Abstract
Background
Schizophrenia is a complex genetic disorder, with multiple putative risk genes and many reports of reduced cortical gray matter. Identifying the genetic loci contributing to these structural alterations in schizophrenia (and likely also to normal structural gray matter patterns) could aid understanding of schizophrenia’s pathophysiology. We used structural parameters as potential intermediate illness markers to investigate genomic factors derived from single nucleotide polymorphism (SNP) arrays.
Method
We used research quality structural magnetic resonance imaging (sMRI) scans from European American subjects including 33 healthy control subjects and 18 schizophrenia patients. All subjects were genotyped for 367 SNPs. Linked sMRI and genetic (SNP) components were extracted to reveal relationships between brain structure and SNPs, using parallel independent component analysis, a novel multivariate approach that operates effectively in small sample sizes.
Results
We identified an sMRI component that significantly correlated with a genetic component (r = −.536, p < .00005); components also distinguished groups. In the sMRI component, schizophrenia gray matter deficits were in brain regions consistently implicated in previous reports, including frontal and temporal lobes and thalamus (p < .01). These deficits were related to SNPs from 16 genes, several previously associated with schizophrenia risk and/or involved in normal central nervous system development, including AKT, PI3K, SLC6A4, DRD2, CHRM2, and ADORA2A.
Conclusions
Despite the small sample size, this novel analysis method identified an sMRI component including brain areas previously reported to be abnormal in schizophrenia and an associated genetic component containing several putative schizophrenia risk genes. Thus, we identified multiple genes potentially underlying specific structural brain abnormalities in schizophrenia.
Keywords: Brain development, genetics, imaging, schizophrenia, structural magnetic resonance imaging (sMRI)
Schizophrenia is a heritable psychiatric disorder (1) associated with both genetic and environmental factors. Many schizophrenia susceptibility genes are hypothesized to influence brain morphology and function via processes including neuronal growth, migration, communication, and pruning (2), so it is important to know how they may be involved in “building” normal and (by extension) abnormal brain structure and function (3).
Regional gray matter (GM) volumes are highly heritable (4). Meta-analyses of brain structure in schizophrenia report reduced GM and increased ventricular volume (4,5); first-episode data suggest that these findings are state-independent and persist throughout the illness (6). Structural studies based on voxel-based morphometry (VBM) and automated regional parcellation confirm nonhomogenous GM reductions, especially in frontal, parietal, and temporal regions (7–10). Although other reports temper this conclusion (8), meta-analytic, twin, and family studies suggest that GM volume abnormalities covary in a dose-dependent manner with schizophrenia risk (11). Overall, these findings support brain structural alterations as robustly associated with schizophrenia (6), possibly as an endophenotype (11).
Identifying genetic loci contributing to normal GM structural patterns or to structural abnormalities in schizophrenia aids our understanding of pathophysiology. Many studies have investigated effects of numerous genetic polymorphisms related to GM differences in schizophrenia. Examples include variation in DISC1 associated with hippocampal GM volume and function (12), COMT variants affecting hippocampal and dorsolateral prefrontal GM volume (13) and altered volumes of left inferior temporal gyrus, pre-frontal cortex and lateral occipital cortex associated with brain-derived neurotrophic factor variants (14–16). Most prior structural magnetic resonance imaging (sMRI) studies were hypothesis guided, focused on the influence of only one or two preselected genes at a time, and were based on data from schizophrenia association and linkage studies.
Because schizophrenia is a complex disorder likely contributed to by multiple genes (6), it is likely that multiple genes also contribute additively or multiplicatively to both normal and schizophrenia-associated regional GM volume patterns. To better understand the product of this putatively interactive process, we wished to analyze simultaneously GM volumes from both control subjects and schizophrenia patients along with their allelic variation data. In analyzing such large data sets (i.e., GM and single nucleotide polymorphisms [SNPs]), dimension and computation are necessarily large scale (17) to model potentially hundreds of thousands of MRI data points (GM voxels) and SNPs. A variety of data-driven approaches have been developed, including partial least squares, local linear discriminant analysis, and support vector machines (18). All such approaches have been applied for first-level processing, that is, calculated using single subject spatiotemporal MRI data and finding the neurocircuitry influence of a specific genotype. They have not been, to our knowledge, applied to the combination of multiple genotypes and imaging data. We therefore used parallel independent component analysis (parallel ICA) (19), a novel approach to analyze multimodal data that employs data reduction steps to minimize the “curse of dimensionality” (17). Parallel ICA uses a blind source separation method that separates high dimensional data to discover patterns associated with, for example, clusters (components) of linked GM regions derived from quantitative brain measures or components of associated SNPs derived from a gene array. This technique can identify and quantify associations between these two sets of components and determine differences in a patient-versus-control context embedded in the components (19). It is a variant of ICA designed for multimodality processing that extracts components using an entropy term based on information theory to maximize independence (20) and enhances the interconnection by maximizing the linkage function in a joint estimation process (19) based on higher-order statistics. This technique requires prior knowledge of neither specified genes nor GM patterns, making this a hypothesis-free approach constituting an unsupervised algorithm, analogous to those used in discovering novel genes to reveal regulatory networks through analyzing large data sets (21). The basic ICA technique has been validated by examining multivariate relationships between GM and functional-MRI-measured activity (22) and relationships between DTI-measured white matter (WM) coherence and whole brain functional connectivity (23). Parallel ICA has been previously applied to gene–brain explorations to find simultaneously independent components from a functional MRI auditory oddball task and from a gene array in schizophrenia and control subjects and from EEG data and SNP array data in healthy control subjects (19,24,25). Importantly, this method has the ability to detect significant associations in modest-sized data sets with a preferred ratio of sample size to SNP size of at least 0.02 (26). In the current study, we applied parallel ICA to a relatively small sample for identifying the linked components simultaneously in GM and SNP array. For the first time, we used this approach to extract information on the intrinsic relationship between GM regions and SNP clusters in healthy control subjects and schizophrenia patients. The SNP cluster was derived from putative schizophrenia risk genes combined with a hypothesis-free strategy for gene selection, a so-called hybrid approach.
Methods and Materials
We used data from two modalities, structural MRI and genotype (SNP), to reveal relationships between them that also differed between schizophrenia-diagnosed and healthy control groups.
