Abstract
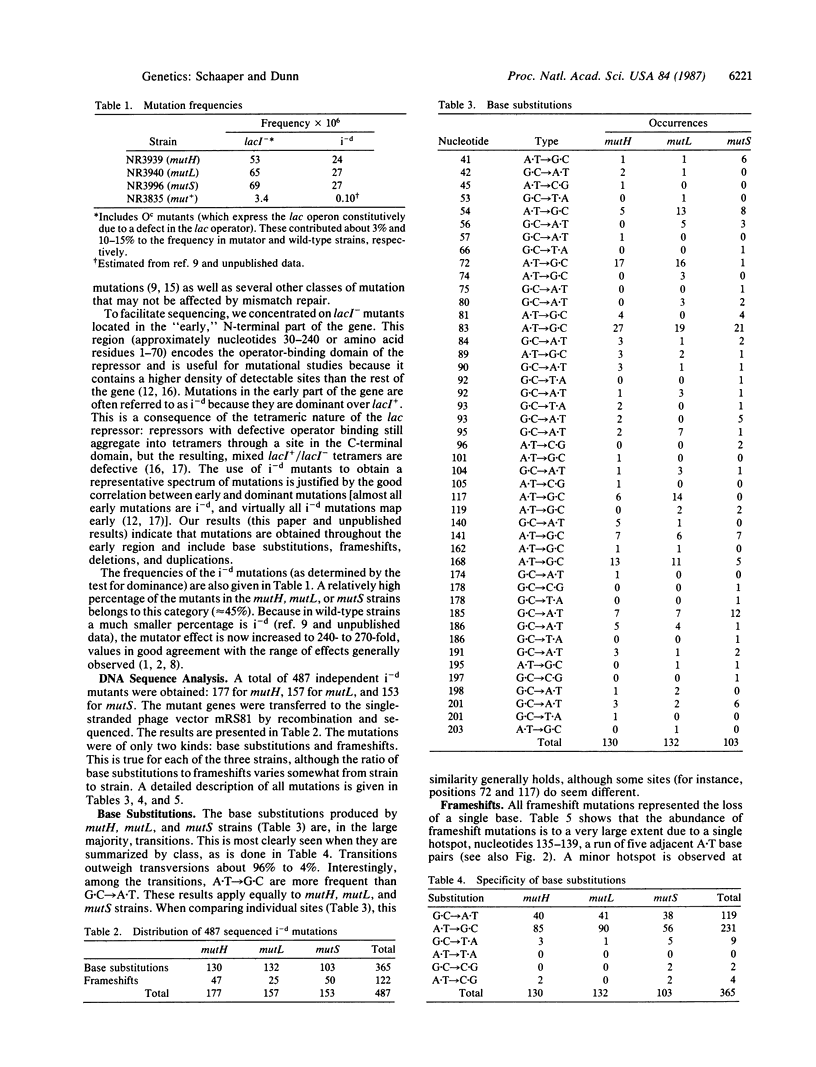
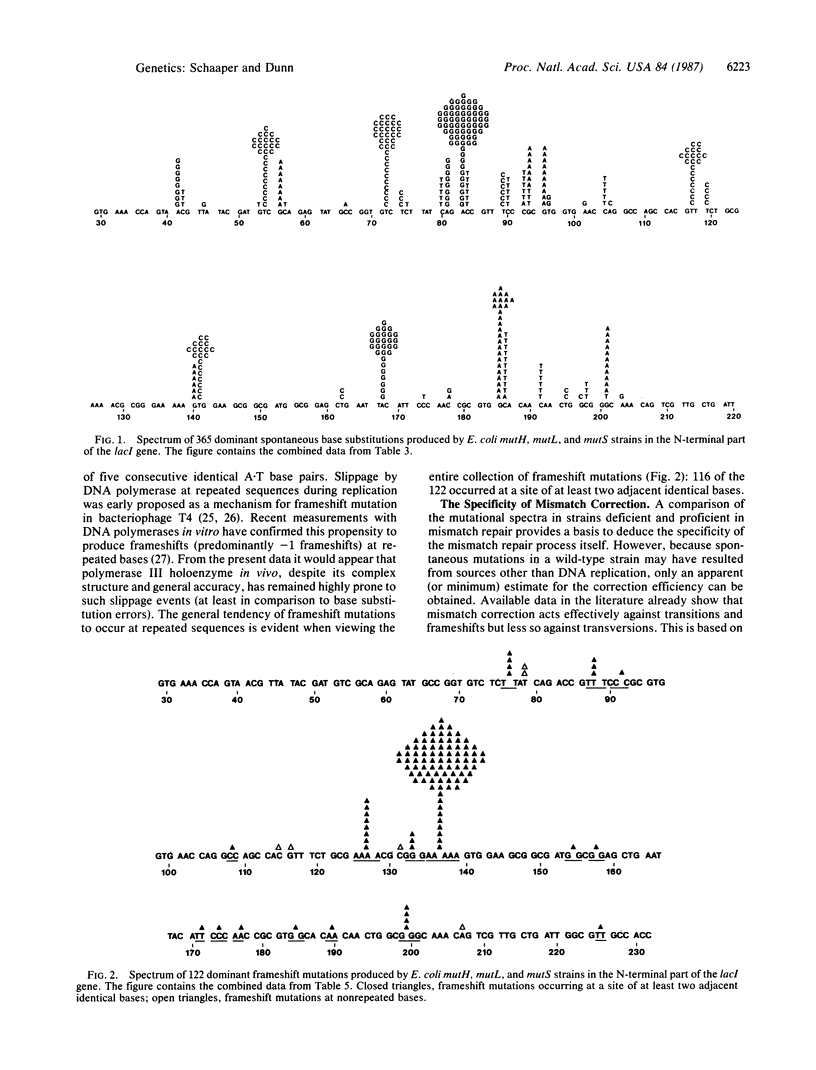
We have determined the DNA sequence changes in 487 spontaneous mutations in the N-terminal part of the lacI gene in mutH, mutL, and mutS strains of Escherichia coli. These strains display elevated spontaneous mutation rates because of a deficiency in the process of postreplicative mismatch correction. As a consequence the mutational spectra reveal the nature of spontaneous DNA replication errors. The spectra consist of base substitutions (75%) and single-base deletions (25%). Among the base substitutions, transitions (both A.T----G.C and G.C----A.T) are strongly favored over transversions (96% versus 4%). Large site-to-site differences are observed among identical base substitutions, presumably reflecting the modulating effects of neighboring bases. The single-base-deletion spectrum is dominated by a large hotspot at a run of adjacent identical base pairs, implying a Streisinger-slippage mechanism. The data, when compared to a previously determined wild-type spectrum, also provide information on the specificity of the mismatch repair system.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Breslauer K. J., Frank R., Blöcker H., Marky L. A. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy H. E., Fowler R. G. The specificity of base-pair substitution induced by the mutL and mutS mutators in E. coli. Mutat Res. 1985 Mar;142(3):93–97. doi: 10.1016/0165-7992(85)90046-6. [DOI] [PubMed] [Google Scholar]
- Claverys J. P., Lacks S. A. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev. 1986 Jun;50(2):133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H. Genetic studies of the lac repressor. IV. Mutagenic specificity in the lacI gene of Escherichia coli. J Mol Biol. 1977 Dec 15;117(3):577–606. doi: 10.1016/0022-2836(77)90059-6. [DOI] [PubMed] [Google Scholar]
- Cox E. C. Bacterial mutator genes and the control of spontaneous mutation. Annu Rev Genet. 1976;10:135–156. doi: 10.1146/annurev.ge.10.120176.001031. [DOI] [PubMed] [Google Scholar]
