Abstract
Complementary techniques that deepen information content and minimize reagent costs are required to realize the full potential of massively parallel sequencing. Here, we describe a resequencing approach that directs focus to genomic regions of high interest by combining hybridization-based purification of multi-megabase regions with sequencing on the Illumina Genome Analyzer (GA). The capture matrix is created by a microarray on which probes can be programmed as desired to target any non-repeat portion of the genome, while the method requires only a basic familiarity with microarray hybridization. We present a detailed protocol suitable for 1–2 µg of input genomic DNA and highlight key design tips in which high specificity (>65% of reads stem from enriched exons) and high sensitivity (98% targeted base pair coverage) can be achieved. We have successfully applied this to the enrichment of coding regions, in both human and mouse, ranging from 0.5 to 4 Mb in length. From genomic DNA library production to base-called sequences, this procedure takes approximately 9–10 d inclusive of array captures and one Illumina flow cell run.
INTRODUCTION
The notion that the personal genome of a human individual could be sequenced in less than a year was virtually unthinkable 5 years ago. Even more unthinkable was the idea of doing a low-coverage sequence survey for <$20,000. The commercial availability and widespread adoption of massively parallel sequencing by synthesis (SBS) platforms has turned our collective focus toward the development of tools and infrastructure to apply their capacity to rapidly and cost-effectively tackle a wide variety of questions related to genome biology. To this end, multiple consortia have assembled to accelerate the generation of sequence catalogs of human genetic variation as exemplified by the 1000 Genomes Project1,2 and The Cancer Genome Atlas3,4. Moreover, a recent surge of publications describing the resequencing of the diploid genomes of Craig Venter5, James Watson6, the first genome of an Asian individual7, and the genome of a malignant tumor8 firmly demonstrates the unprecedented opportunity presented by the use of this technology.
Though many would argue that whole genome sequencing provides the most comprehensive and unbiased approach for the discovery of disease-causing mutations, this method presently has substantial barriers to its routine application. Despite their considerable throughput, the large data sets produced by SBS instruments comprise relatively short sequence read lengths that suffer from higher error rates than conventional methods9. The combined informatic challenges associated with these two characteristics necessitate high levels of sequence sampling to obtain definitive evidence for detection of a variant. In fact, several groups have estimated the minimal required read depth to be around 20-fold coverage for sufficient error compensation to generate accurate base calls10. Furthermore, detecting a heterozygous genotype with high statistical confidence will require even deeper coverage, as adequate sampling of both alleles is needed11. On the basis of these estimates, the cost and sequencing run time required for such endeavors remain too prohibitive for an individual investigator or a small group wishing to undertake a survey of genetic variation in many individuals. Therefore, the ability to fractionate a complex eukaryotic genome for resequencing provides a potential approach to a variety of biological questions.
Recently, we and others showed a means to direct sequencing efforts to sets of defined genomic intervals in order to boost coverage of high-value regions by exclusion of others using a subtractive hybridization strategy12–17. This selection scheme uses either complex libraries of soluble probes or high-density tiling DNA microarrays to purify large continuous or discontinuous genomic regions. After hybridization, the captured material is recovered for direct sequencing on massively parallel SBS platforms. Through array-based hybrid selection and deep sequencing, we achieve significant enrichment and ample coverage of target exon regions, illustrating the potential breadth of this resequencing approach to uncover variations that might otherwise escape detection.
Soluble probes
Earlier versions of solution-based hybrid selection have used BAC/YAC (bacterial/yeast artificial chromosomes) clones as bait for the isolation of specific transcripts from cDNA libraries or regions of total genomic DNA16. More recently, the highly parallel in situ oligonucleotide synthesis characteristic of current microarray printing technologies has enabled the production of complex nucleic acid libraries containing tens of thousands of custom-defined probes. Some synthesis platforms achieve lengths up to 200 nucleotides. In addition, the probes can be cleaved and solubilized by soaking the arrays in an alkaline solution18. This complex pool of probes may then be PCR-amplified to create a renewable source of material for capture, and may be tagged with a biotin label for subsequent affinity purification. Several adaptations of array-released oligo libraries for genomic enrichment have been reported including a ‘molecular inversion probe’ strategy15 and probes transcribed into long (170 bp) RNA baits17. The advantage of hybridization in solution is that high specificity can be achieved with minimal amounts of input material. The disadvantages, however, are that nonuniform representation of probes resulting from their initial molecular preparation and management may produce unequal recovery of targets leading to disparities in coverage of genomic intervals.
In situ, fixed position probes
Tiling arrays consist of chemically synthesized and spatially immobilized oligonucleotides in which predefined sequences reflect regions of the genome at frequent and uniform intervals. Such arrays may be programmed to exclude repetitive elements, thereby optimizing available array capacity and performance. Earlier, we described the genome-wide selection and focal resequencing of more than 200,000 protein coding human exons13. We validated this approach using 385k Nimblegen arrays, which were printed with custom-designed, variable-length probes tiling exons at 20 nucleotide intervals. In brief, genomic DNA (20 µg input)was first sheared to a specified length (~600 bp) and fragments were hybridized to arrays over a period of 3–4 d, followed by stringent array washing and fragment recovery by thermal elution. In the captured material from the initial set of eight arrays, approximately >50% of all sequenced fragments originated from the exonic regions selected by probes. In addition, 25% of the total bases within those intervals were covered by reads. These results are based on the initially described procedure and do not take into account our observation that fragment size and read distribution can influence the interpretation of the data. On the basis of several key lessons gained from this study, we were motivated to develop an improved protocol that better combines array capture with sequencing on short read SBS platforms (Fig. 1).
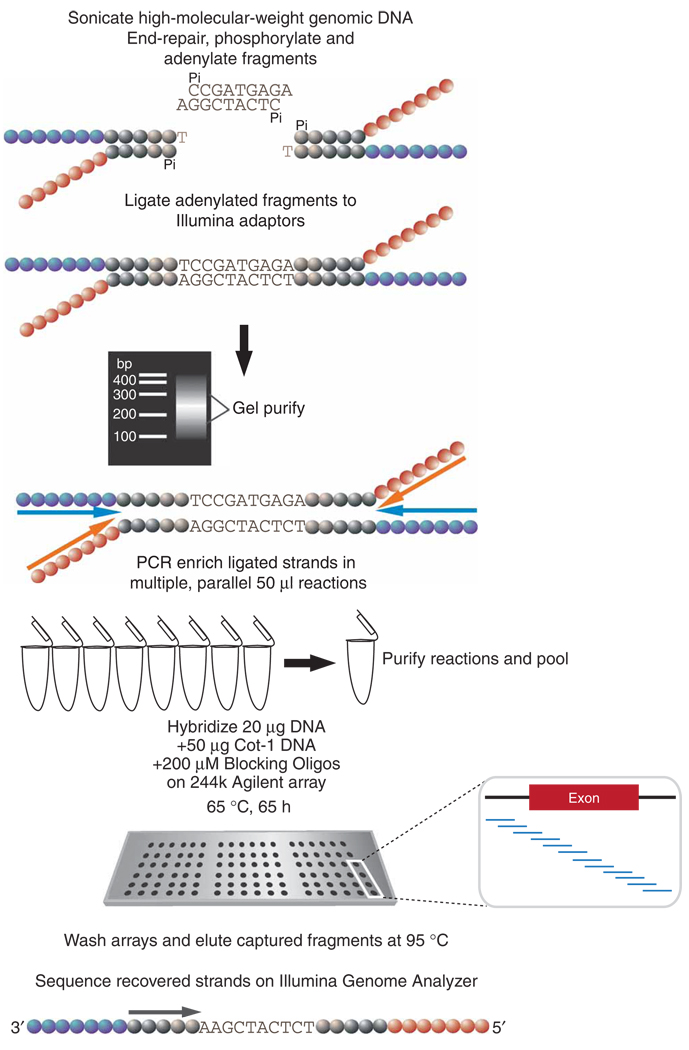
Figure 1.
Schematic diagram of the array capture protocol. High-molecular-weight genomic DNA is fragmented by sonication. The fragments are subjected to a series of enzymatic reactions that repair frayed ends, restore phosphorylation and add a single 3′ adenine overhang. The fragments are symmetrically ligated to an adaptor comprising two partially complementary oligos11 (colored circles indicate distinct adaptor oligo sequences). After ligation, the DNA is gel-purified and size selected for 150–300 bp. Ligated fragments are PCR enriched with primers corresponding to the adaptor ends (colored arrows represent PCR primers). A total of 8–10 reactions are carried out to obtain 20 µg of amplified DNA for hybridization and to ensure that all hybridized fragments contain detectable ends for forming sequence clusters on the Illumina instrument. The PCR products are added to a cocktail of Cot-1 DNA, blocking oligos and hybridization buffer, and the 244k tiling arrays are hybridized for several days. After hybridization, the arrays are stringently washed and captured fragments are eluted in water at 95 °C. The eluted material is lyophilized, and recovered strands either undergo further PCR amplifications or are added directly to the Illumina Genome Analyzer flow cell for cluster formation.
Improvements to the method and their rationale include the following:
Earlier versions of this protocol involved genomic DNA libraries sheared to 600 bp. However, fragment size is a critical consideration for optimal coverage on short-read SBS platforms. Therefore, we reduced the average fragment size of the libraries from 600 to 150–300 bp. The optimal base length may need to be further varied, depending on the platform on which fragments will be analyzed.
-
Hybridization efficiency requires a driver. As probe concentrations on the array are fixed, excess input DNA must drive the hybridization. For rare samples with limited quantities, obtaining such an amount of primary DNA can be difficult. One solution to this limitation is the use of strand displacing enzymes for whole-genome amplification (WGA)14. However, WGA may introduce representational bias that can adversely affect sequence coverage and fragment distribution13,14.
Importantly, the addition of adaptors before selection allows hybridized fragments to be distinguished from contaminants such as oligonucleotides that invariably leach off the arrays and Cot-1 DNA (a repeat-rich additive intended to block nonspecific hybridization). Therefore, as an alternative to WGA, we took advantage of the fact that the DNA is pre-ligated to Illumina compatible adaptors to generate amplifiable libraries using adaptor-specific primers. In this way, even small amounts of sample (approximately 100 ng) could be amplified by high-fidelity PCR to yield material sufficient for multiple captures while minimizing representational bias. Moreover, pre-amplification, coupled with the compressed dynamic range of arrays, in general, tends to normalize the fragment population and maximizes the potential that every captured fragment has the opportunity to be detected by sequencing.