Participants
We assessed 51 self-described European American subjects, whose demographic data are shown in Table 1, comprising 18 schizophrenia patients and 33 healthy control subjects. Individuals belonging to non–European American populations were excluded because of potential population stratification artifacts. Subjects were recruited from outpatient units at the Institute of Living in Hartford, by newspaper, and by word of mouth. Healthy control subjects were screened to exclude current or past history of DSM-IV Axis I diagnosis assessed by the Structured Clinical Interview for DSM-IV Axis I Disorders (SCID) (27) and interviewed to confirm an absent family history of psychosis. Schizophrenia patients met DSM-IV SCID diagnostic criteria on the basis of interview and case file review by a clinician. All participants gave written informed consent approved by Hartford Hospital Internal Review Board. Groups were not statistically different on age, sex, or handedness. IQ scores was estimated from the National Adult Reading Test (28) [t (45) = 3.2, p = .002]. Positive and Negative Syndrome Scale (29) scores for 14 patients were recorded (Table 1). All patients were prescribed antipsychotic medications; 16 took one or more second-generation drugs, three a first-generation drug (one patient received a combination), six took a selective serotonin reuptake inhibitor, six a mood stabilizer, and four a benzodiazepine.
Table 1.
Demographics and Clinical Characteristics of the Sample
| Schizophrenia (n = 18) | Healthy Control Subjects (n = 33) | Statistics | |||
|---|---|---|---|---|---|
| Sex (male/female) | 15/3 | 21/12 | λ2 = 2.17 | ns | |
| Handedness | 15/3 | 31/2 | t(49) = 1.21 | ns | |
| Age (years), Mean (SD) | 35.3 (10.1) | 35.9 (13.5) | t(49)= .14 | ns | |
| Estimated Premorbid Full-Scale IQ (NART), Mean (SD) | 104.09 (9.9) | (n = 29) 111.47 (5.5) | t(45) = 3.2 | .002 | |
| PANSS Total (n = 14) | 58.28 | 14.4 | |||
| PANSS Positive (n = 14) | 16.35 | 4.41 | |||
| PANSS Negative (n = 14) | 12.14 | 5.14 | |||
| PANSS General (n = 14) | 29.78 | 8.06 | |||
NART, National Adult Reading Test; PANSS, Positive and Negative Syndrome Scale.
Structural MRI Parameters and Preprocessing
Structural MRI were obtained with a 3T Siemens Allegra scanner (Siemens, Erlangen, Germany) using a T1-weighted axial magnetization prepared rapid acquisition gradient-echo (repetition time = 2500, echo time = 2.74, inversion time = 900 msec, image matrix = 208 × 256, field of view = 256 mm, voxel size = 1 × 1 × 1 mm3, flip angle = 8°, 176 slices). Data were preprocessed using SPM2 (30) running in MATLAB 6.5. Before preprocessing, images were checked for movement artifacts and the origin on the anterior commissure. We employed optimized VBM methodology (31). Initially, a control-specific template was created by normalizing each structural scan to the Montreal Neurological Institute template. Using the customized template and priors, each participant’s original T1 image was segmented into GM and WM, normalized and smoothed with a 10-mm full width at half maximum Gaussian kernel. The resultant smoothed modulated GM images were input from the sMRI modality to parallel ICA.
SNP Data Collection and Preprocessing
We followed two strategies for genotyping and subsequently combined the two SNP data sets for each subject, yielding 367 SNPs. The first was based on general physiologic factors, and the second specifically targeted schizophrenia candidate genes.
Saliva samples were collected from each subject and DNA extracted using routine procedures. Genotyping performed at Genomas, Inc., used the Illumina BeadArray platform and Goldengate Assay (32,33). Genes were selected from the PG custom SNP array, consisting of 384 SNPs from 222 genes, designed and tested at Genomas, Inc. (Hartford, Connecticut) as a product (34) and service (19). The SNP array covers genes from a variety of brain-related axes including neurotransmitters, their synthetic enzymes, receptors and transporters, and general brain and somatic metabolic processes (35). For the reliability measure, SNPs with a calculated GenCall score of .25 or higher were selected, resulting in 345 SNPs. Genotypes are inherently categorical and codable as either positive or negative numbers. The signs are not important in our test because we consider genotypic variation and not the SNPs themselves. Here the SNP input data were coded as 0 for homozygous, 1 for heterozygous, and 2 for opposite homozygous.
The remainder of the genotyping, performed at Yale University, independently targeted SNPs for 18 schizophrenia candidate genes using a fluorogenic 5′ nuclease assay (the TaqMan method; Applied Biosystems, Foster City, California) (36). All samples were genotyped in duplicate for quality control, with no discrepancies. The same coding procedure described above applied to following 22 SNPs: catechol-O-methyltransferase(rs4680); brain-derived neurotrophic-factor(rs6265); solute carrier family-6(neurotransmitter transporter, serotonin member-4; rs25531); dopamine receptor D2(rs6277, rs1799732); nicotinic cholinergicreceptor, alpha7(rs868437, rs2337506); muscariniccholinergic receptor, alpha5(rs16969968); cannabinoid receptor1(rs1049353); doublecortin domain containing-2(dbSTS BV677278); solute carrier family-24, member-5(rs1426654); dopadecarboxylase (aromatic-l-aminoacid decarboxylase) (rs11238214); dopamine beta-hydroxylase (dopamine beta-monooxygenase) (rs1611115); dopamine receptor D4; calsyntenin-2(rs6439886); KIBRA(rs17070145); solute carrier family-6 (neurotransmitter transporter, serotonin)member-2; Disrupted-in-Schizophrenia-1 (rs751229, rs3738401, rs980989, rs821616); translin-associated factor-X(rs1655285).
