Abstract
Fe-S cluster biogenesis is of interest to many fields, including bioenergetics and gene regulation. The CSD system is one of three Fe-S cluster biogenesis systems in E. coli and is comprised of the cysteine desulfurase CsdA, the sulfur acceptor protein CsdE, and the E1-like protein CsdL. The biological role, biochemical mechanism, and protein targets of the system remain uncharacterized. Here we present that the active site CsdE C61 has a lowered pKa value of 6.5, which is nearly identical to that of C51 in the homologous SufE protein and which is likely critical for its function. We observed that CsdE forms disulfide bonds with multiple proteins and identified the proteins that copurify with CsdE. The identification of Fe-S proteins and both putative and established Fe-S cluster assembly (ErpA, glutaredoxin-3, glutaredoxin-4) and sulfur trafficking (CsdL, YchN) proteins supports the two-pathway model, in which the CSD system is hypothesized to synthesize both Fe-S clusters and other sulfur-containing cofactors. We suggest that the identified Fe-S cluster assembly proteins may be the scaffold and/or shuttle proteins for the CSD system. By comparison with previous analysis of SufE, we demonstrate that there is some overlap in the CsdE and SufE interactomes.
Keywords: iron-sulfur (Fe-S) cluster biogenesis, iron-sulfur cluster assembly, iron-sulfur cluster biosynthesis, cysteine sulfinate desulfinase (CSD), pKa, CsdA, CsdE, CsdL, SufE
Sulfur trafficking pathways are highly conserved, complex protein machineries that are responsible for extracting sulfur from cysteine and synthesizing the sulfur-containing cofactors and thionucleosides, including thiamin, biotin, the molybdenum cofactor, lipoic acid, S-adenosylmethionine, and iron-sulfur (Fe-S) clusters (reviewed in Mueller 2006)1. The first step of sulfur trafficking is removal of inorganic sulfur from free cysteine by a cysteine desulfurase, followed by the transient and covalent binding of the sulfur atom to a reactive cysteine residue sulfhydryl within the desulfurase active site, thereby forming a persulfide (R-S-SH) intermediate. Due to the toxicity of free sulfide, the persulfide intermediate is the universal sulfur donor in these pathways. The sulfur atom is then transferred between nucleophilic cysteine residue sulfhydryls on various protein carriers, ultimately being incorporated into a cofactor or thionucleoside. Despite the biological importance of such sulfur-containing molecules, sulfur trafficking pathways are poorly understood. We therefore developed an in vivo method for the identification of protein-protein interactions within sulfur trafficking systems and applied it successfully to the sulfur mobilization (SUF) Fe-S cluster biogenesis system of E. coli (Bolstad and Wood, 2010, submitted). The goal of the present work was to apply our method to the characterization of another E. coli Fe-S cluster biogenesis system, the cysteine sulfinate desulfinase (CSD) system.
Fe-S clusters are evolutionarily ancient and important cofactors found in all kingdoms of life. They serve as catalysts, electron transfer sites, regulatory sensors (e.g., of iron and oxidants), and structural centers, and are found in proteins involved in processes as diverse as respiration, metabolism, gene regulation, cofactor biosynthesis, and DNA-RNA metabolism 2. Fe-S clusters form spontaneously on apoproteins in vitro under reducing and/or anaerobic conditions in the presence of sulfide and ferrous iron, but in vivo the clusters are formed by multi-protein systems that bind the otherwise toxic free iron and sulfide and accelerate the process. There is tremendous interest in understanding the mechanisms of Fe-S cluster biosynthesis, as several human diseases, including Friedreich’s ataxia, are the result of genetic defects in the protein machinery that makes Fe-S clusters 2. Furthermore, Fe-S cluster biogenesis contributes to bacterial pathogenicity 3–7, and is therefore a potential therapeutic target.
E. coli contains three cysteine desulfurases, IscS, SufS, and CsdA, 8–12 and three associated systems for Fe-S cluster biogenesis, the iron-sulfur cluster assembly (ISC), SUF, and CSD systems (reviewed in Fontecave and Ollagnier-de-Choudens, 2008) 13. The ISC system serves a housekeeping function by synthesizing Fe-S clusters under normal growth conditions 13, and is conserved in higher eukaryotes, including mammals, where it exists in the mitochondria and is essential for cellular Fe-S cluster assembly 2. The E. coli IscS cysteine desulfurase is also responsible for providing sulfur and selenium to the molybdopterin, thiamin, thionucleoside, selenonucleoside, and selenoprotein biosynthetic pathways 1, 14. The SUF system serves in stress response through its induction by iron limitation and oxidative stress 6, 7, 9, 15, 16. Suf proteins are conserved among bacteria and the plastid organelles of higher plants and the malaria parasite. The biological role of the CSD system has yet to be fully characterized, but it appears necessary for bacterial motility 17. The three systems are expressed from the iscRSUA-hscBA-fdx, sufABCDSE, and csdAEL operons (note that csdAE and csdL are transcribed in opposite directions), respectively, and multiple uncharacterized accessory proteins have been identified that interact with the ISC and SUF systems. IscR is a [2Fe-2S]-containing transcription factor that represses isc operon transcription in its holoform 18, whereas in its apoform, formed under conditions of iron limitation and oxidative stress, it stimulates suf operon expression 3, 19–21. IscU is a scaffold protein, that is, Fe-S clusters are assembled upon it and can then be transferred to apoproteins 13. The HscBA chaperone pair catalyzes this transfer process 22. IscA is one of the three A-type carrier proteins (ATCs) in E. coli (the others are SufA and ErpA). Proteins are designated as ATCs based on their sequence. The role of the ATCs is controversial (see Loiseau et al., 2007 and Vinella et al., 2009 for thorough discussions) 23, 24; they have been proposed to serve as scaffold proteins, iron donors, and/or Fe-S cluster shuttles that transfer preformed clusters from scaffold proteins to apoproteins. Recent data, however, supports a shuttle role for ATCs 24, 25. The ferredoxin (fdx) protein is proposed to provide the electrons necessary for Fe-S cluster assembly. SufE and CsdE are so-called sulfur acceptor proteins because a conserved cysteine residue sulfhydryl within these proteins makes a nucleophilic attack on the persulfide sulfur on SufS and CsdA, resulting in a persulfide intermediate on SufE and CsdE and stimulation of the desulfurase activity of SufS and CsdA 26–29. The E1-like (ubiquitin-activating-like) CsdL (formerly YgdL) protein was recently shown to interact with CsdE and to receive sulfur from CsdA in a CsdE-independent manner 17. E. coli contains two other E1-like proteins, MoeB and ThiF, which are necessary for incorporation of sulfur into the precursors of molybdopterin and thiamin, respectively 30–34. It was suggested that CsdE brings CsdA and CsdL together for synthesis of a currently unidentified sulfur-containing molecule, or that CsdE serves as a sulfur relay between CsdA and CsdL, though the biochemical data suggests that this role could be dispensable 17. The CSD system was therefore proposed to engage in two sulfur transfer pathways, one dedicated to the biosynthesis of Fe-S clusters and the other to biosynthesis of a putative thiolated metabolite. SufC is an atypical ABC transporter ATPase 6 that forms a complex with SufB and SufD 28. The purpose of the SufC ATPase activity is unknown. It was recently shown that the SufBCD complex is likely the site of Fe-S cluster formation within the SUF system and that the persulfide sulfur on SufE is transferred to SufB 25, 35, likely explaining the stimulation of SufS cysteine desulfurase activity by SufBCD 28.
The CSD system and components of the SUF system share a high degree of homology, with a shared sequence identity of 45% for the CsdA and SufS proteins and 35% for the CsdE and SufE proteins (Fig. 1A). In fact, CsdA appears to be able to substitute for SufS within the SUF system 17, resulting in the suggestion that CsdAE and SufSE derive from a common heterodimeric cysteine desulfurase ancestor. The CsdE and SufE proteins also exhibit extensive structural homology; the CsdE solution structure (PDB 1NI7) 36 and SufE crystal structure (PDB 1MZG) 37 superimpose with a root-mean-square deviation of 2.0 Angstroms for 132 backbone carbon (Cα) atoms (Fig. 1B). In particular, the structures of the cavities containing the conserved C61 (CsdE) and C51 (SufE) residues are virtually identical (Fig. 1C). Given the sequence conservation between the active sites (Fig. 1A), it is therefore likely that these thiols encounter similar electrostatic environments and therefore have similar pKa values. The pKa of a cysteine sulfhydryl describes its propensity to exist in the protonated form versus the negatively charged thiolate form. A lower pKa, below that of free cysteine (8.3), reflects a tendency towards the thiolate anion form and thus stronger nucleophilic character and reactivity.
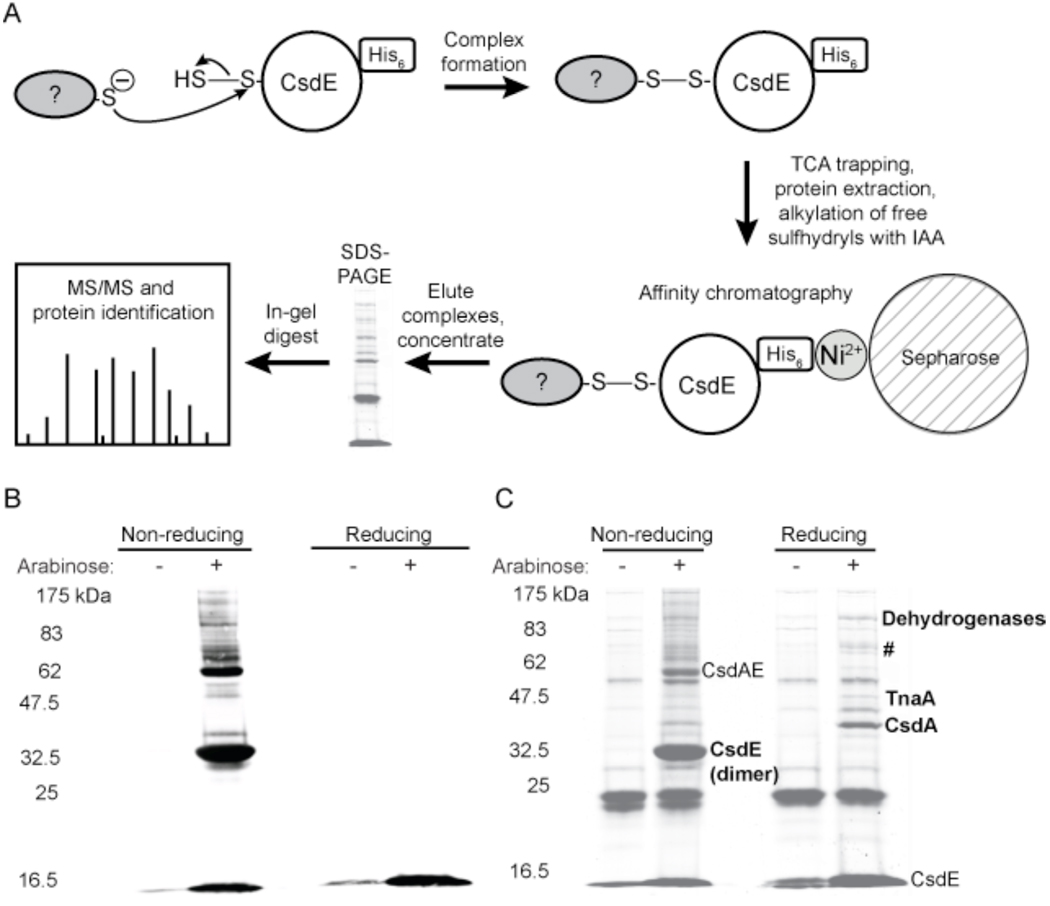
Figure 1. CsdE and SufE display significant sequence and structural homology.