We have continued to develop and optimize this approach in order to streamline the process and to improve target specificity. In the course of this work, we empirically evaluated multiple array platforms. We were compelled to choose a well-established system that is user-friendly with limited opportunity for variability in performance because of user-dependent (or experience-dependent) error. We obtained the most reproducible results on Agilent 244k DNA microarrays. We have also tested, to a limited extent, 1 M-feature Agilent arrays, before their general release, and found that they retain the specificity and selectivity seen with 244k arrays while allowing approximately four-fold greater genomic coverage. Our updated method is essentially a modified version of the standard Agilent aCGH (array comparative genomic hybridization) protocol using stock reagents, and this has eased the transfer of the method to other laboratories familiar with microarray processing.
Here, we describe in detail our array capture protocol. This procedure assumes initial DNA quantities of 1–2 µg. We are applying this protocol to tackle numerous genomic questions in multiple organisms including humans, mouse and rat. In fact, highly comparable enrichment data were obtained from capture experiments probing both human exons from a region below 1 Mb (see ANTICIPATED RESULTS) and from ~1,000 mouse exons for a total of 4 Mb of target sequence. Therefore, we find that the improvements described here yield better, more comparable results that are largely species- and target-independent.
Experimental design
This section outlines key points for the array capture setup providing, in our experience, the best results that we have been able to attain thus far.
Array design
The array configurations currently in use in our lab are designed in a manner similar to earlier described methods13, with several important exceptions. First, Agilent arrays have 243,504 features available for probe design, which are ~140,000 fewer features than the NimbleGen arrays used earlier. However, the Agilent features cover a larger surface area, and thus are likely to contain >50 times more molecules per feature. In addition, we have relied on a standard 60-mer oligonucleotide production format that omits synthesis of variable length probes for Tm matching, reducing complications during probe design and array synthesis. We have designed arrays that target genomic regions ranging from 0.5 to 4 Mb with tiling intervals, meaning the distance in nucleotides that separate the start positions of the probes, from 3 up to 20 bp depending on the size of the areas under selection. Earlier, we noted that tiling across individual regions (in this case exons) generates unequal read depth within the boundaries of that interval. This may be explained by the fact that probes begin to overlap so that the center of the region contains more overlapping probes than the edges of the region (where probes begin to taper off). To compensate for this trend, we begin tiling each interval in flanking regions beginning 60 bp upstream and ending 60 bp downstream of the interval boundaries. Furthermore, if an interval is shorter than 150 bp, we extend the region at both ends to include a minimum of 150 bases of genomic sequence.
Probably the most critical aspect of array design is the filtering of repetitive elements to reduce nonspecific binding. Not only do we exclude nonunique probes but also we eliminate probe sequences that correspond to highly repetitive regions in the genome. There are two methods by which this can be accomplished. The first method, called RepeatMasker filtering (Smit, A.F.A., Hubley, R. & Green, P. RepeatMasker Open-3.0, 1996–2004), results in conservative coverage of the genome with regard to repeat-rich regions. An alternative approach, based on the frequency of 15-mer sequence combinations in the genome, calculates the 15-mer frequency for all 46 potential stretches of contiguous 15 bases within an individual 60-mer probe13,19. If the average genomic frequency for the 46 15-mers is greater than a set threshold, in our case 100, the probe is filtered out. The 15-mer frequency tables that have been generated for both the mouse and human genomes are available upon request. In general, this method is slightly less stringent than the Repeat-Masker filtering, but can be adjusted by setting the threshold lower or higher depending on the desired stringency and the sequence content of the regions of interest. For example, if the desired target is an entire genic region, containing both exons and introns, and a considerable portion of this is repetitive, it may be necessary to relax this threshold to include more of the region or achieve better coverage. On the other hand, if several paralogs exist for a particular gene, a lower, more rigid threshold may be necessary.
To control for performance and reproducibility between experiments, a number of randomly dispersed array features may be reserved for a small subset of informative intervals. These intervals may serve as either positive controls of enrichment (enrichment standards of an expected level), i.e., regions/exons displaying high levels in earlier captured material, or for controlling sample content whereby contamination levels can be estimated by looking at known/expected haplotype regions. The ‘control grid’ can be used to unify results obtained from arrays of different target origin and size (within the same species).
Library construction
The quality of the genomic DNA library strongly influences the success of the array capture. Our library protocol is historically based on the standard Illumina procedure11 for single-end sequencing, but can be easily adapted for paired-end applications. The first step in this process is DNA fragmentation, either by nebulization or by sonication. Nebulization consumes more DNA than sonication owing to the atomizing process and the wide size distributions generated, which is not favorable when size selection is necessary. Thus, for many samples, nebulization is unsuitable because of the large amounts of required input DNA. For this reason, we use sonication as an alternate fragmentation method for most library preparation. Although many options exist, we routinely generate high-quality libraries using a focused acoustics system (Covaris) or a closed system ultrasonic disruptor (Bioruptor, Diagenode). The latter system is lower in cost, while achieving higher energy transfer efficiency and more reproducible performance than standard probe sonicators. In addition, multiple samples can be processed simultaneously in a uniform manner. Subsequently, DNA is subjected to a series of enzymatic reactions that repair frayed ends, phosphorylate the fragments, and add a single nucleotide A overhang (Fig. 1). In these protocols, we have used both the kit reagents purchased from Illumina11 and off-the-shelf reagents as described below with comparable results. After ligating Illumina adaptors, 150–300 bp fragments are selected and purified by gel extraction. At this stage, we carry out multiple parallel PCR amplifications (8–10 per sample) of the ligated fragments after which the amplified products are pooled. By carrying out multiple reactions in parallel and pooling, fewer cycles are required to generate the optimal 20 µg for capture without comprising the complexity or skewing the representation/introducing a representational bias. In addition, barcoding strategies, which enable sample multiplexing, are suitable for array capture and may offset the per sample number of PCRs required. It is also worth noting that significant improvements to the library preparation protocol have been recently reported and could easily be implemented here20.
Blocking repeats
Although the arrays are processed under stringent hybridization and washing conditions, certain factors can influence the specificity of the capture experiment. There are several scenarios in which unintended cross-hybridization of fragments can occur. First, although repetitive probe filtering helps to reduce nonspecific hybridization to the array, it does not prevent repeats from confounding the results altogether. This may be because the average size of the input DNA is generally larger (2.5–5×) than the size of the probes. Therefore, the complementary oligo segment affixed to the array accounts for less than half of the DNA fragment. This leaves an unbound area of single-stranded DNA free to bind to any complement in the applied sample. If a repeat is contained within the unbound segment of the fragment, it may hybridize to complementary repeats in other library fragments (Fig. 2). This is especially challenging on the periphery of interval borders where bound DNA fragments can extend to regions beyond probe coverage into areas not filtered for repeats. To counteract this potential problem, species-specific Cot-1 DNA is added in excess of the input DNA to the hybridization mixture. The Cot-1 DNA is the repeat-rich fraction of genomic DNA that has been isolated on the basis of its re-annealing characteristics. We have used up to five-fold excess Cot-1, achieving our best results at 2.5-fold excess (see ANTICIPATED RESULTS). A second source of cross-hybridization stems from the common adaptor sequences present on the hybridized fragments. When the DNA becomes denatured before hybridization, the complementary adaptor sequences can bind indiscriminately to each other, regardless of the insert sequence (Fig. 2). The Illumina adaptors are also easily long enough to remain annealed under the conditions used for hybridization. Therefore, to compete for adaptor binding, we supplement the hybridization mixture with a molar excess of four distinct ‘blocking oligos’ that complement each strand of the adaptor sequence.
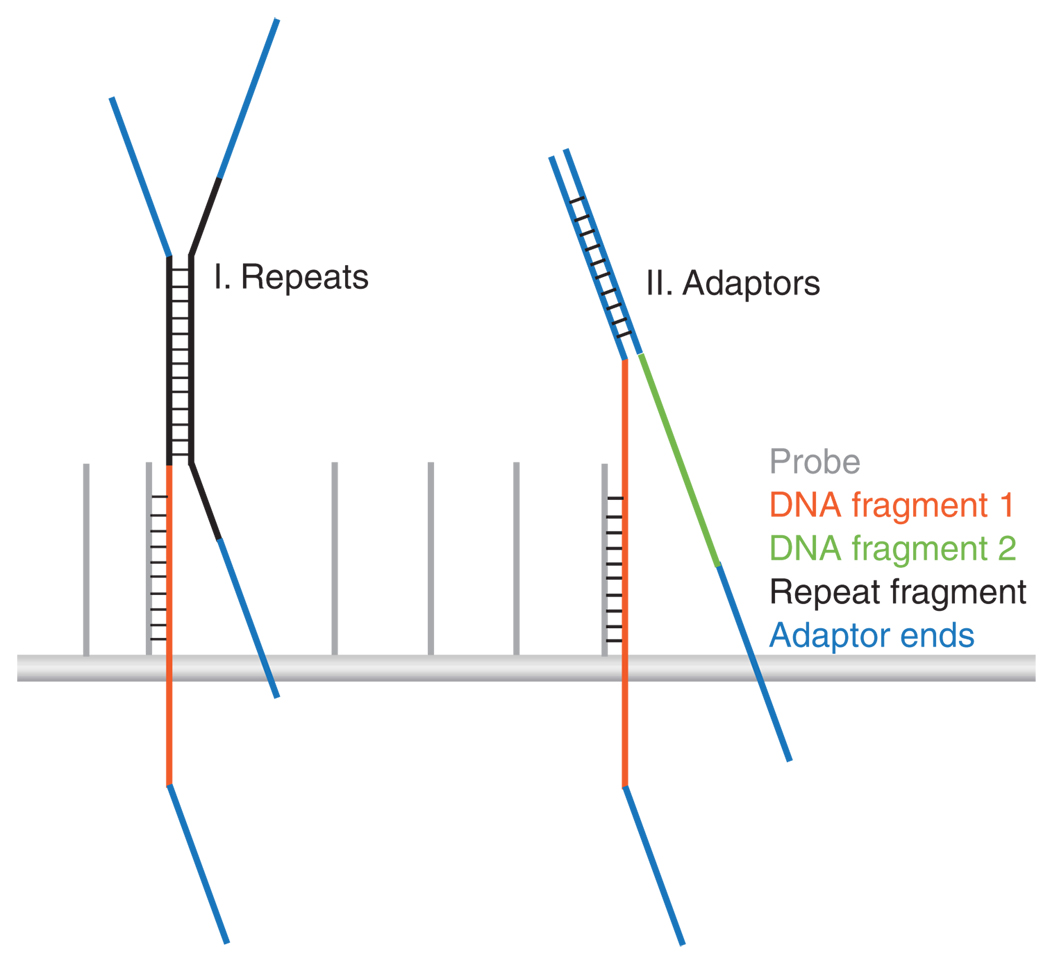
Figure 2.