Data Analysis and Statistics
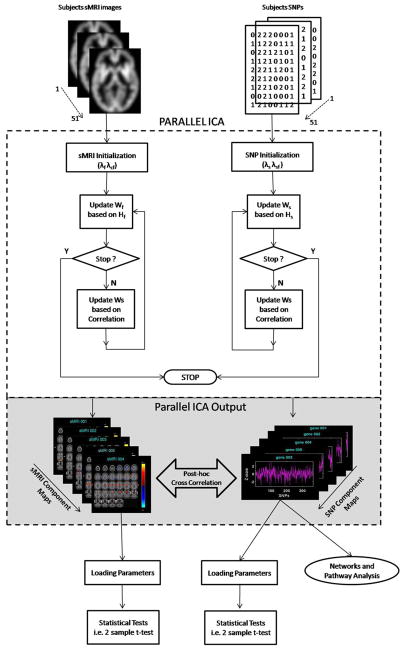
We applied parallel ICA (19) using FIT (Fusion ICA Toolbox, http://icatb.sourceforge.net) in MATLAB-6.5 on the sMRI and SNP data described earlier to identify the structural brain networks, SNP associations, and their interrelationships. A detailed description of the algorithms used in parallel ICA and their validation is found in Liu et al. (19,25) and is depicted in Figure 1.
Figure 1.
Illustration of parallel independent component analysis (ICA) and network/canonical pathways implemented in structural magnetic resonance imaging (sMRI) and single nucleotide polymorphism (SNP) data. sMRI and SNP data are initialized with specified learning rates: sMRI (λf, λcf) and SNP (λs λsf). Parallel ICA identified sMRI and SNP component using an optimization algorithm based on entropy terms (Hf and Hs) and correlation term between Wf−1 (sMRI) and Ws−1 (SNP). Wf−1 and Ws−1 a participant-by-component mixing matrix. The loading parameters of the selected sMRI and SNP component were tested for group differences. SNP component that showed group differences and significant correlation with sMRI component were passed into gene pathway analysis.
The sMRI data including both control and patients group were constructed as a matrix of subjects by smoothed modulated GM images, represented as a set of spatially independent voxels that are linearly mixed (37). The SNP data from both groups were organized as matrix of subjects by SNPs. These two data matrices are the input to the parallel ICA. A dilated GM mask generated using the WFU Pick atlas (http://www.fmri.wfubmc.edu) was used to limit the analysis to GM. The order selection tool in the GIFT software toolbox (38) was used to identify the number of components. The component set size was estimated using both Akaike information criterion (AIC) and minimum description length criteria, which is the standard method for estimating the components from the aggregated data set (39). The order estimated from both sMRI and SNP data were determined to be five components.
Parallel ICA is then applied to identify the latent components and the relationship between the two modalities described earlier. The algorithm identifies the linked components by jointly maximizing the independence between the components within modality as well as enhancing the pairwise correlation between them (19). In Liu et al. (19), it was shown that the parallel ICA algorithm is better able to identify the underlying relationships among SNP/brain features than simple correlation or ICA performed separately on each modality. Components from the sMRI data are maximally independent and measure the localized GM changes and their variation among the subjects. Components extracted from SNP data are distinct, independent, linear combinations of SNPs, and those with a significant linkage are highly associated with brain structure, brain function, or other phenotypes (40). We used a leave-one-out cross-evaluation to test further for consistency of the components and the intermodality linkages. In this method, one subject at a time is randomly omitted and 50 of 51 subjects analyzed repeatedly with the same specification.
The output from the parallel ICA is presented as a pair of sMRI and SNP components with their correlation values expressing the existing relationship between the two modalities. Correlation values were thresholded at p < .05 corrected for multiple comparison using a false discovery rate (41). In addition, each sMRI/SNP component was expressed to different degrees and weights in different subjects, and their influence and expression was captured in the loading parameters (19). Each loading parameter reveals the expression pattern of the correlated components in all subjects and enables testing of group differences (19). Subsequently, we used the loading parameter matrix to test the group difference between normal control subjects and schizophrenia patients in each sMRI and SNP component identified in the significant pairs.
Networks and Canonical Pathway Mapping
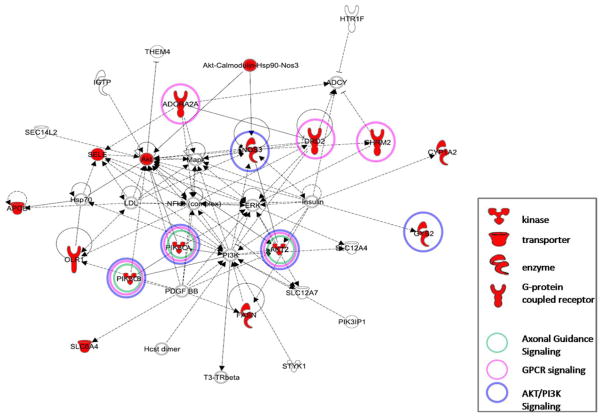
To explore how the genes included in the SNP components identified in the significant correlating pairs were inter-related we used canonical pathway analysis using the Ingenuity Pathway Analysis (IPA) Software (Ingenuity Systems, http://www.ingenuity.com). The analysis uses a network generation algorithm to identify the genes that are highly interconnected and expressed in the data set. These genes were overlaid onto a global molecular network developed from information contained in the IPA knowledge base and a gene network then algorithmically generated based on their connectivity. IPA uses a right-tailed Fisher’s test to calculate the p value for networks. Genes are represented as nodes, and the biological relationship between two nodes is represented as an edge (line). Nodes are displayed using various shapes representing functional classes of gene products (Figure 3 in Results).
Figure 3.
Functional network identified by genes in the single nucleotide polymorphism (SNP) component using independent component analysis. Nodes in the red represent genes in the SNP component. The other molecules found in the network are possible connections and interactions between molecules, based onIngenuity Pathway Analysis knowledge base. Axonal Guidance signaling, G-protein-coupled receptor (GPCR) signaling, and PI3K/AKT signaling were the main signaling networks found. The genes with the colored circles in the network diagram are relevantly expressed in their respective pathways. The shapes of each gene symbol in the network denote the class of that gene as defined by the Ingenuity Pathway Analysis tool.
Results
Parallel ICA identified two sMRI components (sMRI-A and sMRI-B) significantly correlated with a genetic component (SNP). Post hoc correlation analysis on the loading parameters showed both the sMRI component (sMRI-A and sMRI-B) were negatively correlated with the SNP, with the sMRI-A component, r = −.536, p < .00005, and with the sMRI-B component r = −.341, p < .014.
