an uncommon bile salt), although the rollers had additional complexity, with C24 bile acids or C27 bile alcohols or both. The three families of kingfishers analyzed (Alcedinidae, Cerylidae, and Halcyonidae) had substantial variation in bile salts, ranging from C24 bile acids (type VI, Alcedinidae) to a mixture of C24 bile acids and C27 bile alcohols (type III, Cerylidae) to a mixture of C24 and C27 bile acids (type V, Halcyonidae).
Table 1.
Variation in bile salts within selected groups of birds
| Birds | na | Main bile salt(s) b |
|---|---|---|
| Apodiiformes | ||
| Hummingbirds | 4 | 16α-OH-CDCA |
| Ciconiiformes/Pelecaniformes | ||
| Boobies (Sulidae) | 3 | CA, CDCA |
| Cormorants (Phalacrocoracidae) | 4 | CA, CDCA |
| Darters (Anhingidae) | 1 | CA, CDCA |
| Frigatebird (Fregatidae) | 1 | CA, CDCA |
| Egrets, herons (Ardeidae) | 12 | 16α-OH-CDCA |
| Pelicans (Pelecanidae) | 4 | 16α-OH-CDCA |
| Shoebill (Balaeniciptridae) | 1 | 16α-OH-CDCA |
| Coraciiformes | ||
| Bee-eaters (Meropidae) | 6 | 16α-OH-C27 bile acids |
| Kingfishers (3 families) | 12 | Varied (C24 and C27 bile acids) |
| Motmots (Momotidae) | 1 | 16α-OH-C27 bile acids |
| Rollers (Coraciidae) | 7 | 16α-OH-C27 bile acids |
| Falconiformes | ||
| Eagles and hawks (Accipitridae) | 10 | 12-Oxo bile acids |
| Osprey (Accipitridae) | 1 | CA, CDCA |
| Secretarybird (Sagittaridae) | 1 | CA, CDCA |
| New World vultures (Cathartidae) |
4 | 16α-OH-CDCA and C27 bile acids |
| 8 | 16α-OH-CDCA | |
| Falcons and caracaras (Falconidae) | ||
| Gruiformes | ||
| Cranes (Gruidae) | 3 | CA, CDCA, 1β-OH-CDCA |
| Rails (Rallidae) | 7 | CA, CDCA |
| Trumpeters (Psophiidae) | 1 | CDCA, 1β-OH-CDCA |
| Procellariiformes | ||
| Diving petrels (Pelecanoididae) | 1 | 16α-OH-CDCA |
| Petrels, prions (Procellariidae) | 2 | 16α-OH-CDCA |
| Storm petrels (Hydrobatidae) | 1 | 16α-OH-CDCA |
| Sphenisciformes | ||
| Penguins (Spheniscidae) | 3 | 16α-OH-CDCA |
| Strigidae (owls) | ||
| True owls (Strigidae) | 4 | CA, CDCA |
| Tytonidae (barn-owls) | 2 | 16α-OH-CDCA, CA, CDCA |
n = number of species analyzed.
Bile salt abbreviations: CA = cholic acid, CDCA = chenodeoxycholic acid, and OH = hydroxyl.
Bile salts also varied within Falconiformes. The eagles and hawks analyzed within Accipitridae all had 12-oxo bile acids, a type of bile acid not seen in any other species of bird analyzed. The Osprey (Accipitridae: Pandion haliaetus) and the Secretarybird (Sagittaridae: Sagittarius serpentarius) had mainly CA and CDCA and no detectable 12-oxo bile acids. By contrast, all species analyzed within Falconidae (falcons, American Kestrel [Falco sparverius], and caracaras) and Cathartidae (New World vultures) had 16α-hydroxylated bile acids, either 16α-hydroxy-CDCA (Falconidae: Black Vulture [Coragyps atratus]) or 16α-hydroxy-C27 bile acids (California Condor [Gymnogyps californianus], King Vulture [Sarcoramphus papa], and Andean Condor [Vultur gryphus]). Within Strigiformes (owls), the four species analyzed in Strigidae (true owls) had CA and CDCA whereas the two species analyzed in Tytonidae (barn-owls) had 16α-hydroxy-CDCA.
