Abstract
Histone deacetylase (HDAC) inhibitors induce chromatin destabilization. We sought to determine whether HDAC inhibition may amplify alkylator-induced mitotic cell death in multiple myeloma (MM) cells. The combination of SNDX-275, a class I HDAC inhibitor, with melphalan, showed a powerful synergism on growth inhibition with the combination index ranged from 0.27 to 0.75 in MM1.S and RPMI8226 cells. Their combinations as compared with either agent alone promoted much more caspase-dependent apoptosis. Flow cytometry analysis showed that SNDX-275 had minimal effects on cell cycle progression of MM1.S cells, but clearly increased the percentage of S phase in RPMI8226 cells associated with an upregulation in p21waf1 and a reduction in cyclin D1 and E2F1. Melphalan alone significantly arrested both MM1.S and RPMI8226 cells at S phase and enhanced expression of p53 and p21waf1. Furthermore, studies on DNA damage response revealed that phospho-histone H2A.X (γH2A.X), a hall marker of DNA double strand break, along with phosphorylated CHK1 (P-CHK1) and CHK2 (P-CHK2) was dramatically induced by SNDX-275 or melphalan. The increase in γH2A.X and P-CHK1 was considerably higher on combination than either agent alone. These molecular changes correlated well with the significant increase in mitotic catastrophe. Our data indicate that SNDX-275 synergistically enhances melphalan-induced apoptosis in MM cells via intensification of DNA damage, suggesting that SNDX-275 in combination with melphalan may be a novel therapeutic strategy for MM.
Keywords: HDACi, melphalan, DNA damage, apoptosis, multiple myeloma
1. Introduction
Multiple myeloma (MM) is a plasma cell disorder characterized by specific genetic aberrations and abnormal gene expression of several proto-oncogenes and tumor suppressor genes. Despite recent advances in treatment, MM remains incurable in the majority of patients [1; 2]. In addition to autologous stem cell transplantation following high dose melphalan, other chemotherapeutic agents, such as bortezomib, thalidomide, doxorubicin, and dexamethasone, have been the mainstay of treatment for MM [3]. Over the last several years, significant insights into the pathogenesis of MM have identified multiple signaling pathways that have become potential therapeutic targets in tumor cells and its bone marrow microenvironment [4; 5]. Among these, histone acetylation has emerged as one of the potentially important areas in the development of specific inhibitors for MM treatment. Alterations in chromatin structure by histone modification play a vital role in the regulation of gene transcription. Acetylation of core nucleosomal histones is regulated by the opposing behaviors of histone acetyltransferases (HATs) and histone deacetylases (HDACs) [6; 7]. The discovery that drastic changes in histone modifications are commonly found in human cancers, including MM [8], has inspired various laboratories and pharmaceutical industry to develop and study the potential therapeutic activities of HDAC inhibitor (HDACi) [9; 10]. Indeed, numerous studies demonstrate that HDACis possess anti-cancer activity in a variety of tumor cell models via influencing cell cycle progression, apoptosis, differentiation, and tumor angiogenesis [11; 12]. It has been shown that HDACis, such as suberoylanilide hydroxamic acid (SAHA), SNDX-275, sodium butyrate (NaB), and valproic acid (VPA), induce potent apoptosis on both MM cell lines and tumor cells from patients, both sensitive and resistant to conventional chemotherapeutic agents or proteasome inhibitor bortezomib [13; 14; 15]. These data indicate that the use of HDACis, probably in association with classical chemotherapy drugs could be promising for cancer patients [16].
One of the main mechanisms of action of HDACi is the transcriptional reactivation of dormant tumor-suppressor genes [17], however the pro-apoptotic activity of HDACi also comes from their non-transcriptional mechanisms on cell cycle, DNA recombination and repair, extrinsic and intrinsic apoptotic pathways, angiogenesis, autophagy and senescence [11; 18; 19]. Recent studies have shown that several HDACis sensitize cancer cells in either culture or mouse xenograft to DNA damage induced by ionizing radiation [20; 21]. SNDX-275 and SAHA also augment apoptosis by DNA damaging agents, such as mitomycin C, cisplatin, bleomycin, topotecan, doxorubicin, etoposide, 5-fluorouracil and Ara-C [22; 23]. HDACis increase H2A.X phosphorylation-induced by radiation and DNA damaging drugs, alter the global chromatin configuration, and subsequently promote DNA damage signaling pathways [21; 24; 25].
SNDX-275 (Entinostat; formerly MS-275) is a synthetic benzamide derivative class I selective HDACi. It inhibits cancer cell growth with an IC50 in the submicromolar range. The inhibition of cell growth is accompanied by a cell cycle arrest and an induction of the cyclin-dependent kinase (CDK) inhibitor p21waf1, which is one of the most commonly induced genes by HDACis [26]. SNDX-275 exhibits both in vitro and in vivo activities against various cancer types, including colorectal cancer, lung cancer, ovary cancer, and pancreatic cancer [27], pediatric solid tumors [28], leukemia [27; 29; 30; 31], prostate cancer [32], and breast cancer [33; 34; 35]. While other broad spectrum HDACis, such as SAHA, LAQ824 and LBH589 exhibit potent antimyeloma activities [36; 37], SNDX-275’s therapeutic potential and its combinational effects with alkylators on MM remain unclear. In this study, we sought to determine whether SNDX-275 might synergistically enhance melphalan-induced apoptosis in MM cells, and to explore the molecular mechanisms, especially of DNA damage response. The combinations of SNDX-275 and melphalan in MM cells showed a significant synergism. SNDX-275 intensified DNA damage response by melphalan and increased mitotic catastrophe, suggesting a potential role of DNA damage for non-transcriptional induction of cell death. The combinational strategy using an HDAC inhibitor with melphalan expands therapeutic options for patients with MM.
2. Materials and Methods
2.1 Reagents and antibodies
Melphalan (10mg, Sigma Chemical Co., St. Louis, MO) was first dissolved in 100 µl Acid-Ethanol (47 µl concentrated HCl with 1 ml of 100% Ethanol) and then brought up to 1 ml of sterile PBS to make a 33 mM stock solution. SNDX-275 (kindly provided by Syndax Pharmaceuticals, Inc., San Diego, CA) was dissolved in DMSO to make a stock solution at 200 mM. The stock solutions of both Melphalan and SNDX-275 were stored at −20°C.
