Abstract
We demonstrate a hybrid machine learning method to classify schizophrenia patients and healthy controls, using functional magnetic resonance imaging (fMRI) and single nucleotide polymorphism (SNP) data. The method consists of four stages: (1) SNPs with the most discriminating information between the healthy controls and schizophrenia patients are selected to construct a support vector machine ensemble (SNP-SVME). (2) Voxels in the fMRI map contributing to classification are selected to build another SVME (Voxel-SVME). (3) Components of fMRI activation obtained with independent component analysis (ICA) are used to construct a single SVM classifier (ICA-SVMC). (4) The above three models are combined into a single module using a majority voting approach to make a final decision (Combined SNP-fMRI). The method was evaluated by a fully validated leave-one-out method using 40 subjects (20 patients and 20 controls). The classification accuracy was: 0.74 for SNP-SVME, 0.82 for Voxel-SVME, 0.83 for ICA-SVMC, and 0.87 for Combined SNP-fMRI. Experimental results show that better classification accuracy was achieved by combining genetic and fMRI data than using either alone, indicating that genetic and brain function representing different, but partially complementary aspects, of schizophrenia etiopathology. This study suggests an effective way to reassess biological classification of individuals with schizophrenia, which is also potentially useful for identifying diagnostically important markers for the disorder.
Keywords: machine learning, feature selection, support vector machine ensemble, functional magnetic resonance imaging, single nucleotide polymorphisms, gene, schizophrenia
Introduction
Schizophrenia is a severe, chronic, brain disease that disrupts normal thinking, speech, and behavior. Schizophrenia diagnosis currently relies on clinical examination and the illness course, with many subcategories reflecting different aspects of this complex and likely biologically heterogeneous mental disease. Despite the diagnostic reliability achieved by quantifiable examination of overt psychiatric symptoms, researchers have also used biological indices in attempts to classify schizophrenia patients (Murray et al., 1992; Malaspina et al., 1998; Sponheim et al., 2001, 2003). Recently, there have been increasing efforts to utilize brain functional magnetic resonance imaging (fMRI) and examine genetic variation to study potential schizophrenia biomarkers, in order to better understand the pathology of schizophrenia. While most such studies focus on identifying associations between genetics and brain function in schizophrenia, we look at this problem from a different perspective, using biological and genetic information to help classify the disorder. We attempt to improve classification accuracy and provide preliminary data, suggesting that by combining biological and genetic information, we can best reflect the underlying pathophysiology, which ultimately may aid in the diagnosis of schizophrenia and its subcategories. We also predict that by achieving better classification, intrinsic connections between genetic variation and biological function can also be identified.
In the last few years there has been a growing interest in the use of machine learning algorithms for analyzing fMRI data. Machine learning algorithms can be used to train classifiers to decode stimuli, behaviors and other variables of interest from fMRI data (Haynes and Rees, 2006; O'Toole et al., 2007; Pereira et al., 2009). Demirci applied a projection pursuit technique to components obtained via independent component analysis (ICA) of fMRI activation maps, to classify individuals as being either schizophrenia patients or healthy controls (Demirci et al., 2008). Shinkareva et al. (2006) presented a unified feature selection and classification procedure to classify subjects into groups based on four dimensional spatio-temporal data. Zhang et al. (2005) applied the adaptive boosting algorithm (AdaBoost) (Freund and Schapire, 1997) to classify subjects into groups (drug-addicted subjects and healthy non-drug-using controls) based on the observed 3D brain images. Ford et al. (2003) used a Fisher linear discriminant analysis on the fMRI brain activation maps to extract spatial characteristics and to classify healthy controls versus patients with schizophrenia, Alzheimer's disease, and mild traumatic brain injury.
To date, limited work has been done on the use of genotypic information to help classify patients from controls, although Struyf et al. (2008) demonstrated that SVMs can distinguish bipolar and schizophrenia from normal control with a high accuracy by combing gene expression data with demographic and clinical data.
Many researchers now agree that schizophrenia may develop as a result of interplay between genetic predisposition (for example, inheriting certain susceptibility genes) and environmental exposure. While genetic factors play an important role in schizophrenia – persons who have immediate relatives with a history of schizophrenia have a significantly increased risk for developing the disorder over that of the general population. However, even monozygotic twins have only about 42% concordance for the disease (Lee et al., 2005). Environmental factors may well lead to subtle brain alterations that increase the risk of schizophrenia. Thus combining fMRI data (which captures brain function presumably reflecting both genetic and environmental influences) with genetic information, is potentially a useful way to help classify schizophrenia (Hariri and Weinberger, 2003; Pearlson and Folley, 2008; Calhoun et al., 2009; Liu et al., 2009; Potkin et al., 2009).
In this paper we present a supervised machine learning method to classify schizophrenia and control individuals that incorporates fMRI and SNP data. The method to fuse information from both modalities comprises four stages. At the first stage, a support vector machine based classifier ensemble (SVME) is constructed by using signature SNPs selected from a large SNP pool (SNP-SVME). At the second stage, a SVME is trained with a subset of voxels (Voxel-SVME). At the third stage, fMRI activation components obtained with ICA are used to construct a single SVM classifier (ICA-SVMC). Finally, at the fourth state the results obtained from the above three stages are combined into a single module using majority voting (Combined SNP-fMRI). We will first explain the data collection and preparation procedures, and describe the proposed method in detail. Then, we present the experimental results, followed by discussion and conclusion.
Data and Experiments
Subjects
We investigated fMRI and SNP data from 40 subjects, 20 schizophrenia patients (age: 40.2 ± 9.8, three females) and 20 healthy controls (age 42.5 ± 15.5, eight females). All participants provided written, informed, IRB-approved consent at Hartford hospital. Patients met criteria for DSM-IV-TR schizophrenia based on the structured clinical interview for DSM IV (SCID; First et al., 1995) and review of the case file by a clinician. Healthy subjects were screened to ensure they were free from DSMIV Axis I or Axis II psychopathology assessed using the SCID (Spitzer et al., 1996) and also interviewed to determine that there was no history of psychosis in any first-degree relative. All selected subjects were Caucasian/non-Hispanic. Twenty chronic SZ patients were selected and 16 of them had available, contemporaneous positive and negative syndrome scale (PANSS) scores (Kay et al., 1987). For those 16 SZ patients, PANSS total score was 67.6 ± 30.0 (mean ± SD), positive symptom score 15.4 ± 4.1, and negative symptom score 14.5 ± 6.7. Seventeen SZ patients had available medication information. These were taking 26 types of first and second-generation antipsychotics in variable doses, with most patients taking more than one such drug. The most commonly prescribed medicines included olanzapine, risperidone, quetiapine, haloperidol, divalproex, escitalopram, and aripiprazole.