Unusual bile salts
Our study uncovered some unusual bile salt structures in a small number of birds. Bile acids hydroxylated at the 4β- and 5β-positions were found in four species: the Yellow-knobbed Curassow (Crax daubentoni), Chinese Monal (Lophophorus lhuysii), Himalayan Monal (Lophophorus impejanus), and Temminck’s Tragopan (Tragopan temminckii). These four birds are currently classified in either Cracidae (curassows) or Phasianidae, two families robustly grouped into the Galliformes in many phylogenies (e.g., Livezey and Zusi 2007, Hackett et al. 2008). A multigene phylogeny places the genera that include these four birds in a well-supported “erectile clade” with regard to fleshy traits on the head (Kimball and Braun 2008). Bile acids hydroxylated at the 15α position were found in 23 species within Anatidae (specifically the genera Anser, Branta, Bucephala, Cyanochem, Cygnus, Dendrocygna, Malacorhynchus, Neochen, Plectropterus, Sarkidiornis, and Tadorna) and in three species of lapwings (genus Vanellus) within Charadriidae. The Southern Screamer (Chauna torquata) and Hamerkop (Scopus umbretta) were notable in having the highest percentage of allo-bile acids of all the birds analyzed.
Bile salt variation in relation to diet
As described above, we classified birds by diet using two different classification systems and then sorted by bile salts. We additionally classified bile salt profiles as “complex” or “simple” (see footnotes to Table 2). Using this classification, 48.8% of carnivores, 64.6% of herbivores, and 61.0% of omnivores in our sample had complex bile salt profiles. Carnivores thus appear to generally have less complex bile salt profiles than other species (χ2 = 7.88, df = 2, P = 0.020). The most common bile salts in our sample (CA, CDCA, 16α-hydroxy-bile acids, 23R-hydroxy-bile acids, and C27 bile acids) were found in all dietary classes, although there were some differences in distribution. CA and 16α-hydroxy-bile acids were more common in carnivores, whereas 23R-hydroxy-bile acids were more common in herbivores and omnivores. It should be pointed out that the majority of 23R-hydroxy-bile acids in our sample were found in species in Anatidae, a family for which we analyzed a large number of species.
Table 2.
Summary of bile salt variation in various dietary classes
| Diet class a | nb | CDCA c |
CA c | 16α- OH C24 bile acid c |
23R- OH C24 bile acid c |
C27 bile acid c |
Complex profile d |
Simple profile d |
|---|---|---|---|---|---|---|---|---|
| (%) | (%) | (%) | (%) | (%) | (%) | (%) | ||
| 1. Vertebrates | 31 | 96.8 | 77.4 | 29.0 | 0.0 | 9.7 | 48.4 | 3.2 |
| 2. Vertebrates + invertebrates |
83 | 88.0 | 60.2 | 44.6 | 3.6 | 16.9 | 45.8 | 1.2 |
| 3. Invertebrates | 46 | 78.3 | 60.9 | 26.1 | 15.2 | 21.7 | 54.3 | 15.2 |
| 4. Omnivores | 58 | 58.6 | 34.5 | 6.9 | 13.8 | 46.6 | 50.0 | 8.6 |
| 5. Invertebrates + plants |
88 | 86.4 | 43.2 | 21.6 | 26.1 | 17.0 | 68.2 | 10.2 |
| 6. Herbivores | 97 | 87.6 | 43.3 | 17.2 | 22.9 | 10.3 | 64.9 | 9.3 |
| All carnivores | 160 | 86.9 | 63.8 | 36.3 | 6.3 | 16.9 | 48.8 | 5.6 |
| All omnivores | 146 | 75.3 | 39.7 | 15.8 | 21.2 | 28.8 | 61.0 | 9.6 |
Diet classes are defined as follows: (1) diet >90% vertebrate animals; (2) diet >90% vertebrate + invertebrate animals, each >10%; (3) diet >90% invertebrate animals; (4) diet includes vertebrate animals, invertebrate animals, and plants, each >10%; (5) diet >90% plants + invertebrate animals, each >10%; and (6) diet >90% plants. All carnivores = classes 1, 2, and 3. All omnivores = classes 4 and 5.
n = number of species in the category.