Sources of cross-hybridization that influence specificity. Two scenarios are depicted in which unintended fragments can be purified along with targeted sequences. The first illustration shows the potential hybridization between a fragment of repetitive DNA (shown in black) and the probe-bound DNA fragment (shown in orange). The second illustration shows adaptor (shown in blue) complementation between two unrelated DNA inserts.
Elution
The elution step can be challenging without the appropriate equipment. Our goal was to develop a straightforward means of denaturing and recovering the hybridized material without using a specialized elution apparatus. The strategy we use takes advantage of the standard Agilent slide gasket chamber system (Fig. 3). This system comprises a steel chamber base and a rubber gasket-slide that, when assembled, forms a sandwich with the microarray and creates a hybridization compartment for the printed surface of the array. The rubber gasket creates a space through which a small-gauge syringe can be inserted without compromising the integrity of the seal. The procedure we outline requires the use of a hybridization oven that must be capable of reaching 95 °C. The elution mixture is withdrawn from the chamber using a syringe, and the liquid eluate is lyophilized from a 490- to 50-µl volume. Subsequently, an additional PCR step is carried out to accurately quantify the captured material. However, as the captured fragments already contain the full sequences necessary for cluster generation, this step may be eliminated provided the fragments can be quantified by quantitative PCR (qPCR).
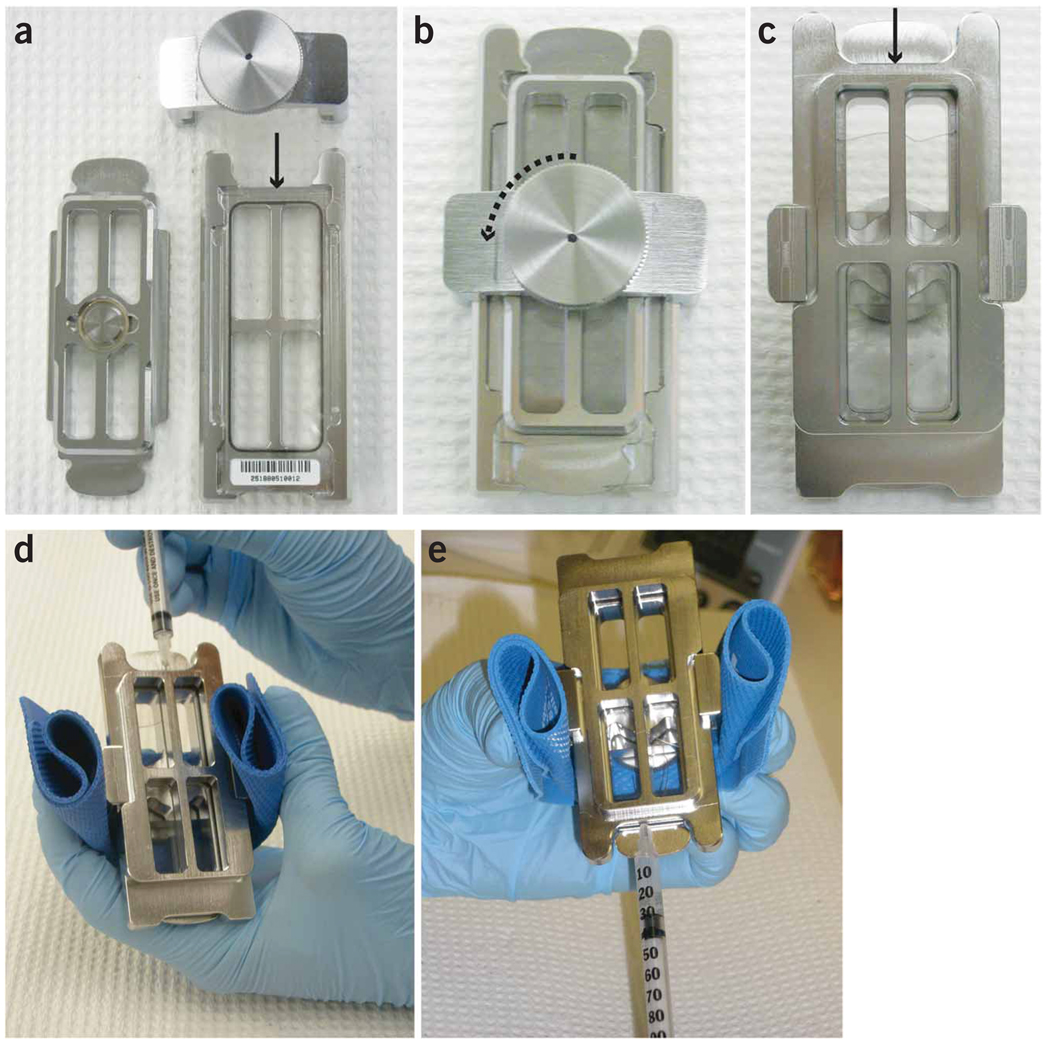
Figure 3.
Elution strategy showing chamber assembly and syringe method. To perform the elution step, nuclease-free water is added to the gasket slide surface and the array is placed with the printed side down (a). The chamber base is assembled, tightened and placed in the rotating oven at 95 °C. After the captured strands are melted, the eluted material is recovered by turning the screw 1/4 turn (b). The chamber is hot and must be handled with care by holding it with a rubber grip (d). There are two different ends to the chamber base, a narrow end (shown by arrows) and the end marked by the array labels (c). The liquid can be seen through the chamber base from the side opposite the screw through which a large air bubble is visible (c). The chamber base is tilted so that the liquid shifts toward the label end and the air bubble is near the narrow end (d). The syringe is inserted through the space between the array and gasket slides into the air bubble at the narrow end (d). The chamber is then tilted back towards the syringe (e). The liquid eluate is carefully withdrawn from the compartment into the syringe and transferred to a 1.7-ml maximum recovery centrifuge tube.
Evaluating the results
Capture performance can be assessed either by qPCR for a defined set of intervals or by sequencing the captured material on an SBS instrument. For qPCR, primer pairs are selected for a small set of target loci and the CT values for each locus are compared between the input DNA and the captured DNA (see ANTICIPATED RESULTS). Larger exons are ideal for designing well-matched pairs. Negative controls should also be included. These can include any genomic region for which there are no selected probes. Each primer should be ~20 bp with a melting temperature of 58–60 °C. Optimal amplicon sizes are around 100 bp. Further instructions on designing primers can be found in the SYBR Green PCR master mix user guide. qPCR is useful for acquiring an initial snapshot of success, and as an experimental quality control for determining whether sequencing the eluate will be informative. However, qPCR does not provide a global sense of enrichment specificity and sensitivity. Therefore, massively parallel sequence analysis from single captured molecules is the only accurate and comprehensive way to estimate performance. The sequence analysis procedure will be described in further detail below.
After the hybrid selection process, enriched samples undergo cluster generation on the Illumina flow cell. Cluster generation is followed by 36 cycles of single base extension and cluster imaging on the Illumina Genome Analyzer (GA). Next, the cluster images are compiled and analyzed, and the called bases are assigned a quality score. Filtered sequences are mapped to the reference human genome (Hg18) using Eland, a built-in mapping algorithm for the Illumina analysis pipeline. Only reads mapping to unique positions in the genome with at most two mismatches to the reference are considered. We evaluate the success of an individual capture experiment by several metrics. First, the specificity is measured by the percentage of reads (the number of reads in target/the number of reads mapped) that overlap with targeted intervals by at least 1 bp (see ANTICIPATED RESULTS). Another way to describe specificity is by generating a ‘fold enrichment’ score that is calculated by dividing the percentage of reads in target by the percentage of bases targeted in the genome.
The sensitivity of the array selection process is evaluated on two levels: (1) sequence coverage of the regions selected and (2) higher resolution coverage at the actual base pair level. The first level of coverage is determined by the number of targeted intervals covered by at least one overlapping read. Similarly, the base pair coverage is measured by the number of total target bases covered by at least one read. Both numbers provide a general estimate of the extensiveness and the complexity of target purification (see ANTICIPATED RESULTS). However, as the overall objective is to enable high-quality variant detection through enhanced coverage, the most valuable measurement will be the median sequencing depth, or the median number of times (X) an individual base within the target interval is covered by a unique read. For confident single nucleotide polymorphism (SNP) detection, we require high-quality base calls (> Q20) with a sequencing depth of at least 20×.
MATERIALS
REAGENTS
1–2 µg purified genomic DNA
Water, nuclease-free (Invitrogen/Gibco, cat. no. 15230)
EB buffer (supplied with Qiagen PCR Purification kits, see below); 10mM Tris-HCl, pH 8.5
Tris, Ultra Pure (MP Biomedicals, cat. no. 103133)
EDTA (Sigma, cat. no. ED2SS)
Sucrose (Fluka, cat. no. 84097)
Bromophenol blue (Sigma-Aldrich, cat. no. B6131)
Xylene cyanol (Sigma-Aldrich, cat. no. G5516)
Glycerol (Fisher Scientific, cat. no. G33-500)
T4 DNA ligase reaction buffer (NEB, cat. no. B0202S), contains 10 mM ATP
dNTPs (Roche, cat. no. 11814362001), supplied as 10 mM each
T4 DNA polymerase (NEB, cat. no. M0203L)
Klenow DNA polymerase I (NEB, cat. no. M0210L)
T4 polynucleotide kinase (NEB, cat. no. M0201L)
dATP (Roche, cat. no. 11934511001), supplied as 100 mM (see REAGENT SETUP)
Klenow fragment (3′ to 5′ exo minus) (NEB, cat. no. M0212L), supplied with 10× Klenow reaction buffer
Rapid DNA Ligation Kit (Roche, cat. no. 11635379001), including 2× T4 DNA ligase buffer, 5× DNA dilution buffer and T4 DNA ligase
DNA Sample Prep Oligo Only Kit (Illumina, cat. no. FC-102-1003), including adapter oligo mix and PCR primers
 Additional PCR primers may be needed to obtain enough initial DNA for hybridization. They can be purchased from (IDT) HPLC purified and lyophilized. Sequences are available in Table 1.
