Abstract
Dicer initiates RNA interference by generating small RNAs involved in various silencing pathways. Dicer participates in centromeric silencing, but its role in the epigenetic regulation of other chromatin domains has not been explored. Here we show that Dicer1 deficiency in Mus musculus leads to decreased DNA methylation, concomitant with increased telomere recombination and telomere elongation. These DNA-methylation defects correlate with decreased expression of Dnmt1, Dnmt3a and Dnmt3b DNA methyltransferases (Dnmts), and methylation levels can be recovered by their overexpression. We identify the retinoblastoma-like 2 protein (Rbl2) as responsible for decreased Dnmt expression in Dicer1-null cells, suggesting the existence of Dicer-dependent small RNAs that target Rbl2. We identify the miR-290 cluster as being downregulated in Dicer1-deficient cells and show that it silences Rbl2, thereby controlling Dnmt expression. These results identify a pathway by which miR-290 directly regulates Rbl2-dependent Dnmt expression, indirectly affecting telomere-length homeostasis.
Dicer is an RNase III family nuclease that generates small RNAs involved in several silencing pathways through a mechanism known as RNA interference (RNAi)1–9. In Schizosaccharomyces pombe, Dicer generates small RNAs from pericentric repeats that initiate histone H3 lysine 9 (H3K9) methylation and heterochromatin formation at these domains3. Dicer and other members of the RNAi machinery have also been involved in telomere clustering and meiotic bouquet formation in yeast10,11. A role for Dicer in heterochromatin assembly in mammals is less well established. In particular, Dicer1-deficient mouse embryonic stem (ES) cells were initially described as having decreased DNA methylation and decreased H3K9 trimethylation (H3K9me3) at pericentric chromatin2. However, more recent reports are less supportive of a putative role for Dicer in the assembly of silenced chromatin domains1,7–9.
DNA methylation is one the best understood epigenetic modifications of the chromatin, with crucial roles in gene expression, imprinting, development and heterochromatin assembly12. In particular, both pericentric and subtelomeric heterochromatin domains are highly methylated13,14, and this epigenetic modification is thought to contribute to the repressive nature of these chromatin domains. DNA-methylation errors are also a common feature of cancer cells and are proposed to contribute to the transformed phenotype15. Three active DNA (cytosine-5-)methyltransferases have been identified in humans and mice—Dnmt1, Dnmt3a and Dnmt3b—all of which participate in the DNA methylation of heterochromatin domains such as pericentric and telomeric chromatin14,16–21. These heterochromatin domains are also enriched in histone modifications characteristic of silenced chromatin, such as H3K9me3 and trimethylated histone H4 lysine 20 (H4K20me3), and show binding of the CBX1, CBX3 and CBX5 homologs of Drosophila melanogaster heterochromatin protein 1 (HP1)13,14,22–26.
Telomeres are repeated elements at the ends of chromosomes that have an essential role in chromosomal stability27,28. Telomere length is maintained by telomerase, a reverse transcriptase that synthesizes telomeres at the chromosome ends27, and by the so-called alternative lengthening of telomeres, or ALT, mechanism29, which relies on homologous recombination events at telomeres30,31. Loss of either DNA methylation or histone trimethylation marks at mammalian telomeres results in increased telomere recombination and aberrant telomere elongation14,22–26. In addition to these epigenetic marks, telomeres originate long noncoding RNAs that can remain associated with the telomeric chromatin32,33 and seem to be important for telomere-length control33. Notably, production of these telomere-associated RNAs is influenced by the status of telomeric heterochromatin33, although a direct role for these RNAs in telomeric silencing is still unknown.
Here we set out to address a putative role for mammalian Dicer and Dicer-dependent small RNAs in the establishment of well-known repressive epigenetic marks such as DNA methylation and histone trimethylation. The findings described here show that abrogation of Dicer results in global DNA-methylation defects resulting from decreased expression of the Dnmts. These DNA-methylation defects include the subtelomeric regions, where they are accompanied by increased telomere recombination and aberrant telomere elongation. Dicer deficiency, however, did not lead to a decreased abundance of H3K9me3 and H4K20me3 marks or HP1 binding at these domains, arguing that Dicer, and therefore Dicer-dependent small RNAs, are not required for the establishment of silenced chromatin domains at these regions. Notably, we identify increased levels of the retinoblastoma-like protein 2 (Rbl2) as responsible for decreased Dnmt expression in Dicer1-null cells, suggesting the existence of microRNAs (miRNAs) that specifically target Rbl2, thereby controlling global DNA methylation. In this regard, we identify the mammalian-specific miR-290 cluster34,35 as being significantly downregulated in Mus musculus Dicer1-null cells and show that miRNAs from this cluster specifically silence Rbl2. Altogether, these results reveal the existence of a previously unknown regulatory pathway by which the highly conserved mammalian miR290 cluster regulates Rbl2 at the post-transcriptional level, leading to a transcriptional repression of Dnmt3a and Dnmt3b and the appearance of DNA-methylation defects. In turn, DNA-methylation defects lead to increased telomere recombination and to aberrant telomere elongation in Dicer1-null cells, demonstrating an indirect link between Dicer and telomere-length homeostasis.
RESULTS
Global DNA-methylation defects in Dicer1-null cells
To explore a role for Dicer in DNA methylation of different chromatin domains, we studied DNA-methylation levels at chromatin regions enriched in either ‘active’ (characterized by the presence of acetylated H3K9, AcH3K9) or ‘inactive’ (or ‘compacted’) chromatin marks (characterized by the presence of HP1, H3K9me3 and H4K20me3) using chromatin immunoprecipitation (ChIP)–immunoblot assays (Methods). As a positive control for DNA-methylation defects, we used Dnmt1−/− ES cells, which show decreased DNA methylation at all different chromatin domains except for the HP1-bound chromatin (Fig. 1a). Notably, DNA methylation was significantly decreased at H3K9me3- and H4K20me3-bound chromatin, as well as at AcH3K9-bound chromatin, in Dicer1-null cells compared to wild-type controls (P < 0.05 for all comparisons, Fig. 1a), suggesting a marked loss of DNA methylation at these chromatin domains in the absence of Dicer. Similarly to Dnmt1-deficient cells, DNA methylation at HP1-bound chromatin was not significantly decreased in Dicer1-null cells (Fig. 1a). These results were confirmed using the B1-SINE Cobra technique, which measures global DNA methylation as determined by methylation of the B1 SINE repeat (Methods). Again, Dicer1-null cells showed a global decrease in DNA methylation, similar to that seen in Dnmt1−/− cells (Fig. 1b). This extensive loss of DNA methylation in Dicer1-null cells suggests a global change in the state of the chromatin as a consequence of Dicer1 deletion.
Figure 1.