SNP data collection and preprocessing
A saliva sample was obtained for each subject and DNA extracted. Genotyping was performed using the Illumina BeadArray™ platform and the GoldenGate™ assay (Oliphant et al., 2002; Fan et al., 2003). The PG Array of Genomas Inc. was used (the detailed composition has been published as a patent application, Ruano, 2006). The SNP array consists of 384 SNPs from 222 genes derived from six physiological systems: neurobiology, metabolism, cell proliferation, cardiovascular, inflammation, and cholesterol biochemistry. Over all systems, the following pathways were represented: insulin resistance, glucose metabolism, energy homeostasis, adiposity, apolipoproteins and receptors, fatty acid and cholesterol metabolism, lipases, receptors, cell signaling and transcriptional regulation, growth factors, drug metabolism, blood pressure, vascular signaling, endothelial dysfunction, coagulation and fibrinolysis, vascular inflammation, cytokines, and behavior (satiety).
Genotyping analysis software, GenCall, was used to cluster the intensities from the genotyping microarray into three clusters: AA, AB, and BB, without assuming dominant or recessive inheritance. On the basis of the GenCall score, a number between 0 and 1 indicating how close to the center of the cluster a sample lies, we chose a threshold to select only reliable genotype results. SNPs with a GenCall score of 0.25 or higher were selected, resulting in 367 SNPs. Genotypes are inherently categorical and can be represented as discrete numbers, e.g., 1 for one type of homozygous, 0 for heterozygous, and −1 for the other type of homozygous. In our study, each subject has a feature vector with 367 discrete numbers.
fMRI data collection and preprocessing
FMRI data were collected during performance of an auditory oddball task (Kiehl and Liddle, 2003), which consists of detecting an infrequent sound within a series of frequent sounds. The same auditory stimuli were used and found to be effective in eliciting fMRI BOLD patterns differentiating healthy controls from schizophrenia subjects (Kiehl et al., 2005). Auditory stimuli were presented to each participant by a computer stimulus presentation system via earphones. Subjects were presented with three types of sounds: target (1000 Hz with probability p = 0.1), novel (non-repeating random digital noises, p = 0.1), and standard (500 Hz, p = 0.8). Subjects were expected to respond and press a button with their right index finger every time they heard a target stimulus and not to respond to standard or novel sounds.
Scans were acquired at the Olin Neuropsychiatry Research Center at the Institute of Living on a Siemens Allegra 3 T dedicated head MRI scanner equipped with 40 mT/m gradients and a standard quadrature head coil. The functional scans were acquired using gradient-echo echo-planar-imaging with the following parameters (repeat time = 1.50 s, echo time = 27 ms, field of view = 24 cm, acquisition matrix = 64 × 64, flip angle = 70°, voxel size = 3.75 × 3.75 × 4 mm3, slice thickness = 4 mm, gap = 1 mm, 29 slices, ascending acquisition).
Six “dummy” scans were performed at the beginning to allow for longitudinal equilibrium, after which the paradigm was automatically triggered to start by the scanner. Data were preprocessed using the software package SPM2 (http://www.fil.ion.ucl.ac.uk/spm/). Images were realigned using INRIalign – a motion correction algorithm unbiased by local signal changes (Freire and Mangin, 2001). Data were spatially normalized into the standard Montreal Neurological Institute space (Friston et al., 1995), resliced to 3 × 3 × 3 mm3, and spatially smoothed with a 10 × 10 × 10 mm3 Gaussian kernel. Data for each participant were analyzed by multiple regression incorporating regressors for the novel, target, and standard and their temporal derivatives plus an intercept term. The target-related contrast images were used in this study. Finally, we used a mask based upon one-sample t-test against zero activation to select meaningful voxels. This results in a size of 7,060 voxels in each fMRI image.
Methods
The hybrid machine learning method
A two-class supervised learning problem can be written as a formula, with m samples (subjects in this study). Each sample xi has d features and a class label yi. From a set of training samples, the machine learning algorithm establishes a classifier, which represents a hypothesis, h. Given unseen samples, the classifier predicts the corresponding y value. An ensemble method constructs a set of classifiers {h1, …, hT}, chooses a set of weights {α1, …, αT} and build a weighted average classifier H(x) = α1h1(x) + … αThT(x). The classification decision of the combined classifier H is +1 if H(x) > and −1 otherwise.
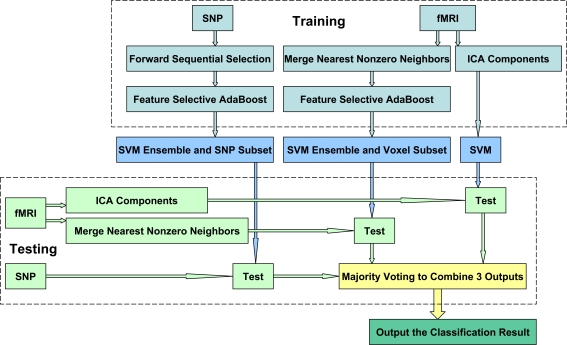
The flowchart of the proposed supervised machine learning method can be seen in Figure 1. There are four stages to fuse fMRI and genetic data, and to classify schizophrenia. The first stage is to select signature SNP loci and construct a SVME for SNPs, termed SNP-SVME. Two steps are involved: (1) to select a subset of candidate SNPs from whole SNPs pool by the forward sequential feature selection method (FSFS) (Liu, 2005); (2) to construct a SVME by a feature selective AdaBoost method (FSA) (Howe, 2003). The second stage is to construct a SVME for fMRI images with the optimal subset of voxels to reach the best classification performance. We first average neighboring voxels to reduce computation complexity, and then construct a SVME using the FSA on the averaged voxels. The third stage is to obtain a SVM classifier using independent components extracted from fMRI activation maps by ICA. The fourth and final stage is to combine the three classification models obtained from above stages into one model using majority voting.
Figure 1.
Flow chart of method.
SNPs subset selection and SVME
Classifying schizophrenia based on genetic data is complicated by small-sample-size classification problems (Fukunaga, 1990). Genetic data have high dimensionality compared to the generally small number of available subject samples. The dimensionality N is often considered large if it is in the range of hundreds. Genetic data, however, can have hundreds of thousands of dimensions (genes or loci). Some genes are related to the schizophrenia classification task, but many are presumably irrelevant. The learning algorithms can be potentially confused by the irrelevant/redundant features and construct poor classifiers (Jain and Chandrasekaran, 1982). To address the small-sample-size classification problem, we propose a two-step algorithm to select informative genes from a high-dimensional space and generate a classifier ensemble through SVM. The first step is a filter that removes most irrelevant features and selects a candidate SNP subset from whole SNP pool using FSFS. The second step combines a SNP selection into AdaBoost SVM ensemble algorithm to construct SVME with signature SNP subset.