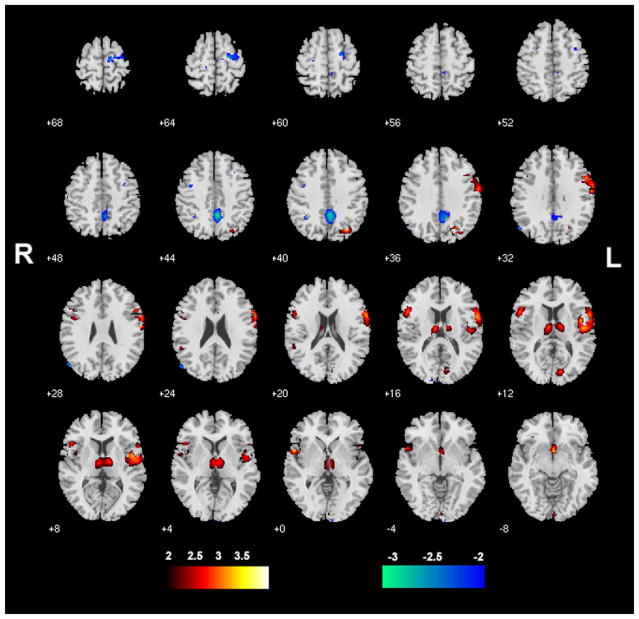
The sMRI Component-A showing the GM distribution thresholded at |Z| >2.0 is shown in Figure 2. The Talairach coordinates for sMRI Component-A is shown in Table 2. Brain regions in this component were identified in previous VBM meta-analyses and large-scale VBM analyses of schizophrenia (5,8,9), and the comparison with prior studies is shown in Table 2.
Figure 2.
The structural magnetic resonance imaging Component-A identified by parallel independent component analysis showing gray matter distributions from all patients and control subjects (n = 51) and thresholded at |z| > 2.0
Table 2.
Coordinates in the Talairach Space of Structural Magnetic Resonance Imaging Component-A of Significant Gray Matter Reduction in Patients Versus Control Subjects
| Regions | Brodmann Area | Volume (cc) |
Random Effects: Max Value (x, y, z) |
Prior Study | |
|---|---|---|---|---|---|
| R/L | R | L | |||
| Positive | |||||
| Anterior Cingulate | 25 | .6/.4 | 3.5 (0, 5, −8) | 3.2 (−3, 5, −10) | (5, 8–10) |
| Precuneus | 7, 19, 39 | 1.9/.0 | 3.5 (27, −65, 36) | NS | (8, 10) |
| Subgenual Cingulate | 25 | .6/.7 | 3.4 (3, 8, −13) | 3.2 (−3, 11, −11) | (9, 10) |
| Inferior Frontal Gyrus | 9, 11, 13, 44, 45, 47 | 5.9/5.3 | 3.2 (53, 13, 27) | 3.4 (−21, 20, −16) | (8–10) |
| Precentral Gyrus | 6, 9, 13, 43, 44 | 6.0/1.0 | 3.3 (59, −4, 31) | 2.7 (−53, 15, 8) | (8–10) |
| Superior Temporal Gyrus | 13, 22, 38 | 3.6/2.4 | 3.3 (45, 19, −24) | 3.3 (−53, 9, 0) | (8–10) |
| Middle Frontal Gyrus | 6, 8, 9, 11, 46 | 1.6/1.0 | 3.2 (45, 13, 30) (13, 30, 45) | 2.6 (−21, 25, −16) | (5, 8, 9) |
| Orbital Gyrus | 11, 47 | 1.1/.4 | 3.2 (12, 31, −24) | 2.7 (−12, 31, −24) | (9, 10) |
| Rectal Gyrus | 11 | 1.6/1.7 | 3.1 (9, 28, −24) | 2.8 (−3, 22, −19) | (8–10) |
| Cuneus | 18 | 1.3/.0 | 3.1 (9, −78, 18) | ns | (8, 10) |
| Insula | 13 | 2.8/.8 | 3.1 (42, 1, 11) | 2.3 (−53, −37, 21) | (5, 8–10) |
| Lingual Gyrus | 17, 18 | .4.4/.3 | 3.0 (0, −91, −8) | 2.7 (0, −90, −3) | |
| Postcentral Gyrus | 1, 3, 43 | .8/.1 | 3.0 (48, −14, 15) | 2.2 (−53, −18, 51) | (5, 8–10) |
| Thalamus | 2.9/3.1 | 2.9 (9, −8, 9) | 3.0 (−9, −11, 14) | (5, 8–10) | |
| Medial Frontal Gyrus | 11, 25 | .3/.8 | 2.7 (3, 11, −16) | 2.9 (−3, 20, −16) | (8–10) |
| Superior Frontal Gyrus | 11 | .4/.1 | 2.8 (24, 37, −22) | 2.2 (−9, 54, −23) | (10) |
| Superior Parietal Lobule | 7 | .6/.0 | 2.6 (27, −68, 45) | NS | |
| Transverse Temporal Gyrus | 41, 42 | .4/.0 | 2.6 (48, −17, 12) | NS | (9, 10) |
| Inferior Parietal Lobule | 40 | .0/.3 | ns | 2.6 (−53, −40, 24) | (8–10) |
| Middle Temporal Gyrus | 20, 38 | .3/.1 | 2.4 (33, 1, −40) | 2.2 (−33, 7, −38) | (8–10) |
| Uncus | 28 | .3/.1 | 2.2 (24, 7, −28) | 2.1 (−21, 4, −30) | (9, 10) |
| Parahippocampal Gyrus | .0/.1 | ns | 2.1 (−12, −4, −17) | (5, 8, 10) | |
| Caudate | .1/.0 | 2.1 (9, −2, 14) | NS | (8–10) | |
| Superior Occipital Gyrus | .1/.0 | 2.0 (33, −77, 26) | NS | (10) | |
| Negative | |||||
| Cingulate Gyrus | 31, 32 | 1.2/.5 | 3.1 (3, −45, 41) | 2.6 (−3, −45, 41) | |
| Superior Frontal Gyrus | 6 | .8/.3 | 3.1 (15, −14, 64) | 2.6 (−15, −14, 64) | (22) |
| Precuneus | 7, 31 | 2.6/.4 | 2.9 (3, −47, 44) | 2.3 (−3, −42, 44) | (22) |
| Middle Temporal Gyrus | 39 | .0/.4 | ns | 2.9 (−56, −69, 28) | (22) |
| Angular Gyrus | 39 | .0/.3 | ns | 2.9 (−53, −68, 31) | (22) |
| Middle Frontal Gyrus | 6 | 1.4/.3 | 2.9 (21, −9, 58) | 2.6 (−42, 2, 39) | (22) |
| Medial Frontal Gyrus | 6 | .4/.1 | 2.9 (12, −11, 64) | 2.2 (−12, −11, 64) | (22) |
| Precentral Gyrus | 6 | .6/.3 | 2.8 (12, −17, 64) | 2.4 (−12, −17, 64) | (22) |
| Cuneus | 18 | .4/.3 | 2.5 (6, −99, 5) | 2.2 (−15, −92, 21) | (22) |
Prior Meta/Voxel-Based Morphometry Studies Showing Similar Regions of Gray Matter Differences Are Listed in the Last Column of the Table.