A, primary sequence alignment of E. coli CsdE and SufE made using ClustalW2. Identical residues are shaded. The bars indicate the residues that line the cavity containing the conserved persulfide-forming Cys61 (CsdE) and Cys51 (SufE) (denoted with a star). B, structural alignment of E. coli CsdE and SufE. The ribbon diagram shows CsdE in cyan and SufE in green, with the side-chains of the conserved persulfide-forming Cys61 (CsdE) and Cys51 (SufE) presented in stick representation with carbon and sulfur atoms colored green and yellow, respectively. The CsdE solution structure (PDB 1NI7) 36 and the SufE crystal structure (PDB 1MZG) 37 were aligned with MacPyMOL. The structures superimpose with a root-mean-square deviation of 2.0 Angstroms for 132 Ca atoms. C, the active site containing the conserved persulfide-forming Cys61 (CsdE) and Cys51 (SufE) is formed by a loop and α-helices 3 and 6.
Unlike the SUF system, however, the CSD system is largely uncharacterized. The biological role of the system and the path of the sulfur atom are unknown. Furthermore, by analogy to the ISC and SUF systems, it is expected that the CSD system utilizes as yet unidentified scaffold and ATC proteins. In this study we sought to characterize the CSD system of E. coli by determining the pKa of C61 in CsdE and by identifying the proteins that interact with CsdE. We found that CsdE C61 has a lowered pKa value of 6.5, which is nearly identical to that of SufE C51 (Bolstad and Wood, 2010, submitted), and which is likely critical for its sulfur acceptor function. Using methodology we previously developed and applied to the SUF system (Bolstad and Wood, 2010, submitted), we observed that CsdE forms disulfide bonds with multiple proteins and identified the proteins that copurify with CsdE. Of these proteins, we predict that a specific subset are heretofore-unknown players within the CSD system and that the Fe-S cluster proteins are among the targets of the CSD system. Our data support the two-pathway model proposed by Trotter et al. 17. Furthermore, by comparison with our previous analysis of SufE (Bolstad and Wood, 2010, submitted), we show that there is some overlap in the CsdE and SufE interactomes.
MATERIALS AND METHODS
Chemicals
Iodoacetamide (IAA) was purchased from Sigma, dithiothreitol (DTT) from Fisher, and beta-mercaptoethanol (BME) from BioRad. (2-Pyridyl)-dithiobimane (PDT-bimane) was purchased from Toronto Research Chemicals. Other chemicals were of the highest quality commercially available.
Strains and Media
Protein was expressed in TOP10 (Invitrogen). All of the bacterial growth was in Lennox Broth (10 g tryptone, 5 g yeast extract, 5 g NaCl/liter) with 100 µg/ml ampicillin.
Plasmid Construction
The csdAE operon was subcloned from E.coli K12 MG1655 genomic DNA and NcoI and HindIII sites were introduced at the 5’ and 3’ ends, respectively. The csdAE product was cloned into the pCR 2.1-TOPO Vector (Invitrogen) using a TOPO TA Cloning Kit (Invitrogen), isolated by digestion with NcoI and HindIII, and ligated into the pBAD/Myc-His C vector (Invitrogen) in frame with the C-terminal Myc-His tag. Electrically competent TOP10 (Invitrogen) cells were transformed with the resulting plasmid, creating the CsdAE strain. The presence of the insert was confirmed by restriction digest.
Protein Expression and Purification
A single colony of TOP10 containing the CsdAE expression construct was used to inoculate 60 ml of Lennox Broth containing 100 µg/ml ampicillin. Cultures were grown overnight with shaking at 37 °C. The following day the cultures were diluted to an OD600 of 0.1 in 4 l fresh medium and grown to mid-log phase. The expression of CsdAE was induced by addition of 0.2% w/v L-arabinose for 3 h. The cells were pelleted by centrifugation and the pellets were frozen at −80 °C. The pellets were defrosted, refrozen, and defrosted again before being resuspended in ice-cold extraction buffer (50 mM sodium phosphate (pH 8), 300 mM NaCl, 5 mM MgCl2, 10% glycerol, Protease Inhibitor Cocktail (EDTA-free, Roche), 10 mM BME). Cells were lysed by sonication and cell debris cleared by centrifugation. The soluble fraction was diluted 1:1 with buffer A (50 mM sodium phosphate (pH 8), 300 mM NaCl, 10 mM imidazole, 10% glycerol, 5 mM BME). Imidazole was added to the extract to give a final concentration of 10 mM imidazole. The extract was applied to a 5 ml HisTrap FF Ni2+-affinity column (GE Healthcare) on an AKTA purifier fast protein liquid chromatography system (FPLC) (Amersham Biosciences) and washed with 10 column volumes buffer A and 10 column volumes 9% buffer B (50 mM sodium phosphate (pH 8), 300 mM NaCl, 250 Mm imidazole, 10% glycerol, 5 mM BME). The bound protein was eluted with 100% buffer B. The eluted protein was yellow due to the pyridoxal-5'-phosphate cofactor on CsdA. 10 mM EDTA was added to the eluted protein to prevent thiol oxidation and the protein was stored overnight at 4 °C. CsdA and CsdE copurify during Ni2+-affinity chromatography, but the proteins can be separated by size exclusion chromatography under reducing conditions. The protein was therefore concentrated with an Amicon Ultra-4 centrifugal device (5-kDa cutoff), treated with 100 mM DTT for 1 h at room temperature, and further purified with a Superdex 75 column (Amersham Biosciences) using a buffer consisting of 25 mM Tris (pH 7.4), 200 mM NaCl, 1 mM EDTA, and 10 mM BME. The CsdE peak was concentrated with an Amicon (5-kDa cutoff) and dialyzed against 25 mM Tris (pH 7.4),100 mM NaCl, 1 mM EDTA. The concentration of the purified protein was determined with the BCA assay (Pierce) and the protein was stored at −80 °C.
pKa Determination of SufE Sulfhydryls with PDT-Bimane
Reaction between PDT-bimane and a cysteine sulfhydryl releases pyridine-2-thione, the formation of which can be monitored by absorbance at 343 nm 38. This reaction has been used previously to determine the pKa of cysteine residue sulfhydryls in proteins 39, 40. All experiments were performed under aerobic conditions. Purified CsdE-(His)6 protein was reduced with 50 mM DTT for 1 h at 25 °C and exchanged into 25 mM potassium phosphate (pH 6.0), 50 mM NaCl,1 mM EDTA using a 5 ml HiTrap FF desalting column (GE Healthcare) operating on an AKTA purifier FPLC (Amersham Biosciences). The reduced protein was concentrated with an Amicon Ultra-4 centrifugal device (10-kDa cutoff) and diluted to 7 µM in sodium citrate or phosphate buffers spanning the pH range 5–9. 150 µl reactions were commenced in a UV-transparent 96-well microplate (Costar) by addition of PDT-bimane to a final concentration of 80 µM followed by rapid mixing, and A343 nm was monitored over 120 min with a Varian Cary 50 Bio UV-visible spectrophotometer and 50 MPR microplate reader. The resulting A343 nm curves were fit to first (Y=Y0 + Ae−x/t¹) or second order (Y=Y0 + Ae−x/t¹ + Be−x/t²) exponential decay functions. At higher pH values, the reaction is biphasic, proceeding rapidly for a brief period and then much more slowly for the remainder of the assay. It is these biphasic curves that are best fit by a second order versus a first order exponential decay function. The t1 values for the first and second order exponential decay functions were plotted against pH. The resulting curve was fit to the Henderson-Hasselbalch equation, from which the pKa value was determined.
Large-Scale Complex Purification and Visualization
A single colony of TOP10 carrying the CsdAE expression construct was used to inoculate 30–40 ml of Lennox Broth containing 100 µg/ml ampicillin. The culture was grown overnight with shaking at 37 °C. The following day the culture was diluted to an OD600 of 0.07–0.13 in 0.5–2 l fresh medium and grown to mid-log phase. Protein expression was induced by addition of 0.2% w/v L-arabinose for 1–2 h. The cells were pelleted by centrifugation at 4 °C. Disulfide-linked protein complexes were rapidly trapped by resuspension of the pellets in 20–30 ml ice-cold 20% w/v trichloroacetic acid (TCA). Safety note: when using TCA, eye protection, lab coat, and gloves should be worn and bench surface should be protected; save media containing TCA as hazardous waste for collection by Environmental Health and Safety personnel. The cells were pelleted again and the pellets were frozen at –80 °C. The pellets were defrosted, and the cells lysed by agitation with 5–20 ml 20% w/v TCA and 1.5–3 ml glass beads. The beads were washed with 5% w/v TCA and the wash solutions were combined with the protein solution. The TCA-precipitated protein was pelleted by centrifugation and the pellet washed three times with 5 ml acetone. The pellets were air-dried and then resuspended in 5–10 ml buffer containing 100 mM Tris (pH 8), 1% SDS, and 75 mM IAA and reacted in the dark for 20–90 min at 37 °C. The insoluble protein was pelleted by centrifugation, resuspended in 3–5 ml IAA buffer, incubated in the dark for 10–20 min at 37 °C, spun, and the soluble fractions were combined. The protein was diluted 1:3–1:5 with buffer A (50 mM sodium phosphate (pH 8), 300 mM NaCl, 10 mM imidazole, 10% glycerol) and imidazole was added to the diluted extract to give a final concentration of 10 mM imidazole. The sample was applied to a 5 ml HisTrap FF Ni2+-affinity column (GE Healthcare) on an AKTA purifier FPLC system (Amersham Biosciences) and washed with 10 column volumes buffer A and 10 column volumes 9% buffer B (50 mM sodium phosphate (pH 8), 300 mM NaCl, 250 mM imidazole, 10% glycerol). The bound protein was eluted with 100% buffer B. Protein concentration was determined by the BCA assay (Pierce). The protein samples were concentrated with an Amicon Ultra-4 centrifugal device (5–10–kDa cutoff), and when necessary, further concentrated by precipitation with methanol and chloroform. The purified samples were analyzed with reducing and nonreducing SDS-PAGE (8% (37.5:1) acrylamide gels were constructed for Fig. 3B and C whereas a pre-cast acrylamide gradient gel (Bio-Rad Ready Gel, 4–15% Tris-HCl) was used for Fig. 4A), and the gels were either stained with Coomassie Brilliant Blue (CBB) (GelCode Blue, Pierce) or transferred to nitrocellulose membranes. CsdE-(His)6 was visualized with HRP-linked anti-His6 antibodies (Clontech) and a FujiFilm LSA-3000 imaging system.
Figure 3. Determination of protein interactions involving CsdE.