Forward sequential feature selection (FSFS) method
The FSFS algorithm is a good choice for irrelevancy removal. It applies independent evaluation criteria without involving any learning algorithm. It does not inherit any bias of a learning algorithm and it is also computationally efficient (Liu, 2005). The FSFS algorithm starts the search from an empty SNP set. As the search proceeds, SNPs are added into the SNP subset one at a time. On each round, the best SNP for classification among unselected ones is chosen based on a distance measure. Distance measures are also known as separability, divergence, or discrimination measures. We try to find the SNP that can separate the patients and healthy controls as far as possible. The distance measure used in this paper is Mahalanobis distance. The SNP subset grows until it reaches the full set of original SNPs. A rank list is computed according to how early a SNP is added into the list. Then a certain number of SNPs are selected to construct a candidate SNP subset for second step. Both the prior knowledge of the SNP dataset and experience are used to decide how many SNPs are selected. In order to keep more informative SNPs, we select about top 40% SNPs in the rank list to construct a candidate SNP subset. The candidate SNP subset is much smaller than original SNP set, but still contains unrelated SNPs which need to be removed.
Feature selective adaboost (FSA) method
The second step is constructing a SVME by the FSA method. AdaBoost proposed by Freund and Schapire (1997) can be used in conjunction with any other iterative learning algorithms to improve their performance. Here, we use AdaBoost with SVM to build a SVM classifier ensemble. In addition, we modify AdaBoost to add a feature selection function, then propose a feature selective AdaBoost method. The FSA algorithm aims at training classifiers to get the best performance and selecting features with the best discriminating power simultaneously. The FSA algorithm is given below
Given a training set X from two classes, including m samples and d features per sample.
Initialize weights for the m samples: D1(i) = 1/m.
-
For t = 1:1:T iteration, do
-Feature selection
Train a classifier for each feature on the weighted samples.
Rank each feature based on training error rate of each classifier .
Select l features with the lowest , and form a new training dataset
-Train a classifier on the weighted samples X*.
-Compute which weighs by its classification performance.
-Update and normalize the weighted distribution to be
, where Zt is a normalization factor.
Output the final classifier ensemble:
As shown above, the FSA algorithm runs for T iterations, and the final classification output of H is a weighted T individual classifiers. Initially, all weights of training samples are set equally. On each round the weights of misclassified samples are increased so that the algorithm forces classifiers to focus on those samples in the training set.
Furthermore, within each iteration cycle, the FSA algorithm ranks all features with training error rate, and selects l features with the lowest training error rate. The number l is decided based on the leave one out (LOO) SVMs performance with the weighted training samples used in this iteration. Thus the FSA algorithm selects the feature subset that contains the most discriminating information on each round and trains a classifier based on weighted training samples with the selected features. Accuracy and diversity of individual classifiers critically influence the classification performance of ensemble methods. The FSA increases the diversity among the classifiers by allowing a flexible feature space, which in turn enhances the overall performance of SVME.
Valentini and Dietterich (2002) analyzed bias-variance decomposition of the error in SVM, and showed that the bias-variance decomposition offers a rationale to develop ensemble methods using SVMs as base learners. In this paper, the kernel function of SVM is the radial basis function (RBF) kernel. SVM is a statistical learning method based on the structure risk minimization principle that has been shown to be very efficient in pattern recognition applications (Vapnik, 2000). However, the classification performance of SVM heavily depends on a proper setting of parameters. The RBF-SVM has two parameters: one is the RBF kernel parameter σ, and the other is C, which controls the trade-off between training error and the margin. On each round of the FSA algorithm, we compute the optimal parameters of RBF-SVM by evaluating its accuracy and diversity with the weighted training dataset through the bias-variance decomposition of the error in SVM (Valentini and Dietterich, 2002).
Voxels selection and SMVE
The goal of this stage is to select informative voxels to aid in diagnostic classification. As mentioned above, the fMRI image has voxels with 7060 non-zero meaningful voxels. The amount of non-zero voxels is very large compared to the number of samples. It is necessary to decrease the dimensionality while retaining the group discrimination information. First, we merge the 3 × 3 × 3 non-zero neighboring voxels by averaging. Thus the resultant images have 261 large voxels. In the second step, we apply FSA algorithm described in section “SNPs subset selection and SVME” to further select informative voxels and construct SVME. At each FSA iteration, voxels ranked with high discriminative values are used for training a SVM classifier. The final decision is a weighted ensemble of individual classifiers.
ICA component extraction
In prior research, ICA has been applied to the analysis of fMRI data to discover hidden components presenting brain activation and characterize their spatial locations in healthy control subjects and patients with schizophrenia (Calhoun et al., 2004; Sui et al., 2009). The basic ICA model defines a generative model for the observed data, with a goal of identifying hidden independent components from linearly mixed observations.
In above equation, O is an observation matrix that can be composed of measurements from MRI images. S contains the independent components, which consists of unknown sources such as brain activation networks. A is a linear mixing matrix, relating the sources to the mixed measurements. W is an unmixing matrix. If W equals the inverse of A, then the Z, the estimated component matrix, is equivalent to S, the source matrix. There are many ICA algorithms based on different independence criteria. The ICA algorithm we use here is the infomax algorithm which attempts to find the W matrix through maximizing an entropy function (Bell and Sejnowski, 1995; Cardoso, 1997). And we use modified Akaike information criterion (AIC) method proposed by Li et al. to estimate the correct number of components (Akaike, 1974; Li et al., 2007). At this stage, there are five components extracted from the fMRI image of each sample. These five components are used as classification features to train a linear SVM classifier.
Classification combination
The fourth and final stage combines the results from the above three stages and makes a final decision via majority voting.
Classification Experiments and Results
We next applied the hybrid machine learning method to the problem of separating patients from controls. All statistical results of our experiments are based on the LOO cross-validation method. Thirty-nine subjects were used for training, while one subject was used for testing. A total of 40 training-testing sets were implemented. The performance measures used in this paper are specificity, sensitivity, and accuracy. The test output of our method can be positive (patient) and negative (control). A true positive means a patient correctly diagnosed as a patient, a false positive means healthy people wrongly identified as sick. True negative means healthy people correctly identified as healthy. A false negative means sick people wrongly identified as healthy. The specificity, sensitivity and accuracy are defined as below:
Results
Taking into account of the stochastic property of the algorithm, we performed the algorithm 20 times for each pair of training-testing dataset and the average classification results in each stage and final combined classification model are shown in Table 1. For comparison, we also trained the SVMC with all 367 SNPs and 7060 Voxels. The LOO accuracy are 0.4 (367 SNPs) and 0.675 (7060 Voxels). The experiment results suggest that SNP and voxel selection is necessary.