Percentage of animals that have this bile salt in their top three most-abundant biliary bile salts. Abbreviations: CA = cholic acid, CDCA = chenodeoxycholic acid, and −OH = hydroxyl.
Complex profile is defined as (1) bile salt class II, III, or V; and/or (2) presence of 3 or more bile salts in biliary bile, each at ≥10%. Simple profile is defined as the presence of a single bile salt that accounts for >95% of total biliary bile salts.
Analysis of fecal bile salts in birds
We examined the fecal bile salts of three birds—the Snowy Owl (Bubo scandiaca), Taveta Golden Weaver (Ploceus castaneiceps), and Red-legged Seriema (Cariama cristata)—that secrete taurine-conjugated CDCA as a primary bile acid. In all three birds, the potentially toxic secondary bile acid LCA was detected as a component of fecal bile salts (Supplemental Fig. 2N–Q).
Discussion
We have presented an extensive survey of bile salts in birds, and we use these data for comparative purposes within Aves and other vertebrates. Elsewhere, we report on the bile salts of 748 nonavian species, including 175 fish (38 orders), 29 amphibians (9 families), 218 reptiles (37 families), and 326 mammals (63 families) (Hagey et al. 2010a, b; Hofmann et al. 2010). One of the most striking observations to emerge is the variety of bile salts in neognath birds. Neognath birds are remarkable in comparison to other vertebrates in the extent of variation in bile salt profiles within bird orders and families, bearing in mind that the classification of some neognath orders and families remains controversial and subject to change in the future. The structural diversity of bird bile salts is at least as extensive as that found in our survey of 218 reptile species (Hagey et al. 2010b, Hofmann et al. 2010).
Our analysis showed that simpler bile salt profiles in birds were more common in carnivores than in herbivores and omnivores. A similar trend was noted in a survey of mammalian bile salts (Hagey et al. 2010b). However, there was no correlation between bile salt diversity and diet in surveys of bile salts in fish or reptiles (Hagey et al. 2010a, b). However, there are groups of birds that have very similar bile salt profiles but different diets. An example is Anatidae, which has carnivores (e.g., Common Merganser [Mergus merganser]), herbivores, and omnivores that have essentially identical bile salt profiles. Overall, common bile acids such as CA and CDCA are found in species with a wide variety of diets. With uncommon bile acids, there are insufficient data to make generalizations. For example, the unusual 4β- and 5β-hydroxy bile acids of the Chinese Monal and Himalayan Monal could theoretically be advantageous to the diets of these particular species, pending more extensive surveys of birds.
One frequently encountered bile salt in birds is 16α-hydroxy-CDCA, a bile acid that differs from CA (common bile acid) in the arrangement of one hydroxyl group (3α,7α,16α for 16α-hydroxy-CDCA vs. 3α,7α,12α for CA). 16α-hydroxy-CDCA has been chemically synthesized and the trivial name avicholic acid proposed (Iida et al. 2002, Mukhopadhyay and Maitra 2004). Bile acids with 16α-hydroxylation are otherwise rare in vertebrates, being found so far only in the bile of snakes within three families (Boidae, Cylindrodrophiidae, and Pythonidae; Hofmann et al. 2010) and in hagfish (Haslewood 1966, Hagey et al. 2010a).
Bile acids hydroxylated at C-23 are also common in birds but relatively uncommon in other vertebrates, although they are also found in pinniped mammals (seals, sea lions, and walruses) and some snakes within the family Viperidae (Hofmann et al. 2010). One possible physiological advantage of bile acids with a hydroxyl group at C-23 is that the taurine conjugates of these acids are more resistant to deconjugation by intestinal bacteria than the conjugate of the corresponding bile acid without a hydroxyl group at C-23 (Merrill et al. 1996). The character states of 16α- and 23-hydroxylation in unrelated groups of vertebrates likely represent examples of homoplasy.