Antibodies for western blot analysis were from following sources: caspase-8 mouse mAb (1C12), caspase-9 polyclonal antibody, caspase-3 rabbit mAb (8G10), Ac-Histone H3 (Lys9), Histone H3, p53 rabbit polyclonal antibody, γH2A.X (Ser139) rabbit antibody, H2A.X rabbit polyclonal antibody, P-CHK1 (Ser345) (133D3) rabbit mAb, CHK1 rabbit antibody, P-CHK2 (Thr68) rabbit polyclonal antibody, and CHK2 rabbit polyclonal antibody (Cell Signaling Technology, Inc., Beverly, MA); Cyclin D1 (M-20), E2F1 (KH95), p21waf1 (F-5) (Santa Cruz Biotechnology Inc., Santa Cruz, CA); Poly (ADP-ribose) polymerase (PARP) mAb (C-2-10) (BIOMOL Research Laboratories Inc., Plymouth Meeting, PA); β-actin mouse mAb (clone AC-75) (Sigma Chemical Co., St. Louis, MO). All other reagents were purchased from Sigma unless otherwise specified.
2.2 Cells and cell culture
Human MM cell line RPMI8226 was purchased from the American Type Culture Collection (ATCC, Manassas, VA). Human MM cell line MM1.S [38] was kindly provided by Dr. Steven Rosen (Department of Medicine, Robert H. Lurie Comprehensive Cancer Center, Northwestern University, Chicago, IL). All cell lines were maintained in RPMI1640 cell culture medium supplemented with 10% fetal bovine serum (FBS) at a 37°C humidified atmosphere containing 95% air and 5% CO2 and were split twice a week.
2.3 Cell proliferation assay
The CellTiter96™ AQ non-radioactive cell proliferation kit (Thermo Fisher Scientific Inc., Waltham, MA) was used to determine cell viability as we previously described [33]. In brief, cells were plated onto 96-well plates with either 0.1 ml complete medium with 5% FBS as control, or 0.1 ml of the same medium containing a series doses of melphalan or SNDX-275 alone or combination of melphalan and SNDX-275, and incubated for 72 hrs in a 37°C humidified atmosphere containing 95% air and 5% CO2. After reading all wells at 490 nM with a micro-plate reader, the percentages of surviving cells from each group relative to controls, defined as 100% survival, were determined by reduction of MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt).
2.4 Flow cytometric analysis of cell cycle
Flow cytometric analyses were performed as described previously [33] to define the cell cycle distribution for treated and untreated cells. In brief, cells grown in 100-mm culture dishes were harvested and fixed with 70% ethanol. Cells were then stained for total DNA content with a solution containing 50 µg/ml propidium iodide and 100 µg/ml RNase I in PBS for 30 min at 37°C. Cell cycle distribution was analyzed at the Flow Cytometry Core Facility of University of Colorado Cancer Center with a FACScan flow cytometer (BD Biosciences, San Jose, CA).
2.5 Quantification of apoptosis
An apoptosis ELISA kit (Roche Diagnostics Corp., Indianapolis, IN) was used to quantitatively measure cytoplasmic histone-associated DNA fragments (mononucleosomes and oligonucleosomes) as previously reported [33]. This photometric enzyme immunoassay was performed according to the manufacturer’s instructions.
2.6 Morphologic evaluation of mitotic catastrophe
Cultured MM.1S (or RPMI8226) MM cells were harvested and cytocentrifuged for 2 min at 400 rpm. Cells were fixed in acetone for 10 min and then in chloroform for 5 min and processed. Cells were rehydrated once in 100% alcohol for 2 min, in 90% alcohol for 2 min, in 70% alcohol for 2 min and rinsed twice in deionized water for 2 min. Samples were then incubated for 1 hr in Giemsa's solution diluted 1:5 in deionized water at room temperature, rinsed in acetic acid diluted 1:400 in deionized water for 30 sec and dehydrated in 90% and 100% alcohols and in xylene. Slides were examined under a photomicroscope (Olympus). Pathologist was blinded on each slide set regarding the treatment groups. Cells that showed abnormal mitotic figures, chromatin condensation and fragmentation were counted against normal cells, and reported in percentage.
2.7 Western blotting analysis
Protein expression levels were determined by western blot analysis as previously described [33]. Briefly, cells were lysed in a buffer containing 50 mM Tris, pH 7.4, 50 mM NaCl, 0.5% NP-40, 50 mM NaF, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 25 µg/ml leupeptin, and 25 µg/ml aprotinin. The lysates were centrifuged at full speed in a microcentrifuge for 20 min and the supernatants were collected for protein concentration determination by the Coomassie Plus protein assay reagent (Pierce Chemical Co., Rockford, IL). Equal amounts of cell lysates were boiled in Laemmli SDS-sample buffer, resolved by SDS-PAGE, and western blot analysis with specific antibodies as described in the figure legends.
2.8 Statistical analysis
Assessment of synergy versus antagonism between SNDX-275 and melphalan was performed by the Calcusyn software program (Biosoft, Ferguson, MO). Combination index (CI) was calculated by the Chou-Talalay method [39; 40]. The following equation was used to determine the value of CI. Drug synergism and antagonism are defined by CI of less than 1 and of above 1, respectively. CI of 1 represents additive effects.
3. Results
3.1 SNDX-275 synergistically enhances the inhibitory effects of melphalan on MM cells
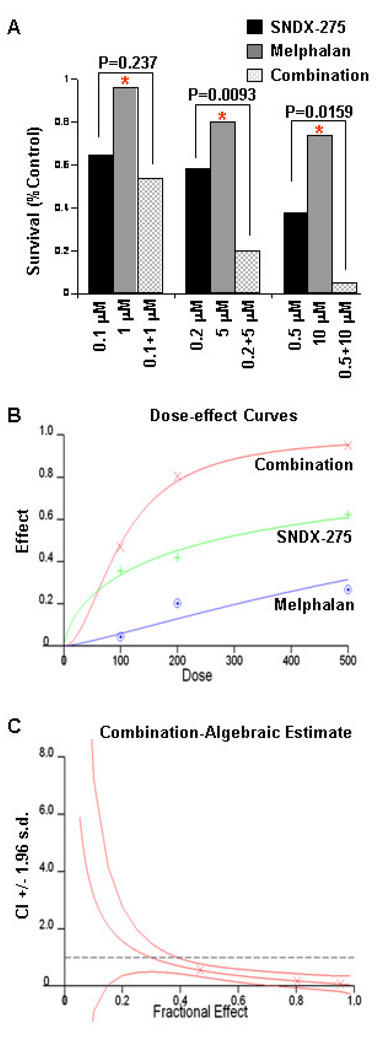
To explore whether SNDX-275 might be a potential therapeutic agent against MM, we investigated the anti-proliferative/anti-survival activities of SNDX-275 as a single agent or in combination with melphalan in MM cell lines. In 72 hrs cell proliferation MTS assays, either SNDX-275 or melphalan alone inhibited proliferation of RPMI 8226 cells in a dose-dependent manner, although SNDX-275 exhibited a much potent inhibitory effect than melphalan (Fig. 1A & B). The combinations of SNDX-275 and melphalan revealed significant anti-proliferative/anti-survival effects on RPMI 8226 cells as compared with SNDX-275 at higher doses (0.2 µM and 0.5 µM) and melphalan at all three doses tested (P<0.002). The combination index (CI) curves were calculated according to the Chou-Talalay equation [39; 40]. The curves represent CI + 1.96 × standard deviation (Fig. 1C). The CIs were 0.573 between 100 nM of SNDX-275 and 1 µM of melphalan, 0.668 between 200 nM of SNDX-275 and 5 µM of melphalan, and 0.073 between 500 nM of SNDX-275 and 10 µM of melphalan, suggesting a powerful synergy. Similar results were obtained with another cell line MM1.S (data not shown). Thus, SNDX-275 synergistically enhances melphalan-induced growth inhibition in MM cells.