Table 1.
Performance of the classification model.
| Measures of performance | ||||||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Accuracy | ||||
| CLASSIFICATION MODEL | ||||||
| SVMC with all 367 SNPs | 0.4000 | 0.4000 | 0.4000 | |||
| SVMC with all 7060 Voxels | 0.6500 | 0.7000 | 0.6750 | |||
| THE PROPOSED CLASSIFICATION MODEL | ||||||
| SNP-SVME | 0.7175 | 0.7600 | 0.7388 | |||
| Voxel-SVME | 0.7875 | 0.8450 | 0.8163 | |||
| ICA-SVMC | 0.8000 | 0.8500 | 0.8250 | |||
| Combination | 0.8575 | 0.8875 | 0.8725 | |||
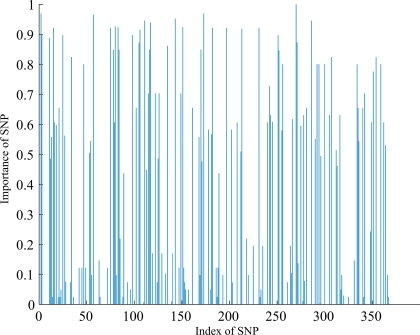
At the first stage, we examined the SNPs database using the two-step method described in section “The hybrid machine learning method”. After the most irrelevant SNPs filtered out from whole SNPs dataset using FSFS, 150 SNPs were selected. These 150 SNPs were then used as input features of the FSA algorithm. The number of iterations for FSA was set to 20 empirically since the performance was saturated after 20 classifiers. At each iteration the algorithm selected a certain number of SNPs from 150 SNPs and trained a SVM classifier. The number of SNPs selected in each iteration was estimated by the LOO algorithm on weighted training dataset. Those SNPs having more discrimination information are expected to have a high frequency of being selected. The importance of each SNP to the classification task can be denoted by the ratio of the number of times each SNPs selected over the number of iterations of FSA. Figure 2 shows the importance of individual SNP, and the most important 15 SNPs are listed in Table 2.
Figure 2.
Importance of individual SNP.
Table 2.
Top 15 SNPs.
| SNP | Gene |
|---|---|
| rs6136 | SELP: selectin P (granule membrane protein 140 kDa, antigen CD62) |
| rs737865 | COMT: catechol-O-methyltransferase |
| rs7072137 | GAD2: glutamic acid decarboxylase 2 |
| rs1176744 | HTR3B: 5-hydroxytryptamine (serotonin) receptor 3B |
| rs821616 | DISC1: disrupted in schizophrenia 1 |
| rs11188092 | CYP2C19: cytochrome P450, family 2, subfamily C, polypeptide 19 |
| rs3771892 | TNFAIP6: tumor necrosis factor alpha-induced protein 6 |
| rs1128503 | ABCB1: ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| rs2066470 | MTHFR: 5,10-methylenetetrahydrofolate reductase (NADPH) |
| rs2020933 | SLC6A4: solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 |
| rs2192752 | IL1R1: interleukin 1 receptor, type I |
| rs2298122 | DRD1IP: dopamine receptor D1 interacting protein |
| rs2276307 | HTR3B: 5-hydroxytryptamine (serotonin) receptor 3B |
| rs3758947 | ABCC8: ATP-binding cassette, sub-family C, member 8 |
| rs11212515 | ACAT1: acetyl-coenzyme A acetyl transferase 1 |
At the second stage, the FSA selected a certain number of voxels that containing the most discriminating information from 261 large voxels and trained a SVM at each iteration. The number of voxels to be selected at each iteration was estimated by the LOO algorithm with weighted training dataset used in that iteration. The importance of each voxel to the classification task can be denoted by the ratio of the number of times each voxel selected over the number of iterations of FSA. Figure 3 shows the location of selected voxels in the brain and their importance. The volume of each region represents the importance of voxels. Yellow indicates the highly important region, followed by orange and red. Table 3 lists the anatomical brain regions of selected voxels.
Figure 3.
The location of selected voxels.
Table 3.
The detail region of selected voxels.
| Area | Broadmann area | L/R volume (cc) | L/R importance: value (x,y,z) |
|---|---|---|---|
| Postcentral gyrus | : 3: 5: 2: 7 | 0.7/0.6 | 1 (−24,−29,71)/1 (18,−34,71) |
| Precentral gyrus | : 4: 6: 44: 9 | 0.9/1.0 | 1 (−12,−29,71)/1 (18,−29,71) |
| Paracentral lobule | : 6: 4: 5: 31 | 0.2/0.2 | 1 (0,−34,71)/1 (6,−29,71) |
| Cingulate gyrus | : 31: 32: 24 | 1.8/1.6 | 0.341 (−6,−42,44)/0.341 (12,−42,44) |
| Superior parietal lobule | : 7 | 0.3/0.2 | 0.341 (−30,−47,44)/0.341 (30,−53,44) |
| Inferior parietal lobule | : 40 | 0.6/0.4 | 0.341 (−30,−42,44)/0.341 (48,−42,44) |
| Precuneus | : 7: 31 | 1.0/0.9 | 0.341 (0,−42,44)/0.341 (30,−42,44) |
| Medial frontal gyrus | : 11: 32: 10: 6: 9 | 1.1/1.1 | 0.268 (−6,49,−15)/0.268 (6,49,−15) |
| Superior temporal gyrus | : 38: 22:*: 41: 42 | 1.0/1.0 | 0.268 (−48,20,−14)/0.268 (48,20,−14) |
| Middle frontal gyrus | : 11: 10: 47: 6: 46: 9 | 3.9/2.3 | 0.268 (−42,49,−15)/0.268 (24,37,−14) |
| Inferior frontal gyrus | : 47: 11:*: 46: 45: 9: 13 | 3.2/1.9 | 0.268 (−36,14,−8)/0.268 (42,20,−14) |
| Superior frontal gyrus | : 11: 10: 9 | 1.0/0.6 | 0.268 (−18,60,−16)/0.268 (18,60,−16) |
| Anterior cingulate | : 32: 24 | 0.2/0.3 | 0.036 (−6,39,23)/0.121 (12,43,−10) |
| Middle temporal gyrus | : 22: 19: 21: 20 | 0.8/0.6 | 0.024 (−53,−32,4)/0.024 (65,−32,4) |
| Caudate | : | 0.1/0.2 | 0.024 (−36,−32,4)/0.024 (36,−32,4) |
| Transverse temporal gyrus | : 42: 41 | 0.2/0.3 | 0.012 (−59,−14,9)/0.012 (59,−14,9) |
| Posterior cingulate | : 31:*: 30 | 0.2/0.1 | 0.012 (−30,−60,17)/0.012 (30,−66,17) |
| Insula | : 13 | 0.1/0.2 | 0.012 (−30,27,18)/0.073 (30,26,1) |
| Cuneus | : 19: 18: 30: 17: 23 | 0.8/0.7 | 0.012 (−18,−89,24)/0.012 (12,−95,24) |
Discussion
Classification results