A, Schematic representation of the methodology used to trap, purify, and identify protein complexes containing CsdE-(His)6. Disulfide-bound protein complexes form when a cysteine thiol on an interacting protein makes a nucleophilic attack on the bridging sulfur of the CsdE persulfide. Complexes formed within E. coli cells expressing CsdAE-(His)6 are trapped and extracted in the presence of TCA, free sulfhydryls are alkylated with IAA, and the extract is applied to a Ni2+ affinity column. The eluted proteins are concentrated, separated by reducing and nonreducing SDS-PAGE, digested by in-gel proteolysis, and analyzed with either MALDI-TOF/TOF or LC-MS/MS. The mass spectrometry data is searched against an E. coli proteomic database to identify the proteins that interact with CsdE. B and C, characterization of protein complex formation and identification of proteins that interact with CsdE. Arabinose was used to induce protein expression in the CsdAE-(His)6 E. coli strain and protein extracts were prepared as described above, resolved by nonreducing and reducing SDS-PAGE, and then transferred to nitrocellulose for immunoblotting with anti-His6 antibodies (B) or stained for total protein (C). The CsdE monomer is observed at ~15 kDa, and under nonreducing conditions the CsdE dimer can be observed at ~32.5 kDa and the predicted CsdE-CsdA complex can be observed at ~62 kDa. Under nonreducing conditions, complexes containing CsdE are observed in the immunoblot (B). These complexes disappear under reducing conditions, indicating that the complexes are bound by disulfide bonds. When stained for total protein (C), the proteins that copurify with CsdE are observed. These proteins were identified by in-gel digest and MALDI-TOF/TOF mass spectrometry (bold). #=pyruvate dehydrogenase (AceF), threonine-tRNA ligase (ThrS), transketolase 1 (TktA), BolA, YdfW, and Q1RE36_ECOUT. The dehydrogenases include SucA, AdhE, AceE, hypothetical proteins ECs3276, Q8FGG9_ECOL6, Q1RBX2_ECOUT, and Q1RCI6_ECOUT.
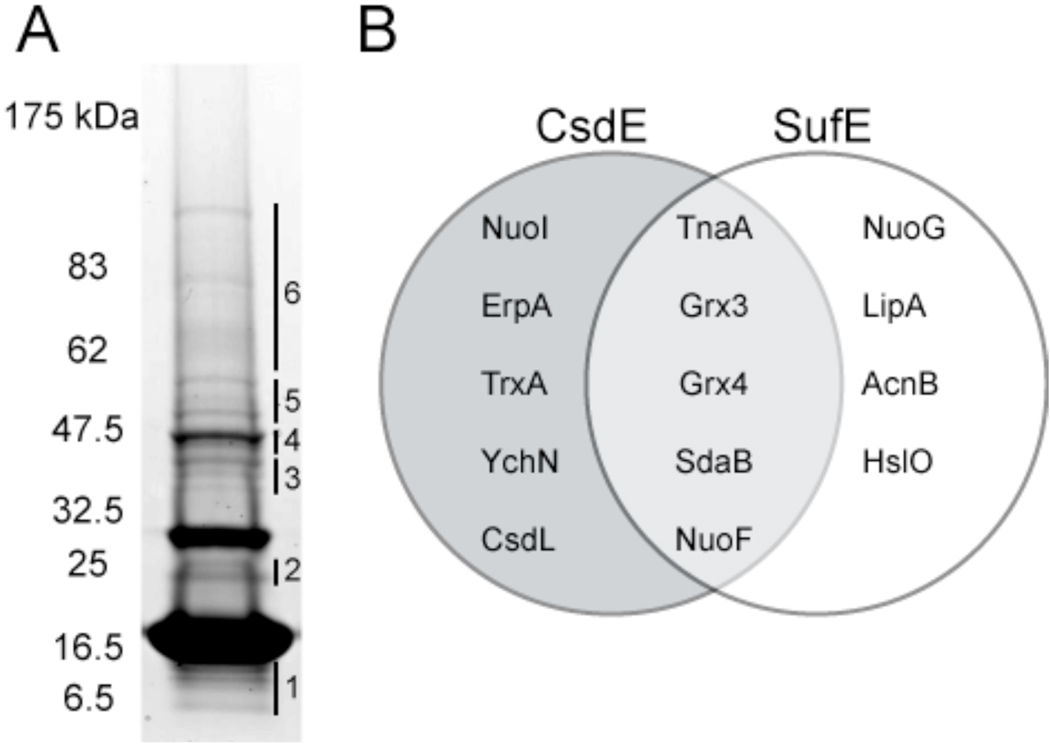
Figure 4. Identification of CsdE interactome by LC-MS/MS mass spectrometry and comparison with SufE interactome.
A, TCA-prepared protein extracts from cells expressing CsdAE-(His)6 were purified by Ni2+ affinity chromatography, resolved by reducing SDS-PAGE, and stained with CBB. The CsdE monomer is observed at ~16.5 kDa. The gel was subsectioned and in-gel proteolysis followed by LC-MS/MS was performed on each section. CsdA was identified in section #4. SI Tables S2 and S3 list all 116 proteins that were identified with a minimum of 2 peptides and 95% confidence in the subsections. Table 1 presents the proteins of particular interest that were identified. B, Comparison between the CsdE and SufE interactomes (SufE interactome presented in Bolstad and Wood, 2010, submitted). Proteins were identified by LC-MS/MS. Only those proteins that represent interactions of predicted significance are shown.
In-gel Proteolysis
Protein bands of interest were excised from the SDS-PAGE gel using a sterile blade. Bands were destained with two washes of 0.5 ml 50/50 0.1 M NH4HCO3/methanol over a period of 1 h at 30 °C. Bands were washed with 0.5 ml 0.1 M NH4HCO3 for 1 h at 30 °C followed by 0.5 ml 50/50 acetonitrile (ACN)/0.1 M NH4HCO3 for 60–80 min at 30 °C. Bands were shrunk with 50–100 µl ACN for 10–15 min at 25 °C and then dried at 50 °C. The bands were reswelled with 10–20 µl 25 mM NH4HCO3 containing 0.02 µg/µl reductively methylated 41 trypsin (Worthington Biochemical Co.) for 10 min at 25 °C. 25 mM NH4HCO3 was added to completely cover the bands, and they were incubated overnight at 37 °C. The following day, the supernatant (SUP1) was transferred to a clean tube. Peptides were extracted by addition of 30–60 µl 50/50 ACN/5% trifluoroacetic acid (TFA) to the bands followed by 10 min of sonication. This extraction solution was combined with SUP1. 30–60 µl 95/5 ACN/5% TFA was added to the bands and sonicated as before. This second extraction solution was added to SUP1, and the bulk solution concentrated with a SpeedVac to near-dryness (~5 µl). The peptides were resuspended in 20–30 µl 0.1% TFA.
Protein Identification by MALDI-TOF/TOF MS
Peptides were desalted with C18 ZipTips (Millipore). ZipTips were wetted with 50/50 ACN/0.1% TFA and equilibrated with 0.1% TFA, after which the peptides were bound to the tip with five passes of the peptide solution through the ZipTip. The ZipTip was washed 2X with 0.1% TFA, and the peptides were eluted with 3–5 passes of 3 µl 50/50 ACN/0.1% TFA. Two 0.5 µl aliquots of the peptide solution were spotted on a MALDI target (Applied Biosystems), with the first aliquot left to dry before the next was spotted, and then 0.5 µl of 6 mg/ml α-cyano-4-hydroxycinnamic acid (Applied Biosystems 4700 Proteomics Analyzer Mass Standards Kit) in 50/50 ACN/0.1% TFA was spotted. Applied Biosystems 4700 Proteomics Analyzer Mass Standards Kit calibration standard was diluted 1:10 with 6 mg/ml α-cyano-4-hydroxycinnamic acid (Applied Biosystems 4700 Proteomics Analyzer Mass Standards Kit) in 50/50 ACN/0.1% TFA, and 2 × 0.5 µl was spotted on each calibration spot. The MALDI target was analyzed using Applied Biosystems 4000 Series Explorer software and an Applied Biosystems 4700 Proteomics Analyzer (Applied Biosystems Inc., Foster City, CA) with TOF/TOF Optics. MALDI-MS data were collected in the m/z range of 700–4000 using 1125 shots per spectrum, and where possible, trypsin autolysis peaks of m/z 842.510, 1045.600, 2211.105, and 2239.136 Da were used as internal calibrants. The top 10 precursor peptides (excluding trypsin autolysis peaks) above a S/N threshold of 10 were selected for MS/MS. Data were analyzed with Applied Biosystems GPS Explorer Software v 3.6 using a Gel-based Workflow, with a precursor tolerance of 100 ppm, MS/MS fragment tolerance 0.3 Da, variable modifications: carbamidomethyl (C) and oxidation (M), a peptide charge of +1 and a maximum of two missed trypsin cleavages. Precursor peptides resulting from trypsin autolysis were excluded from the analysis. MSDB (September 2006, 7078992 total entries) database searches of the E. coli proteome were performed on all MS and MS/MS data sets using GPS Explorer Workstation v3.6 software, and a probability-based Mascot score >57 (equivalent to p ≤ 0.05) was considered statistically significant.
Protein Identification by LC-MS/MS
Peptides were submitted to the Genome Center Proteomics Core at the University of California, Davis, for mass spectrometry (LC-MS/MS) based protein identification. Protein identification was performed using a Michrom Paradigm HPLC and CTC Pal autosampler (Michrom, Auburn, CA) coupled to an LTQ ion trap mass spectrometer (Thermo-Scientific, San Jose, CA) through a Michrom ADVANCE Plug and Play Nano Spray Source. Peptides were desalted onto an Agilent nanotrap (Zorbax 300SB-C18, Agilent Technologies) and then eluted from the trap and separated by a 200 mm × 15 cm Michrom Magic C18 AQ column. Peptides were eluted using a 40 min gradient of 2–80% buffer B (buffer A = 0.1% formic acid, buffer B = 95% ACN/0.1% formic acid). The top 10 ions in each survey scan were subjected to automatic low-energy CID. Tandem mass spectra were extracted by BioWorks version 3.3. Charge state deconvolution and deisotoping were not performed. All MS/MS samples were analyzed using X! Tandem (www.thegpm.org; version TORNADO (2008.02.01.2)). X! Tandem was set up to search Uniprot Protein Knowledgebase Organism# 83333 (E-coli Strain K12) (May 2009, 4383 entries) assuming the digestion enzyme trypsin. X! Tandem was searched with a fragment ion mass tolerance of 0.4 Da and a parent ion tolerance of 1.8 Da. Iodoacetamide derivative of cysteine was specified in X! Tandem as a fixed modification. Deamidation of asparagine and glutamine, oxidation of methionine and tryptophan, sulphone of methionine, tryptophan oxidation to formylkynurenin of tryptophan and acetylation of the N-terminus were specified in X! Tandem as variable modifications. Scaffold (version Scaffold_2_02_03, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm 42. Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm 43. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
RESULTS AND DISCUSSION
Analysis of the pKa of the CsdE Cysteine Sulfhydryl
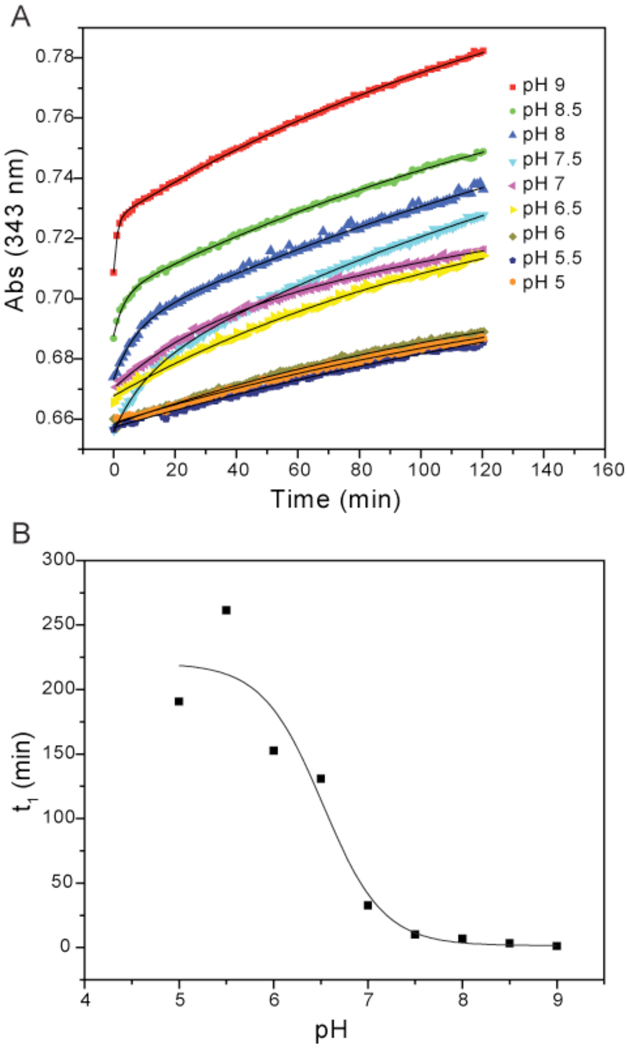
Since CsdE C61 accepts sulfur via nucleophilic attack on the persulfide intermediate of CsdA 29, we hypothesized that it would have a lowered pKa. To determine the pKa of the CsdE C61 sulfhydryl we used a thiol reactivity assay described previously for cysteine sulfhydryl pKa determination 39, 40. Briefly, the chemical PDT-bimane is reacted with a protein containing a single cysteine residue over a range of pH values 38. The course of the reaction is monitored at 343 nm and the resulting curves are fit to a first or second order exponential decay function. We performed this analysis on purified CsdE protein over a range of pH values from 5 to 9 (Fig. 2A). These data show that the initial rate of the reaction between C61 and PDT-bimane increases as the pH increases and C61 is deprotonated. The t1 values for the curves were plotted as a function of pH (Fig. 2B), fit to the Henderson-Hasselbalch equation, and a pKa of 6.5 was determined. This value is almost two pH units lower than the pKa of free cysteine and shows that >88% of C61 would be in the negatively charged and highly nucleophilic thiolate form at physiological pH. Given the high degree of sequence and structural conservation in the active sites of CsdE and SufE, it is not surprising that this value is nearly identical to the pKa value of 6.3 determined for the homologous SufE C51 (Bolstad and Wood, 2010, submitted). These lowered pKa values serve the basis for the propensity of the active site cysteines to make a nucleophilic attack on the persulfide of the corresponding cysteine desulfurase. Several factors can contribute to a lowered pKa. It was hypothesized that an ion pair between SufE C51 and the sole charged residue in the SufE active site - R119 (R129 in CsdE) – serves to stabilize the thiolate anion 37. However, mutation of R119 did not affect the pKa of SufE C51 (Bolstad and Wood, 2010, submitted), supporting the idea that this conserved arginine residue instead mediates binding between the active site surfaces of CsdE/SufE and CsdA/SufS through charge-charge interactions 36, 37.