Figure 1. SNDX-275 and melphalan synergistically inhibit proliferation/survival of MM cells.
A, Human MM cells (RPMI8226) were plated onto 96-well plates with complete culture medium (RPMI1640, 10% FBS). After 24 hrs, the medium was replaced with control medium (fresh RPMI1640, 0.5% FBS) or same medium containing indicated concentrations of SNDX-275, melphalan, or combinations of SNDX-275 and melphalan for another 72 hrs incubation. The percentages of surviving cells from each treatment to controls, defined as 100% survival, were determined by reduction of MTS. Data shows the representative of at least three independent experiments. * P<0.002 vs combination of SNDX-275 and melphalan. B, Dose-effect curves were generated with Chou-Talalay equation of statistical analysis. C, The combination index (CI) curves were calculated according to the Chou-Talalay equation.
3.2 SNDX-275 potentiates melphalan-induced apoptosis and cell cycle arrest in MM cells
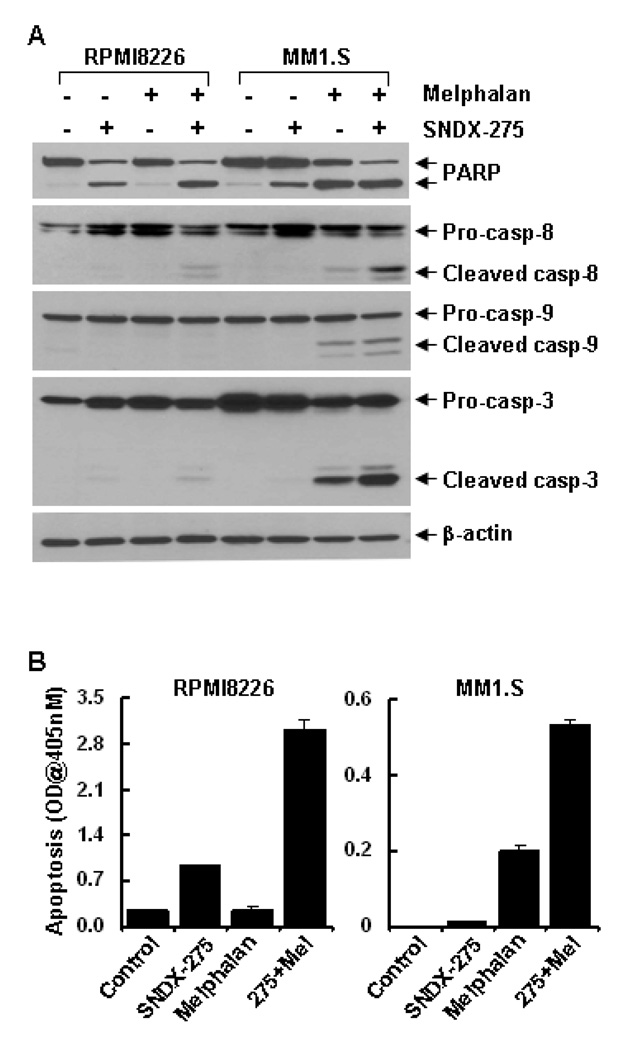
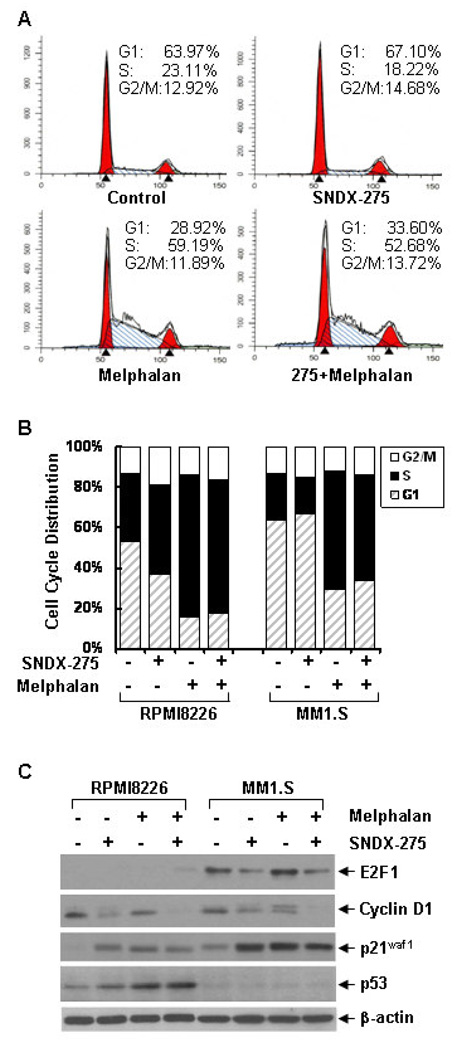
To determine the molecular mechanisms by which SNDX-275 enhances melphalan-mediated anti-proliferative/anti-survival effects, we first studied whether SNDX-275 might augment melphalan-induced apoptosis in MM cells. Apoptosis specific ELISA revealed that SNDX-275 (0.5 µM) alone induced minor apoptosis in RPMI8226 cells, but had no effect on MM1.S cells, whereas melphalan alone induced minor apoptosis in MM1.S cells, but had minimum effect on RPMI8226 cells. However, their combinations significantly induced apoptosis in both RPMI8226 and MM1.S cells (Fig. 2B). Furthermore, western blot analyses indicated that treatment with either SNDX-275 or melphalan alone induced some PARP cleavage in RPMI8226 and MM1.S cells, respectively. Consistent with the ELISA data, the combinations of SNDX-275 and melphalan strongly resulted in PARP cleavage and activation of caspase-8, -9, and -3 evidenced by the increases of cleaved caspase-8, -9, and -3 (Fig. 2A), suggesting that SNDX-275 potentiates melphalan-induced caspase-dependent apoptosis in MM cells. Next we investigated whether SNDX-275 and melphalan might have different modulation on cell cycle progression. Cell cycle assays showed that SNDX-275 induced a minor increase of the cells in G1 phase, whereas melphalan dramatically increased the MM1.S cells in S phase and both agents together led to the majority of cells still staying at S phase (Fig. 3A). In contrast, either SNDX-275 or melphalan alone, or their combinations mainly arrested RPMI8226 cells at S phase (Fig. 3B). These data correlated well with the western blot analyses on several key molecular markers regulating cell cycle progression. Consistent with a previous report showing that the CDK inhibitor p21waf1 is one of the most commonly induced genes by HDACi [26], we did discover a significant induction of p21waf1 by SNDX-275 (Fig. 3C). Cyclin D1 and E2F1 are two important positive regulators at the G1-S checkpoint of cell cycle. Treatment with SNDX-275 reduced the levels of cyclin D1 and E2F1 in both RPMI8226 and MM1.S cells cells (Fig. 3C). Taken together, the effects of SNDX-275 on p21waf1, cyclin D1, and E2F1 may contribute to its influence on the cell cycle G1-S transition. In contrast, although melphalan also upregulated p21waf1, but had no effect on E2F1, and might induced Cyclin D1 phosphorylation in MM1.S cells (Fig. 3C). Thus, melphalan alone arrested the cells at S phase. It appeared that the combinations of SNDX-275 and melphalan antagonistically compromised their individual effects on cell cycle progression (Fig. 3A). Collectively, our results demonstrate that SNDX-275 enhances melphalan-induces apoptosis and both SNDX-275 and melphalan blocks cell cycle progression in MM cells.