The sMRI component-B’s GM distribution and Talairach coordinates are detailed in Supplement 1 because subsequent analyses (described below) indicated that this component did not show diagnostic group differences that were of greatest interest in this study.
The genetic component with its contributing polymorphisms is listed in Table 3. The identified component consists of an association of 367 linearly weighted SNP genotypes, which together is assumed to influence an independent structural factor. The relative weightings for each SNP is calculated and ranked based on |z| > 2. The thresholding we use is invariant to the sign of the association. We discuss those SNPs that most heavily contribute to the identified SNP factor. The SNP component comprised 18 SNPs corresponding to the following 16 genes: AKT2, PIK3CA, PIK3CB, DRD2, CHRM2, ADORA2A, SLC6A4, GYS2, CYP1A2, NOS3, Sele, APOL5, APOB, OLR1, FASN, and GNB3.
Table 3.
The SNP Component Identified by Parallel Independent Component Analysis Linked with the Structural Components (A and B)
| SNP RS# | Z Score | Gene | Gene Name |
|---|---|---|---|
| rs762551 | −3.2336605 | CYP1A2 | Cytochrome P450, family 1, subfamily A, polypeptide 2 |
| rs5361 | 3.1389192 | Sele | Selectin E |
| rs2470890 | −2.8897406 | CYP1A2 | Cytochrome P450, family 1, subfamily A, polypeptide 2 |
| rs2228309 | −2.8814566 | FASN | Fatty-acid synthase |
| rs4802071 | 2.7635226 | AKT2 | v-akt murine thymoma viral oncogene homologue 2 |
| rs3761422 | −2.70295 | ADORA2A | Adenosine A2a receptor |
| rs2742115 | −2.4589744 | OLR1 | Oxidized low-density lipoprotein (lectin-like) receptor 1 |
| rs2471857 | −2.4375109 | DRD2 | Dopamine receptor D2 |
| rs2076672 | −2.4218938 | APOL5 | Apolipoprotein L, 5 |
| rs10513055 | −2.320138 | PIK3CB | Phosphoinositide-3-kinase, catalytic, beta polypeptide |
| rs870995 | −2.299162 | PIK3CA | Phosphoinositide-3-kinase, catalytic, alpha polypeptide |
| rs7641983 | −2.2806524 | PIK3CA | Phosphoinositide-3-kinase, catalytic, alpha polypeptide |
| rs1478290 | −2.2346463 | GYS2 | Glycogen synthase 2 |
| rs2020933 | −2.1976008 | SLC6A4 | Serotonin neurotransmitter transporter, solute carrier family 6, member 4 |
| rs1800783 | −2.1707895 | NOS3 | Nitric oxide synthase 3 (endothelial cell) |
| rs676210 | 2.1390609 | APOB | apolipoprotein B (including Ag(x) antigen) |
| rs324651 | −2.1153049 | CHRM2 | Cholinergic receptor, muscarinic 2 |
| rs6489738 | −2.0053828 | GNB3 | Guanine nucleotide binding protein (G protein), beta polypeptide 3 |
Group Differences
The loading parameters of the selected sMRI/SNP components were compared for the patient versus normal control groups. Two-sample t tests were computed on the loading parameters. For MRI, only the sMRI-A component showed a group difference (t = 2.53, p < .01). Some regions coded as negative in sMRI-A (Table 2) such that in these regions, GM values for the extracted component are larger in schizophrenia. The sMRI-B component did not show a group difference (t = 1.2, p = .2). The SNP component also differed between diagnostic groups (t = −2.6, p < .01).
Pathway Analysis
The neurologic significance and relationships among the genes in the SNP component was further analyzed using biological network–pathway analysis identified within Ingenuity, as described earlier. Those significant networks were associated with the categories “psychologic disorder,” “cell signaling,” and “neurologic disease.” All genes in the network are upregulated or highly expressed in the central nervous system. Figure 3 depicts the network containing the 14 key gene products. Pathway analysis indicated three signaling pathways based on their significance (p < .05) including Axonal Guidance, G-protein coupled receptor (GPCR), and PI3K/AKT signaling.
Discussion
This is the first study using parallel ICA to investigate genetic networks associated with structural brain volumes in schizophrenia and healthy control subjects, based on the overarching hypothesis that multiple genes promote relevant structural brain changes (42). We reasoned that a method comparing two modalities simultaneously might provide more insight into how multiple genes may influence structural GM patterns in control subjects and schizophrenia. Considering the limited sample and different data dimensionality, parallel ICA is robust in finding the connection strength between different modalities, thus avoiding overfitting and underfitting issues (43). We identified two structural components that were associated with a single genetic component in the entire sample.
The structural component (sMRI-A) with the stronger relationship to the identified genes included multiple frontal lobe cortices (e.g., anterior cingulate), temporal cortices (insula, middle, inferior and superior gyri), precuneus, thalamus, and striatum. Importantly, sMRI-A (and SNP) also showed significant group differences, such that patients showed GM deficits compared with control subjects. The sMRI-A component also identified more GM in particular brain regions of SZ patients consistent with some prior reports (22). The negative association was seen in regions including precuneus and frontal lobe cortices (middle, medial, and superior gyri). Although most studies report more GM in control subjects, a small number have reported more local GM in specific SZ brains regions. One possible explanation is that higher regional cortical GM in patients might result from antipsychotic treatment (44), altered pruning processes (45), or compensatory synaptic increases secondary to reduced inputs from other deficient brain areas (46). The significant negative correlation between GM regions and the SNP component indicated that genetic signaling differences were associated with less GM.