Figure 2. pKa determination of CsdE C61 with PDT-bimane.
A, reaction of CsdE with PDT-bimane was monitored at 343 nm at pH values ranging from 5 to 9. The increase at 343 nm results from the release of pyridyl-2-thione from PDT-bimane. Each curve was fit to either a first or second order exponential function (solid black line), and the rate constants were determined. B, inverse of the initial rate constant (t1) was plotted as a function of pH. The results were fit to the Henderson-Hasselbalch equation, and from these curve fits a sulfhydryl pKa value of 6.5 was determined for CsdE C61.
In Vivo Characterization of Protein-Protein Interactions Involving CsdE and Identification by MALDI-TOF/TOF
Disulfide-mediated complex formation between CsdE and other proteins was examined in vivo with a large-scale trapping and purification procedure (Fig. 3A) that was previously used to identify protein interactions within the SUF system of E. coli (Bolstad and Wood, 2010, submitted). This procedure allows for the identification of proteins that bind to a His6-tagged protein of interest and potentially members of multiprotein complexes containing the protein of interest. In the aforementioned manuscript we demonstrate the method’s ability to identify physiologically relevant interactions, though interactions that are likely nonspecific are identified as well. It is important to note, however, that the method does not reveal whether the identified interactions are functional, inhibitory, or nonspecific. The value of the method is therefore in its use as a screening tool to reveal potentially novel interactions, which must then be further characterized by biochemical methods.
As CsdA and CsdE are expressed from an operon, and CsdE is likely to interact with other proteins via transfer of the transiently bound sulfur atom derived from CsdA 29, we created an E. coli strain in which both proteins are expressed and CsdE bears a C-terminal myc-His6 tag. Note that since CsdA is also overexpressed, it may transfer multiple sulfur atoms to CsdE C51, and the subsequent reaction of this polysulfide CsdE species with another Cys-containing protein may result in a polysulfide linkage rather than a disulfide bond. L-arabinose was used to induce overexpression of CsdAE in E. coli and disulfide-bonded protein complexes were trapped by treatment of the cells with TCA. TCA-prepared protein extracts were purified with Ni2+ affinity chromatography and resolved by nonreducing and reducing SDS-PAGE, after which gels were transferred to nitrocellulose for immunoblotting with anti-His6 antibodies (Fig. 3B) or stained with CBB (Fig. 3C). The fully reduced CsdE monomer is observed at ~16.5 kDa and the disulfide-bonded CsdE dimer can be observed at ~32.5 kDa under nonreducing conditions. The immunoblot shows the complexes containing CsdE (Fig. 3B). Trace amounts of CsdE monomer are evident in the uninduced lanes, evidence that minimal ara promoter activity exists under basal conditions. In the induced lanes under nonreducing conditions, there were observable disulfide-bonded complexes between CsdE and unknown proteins (Fig. 3B). These complexes disappeared under reducing conditions, demonstrating that the protein complexes were formed via disulfide bonds. The complex composed of native CsdA and CsdE-myc-His6 is predicted to be ~62 kDa, and thus the intense band at ~62 kDa is predicted to be the CsdA-CsdE complex. The presence of the other complexes suggests that CsdE interacts with other unknown proteins. One of these is predicted to be the recently identified 29 kDa CsdL protein, which was shown to interact with CsdE and to receive sulfur from CsdA 17. Whether CsdL participates in a CsdAEL complex, as proposed by Trotter et al. 17, or a CsdEL complex, is unknown, and thus the complex containing CsdL may be ~48 kDa (CsdEL) or ~91 kDa (CsdAEL). The CBB-stained gel (Fig. 3C) shows the proteins that copurify with CsdE. The bands in the uninduced lanes represent proteins that bind nonspecifically to the column. The dark band at ~16.5 kDa in the uninduced lanes appears to be the CsdE monomer, but is in fact the dye front and contains unresolved low molecular weight proteins (<16.5 kDa). This is confirmed by the immunoblot (Fig. 3B), in which the uninduced lanes contain only a slight amount of CsdE monomer. Upon induction, the intensity of the 16.5 kDa band increases, confirming the expression of CsdE. The 32.5 kDa CsdE dimer is also evident in the induced lane under nonreducing conditions, as is the predicted CsdA-CsdE complex at ~57 kDa. The bands that are unique to the induced, nonreducing lane or found at greater levels when compared to the uninduced lane represent complexes containing CsdE. This is demonstrated by the fact that the unique and upregulated bands in the induced, nonreducing lane are also present in the induced, nonreducing lane of the immunoblot (Fig. 3B). The only exception is the band at ~40 kDa, which is not observed on the immunoblot. The complexes disappear under reducing conditions, indicating that the complexes are formed by disulfide bonds. Similarly, those bands that are unique to the induced, reducing lane or found at greater levels when compared to the uninduced lane represent proteins that copurify with CsdE when CsdAE is expressed. The latter proteins were identified (p ≤ 0.05) by in-gel proteolysis and MALDI-TOF/TOF mass spectrometry and are shown in bold (see SI Table S1 for more information). As a positive control, the 32.5 kDa band, predicted to be the CsdE dimer, was indeed identified as CsdE. CsdA was identified at ~40 kDa, and therefore it was CsdA that was present at ~40 kDa in the induced, nonreducing CBB lane but was not evident on the immunoblot. Due to reduction of the CsdA-CsdE complex, the intensity of the CsdA band is greater in the reducing lane as compared to the nonreducing lane. Tryptophanase (TnaA) was identified at ~47.5 kDa (see discussion below). The ~70 kDa band (marked with an #) contained the following proteins: dihydrolipoyllysine-residue acetyltransferase component E2 of the pyruvate dehydrogenase complex (AceF), threonyl-tRNA synthetase (ThrS), transketolase 1 (TktA), the general stress response gene protein BolA (see discussion below) 44, the uncharacterized protein YdfW in the Qin prophage region, and hypothetical protein Q1RE36_ECOUT. The band containing these proteins is not believed to represent a single multiprotein complex but rather contains multiple unresolved bands. The copurification of most of these proteins likely results from nonspecific interactions with CsdE. Multiple proteins which also likely represent nonspecific interactions were identified at ~114 kDa, including oxoglutarate dehydrogenase (SucA), aldehyde-alcohol dehydrogenase (AdhE), pyruvate dehydrogenase subunit E1 (AceE), the putative virulence protein/transposase hypothetical protein ECs3276, hypothetical protein Q8FGG9_ECOL6, hypothetical protein Q1RBX2_ECOUT, and hypothetical protein Q1RCI6_ECOUT. SucA is a recognized contaminant of immobilized metal affinity chromatography 45, and nonspecific interactions with CsdE C61 may be explained by the presence of a pair of redox active cysteines in the active site of AceF 46 and the nucleophilic active site cysteine of aldehyde dehydrogenase.
Identification of Protein-Protein Interactions Involving CsdE by LC-MS/MS
To enhance our ability to identify interactions between CsdE and low abundance proteins, we used LC-MS/MS for protein identification instead of MALDI-TOF/TOF because the former provides greater sensitivity. A large-scale purification was performed with cells expressing CsdAE. The affinity purified extract was resolved by reducing SDS-PAGE and stained with CBB (Fig. 4A). The CsdE monomer is observed at ~16.5 kDa. Two bands of strong intensity and at least twelve bands of weak intensity were observed, just as in the identically-prepared sample in Fig. 3C (reducing, + arabinose lane). However, the separation among the bands is different between the figures because the sample in Fig. 4A was resolved on an acrylamide gradient gel, whereas those in Fig. 3C were resolved on a fixed-percentage gel. The gel was subsectioned (Fig. 4A), and in-gel proteolysis followed by LC-MS/MS was performed on each section. SI Tables S2 and S3 list all 116 proteins that were identified with a minimum of 2 peptides and 95% confidence in the subsections. Table 1 presents the proteins of particular interest that were identified. As evident in the tables, the proteins identified perform a multitude of functions. The identification of CsdE, CsdA, TnaA, AceF, ThrS, and AdhE by LC-MS/MS replicates the set of proteins identified by MALDI-TOF/TOF (Fig. 3C), further confirming that CsdE interacts with CsdA (band 4) and tryptophanase (band 5) in vivo. As summarized in Fig. 4B, some of the proteins identified were also found to interact with SufE (Bolstad and Wood, 2010, submitted), whereas others are unique to CsdE. The differences in the CsdE and SufE interactomes and the associated functions of the proteins may extend from their structural differences; though their structures align very closely, the polypeptide segment of CsdE helix II' does not adopt a helical conformation in SufE 36. Moreover, helices I and VI have different orientations in the two proteins.
TABLE 1.