Figure 2. Combinations of SNDX-275 and melphalan significantly induce PARP cleavage, activation of caspase-8, -9, -3, and apoptosis in MM cells.
RPMI8226 and MM1.S cells were cultured with RPMI1640 (0.5% FBS) in the absence or presence of SNDX-275 (0.5 µM), melphalan (10 µM) alone or combinations of SNDX-275 and melphalan for 24 hrs. Cells were collected and subjected to western blot analyses with specific antibodies directed against PARP, caspase-8, caspase-9, caspase-3, or β-actin (A) or apoptosis ELISA (B).
Figure 3. SNDX-275 and melphalan block cell cycle progression and modulate expression of several key cell cycle regulators.
A, MM1.S cells were cultured with RPMI1640 (0.5% FBS) in the absence or presence of SNDX-275 (0.5 µM), melphalan (10 µM) alone or combinations of SNDX-275 and melphalan for 24 hrs. Cells were harvested and subjected to flow cytometry analysis of cell cycle distribution. B, Similar experiments were also performed with RPMI8226 cells. The bar graph reflects the percentage of cells in G1, S, G2/M phase of the cell cycle. Data shows the representative of at least three independent experiments. C, RPMI8226 and MM1.S cells similarly treated as in (A) were collected and subjected to western blot analyses with specific antibodies directed against E2F1, cyclin D1, p21waf1, p53, or β-actin.
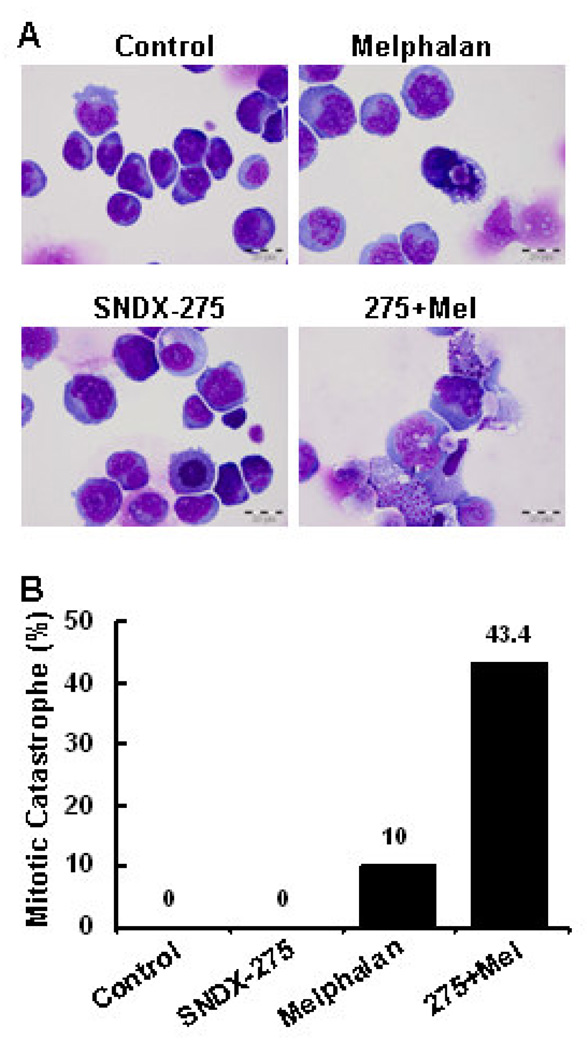
3.3 Combination of SNDX-275 and melphalan results in strong DNA damage responses associated with enhanced mitotic catastrophe
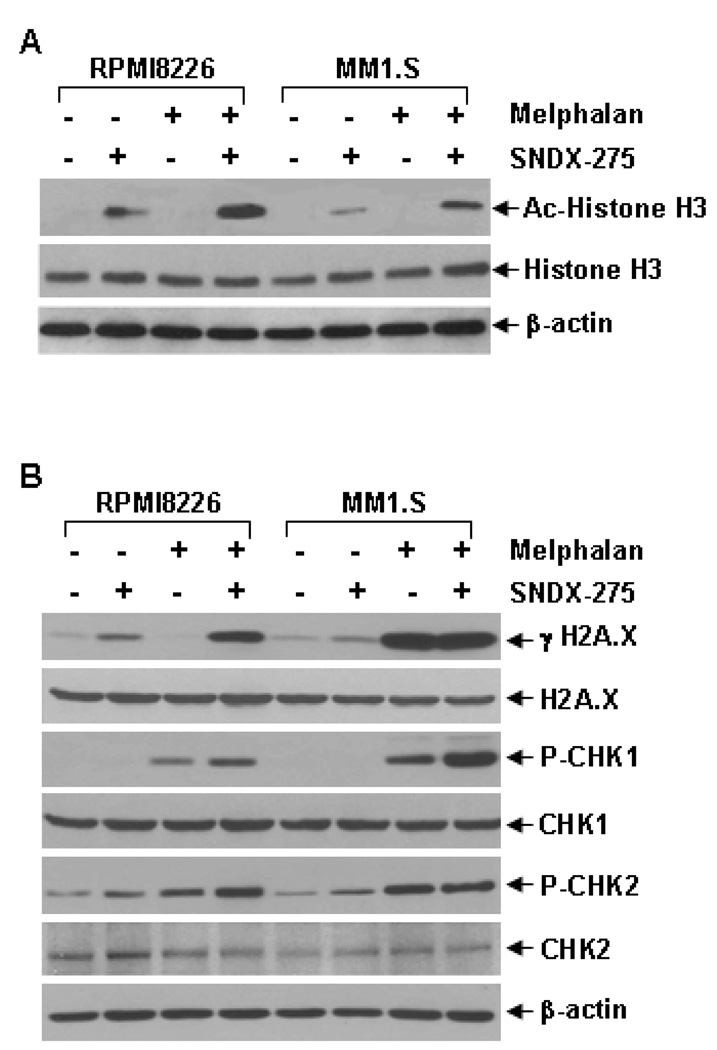
Our subsequent studies showed that while SNDX-275 alone clearly induced acetylation of histone H3 in both RPMI8226 and MM1.S cells, the combinations of SNDX-275 and melphalan significantly increased the levels of histone H3 acetylation (Fig. 4A), suggesting that SNDX-275 and melphalan might synergistically induce gene expression/modification in MM cells. As one of the major alkylators currently used in the treatment of MM, melphalan induces mitotic cell death via DNA damage. Thus, we next focused on investigating if SNDX-275 may enhance modification of genes associated with melphalan-mediated DNA damage responses in RPMI8226 and MM1.S cells. Phosphorylation of H2A.X (γH2A.X), a hall marker of DNA double strand break [41], was increased upon treatment with SNDX-275 alone in RPMI8226 cells and with melphalan alone in MM1.S cells (Fig. 4B). Combinations of SNDX-275 and melphalan significantly increased the levels of γH2A.X in both MM cells. Similar results were also observed on phosphorylation of CHK1, indicative of initiation of DNA damage response [41]. The combinations of two agents induced considerably higher levels of P-CHK1 than either agent alone in both RPMI8226 and MM1.S cells. While melphalan alone clearly increased P-CHK2 levels, its combination with SNDX-275 did not further increase the levels of P-CHK2 in MM1.S cells. The combinations of SNDX-275 and melphalan induced much higher P-CHK2 levels than either agent alone in RPMI8226 cells (Fig. 4B). These data suggest that SNDX-275 synergistically enhances melphalan-mediated DNA damage responses mainly through induction of γH2A.X, P-CHK1, and/or P-CHK2 in MM cells. Furthermore, morphologic assessment of MM1.S cells treated with either agent alone or their combinations in cell culture revealed a significant increase in mitotic catastrophe upon combinational treatment (Fig. 5). Taken together, our studies indicate that HDAC inhibition synergizes with melphalan to induce apoptosis in MM cells, and that intensification of DNA damage is one of the mechanisms.