In the method described, three kinds of classification information were extracted from genetic and fMRI data in order to classify schizophrenia and healthy control subjects using three models: SNP-SVME, Voxel-SVME, and ICA-SVMC. Among them, Voxel-SVME and ICA-SVMC both extract information from fMRI data, while only SNP-SVME extracts classification information from SNP data. FMRI data have more weight than SNP data in the proposed method for two reasons: (a) fMRI images contain more discriminating information than SNP data, due to the fact that brain function is logically closer to the expression of mental illness symptoms and as expected the fMRI classification models performed better than SNP classification model in our experiments; (b) although Voxel-SVME and ICA-SVMC models are both constructed with fMRI data, the two models present discriminating information from different perspectives. This does not imply that the SNP classification model is unnecessary. In fact, when the two fMRI models disagree with each other, the decision of SNP-SVME is especially important because this model makes the decision based on a totally different data source. A necessary and sufficient condition for an ensemble of classifiers to be more accurate than any of its individual members is that the classifiers are accurate (better than random guessing) and the errors are at least somewhat uncorrelated (Dietterich, 2000). The proposed method meets the requirement by constructing individual classification models from different perspectives and different data source.
From data shown in Table 1, we know that the proposed four-stage method achieves better classification accuracy by combining genetic data and fMRI data than using either alone. The results indicate that even though abnormal brain function and genetic variation are both related to a clinical diagnosis of schizophrenia, they reflect different aspects of schizophrenia etiopathology, and cannot replace each other in terms of reflecting the disease. Overall, 87% accuracy was achieved, suggesting that combining genetic and brain functional information best represents the majority of symptomatic information used currently to arrive at a clinical diagnosis. For misclassified cases, many reasons may be involved including the small size of the SNP array, the rather simple and non-specific brain activation patterns reflected in the auditory stimuli paradigm and the sub-optimal sensitivity of the model. One observation worthy of note is that two patients were consistently misclassified by all classification models. This may be due to inaccuracy in all models, or the fact that there is discrepancy between biological/genetic and clinical interview-based diagnosis.
The schizophrenia patients used in this study were chronic and all taking antipsychotic medication. Aware of the potential effects of such medication on brain function, we assume that these drugs had a common, general effect on all 20 patients, since most patients were using multiple medicines (1–5 types of medicines, and a total of 26 types of medicines were prescribed) at various dosages. This study is a proof-of-concept with a small sample size and limited numbers of SNPs, to demonstrate the power of combining genetics with brain function applied in the classification framework. For a full validation, the proposed method will need to be applied to a much larger group of subjects, including multiple SZ subcategories (including schizo-affective disorder), multiple clinical treatment group (including current-naïve subjects), and using more SNPs. Future work will also focus on early differentiation of sub-groups (which in the case of prodromal subjects can take weeks to months), prediction of treatment response, or early diagnosis at the time of first presentation.
Gene selection
As shown in Table 2, the top 15 SNPs ranked by the proposed method were located in 14 genes: Among them, some are well-known putative schizophrenia susceptibility genes, such as COMT (Handoko et al., 2005; Shifman et al., 2006; Nicodemus et al., 2007), DISC1 (St Clair et al., 1990; Hodgkinson et al., 2004; Cannon et al., 2005; Callicott et al., 2005; Nicodemus et al., 2007; Saetre et al., 2008; Liu et al., 2009), MTHFR (Godfrey et al., 1990; Zintzaras, 2006; Gilbody et al., 2007; Jönsson et al., 2008; Roffman et al., 2008), and HTR3B (Maziade et al., 1995; Levinson et al., 1998; Gurling et al., 2001; Frank et al., 2004; Yamada et al., 2006). Some are brain related genes including GAD2 (De et al., 2004; Arai et al., 2009), SLC6A4 (Shi et al., 2008; Zaboli et al., 2008), and ABCB1 and ABCC (Bozina et al., 2008), possible candidates for schizophrenia susceptibility.
Voxel selection
From Table 3, the brain regions contributing most to classify schizophrenia patients and healthy controls consist of inferior, middle and medial frontal gyri, cingulate gyrus, superior temporal gyrus, and precuneus. Since our input fMRI data were contrast images (target stimulus vs. standard stimulus) collected in the auditory oddball test, it is reasonable that voxels in these regions were selected. The results are in accordance with previous structural and functional brain findings (Barta et al., 1990; Pearlson et al., 1996; Calhoun et al., 2004; Cavanna and Trimble, 2006; Garrity et al., 2007).