Proteins of interest that interact with CsdE-(His)6a
| Number of Unique Peptidesb Gel Section |
||||||||
|---|---|---|---|---|---|---|---|---|
| Function | MW | Fe-S (Ref) | 1 | 2 | 3 | 4 | 5 | 6 |
| Iron-sulfur cluster assembly | ||||||||
| ErpA | 12 kDa | 3 | ||||||
| SufE | 16 kDa | 4 | 4 | 3 | 2 | 2 | ||
| CsdA | 43 kDa | 4 | 18 | 12 | 30 | 20 | 8 | |
| CsdE | 16 kDa | 65 | 59 | 43 | 25 | 32 | 11 | |
| IscS | 45 kDa | 4 | ||||||
| Porphyrin metabolism | ||||||||
| HemL | 45 kDa | 6 | ||||||
| Cellular redox homeostasis | ||||||||
| GrxC | 9 kDa | 2 | ||||||
| GrxD | 13 kDa | Yes (64,65) | 4 | |||||
| TrxA | 12 kDa | 3 | ||||||
| AhpC | 21 kDa | 2 | 16 | |||||
| Oxidative phosphorylation | ||||||||
| NuoI | 21 kDa | Yes (52) | 2 | |||||
| NuoF | 49 kDa | Yes (51) | 2 | |||||
| Gluconeogenesis | ||||||||
| SdaB | 49 kDa | Yes (53) | 6 | |||||
| Amino acid metabolism | ||||||||
| TnaA | 53 kDa | 2 | 4 | 5 | 27 | 3 | ||
| Uncharacterized | ||||||||
| YchN | 13 kDa | 2 | ||||||
| CsdL | 29 kDa | 2 | ||||||
Protein identification probability is 100% for all proteins except for CsdL, gel section 3, where it is 98%
All peptides identified with ≥95% confidence
One unique interaction that we identified is that between CsdE and the E1-like (ubiquitin-activating-like) CsdL (formerly YgdL) protein. Interestingly, csdL lies directly downstream of csdAE, yet is transcribed in the opposite direction. CsdL was completely uncharacterized until, during preparation of this manuscript, it was independently shown to interact with CsdE by both copurification and yeast two-hybrid procedures and to receive sulfur from CsdA in a CsdE-independent manner 17. E. coli contains two other E1-like proteins, MoeB and ThiF, which are necessary for incorporation of sulfur into the precursors of molybdopterin and thiamin, respectively 30–34. It was therefore suggested that CsdE brings CsdA and CsdL together for synthesis of a currently unidentified sulfur-containing molecule, or that CsdE serves as a sulfur relay between CsdA and CsdL, though the biochemical data suggests that this role could be dispensable 17. A model was proposed in which the CSD system engages in two sulfur transfer pathways, one dedicated to the biosynthesis of Fe-S clusters and the other to biosynthesis of a putative thiolated metabolite 17. Our identification of the interaction between CsdE and CsdL – an interaction independently confirmed and coupled with biochemical evidence for a functional interaction between CsdL and CsdA – further demonstrates that our method has the ability to capture physiologically relevant interactions.
Another protein that is unique to the CsdE interactome is the uncharacterized YchN protein. YchN contains a probable active-site region that harbors two conserved cysteine residues in a C-X-X-C motif47. YchN also shares the same three dimensional fold as the DsrE protein, which is essential for the oxidation of reduced sulfur compounds in photo- and chemolithoautotrophic bacteria 48, and the TusD protein, which is part of the sulfur relay system that synthesizes 2-thiouridine in bacterial tRNAs 49. The Dsr proteins have been suggested to be part of a sulfur trafficking system, in which the cysteine residue of DsrE that is conserved in both YchN and TusD plays a pivotal role 48, 50. In the Tus system, IscS provides the sulfur atom to TusA, which transfers it to TusD in the TusBCD complex 49. YchN may therefore play a sulfur transfer role within the CSD system, and by analogy with the Tus system, the sulfur atom would be transferred from CsdA to CsdE and then to YchN. The identification of YchN provides further credence to the proposal by Trotter et al. that the CSD system participates not only in Fe-S cluster biogenesis but also in a sulfur transfer pathway dedicated to biosynthesis of an as yet unknown thiolated metabolite 17.
Several Fe-S and Fe-S cluster assembly proteins were identified, supporting the other arm of the two-pathway hypothesis. The Fe-S proteins identified are NADH:ubiquinone oxidoreductase, chain F (NuoF) 51, NADH:ubiquinone oxidoreductase, chain I (NuoI) 52, and the predicted Fe-S protein L-serine dehydratase 2 (SdaB) 53. NuoF and SdaB were also found to interact with SufE (Bolstad and Wood, 2010, submitted), whereas NuoI was not. The identification of these proteins may suggest a role for the CSD system in the biosynthesis of their Fe-S clusters. As a member of the dehydratase enzyme family, SdaB contains a particularly labile [4Fe-4S] cluster, in which a solvent-exposed, noncysteine-liganded Fe atom binds and activates substrate. This makes the cluster unusually vulnerable to oxidation and loss of the Fe atom 54–57, and would necessitate cluster repair and/or de novo synthesis under both oxidative stress and basal conditions, possibly explaining an interaction with the Csd proteins. Another protein that we identified, and which is unique to CsdE, is the recently discovered A-type Fe-S protein ErpA 23. ErpA is essential for E. coli growth under both aerobiosis and anaerobiosis due to its role in the maturation of the Fe-S proteins IspG/H 23, which synthesize the quinone precursor molecule isopentenyl diphosphate 58, 59. ErpA was found to assemble [2Fe-2S] and [4Fe-4S] clusters and to transfer the clusters to apoIspG. Unlike the other ATCs of E. coli (IscA and SufA), ErpA maps at a distance from any other Fe-S cluster biogenesis-related gene, and thus there is interest in determining the Fe-S cluster assembly system with which it associates. Furthermore, the erpA gene is in a chromosomal region that is particularly rich in iron uptake-related genes and IscR represses its expression under anaerobic conditions 20, suggesting a role for ErpA in iron homeostasis. ErpA has been proposed to act as an Fe-S cluster shuttle for the SUF and ISC systems, transferring formed Fe-S clusters from these systems to apoproteins 24. This model, however, does not consider the role of the CSD system nor the potential for interaction between ErpA and this system. The present results lead us to suggest that in addition to a role in the SUF and ISC systems, ErpA may serve as a scaffold or shuttle protein for the CSD system. SufE and IscS were also identified. The identification of SufE may be due to a proposed interaction between SufE and CsdA 17 and the fact that CsdA forms a dimer 29. The CsdA dimer may bind one molecule each of SufE and CsdE, the latter of which would bind to the Ni2+ column. CsdE does not alter the activity of IscS 29 and thus any interaction between the two is likely nonspecific.
Glutaredoxin 3 (Grx3) and glutaredoxin 4 (Grx4), which were also found to interact with SufE (Bolstad and Wood, 2010, submitted), and thioredoxin-A (TrxA), were identified. Until recently, the observed stimulation of Fe-S cluster assembly and transfer by Grx and Trx was thought to be via their reduction of protein and mixed protein-glutathione disulfides, which would free up key cysteine residues so that they may bind iron, sulfur, and intact clusters 60, 61, 62. It was recently discovered, however, that homodimers of some prokaryotic and eukaryotic Grx isoforms, including EcGrx4 and its homologs, coordinate Fe-S clusters, and the eukaryotic EcGrx4 homolog Grx5 is necessary for Fe-S cluster assembly 63–70 (also reviewed in Rouhier et al., 2009; Bandyopadhyay et al., 2008; Herrero and de la Torre-Ruiz, 2007) 71–73. Data suggests that Grx5 has a role in the mitochondrial ISC system, perhaps as a scaffold or Fe-S cluster transfer protein, and that its function is conserved among its homologs, including EcGrx4 63, 66, 69, 70, 74–77. Among prokaryotes, the phylogenetic profile of Grx5 homologues is similar to that of some ISC system proteins, implying that prokaryotic Grx5 orthologues are also required for Fe-S cluster assembly 76. Monothiol Grx like Grx5 are also found in or near putative microbial nif, suf, isc, and molybdenum cofactor synthesis operons 73. Currently, the working hypothesis is that monothiol Grx (e.g., EcGrx4) act as Fe-S cluster assembly scaffolds and/or transfer proteins, whereas the clusters on dithiol Grx (e.g., EcGrx3) are sensors of oxidative stress, releasing active monomeric Grx upon oxidative degradation of the cluster. Our results may therefore suggest that EcGrx4 serves as a previously unidentified scaffold and/or transfer protein for the SUF and CSD systems, and that the latter systems assemble a cluster on EcGrx3. Interestingly, a genetic interaction was recently observed between EcGrx4 and ISC system proteins, suggesting a role for EcGrx4 in the ISC system 78. EcGrx4 may therefore have a general role in Fe-S cluster assembly within E. coli, a role that is consistent with the synthetic lethality of the grx4 null mutation and its upregulation by iron depletion 79, 80. Interestingly, the uncharacterized protein BolA was identified by MALDI-TOF/TOF (Fig. 3C). Bioinformatic data, including gene proximity and cooccurrence, predicts that BolA proteins interact with monothiol Grxs, 73, 81, 82 and indeed, yeast Grx3 and Grx4 were found to interact with BolA 83. Chloroplast SufE contains a C-terminal sequence homologous to the conserved BolA sequence 84 and several proteobacteria contain a protein made up of three domains: a Grx, an ATC-like domain that lacks the conserved Cys that bind Fe-S clusters, and a sulfurtransferase 85. Collectively, the available data points to a functional interaction between BolA, Grx, and SufE-type proteins such as CsdE.
Tryptophanase (TnaA), which was also found to interact with SufE (Bolstad and Wood, 2010, submitted), was identified. Metabolism of cysteine by tryptophanase produces hydrogen sulfide 86, which could be transferred to the CsdE and SufE sulfur acceptor proteins. Tryptophanase may therefore serve as an alternate source of sulfur for the CSD and SUF systems. Like the cysteine desulfurases that provide sulfur for sulfur trafficking systems, tryptophanase is a PLP-dependent enzyme, and based on its sequence, belongs to the same family of PLP-dependent enzymes as NifS, the cysteine desulfurase responsible for biosynthesis of the nitrogenase Fe-S clusters 87, 88. Furthermore, tnaA is one of the genes most upregulated upon superoxide and hydrogen peroxide exposure 15, 89, making it plausible that tryptophanase serves to augment the activity of the CsdA and SufS desulfurases during oxidative stress, when demand for new Fe-S clusters is increased. We are currently pursuing biochemical confirmation of this hypothesis.