The genetic component comprised SNPs from 16 genes, several of which have been previously associated with schizophrenia risk or involved in normal central nervous system development. Here, we discuss both aspects of these gene roles for AKT, PI3K, SLC6A4, DRD2, CHRM2, and ADORA2A, all of which have been identified as schizophrenia or major mental illness susceptibility genes (47–51). In addition, several of them are involved in synaptic vesicle trafficking, synaptogenesis, or neurotransmitter release.
We wished to explore how genes identified in the SNP component were interrelated, using canonical pathway analysis (Figure 3) and to interpret how these relationships might be associated with the structural brain abnormalities identified in schizophrenia. Also, pathway analysis of genes illustrates how brain morphology and function may be influenced by processes including neuronal growth, migration, communication, and pruning. In this regard, axonal guidance signaling establishes normal connectivity between developing neurons and helps orient and promote axonal outgrowth (52,53). AKT2, PIK3CA, PIK3CB, and GNB3 were the relevant genes expressed in this signaling pathway. Nerve growth factor activates the AKT/PI3K pathway, which, among many functions, contributes to cell survival. To promote neuronal survival, neurotrophins require a functional AKT/PI3K pathway in cell body and distal axons (54) and to polarize axonal outgrowth (55). Guanine nucleotide protein (GNB3), another gene in the pathway, has essential roles in cell migration, proliferation, and differentiation. The G-protein beta subunit regulates axonal growth (56,57).
The next relevant gene pathway we identified was GPCR signaling. GPCRs bind and regulate most neurotransmitters; approximately 90% of them are located in the human brain (58). Our pathway analyses revealed DRD2, ADORA2A, CHRM2, AKT2, PIK3CA, and PIK3CB. DRD2 are widely expressed in postsynaptic dopaminergic neurons (59) and are relevant to working memory function (48). Neuronal outgrowth is induced in cultured cortical neurons by D2 receptor activation (60). The dephosphorylation/inactivation of AKT is associated with DRD2 expression. There is a well-established antagonist interaction between DRD2 and ADORA2A at the second messenger level, through their stimulating and inhibiting coupling to adenylyl cyclase activity (61). For CHRM2, pharmacologic studies indicate that this presynaptic receptor on cholinergic terminals plays a key role in regulating acetylcholine. In addition, CHRM2 participates in modulating neuronal excitability, synaptic plasticity, and feedback regulation of acetylcholine (ACh) release (62).
The PI3K/AKT pathway plays an important role in cell growth, apoptosis, inhibition, and glucose uptake. AKT2, PIK3CA, PIK3CB, NOS3, and GYS2 are represented in this pathway. AKT is highly expressed in brain and is important in adult nervous system plasticity (63). We identified AKT2 in our SNP component. Postmortem schizophrenia brains showed no differences in AKT2 levels compared with control subjects (64). Although AKT2, another AKT isoform, may play a secondary role in AKT signaling. Activation of AKT may begin with multiple events, including GPCRs, which activate PI3K through associated G proteins. PI3K promotes AKT phosphorylation and activation, a general mediator of cell survival (65). PI3K activity is required for nerve growth factor–induced suppression of distal axon growth (66). PI3K/AKT signaling phosphorylates endothelial nitric oxide synthase (eNOS/NOS3), which plays a major role in neural transmission through axons. NOS3 is widely expressed in brain endothelial cells, and its mRNA is also expressed in the basal ganglia (67). Excessive formation or inadequate degradation of nitric oxide may be an important factor in the etiology of several neurological disorders (68). AKT is also involved in glycogen synthesis by phosphorylating and inactivating GSK3 (glycogen synthase kinase 3), leading to the activation of glycogen synthase. GYS2 catalyzes glycogen, and its activity is regulated by a complex phosphorylation–dephosphorylation mechanism (69,70). Altered regulation of these genes may lead to disrupted AKT/PI3K signaling, which has been identified as a potentially altered pathway in schizophrenia (47,71). In summary, all these genes may play key roles in neurogenesis and neocortical signal functions.
We identified several other genes apart from the major networks/pathways just discussed. SLC6A4 encodes the serotonin transporter, which plays an important role in synaptic modulation of serotonin (5-HT) by controlling its uptake in presynaptic terminals (72). In addition, serotonin acts in neuronal division, differentiation, migration, synaptogenesis, and adult neurogenesis (73). The association of SLC6A4 with schizophrenia remains uncertain (72). CYP1A2 is regulated by various endogenous hormones and immune factors and is distributed in neurons and glial cells and at the blood–brain interface. Dopaminergic pathways may play a role in regulating CYP isoforms in schizophrenia (74). SEL E is mainly expressed in vascular endothelial cells, plays a role in immune response and showed no differences in unmedicated schizophrenia patients (75). In cultured glioma cells, FASN expression significantly increases with time for clozapine (76). APOL 5 plays a role in cholesterol transport and has no known association with schizophrenia. The other members of apolipoprotein family—APOL1, L2, and L4—showed significant upregulation in prefrontal cortex in schizophrenia (77). Serum levels of ApoB were higher in schizophrenia patients taking phenothiazine drugs for longer periods (78). GNB3 plays a role in integrating signals between receptor and effector proteins, acting as a cellular switch (79). No association is reported with schizophrenia, but there was a significant association of T allele in depression (80). OLR1 is regulated through the cyclic AMP pathway, and a mutation of this gene may increase risk for Alzheimer’s disease (81) but to date has no reported association with schizophrenia.
In conclusion, parallel ICA is a novel method for detecting clusters of genes related to a particular biological modality (in this case, sMRI measurements) that can detect associations in modest-sized samples (26,82). Our data not only suggest networks and pathways in which the identified genes work together but may help to clarify their function underlying aspects of normal brain structure and, by extension, their role in schizophrenia. In addition to rare copy number variant effects (83), schizophrenia is seen as a complex genetic disorder caused in part by concurrent inheritance of multiple common, ordinarily nonpathogenic SNP variants, that interact, perhaps epistatically, at molecular bottlenecks to increase disease risk (9,84). It is not possible to determine whether our SNP component represents one or more epistatic interactions. However, several of the genes identified, or their variants, have been implicated as putative schizophrenia risk genes, and a number act together in previously identified, well-characterized physiologic pathways. For example, ADORA2 interacts with DRD2 at a GCPR level and via PI3K affects AKT, one effect of which is on eNOS. This genomic pathway–guided approach can help clarify the pathology of a complex disease such as schizophrenia, and the gene component we identified may warrant investigation for further evidence of epistasis. Both the structural and genetic components seen in our results are schizophrenia-relevant.