Defective DNA methylation in Dicer1-null cells. (a) 5-methyl cytosine (5MetCyt) abundance after chromatin immunoprecipitation (ChIP) with antibodies to the substrates indicated above the blots. Values were corrected by the abundance of these marks at pericentric repeats (left). Dnmt1−/− cells were used as a control. AcH3K9, acetylated H3 lysine 9; H4K20me3, trimethylated histone H3 lysine 20; H3K9me3, trimethylated histone H3 lysine 9; Het., heterozygous; HP1, heterochromatin protein 1. (b) Fraction of methylated B1 SINE repeat element in the indicated genotypes. Dnmt1−/− cells were used as control. WT, wild type. (c) Abundance of methylated CpG dinucleotides as determined by bisulfite sequencing. We analyzed 9–16 clones per genotype. Yellow and blue represent the frequency of methylated and unmethylated CpG dinucleotides, respectively. Grey corresponds to undetermined methylation. CpG, CpG position; U, unmethylated; M, methylated; NP, not present. Statistically significant differences are indicated with an asterisk.
We next studied DNA methylation specifically at heterochromatic domains such as subtelomeric repeats, where heterochromatin has a role in controlling telomere recombination and telomere length14. To this end, we quantified CpG methylation at two subtelomeric regions in mouse chromosomes 1 and 2 (refs. 25,26) using bisulfite sequencing (Methods). Whereas both subtelomeric regions are heavily methylated in wild-type cells, this modification is significantly decreased at both subtelomeric regions in the case of 27G5 Dicer1-null cells and at one subtelomeric region in the case of 27H10 Dicer1-null cells (Fig. 1c), suggesting DNA-methylation defects at subtelomeric regions in the absence of Dicer. Such defects, however, were not detected at these subtelomeric regions in Dicer1-heterozygous cells. Similarly, we did not detect obvious DNA-methylation defects at pericentric chromatin when using nonquantitative methylation-sensitive restriction enzyme analysis with HpaII and MspI isoschizomers8 (Supplementary Fig. 1a,b), although we cannot rule out differences in DNA methylation at these regions when using more sensitive techniques.
Next, we studied whether decreased DNA methylation in Dicer1-null cells was the direct consequence of decreased Dnmt levels. Real-time quantitative RT-PCR analysis showed a significant decrease in Dnmt1, Dnmt3a and Dnmt3b mRNA levels in Dicer1-null cells (Fig. 2a and Supplementary Methods). Western blot analysis confirmed decreased Dnmt1, Dnmt3a2 and Dnmt3b protein levels in Dicer1-null cells (Fig. 2b,c). Dnmt3a protein levels were undetectable by western blot in all genotypes (Fig. 2b; Dnmt3a,3b−/− cells reconstituted with the Dnmt3a enzyme were used as a control for mobility14). As a negative control, we used Dnmt1−/− and Dnmt3a,3b−/− cells (Fig. 2b,c). Altogether, these results indicate that Dicer abrogation results in decreased mRNA and protein levels of the three main Dnmts.
Figure 2.
Defective DNA methylation of Dicer1-null cells is corrected by Dnmt overexpression. (a) Quantification of Dnmt mRNA levels. (b) Representative western blots. Dnmt1- and Dnmt3a,3b-deficient cells were used as negative controls and Dnmt3a,3b-deficient cells were reconstituted with the Dnmt3a enzyme (Dnmt3a,3b−/−* Dnmt3a) as a control for Dnmt3a mobility in the gel. (c) Quantification of western blot results shown in part b. (d) Dnmt mRNA levels before or after expression of the indicated enzymes. (e) Fraction of methylated B1 SINE repeat element in two Dicer1-null cultures (27H10 and 27G5) before and after Dnmt overexpression. (f) Percentage of CpG methylation at the indicated subtelomeres before and after overexpression of the indicated Dnmts. Bisulfite sequencing of 2–16 individual clones was performed.
Dnmts rescues DNA methylation in Dicer1-null cells
Next, we addressed whether overexpression of Dnmt1, or both Dnmt3a and Dnmt3b, in Dicer1-null cells was able to recover DNA-methylation defects in these cells (Methods). Overexpression of Dnmt enzymes in Dicer1-null cells was confirmed by quantitative RT-PCR (Fig. 2d), and was sufficient to largely recover global DNA methylation as well as subtelomeric DNA methylation in these cells (Fig. 2e,f), suggesting that defective DNA methylation in Dicer1-null cells is the consequence of decreased Dnmt expression. Notably, expression profiling using the Agilent 4×44K mouse 60-mer oligonucleotide microarrays showed that the global pattern of differentially expressed genes in Dicer1-null cells was inverted upon introduction of either Dnmt1 or Dnmt3a and Dnmt3b (Supplementary Fig. 2), suggesting that defective DNA methylation may account for a large part of the gene-expression changes associated with Dicer ablation.
Dicer abrogation increases telomere recombination and length
Defective subtelomeric DNA methylation has been previously shown to result in increased telomere recombination and aberrant telomere elongation14. To determine whether subtelomeric DNA-methylation defects in Dicer1-null cells were sufficiently severe to trigger these phenotypes, we determined the frequency of telomeric sister chromatid exchange events (T-SCE) using chromosome orientation fluoresecence in situ hybridization (CO-FISH; Methods and Fig. 3a). Two-color CO-FISH showed that T-SCE is significantly increased in Dicer1-heterozygous cells compared to wild-type controls (Fig. 3a; χ2-test P < 0.0001) and even further increased in Dicer1-null cells (Fig. 3a; χ2-test P < 0.0001). These results demonstrate that abrogation of Dicer leads to augmented T-SCE events at telomeres concomitant with defective DNA methylation at subtelomeric repeats. Notably, overexpression of Dnmt1 or Dnmt3a and Dnmt3b enzymes in Dicer1-null cells restored telomere recombination to normal levels (Fig. 3b), indicating that defective DNA methylation is responsible for this phenotype.
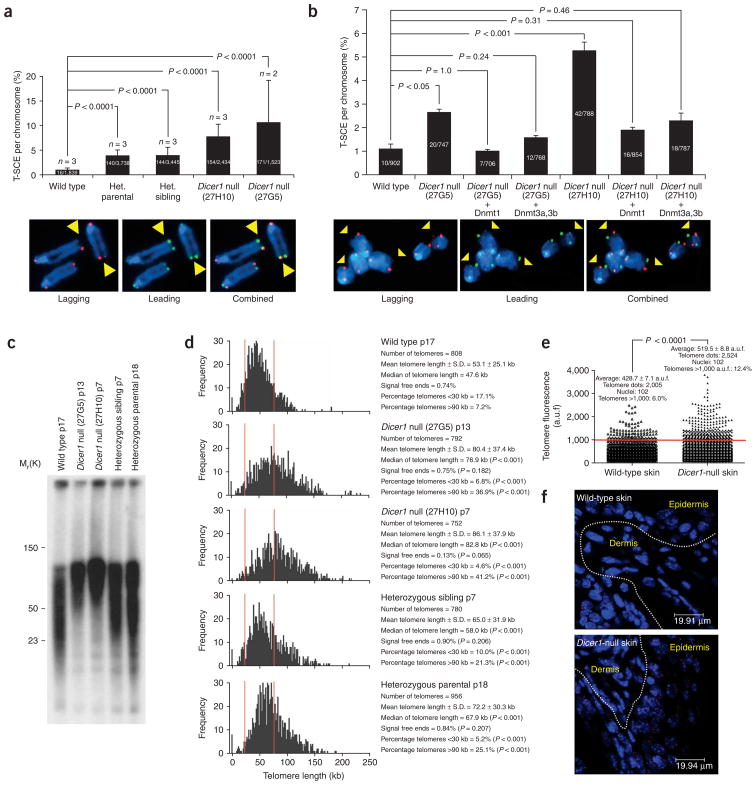
Figure 3.