It is possible that any accessible cysteine residue will react with the persulfide on CsdE, forming a disulfide-bonded protein complex that would be purified using our method. For those proteins that do not contribute to the function of the CSD system, this interaction is considered nonspecific and likely accounts for the identification of some of the proteins reported in this study, including the following proteins that contain redox- or catalytically-active cysteine residues: alkyl hydroperoxide reductase subunit C (AhpC) 90, methionine sulfoxide reductase (MsrB) 91, Hsp31 (HchA) 92, glyceraldehyde-3-phosphate dehydrogenase A (GapA) 93, and dihydrolipoyllysine-residue acetyltransferase component E2 of the pyruvate dehydrogenase complex (AceF) 46. However, the fact that we identified some proteins that interact only with CsdE and not SufE and vice versa is evidence that our method is capable of identifying functional interactions and not just those between the protein of interest and abundant proteins with accessible thiols. Furthermore, published literature and homology helps to identify the interactions that are likely functional, weeding out the interactions that are likely nonspecific and increasing the value of the method as a screening tool for protein interactions within sulfur trafficking systems. Nevertheless, indiscriminate disulfide bond formation may occur, but if this were the case we would predict that more proteins with reactive cysteine residues would have been identified. Examples include the peroxiredoxin Bcp, the glutathione peroxidase homolog BtuE, and the organic peroxide sensor OhrR 94, none of which were identified in our study. It may be that the expression level of these proteins precluded identification, whereas the abundance of the peroxiredoxin AhpC facilitated its identification. In contrast, we predict that the protein(s) that receives the persulfide sulfur on CsdE exhibits significant affinity for CsdE and would effectively compete with proteins that do not contribute to the function of the CSD system yet harbor an accessible cysteine thiol. For example, the homologous SufE protein specifically interacts with SufB – the sulfur recipient - and not SufA, even though SufA also has reactive cysteine residues 35. Furthermore, the surface surrounding the active site thiol of CsdE/SufE homologs is highly conserved, as is the corresponding surface on CsdA/SufS homologs 36, suggesting that the interacting surface on the recipient protein is also conserved and mediates a tight interaction with CsdE. In the crystal structure of SufE, which Fig. 1 shows overlaps significantly with the solution structure of CsdE, it was observed that the cavity containing the active site thiol is large enough to accommodate the perfsulfide group in a solvent-sequestered environment 37. Therefore, sulfur transfer to the recipient protein is likely mediated by a conformational change in SufE/CsdE that would likely be triggered only by a specific interaction with the recipient protein. Such a conformational change was predicted to to be required for SufE to receive sulfur from SufS 37.
Proteins that are known to bind nonspecifically to Ni2+ columns were also identified 45, including carbonic anhydrase 2 (Can), catabolite gene activator protein (Crp), host factor-I protein (Hfq), FKBP-type peptidyl-prolyl cis-trans isomerase (SlyD), 30S ribosomal protein S15 (RpsO), 60 kDa chaperonin (GroL), and dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex (SucB). These proteins bind to the Ni2+ column via native metal binding sites, surface histidine residues, or through an uncharacterized mechanism. We previously found that the YqjI protein - which was also identified - binds nonspecifically to Ni2+ columns (Bolstad and Wood, 2010, submitted), most likely as a result of its predicted affinity for positively charged metalloids and significant histidine content. The bands containing YqjI (band 2) are present in both the uninduced and induced samples (Fig. 3C), further evidence that YqjI binds nonspecifically to Ni2+ columns. A heat shock-like response occurs during overexpression of recombinant proteins 95, inducing the expression of chaperone proteins, which are common contaminants of Ni2+ affinity chromatography 45. The chaperones identified therefore likely represent nonspecific interactions, though the ISC system contains the HscBA chaperone pair, which catalyzes the transfer of Fe-S clusters to apoproteins 22, and thus it’s possible that the CSD system also employs chaperone proteins. Translation elongation factors, ribosomal proteins, and aminoacyl-tRNA synthetases are often identified in proteomics work, and are likely an artifact of protein overexpression.
As mentioned previously, Trotter et al. first demonstrated the interaction between CsdE and CsdL 17. In the same analysis, they also found that CsdE interacts, unsurprisingly, with CsdA, as well as ribose-phosphate pyrophosphokinase (Prs) and DnaK (Hsp 70). Of these four proteins, we identified all but DnaK, which may be due to differences in experimental design. Trotter et al. expressed CsdE with an N-terminal TAP tag whereas we expressed CsdAE in which CsdE had a C-terminal myc-His6 tag. Thus, purification of the protein complexes required different methods. Furthermore, Trotter et al. did not trap the cells with acid nor alkylate free cysteine residues prior to purification, as we did. It is possible that CsdE and DnaK interact via some mechanism other than a disulfide bond, and that the acid, or the SDS in the alkylation buffer, disrupt this interaction, thereby preventing purification of the CsdE-DnaK complex.
In conclusion, we have discovered potential new players in the CSD Fe-S cluster biogenesis system of E. coli. Our results point to the value of our approach, in which recombinant protein systems are screened in a high throughput manner for protein-protein interactions, thereby revealing novel interactions that may then be confirmed by in vitro and in vivo methods. Along this vein, we are currently pursuing biochemical confirmation of the interactions involving tryptophanase and ErpA. The cloning and characterization of the YchN protein, as well as confirmation of the interactions of YchN, Grx3, and Grx4 with CsdE (and SufE where applicable) would help to further characterize the CSD system. Though we have demonstrated that there is some overlap in the CsdE and SufE interactomes, it is the differences that likely enable the CSD and SUF systems to execute unique functions and to possibly mature unique subsets of Fe-S proteins. Defining the mechanism and biological role of the CSD system and the recipients of the Fe-S clusters produced by the ISC, CSD, and SUF systems remain significant challenges.
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded by a grant from the American Heart Association (0635328N). H. M. Bolstad was funded by a NIEHS Training Grant in Environmental Toxicology (ES007059). We thank Rudy Alvarado and Brett Phinney of the UC Davis Proteomics Core for LC-MS/MS analysis and help with data analysis. We thank William Jewell of the UC Davis Molecular Structure Facility and Page Spiess for help with MALDI-TOF/TOF analysis. We thank Christina Takanishi and Nicholas Kettenhofen for their insightful comments and Gopal Lalchandani for his contributions.
Abbreviations
- CSD
cysteine sulfinate desulfinase
- Fe-S
iron-sulfur
- ATC
A-type carrier protein
- IAA
iodoacetamide
- DTT
dithiothreitol
- BME
beta-mercaptoethanol
- PDT-bimane
(2-Pyridyl)-dithiobimane
- FPLC
fast protein liquid chromatograph
- TCA
trichloroacetic acid
- HRP
horseradish peroxidase
- CBB
Coomassie Brilliant Blue
- ACN
acetonitrile
- TFA
trifluoroacetic acid
Footnotes
SUPPORTING INFORMATION AVAILABLE
Table S1 (Fig. 3C), Table S2 (Fig. 4A), and Table S3 (Fig. 4A) contain proteomic parameters for the proteins identified. Table S1 (Fig. 3C) contains accession numbers, molecular weight, protein score, number nonredundant peptides, protein score confidence interval, and sequence coverage. Table S2 (Fig. 4A) groups the proteins by function, presents protein molecular weight, and identifies the proteins containing Fe-S clusters. Table S3 (Fig. 4A) contains accession numbers, molecular weight, protein identification probability, quantitative value, sequence coverage, and number unique peptides. This information is available free of charge via the Internet at http://pubs.acs.org/.
REFERENCES
- 1.Mueller EG. Trafficking in persulfides: delivering sulfur in biosynthetic pathways. Nature chemical biology. 2006;2(4):185–194. doi: 10.1038/nchembio779. [DOI] [PubMed] [Google Scholar]
- 2.Lill R, Muhlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annual review of biochemistry. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 3.Rincon-Enriquez G, Crete P, Barras F, Py B. Biogenesis of Fe/S proteins and pathogenicity: IscR plays a key role in allowing Erwinia chrysanthemi to adapt to hostile conditions. Molecular microbiology. 2008;67(6):1257–1273. doi: 10.1111/j.1365-2958.2008.06118.x. [DOI] [PubMed] [Google Scholar]
- 4.Runyen-Janecky L, Daugherty A, Lloyd B, Wellington C, Eskandarian H, Sagransky M. Role and regulation of iron-sulfur cluster biosynthesis genes in Shigella flexneri virulence. Infect Immun. 2008;76(3):1083–1092. doi: 10.1128/IAI.01211-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huet G, Daffe M, Saves I. Identification of the Mycobacterium tuberculosis SUF machinery as the exclusive mycobacterial system of [Fe-S] cluster assembly: evidence for its implication in the pathogen's survival. Journal of bacteriology. 2005;187(17):6137–6146. doi: 10.1128/JB.187.17.6137-6146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nachin L, Loiseau L, Expert D, Barras F. SufC: an unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. The EMBO journal. 2003;22(3):427–437. doi: 10.1093/emboj/cdg061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nachin L, El Hassouni M, Loiseau L, Expert D, Barras F. SoxR-dependent response to oxidative stress and virulence of Erwinia chrysanthemi: the key role of SufC, an orphan ABC ATPase. Molecular microbiology. 2001;39(4):960–972. doi: 10.1046/j.1365-2958.2001.02288.x. [DOI] [PubMed] [Google Scholar]
- 8.Flint DH. Escherichia coli contains a protein that is homologous in function and N-terminal sequence to the protein encoded by the nifS gene of Azotobacter vinelandii and that can participate in the synthesis of the Fe-S cluster of dihydroxy-acid dehydratase. J Biol Chem. 1996;271(27):16068–16074. [PubMed] [Google Scholar]
- 9.Patzer SI, Hantke K. SufS is a NifS-like protein, and SufD is necessary for stability of the [2Fe-2S] FhuF protein in Escherichia coli. Journal of bacteriology. 1999;181(10):3307–3309. doi: 10.1128/jb.181.10.3307-3309.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mihara H, Maeda M, Fujii T, Kurihara T, Hata Y, Esaki N. AnifS-like gene, csdB encodes an Escherichia coli counterpart of mammalian selenocysteine lyase Gene cloning, purification, characterization and preliminary x-ray crystallographic studies. J Biol Chem. 1999;274(21):14768–14772. doi: 10.1074/jbc.274.21.14768. [DOI] [PubMed] [Google Scholar]