Additional PCR primers may be needed to obtain enough initial DNA for hybridization. They can be purchased from (IDT) HPLC purified and lyophilized. Sequences are available in Table 1.2× Phusion High-Fidelity PCR Master Mix (Finnzymes, cat. no. F-531); contains 0.04 U µl−1 Phusion DNA polymerase, 2× Phusion HF buffer and 400 µM of each dNTP. The master mix provides 1.5 mM MgCl2 and 200 µM dNTP in final reaction concentration
Agarose gel (Roche, cat. no. 11685660001) (see REAGENT SETUP)
Ethidium bromide (Sigma, cat. no. 46067)
 Mutagen and potential carcinogen.
Mutagen and potential carcinogen.MinElute PCR Purification Kit (Qiagen, cat. no. 28004)
 Buffer PBI contains materials that cause damage to the skin and eyes; may be harmful if swallowed; flammable.
Buffer PBI contains materials that cause damage to the skin and eyes; may be harmful if swallowed; flammable.QIAquick PCR Purification Kit (Qiagen, cat. no. 28104)
 Buffer PBI contains materials that cause damage to the skin and eyes; may be harmful if swallowed; flammable.
Buffer PBI contains materials that cause damage to the skin and eyes; may be harmful if swallowed; flammable.QIAquick Gel Extraction Kit (Qiagen, cat. no. 28704)
 Buffer QG contains materials that cause damage to the skin; may be harmful if swallowed or inhaled.
Buffer QG contains materials that cause damage to the skin; may be harmful if swallowed or inhaled.Agilent 244k DNA microarray (Agilent, see REAGENT SETUP)
 Microarrays are shipped vacuum packed in foil. If microarrays are not immediately used after opening the foil pack, store them at 22 °C (room temperature, RT) inside a vacuum desiccator.
Microarrays are shipped vacuum packed in foil. If microarrays are not immediately used after opening the foil pack, store them at 22 °C (room temperature, RT) inside a vacuum desiccator.Blocking oligonucleotide mix (IDT), desalted and lyophilized. Sequences available in Table 1
Cot-1 DNA (Invitrogen or Applied Genetics); supplied as 1 mg ml−1
 Order appropriate species-specific Cot-1 DNA.
Order appropriate species-specific Cot-1 DNA.Oligo aCGH Hybridization Kit (Agilent, cat. no. 5188-5220), including 10× blocking agent and 2× hybridization buffer
 Contains materials that cause damage to the skin and the central nervous system. May be harmful if swallowed
Contains materials that cause damage to the skin and the central nervous system. May be harmful if swallowed  10× blocking agent is supplied as a lyophilized pellet that must be reconstituted (see REAGENT SETUP).
10× blocking agent is supplied as a lyophilized pellet that must be reconstituted (see REAGENT SETUP).Oligo aCGH Wash Buffer Kit (Agilent, cat. no. 5188-5226), including two 4-liter containers of wash buffer 1 and one 4-liter container of wash buffer 2
 Contains material that causes damage to the skin and the central nervous system; may be harmful if swallowed
Contains material that causes damage to the skin and the central nervous system; may be harmful if swallowed  Shake well before using.
Shake well before using.50 bp DNA Ladder (Crystalgen, cat. no. 65-0371)
SYBR Green PCR Master Mix (Applied Biosystems, cat. no. 4309155)
Real-time PCR primers (IDT), desalted and lyophilized (see REAGENT SETUP)
TABLE 1.
Genomic DNA oligonucleotide sequences.
| PCR primers (50 µM each) | |
| PCR primer 1.1 | 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT–3′ |
| PCR primer 2.1 | 5′-CAAGCAGAAGACGGCATACGAGCTCTTCCGATCT-3′ |
| Blocking oligonucleotide (BO) mix (200 µM each) | |
| BO 1 (PCR primer 1.1) | 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′ |
| BO 2 (PCR primer 2.1) | 5′-CAAGCAGAAGACGGCATACGAGCTCTTCCGATCT-3′ |
| BO 3 (reverse complement 1.1) | 5′-AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGCCGTATCATT 3′ |
| BO 4 (reverse complement 2.1) | 5′-AGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG-3′ |
Oligonucleotides sequences © 2006 and 2008 Illumina Inc. All rights reserved.
EQUIPMENT
1.5 ml centrifuge tubes (Eppendorf)
1.7 ml maximum recovery centrifuge tubes (Axygen)
0.2 ml 8-well PCR strip tubes or individual 0.2 ml PCR tubes (VWR)
Sonicator (e.g., Diagenode Model Bioruptor UCD-200, Diagenode; equipped with 1.5-ml microtube unit) (see EQUIPMENT SETUP)
 Genomic DNA can be fragmented using other sonication systems or other methods of shearing such as nebulization. However, sonication is recommended as it results in a higher recovery of input DNA.
Genomic DNA can be fragmented using other sonication systems or other methods of shearing such as nebulization. However, sonication is recommended as it results in a higher recovery of input DNA.Vortex (Fisher Scientific, cat. no. 02215365)
Centrifuge (Model 5417C, Eppendorf)
Minicentrifuge (Fisher Scientific, cat. no. 05090100)
Thermal cycler (Model PTC-225, MJ Research)
Agarose Gel Electrophoresis Unit (Bio-Rad)
UV transilluminator
 UV radiation can cause damage to unprotected eyes and skin.
UV radiation can cause damage to unprotected eyes and skin.NanoDrop ND-1000 spectrophotomoter (NanoDrop Technologies)
Heating blocks (95, 50 and 37 °C)
Incubator (37 °C)
Vacuum desiccator (Nalgene, cat. no. 5311-0250)
 It is important to store DNA microarray slides in a desiccator to keep them as dry as possible when not in use.
It is important to store DNA microarray slides in a desiccator to keep them as dry as possible when not in use.Microarray gasket slides (Agilent, cat. no. G2534-60003)
SureHyb DNA Microarray Chamber Kit (Agilent, cat. no. G2534A), including a stainless steel chamber base and top, thumb clamp and screw, and plastic tweezers (see EQUIPMENT SETUP)
Microarray hybridization oven (SciGene Model 700 Microarray Oven, SciGene, cat. no. 1070-00-1)
 This particular oven is ideal because it is compatible with Agilent SureHyb chambers and is capable of heating up to 99 °C.
This particular oven is ideal because it is compatible with Agilent SureHyb chambers and is capable of heating up to 99 °C.Sterile disposable bottles (Nalgene, cat. no. 455-0500)
Glass slide-staining dishes with lids (Electron Microscopy Sciences, cat. no. 71420-DL), protocol requires at least three dishes
Metal slide racks with handles (Electron Microscopy Sciences, cat. no. 71420-SR)
Glass drying trays (Corning, cat. no. 3175-7)
Magnetic stir bars (Fisher Scientific, cat. no. 14-513-51)
Magnetic stirring hotplate (Thermo Scientific Model Thermolyne Nuova, Thermo Scientific)
Centrifuge fitted with microplate adapters (Sorvall Legend T, Sorvall)
SpeedVac Concentrator (Model SVC 100, Savant)
30-G syringes (BD Medical, cat. no. 328411)
Skirted real-time PCR plates (Bio-Rad Laboratories, cat. no. HSP-9655)
Flat-cap strips for real-time PCR plates (Bio-Rad Laboratories, cat. no. TCS0803)
Real-time PCR thermal cycler (Model PTC-0221G, Bio-Rad Laboratories)
Illumina Genome Analyzer and associated equipment
REAGENT SETUP
6× DNA-loading dye
0.25% (wt/vol) bromophenol blue, 0.25% (wt/vol) xylene cyanol FF, 30% (vol/vol) glycerol in water. Add 1 vol of 6× DNA-loading dye to 5 vol of DNA sample. Store at RT for regular use or −20 °C for long-term storage.
Sucrose loading buffer
50 mM Tris (pH 8.0), 40 mM EDTA, 40% (wt/vol) sucrose: can be stored at RT (20–25 °C) for several months.
20× Tris–acetate–EDTA (TAE) electrophoresis buffer
Dissolve 96.8 g Tris in 250 ml water. Add 22.8 ml acetic acid and 40 ml of 0.5 M EDTA (pH 8.0). Adjust to a final volume of 1 liter. Vacuum filter the buffer before use. Prepare a 1× TAE working dilution; can be stored at RT for several months.
2% agarose gel
Add 3 g agarose to 150 ml 1× TAE electrophoresis buffer. Heat in a microwave oven until completely melted. Add ethidium bromide (final concentration 0.4–0.5 µg ml−1) to facilitate visualization of DNA after electrophoresis. After cooling to 50–60 °C, pour gel into a casting tray containing a gel comb and allow it to solidify at RT. Make a fresh gel before loading samples.
1 mM dATP
Make 1 mM working dilutions from 100 mM stock. Store at −20 °C until further usage.
Preparing PCR primers
Dissolve the primers in nuclease-free water to obtain 100 µM stock solutions. Prepare 50 µM working dilutions. Freeze at −20 °C until further usage.
Preparing blocking oligonucleotide mix
Dissolve the primers in nuclease-free water to obtain 200 µM working dilutions for the hybridization master mix. Be sure to vortex thoroughly. Freeze at −20 °C until further usage.
10× blocking agent
Add 1,350 µl nuclease-free water to the lyophilized pellet. Leave at RT for 60 min. Mix gently by vortexing. Aliquot ~110 µl into several 1.5-ml centrifuge tubes to avoid freeze–thaw cycles. Store at −20 °C until further usage.
Preparing real-time PCR primers
Dissolve the real-time primers in nuclease-free water to obtain 100 µM stock solutions. Prepare 10 µM working dilutions for real-time PCR master mix. Freeze at −20 °C until further usage.
EQUIPMENT SETUP
Sonicator
We use the Diagenode Bioruptor UCD-200 equipped with a 1.5-ml microtube unit, which holds up to six 1.5-ml centrifuge tubes. Set the power to high (200 W) with alternating cycles of a 30-s burst of sonication followed by a 30-s pause.
SureHyb DNA microarray chambers
Refer to the Agilent microarray chamber user guide (G2354-90001) for detailed instructions on how to load samples, assemble and disassemble chambers, as well as for other useful tips. The user guide is also available with the purchase of the Agilent microarray hybridization chamber kit (G2534A).
PROCEDURE
Sonicate genomic DNA to generate fragments between 100 and 500 bp  1.5 h
1.5 h
-
1|
Dissolve 2 µg of each purified genomic DNA sample in a total volume of 80 µl using EB buffer in a 1.5-ml centrifuge tube.