Increased telomere recombination and aberrantly elongated telomeres in Dicer1-null cells. (a) Quantification of telomere recombination events (T-SCE) in the indicated genotypes. Error bars correspond to two or three independent experiments (n). The total number of T-SCE out of the total number of chromosomes analyzed per genotype is indicated. Representative chromosome orientation fluorescence in situ hybridization (CO-FISH) images after labeling leading (green) and lagging (red) strand telomeres are shown below. A T-SCE was considered positive only when observed with both probes and involving an unequal exchange of fluorescence (yellow arrows). Het., heterozygous (b) Quantification of T-SCE in the indicated genotypes before and after overexpression of Dnmt1 or Dnmt3a,3b. The experiment was performed in duplicate using two independent Dicer1-null cultures (27G5 and 27H10). The total number of T-SCE out of the total number of chromosomes analyzed per genotype is indicated. The results are not directly comparable to those shown in part a, as they correspond to different experiments. T-SCE events are indicated with yellow arrows. (c) Telomere restriction fragment (TRF) analysis in the indicated ES cells. (d) Telomere-length distribution in the indicated ES cells as determined by quantitative FISH (Q-FISH). p refers to the passage number of the ES cells. The vertical red lines highlight the longer and more widely scattered telomeres of Dicer1-heterozygous and Dicer1-null cells compared to wild-type controls. (e) Quantification of telomere fluorescence in tail skin sections of the indicated genotypes. More than 100 keratinocyte nuclei and 2,000 telomere dots per genotype were analyzed. a.u.f., arbitrary units of fluorescence. (f) Representative images of telomere fluorescence in skin sections from wild-type and Dicer1-null mice.
Activation of recombination-based ALTmechanisms have previously been shown to be accompanied by aberrant telomere elongation14,29–31. To address this, we next measured telomere length using Southern blot terminal restriction fragment (TRF) analysis in wild-type cells and in two independent Dicer1-null ES cell lines, as well as in parental and sibling heterozygous cultures (Methods). Digestion of genomic DNA with MboI releases a TRF that contains the telomeric TTAGGG repeats and part of the subtelomeric region. Dicer1-heterozygous ES cell cultures presented higher molecular weight TRFs than wild-type cells, and this was exacerbated in the case of Dicer1-null ES cells (Fig. 3c), suggesting that aberrant telomere elongation occurs in the absence of Dicer. To confirm TRF results with an independent method, we measured telomere length by quantitative telomere fluorescence in situ hybridization (Q-FISH) on metaphases, which allows accurate measurement of all individual telomeres (Methods). Again, Dicer1-heterozygous ES cells showed significantly longer telomeres than wild-type cells, and this telomere elongation was further increased in Dicer1-null ES cells (Fig. 3d and representative Q-FISH images in Supplementary Fig. 3a). The differences in telomere length were highly significant for all comparisons (P < 0.001, Fig. 3d). Of note, Q-FISH telomere frequency histograms revealed that Dicer1-null telomeres are widely scattered, in agreement with the possible activation of recombination-based ALT pathways in these cells29–31. We found it interesting that the aberrantly elongated telomeres in Dicer1-null cells retained their capping function27,28, as we did not detect a significant increase in chromosomal aberrations involving telomeres in Dicer1-null cells compared to wild-type controls (Supplementary Fig. 3b). Notably, the long-telomere phenotype observed in cultured Dicer1-null ES cells was confirmed in conditionally deleted adult mouse skin36 (P < 0.0001, Fig. 3e,f). Dnmt overexpression in Dicer1-null cells, however, did not lead to a detectable shortening of their aberrantly long telomeres (Supplementary Fig. 4), probably owing to the slow rate of telomere shortening associated with cell division (100–200 base pairs (bp) per cell division) as well as to the fact that Dicer1-null cells are positive for telomerase activity (see below).
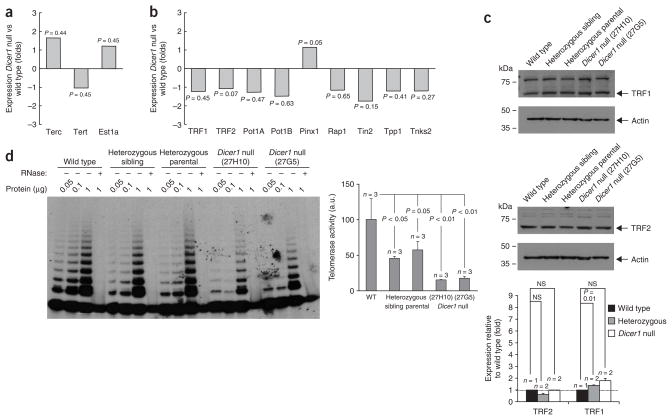
Finally, we addressed whether the abnormally elongated telomeres of Dicer1-null cells could be the consequence of altered expression of well-known telomere-length regulators, such as telomerase or telomere-binding proteins27,28. To this end, we studied the expression of telomerase components and telomere-binding proteins using the Agilent 4×44K mouse 60-mer oligonucleotide microarray (Methods). We did not detect significant changes in the expression of the telomerase RNA component (Terc) and telomerase reverse transcriptase (Tert) telomerase core components, nor in the expression of the telomerase-interacting factor Est1a (also known as Smg6)37 (Fig. 4a). Similarly, we did not find significant changes in the expression of various telomere-binding factors (Figure 4b shows gene-expression changes and Figure 4c shows protein levels of telomere repeat binding factors 1 and 2, Trf1 and Trf2), except for increased expression of the Pin2-interacting protein X1 (Pinx1; P = 0.05, Fig. 4b), a Trf1-interacting protein previously suggested to inhibit telomerase activity38. Notably, telomerase activity was decreased in Dicer1-null cells compared to wild-type controls, and Dicer1-heterozygous cells showed an intermediate activity (Fig. 4d and Methods). Altogether, these results suggest that altered expression of telomerase or telomere-binding proteins is not likely to be the cause of elongated telomeres in Dicer1-null cells. Instead, subtelomeric DNA-methylation defects and increased telomere recombination could be associated with the aberrant telomere elongation shown by Dicer1-null cells.
Figure 4.
Decreased telomerase activity and normal expression of telomere-binding proteins in the absence of Dicer. (a) Expression levels of the indicated telomerase components in wild-type and Dicer1-null ES cells using Agilent 4×44K mouse 60-mer oligonucleotide microarrays. Data are mean values from two experiments using cells derived from two independent Dicer1-null cultures (27G5 and 27H10). Statistical significance using the Student’s t-test is also indicated. Note the absence of significant differences between genotypes. (b) Expression levels of the indicated telomere-binding proteins determined as in part a. Statistical significance using the Student’s t-test is indicated. (c) Representative western blots showing the abundance of telomere repeat binding factors 1 and 2 (TRF1 and TRF2) in the indicated genotypes. Quantification is shown below. Statistical significance is indicated. NS, not significant. (d) Representative images of telomerase telomere repeat amplification protocol (TRAP) activity in the indicated genotypes are shown on the left. (+), treated with RNase; (–), not treated with RNase. Quantification of telomerase TRAP activity levels in the indicated genotypes is shown on the right. a.u., arbitrary units.