Thirteen bird families had species with different bile salt profiles. By contrast, in our survey of nonavian species, only three reptile families (Chelidae and Testudinae within Testudines [turtles and tortoises] and Scincidae [skinks]), one mammalian family (Cebidae [New World monkeys]), and no fish families showed any variation among species in bile salt profiles (Hagey et al. 2010a, b; Hofmann et al. 2010). Five bird families even had species with non-overlapping bile salt profiles. Such variation in bile salts within families has not been found in any other vertebrate orders and may result from the rapid and extensive diversification of neognath birds during evolution (Livezey and Zusi 2007, Hackett et al. 2008, van Tuinen 2009). The last common ancestor of extant neognath birds has been estimated to have lived ~100 mya (van Tuinen 2009). In this interval, there have been substantial modifications to the bile salt synthetic pathway in birds, in terms of hydroxylation patterns and bile salt category. In contrast to neognath birds, we have found examples of vertebrates for whom bile salt profiles have likely stayed essentially unchanged for at least 100 million years. Examples include Crocodylia (with three extant families, Alligatoridae, Crocodylidae, and Gavialidae; Brochu 2009) and hagfishes (two extant subfamilies, Myxininae and Eptatretinae; Kuraku and Kuratani 2006).
Birds demonstrate a number of bile salt modifications that are unusual in relation to other vertebrates. These include allo-bile acids (“Gruiformes”), 1β-hydroxy C27 bile acids (tinamous), 15α-hydroxy bile acids (Anatidae, Charadriidae), 4β- and 5β-hydroxy bile acids (Chinese Monal, Himalayan Monal, monal pheasants, and Yellow-knobbed Curassow), and 12-oxo bile acids (Accipitridae). Allo-bile acids are generally uncommon in vertebrates, being very rare in mammals, fish, and amphibians (Hofmann et al. 2010). Other than birds, the only other vertebrates that commonly use allo-bile acids are iguanid lizards (Hofmann et al. 2010). 1β-hydroxylated C24 bile acids are uncommon in vertebrates outside birds, being found so far only in marsupials (Hofmann et al. 2010). 15α-hydroxy bile acids, common in Anatidae, have been described outside birds only in the Common Wombat (Vombatus ursinus), a marsupial (Kakiyama et al. 2007). 4β- and 5β-hydroxylated bile acids are so far unique to birds. We also found in many birds a complex mixture of bile alcohols at low concentration, even in species whose biliary bile salts are >95% C24 bile acids; the minor fraction of bile alcohols may even include unusual C25 and C26 compounds. The physiological importance (if any) of these trace bile salts is as yet unknown.
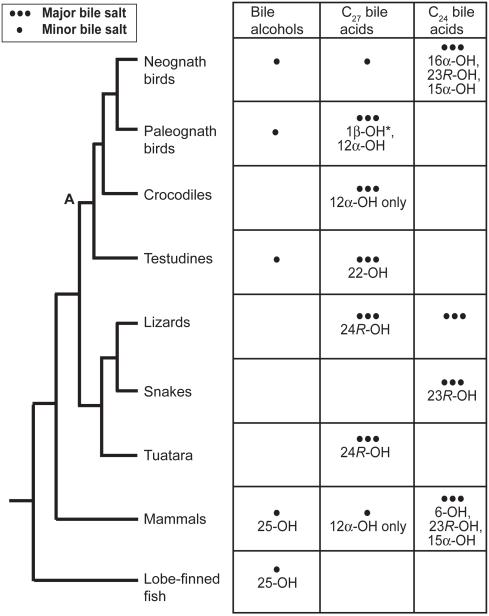
Figure 4 shows a tree of reptiles, birds, and mammals, with lobe-finned fish used as an outgroup. This tree fits the current molecular data and places Testudines as a sister group to Crocodylia and Aves (Meyer and Zardoya 2003). The observed bile salt phenotypes of reptiles and birds are consistent with this phylogeny. The bile salt profiles of crocodilians and paleognath birds are very similar, with both groups predominantly using C27 bile acids with either a stem hydroxylation pattern (3α,7α) or with an additional 12α-hydroxyl group (same hydroxylation pattern as in the most common bile acid, CA). By contrast, Testudines predominantly use C27 bile acids with 22S-hydroxylation, a modification to C27 bile acids that may represent an autapomorphy, having not been found in any other vertebrate group analyzed to date (Kakiyama et al. 2007). From these relationships, one can speculate about the bile salt profiles of the most recent common ancestor to Testudines–Crocodylia–Aves. This ancestor can be inferred to have had a bile salt profile consisting mainly of C27 bile alcohols and dihydroxy- and trihydroxy-C27 bile acids (3α,7α,12α-trihydroxy-5β-cholestan-27-oic acid). Crocodylia retain the trihydroxy-C27 bile acid as their main primary bile salt but now produce only trace amounts of C27 bile alcohols. Testudines differ by having a (so far) unique modification to C27 bile acids, namely 22-hydroxylation (Hofmann et al. 2010).