-
2|
Fill the Bioruptor water tank with distilled cold water (4 °C) and supplement with 0.5-cm crushed ice.
 The level of the water and ice should always reach the blue line on the wall of the water tank. The ultrasonic waves generated by the sonicator produce a considerable amount of heat. Using cold water and ice helps regulate the temperature of the samples.
The level of the water and ice should always reach the blue line on the wall of the water tank. The ultrasonic waves generated by the sonicator produce a considerable amount of heat. Using cold water and ice helps regulate the temperature of the samples. -
3|
Assemble sample tubes into the 1.5-ml microtube unit. Align the microtube unit with the motorized lid with the samples immersed in the water bath.
-
4|
Sonicate for 7 min (see EQUIPMENT SETUP for sonicator settings).
-
5|
Remove the water in the tank. Re-fill with cold water and crushed ice as above.
-
6|
Follow Steps 3–5, two additional times making sure to change the water between 7-min intervals. The total sonication time is 21 min, which is equivalent to 21 cycles with a cumulative sonication time of 10.5 min.
 Samples can be spun down and stored at −20 °C overnight.
Samples can be spun down and stored at −20 °C overnight. -
7|
Add 1 µl 6× DNA-loading dye to 5 µl (125 ng) of each fragmented sample. Load 6 µl on a 2% (wt/vol) agarose gel (see REAGENT SETUP), reserving one lane for 6 µl of the 50-bp ladder. Run gel at ~100 V for 30 min to validate that the fragment size is between 100 and 500 bp.
 If the majority of the fragments for a particular sample are >500 bp, sonicate for an additional few minutes. (Agarose gel can be prepared earlier to save time.)
If the majority of the fragments for a particular sample are >500 bp, sonicate for an additional few minutes. (Agarose gel can be prepared earlier to save time.)
Carry out end repair on fragmented genomic DNA to generate blunt ends  0.75 h
0.75 h
-
8|Prepare the following master mix in a 1.5-ml centrifuge tube for each sample. Mix carefully by pipetting up and down or flicking the tube, and spin down.
Reagent Volume (µl) per sample Final concentration in reaction T4 DNA ligase buffer with 10 mM ATP (10×) 10 1 mM ATP (1×) dNTP mix (10 mM each) 4 400 µM each T4 DNA polymerase (3 U µl−1) 5 0.15 U µl−1 Klenow DNA polymerase (5 U µl−1) 1 0.05 U µl−1 T4 polynucleotide kinase (10 U µl−1) 5 0.5 U µl−1  All reagents are stored at −20 °C. Prepare the master mix on ice.
All reagents are stored at −20 °C. Prepare the master mix on ice. -
9|
Add 25 µl of master mix to each fragmented DNA sample to obtain a total reaction volume of 100 µl. Mix and spin down. Incubate in a thermal cycler for 30 min at 20 °C.
-
10|
Purify each sample on one column using reagents from the QIAquick PCR Purification Kit following the manufacturer’s instructions. Elute in 32 µl EB buffer.
 Samples can be stored at 4 °C overnight or −20 °C for longer storage.
Samples can be stored at 4 °C overnight or −20 °C for longer storage.
Add 3′ A-overhangs to the ends of the blunted DNA fragments  0.75 h
0.75 h
-
11|Prepare the following master mix in a 1.5-ml centrifuge tube for each sample. Mix carefully and spin down.
Reagent Volume (µl) per sample Final concentration in reaction Klenow buffer (10×) 5 1× dATP (1 mM) 10 0.2 mM Klenow fragment exo− (5 U µl−1) 3 0.3 U µl−1  All these reagents are stored at −20 °C. Prepare the master mix on ice to maintain full enzyme activity.
All these reagents are stored at −20 °C. Prepare the master mix on ice to maintain full enzyme activity. -
12|
Add 18 µl of master mix to each sample to obtain a total reaction volume of 50 µl. Mix and spin down. Incubate for 30 min at 37 °C.
-
13|
Purify each sample on one column using the reagents from the MinElute PCR Purification Kit following the manufacturer’s instructions. Elute in 10 µl EB buffer.
 MinElute columns should be stored at 4 °C before use to maximize recovery.
MinElute columns should be stored at 4 °C before use to maximize recovery. Samples can be stored at 4 °C overnight or −20 °C for longer storage.
Samples can be stored at 4 °C overnight or −20 °C for longer storage.
Ligate adapters to DNA fragments  0.5 h
0.5 h
-
14|Prepare the following master mix in a 1.5-ml centrifuge tube for each sample. Mix thoroughly and spin down.
Reagent Volume (µl) per sample Final concentration in reaction T4 DNA ligase buffer (2×) 25 ~1× DNA dilution buffer (5×) 5 0.5× Adapter oligo mix 10 µM 10 2 µM T4 DNA ligase (5 U µl−1) 1 0.1 U µl−1  All these reagents are stored at −20 °C. Prepare the master mix on ice to maintain full enzyme activity.
All these reagents are stored at −20 °C. Prepare the master mix on ice to maintain full enzyme activity. -
15|
Add 40 µl of master mix to each ligated DNA sample to obtain a total reaction volume of 51 µl. Mix gently and spin down. Incubate at RT for 15 min.
 Proceed immediately to gel purification.
Proceed immediately to gel purification.
Size-select and gel-purify ligated products  1.5 h
1.5 h
-
16|
Prepare a 2% agarose gel using 1× TAE (see REAGENT SETUP). Use a gel comb that is wide enough to hold 71 µl of each sample. (Agarose gel can be prepared earlier to save time.)
-
17|
Add 20 µl of sucrose loading buffer to 51 µl of each purified adaper-ligated DNA sample.
-
18|
Load 5 µl of the 50-bp DNA ladder to one lane of the gel.
-
19|
Load the entire sample in another lane of the gel, leaving at least one empty well between the ladder and the sample, and between samples.
 It is essential to leave a well between the ladder and the sample, and between multiple samples to prevent cross-contamination.
It is essential to leave a well between the ladder and the sample, and between multiple samples to prevent cross-contamination. -
20|
Run the gel at 100–110 V for 30–40 min. View the gel on a UV transilluminator.
 Prolonged exposure to UV light can damage DNA and cause harm to the skin and eyes.
Prolonged exposure to UV light can damage DNA and cause harm to the skin and eyes.
-
21|
Using a clean scalpel for each slice, excise a gel slice between 150 and 300 bp and place in a 1.5-ml centrifuge tube.
 Use a clean scalpel for each sample to minimize cross-contamination.
Use a clean scalpel for each sample to minimize cross-contamination. -
22|
Use the Qiagen gel extraction kit to purify the DNA from each agarose slice following the manufacturer’s instructions. Use one column per sample. Elute in 1.7-ml maximum recovery centrifuge tubes using 30 µl EB buffer.
 Up to 400 mg agarose can be processed per spin column with a maximum volume of 750 µl per spin cycle. If the gel slice is large, it may be necessary to spin down the entire gel slice solution using several spin cycles or on several columns and pooling the eluates.
Up to 400 mg agarose can be processed per spin column with a maximum volume of 750 µl per spin cycle. If the gel slice is large, it may be necessary to spin down the entire gel slice solution using several spin cycles or on several columns and pooling the eluates. Samples can be stored at 4 °C overnight or −20 °C for longer storage.
Samples can be stored at 4 °C overnight or −20 °C for longer storage.
Enrich the adapter-modified DNA fragments by PCR  2.5 h
2.5 h
-
23|Prepare the following master mix in a 1.5-ml centrifuge tube for each sample. Mix and spin down.
Reagent Volume (µl) per sample Final concentration in reaction Phusion DNA polymerase (2×) 25 1× PCR primer 1.1 (50 µM) 1 1 µM PCR primer 2.1 (50 µM) 1 1 µM Water 22  All reagents are stored at −20 °C. Prepare master mix on ice.
All reagents are stored at −20 °C. Prepare master mix on ice. -
24|
Add 49 µl of master mix to 1 µl of each sample to obtain a total reaction volume of 50 µl. Carry out 8–12 parallel reactions for each sample. Mix gently and spin down.
 Multiple parallel reactions are required to obtain at least 21 µg of each library after PCR purification for hybridization and downstream real-time PCR. If <21 µg is obtained, carry out additional rounds of PCR.
Multiple parallel reactions are required to obtain at least 21 µg of each library after PCR purification for hybridization and downstream real-time PCR. If <21 µg is obtained, carry out additional rounds of PCR. -
25|Amplify using the following PCR conditions:
- Step 1: 98 °C for 30 s
- Step 2: 98 °C for 10 s
- Step 3: 65 °C for 30 s
- Step 4: 72 °C for 30 s
- Step 5: Repeat Steps 2–4, 17 times for a total of 18 cycles
- Step 6: 72 °C for 5 min
- Step 7: Hold at 4 °C
-
26|
Purify 2.5–3 reactions of each sample on one column using reagents from the QIAquick PCR purification kit following the manufacturer’s instructions. Elute each column in 30 µl EB buffer. Pool the eluted material for each sample into one tube.
-
27|
Use a NanoDrop spectrophotometer to quantify the purified enriched adapter-modified DNA.
 Samples can be stored at −20 °C.
Samples can be stored at −20 °C.
-
28|
Analyze 5 µl on a 2% agarose gel as described in Step 7 to validate that the fragment size is between 150 to 300 bp.
 The size range should be similar to the size range excised during the gel purification step (Step 21). (Agarose gel can be prepared earlier to save time.)
The size range should be similar to the size range excised during the gel purification step (Step 21). (Agarose gel can be prepared earlier to save time.)
Prepare enriched adapter-modified genomic DNA for hybridization  1 h
1 h
-
29|
Dissolve 20 µg of adapter-modified genomic DNA in a total volume of 138 µl in a 1.5-ml centrifuge tube using nuclease-free water.
-
30|Prepare the hybridization mixture in a 1.5-ml centrifuge tube for each sample. The final hybridization mixture volume is 520 µl.
Reagent Volume (µl) per sample Final concentration in reaction 20 µg adapter-modified genomic DNA 138 ~38.5 ng µl−1 BO 1 (200 µM) 5 ~2 µM BO 2 (200 µM) 5 ~2 µM BO 3 (200 µM) 5 ~2 µM BO 4 (200 µM) 5 ~2 µM Cot-1 DNA (1 mg ml−1) 50 ~0.01 mg ml−1 Agilent blocking agent (10×) 52 1× Agilent hybridization buffer (2×) 260 1× BO, blocking oligonucleotide. It is important to add the components in the order listed to prevent precipitation of DNA. Avoid creating bubbles in the hybridization mixture. Mix the 2× hybridization buffer by inverting gently before adding it to the hybridization mixture.