A potential limitation of our study is the relatively small number of subjects, and thus this effort must be seen as preliminary. Another limitation is that we focused on the variation of genoytpes but not the SNPs themselves. Because we did not have sufficient number of subjects to recode the less frequent alleles, the results should be considered cautiously until replicated.
We note that parallel ICA has previously produced significant findings in similar-sized data sets, identifying for example genes for neurotransmitters known to be associated with the P300 event-related potential complex in healthy control subjects and the putative schizophrenia risk genes DISC1, BDNF, and DAT in functional MRI investigations in schizophrenia (19,25,82). Strengths of the current study are that we maximized uniformity with regard to data collection, population ethnicity, and image and genetic analysis. Although the sample size was small, data from the structural component agree substantially with those of previous schizophrenia studies, and several of the identified genes are previously identified with possible schizophrenia risk and/or are known to interact with each other in pathways concerned with brain development. However, these results should be considered cautiously until replication in a larger SNP set with more schizophrenia subjects including those of different ethnicity.
Acknowledgments
This research was supported by the National Institutes of Health under Grant Nos. R37 MH43775, R01 MH074797, and R01 MH077945 (to GP); R01 EB005846 and R01 EB006841 (to VDC); and R43 MH075481 (to GR). We thank the research staff at the Olin Neuropsychiatry Research Center who helped to collect the data.
Footnotes
All authors reported no biomedical financial interests or potential conflicts of interest.
Supplementary material cited in this article is available online.
References
- 1.Goldman AL, Pezawas L, Mattay VS, Fischl B, Verchinski BA, Zoltick B, et al. Heritability of brain morphology related to schizophrenia: a large-scale automated magnetic resonance imaging segmentation study. Biol Psychiatry. 2008;63:475–483. doi: 10.1016/j.biopsych.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Cicchetti D, Cannon TD. Neurodevelopmental processes in the ontogenesis and epigenesis of psychopathology. Dev Psychopathol. 1999;11:375–393. doi: 10.1017/s0954579499002114. [DOI] [PubMed] [Google Scholar]
- 3.DeLisi LE. The concept of progressive brain change in schizophrenia: Implications for understanding schizophrenia. Schizophr Bull. 2008;34:312–321. doi: 10.1093/schbul/sbm164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright IC, Sham P, Murray RM, Weinberger DR, Bullmore ET. Genetic contributions to regional variability in human brain structure: Methods and preliminary results. Neuroimage. 2002;17:256–271. doi: 10.1006/nimg.2002.1163. [DOI] [PubMed] [Google Scholar]
- 5.Glahn DC, Laird AR, Ellison-Wright I, Thelen SM, Robinson JL, Lancaster JL, et al. Meta-analysis of gray matter anomalies in schizophrenia: application of anatomic likelihood estimation and network analysis. Biol Psychiatry. 2008;64:774–781. doi: 10.1016/j.biopsych.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keshavan MS, Prasad KM, Pearlson G. Are brain structural abnormalities useful as endophenotypes in schizophrenia? Int Rev Psychiatry. 2007;19:397–406. doi: 10.1080/09540260701486233. [DOI] [PubMed] [Google Scholar]
- 7.Giuliani NR, Calhoun VD, Pearlson GD, Francis A, Buchanan RW. Voxel-based morphometry versus region of interest: A comparison of two methods for analyzing gray matter differences in schizophrenia. Schizophr Res. 2005;74:135–147. doi: 10.1016/j.schres.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Honea R, Crow TJ, Passingham D, Mackay CE. Regional deficits in brain volume in schizophrenia: A meta-analysis of voxel-based morphometry studies. Am J Psychiatry. 2005;162:2233–2245. doi: 10.1176/appi.ajp.162.12.2233. [DOI] [PubMed] [Google Scholar]
- 9.Meda SA, Giuliani NR, Calhoun VD, Jagannathan K, Schretlen DJ, Pulver A, et al. A large scale (N=400) investigation of gray matter differences in schizophrenia using optimized voxel-based morphometry. Schizophr Res. 2008;101:95–105. doi: 10.1016/j.schres.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segall JM, Turner JA, van Erp TG, White T, Bockholt HJ, Gollub RL, et al. Voxel-based morphometric multisite collaborative study on schizophrenia. Schizophr Bull. 2009;35:82–95. doi: 10.1093/schbul/sbn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prasad KM, Keshavan MS. Structural cerebral variations as useful endophenotypes in schizophrenia: Do they help construct “extended endophenotypes? Schizophr Bull. 2008;34:774–790. doi: 10.1093/schbul/sbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Giorgio A, Blasi G, Sambataro F, Rampino A, Papazacharias A, Gambi F, et al. Association of the SerCys DISC1 polymorphism with human hippocampal formation gray matter and function during memory encoding. Eur J Neurosci. 2008;28:2129–2136. doi: 10.1111/j.1460-9568.2008.06482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Honea R, Verchinski BA, Pezawas L, Kolachana BS, Callicott JH, Mattay VS, et al. Impact of interacting functional variants in COMT on regional gray matter volume in human brain. Neuroimage. 2009;45:44–51. doi: 10.1016/j.neuroimage.2008.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho BC, Milev P, O’Leary DS, Librant A, Andreasen NC, Wassink TH. Cognitive and magnetic resonance imaging brain morphometric correlates of brain-derived neurotrophic factor Val66Met gene polymorphism in patients with schizophrenia and healthy volunteers. Arch Gen Psychiatry. 2006;63:731–740. doi: 10.1001/archpsyc.63.7.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pezawas L, Verchinski BA, Mattay VS, Callicott JH, Kolachana BS, Straub RE, et al. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci. 2004;24:10099–10102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan YL, Zhou DF, Cao LY, Zou YZ, Wu GY, Zhang XY. Effect of the BDNF Val66Met genotype on episodic memory in schizophrenia. Schizophr Res. 2005;77:355–356. doi: 10.1016/j.schres.