- Cox E. C., Degnen G. E., Scheppe M. L. Mutator gene studies in Escherichia coli: the mutS gene. Genetics. 1972 Dec;72(4):551–567. doi: 10.1093/genetics/72.4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohet C., Wagner R., Radman M. Methyl-directed repair of frameshift mutations in heteroduplex DNA. Proc Natl Acad Sci U S A. 1986 May;83(10):3395–3397. doi: 10.1073/pnas.83.10.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohet C., Wagner R., Radman M. Repair of defined single base-pair mismatches in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jan;82(2):503–505. doi: 10.1073/pnas.82.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Glickman B. W., Fix D. F., Yatagai F., Burns P. A., Schaaper R. M. Mechanisms of spontaneous mutagenesis: clues from mutational specificity. Basic Life Sci. 1986;38:425–437. doi: 10.1007/978-1-4615-9462-8_45. [DOI] [PubMed] [Google Scholar]
- Glickman B. W., Radman M. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc Natl Acad Sci U S A. 1980 Feb;77(2):1063–1067. doi: 10.1073/pnas.77.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman B. W. Spontaneous mutagenesis in Escherichia coli strains lacking 6-methyladenine residues in their DNA: an altered mutational spectrum in dam- mutants. Mutat Res. 1979 Jul;61(2):153–162. doi: 10.1016/0027-5107(79)90122-2. [DOI] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Frameshift mutagenesis by eucaryotic DNA polymerases in vitro. J Biol Chem. 1986 Oct 15;261(29):13581–13587. [PubMed] [Google Scholar]
- Leong P. M., Hsia H. C., Miller J. H. Analysis of spontaneous base substitutions generated in mismatch-repair-deficient strains of Escherichia coli. J Bacteriol. 1986 Oct;168(1):412–416. doi: 10.1128/jb.168.1.412-416.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Clark S., Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H., Calos M. P., Combépine C., Hofer M. Further correlations of the lacI genetic map with the DNA sequence. J Mol Biol. 1981 Nov 25;153(1):65–66. doi: 10.1016/0022-2836(81)90526-x. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Ganem D., Lu P., Schmitz A. Genetic studies of the lac repressor. I. Correlation of mutational sites with specific amino acid residues: construction of a colinear gene-protein map. J Mol Biol. 1977 Jan 15;109(2):275–298. doi: 10.1016/s0022-2836(77)80034-x. [DOI] [PubMed] [Google Scholar]