It is important to add the components in the order listed to prevent precipitation of DNA. Avoid creating bubbles in the hybridization mixture. Mix the 2× hybridization buffer by inverting gently before adding it to the hybridization mixture. -
31|
Denature samples for 3 min at 95 °C.
-
32|
Immediately transfer sample tubes to a 37 °C heat block. Incubate at 37 °C for at least 30 min.
-
33|
Remove sample tubes from the heat block and spin for 1 min at 17,800g to collect the sample at the bottom of the tube.
Set up microarray hybridization  65 h
65 h
-
34|
Preheat the microarray hybridization oven to 65 °C.
-
35|
Load a clean gasket into the Agilent SureHyb chamber base with the rubber seal facing up. Align the gasket label with the rectangular section of the chamber base.
 Ensure that the gasket slide is level with the chamber base.
Ensure that the gasket slide is level with the chamber base. -
36|
Carefully dispense 490 µl of the hybridization mixture (from Step 33) onto the gasket surface. Start close to one end of the inner rubber seal and slowly dispense the mixture while moving the pipette to the opposite end of the well moving up and down in a zigzag manner.
 Avoid pipetting the hybridization mixture too close to the rubber seal of the gasket to prevent leakage.
Avoid pipetting the hybridization mixture too close to the rubber seal of the gasket to prevent leakage.
-
37|
Place the desired microarray slide onto the gasket slide with the ‘Agilent’-labeled barcode facing down and the numeric barcode facing up. Make sure the array-gasket sandwich is properly aligned with the gasket and microarray labels facing each other on the same side.
 Each microarray is printed on the side containing the ‘Agilent’-labeled barcode. This is the ‘active side.’ Avoid touching the active surface of the array by carefully handling the array by its edges.
Each microarray is printed on the side containing the ‘Agilent’-labeled barcode. This is the ‘active side.’ Avoid touching the active surface of the array by carefully handling the array by its edges. -
38|
Place the SureHyb chamber cover onto the sandwiched slides. Slide the clamp assembly onto the chamber and hand-tighten.
-
39|
Vertically rotate the assembled chamber to assess the mobility of bubbles. Gently tap the assembly corners on a hard surface to dislodge any stationary bubbles.
 Multiple bubbles are acceptable as long as they move when the chamber is rotated.
Multiple bubbles are acceptable as long as they move when the chamber is rotated. -
40|
Place assembled slide chamber in the rotator rack of the hybridization oven set to 65 °C.
 Make sure the hybridization chambers on the rotator rack are balanced.
Make sure the hybridization chambers on the rotator rack are balanced. -
41|
Hybridize at 65 °C for 65 h at 12 r.p.m.
Pre-warm oligo aCGH wash buffer 2  overnight
overnight
-
42|
Add 500 ml of oligo aCGH wash buffer 2 to a sterile disposable bottle. Warm overnight in an incubator set to 37 °C along with a slide-staining dish, a 1.5- to 1.75-ml glass drying tray, and 1-liter bottle of distilled water.
 Wash buffer 2 is carried out optimally at 37 °C. Using pre-heated glassware ensures that the wash buffer 2 is maintained at 37 °C during the wash. Up to five slides can be washed in 500 ml wash buffer 2. Heat the appropriate amount of wash buffer 2 and the glassware. Perform this step the night before the hybridization is completed.
Wash buffer 2 is carried out optimally at 37 °C. Using pre-heated glassware ensures that the wash buffer 2 is maintained at 37 °C during the wash. Up to five slides can be washed in 500 ml wash buffer 2. Heat the appropriate amount of wash buffer 2 and the glassware. Perform this step the night before the hybridization is completed.
Post-hybridization microarray wash  0.5 h
0.5 h
-
43|
Fill a slide-staining dish with ~500 ml oligo aCGH wash buffer 1 at RT. (This dish will be used to disassemble the array-gasket sandwich.)
-
44|
Place a slide rack and a magnetic stir bar into a second slide-staining dish. Fill the dish with enough wash buffer 1 at RT to cover the slide rack. (This dish will be used to wash the array.)
 Up to five slides can be washed in 500 ml wash buffer 1.
Up to five slides can be washed in 500 ml wash buffer 1. -
45|
Remove the hybridization chambers from the hybridization oven and raise the temperature of the oven to 95 °C.
-
46|
On a flat surface, loosen and slide off the clamp assembly. Remove the chamber cover by holding the slide sandwich in place with plastic tweezers.
-
47|
Remove the array-gasket sandwich from the chamber base by pulling up from the ends. Quickly submerge the sandwich with the numeric barcode side facing up into the disassembly slide-staining dish with wash buffer 1 (from Step 43) at RT.
-
48|
With the sandwich completely submerged, pry the sandwich open from the numeric barcode end using plastic tweezers for leverage. Let the gasket drop to the bottom of the dish. Remove the microarray slide and place it into the slide rack in the second wash buffer 1 slide-staining dish (from Step 44) and stir at medium speed for 10 min.
 Minimize exposure of the slide to air by transferring slides as quickly and carefully as possible from one slide-staining dish to another.
Minimize exposure of the slide to air by transferring slides as quickly and carefully as possible from one slide-staining dish to another. -
49|
When 1–2 min are left for washing in buffer 1, remove wash buffer 2 and the heated glassware from the 37 °C incubator. Fill the glass drying tray three-quarters of the way up with the heated water. Place the slide-staining dish in the glass drying tray. Add a magnetic stir bar to the slide-staining dish and fill the dish with ~500 ml prewarmed wash buffer 2.
-
50|
Transfer the slide rack from wash buffer 1 (see Step 48) to wash buffer 2 (prepared in Step 49). Stir at medium speed for 5 min.
-
51|
Slowly remove the slide rack and spin in a microplate centrifuge at 75g (600 r.p.m.) for 1 min at RT (20–25 °C).
 Make sure the speed of the microplate centrifuge is not set higher than 75g. High speeds may crack the arrays.
Make sure the speed of the microplate centrifuge is not set higher than 75g. High speeds may crack the arrays.
Elute hybridized genomic DNA  0.5 h
0.5 h
-
52|
Load a clean gasket into the chamber base and dispense 490 µl nuclease-free water onto the center of the gasket surface.
 Water does not spread as easily as the hybridization mixture. Try to dispense the water as one large bubble avoiding the edges of the gasket’s rubber seal.
Water does not spread as easily as the hybridization mixture. Try to dispense the water as one large bubble avoiding the edges of the gasket’s rubber seal. -
53|
Place the dried microarray slide (from Step 51) onto the gasket slide (from Step 52) with the ‘Agilent’-labeled barcode facing down and the numeric barcode facing up. Make sure the array-gasket sandwich is properly aligned before reassembling the chamber cover and clamp.
-
54|
Hand-tighten the clamp and vertically rotate the assembled chamber to assess the mobility of bubbles. Tap the assembly on a hard surface to dislodge any stationary bubbles.
-
55|
Place assembled slide chamber in the rotator rack of the hybridization oven set to 95 °C.
-
56|
Elute at 90–95 °C for 10 min at 12 r.p.m.
 The oven temperature may decrease owing to the release of heat when the door is opened. Once the oven reaches 90 °C, start timing the elution.
The oven temperature may decrease owing to the release of heat when the door is opened. Once the oven reaches 90 °C, start timing the elution. -
57|
Carefully remove the chamber from the oven and loosen the clamp approximately half a turn.
 The chambers are extremely hot. Handle with paper towels or rubber grip to avoid burns.
The chambers are extremely hot. Handle with paper towels or rubber grip to avoid burns. -
58|
Loosen the clamp screw to 1/4 turn. Tilt the chamber so that the liquid pools on the labeled end of the array. Slowly and carefully insert the tip of a 30-gauge needle per 1 ml syringe through the rubber seal on the non-labeled end of the array-gasket sandwich where there should be an air pocket. Reverse the tilt of the chamber so that the liquid moves toward the syringe tip. Remove as much of the eluate as possible with the syringe and dispense the contents of the syringe into a 1.7 ml maximum recovery centrifuge tube.
 It may be necessary to re-insert the syringe to remove all the eluate. Close to 490 µl should be recovered.
It may be necessary to re-insert the syringe to remove all the eluate. Close to 490 µl should be recovered. Samples can be stored at 4 °C overnight.
Samples can be stored at 4 °C overnight.
Concentrate the eluted DNA  2–4 h
2–4 h
-
59|
Separate the eluted material into two 1.7-ml maximum recovery centrifuge tubes with equal volumes (or siliconized tubes). Each tube should have ~245 µl.
-
60|
Concentrate the eluted material in a SpeedVac set to medium or high for 2–4 h. Lyophilize each tube down to ~25 µl each.
 Periodically check the volume of the tubes to avoid forming a pellet.
Periodically check the volume of the tubes to avoid forming a pellet. -
61|
Combine the contents of the separated tubes from the same microarray. If <50 µl is obtained, bring the volume of the sample up to 50 µl using nuclease-free water.
 Samples can be stored at 4 °C overnight or −20 °C for longer storage.
Samples can be stored at 4 °C overnight or −20 °C for longer storage.
Amplify the eluted DNA  2–2.5 h
2–2.5 h
-
62|Prepare the following master mix in a 1.5-ml centrifuge tube for each sample. Mix and spin down.
Reagent Volume (µl) per sample Final concentration Phusion DNA polymerase (2×) 25.0 1× PCR primer 1.1 (50 µM) 1.0 1 µM PCR primer 2.1 (50 µM) 1.0 1 µM Water 18.0  All reagents are stored at −20 °C. Prepare the master mix on ice.
All reagents are stored at −20 °C. Prepare the master mix on ice. -
63|
Add 45 µl of master mix to 5 µl of each sample to obtain a total reaction volume of 50 µl. Carry out five parallel reactions for each sample. Mix gently and spin down.
-
64|Amplify using the following PCR conditions:
- Step 1: 98 °C for 30 s
- Step 2: 98 °C for 10 s
- Step 3: 65 °C for 30 s
- Step 4: 72 °C for 30 s
- Step 5: Repeat Steps 2–4, 17 times for a total of 18 cycles
- Step 6: 72 °C for 5 min
- Step 7: Hold at 4 °C
-
65|
Purify 2.5 reactions of each sample on one column (two columns total per sample) using reagents from the QIAquick PCR Purification Kit following the manufacturer’s instructions. Elute each column in 30 µl EB buffer. Pool the eluted material from each sample in one tube.