Dicer1 is not required for histone trimethylation at telomeres
We have previously shown that either abrogation of histone methyltransferase (HMT) activities, such as Suppressor of variegation 3–9 homologs 1 and 2 (Suv39h1 and Suv39h2) and Suppressor of variegation 4–20 homologs 1 and 2 (Suv4-20h1 and h2), or abrogation of Dnmt1 or Dnmt3a and Dnmt3b leads to abnormal telomere elongation and increased telomere recombination22–26. To address whether Dicer1 deficiency also provoked these telomere phenotypes as a result of defective establishment of histone and DNA methylation at telomeric domains, we carried out ChIP analysis of both subtelomeric and telomeric chromatin in wild-type, Dicer1-heterozygous and Dicer1-null ES cells (Supplementary Methods). First, we confirmed our previous observations that Dicer1-null cells have a normal density of H3K9me3 and H4K20me3 marks and HP1 binding at pericentric repeats8 (Fig. 5a,b). These heterochromatic histone marks were significantly increased at telomeric chromatin in Dicer1-null cells compared to what was observed in wild-type controls (P ≤ 0.01, Fig. 5a,b), with Dicer1-heterozygous cells showing an intermediate phenotype (Fig. 5a,b). In turn, active chromatin marks such as AcH3K9 were decreased at Dicer1-null telomeres compared to those of wild-type cells (P = 0.08, Fig. 5a,b), although the density of this mark was not significantly decreased at pericentric chromatin (Fig. 5a,b). These results suggest a higher degree of chromatin compaction and silencing associated with the longer telomeres of Dicer1-null cells (Fig. 5a,b), which is in agreement with recent data from our group showing decreased telomere transcription in Dicer1-null cells33. Moreover, these findings indicate that the long-telomere phenotype of Dicer1-null cells is not initially triggered by loss of heterochromatic marks at telomeric chromatin, similarly to what has previously been shown for Dnmt-deficient cells14. Finally, the fact that Dicer is not required to establish histone heterochromatic marks at telomeres suggests that heterochromatinization and silencing of these domains is not mediated by Dicer-dependent small RNAs. This is consistent with recent in-depth sequencing and computational studies showing no evidence for the generation of small RNAs at subtelomeric or telomeric regions39,40.
Figure 5.
Dicer is not required to direct heterochromatic histone marks and HP1 binding at mammalian telomeres and subtelomeres. (a) Representative chromatin immunoprecipitation (ChIP) data of wild-type, Dicer1-heterozygous and Dicer1-null ES cells with the indicated antibodies. AcH3K9, acetylated H3 lysine 9; H4K20me3, trimethylated histone H3 lysine 20; H3K9me3, trimethylated histone H3 lysine 9; Het., heterozygous; HP1, heterochromatin protein 1. (b) Quantification of immunoprecipitated telomeric and pericentric repeats. In the case of telomeric chromatin, quantification was done after normalization to both telomeric and pericentric input signals. Error bars correspond to two to eight independent experiments (n = 2–8). Dicer1-null ES cells showed a significant increase in heterochromatic features at telomeric chromatin compared to wild-type controls, which was not detected at pericentric chromatin. Statistical significance values are shown.
Rb proteins decrease Dnmt expression in Dicer1-null cells
To gain insight into the mechanisms by which Dicer regulates DNA methylation and Dnmt levels, we further analyzed Agilent 4×44K mouse 60-mer oligonucleotide microarray data (Methods). First, we confirmed that ablation of Dicer caused a highly significant repression of Dnmt3a and Dnmt3b gene expression (P = 0.017 and P = 0.001, respectively; Fig. 6a), in agreement with quantitative RT-PCR and western blot results (Fig. 2a–c). Expression of the Dnmt1 gene was not detectable (n.d.) in the microarray because of an extensive mismatch between the oligonucleotide probe on the array and the Dnmt1 mRNA (Fig. 6a). Notably, downregulation of Dnmt3a and Dnmt3b gene expression in Dicer1-null cells was paralleled by a significant induction of the Rb family genes Rbl1 and Rbl2, the Rb regulator gene Rb1cc1 (ref. 41) and Rbak, which encodes a putative mediator of the Rb-dependent suppression of E2f-mediated transcriptional activation42 (Fig. 6b). Ablation of Dicer also increased the expression of Rbbp1 (Fig. 6b), also known as Rbp1 or Arid4a. The gene product of Rbbp1 interacts with Rb proteins and is thought to mediate some of the epigenetic effects of Rb, such as recruitment of histone deacetylases (HDACs), bringing about transcriptional repression of E2f-dependent promoters43.
Figure 6.
Increased expression of Retinoblastoma (Rb) family proteins in Dicer1-null cells is responsible of the decreased expression of Dnmt1, Dnmt3a and Dnmt3b. (a) Gene-expression changes in the indicated genotypes using Agilent 4×44K mouse 60-mer oligonucleotide microarrays. Data are mean values from two experiments using two independent Dicer1-null cultures (27G5 and 27H10). Ablation of Dicer results in decreased Dnmt3a,3b expression. Expression of Dnmt1 was not detectable (n.d.) because of an extensive mismatch between the oligonucleotide probe on the array and Dnmt1 mRNA. (b) Downregulation of Dnmt3a,3b expression was paralleled by a significant increase in the expression of Rb family genes Rbl1 and Rbl2, and of the Rb regulators Rb1cc1, Rbak and Rbbp1. (c) Representative RT-PCR gel confirming overexpression of TAg (T121) in Dicer1-null cells. β-Actin is used as a control. (d) Dnmt1, Dnmt3a and Dnmt3b mRNA levels as determined by quantitative RT-PCR in Dicer1-null ES cells transfected with a control vector or a truncated form of the SV40 large T-antigen (T121). Inhibition of Rb function by TAg T121 in Dicer1-null cells significantly increases Dnmt1 and Dnmt3b expression. (e) Expression of TAg T121 in Dicer1-null ES cells rescues subtelomeric DNA-methylation defects as determined by bisulfite sequencing of 4–16 individual clones. (f) Histone acetylation (AcH3K9) as determined by ChIP at the promoters of Dnmt genes in the indicated genotypes. Dnmt1-deficient cells were used as negative control.
Rb proteins were previously described to transcriptionally inactivate Dnmts, and this effect was shown to be reversible by Rb-inactivating viral oncoproteins such as the T-antigen44–48. To directly address whether Rb proteins were responsible for decreased Dnmt expression in Dicer1-null cells, we expressed a truncated form of the T-antigen, which specifically binds and inhibits Rb family proteins (T121)49 (Methods and Fig. 6c). Expression of T121 in Dicer1-null cells resulted in increased Dnmt1 and Dnmt3b mRNA levels as determined by quantitative RT-PCR (Fig. 6c,d). Furthermore, T121 was also able to partially rescue subtelomeric DNA-methylation defects in Dicer1-null cells (Fig. 6e), demonstrating that Rb proteins are responsible for DNA-methylation defects in these cells. Rb proteins have previously been shown to epigenetically repress promoters by associating with histone deacetylases and decreasing promoter acetylation50. In support of a role for the Rb proteins in epigenetically repressing Dnmt promoters, ChIP analysis showed that Dicer1-null cells have a decreased abundance of AcH3K9 at the promoter regions of the Dnmt1, Dnmt3a and Dnmt3b genes compared to wild-type cells (Fig. 6f). As negative control, promoter acetylation did not change in Dnmt1-deficient cells (Fig. 6f). Altogether, these results indicate that increased levels of Rb proteins in Dicer1-null cells are responsible for decreased Dnmt expression and DNA-methylation defects in these cells.