Conclusion
We propose a hybrid machine learning method for fusing fMRI and genetic data to separate individuals with schizophrenia from healthy controls. Experimental results showed that better classification accuracy is achieved by combining genetic and fMRI data than using either alone, suggesting that genetic and brain function represent different aspects, partially complementary to each other, of schizophrenia beauty of pathology. The method is able to extract the discriminating information to classify schizophrenia effectively and is potentially useful to identify important diagnostic markers for schizophrenia. Given the limited sample size and relatively small SNP array, this manuscript presents preliminary results and we are now in the process of attempting to replicate these findings in an independent sample.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Akaike H. (1974). A new look at statistical model identification. IEEE Trans. Automatic Control 19, 716–723 10.1109/TAC.1974.1100705 [DOI] [Google Scholar]
- Arai S., Shibata H., Sakai M., Ninomiya H., Iwata N., Ozaki N., Fukumaki Y. (2009). Association analysis of the glutamic acid decarboxylase 2 and the glutamine synthetase genes (GAD2, GLUL) with schizophrenia. Psychiatr. Genet. 19, 6–13 10.1097/YPG.0b013e328311875d [DOI] [PubMed] [Google Scholar]
- Barta P. E., Pearlson G. D., Powers R. E., Richards S. S., Tune L. E. (1990). Auditory hallucinations and smaller superior temporal gyral volume in schizophrenia. Am. J. Psychiatry 147, 1457–1462 [DOI] [PubMed] [Google Scholar]
- Bell A. J., Sejnowski T. J. (1995). An information-maximization approach to blind separation and blind deconvolution. Neural. Comput. 7, 1129–1159 10.1162/neco.1995.7.6.1129 [DOI] [PubMed] [Google Scholar]
- Bozina N., Kuzman M. R., Medved V., Jovanovic N., Sertic J., Hotujac L. (2008). Associations between MDR1 gene polymorphisms and schizophrenia and therapeutic response to olanzapine in female schizophrenic patients. J. Psychiatr. Res. 42, 89–97 10.1016/j.jpsychires.2006.10.002 [DOI] [PubMed] [Google Scholar]
- Calhoun V. D., Kiehl K. A., Liddle P. F., Pearlson G. D. (2004). Aberrant localization of synchronous hemodynamic activity in auditory cortex reliably characterizes schizophrenia. Biol. Psychiatry 55, 842–849 10.1016/j.biopsych.2004.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun V. D., Liu J., Adali T. (2009). A review of group ICA for fMRI data and ICA for joint inference of imaging, genetic, and ERP data. Neuroimage 45, 163–172 10.1016/j.neuroimage.2008.10.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callicott J. H., Straub R. E., Pezawas L., Egan M. F., Mattay V. S., Hariri A. R., Verchinski B. A., Meyer L. A., Balkissoon R., Kolachana B., Goldberg T. E., Weinberger D. R. (2005). Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc. Natl. Acad. Sci. U.S.A. 102, 8627–8632 10.1073/pnas.0500515102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon T. D., Hennah W., Van E. T., Thompson P. M., Lonnqvist J., Huttunen M., Gasperoni T., Tuulio H. A., Pirkola T., Toga A. W., Kaprio J., Mazziotta J., Peltonen L. (2005). Association of DISC1/TRAX haplotypes with schizophrenia, reduced prefrontal gray matter, and impaired short- and long-term memory. Arch. Gen. Psychiatry 62, 1205–1213 10.1001/archpsyc.62.11.1205 [DOI] [PubMed] [Google Scholar]
- Cardoso J. F. (1997). Infomax and maximum likelihood for blind source separation. IEEE Signal Proc. Lett. 4, 112–114 10.1109/97.566704 [DOI] [Google Scholar]
- Cavanna A. E., Trimble M. R. (2006). The precuneus: a review of its functional anatomy and behavioural correlates. Brain 129, 564–583 10.1093/brain/awl004 [DOI] [PubMed] [Google Scholar]
- De L. V., Muglia P., Masellis M., Jane D. E., Wong G. W., Kennedy J. L. (2004). Polymorphisms in glutamate decarboxylase genes: analysis in schizophrenia. Psychiatr. Genet. 14, 39–42 10.1097/00041444-200403000-00006 [DOI] [PubMed] [Google Scholar]
- Demirci O., Clark V. P., Calhouna V. D. (2008). A projection pursuit algorithm to classify individuals using fMRI data: application to schizophrenia. Neuroimage 39, 1774–1782 10.1016/j.neuroimage.2007.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietterich T. G. (2000). Ensemble methods in machine learning. Proc. MCS’2000, Lect. Notes Comput. Sci. 1857, 1–15 10.1007/3-540-45014-9_1 [DOI] [Google Scholar]
- Fan J. B., Oliphant A., Shen R., Kermani B. G., Garcia F., Gunderson K. L., Hansen M., Steemers F., Butler S. L., Deloukas P., Galver L., Hunt S., McBride C., Bibikova M., Rubano T., Chen J., Wickham E., Doucet D., Chang W., Campbell D., Zhang B., Kruglyak S., Bentley D., Haas J., Rigault P., Zhou L., Stuelpnagel J., Chee M. S. (2003). Highly parallel SNP genotyping. Cold Spring Harbor Symp. Quant. Biol. 68, 69–78 10.1101/sqb.2003.68.69 [DOI] [PubMed] [Google Scholar]
- First M. B., Spitzer R. L., Gibbon M., Williams J. B. (1995). Structured Clinical Interview for DSM-IV Axis I Disorders, Patient Edition (SCID-I/P, version 2.0). New York: Biometrics Research Department, New York State Psychiatric Institute [Google Scholar]
- Ford J., Farid H., Makedon F., Flashman L., McAllister T., Megalooikonomou V., Saykin A. (2003). Patient classification of Fmri activation maps. Medical Image Computing and Computer Assisted Intervention (MICCAI 03), 58–65 [Google Scholar]
- Frank B., Niesler B., Nöthen M. M., Neidt H., Propping P., Bondy B., Rietschel M., Maier W., Albus M., Rappold G. (2004). Investigation of the human serotonin receptor gene HTR3B in bipolar affective and schizophrenic patients. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 131, 1–5 10.1002/ajmg.b.30070 [DOI] [PubMed] [Google Scholar]
- Freire L., Mangin J. F. (2001). Motion correction algorithms may create spurious brain activations in the absence of subject motion. Neuroimage 14, 709–722 10.1006/nimg.2001.0869 [DOI] [PubMed] [Google Scholar]
- Freund Y., Schapire R. E. (1997). A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 55, 119–139 10.1006/jcss.1997.1504 [DOI] [Google Scholar]