-
66|
Use a NanoDrop spectrophotometer to quantify the amplified captured DNA.
 Samples can be stored at −20 °C.
Samples can be stored at −20 °C.
-
67|
Analyze 5 µl on a 2% agarose gel as described in Step 7 to validate that the fragment size is between 150 and 300 bp.
 The size range should be similar to the size range excised during the gel purification step (from Step 21).
The size range should be similar to the size range excised during the gel purification step (from Step 21). -
68|
Samples are now ready for analysis using the Illumina Cluster Station and Genome Analyzer. Illumina recommends storing prepped DNA at a concentration of 10 nM. The prepped DNA can be adjusted to 10 nM using EB buffer and stored in a 1.7-ml maximum recovery centrifuge tube at −20 °C for several weeks or until the samples are sequenced.
Validate enrichment with real-time PCR  3.5 h
3.5 h
-
69|Prepare the following master mix in a 1.5-ml centrifuge tube. Mix and spin down.
Reagent Volume (µl) per sample Final concentration SYBR Green PCR master mix (2×) 10.0 1× Forward primer (10 µM) 0.5 0.25 µM Reverse primer (10 µM) 0.5 0.25 µM Template (20 ng µl−1) 1.0 (1 ng µl−1) Water 8.0  Prepare master mix on ice to maintain full enzyme activity. The SYBR Green PCR Master Mix is light sensitive. After adding the PCR master mix to the sample tubes, avoid exposing them to light for prolonged periods of time.
Prepare master mix on ice to maintain full enzyme activity. The SYBR Green PCR Master Mix is light sensitive. After adding the PCR master mix to the sample tubes, avoid exposing them to light for prolonged periods of time. -
70|
The final volume per well is 20 µl. Carry out each reaction in triplicate for both the pre-hybridized amplified template (the product generated after PCR enrichment in Step 26) and the post-hybridized amplified template (generated in Step 65). Include a triplicate of blank wells (no template).
-
71|Use the following real-time PCR conditions:
- Step 1: 95 °C for 10 min
- Step 2: 95 °C for 15 s
- Step 3: 60 °C for 1 min
- Step 4: Repeat Steps 2–3, 39 times for a total of 40 cycles
- Step 5: Hold at 4 °C
-
72|
Evaluate the ΔCT between the pre- and post-hybridized DNA to determine the level of enrichment (helpful information is available at http://www3.appliedbiosystems.com/applicationstechnologies/real-timepcr/index.htm).
![]()
SUMMARY 5–6 d
Steps 1–22: 5 h (1 d)
Fragment genomic DNA, repair ends and add 3′A-overhangs (Steps 1–13): 3 h
Ligate adapters (Steps 14 and 15): 0.5 h
Size-select and gel-purify adapted DNA fragments (Steps 16–22): 1.5 h
Steps 23–42: 68.5 h (3 d)
Enrich adapter-modified DNA and prepare it for hybridization (Steps 23–33): 3.5 h
Set up hybridization and pre-warm wash buffer (Steps 34–42): 65 h
Steps 43–68: 1 d
Post-hybridization wash and elution (Steps 43–58): 1 h
Concentrate and amplify eluted material (Steps 59–68): 4.5–6.5 h
Steps 69–72: 3.5 h (0.5 d)
Validate enrichment with real-time PCR (Steps 69–72): 3.5 h
![]()
Troubleshooting advice can be found in Table 2.
TABLE 2.
Troubleshooting table.
| Problem | Possible reason | Solution |
|---|---|---|
| Adaptor dimer formation (Step 20) | Less DNA than expected in ligation reaction, i.e., <500 ng (Step 14) | Reduce adaptor concentration in ligation reaction |
| Insufficient DNA for hybridization (Step 27) | Possible loss during purification steps (Step 21) | Ensure ethanol is added properly to wash buffers and check pH of elution buffer |
| Inadequate number of PCR reactions were carried out (Step 24) | Set up 2–3 more reactions. Alternatively, increase the number of cycles to 20 | |
| Precipitate forms in hybridization solution before loading the gasket (Step 36) | Hybridization mixture components were not added in the order indicated (Step 30) | Ensure that all nucleic acid components are combined first before adding the hybridization buffer |
| Hybridization mixture was not immediately loaded on the gasket after centrifugation (Step 33) | Avoid pipetting the mixture from bottom of the tube and load onto the gasket immediately | |
| Concentration of nucleic acid too high | Total amount of nucleic acid should not exceed 500 ng µl−1 | |
| Low yield after amplification of recovered material (Step 66) | Wash buffer 2 may not have reached optimal temperature (Step 42) | Ensure temperature of wash buffer 2 does not exceed 37 °C |
| Samples were not held at 90–95 °C for at least 5 min (Step 56) | Elution steps can be repeated | |
| Check fragment recovery by qPCR | ||
| Extremely low amounts of DNA, such as the amount recovered from arrays, can stick to polypropylene tubes | Use low-binding or siliconized tubes. Alternatively, 0.1% Tween can be used in the EB buffer |
qPCR, quantitative PCR.
ANTICIPATED RESULTS
After elution and amplification of the captured material, qPCR may be carried out for a set of selected and depleted exons. The exons depleted from the captured genomic DNA are negative controls containing regions not selected by probes (Fig. 4). For example, in Figure 4, a difference of ~10 cycles is observed between the pre- and post-hybridization templates from a single experiment, indicating >1,000× copies of the selected exon are present in the enriched sample. If the cycle threshold is lower in the captured library than in the input library, the selection has passed an initial quality control and may proceed through sequencing.
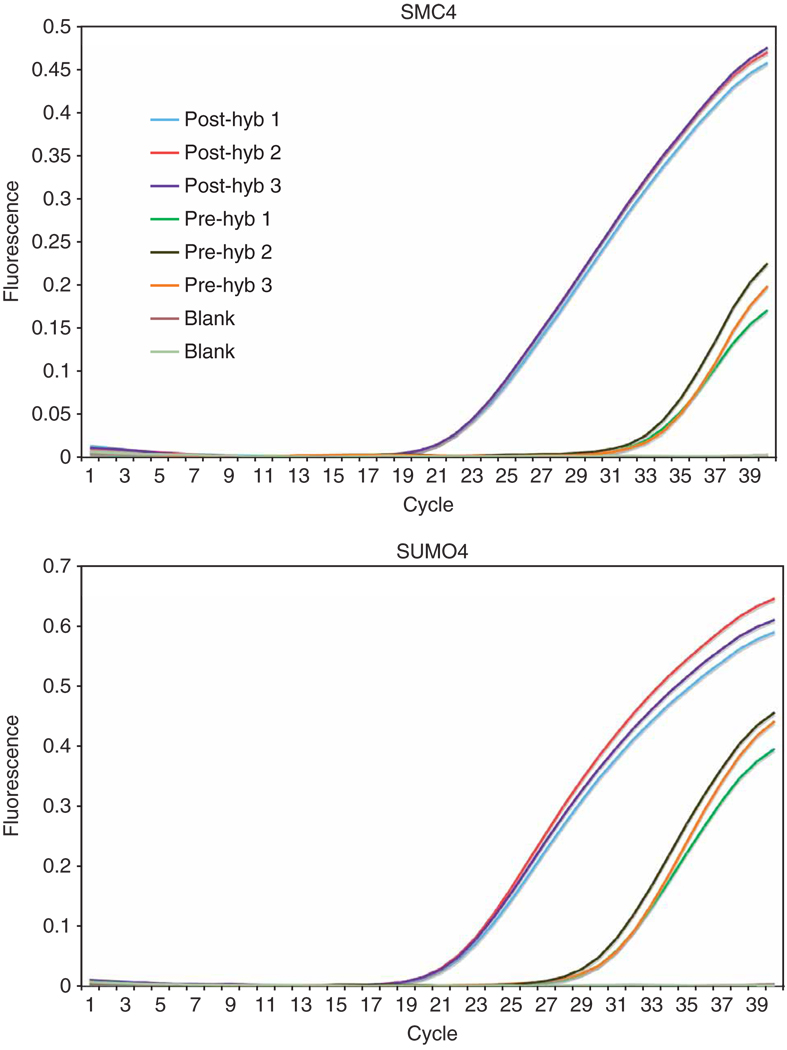
Figure 4.
qPCR analysis of two selected exons. qPCRs were carried out in triplicate for individually selected exons on both the input material (pre-hyb genomic DNA) and the enriched material (post-hyb). The differential CT values between ‘pre’ and ‘post’ array capture give a clear indication that the purification of the exons has succeeded. Non-selected exons may also be included (data not shown) in this analysis to confirm depletion of unselected targets.
To show how various components of hybridization influence capture efficiency, we performed a series of experiments shown in Table 3. These results stem from iterative optimization trials carried out on 244k arrays designed to select ~0.8 Mb of human exonic sequence from three independent chromosomal regions. The genomic DNA used for all captures was obtained from a single HapMap individual from the CEPH group (Coriell NA12762). It should be noted that the results described here are based on sequence data obtained from one lane of the eight-lane flow cell for each experiment/array capture. For the fourth experiment, two replicates (4A and 4B) are shown. These captures were performed and sequenced independently with two genomic DNA libraries generated separately from the same HapMap sample. Although there were differences in the number of total and mappable reads produced (a normal variation between sequencing runs), the results are highly comparable, indicating the reproducibility of the procedure given the optimal components and conditions.
TABLE 3.
Sequencing results from five array capture experiments.
| Experiment nos. | 1 | 2 | 3 | 4A | 4B |
|---|---|---|---|---|---|
| Experimental conditions | 10 µg − Cot-1 | 10 µg + Cot-1 | 20 µg + Cot-1 | 20 µg + Cot-1 + Oligos | 20 µg + Cot-1 + Oligos |
| Total reads | 6,812,441 | 5,459,328 | 9,279,439 | 15,575,292 | 17,352,851 |
| Total mappable reads | 3,326,872 | 2,835,630 | 4,745,834 | 9,136,698 | 7,985,886 |
| Reads in target | 458,496 | 827,676 | 1,539,894 | 5,999,669 | 4,963,481 |
| % Reads in target | 13.78 | 29.19 | 32.45 | 65.67 | 62.15 |
| Enrichment | 548 | 1,161 | 1,290 | 2,612 | 2,472 |
| No. of regions with read | 1,342 | 1,342 | 1,342 | 1,342 | 1,342 |
| % Regions with read | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| Target length (bp)a | 829,564 | 829,564 | 829,564 | 829,564 | 829,564 |
| Mean depth (X) | 20.68 | 36.98 | 69.22 | 266.05 | 220.46 |
| Median depth (X) | 19 | 36 | 66 | 189 | 201 |
| bp coverage (%)b | 98.01 | 98.12 | 98.74 | 98.79 | 99.21 |
The target length includes 60 bp probe regions that flank the target intervals.