Fig. 4.
Overall bile salt variation in birds, reptiles, and mammals. The phylogeny depicted has Testudines as sister group to Aves–Crocodylia. Lizards are paraphyletic but are indicated here as a single group for comparative purposes. Amphibians are not included in the figure, but analyses of biliary bile salts from Anura and Caudata have revealed extensive structural diversity of bile salts, including C24 bile acids, C27 bile acids, and C27 bile alcohols (some species show all three bile salt classes), in addition to unusual C26, C28, and C29 bile salts.
There is still little understanding of what may have driven variation in bile salt diversity throughout vertebrate evolution (Hofmann et al. 2010). Diet may be one factor, given our observation that complex bile salt profiles are more common in herbivores and omnivores than in carnivores. However, typical bile acids such as CDCA and CA are found in birds, reptiles, and mammals with a wide variety of diets, which suggests that additional factors influence bile salt structural diversity. It is also unclear what benefit may be conferred to those birds (e.g., Southern Screamer and Hamerkop) that have a high percentage of allo-bile salts. The complicated structural variation of bile salts may also relate to other functions of bile salts that are not well understood and in need of further research, including communication (e.g., as pheromones or territory markers), influencing of the microbial environment in the gut, and regulation of hepatobiliary development and regeneration (Li et al. 2002, Hofmann and Eckmann 2006, Hofmann and Hagey 2008).
We have speculated that one benefit of bile salt diversity is to avoid or mitigate damage to bile salts by the action of intestinal bacteria (Hofmann 2004, Hofmann et al. 2010). Our analyses of the fecal bile salts in three bird species showed the presence of the potentially toxic secondary bile acid LCA, a compound typically formed by enzymatic alterations to primary bile acids mediated by intestinal bacteria. If birds generally have intestinal flora capable of damaging bile salts, then bile salt structural diversity could in part be an evolutionary adaptation to minimize bacterial alterations to primary bile acids, as has been speculated with regard to mammals (Hofmann 2004, Hofmann et al. 2010). Our initial, limited study of bird fecal bile salts will obviously need verification by a wider survey of fecal bile salts in birds and other nonmammalian species.
Finally, the extensive variation in bile salt structures across birds predicts that protein receptors that bind bile salts and bile salt synthetic enzymes will also show variation in structure and specificity for ligands (receptors) or substrates (enzymes), as we have shown for three nuclear hormone receptors involved in bile salt homeostasis and detoxification (Krasowski et al. 2005a, b; Reschly et al. 2007, 2008). Analysis of these receptors in birds with different bile salt profiles may be particularly interesting to study as model systems for investigating protein evolution. Analysis of species with plesiomorphic bile salt profiles (e.g., paleognath birds) may also be valuable in trying to understand key evolutionary changes in bile salt synthetic enzymes, especially as genome sequence information becomes available for more birds and other vertebrate species.
Supplementary Material
Acknowledgments
M.D.K. is supported by K08-GM074238 from the National Institutes of Health (NIH). A.F.H. and L.R.H. were supported by NIH grant DDK 64891 (to A.F.H.). We thank the Pathology Laboratory of the San Diego Zoo for samples. Samples were also provided by G. A. D. Haslewood (now deceased), professor of biochemistry at Guy’s Hospital (London); V. Lance (Department of Biology, San Diego State University); J. Barrett and M. Becker (University of Memphis); H. Johnston (Veterinarian of the County of San Diego); R. Norman (University of Melbourne); and S. Sarro (National Aviary, Pittsburgh, Pennsylvania). Avian Web is available at www.avianweb.com, Biodiversity Explorer at www.biodiversityexplorer.org, and the IUCN Red List of Threatened Species at www.iucnredlist.org.