- 11.Mihara H, Kurihara T, Yoshimura T, Soda K, Esaki N. Cysteine sulfinate desulfinase a NIFS-like protein of Escherichia coli with selenocysteine lyase and cysteine desulfurase activities Gene cloning, purification, and characterization of a novel pyridoxal enzyme. J Biol Chem. 1997;272(36):22417–22424. doi: 10.1074/jbc.272.36.22417. [DOI] [PubMed] [Google Scholar]
- 12.Mihara H, Kurihara T, Yoshimura T, Esaki N. Kinetic and mutational studies of three NifS homologs from Escherichia coli: mechanistic difference between L-cysteine desulfurase and L-selenocysteine lyase reactions. J Biochem. 2000;127(4):559–567. doi: 10.1093/oxfordjournals.jbchem.a022641. [DOI] [PubMed] [Google Scholar]
- 13.Fontecave M, Ollagnier-de-Choudens S. Iron-sulfur cluster biosynthesis in bacteria: Mechanisms of cluster assembly and transfer. Archives of biochemistry and biophysics. 2008;474(2):226–237. doi: 10.1016/j.abb.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Urban A, Mihara H, Leimkuehler S, Kurihara T, Esaki N. IscS functions as a primary sulfur-donating enzyme by interacting specifically with MoeB and MoaD in the biosynthesis of molybdopterin in Escherichia coli. J Biol Chem. 2009 doi: 10.1074/jbc.M109.082172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. Journal of bacteriology. 2001;183(15):4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Outten FW, Djaman O, Storz G. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Molecular microbiology. 2004;52(3):861–872. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- 17.Trotter V, Vinella D, Loiseau L, Ollagnier de, Choudens S, Fontecave M, Barras F. The CsdA cysteine desulphurase promotes Fe/S biogenesis by recruiting Suf components and participates to a new sulphur transfer pathway by recruiting CsdL (ex-YgdL), a ubiquitin-modifying-like protein. Molecular microbiology. 2009;74(6):1527–1542. doi: 10.1111/j.1365-2958.2009.06954.x. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz CJ, Giel JL, Patschkowski T, Luther C, Ruzicka FJ, Beinert H, Kiley PJ. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(26):14895–14900. doi: 10.1073/pnas.251550898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee KC, Yeo WS, Roe JH. Oxidant-responsive induction of the suf operon, encoding a Fe-S assembly system, through Fur and IscR in Escherichia coli. Journal of bacteriology. 2008;190(24):8244–8247. doi: 10.1128/JB.01161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giel JL, Rodionov D, Liu M, Blattner FR, Kiley PJ. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Molecular microbiology. 2006;60(4):1058–1075. doi: 10.1111/j.1365-2958.2006.05160.x. [DOI] [PubMed] [Google Scholar]
- 21.Yeo WS, Lee JH, Lee KC, Roe JH. IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Molecular microbiology. 2006;61(1):206–218. doi: 10.1111/j.1365-2958.2006.05220.x. [DOI] [PubMed] [Google Scholar]
- 22.Bonomi F, Iametti S, Morleo A, Ta D, Vickery LE. Studies on the mechanism of catalysis of iron-sulfur cluster transfer from IscU[2Fe2S] by HscA/HscB chaperones. Biochemistry. 2008;47(48):12795–12801. doi: 10.1021/bi801565j. [DOI] [PubMed] [Google Scholar]
- 23.Loiseau L, Gerez C, Bekker M, Ollagnier-de Choudens S, Py B, Sanakis Y, Teixeira de, Mattos J, Fontecave M, Barras F. ErpA, an iron sulfur (Fe S) protein of the A-type essential for respiratory metabolism in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(34):13626–13631. doi: 10.1073/pnas.0705829104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vinella D, Brochier-Armanet C, Loiseau L, Talla E, Barras F. Iron-sulfur (Fe/S) protein biogenesis: phylogenomic and genetic studies of A-type carriers. PLoS Genet. 2009;5(5):e1000497. doi: 10.1371/journal.pgen.1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chahal HK, Dai Y, Saini A, Ayala-Castro C, Outten FW. The SufBCD Fe-S scaffold complex interacts with SufA for Fe-S cluster transfer. Biochemistry. 2009;48(44):10644–10653. doi: 10.1021/bi901518y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loiseau L, Ollagnier-de-Choudens S, Nachin L, Fontecave M, Barras F. Biogenesis of FeS cluster by the bacterial Suf system: SufS and SufE form a new type of cysteine desulfurase. J Biol Chem. 2003;278(40):38352–38359. doi: 10.1074/jbc.M305953200. [DOI] [PubMed] [Google Scholar]
- 27.Ollagnier-de-Choudens S, Lascoux D, Loiseau L, Barras F, Forest E, Fontecave M. Mechanistic studies of the SufS-SufE cysteine desulfurase: evidence for sulfur transfer from SufS to SufE. FEBS letters. 2003;555(2):263–267. doi: 10.1016/s0014-5793(03)01244-4. [DOI] [PubMed] [Google Scholar]
- 28.Outten FW, Wood MJ, Munoz FM, Storz G. The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe-S cluster assembly in Escherichia coli. J Biol Chem. 2003;278(46):45713–45719. doi: 10.1074/jbc.M308004200. [DOI] [PubMed] [Google Scholar]
- 29.Loiseau L, Ollagnier-de Choudens S, Lascoux D, Forest E, Fontecave M, Barras F. Analysis of the heteromeric CsdA-CsdE cysteine desulfurase, assisting Fe-S cluster biogenesis in Escherichia coli. J Biol Chem. 2005;280(29):26760–26769. doi: 10.1074/jbc.M504067200. [DOI] [PubMed] [Google Scholar]
- 30.Sambasivarao D, Turner RJ, Bilous PT, Rothery RA, Shaw G, Weiner JH. Differential effects of a molybdopterin synthase sulfurylase (moeB) mutation on Escherichia coli molybdoenzyme maturation. Biochem Cell Biol. 2002;80(4):435–443. doi: 10.1139/o02-131. [DOI] [PubMed] [Google Scholar]
- 31.Lake MW, Wuebbens MM, Rajagopalan KV, Schindelin H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature. 2001;414(6861):325–329. doi: 10.1038/35104586. [DOI] [PubMed] [Google Scholar]
- 32.Leimkuhler S, Wuebbens MM, Rajagopalan KV. Characterization of Escherichia coli MoeB and its involvement in the activation of molybdopterin synthase for the biosynthesis of the molybdenum cofactor. J Biol Chem. 2001;276(37):34695–34701. doi: 10.1074/jbc.M102787200. [DOI] [PubMed] [Google Scholar]
- 33.Taylor SV, Kelleher NL, Kinsland C, Chiu HJ, Costello CA, Backstrom AD, McLafferty FW, Begley TP. Thiamin biosynthesis in Escherichia coli. Identification of this thiocarboxylate as the immediate sulfur donor in the thiazole formation. J Biol Chem. 1998;273(26):16555–16560. doi: 10.1074/jbc.273.26.16555. [DOI] [PubMed] [Google Scholar]
- 34.Xi J, Ge Y, Kinsland C, McLafferty FW, Begley TP. Biosynthesis of the thiazole moiety of thiamin in Escherichia coli: identification of an acyldisulfide-linked protein--protein conjugate that is functionally analogous to the ubiquitin/E1 complex. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(15):8513–8518. doi: 10.1073/pnas.141226698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Layer G, Gaddam SA, Ayala-Castro CN, Ollagnier-de Choudens S, Lascoux D, Fontecave M, Outten FW. SufE transfers sulfur from SufS to SufB for iron-sulfur cluster assembly. J Biol Chem. 2007;282(18):13342–13350. doi: 10.1074/jbc.M608555200. [DOI] [PubMed] [Google Scholar]
- 36.Liu G, Li Z, Chiang Y, Acton T, Montelione GT, Murray D, Szyperski T. High-quality homology models derived from NMR, X-ray structures of E. coli proteins YgdK and Suf E suggest that all members of the YgdK/Suf E protein family are enhancers of cysteine desulfurases. Protein Sci. 2005;14(6):1597–1608. doi: 10.1110/ps.041322705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldsmith-Fischman S, Kuzin A, Edstrom WC, Benach J, Shastry R, Xiao R, Acton TB, Honig B, Montelione GT, Hunt JF. The SufE sulfur-acceptor protein contains a conserved core structure that mediates interdomain interactions in a variety of redox protein complexes. Journal of molecular biology. 2004;344(2):549–565. doi: 10.1016/j.jmb.2004.08.074. [DOI] [PubMed] [Google Scholar]
- 38.Mansoor SE, Farrens DL. High-throughput protein structural analysis using site-directed fluorescence labeling and the bimane derivative (2-pyridyl)dithiobimane. Biochemistry. 2004;43(29):9426–9438. doi: 10.1021/bi036259m. [DOI] [PubMed] [Google Scholar]
- 39.Hong M, Fuangthong M, Helmann JD, Brennan RG. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Molecular cell. 2005;20(1):131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 40.Ma LH, Takanishi CL, Wood MJ. Molecular mechanism of oxidative stress perception by the Orp1 protein. J Biol Chem. 2007;282(43):31429–31436. doi: 10.1074/jbc.M705953200. [DOI] [PubMed] [Google Scholar]
- 41.Rice RH, Means GE, Brown WD. Stabilization of bovine trypsin by reductive methylation. Biochim Biophys Acta. 1977;492(2):316–321. doi: 10.1016/0005-2795(77)90082-4. [DOI] [PubMed] [Google Scholar]
- 42.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 43.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 44.Santos JM, Freire P, Vicente M, Arraiano CM. The stationary-phase morphogene bolA from Escherichia coli is induced by stress during early stages of growth. Molecular microbiology. 1999;32(4):789–798. doi: 10.1046/j.1365-2958.1999.01397.x. [DOI] [PubMed] [Google Scholar]
- 45.Bolanos-Garcia VM, Davies ORStructural analysis, classification of native proteins from E. coli commonly co-purified by immobilised metal affinity chromatography. Biochim Biophys Acta. 2006;1760(9):1304–1313. doi: 10.1016/j.bbagen.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 46.Wilkinson KD, Williams CH., Jr Evidence for multiple electronic forms of two-electron-reduced lipoamide dehydrogenase from Escherichia coli. J Biol Chem. 1979;254(3):852–862. [PubMed] [Google Scholar]
- 47.Shin DH, Yokota H, Kim R, Kim SH. Crystal structure of a conserved hypothetical protein from Escherichia coli. J Struct Funct Genomics. 2002;2(1):53–66. doi: 10.1023/a:1014450817696. [DOI] [PubMed] [Google Scholar]
- 48.Dahl C, Schulte A, Stockdreher Y, Hong C, Grimm F, Sander J, Kim R, Kim SH, Shin DH. Structural and molecular genetic insight into a widespread sulfur oxidation pathway. Journal of molecular biology. 2008;384(5):1287–1300. doi: 10.1016/j.jmb.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 49.Ikeuchi Y, Shigi N, Kato J, Nishimura A, Suzuki T. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Molecular cell. 2006;21(1):97–108. doi: 10.1016/j.molcel.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 50.Cort JR, Selan U, Schulte A, Grimm F, Kennedy MA, Dahl C. Allochromatium vinosum DsrC: solution-state NMR structure, redox properties, and interaction with DsrEFH, a protein essential for purple sulfur bacterial sulfur oxidation. Journal of molecular biology. 2008;382(3):692–707. doi: 10.1016/j.jmb.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohnishi T. Iron-sulfur clusters/semiquinones in complex I. Biochim Biophys Acta. 1998;1364(2):186–206. doi: 10.1016/s0005-2728(98)00027-9. [DOI] [PubMed] [Google Scholar]
- 52.Yakovlev G, Reda T, Hirst J. Reevaluating the relationship between EPR spectra and enzyme structure for the iron sulfur clusters in NADH:quinone oxidoreductase. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(31):12720–12725. doi: 10.1073/pnas.0705593104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hofmeister AE, Textor S, Buckel W. Cloning and expression of the two genes coding for L-serine dehydratase from Peptostreptococcus asaccharolyticus: relationship of the iron-sulfur protein to both L-serine dehydratases from Escherichia coli. Journal of bacteriology. 1997;179(15):4937–4941. doi: 10.1128/jb.179.15.4937-4941.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Imlay JA. Iron-sulphur clusters and the problem with oxygen. Molecular microbiology. 2006;59(4):1073–1082. doi: 10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- 55.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annual review of biochemistry. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 56.Keyer K, Imlay JA. Inactivation of dehydratase [4Fe-4S] clusters and disruption of iron homeostasis upon cell exposure to peroxynitrite. J Biol Chem. 1997;272(44):27652–27659. doi: 10.1074/jbc.272.44.27652. [DOI] [PubMed] [Google Scholar]