Figure 4. Combinations of SNDX-275 and melphalan significantly enhance acetylation of histone H3, and exhibit super-induction of molecular markers-associated with DNA damage response.
RPMI8226 and MM1.S cells were cultured with RPMI1640 (0.5% FBS) in the absence or presence of SNDX-275 (0.5 µM), melphalan (10 µM) alone or combinations of SNDX-275 and melphalan for 24 hrs. Cells were collected and subjected to western blot analyses with specific antibodies directed against (acetylated) Ac-Histone H3, Histone H3, or β-actin (A) or γH2A.X, H2A.X, P-CHK1, CHK1, P-CHK2, CHK2, or β-actin (B).
Figure 5. SNDX-275 enhances melphalan-induced mitotic catastrophe in MM cells.
MM1.S cells were cultured with RPMI1640 (0.5% FBS) in the absence or presence of SNDX-275 (0.5 µM), melphalan (10 µM) alone or combinations of SNDX-275 and melphalan for 24 hrs. Cells were harvested and subjected to cytospin onto cell slides. A, Slides were stained with HE, examined, and pictures were taken under a photomicroscope. B, Pathologist was blinded on each slide set regarding the treatment groups. Cells that showed atypical mitotic figures, multi-nucleation, atypical chromosome clusters, and apoptosis were counted against normal cells, and reported in percentage.
4. Discussion
Recent advances in identifying novel therapeutics against myeloma have provided new hope for this incurable disease. HDACi is one of those promising agents for treatment of MM [42; 43]. It has been shown that several HDACis, such as SAHA, LAQ824, and LBH589, exhibit potent anti-myeloma activities in cell culture model and are currently in clinical trials for MM patients [4; 37]. While all of these HDACis are pan-inhibitors towards all classes of HDACs, SNDX-275 is a specific class I HDACi [11; 12]. Numerous studies have reported that SNDX-275 is a more selective anti-cancer agent and has gone under clinical trials of many human cancers (http://www.clinicaltrials.gov/ct2/results?term=SNDX-275). However, SNDX-275’s potential therapeutic activity in MM remains unclear. Here we provide strong evidence indicating that SNDX-275 not only shows anti-myeloma activities when used as a single agent, it also synergistically potentiates melphalan-induced apoptosis via enhanced DNA damage response. Thus, SNDX-275 may be a valuable option for the treatment of MM patients.
We have found that SNDX-275’s inhibitory effects on cell cycle progression may be cell type-dependent. SNDX-275 alone mainly induced S phase arrest in RPMI8226 cells, it had only minimal induction of G1 phase in MM1.S cells (Fig. 3A & 3B). This disparity effects of SNDX-275 on cell cycle progression have been reported in our previous studies on breast cancer cells with distinct subtypes [33]. Further studies on the key regulators involving in G1-S checkpoint of cell cycle revealed that treatment with SNDX-275 resulted in a significant reduction in the levels of E2F1 and Cyclin D1, and a dramatic increase in p21waf1. These data are consistent not only with our observations in erbB2-overexpressing breast cancer cells upon SNDX-275 treatment [33], but also with a number of reports indicating that p21waf1 is one of the most commonly induced genes by HDACis [15; 26; 44]. SNDX-275 either alone or in combination with melphalan induced apoptosis through caspase-dependent mechanisms, similar results were also observed in studies with LBH589 and LAQ824 [44; 45]. It appeared SAHA-induced apoptosis in MM cells might be via caspase-independent pathways [14]. The molecular mechanisms of this difference remain to be elucidated.