Literature Cited
- Anderson IG, Haslewood GAD. Comparative studies of ‘bile salts’. 20. Bile salts of the coelacanth, Latimeria chalumnae Smith. Biochemical Journal. 1964;93:34–39. doi: 10.1042/bj0930034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochu CA. Crocodylians (Crocodylia) In: Hedges SB, Kumar S, editors. The Timetree of Life. Oxford University Press; Oxford, United Kingdom: 2009. pp. 402–406. [Google Scholar]
- Brown JW, Rest JS, García-Moreno J, Sorenson MD, Mindell DP. Strong mitochondrial DNA support for a Cretaceous origin of modern avian lineages. BMC Biology. 2008;6:6. doi: 10.1186/1741-7007-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cracraft J, Barker FK, Braun M, Harshman J, Dyke GJ, Feinstein J, Stanley S, Cibois A, Schikler P, Beresford P. Phylogenetic relationships among modern birds (Neornithes): Toward an avian tree of life. In: Cracraft J, Donoghue MJ, editors. Assembling the Tree of Life. Oxford University Press; Oxford, United Kingdom: 2004. pp. 468–489. others. [Google Scholar]
- Doornbos G. Winter food habits of smew (Mergus albellus L.) on Lake Yssel, The Netherlands: Species and size selection in relation to fish stocks. Ardea. 1979;67:42–48. [Google Scholar]
- Fuentes C, Sánchez MI, Selva N, Green AJ. The diet of the Marbled Teal Marmaronetta angustirostris in southern Alicante, eastern Spain. Revue d’Ecologie, Terre et Vie. 2004;59:475–490. [Google Scholar]
- Gass J, Vora H, Hofmann AF, Gray GM, Khosla C. Enhancement of dietary protein digestion by conjugated bile acids. Gastroenterology. 2007;133:16–23. doi: 10.1053/j.gastro.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Hackett SJ, Kimball RT, Reddy S, Bowie RCK, Braun EL, Braun MJ, Chojnowski JL, Cox WA, Han K-L, Harshman J. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. others. [DOI] [PubMed] [Google Scholar]
- Hagey LR, Kakiyama G, Muto A, Iida T, Mushiake K, Goto T, Mano N, Goto J, Oliveira CA, Hofmann AF. A new, major C27 biliary bile acid in the Red-winged Tinamou (Rhynchotus rufescens):(25R)-1β,3α,7α-trihydroxy-5β-cholestan-27-oic acid. Journal of Lipid Research. 2009;50:651–657. doi: 10.1194/jlr.M800521-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagey LR, Møller PR, Hofmann AF, Krasowski MD. Diversity of bile salts in fish and amphibians: Evolution of a complex biochemical pathway. Physiological and Biochemical Zoology. 2010a;83:308–321. doi: 10.1086/649966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagey LR, Vidal N, Hofmann AF, Krasowski MD. Evolutionary diversity of bile salts in reptiles and mammals, including analysis of ancient human and extinct giant ground sloth coprolites. BMC Evolutionary Biology. 2010b;10:133. doi: 10.1186/1471-2148-10-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BD, Baron LA, Somers CM. Mercury concentrations in surface water and harvested waterfowl from the prairie pothole region of Saskatchewan. Environmental Science and Technology. 2009;43:8759–8766. doi: 10.1021/es9024589. [DOI] [PubMed] [Google Scholar]
- Harshman J. Classification and phylogeny of birds. In: Jamieson BGM, editor. Reproductive Biology and Phylogeny of Birds. Part A: Phylogeny, Morphology, Hormones, Fertilization. Science Publishers; Enfield, New Hampshire: 2007. pp. 1–35. [Google Scholar]
- Haslewood GAD. Comparative studies of bile salts. Myxinol disulphate, the principal bile salt of hagfish (Myxinidae) Biochemical Journal. 1966;100:233–237. doi: 10.1042/bj1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslewood GAD. Bile salt evolution. Journal of Lipid Research. 1967;8:535–550. [PubMed] [Google Scholar]