- Friston K., Ashburner J., Frith C. D., Poline J. P., Heather J. D., Frackowiak R. S. (1995). Spatial registration and normalization of images. Hum. Brain Mapp. 2, 165–189 10.1002/hbm.460030303 [DOI] [Google Scholar]
- Fukunaga K. (1990). Introduction to Statistical Pattern Recognition. New York: Academic Press [Google Scholar]
- Garrity A. G., Pearlson G. D., McKiernan K., Lloyd D., Kiehl K. A., Calhoun V. D. (2007). Aberrant “default mode” functional connectivity in schizophrenia. Am. J. Psychiatry 164, 450–457 10.1176/appi.ajp.164.3.450 [DOI] [PubMed] [Google Scholar]
- Gilbody S., Lewis S., Lightfoot T. (2007). Methylenetetrahydrofolate reductase (MTHFR) genetic polymorphisms and psychiatric disorders: a HuGE review. Am. J. Epidemiol. 165, 1–13 10.1093/aje/kwj347 [DOI] [PubMed] [Google Scholar]
- Godfrey P. S., Crellin R., Toone B. K., Carney M. W., Flynn T. G., Bottiglieri T., Laundy M., Chanarin I., Reynolds E. H. (1990). Enhancement of recovery from psychiatric illness by methylfolate. Lancet 336, 392–395 10.1016/0140-6736(90)91942-4 [DOI] [PubMed] [Google Scholar]
- Gurling H. M., Kalsi G., Brynjolfson J., Sigmundsson T., Sherrington R., Mankoo B. S., Read T., Murphy P., Blaveri E., McQuillin A., Petursson H., Curtis D. (2001). Genomewide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21-22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3-24 and 20q12.1-11.23. Am. J. Hum. Genet. 68, 661–673 10.1086/318788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoko H. Y., Nyholt D. R., Hayward N. K., Nertney D. A., Hannah D. E., Windus L. C., McCormack C. M., Smith H. J., Filippich C., James M. R., Mowry B. J. (2005). Separate and interacting effects within the catechol-O-methyltransferase (COMT) are associated with schizophrenia. Mol. Psychiatry 10, 589–597 10.1038/sj.mp.4001606 [DOI] [PubMed] [Google Scholar]
- Hariri A. R., Weinberger D. R. (2003). Imaging genomics. Br. Med. Bull. 65, 259–270 10.1093/bmb/65.1.259 [DOI] [PubMed] [Google Scholar]
- Haynes J., Rees G. (2006). Decoding mental states from brain activity in humans. Nat. Rev. Neurosci. 7, 523–534 10.1038/nrn1931 [DOI] [PubMed] [Google Scholar]
- Hodgkinson C. A., Goldman D., Jaeger J., Persaud S., Kane J. M., Lipsky R. H., Malhotr A. K. (2004). Disrupted in schizophrenia 1 (DISC1): association with schizophrenia, schizoaffective disorder, and bipolar disorder. Am. J. Hum. Genet. 75, 862–872 10.1086/425586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe N. R. (2003). “A closer look at boosted image retrieval,” Proceedings of Second International Conference on Image and Video Retrieval, 61–70 [Google Scholar]
- Jain A. K., Chandrasekaran B. (1982). Dimensionality and sample size considerations. Pattern Recogn. Pract. 2, 835–855 [Google Scholar]
- Jönsson E. G., Larsson K., Vares M., Hansen T., Wang A. G., Djurovic S., Rønningen K. S., Andreassen O. A., Agartz I., Werge T., Terenius L., Hall H. (2008). Two methylenetetrahydrofolate reductase gene (MTHFR) polymorphisms, schizophrenia and bipolar disorder: an association study. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 147B, 976–982 10.1002/ajmg.b.30671 [DOI] [PubMed] [Google Scholar]
- Kay S. R., Fiszbein A., Opler L. A. (1987). The positive and negative syndrome scale (PANSS) for schizoprenia. Schizophr. Bull. 13, 261–276 [DOI] [PubMed] [Google Scholar]
- Kiehl K. A., Liddle P. F. (2003). Reproducibility of the hemodynamic response to auditory oddball stimuli: a six-week test–retest study. Hum. Brain Mapp. 18, 42–52 10.1002/hbm.10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehl K. A., Stevens M. C., Laurens K. R., Pearlson G., Calhoun V. D., Liddle P. F. (2005). An adaptive reflexive processing model of neurocognitive function: supporting evidence from a large scale (n = 100) fMRI study of an auditory oddball task. Neuroimage 25, 899–915 10.1016/j.neuroimage.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Lee C., McGlashan T. H., Woods S. W. (2005). Prevention of schizophrenia: can it be achieved? CNS Drugs 19, 193–206 10.2165/00023210-200519030-00002 [DOI] [PubMed] [Google Scholar]
- Levinson D. F., Mahtani M. M., Nancarrow D. J., Brown D. M., Kruglyak L., Kirby A. (1998). Genome scan of schizophrenia. Am. J. Psychiatry 155, 741–750 [DOI] [PubMed] [Google Scholar]
- Li Y., Adali T., Calhoun V. D. (2007). Estimating the number of independent components for functional magnetic resonance imaging data. Hum. Brain Mapp. 28, 1251–1266 10.1002/hbm.20359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. (2005). Toward integrating feature selection algorithms for classification and clustering. IEEE Trans. Knowl. Data Eng. 17, 491–502 10.1109/TKDE.2005.66 [DOI] [Google Scholar]
- Liu J., Pearlson G., Windemuth A., Ruano G., Perrone-Bizzozero N. I., Calhoun V. D. (2009). Combining fMRI and SNP data to investigate connections between brain function and genetics using parallel ICA. Hum. Brain Mapp. 30, 241–255 10.1002/hbm.20508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaspina D., Friedman J. H., Kaufmann C., Bruder G., Amador X., Strauss D., Clark S., Yale S., Lukens E., Thorning H., Goetz R., Gorman J. (1998). Psychobiological heterogeneity of familial and sporadic schizophrenia. Biol. Psychiatry 43, 489–496 10.1016/S0006-3223(97)00527-1 [DOI] [PubMed] [Google Scholar]
- Maziade M., Raymond V., Cliché D., Fournier J. P., Caron C., Garneau Y. (1995). Linkage results on 11Q21-22 in Eastern Quebec pedigrees densely affected by schizophrenia. Am. J. Med. Genet. 60, 522–528 10.1002/ajmg.1320600607 [DOI] [PubMed] [Google Scholar]
- Murray R. M., O'Callaghan E., Castle D. J., Lewis S. W. (1992). A neurodevelopmental approach to the classification of schizophrenia. Schizophr. Bull. 18, 319–332 [DOI] [PubMed] [Google Scholar]
- Nicodemus K. K., Kolachana B. S., Vakkalanka R., Straub R. E., Giegling I., Egan M. F., Rujescu D., Weinberger D. R. (2007). Evidence for statistical epistasis between catechol-O-methyltransferase (COMT) and polymorphisms in RGS4, G72 (DAOA), GRM3, and DISC1: influence on risk of schizophrenia. Hum. Genet. 120, 889–906 10.1007/s00439-006-0257-3 [DOI] [PubMed] [Google Scholar]