Although HDACi using as a single agent holds promising for treatment of MM patients, more studies are carried out by combinations of an HDACi and conventional chemotherapeutic drugs or newly introduced agents, such as thalidomide, its immunomodulatory derivative lenalidomide and the proteasome inhibitor bortezomib [43], with a hope of overcoming drug resistance and reduced side effects. It has been reported that SAHA enhances cytotoxicity of dexamethasone, doxorubicin, thalidomide, or bortezomib in MM1.1S cells [15; 46]. LBH589 also shows synergistic anti-myeloma activity in combinations with dexamethasone, bortezomib, or melphalan [44; 47]. While majority of these combinational studies focus on increased induction of apoptosis and cell cycle regulators, such as p21waf1, in MM.1S cell line, our data indicate that SNDX-275 synergistically potentiates melphalan-induced apoptosis via enhanced DNA damage responses in both RPMI8226 and MM.1S cells. This augmentation might be due to SNDX-275’s strong inhibition on PI-3K/Akt signaling as we observed in breast cancer cells [33], since Akt activation protects melphalan-mediated DNA damage-induced apoptosis in breast epithelial cells via suppression of the ASK1/JNK pathway [48]. It will be very interesting to study if SNDX-275 may inhibit Akt signaling and subsequently enhance melphalan-induced activation of ASK1/JNK pathway in MM cells. To further explore its therapeutic potential in MM, we are currently testing whether SNDX-275 may also exhibit synergistic inhibitory effects with dexamethasone, doxorubicin, thalidomide, or bortezomib on MM cells.
Recent advances in identifying novel therapeutic targets for the treatment of MM have discovered that the IGF-1 as a major growth factor promotes cell proliferation and survival, and plays a critical role in myeloma development [49; 50]. Indeed, two strategies targeting IGF-1R – blocking antibodies and small molecule inhibitors – show very encouraging preclinical results against MM cells, and both strategies are now in clinical trials [49]. Since PI-3K/Akt signaling is one of the major downstream pathways under IGF-1/IGF-1R, IGF-1R-targeted therapies may enhance melphalan-mediated DNA damage response via inactivation of Akt. In addition, SNDX-275, because of its strong inhibitory effects on PI-3K/Akt signaling, may also synergistically increase the efficacy of IGF-1R-targeted therapies in MM. It has been recently reported that the death ligand TRAIL may be a good candidate for the treatment of MM because of its low toxic for normal human cells [51; 52]. Interestingly, while HDACis, such as SAHA and trichostatin (TSA), were shown to enhance TRAIL-induced apoptosis [53], another HDACi, VPA, was able to overcome TRAIL resistance in MM cells [51]. Moreover, studies have also found SNDX-275’s capability of inducing TRAIL expression in breast cancer cells [34; 35]. It is therefore interesting and necessary to determine if SNDX-275 may enhance TRAIL-induced apoptosis in MM cells via induction of TRAIL expression.
In summary, we provide compelling evidence indicating that the class I HDACi SNDX-275 synergistically enhances melphalan-mediated DNA damage-induced apoptosis in MM cells. Our data suggest that SNDX-275 in combination with conventional chemotherapeutic drugs or newly discovered agents may be promising for the treatment of patients with MM.
Acknowledgements
The authors are grateful to Dr. Peter Ordentlich (Syndax Pharmaceuticals, Inc., San Diego, CA) for providing SNDX-275. This work was supported in part by a PO1 Aging and Cancer Grant (P20-CA103680) from University of Colorado Cancer Center (to CKL), a Susan G. Komen for the Cure Research Grant (BCTR0707449) and ACS IRG #57-001-50 from University of Colorado Cancer Center (to BL).
Abbreviations
- MM
multiple myeloma
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HDACi
inhibitor of HDAC
- SAHA
suberoylanilide hydroxamic acid
- VPA
valproic acid
- TRAIL
tumor necrosis factor-related apoptosis-inducing ligand
- MAPK
mitogen-activated protein kinase
- MEK
MAPK kinase
- PI-3K
phosphoinositide 3-kinase
- IGF-1
insulin-like growth factor-1
- IGF-1R
IGF-1 receptor
- PARP
poly(ADP-ribose) polymerase
- ELISA
enzyme-linked immunosorbent assay
- PAGE
polyacrylamide gel electrophoresis
- SNDX-275
N-(2-Aminophenyl)-4-[N-(pyridine-3-ylmethoxycarbonyl)aminomethyl]benzamide
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium,inner salt
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest Statement
None Declared
References
- 1.Kyle RA, Rajkumar SV. Multiple myeloma. N. Engl. J. Med. 2004;351:1860–1873. doi: 10.1056/NEJMra041875. [DOI] [PubMed] [Google Scholar]
- 2.Naumann F, Weingart O, Kruse E, Schulz H, Bohlius J, Hulsewede H, Engert A. Fifth biannual report of the cochrane haematologic malignancies group--focus on multiple myeloma. J. Natl. Cancer Inst. 2006;98 doi: 10.1093/jnci/djj328. E2-E. [DOI] [PubMed] [Google Scholar]
- 3.Diagnosis and management of multiple myeloma. Br. J. Haematol. 2001;115:522–540. doi: 10.1046/j.1365-2141.2001.03206.x. [DOI] [PubMed] [Google Scholar]
- 4.Bruno B, Giaccone L, Rotta M, Anderson K, Boccadoro M. Novel targeted drugs for the treatment of multiple myeloma: from bench to bedside. Leukemia. 2005;19:1729–1738. doi: 10.1038/sj.leu.2403905. [DOI] [PubMed] [Google Scholar]
- 5.Hideshima T, Chauhan D, Richardson P, Anderson KC. Identification and validation of novel therapeutic targets for multiple myeloma. J. Clin. Oncol. 2005;23:6345–6350. doi: 10.1200/JCO.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 6.Archer SY, Hodin RA. Histone acetylation and cancer. Curr. Opin. Genet. Dev. 1999;9:171–174. doi: 10.1016/s0959-437x(99)80026-4. [DOI] [PubMed] [Google Scholar]