Control of DNA methylation by the miR-290 cluster
Increased Rb protein levels in Dicer1-null cells may suggest the existence of Dicer-dependent miRNAs that specifically silence these genes and are downregulated upon Dicer deletion. To identify such miRNAs, we performed microarray analysis of Dicer1-null and wild-type ES cells using a Spanish National Cancer Centre (CNIO) custom-made miRNA microarray (Methods). We identified several miRNAs with predicted targets in the Rb family proteins (Rb, Rbl1 and Rbl2) whose expression was significantly downregulated in Dicer1-null ES cells compared to wild-type controls (Fig. 7a) Most notably, a family of miRNAs from the miR-290 miRNA cluster showed substantially downregulated expression in Dicer1-null cells compared to wild-type controls (Fig. 7a,b). The miR-290 family is restricted to placental mammals35,36 and encompasses miR-290, miR-291-3p, miR-291-5p, miR-292-3p, miR-292-5p, miR-293, miR-294 and miR-295, all of which show decreased expression in Dicer1-null cells (Fig. 7b). Notably, all miRNAs of the miR-290 cluster except for miR-290 had Rbl2 among its predicted targets in both mouse and human cells (Fig. 7c), and Rbl2 was also the most significantly upregulated Rb protein in Dicer1-null cells (Fig. 6b).
Figure 7.
The miR-290 cluster targets Rbl2 and controls Dnmt3a,3b expression. (a) Each data point represents the expression level of miRNAs in Dicer1-null compared to wild-type ES cells. miRNAs of the miRNA-290 cluster are indicated in orange; other miRNAs predicted to target Rb family members are shown in green. MicroRNAs targeting Rbl2, Rbl1 and Rb1 whose expression is significantly changed in the absence of Dicer are shown. (b) Ablation of Dicer causes decreased expression of all members of the miR-290 cluster. (c) Rb-related targets of the miR-290 cluster in mouse and human cells. Arid4a, ARID domain–containing protein 4A; Rbbp, retinoblastoma binding protein. (d) Representative western blot showing Rbl2 expression after transfection with the indicated miRNAs. (Rb1/Rbl1/Rbl2)−/− mouse embryonic fibroblasts were included as a negative control. (e) Quantification of the western blot shown in part d. Values were corrected by actin levels. (f) Decreased Rbl2 mRNA levels in C2C12 cells after transfection with the indicated miRNAs. No changes were observed in Rb1 or Rbl1 mRNAs. (g) Dnmt levels in C2C12 cells after transfection with the indicated miRNAs. (h) Expression of the indicated Rb proteins after transfection of the miR-290 cluster in Dicer1-null cells. (i) Dnmt levels in Dicer1-null cells after transfection with the indicated miRNAs. Only Dnmt3a and Dnmt3b were affected. Values were normalized to actin expression in parts f–i.
To address whether the miR-290 cluster specifically silences Rbl2, we overexpressed each of the miRNAs from this cluster in mouse C2C12 cells (Methods). Transfection of the entire miR-290 cluster, or single transfection of miR-291-3p, miR-292-3p, miR-294 and miR-295 decreased Rbl2 expression to undetectable levels compared to a control miRNA (the MiRIDIAN miRNA mimic; Fig. 7d,e and Supplementary Methods). As a negative control for Rbl2 expression, we included mouse embryonic fibroblasts (MEFs) that were triple deficient for the Rb family of proteins, (Rb1/Rbl1/Rbl2)−/− cells23,24 (Fig. 7d,e). Notably, single transfection of miR-290 did not affect Rbl2 protein levels, in agreement with the fact that Rbl2 is not a predicted target of this particular miRNA (Fig. 7c–e). In the case of miR-293, we saw an intermediate decrease in Rbl2 mRNA levels (Fig. 7d,e). These results were confirmed by quantitative RT-PCR, further demonstrating a role for miR-290 cluster miRNAs in specifically regulating Rbl2 expression (Fig. 7f). Of note, Rb1 and Rbl1 mRNA levels were not affected by any of the members of the miR-290 cluster, indicating the specificity of this cluster for Rbl2 (Fig. 7f). Altogether, these results suggest that the miR-290 cluster is directly involved in silencing the expression of Rbl2. Furthermore, these data suggest that decreased levels of Dnmts and decreased DNA methylation in Dicer1-null cells are the consequence of downregulated expression of miRNAs of the miR-290 cluster, via loss of Rbl2 silencing.
To directly test this hypothesis, we analyzed Dnmt1, Dnmt3a and Dnmt3b mRNA levels in cells transfected with different members of the miR-290 cluster. Interestingly, decreased Rbl2 protein and mRNA levels in cells transfected with miR-291-3p, miR-292-3p, miR-294 and miR-295 were paralleled by a simultaneous increases in Dnmt3a and Dnmt3b mRNA expression (Fig. 7g). Of note, Dnmt1 expression levels were not significantly altered by transfection of the entire miR-290 cluster, indicating that Rbl2 specifically targets the Dnmt3a and Dnmt3b enzymes. Altogether, these findings suggest that decreased levels of Dnmt3a and Dnmt3b in Dicer1-null cells are the consequence of downregulation of the miR-290 cluster, which in turn results in increased Rbl2-mediated silencing of these genes.
Finally, to directly address whether decreased Dnmt3a and Dnmt3b levels in Dicer1-null ES cells are the consequence of an absence of post-transcriptional repression of Rbl2 by the miR-290 cluster, we co-transfected Dicer1-null ES cells with a vector encoding the enhanced green fluorescent protein (EGFP) and the whole miR-290 cluster, or with a control miRNA (miR-control). Transfected ES cells were sorted on the basis of their EGFP expression, and Rb family and Dnmt mRNA levels were determined by quantitative RT-PCR. Consistent with the results obtained with mouse C2C12 cells, transfection of the miR-290 cluster into Dicer1-null cells resulted in a specific reduction in Rbl2 mRNA levels without affecting Rb1 and Rbl1 expression (Fig. 7h). Decreased Rbl2 protein and mRNA levels in Dicer1-null cells transfected with the miR-290 cluster correlated with significantly rescued expression of the Dnmt3a and Dnmt3b enzymes but not of Dnmt1 (Fig. 7i).