- Oliphant A., Barker D. L., Stuelpnagel J. R., Chee M. S. (2002). BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques Suppl. 56–58, 60–61 [PubMed] [Google Scholar]
- O'Toole A. J., Jiang F., Abdi H., Penard N., Dunlop J. P., Parent M. A. (2007). Theoretical, statistical, and practical perspectives on pattern-based classification approaches to functional neuroimaging analysis. J. Cogn. Neurosci. 19, 1735–1752 10.1162/jocn.2007.19.11.1735 [DOI] [PubMed] [Google Scholar]
- Pearlson G. D., Folley B. S. (2008). Endophenotypes, dimensions, risks: is psychosis analogous to common inherited medical illnesses? Clin. EEG Neurosci. 39, 73–77 [DOI] [PubMed] [Google Scholar]
- Pearlson G. D., Petty R. G., Ross C. A., Tien A. Y. (1996). Schizophrenia: a disease of heteromodal association cortex? Neuropsychopharmacology 14, 1–17 10.1016/S0893-133X(96)80054-6 [DOI] [PubMed] [Google Scholar]
- Pereira F., Mitchell T., Botvinick M. (2009). Machine learning classifiers and fMRI: a tutorial overview. Neuroimage 45, 199–209 10.1016/j.neuroimage.2008.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potkin S. G., Turner J. A., Guffanti G., Lakatos A., Fallon J. H., Nguyen D. D., Mathalon D., Ford J., Lauriello J., Macciardi F., FBIRN (2009). A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophr. Bull. 35, 96–108 10.1093/schbul/sbn155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roffman J. L., Gollub R. L., Calhoun V. D., Wassink T. H., Weiss A. P., Ho B. C., White T., Clark V. P., Fries J., Andreasen N. C., Goff D. C., Manoach D. S. (2008). MTHFR 677C → T genotype disrupts prefrontal function in schizophrenia through an interaction with COMT 158Val → Met.Proc. Natl. Acad. Sci. U.S.A. 105, 17573–17578 10.1073/pnas.0803727105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruano G. (2006). Physiogenomic method for predicting clinical outcomes of treatments in patients. Patent USPTO #20060278241. [Google Scholar]
- Saetre P., Agartz I., De F. A., Lundmark P., Djurovic S., Kähler A., Andreassen O. A., Jakobsen K. D., Rasmussen H. B., Werge T., Hall H., Terenius L., Jönsson E. G. (2008). Association between a disrupted-in-schizophrenia 1 (DISC1) single nucleotide polymorphism and schizophrenia in a combined Scandinavian case-control sample. Schizophr. Res. 106, 237–241 10.1016/j.schres.2008.08.024 [DOI] [PubMed] [Google Scholar]
- Shi J., Gershon E. S., Liu C. (2008). Genetic associations with schizophrenia: meta-analyses of 12 candidate genes. Schizophr. Res. 104, 96–107 10.1016/j.schres.2008.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman S., Levit A., Chen M. L., Chen C. H., Bronstein M., Weizman A., Yakir B., Navon R., Darvasi A. (2006). A complete genetic association scan of the 22q11 deletion region and functional evidence reveal an association between DGCR2 and schizophrenia. Hum. Genet. 120, 160–170 10.1007/s00439-006-0195-0 [DOI] [PubMed] [Google Scholar]
- Shinkareva S. V., Ombao H. C., Sutton B. P., Mohanty A., Miller G. A. (2006). Classification of functional brain images with a spatio-temporal dissimilarity map. Neuroimage 33, 63–71 10.1016/j.neuroimage.2006.06.032 [DOI] [PubMed] [Google Scholar]
- Spitzer R. L., Williams J. B., Gibbon M. (1996). Structured Clinical Interview for DSM-IV: Non-patient Edition (SCID-NP). New York: Biometrics Research Department, New York State Psychiatric Institute [Google Scholar]
- Sponheim S. R., Iacono W. G., Thuras P. D., Beiser M. (2001). Using biological indices to classify schizophrenia and other psychotic patients. Schizophr. Res. 50, 139–150 10.1016/S0920-9964(00)00160-2 [DOI] [PubMed] [Google Scholar]
- Sponheim S. R., Iacono W. G., Thuras P. D., Nugent S. M., Beiser M. (2003). Sensitivity and specificity of select biological indices in characterizing psychotic patients and their relatives. Schizophr. Res. 63, 27–38 10.1016/S0920-9964(02)00385-7 [DOI] [PubMed] [Google Scholar]
- St Clair C. D., Blackwood D., Muir W., Carothers A., Walker M., Spowart G., Gosdeen C., Evans H. J. (1990). Association within a family of a balanced autosomal translocation with major mental illness. Lancet 336, 13–16 10.1016/0140-6736(90)91520-K [DOI] [PubMed] [Google Scholar]
- Struyf J., Dobrin S., Page D. (2008). Combining gene expression, demographic and clinical data in modeling disease: a case study of bipolar disorder and schizophrenia. BMC Genomics 9, 531. 10.1186/1471-2164-9-531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Adali T., Pearlson G. D., Clark V. P., Calhoun V. D. (2009). A method for accurate group difference detection by constraining the mixing coefficients in an ICA framework. Hum. Brain Mapp. 30, 2953–2970 10.1002/hbm.20721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini G., Dietterich T. G. (2002). Bias-variance analysis and SVM ensemble. Lect. Notes Comput. Sci. 27–38 [Google Scholar]
- Vapnik V. N. (2000). The Nature of Statistical Learning Theory. New York: Springer-Verlag Press [Google Scholar]
- Yamada K., Hattori E., Iwayama Y., Ohnish T., Ohba H., Toyota T., Takao H., Minabe Y., Nakatani N., Higuchi T., Detera-Wadleigh S. D., Yoshikawa T. (2006). Distinguishable haplotype blocks in the HTR3A and HTR3B region in the Japanese reveal evidence of association of HTR3B with female major depression. Biol. Psychiatry 60, 192–201 10.1016/j.biopsych.2005.11.008 [DOI] [PubMed] [Google Scholar]
- Zaboli G., Jönsson E. G., Gizatullin R., De F. A., Asberg M., Leopardi R. (2008). Haplotype analysis confirms association of the serotonin transporter (5-HTT) gene with schizophrenia but not with major depression. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 147, 301–307 10.1002/ajmg.b.30597 [DOI] [PubMed] [Google Scholar]
- Zhang L., Samaras D., Tomasi D., Volkow N., Goldstein R. (2005). Machine learning for clinical diagnosis from functional magnetic resonance imaging. IEEE Proc. Comput. Vision Pattern Recogn. 1, 1211–1217 [Google Scholar]
- Zintzaras E. (2006). C677T and A1298C methylenetetrahydrofolate reductase gene polymorphisms in schizophrenia, bipolar disorder and depression: a meta-analysis of genetic association studies. Psychiatr. Genet. 16, 105–115 10.1097/01.ypg.0000199444.77291.e2 [DOI] [PubMed] [Google Scholar]