- 57.Flint DH, Allen RM. Ironminus signSulfur Proteins with Nonredox Functions. Chem Rev. 1996;96(7):2315–2334. doi: 10.1021/cr950041r. [DOI] [PubMed] [Google Scholar]
- 58.Grawert T, Kaiser J, Zepeck F, Laupitz R, Hecht S, Amslinger S, Schramek N, Schleicher E, Weber S, Haslbeck M, Buchner J, Rieder C, Arigoni D, Bacher A, Eisenreich W, Rohdich F. IspH protein of Escherichia coli: studies on iron-sulfur cluster implementation and catalysis. J Am Chem Soc. 2004;126(40):12847–12855. doi: 10.1021/ja0471727. [DOI] [PubMed] [Google Scholar]
- 59.Wolff M, Seemann M, Tse Sum, Bui B, Frapart Y, Tritsch D, Garcia Estrabot A, Rodriguez-Concepcion M, Boronat A, Marquet A, Rohmer M. Isoprenoid biosynthesis via the methylerythritol phosphate pathway: the (E)-4-hydroxy-3-methylbut-2-enyl diphosphate reductase (LytB/IspH) from Escherichia coli is a [4Fe-4S] protein. FEBS letters. 2003;541(1–3):115–120. doi: 10.1016/s0014-5793(03)00317-x. [DOI] [PubMed] [Google Scholar]
- 60.Achebach S, Tran QH, Vlamis-Gardikas A, Mullner M, Holmgren A, Unden G. Stimulation of Fe-S cluster insertion into apoFNR by Escherichia coli glutaredoxins 1, 2 and 3 in vitro. FEBS letters. 2004;565(1–3):203–206. doi: 10.1016/j.febslet.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 61.Ding H, Demple B. Thiol-mediated disassembly and reassembly of [2Fe-2S] clusters in the redox-regulated transcription factor SoxR. Biochemistry. 1998;37(49):17280–17286. doi: 10.1021/bi980532g. [DOI] [PubMed] [Google Scholar]
- 62.Ding H, Harrison K, Lu J. Thioredoxin reductase system mediates iron binding in IscA and iron delivery for the iron-sulfur cluster assembly in IscU. J Biol Chem. 2005;280(34):30432–30437. doi: 10.1074/jbc.M504638200. [DOI] [PubMed] [Google Scholar]
- 63.Kim KD, Chung WH, Kim HJ, Lee KC, Roe JH. Monothiol glutaredoxin Grx5 interacts with Fe-S scaffold proteins Isa1 and Isa2 and supports Fe-S assembly and DNA integrity in mitochondria of fission yeast. Biochem Biophys Res Commun. 392(3):467–472. doi: 10.1016/j.bbrc.2010.01.051. [DOI] [PubMed] [Google Scholar]
- 64.Iwema T, Picciocchi A, Traore DA, Ferrer JL, Chauvat F, Jacquamet L. Structural basis for delivery of the intact [Fe2S2] cluster by monothiol glutaredoxin. Biochemistry. 2009;48(26):6041–6043. doi: 10.1021/bi900440m. [DOI] [PubMed] [Google Scholar]
- 65.Picciocchi A, Saguez C, Boussac A, Cassier-Chauvat C, Chauvat F. CGFS-type monothiol glutaredoxins from the cyanobacterium Synechocystis PCC6803 and other evolutionary distant model organisms possess a glutathione-ligated [2Fe-2S] cluster. Biochemistry. 2007;46(51):15018–15026. doi: 10.1021/bi7013272. [DOI] [PubMed] [Google Scholar]
- 66.Bandyopadhyay S, Gama F, Molina-Navarro MM, Gualberto JM, Claxton R, Naik SG, Huynh BH, Herrero E, Jacquot JP, Johnson MK, Rouhier N. Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. The EMBO journal. 2008;27(7):1122–1133. doi: 10.1038/emboj.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Camaschella C, Campanella A, De Falco L, Boschetto L, Merlini R, Silvestri L, Levi S, Iolascon A. The human counterpart of zebrafish shiraz shows sideroblastic-like microcytic anemia and iron overload. Blood. 2007;110(4):1353–1358. doi: 10.1182/blood-2007-02-072520. [DOI] [PubMed] [Google Scholar]
- 68.Rodriguez-Manzaneque MT, Ros J, Cabiscol E, Sorribas A, Herrero E. Grx5 glutaredoxin plays a central role in protection against protein oxidative damage in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19(12):8180–8190. doi: 10.1128/mcb.19.12.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rodriguez-Manzaneque MT, Tamarit J, Belli G, Ros J, Herrero E. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol Biol Cell. 2002;13(4):1109–1121. doi: 10.1091/mbc.01-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wingert RA, Galloway JL, Barut B, Foott H, Fraenkel P, Axe JL, Weber GJ, Dooley K, Davidson AJ, Schmid B, Paw BH, Shaw GC, Kingsley P, Palis J, Schubert H, Chen O, Kaplan J, Zon LI. Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature. 2005;436(7053):1035–1039. doi: 10.1038/nature03887. [DOI] [PubMed] [Google Scholar]
- 71.Bandyopadhyay S, Chandramouli K, Johnson MK. Iron-sulfur cluster biosynthesis. Biochem Soc Trans. 2008;36(Pt 6):1112–1119. doi: 10.1042/BST0361112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Herrero E, de la, Torre-Ruiz MA. Monothiol glutaredoxins: a common domain for multiple functions. Cell Mol Life Sci. 2007;64(12):1518–1530. doi: 10.1007/s00018-007-6554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rouhier N, Couturier J, Johnson MK, Jacquot JP. Glutaredoxins: roles in iron homeostasis. Trends Biochem Sci. 2009 doi: 10.1016/j.tibs.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alves R, Herrero E, Sorribas A. Predictive reconstruction of the mitochondrial iron-sulfur cluster assembly metabolism II Role of glutaredoxin Grx5. Proteins. 2004;57(3):481–492. doi: 10.1002/prot.20228. [DOI] [PubMed] [Google Scholar]
- 75.Muhlenhoff U, Gerber J, Richhardt N, Lill R. Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. The EMBO journal. 2003;22(18):4815–4825. doi: 10.1093/emboj/cdg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vilella F, Alves R, Rodriguez-Manzaneque MT, Belli G, Swaminathan S, Sunnerhagen P, Herrero E. Evolution and cellular function of monothiol glutaredoxins: involvement in iron-sulphur cluster assembly. Comp Funct Genomics. 2004;5(4):328–341. doi: 10.1002/cfg.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Molina-Navarro MM, Casas C, Piedrafita L, Belli G, Herrero E. Prokaryotic and eukaryotic monothiol glutaredoxins are able to perform the functions of Grx5 in the biogenesis of Fe/S clusters in yeast mitochondria. FEBS letters. 2006;580(9):2273–2280. doi: 10.1016/j.febslet.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 78.Butland G, Babu M, Diaz-Mejia JJ, Bohdana F, Phanse S, Gold B, Yang W, Li J, Gagarinova AG, Pogoutse O, Mori H, Wanner BL, Lo H, Wasniewski J, Christopolous C, Ali M, Venn P, Safavi-Naini A, Sourour N, Caron S, Choi JY, Laigle L, Nazarians-Armavil A, Deshpande A, Joe S, Datsenko KA, Yamamoto N, Andrews BJ, Boone C, Ding H, Sheikh B, Moreno-Hagelseib G, Greenblatt JF, Emili A. eSGA: E. coli synthetic genetic array analysis. Nat Methods. 2008;5(9):789–795. doi: 10.1038/nmeth.1239. [DOI] [PubMed] [Google Scholar]
- 79.Fernandes AP, Fladvad M, Berndt C, Andresen C, Lillig CH, Neubauer P, Sunnerhagen M, Holmgren A, Vlamis-Gardikas A. A novel monothiol glutaredoxin (Grx4) from Escherichia coli can serve as a substrate for thioredoxin reductase. J Biol Chem. 2005;280(26):24544–24552. doi: 10.1074/jbc.M500678200. [DOI] [PubMed] [Google Scholar]
- 80.Gerdes SY, Scholle MD, Campbell JW, Balazsi G, Ravasz E, Daugherty MD, Somera AL, Kyrpides NC, Anderson I, Gelfand MS, Bhattacharya A, Kapatral V, D’Souza M, Baev MV, Grechkin Y, Mseeh F, Fonstein MY, Overbeek R, Barabasi AL, Oltvai ZN, Osterman AL. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. Journal of bacteriology. 2003;185(19):5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huynen MA, Spronk CA, Gabaldon T, Snel B. Combining data from genomes, Y2H and 3D structure indicates that BolA is a reductase interacting with a glutaredoxin. FEBS letters. 2005;579(3):591–596. doi: 10.1016/j.febslet.2004.11.111. [DOI] [PubMed] [Google Scholar]
- 82.Couturier J, Jacquot JP, Rouhier N. Evolution and diversity of glutaredoxins in photosynthetic organisms. Cell Mol Life Sci. 2009;66(15):2539–2557. doi: 10.1007/s00018-009-0054-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kumanovics A, Chen OS, Li L, Bagley D, Adkins EM, Lin H, Dingra NN, Outten CE, Keller G, Winge D, Ward DM, Kaplan J. Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J Biol Chem. 2008;283(16):10276–10286. doi: 10.1074/jbc.M801160200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ye H, Abdel-Ghany SE, Anderson TD, Pilon-Smits EA, Pilon M. CpSufE activates the cysteine desulfurase CpNifS for chloroplastic Fe-S cluster formation. J Biol Chem. 2006;281(13):8958–8969. doi: 10.1074/jbc.M512737200. [DOI] [PubMed] [Google Scholar]
- 85.Alves R, Vilaprinyo E, Sorribas A, Herrero E. Evolution based on domain combinations: the case of glutaredoxins. BMC Evol Biol. 2009;9:66. doi: 10.1186/1471-2148-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Newton WA, Snell EE. Catalytic Properties of Tryptophanase, a Multifunctional Pyridoxal Phosphate Enzyme. Proceedings of the National Academy of Sciences of the United States of America. 1964;51:382–389. doi: 10.1073/pnas.51.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alexander FW, Sandmeier E, Mehta PK, Christen P. Evolutionary relationships among pyridoxal-5'-phosphate-dependent enzymes. Regio-specific alpha, beta and gamma families. European journal of biochemistry / FEBS. 1994;219(3):953–960. doi: 10.1111/j.1432-1033.1994.tb18577.x. [DOI] [PubMed] [Google Scholar]
- 88.Zheng L, Dean DR. Catalytic formation of a nitrogenase iron-sulfur cluster. J Biol Chem. 1994;269(29):18723–18726. [PubMed] [Google Scholar]
- 89.Pomposiello PJ, Bennik MH, Demple B. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. Journal of bacteriology. 2001;183(13):3890–3902. doi: 10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ellis HR, Poole LB. Novel application of 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole to identify cysteine sulfenic acid in the AhpC component of alkyl hydroperoxide reductase. Biochemistry. 1997;36(48):15013–15018. doi: 10.1021/bi972191x. [DOI] [PubMed] [Google Scholar]
- 91.Olry A, Boschi-Muller S, Branlant G. Kinetic characterization of the catalytic mechanism of methionine sulfoxide reductase B from Neisseria meningitidis. Biochemistry. 2004;43(36):11616–11622. doi: 10.1021/bi049306z. [DOI] [PubMed] [Google Scholar]
- 92.Malki A, Caldas T, Abdallah J, Kern R, Eckey V, Kim SJ, Cha SS, Mori H, Richarme G. Peptidase activity of the Escherichia coli Hsp31 chaperone. J Biol Chem. 2005;280(15):14420–14426. doi: 10.1074/jbc.M408296200. [DOI] [PubMed] [Google Scholar]
- 93.D'Alessio G, Josse J. Glyceraldehyde phosphate dehydrogenase of Escherichia coli Structural and catalytic properties. J Biol Chem. 1971;246(13):4326–4333. [PubMed] [Google Scholar]
- 94.Poole LB, Karplus PA, Claiborne A. Protein sulfenic acids in redox signaling. Annu Rev Pharmacol Toxicol. 2004;44:325–347. doi: 10.1146/annurev.pharmtox.44.101802.121735. [DOI] [PubMed] [Google Scholar]
- 95.Durrschmid K, Marzban G, Durrschmid E, Striedner G, Clementschitsch F, Cserjan-Puschmann M, Bayer K. Monitoring of protein profiles for the optimization of recombinant fermentation processes using public domain databases. Electrophoresis. 2003;24(1–2):303–310. doi: 10.1002/elps.200390027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.