In summary, re-introduction of the miR-290 cluster into Dicer1-deficient ES cells rescues decreased expression of Dnmt3a and Dnmt3b enzymes in these cells, with coincidentally decreased Rbl2 mRNA levels. This indicates an important role for the miR-290 cluster in mammalian cells in regulating global DNA methylation, which in turn controls telomere length and telomere recombination. Finally, it is unlikely that telomere phenotypes in Dicer1-null cells could be the result of a direct role for the miR-290 cluster in targeting mouse sub-telomeric sequences, as we did not find a preferential accumulation of predicted miR-290 targets in these regions compared to the rest of the genome (Supplementary Fig. 5). In particular, we compared the seed and mature miRNAs of the miR-290 cluster with 10-kb subtelomeric DNA fragments derived from each of the 19 mouse autosomes and with 1,000 randomly chosen 10-kb genomic DNA fragments (Supplementary Methods). We were not able to detect matches for mature miRNAs of the miR-290 cluster in any of the subtelomeric regions tested; moreover, miR-290 seed matches were not significantly enriched at subtelomeric sequences compared to the rest of the genome39,40 (Supplementary Fig. 5).
DISCUSSION
Here we identify a previously unknown regulatory pathway involving the mammalian Dicer-dependent miR-290 cluster as an important post-transcriptional regulator of Rbl2, which in turn acts as a transcriptional repressor of the Dnmt3a and Dnmt3b enzymes (Fig. 8). In the absence of Dicer, downregulation of the miR-290 cluster leads to increased mRNA levels of the miR-290 cluster target gene Rbl2, whose product in turn inhibits Dnmt3a,3b expression. Decreased Dnmt expression, in part mediated by Rbl2, leads to a significant hypomethylation of the genome, including the subtelomeric regions, and to the appearance of telomeric phenotypes such as increased telomere recombination and increased telomere length (Fig. 8).
Figure 8.
A previously unknown miR-290–dependent pathway controls DNA methylation via Rbl2 and affects telomere integrity and telomere-length homeostasis. We identify here a regulatory pathway involving the highly conserved mammalian miR-290 cluster as an important post-transcriptional regulator of Rbl2, which in turn acts as a transcriptional repressor of Dnmt3a and Dnmt3b (a). In the absence of Dicer (b), miR-290 downregulation leads to increased levels of the miR-290 target Rbl2, which in turn acts as a transcriptional repressor of Dnmt3a,3b expression. Decreased Dnmt expression leads to a hypomethylation of the genome, including the subtelomeric regions, and to the appearance of telomere phenotypes such as increased telomere recombination and aberrantly long telomeres.
Notably, Dicer1-null cells did not show a decreased abundance of histone trimethylation marks (H3K9me3 and H4K20me3) or HP1 binding at telomeric regions, arguing that Dicer, and therefore Dicerdependent small RNAs, are not required for the establishment of histone heterochromatic marks at these domains. These findings are in agreement with recent reports also showing that Dicer-dependent small RNAs are not required for the assembly of histone trimethylation marks at other repeated elements such as pericentric repeats and transgene repeats8,9 (see also this paper). This situation is in marked contrast to the well-established role of the small interfering RNA (siRNA) machinery in directing these marks at heterochromatic domains in fission yeast and D. melanogaster3–5. In addition, the fact that Dicer-dependent small RNAs are not required for the assembly of histone heterochromatic marks and HP1 binding at telomeric domains opens the possibility that the recently described long noncoding telomeric RNAs may be involved in heterochromatin formation at telomeres32,33.
Finally, we anticipate that regulation of DNA methylation by the miR-290 cluster may have important roles in fundamental processes such as development, tumorigenesis and epigenetic gene regulation. In this regard, the miRNA-290 family is highly expressed in pluripotent ES cells and repressed upon differentiation35. Furthermore, in vitro differentiation of ES cells recapitulates some of the global genomic methylation that takes place shortly after implantation51,52, in support of a putative role for the miR-290 family in these processes34,35. We anticipate that control of DNA methylation by the miR-290 cluster via Rbl2 could represent a new important aspect of the epigenetic regulation of embryonic development. In particular, miR-290 cluster–dependent regulation of DNA methylation could have an important impact on the regulation of telomere recombination and telomere length during early embryonic development53, thus contributing to the establishment of telomere-length homeostasis.
METHODS
ES cell culture
Different ES cells were generated and maintained as described8. Mouse C2C12 cells and (Rb1/Rbl1/Rbl2)−/− (ref. 24) were cultured in DMEM with 10% (v/v) FCS.
Terminal restriction fragment analysis
We prepared cells in agarose plugs and carried out TRF analysis as described23.
Telomere quantitative fluorescence in situ hybridization on metaphases
Metaphases were prepared and hybridized as described54. To correct for lamp intensity and alignment, images from fluorescent beads were analyzed in parallel (Molecular Probes, Invitrogen) using the TFL-Telo program55 (a gift from P. Lansdorp, University of British Columbia). Images and telomere fluorescence values were obtained from at least ten metaphases for each data point as described54.
Telomere quantitative fluorescence in situ hybridization on skin sections
Measurement of telomere length on skin sections from mice conditionally deleted for Dicer36 was performed as described56.
Methylation-sensitive restriction-enzyme analysis
ES cells (1×106) were embedded in agarose plugs and digested with Msp I or HpaII enzymes (Fermentas) at 37 °C overnight. After washing, genomic DNA was separated by pulse-field electrophoresis and transferred to a nitrocellulose membrane. Southern hybridization was performed with a major satellite probe as described14.
Telomerase assay
Telomerase activity was measured with a modified telomere repeat amplification protocol (TRAP) as described14. An internal control for PCR efficiency was included (TRAPeze kit, Oncor).
Chromosome orientation fluorescence in situ hybridization
Confluent mouse ES cells were subcultured in the presence of 5′-bromo-2′-deoxyuridine (BrdU; Sigma) at a final concentration of 1×10−5 M and then allowed to replicate their DNA once at 37 °C overnight. Colcemid was added at a concentration of 0.1% (w/v) during the last hour. Cells were then recovered and metaphases prepared as described54. CO-FISH was performed as described14,57 using first a (TTAGGG)7 PNA probe labeled with Cy3 and then a second (CCCTAA)7 PNA probe labeled with Fluorescein (Applied Biosystems). Metaphase spreads were captured on a Leitz Leica DMRB fluorescence microscope. For chromosomal aberrations, 42–92 metaphases from each ES culture were analyzed as described54.
Analysis of genomic subtelomeric DNA methylation
DNA methylation of subtelomeric genomic DNA regions was established by PCR analysis after bisulfite modification as described14. Automatic sequencing of 2–16 colonies for each sequence was performed to obtain the methylation status of the subtelomeric CpG island. Bisulfite genomic sequencing primers were designed against subtelomeric regions in chromosomes 1 and 2 as described25,26. Results were analyzed with BiQ Analyzer Software for DNA methylation analysis (Applied Biosystem Software Community Program).
Immuno-dot-blot with 5-MetCyt
To detect DNA methylation at specific chromatin regions, we performed ChIP experiments with specific antibodies: 0.1% (v/v) of anti–H3K9me3 antibody (#07-442, Upstate); or 0.1% (v/v) rabbit polyclonal anti–H4K20me3 antibody (#07-463, Uupstate); 0.1% (v/v) polyclonal anti–H3K9Ac antibody (#H9286-072K4824, Sigma); or 0.1% (v/v) of monoclonal anti-HP1γ antibody (MAB 3450 #25010489, Chemicon). This was followed by immunoblot analysis using 0.5% (v/v) 5-MetCyt (BI-MECY-0100, Eurogentec; purified ascites) and the specific secondary antibody coupled to horseradish peroxidase. For total DNA samples, cell lysates were diluted 1 in 10, 1 in 100 and 1 in 1000 after reversal of cross-linking and processed along with the immunoprecipitates. Quantification of the signals was done with ImageJ (http://rsb.info.nih.gov/ij/index.html) software. The immunoblot signals were normalized to the ChIP signals of major satellite centromeric repeats.