Coverage of bp flanking interval boundaries is not considered.
Optimal results were achieved in the presence of excess Cot-1 DNA and adaptor blocking oligos. To show how enrichment specificity is determined, if 65.37% of the reads overlap with targeted intervals and the total target length is 829,564 bp, or 0.025% of the 3.3 × 109 bp human genome, the enrichment score will be 65.37%/0.025% or ~2,600, as indicated in Table 3. In other words, of the 328 Mb of sequence output generated in experiment 4A, 215 Mb are potentially informative. Importantly, the enrichment score is inversely proportional to the target length printed on the array. Therefore, it should only be used as a parallel comparison between two arrays consisting of comparable target lengths, and not a cross-comparison between two arrays directed to markedly different genomic sizes.
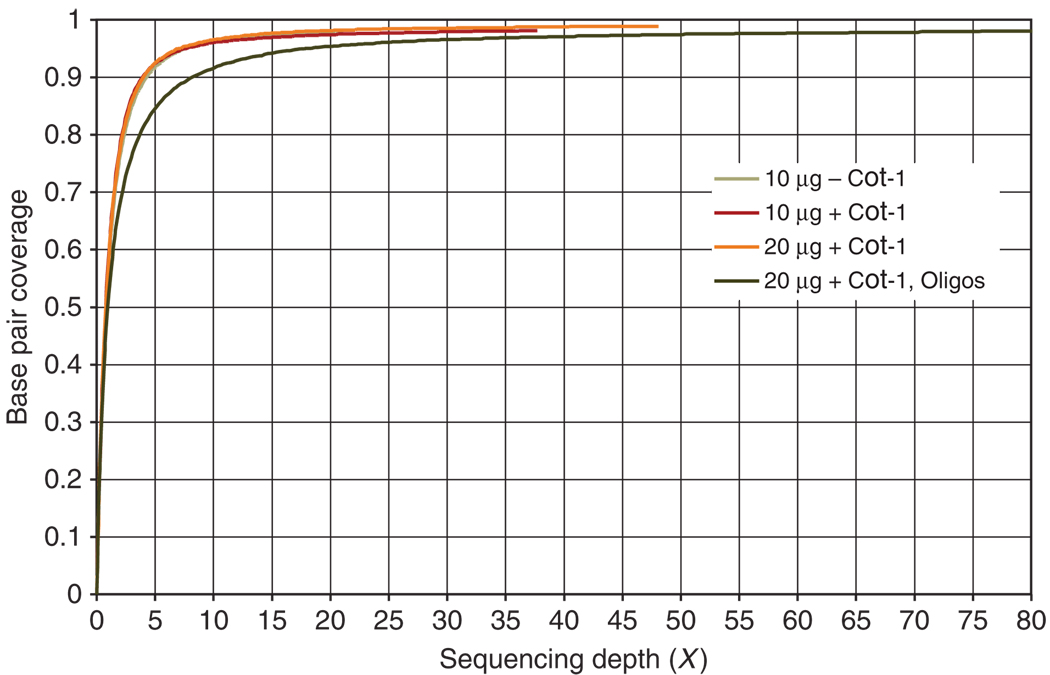
For many assays of this kind, specificity and sensitivity are often at odds with one another. Skewing the representation disproportionately in favor of some regions over others can result in some target regions being missed while others become overrepresented in the data. However, we show here that high sensitivity may also be achieved by this method, as all of the targeted exons are occupied by at least one captured fragment (% regions with read) and >98% of the targeted bases are covered by at least one read (Fig. 5).
Figure 5.
Coverage plot. Target coverage at the base pair level versus sequencing depth is plotted for each of the experiments detailed in Table 3. For all four experiments, significant breadth of coverage is achieved at or around 5×, while similarly high sequence complexity is maintained as exemplified by the corresponding curve behaviors that follow characteristics reasonably reflective of the classic Lander and Waterman21 curve. In all cases, 98% base pair coverage is achieved. The 2% bases not covered by reads likely reflect regions of the genome that are inaccessible or ‘unmappable’ at 36 bp.
Importantly, both the total number of reads obtained and the percentage of those reads that can be mapped to the genome for any given sequencing run directly influence sequencing depth. After recent upgrades to the reagent chemistry of the GA, imaging hardware and accompanying software, we achieve nearly 3× higher read depth than what was generated by the earlier GA. The upgrade from GA1 to GA2 is exemplified by the discrepancy in total read depth reported in Table 3 between experiments 1 and 4. At present, we typically see 14–15 million reads per lane, of which 50–60% map uniquely to the reference genome with ≤2 mismatches. A majority of ‘background’ reads are likely derived from the genome as well, but are cast aside as a result of either ambiguous mapping positions or lower read quality stemming from high rates of erroneous base calls/high per read error rates. In our experience, one GA flow cell lane of array-captured sequence is sufficient to achieve >20× sequencing depth for >93% of the bases inside a 0.7–0.8-Mb target. Consequently, for larger genomic regions, the specificity is only marginally affected, while the sequencing depth will be reduced. Thus, it is necessary to sequence more lanes in order to achieve deeper coverage.
The ultimate and most informative measure of sensitivity is the ability to effectively determine the full extent of polymorphisms present in a sample. To test this we compared our sequence data with the set of known SNPs registered within the genomic intervals captured from the HapMap library (Table 4). From this data, >90% of the known SNPs, both homozygous and heterozygous, were correctly called, whereas <3% were not called due to lack of coverage. Table 4 also illustrates the accuracy of SNP detection when read depth is considered as a criterion/cut-off for calling SNPs. In other words, a SNP must be supported by a minimal number of reads covering that base position in order to be considered a confident call. We performed SNP analysis using a minimal requirement of either 20× or 50× read depth. The more stringent requirement of at least 50× read depth reduces the rate of false positives, but the total number of identified SNPs is also significantly reduced, especially in experiment 3, where three times fewer reads in target were obtained compared with 4A and 4B. Our data show that a sufficient compromise between accuracy and coverage can be achieved when the read depth cutoff is set to 20×.
TABLE 4.
HapMap genotype calls for known SNPs.
| Homozygous SNPs (Total 124) | Heterozygous SNPs (Total 112) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Read depth | Experiment no.a | Good coverage | Low coverageb | No coverage | Good coverage | Low coverageb | No coverage | ||||
| Correctc | False | Correct | False | Correctc | False | Correct | False | ||||
| ≥20× | 3 | 72 | 1 | 43 | 4 | 4 | 57 | 7 | 38 | 8 | 2 |
| 4A | 103 | 2 | 13 | 3 | 3 | 81 | 21 | 7 | 2 | 1 | |
| 4B | 105 | 2 | 12 | 1 | 4 | 94 | 9 | 4 | 4 | 1 | |
| ≥50× | 3 | 15 | 0 | 100 | 5 | 4 | 14 | 1 | 81 | 14 | 2 |
| 4A | 73 | 0 | 43 | 5 | 3 | 64 | 16 | 24 | 7 | 1 | |
| 4B | 79 | 0 | 38 | 3 | 4 | 77 | 4 | 21 | 9 | 1 | |
SNP, single nucleotide polymorphism.
The experiment number listed in Table 3.
Read depth lower than 20× (top half of table) or lower than 50× (including SNPs that may have had good coverage at 20×, bottom half of table).
Non-reference allele frequency for homozygous ≥0.9 and heterozygous 0.3–0.8.
ACKNOWLEDGMENTS
We are thankful to Danea Rebolini, Laura Cardone and Melissa Kramer for sequencing and informatic support. We also thank Mona Spector for helpful discussions. This work was supported by an NIH postdoctoral training grant (E.H.) and by kind gifts from the Stanley Foundation and Kathryn W. Davis (G.J.H.). G.J.H. is an investigator of the Howard Hughes Medical Institute.
Footnotes
COMPETING FINANCIAL INTERESTS The authors declare competing financial interests (see the HTML version of this article for details).
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Kaiser J. DNA sequencing. A plan to capture human diversity in 1000 genomes. Science. 2008;319:395. doi: 10.1126/science.319.5862.395. [DOI] [PubMed] [Google Scholar]
- 2.Siva N. 1000 Genomes Project. Nat. Biotechnol. 2008;26:256. doi: 10.1038/nbt0308-256b. [DOI] [PubMed] [Google Scholar]
- 3.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins FS, Barker AD. Mapping the cancer genome. Pinpointing the genes involved in cancer will help chart anew course across the complex landscape of human malignancies. Sci. Am. 2007;296:50–57. [PubMed] [Google Scholar]
- 5.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erlich Y, Mitra PP, delaBastide M, McCombie WR, Hannon GJ. Alta-Cyclic: a self-optimizing base caller for next-generation sequencing. Nat. Methods. 2008;5:679–682. doi: 10.1038/nmeth.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albert TJ, et al. Direct selection of human genomic loci by microarray hybridization. Nat. Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 13.Hodges E, et al. Genome-wide in situ exon capture for selective resequencing. Nat. Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 14.Okou DT, et al. Microarray-based genomic selection for high-throughput resequencing. Nat. Methods. 2007;4:907–909. doi: 10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- 15.Porreca GJ, et al. Multiplex amplification of large sets of human exons. Nat. Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 16.Bashiardes S, et al. Direct genomic selection. Nat. Methods. 2005;2:63–69. doi: 10.1038/nmeth0105-63. [DOI] [PubMed] [Google Scholar]
- 17.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 2009 doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cleary MA, et al. Production of complex nucleic acid libraries using highly parallel in situ oligonucleotide synthesis. Nat. Methods. 2004;1:241–248. doi: 10.1038/nmeth724. [DOI] [PubMed] [Google Scholar]
- 19.Morgulis A, Gertz EM, Schaffer AA, Agarwala R. WindowMasker: window-based masker for sequenced genomes. Bioinformatics. 2006;22:134–141. doi: 10.1093/bioinformatics/bti774. [DOI] [PubMed] [Google Scholar]
- 20.Quail MA, et al. A large genome center’s improvements to the Illumina sequencing system. Nat. Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lander ES, Waterman MS. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics. 1988;2:231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]