- Miller J. H. Genetic studies of the lac repressor. XII. Amino acid replacements in the DNA binding domain of the Escherichia coli lac repressor. J Mol Biol. 1984 Nov 25;180(1):205–212. doi: 10.1016/0022-2836(84)90438-8. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Schmeissner U. Genetic studies of the lac repressor. X. Analysis of missense mutations in the lacI gene. J Mol Biol. 1979 Jun 25;131(2):223–248. doi: 10.1016/0022-2836(79)90074-3. [DOI] [PubMed] [Google Scholar]
- Modrich P. DNA mismatch correction. Annu Rev Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- Müller-Hill B. Lac repressor and lac operator. Prog Biophys Mol Biol. 1975;30(2-3):227–252. doi: 10.1016/0079-6107(76)90011-0. [DOI] [PubMed] [Google Scholar]
- Ornstein R. L., Fresco J. R. Correlation of Tm and sequence of DNA duplexes with delta H computed by an improved empirical potential method. Biopolymers. 1983 Aug;22(8):1979–2000. doi: 10.1002/bip.360220811. [DOI] [PubMed] [Google Scholar]
- Petruska J., Goodman M. F. Influence of neighboring bases on DNA polymerase insertion and proofreading fidelity. J Biol Chem. 1985 Jun 25;260(12):7533–7539. [PubMed] [Google Scholar]
- Pukkila P. J., Peterson J., Herman G., Modrich P., Meselson M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics. 1983 Aug;104(4):571–582. doi: 10.1093/genetics/104.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman M., Wagner R. Mismatch repair in Escherichia coli. Annu Rev Genet. 1986;20:523–538. doi: 10.1146/annurev.ge.20.120186.002515. [DOI] [PubMed] [Google Scholar]
- Schaaper R. M., Danforth B. N., Glickman B. W. Mechanisms of spontaneous mutagenesis: an analysis of the spectrum of spontaneous mutation in the Escherichia coli lacI gene. J Mol Biol. 1986 May 20;189(2):273–284. doi: 10.1016/0022-2836(86)90509-7. [DOI] [PubMed] [Google Scholar]
- Schaaper R. M., Danforth B. N., Glickman B. W. Rapid repeated cloning of mutant lac repressor genes. Gene. 1985;39(2-3):181–189. doi: 10.1016/0378-1119(85)90312-9. [DOI] [PubMed] [Google Scholar]
- Siegel E. C., Kamel F. Reversion of frameshift mutations by mutator genes in Escherichia coli. J Bacteriol. 1974 Mar;117(3):994–1001. doi: 10.1128/jb.117.3.994-1001.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Owen J. Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics. 1985 Apr;109(4):633–659. doi: 10.1093/genetics/109.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S. S., Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]