- 7.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Espino PS, Drobic B, Dunn KL, Davie LR. Histone modifications as a platform for cancer therapy. J. Cell Biochem. 2005;94:1088–1102. doi: 10.1002/jcb.20387. [DOI] [PubMed] [Google Scholar]
- 9.Mottet D, Castronovo V. Histone deacetylases: target enzymes for cancer therapy. Clin. Exp. Metastasis. 2008;25:183–189. doi: 10.1007/s10585-007-9131-5. [DOI] [PubMed] [Google Scholar]
- 10.Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26:5310–5318. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- 11.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug. Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 12.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 13.Kaiser M, Zavrski I, Sterz J, Jakob C, Fleissner C, Kloetzel PM, Sezer O, Heider U. The effects of the histone deacetylase inhibitor valproic acid on cell cycle, growth suppression and apoptosis in multiple myeloma. Haematologica. 2006;91:248–251. [PubMed] [Google Scholar]
- 14.Mitsiades N, Mitsiades CS, Richardson PG, McMullan C, Poulaki V, Fanourakis G, Schlossman R, Chauhan D, Munshi NC, Hideshima T, Richon VM, Marks PA, Anderson KC. Molecular sequelae of histone deacetylase inhibition in human malignant B cells. Blood. 2003;101:4055–4062. doi: 10.1182/blood-2002-11-3514. [DOI] [PubMed] [Google Scholar]
- 15.Pei XY, Dai Y, Grant S. Synergistic induction of oxidative injury and apoptosis in human multiple myeloma cells by the proteasome inhibitor bortezomib and histone deacetylase inhibitors. Clin. Cancer Res. 2004;10:3839–3852. doi: 10.1158/1078-0432.CCR-03-0561. [DOI] [PubMed] [Google Scholar]
- 16.Villar-Garea A, Esteller M. Histone deacetylase inhibitors: understanding a new wave of anticancer agents. Int. J. Cancer. 2004;112:171–178. doi: 10.1002/ijc.20372. [DOI] [PubMed] [Google Scholar]
- 17.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: overview and perspectives. Mol. Cancer Res. 2007;5:981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 19.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 20.Camphausen K, Scott T, Sproull M, Tofilon PJ. Enhancement of xenograft tumor radiosensitivity by the histone deacetylase inhibitor MS-275 and correlation with histone hyperacetylation. Clin. Cancer Res. 2004;10:6066–6071. doi: 10.1158/1078-0432.CCR-04-0537. [DOI] [PubMed] [Google Scholar]
- 21.Geng L, Cuneo KC, Fu A, Tu T, Atadja PW, Hallahan DE. Histone deacetylase (HDAC) inhibitor LBH589 increases duration of gamma-H2AX foci and confines HDAC4 to the cytoplasm in irradiated non-small cell lung cancer. Cancer Res. 2006;66:11298–11304. doi: 10.1158/0008-5472.CAN-06-0049. [DOI] [PubMed] [Google Scholar]
- 22.Kim MS, Blake M, Baek JH, Kohlhagen G, Pommier Y, Carrier F. Inhibition of histone deacetylase increases cytotoxicity to anticancer drugs targeting DNA. Cancer Res. 2003;63:7291–7300. [PubMed] [Google Scholar]
- 23.Ozaki K, Kishikawa F, Tanaka M, Sakamoto T, Tanimura S, Kohno M. Histone deacetylase inhibitors enhance the chemosensitivity of tumor cells with cross-resistance to a wide range of DNA-damaging drugs. Cancer Sci. 2008;99:376–384. doi: 10.1111/j.1349-7006.2007.00669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 25.Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, Yang SM, Blasco MA, Skoultchi AI, Fernandez-Capetillo O. Global chromatin compaction limits the strength of the DNA damage response. J. Cell Biol. 2007;178:1101–1108. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gui CY, Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc. Natl. Acad. Sci. U S A. 2004;101:1241–1246. doi: 10.1073/pnas.0307708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito A, Yamashita T, Mariko Y, Nosaka Y, Tsuchiya K, Ando T, Suzuki T, Tsuruo T, Nakanishi O. A synthetic inhibitor of histone deacetylase, MS-27-275, with marked in vivo antitumor activity against human tumors. Proc. Natl. Acad. Sci. U S A. 1999;96:4592–4597. doi: 10.1073/pnas.96.8.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaboin J, Wild J, Hamidi H, Khanna C, Kim CJ, Robey R, Bates SE, Thiele CJ. MS-27-275, an inhibitor of histone deacetylase, has marked in vitro and in vivo antitumor activity against pediatric solid tumors. Cancer Res. 2002;62:6108–6115. [PubMed] [Google Scholar]
- 29.Gojo I, Jiemjit A, Trepel JB, Sparreboom A, Figg WD, Rollins S, Tidwell ML, Greer J, Chung EJ, Lee MJ, Gore SD, Sausville EA, Zwiebel J, Karp JE. Phase 1 and pharmacologic study of MS-275, a histone deacetylase inhibitor, in adults with refractory and relapsed acute leukemias. Blood. 2007;109:2781–2790. doi: 10.1182/blood-2006-05-021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lucas DM, Davis ME, Parthun MR, Mone AP, Kitada S, Cunningham KD, Flax EL, Wickham J, Reed JC, Byrd JC, Grever MR. The histone deacetylase inhibitor MS-275 induces caspase-dependent apoptosis in B-cell chronic lymphocytic leukemia cells. Leukemia. 2004;18:1207–1214. doi: 10.1038/sj.leu.2403388. [DOI] [PubMed] [Google Scholar]
- 31.Rosato RR, Almenara JA, Grant S. The histone deacetylase inhibitor MS-275 promotes differentiation or apoptosis in human leukemia cells through a process regulated by generation of reactive oxygen species and induction of p21CIP1/WAF1 1. Cancer Res. 2003;63:3637–3645. [PubMed] [Google Scholar]
- 32.Yang J, Wezeman M, Zhang X, Lin P, Wang M, Qian J, Wan B, Kwak LW, Yu L, Yi Q. Human C-reactive protein binds activating Fcgamma receptors and protects myeloma tumor cells from apoptosis. Cancer Cell. 2007;12:252–265. doi: 10.1016/j.ccr.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 33.Huang X, Gao L, Wang S, Lee CK, Ordentlich P, Liu B. HDAC inhibitor SNDX-275 induces apoptosis in erbB2-overexpressing breast cancer cells via down-regulation of erbB3 expression. Cancer Res. 2009;69:8403–8411. doi: 10.1158/0008-5472.CAN-09-2146. [DOI] [PubMed] [Google Scholar]