- Hofmann AF. Thin layer absorption chromatography of free and conjugated bile acids on silicic acid. Journal of Lipid Research. 1962;3:127–128. [Google Scholar]
- Hofmann AF. Detoxification of lithocholic acid, a toxic bile acid: Relevance to drug hepatotoxicity. Drug Metabolism Reviews. 2004;36:703–722. doi: 10.1081/dmr-200033475. [DOI] [PubMed] [Google Scholar]
- Hofmann AF, Eckmann L. How bile acids confer gut mucosal protection against bacteria. Proceedings of the National Academy of Sciences USA. 2006;103:4333–4334. doi: 10.1073/pnas.0600780103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann AF, Hagey LR. Bile acids: Chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cellular and Molecular Life Sciences. 2008;65:2461–2483. doi: 10.1007/s00018-008-7568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann AF, Hagey LR, Krasowski MD. Bile salts of vertebrates: Structural variation and possible evolutionary significance. Journal of Lipid Research. 2010;51:226–246. doi: 10.1194/jlr.R000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hylemon PB, Zhou H, Pandak WM, Ren S, Gil G, Dent P. Bile acids as regulatory molecules. Journal of Lipid Research. 2009;50:1509–1520. doi: 10.1194/jlr.R900007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida T, Hikosaka M, Kakiyama G, Shiraishi K, Schteingart CD, Hagey LR, Ton-Nu HT, Hofmann AF, Mano N, Goto J, Nambara T. Potential bile acid metabolites. 25. Synthesis and chemical properties of stereoisomeric 3alpha,7alpha,16- and 3alpha,7alpha,15-trihydroxy-5beta-cholan-24-oic acids. Chemical & Pharmaceutical Bulletin (Tokyo) 2002;50:1327–1334. doi: 10.1248/cpb.50.1327. [DOI] [PubMed] [Google Scholar]
- Kakiyama G, Tamegai H, Iida T, Mitamura K, Ikegawa S, Goto T, Mano N, Goto J, Holz P, Hagey LR, Hofmann AF. Isolation and chemical synthesis of a major, novel biliary bile acid in the common wombat (Vombatus ursinus): 15α-hydroxylithocholic acid. Journal of Lipid Research. 2007;48:2682–2692. doi: 10.1194/jlr.M700340-JLR200. [DOI] [PubMed] [Google Scholar]
- Kimball RT, Braun EL. A multigene phylogeny of Galliformes supports a single origin of erectile ability in non-feathered facial traits. Journal of Avian Biology. 2008;39:438–445. [Google Scholar]
- Krasowski MD, Yasuda K, Hagey LR, Schuetz EG. Evolution of the pregnane X receptor: Adaptation to cross-species differences in biliary bile salts. Molecular Endocrinology. 2005a;19:1720–1739. doi: 10.1210/me.2004-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasowski MD, Yasuda K, Hagey LR, Schuetz EG. Evolutionary selection across the nuclear hormone receptor superfamily with a focus on the NR1I subfamily (vitamin D, pregnane X, and constitutive androstane receptors) Nuclear Receptor. 2005b;3:2. doi: 10.1186/1478-1336-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S, Kuratani S. Time scale for cyclostome evolution inferred with a phylogenetic diagnosis of hagfish and lamprey cDNA sequences. Zoological Science. 2006;23:1053–1064. doi: 10.2108/zsj.23.1053. [DOI] [PubMed] [Google Scholar]
- Li W, Scott AP, Siefkes MJ, Yan H, Liu Q, Yun S-S, Gage DA. Bile acid secreted by male sea lamprey that acts as a sex pheromone. Science. 2002;296:138–141. doi: 10.1126/science.1067797. [DOI] [PubMed] [Google Scholar]
- Livezey BC, Zusi RL. Higher-order phylogeny of modern birds (Theropoda, Aves: Neornithes) based on comparative anatomy. II. Analysis and discussion. Zoological Journal of the Linnean Society. 2007;149:1–95. doi: 10.1111/j.1096-3642.2006.00293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill JR, Schteingart CD, Hagey LR, Peng Y, Ton-Nu HT, Frick E, Jirsa M, Hofmann AF. Hepatic biotransformation in rodents and physicochemical properties of 23(R)-hydroxychenodeoxycholic acid, a natural a-hydroxy bile acid. Journal of Lipid Research. 1996;37:98–112. [PubMed] [Google Scholar]