B1-SINE Cobra analyisis for global DNA methylation
Global DNA-methylation levels were determined using the B1-SINE Cobra method as previously described25,26. Estimation of the fraction of methylated B1 elements for each genotype was done using the formula: ((Molarity of 45-bp band)/2)/((Molarity of 45-bp band) + (Molarity of 100-bp band)).
Expression of DNA methyltransferases in Dicer1-null cells
Expression vectors encoding Dnmt3a and Dnmt3b isoforms or Dnmt1 were electroporated into Dicer1-null ES cells, which were subsequently selected in blasticidin-containing medium for 7 d as described20. Blasticin-resistant cells were examined for Dnmt1, Dnmt3a and Dnmt3b mRNA expression as described above. Protein expression of different Dnmts was studied by immunoblotting analysis with the following antibodies: monoclonal anti-Dnmt3a (clone 64B1446-IMG268, Imgenex) or polyclonal anti-Dnmt1 (a gift from S. Tajima, Osaka University, Japan).
Transient transfection of miR-290 cluster into C2C12 mouse cells
C2C12 cells were transiently transfected according to manufacturer’s suggestions with 70 nM each of mmu-mir-290, mmu-miR-291-3b, mmu-miR-292-3b, mmu-miR293, mmu-miR-294 or mmu-miR-295 mimics (Dharmacon), or a 70-nM mix of all miR-290 cluster mimics, using DarmaFECT (Dharmacon). MiR-IDIAN miRNA mimic (CN-001000-01-05, Dharmacon) was used as a negative control. Total RNA and protein was prepared 72 h after transfection (see above).
Expression of a truncated form of T-antigen in Dicer1-null cells
Expression vectors encoding a fragment of the SV40 T antigen (T121), which binds to and inhibits the Rb family proteins, provided by M. Barbacid (CNIO), was derived from the original construct described in ref. 49 after subcloning into a pBabe vector by PCR amplification using 5′-CGCGGATCCATGGATAAAGTTTT AAACAGAGAG-3′ and 5′-CGCGAATTCTTAGCAATTCTGAAGGAAAGTCC-3′ oligonucleotides. pBabeT121 was electroporated together with the pHygro vector (ratio 10:1) into Dicer1-null ES cells, which were subsequently selected in hygromycin-containing medium (final concentration 0.01% (w/v) for 28 d. Hygromycin-resistant cells were examined for Dnmt1, Dnmt3a and Dnmt3b mRNA expression as described above and for T121 expression using the primer pair 5′-CGCGGATCCATGGATAAAGTTTTAAACAGAGAG-3′ and 5′-CGCGAATTCTTAGCAATTCTGAAGGAAAGTCC-3′.
Scanning and analysis of oligonucleotide microarray results
Signal quanti-fication and data normalization were carried out with Feature Extraction 9.1 software (Agilent Technologies) using default analysis parameters for 4×44K gene-expression arrays (feature extraction protocol GE2-v5_91_0806). Gene-Spring GX 7.3.1 Expression Analysis software (Agilent Technologies) was used to identify differentially expressed genes; Welch ANOVA was run on hybridizations data regrouped in three different condition groups: (i) Dicer1-null 27G5 versus wild-type ES cells and Dicer1-null 27H10 versus wild-type ES cells; (ii) Dicer1-null 27G5 cells overexpressing Dnmt1 versus Dicer1-null 27G5 cells and Dicer1-null 27H10 cells overexpressing Dnmt1 versus Dicer1-null 27H10 cells; and (iii) Dicer1-null 27G5 cells overexpressing Dnmt3a,3b versus Dicer1-null 27G5 cells and Dicer1-null 27H10 overexpressing Dnmt3a,3b versus Dicer1-null 27H10 cells. In parallel, a described multiple-testing correction procedure58 was applied to keep the rate of false discoveries (FDR) under 10%, and the Tukey post hoc test was used to determine which specific group pairs were statistically different (P < 0.001) from each other.
MicroRNA arrays
Small RNA fractions containing the miRNA population were isolated using the PureLink miRNA isolation kit (Invitrogen). Labeling and hybridization to CNIO miRNA arrays were performed with the NCode labeling system (Invitrogen) following manufacturer recommendations. CNIO miRNA array v.1 contained the NCode miRNA multispecies probe set (Invitrogen) spotted on epoxy slides. The array contains probes directed to mature miRNAs from five species: human, mouse, rat, D. melanogaster and Caenorhabditis elegans. Probes were designed from the miRNA sequences in Sanger miRBase 7.0 (http://microrna.sanger.ac.uk) and also include predicted miRNAs. Probe oligonucleotides are dimeric (head-to-tail repeats) and are designed to maximize hybridization specificity59.
Supplementary Material
Acknowledgments
We are indebted to O. Dominguez and D. Pisano from the Genomics and Bioinformatics Units of the Spanish National Cancer Centre (CNIO), respectively, for help with bisulfite sequencing analysis and various microarray and genome-sequence analyses. R.B. is a staff investigator of the CNIO, S.G. is a Fondo de Investigaciones Sanitarias (FIS) senior scientist. P.M. is a ‘Ramon y Cajal’ senior scientist. I.J. is a student of the Gulbenkian Ph.D. Program in Biomedicine, supported by FCT/MCT (Portugal). M.A.B.’s laboratory is funded by the MCyT (SAF2005-00277, GEN2001-4856-C13-08), by the Regional Government of Madrid (GR/SAL/0597/2004), European Union (TELOSENS FIGH-CT-2002-00217, INTACT LSHC-CT-2003-506803, ZINCAGE FOOD-CT-2003-506850, RISC-RAD FI6R-CT-2003-508842, MOL CANCER MED LSHC-CT-2004-502943) and the Josef Steiner Cancer Research Award, 2003.
Footnotes
Accession codes. NCBI-GEO. Microarray data were deposited under the accession code GSE11229.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
AUTHOR CONTRIBUTIONS
R.B. performed experiments shown in all manuscript figures as well as designed the experiments together with M.A.B. and S. Gonzalo. S. Gonzalo performed part of the experiments shown in Figure 3c,d and Figure 5. I.J. performed some of the experiments shown in Figure 3a,b. P.M. performed the experiments shown in Figure 3e,f. S. Gonzalez and M.S. generated the miRNA data shown in Figure 7a. S.S. contributed to initial characterization of Dicer1-null telomeric chromatin. E.M. and G.H. provided the Dicer1-null cells and helped with the writing of the manuscript. T.A. and S.M. provided the Dicer1-null skin samples. T.C. and E.L. provided the Dnmt-deficient cells and the Dnmt constructs. P.K. and M.A.B. prepared the manuscript figures. M.A.B., R.B. and S. Gonzalo wrote the paper.