- 34.Singh TR, Shankar S, Srivastava RK. HDAC inhibitors enhance the apoptosis-inducing potential of TRAIL in breast carcinoma. Oncogene. 2005;24:4609–4623. doi: 10.1038/sj.onc.1208585. [DOI] [PubMed] [Google Scholar]
- 35.Xu J, Zhou JY, Wei WZ, Philipsen S, Wu GS. Sp1-mediated TRAIL induction in chemosensitization. Cancer Res. 2008;68:6718–6726. doi: 10.1158/0008-5472.CAN-08-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marks PA, Xu WS. Histone deacetylase inhibitors: Potential in cancer therapy. J. Cell Biochem. 2009;107:600–608. doi: 10.1002/jcb.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitsiades CS, Hayden PJ, Anderson KC, Richardson PG. From the bench to the bedside: emerging new treatments in multiple myeloma. Best Pract. Res. Clin. Haematol. 2007;20:797–816. doi: 10.1016/j.beha.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greenstein S, Krett NL, Kurosawa Y, Ma C, Chauhan D, Hideshima T, Anderson KC, Rosen ST. Characterization of the MM.1 human multiple myeloma (MM) cell lines: a model system to elucidate the characteristics, behavior, and signaling of steroid-sensitive and -resistant MM cells. Exp. Hematol. 2003;31:271–282. doi: 10.1016/s0301-472x(03)00023-7. [DOI] [PubMed] [Google Scholar]
- 39.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 40.Chou TC, Talaly P. A simple generalized equation for the analysis of multiple inhibitions of Michaelis-Menten kinetic systems. J. Biol. Chem. 1977;252:6438–6442. [PubMed] [Google Scholar]
- 41.Peng A, Lewellyn AL, Schiemann WP, Maller JL. Repo-man controls a protein phosphatase 1-dependent threshold for DNA damage checkpoint activation. Curr. Biol. 2010;20:387–396. doi: 10.1016/j.cub.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hideshima T, Mitsiades C, Tonon G, Richardson PG, Anderson KC. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat. Rev. Cancer. 2007;7:585–598. doi: 10.1038/nrc2189. [DOI] [PubMed] [Google Scholar]
- 43.Podar K, Chauhan D, Anderson KC. Bone marrow microenvironment and the identification of new targets for myeloma therapy. Leukemia. 2009;23:10–24. doi: 10.1038/leu.2008.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maiso P, Carvajal-Vergara X, Ocio EM, Lopez-Perez R, Mateo G, Gutierrez N, Atadja P, Pandiella A, San Miguel JF. The histone deacetylase inhibitor LBH589 is a potent antimyeloma agent that overcomes drug resistance. Cancer Res. 2006;66:5781–5789. doi: 10.1158/0008-5472.CAN-05-4186. [DOI] [PubMed] [Google Scholar]
- 45.Catley L, Weisberg E, Tai YT, Atadja P, Remiszewski S, Hideshima T, Mitsiades N, Shringarpure R, LeBlanc R, Chauhan D, Munshi NC, Schlossman R, Richardson P, Griffin J, Anderson KC. NVP-LAQ824 is a potent novel histone deacetylase inhibitor with significant activity against multiple myeloma. Blood. 2003;102:2615–2622. doi: 10.1182/blood-2003-01-0233. [DOI] [PubMed] [Google Scholar]
- 46.Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Hideshima T, Akiyama M, Chauhan D, Munshi N, Gu X, Bailey C, Joseph M, Libermann TA, Richon VM, Marks PA, Anderson KC. Transcriptional signature of histone deacetylase inhibition in multiple myeloma: biological and clinical implications. Proc. Natl. Acad. Sci. U S A. 2004;101:540–545. doi: 10.1073/pnas.2536759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Catley L, Weisberg E, Kiziltepe T, Tai YT, Hideshima T, Neri P, Tassone P, Atadja P, Chauhan D, Munshi NC, Anderson KC. Aggresome induction by proteasome inhibitor bortezomib and alpha-tubulin hyperacetylation by tubulin deacetylase (TDAC) inhibitor LBH589 are synergistic in myeloma cells. Blood. 2006;108:3441–3449. doi: 10.1182/blood-2006-04-016055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meurette O, Stylianou S, Rock R, Collu GM, Gilmore AP, Brennan K. Notch activation induces Akt signaling via an autocrine loop to prevent apoptosis in breast epithelial cells. Cancer Res. 2009;69:5015–5022. doi: 10.1158/0008-5472.CAN-08-3478. [DOI] [PubMed] [Google Scholar]
- 49.Menu E, van Valckenborgh E, van Camp B, Vanderkerken K. The role of the insulin-like growth factor 1 receptor axis in multiple myeloma. Arch. Physiol. Biochem. 2009;115:49–57. doi: 10.1080/13813450902736583. [DOI] [PubMed] [Google Scholar]
- 50.Sprynski AC, Hose D, Caillot L, Reme T, Shaughnessy JD, Barlogie B, Jr, Seckinger A, Moreaux J, Hundemer M, Jourdan M, Meissner T, Jauch A, Mahtouk K, Kassambara A, Bertsch U, Rossi JF, Goldschmidt H, Klein B. The role of IGF-1 as a major growth factor for myeloma cell lines and the prognostic relevance of the expression of its receptor. Blood. 2009;113:4614–4626. doi: 10.1182/blood-2008-07-170464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gomez-Benito M, Martinez-Lorenzo MJ, Anel A, Marzo I, Naval J. Membrane expression of DR4, DR5 and caspase-8 levels, but not Mcl-1, determine sensitivity of human myeloma cells to Apo2L/TRAIL. Exp. Cell Res. 2007;313:2378–2388. doi: 10.1016/j.yexcr.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 52.Labrinidis A, Diamond P, Martin S, Hay S, Liapis V, Zinonos I, Sims NA, Atkins GJ, Vincent C, Ponomarev V, Findlay DM, Zannettino AC, Evdokiou A. Apo2L/TRAIL inhibits tumor growth and bone destruction in a murine model of multiple myeloma. Clin. Cancer Res. 2009;15:1998–2009. doi: 10.1158/1078-0432.CCR-08-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fandy TE, Shankar S, Ross DD, Sausville E, Srivastava RK. Interactive effects of HDAC inhibitors and TRAIL on apoptosis are associated with changes in mitochondrial functions and expressions of cell cycle regulatory genes in multiple myeloma. Neoplasia. 2005;7:646–657. doi: 10.1593/neo.04655. [DOI] [PMC free article] [PubMed] [Google Scholar]