- Meyer A, Zardoya R. Recent advances in the (molecular) phylogeny of vertebrates. Annual Review of Ecology, Evolution, and Systematics. 2003;34:311–308. [Google Scholar]
- Moschetta A, Xu F, Hagey LR, van Berge Henegouwen GP, van Erpecum KJ, Brouwers JF, Cohen JC, Bierman M, Hobbs HH, Steinbach JH, Hofmann AF. A phylogenetic survey of biliary lipids in vertebrates. Journal of Lipid Research. 2005;46:2221–2232. doi: 10.1194/jlr.M500178-JLR200. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Maitra U. Facile synthesis, aggregation behavior, and cholesterol solubilization ability of avicholic acid. Organic Letters. 2004;6:31–34. doi: 10.1021/ol036073f. [DOI] [PubMed] [Google Scholar]
- Mwangomo EA, Hardesty LH, Sinclair ARE, Mduma SAR, Metzger KL. Habitat selection, diet and interspecific associations of the Rufous-tailed Weaver and Fischer’s Lovebird. African Journal of Ecology. 2007;46:267–275. [Google Scholar]
- Norlin M, Wikvall K. Enzymes in the conversion of cholesterol into bile acids. Current Molecular Medicine. 2007;7:199–218. doi: 10.2174/156652407780059168. [DOI] [PubMed] [Google Scholar]
- Oude Elferink RPJ, de Haan J, Lambert KJ, Hagey LR, Hofmann AF, Jansen PLM. Selective hepatobiliary transport of nordeoxycholate side chain conjugates in mutant rats with a canalicular transport defect. Hepatology. 1989;9:861–865. doi: 10.1002/hep.1840090612. [DOI] [PubMed] [Google Scholar]
- Reschly EJ, Ai N, Ekins S, Welsh WJ, Hagey LR, Hofmann AF, Krasowski MD. Evolution of the bile salt nuclear receptor FXR in vertebrates. Journal of Lipid Research. 2008;49:1577–1587. doi: 10.1194/jlr.M800138-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reschly EJ, Bainy ACD, Mattos JJ, Hagey LR, Bahary N, Mada SR, Ou J, Venkataramanan R, Krasowski MD. Functional evolution of the vitamin D and pregnane X receptors. BMC Evolutionary Biology. 2007;7:222. doi: 10.1186/1471-2148-7-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi SS, Converse JL, Hofmann AF. High pressure liquid chromatography analysis of conjugated bile acids in human bile: Simultaneous resolution of sulfated and unsulfated lithocholyl amidates and the common conjugated bile acids. Journal of Lipid Research. 1987;28:589–595. [PubMed] [Google Scholar]
- Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annual Review of Biochemistry. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- Sibley CG, Ahlquist JE. Phylogeny and Classification of Birds: A Study in Molecular Evolution. Yale University Press; New Haven, Connecticut: 1990. [Google Scholar]
- Tella JL, Figuerola J, Negro JJ, Blanco G, Rodríguez-Estrella R, Forero MG, Blázquez MC, Green AJ, Hiraldo F. Ecological, morphological and phylogenetic correlates of interspecific variation in plasma carotenoid concentration in birds. Journal of Evolutionary Biology. 2004;17:156–164. doi: 10.1046/j.1420-9101.2003.00634.x. [DOI] [PubMed] [Google Scholar]
- Une M, Hoshita T. Natural occurrence and chemical synthesis of bile alcohols, higher bile acids, and short side chain bile acids. Hiroshima Journal of Medical Sciences. 1994;43:37–67. [PubMed] [Google Scholar]
- van Tuinen M. Advanced birds (Neoaves) In: Hedges SB, Kumar S, editors. The Timetree of Life. Oxford University Press; Oxford, United Kingdom: 2009. pp. 419–422. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.