References
- 1.Fukagawa T, et al. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- 2.Kanellopoulou C, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Volpe TA, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 4.Hall IM, et al. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 5.Pal-Bhadra M, et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 6.Verdel A, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cobb BS, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murchison EP, et al. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang F, et al. The assembly and maintenance of heterochromatin initiated by transgene repeats are independent of the RNA interference pathway in mammalian cells. Mol Cell Biol. 2006;26:4028–4040. doi: 10.1128/MCB.02189-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall IM, Noma K, Grewal SI. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci USA. 2003;100:193–198. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci USA. 2005;102:152–157. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 13.Lehnertz B, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalo S, et al. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 15.Esteller M. Relevance of DNA methylation in the management of cancer. Lancet Oncol. 2003;4:351–358. doi: 10.1016/s1470-2045(03)01115-x. [DOI] [PubMed] [Google Scholar]
- 16.Dodge JE, et al. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J Biol Chem. 2005;280:17986–17991. doi: 10.1074/jbc.M413246200. [DOI] [PubMed] [Google Scholar]
- 17.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 18.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 19.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 20.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen T, Tsujimoto N, Li E. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol Cell Biol. 2004;24:9048–9058. doi: 10.1128/MCB.24.20.9048-9058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Cao M, O’Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- 23.García-Cao M, Gonzalo S, Dean D, Blasco MA. Role of the Rb family members in controlling telomere length. Nat Genet. 2002;32:415–419. doi: 10.1038/ng1011. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalo S, et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol. 2005;7:420–428. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- 25.Benetti R, Garcia-Cao M, Blasco MA. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat Genet. 2007;39:243–250. doi: 10.1038/ng1952. [DOI] [PubMed] [Google Scholar]
- 26.Benetti R, et al. Suv4–20h deficiency results in telomere elongation and de-repression of telomere recombination. J Cell Biol. 2007;178:925–936. doi: 10.1083/jcb.200703081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 28.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 29.Muntoni A, Reddel RR. The first molecular details of ALT in human tumor cells. Hum Mol Genet. 2005;14:R191–R196. doi: 10.1093/hmg/ddi266. [DOI] [PubMed] [Google Scholar]
- 30.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 31.Bailey SM, Brenneman MA, Goodwin EH. Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res. 2004;32:3743–3751. doi: 10.1093/nar/gkh691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 33.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA dependent RNA polymerase II. Nat Cell Biol. 2007;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 34.Houbaviy HB, Dennis L, Jaenisch R, Sharp PA. Characterization of a highly variable eutherian microRNA gene. RNA. 2005;11:1245–1257. doi: 10.1261/rna.2890305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 36.Andl T, et al. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reichenbach P, et al. A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr Biol. 2003;13:568–574. doi: 10.1016/s0960-9822(03)00173-8. [DOI] [PubMed] [Google Scholar]
- 38.Zhou XZ, Lu KP. The Pin2/TRF1-interacting protein PinX1 is a potent telomerase inhibitor. Cell. 2001;107:347–359. doi: 10.1016/s0092-8674(01)00538-4. [DOI] [PubMed] [Google Scholar]
- 39.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berezikov E, et al. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006;16:1289–1298. doi: 10.1101/gr.5159906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chano T, et al. Identification of RB1CC1, a novel human gene that can induce RB1 in various human cells. Oncogene. 2002;21:1295–1298. doi: 10.1038/sj.onc.1205178. [DOI] [PubMed] [Google Scholar]
- 42.Skapek SX, et al. Cloning and characterization of a novel Kruppel-associated box family transcriptional repressor that interacts with the retinoblastoma gene product, RB. J Biol Chem. 2000;275:7212–7223. doi: 10.1074/jbc.275.10.7212. [DOI] [PubMed] [Google Scholar]
- 43.Lai A, et al. RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol Cell Biol. 2001;21:2918–2932. doi: 10.1128/MCB.21.8.2918-2932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCabe MT, Low JA, Imperiale MJ, Day ML. Human polyomavirus BKV transcriptionally activates DNA methyltransferase 1 through the pRb/E2F pathway. Oncogene. 2006;25:2727–2735. doi: 10.1038/sj.onc.1209266. [DOI] [PubMed] [Google Scholar]
- 45.McCabe MT, et al. Inhibition of DNA methyltransferase activity prevents tumorigenesis in a mouse model of prostate cancer. Cancer Res. 2006;66:385–392. doi: 10.1158/0008-5472.CAN-05-2020. [DOI] [PubMed] [Google Scholar]
- 46.McCabe MT, Davis JN, Day ML. Regulation of DNA methyltransferase 1 by the pRb/E2F1 pathway. Cancer Res. 2005;65:3624–3632. doi: 10.1158/0008-5472.CAN-04-2158. [DOI] [PubMed] [Google Scholar]
- 47.Kimura H, Nakamura T, Ogawa T, Tanaka S, Shiota K. Transcription of mouse DNA methyltransferase 1 (Dnmt1) is regulated by both E2F-Rb-HDAC-dependent and -independent pathways. Nucleic Acids Res. 2003;31:3101–3113. doi: 10.1093/nar/gkg406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pradhan S, Kim GD. The retinoblastoma gene product interacts with maintenance human DNA (cytosine-5) methyltransferase and modulates its activity. EMBO J. 2002;21:779–788. doi: 10.1093/emboj/21.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simin K, et al. pRb inactivation in mammary cells reveals common mechanisms for tumor initiation and progression in divergent epithelia. PLoS Biol. 2004;2:e22. doi: 10.1371/journal.pbio.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brehm A, et al. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 51.Shiota K, Yanagimachi R. Epigenetics by DNA methylation for development of normal and cloned animals. Differentiation. 2002;69:162–166. doi: 10.1046/j.1432-0436.2002.690406.x. [DOI] [PubMed] [Google Scholar]
- 52.Ohgane J, Hattori N, Oda M, Tanaka S, Shiota K. Differentiation of trophoblast lineage is associated with DNA methylation and demethylation. Biochem Biophys Res Commun. 2002;290:701–706. doi: 10.1006/bbrc.2001.6258. [DOI] [PubMed] [Google Scholar]
- 53.Liu L, et al. Telomere lengthening early in development. Nat Cell Biol. 2007;9:1436–1441. doi: 10.1038/ncb1664. [DOI] [PubMed] [Google Scholar]
- 54.Samper E, Goytisolo FA, Slijepcevic P, van Buul PP, Blasco MA. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 2000;1:244–252. doi: 10.1093/embo-reports/kvd051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zijlmans JM, et al. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc Natl Acad Sci USA. 1997;94:7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muñoz P, Blanco R, Flores JM, Blasco MA. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat Genet. 2005;37:1063–1071. doi: 10.1038/ng1633. [DOI] [PubMed] [Google Scholar]
- 57.Bailey SM, et al. Strand-specific postreplicative processing of mammalian telomeres. Science. 2001;293:2462–2465. doi: 10.1126/science.1062560. [DOI] [PubMed] [Google Scholar]
- 58.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 59.Goff LA, et al. Rational probe optimization and enhanced detection strategy. RNA Biol. 2005;2:93–100. doi: 10.4161/rna.2.3.2059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.