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 17.Pearlson G. Multisite collaborations and large databases in psychiatric neuroimaging: Advantages, problems, and challenges. Schizophr Bull. 2009;35:1–2. doi: 10.1093/schbul/sbn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sui J, Adali T, Pearlson G, Yang H, Sponheim SR, White T, Calhoun VD. 15) A CCA+ICA based model for multi-task brain imaging data fusion and its application to schizophrenia. Neuroimage. 2010;51:123–134. doi: 10.1016/j.neuroimage.2010.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Pearlson G, Windemuth A, Ruano G, Perrone-Bizzozero NI, Calhoun V. Combining fMRI and SNP data to investigate connections between brain function and genetics using parallel ICA. Hum Brain Mapp. 2009;30:241–255. doi: 10.1002/hbm.20508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bell AJ, Sejnowski TJ. An information-maximization approach to blind separation and blind deconvolution. Neural Comput. 1995;7:1129–1159. doi: 10.1162/neco.1995.7.6.1129. [DOI] [PubMed] [Google Scholar]
- 21.Lee SI, Batzoglou S. Application of independent component analysis to microarrays. Genome Biol. 2003;4:R76. doi: 10.1186/gb-2003-4-11-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calhoun VD, Adali T, Giuliani NR, Pekar JJ, Kiehl KA, Pearlson GD. Method for multimodal analysis of independent source differences in schizophrenia: Combining gray matter structural and auditory oddball functional data. Hum Brain Mapp. 2006;27:47–62. doi: 10.1002/hbm.20166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stevens MC, Skudlarski P, Pearlson GD, Calhoun VD. Age-related cognitive gains are mediated by the effects of white matter development on brain network integration. Neuroimage. 2009;48:738–746. doi: 10.1016/j.neuroimage.2009.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calhoun VD, Liu J, Adali T. A review of group ICA for fMRI data and ICA for joint inference of imaging, genetic, and ERP data. Neuroimage. 2009;45:S163–S172. doi: 10.1016/j.neuroimage.2008.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Kiehl KA, Pearlson G, Perrone-Bizzozero NI, Eichele T, Calhoun VD. Genetic determinants of target and novelty-related event-related potentials in the auditory oddball response. Neuroimage. 2009;46:809–816. doi: 10.1016/j.neuroimage.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Bixler JN, Calhoun VD. A multimodality ICA study—integrating genomic single nucleotide polymorphisms with functional neuroimaging data. Presented at the Bioinformatics and Biomedicine Workshops, 2008 BIBMW. 2008 IEEE International Conference; Philadelphia, PA. 2008. pp. 151–157. [Google Scholar]
- 27.First MB, Spitzer RL, Gibbon M, Williams JBW. Structured Clinical Interview for DSM-IV Axis I Disorders—Clinical Version (SCID-IV) Washington, DC: Psychiatric Publishing Group; 2002. [Google Scholar]
- 28.Blair JR, Spreen O. Predicting premorbid IQ: A revision of the National Adult Reading Test. Clin Neuropsychol. 1989;3:129–136. [Google Scholar]
- 29.Kay SR, Fiszbein A, Opler LA. The Positive and Negative Syndrome Scale (PANSS) for schizophrenia. Schizophr Bull. 1987;13:261–276. doi: 10.1093/schbul/13.2.261. [DOI] [PubMed] [Google Scholar]
- 30.SPM2. Statistical Parametric Mapping Software. London: Wellcome Department of Cognitive Neurology; 1994–2007. [Google Scholar]
- 31.Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage. 2001;14:21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- 32.Fan JB, Oliphant A, Shen R, Kermani BG, Garcia F, Gunderson KL, et al. Highly parallel SNP genotyping. Cold Spring Harb Symp Quant Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- 33.Oliphant A, Barker DL, Stuelpnagel JR, Chee MS. BeadArray technology: Enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques. 2002;56–58(suppl):60–61. [PubMed] [Google Scholar]
- 34.Ruano G. Physiogenomic method for predicting clinical outcomes of treatments in patients 2006 [Google Scholar]
- 35.Ruano G, Bernene J, Windemuth A, Bower B, Wencker D, Seip RL, et al. Physiogenomic comparison of edema and BMI in patients receiving rosiglitazone or pioglitazone. Clin Chim Acta. 2009;400:48–55. doi: 10.1016/j.cca.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 36.Shi MM, Myrand SP, Bleavins MR, de la Iglesia FA. High throughput genotyping for the detection of a single nucleotide polymorphism in NAD(P)H quinone oxidoreductase (DT diaphorase) using TaqMan probes. Mol Pathol. 1999;52:295–299. doi: 10.1136/mp.52.5.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calhoun VD, Adali T, Pearlson GD, Pekar JJ. Spatial and temporal independent component analysis of functional MRI data containing a pair of task-related waveforms. Hum Brain Mapp. 2001;13:43–53. doi: 10.1002/hbm.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calhoun VD, Adali T, McGinty VB, Pekar JJ, Watson TD, Pearlson GD. fMRI activation in a visual-perception task: Network of areas detected using the general linear model and independent components analysis. Neuroimage. 2001;14:1080–1088. doi: 10.1006/nimg.2001.0921. [DOI] [PubMed] [Google Scholar]
- 39.Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp. 2001;14:140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dawy ZS, Hagenauer M, Mueller JC. Fine-scale genetic mapping using independent component analysis. IEEE-ACM Trans Comput Biol Bioinform. 2008;5:448–460. doi: 10.1109/TCBB.2007.1072. [DOI] [PubMed] [Google Scholar]
- 41.Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 2002;15:870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- 42.Pearlson GD, Folley BS. Schizophrenia, psychiatric genetics, and Darwinian psychiatry: An evolutionary framework. Schizophr Bull. 2008;34:722–733. doi: 10.1093/schbul/sbm130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J, Demirci O, Calhoun VD. A parallel independent component analysis approach to investigate genomic influence on brain function. IEEE Signal Process Lett. 2008;15:413–416. doi: 10.1109/LSP.2008.922513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keshavan MS, Haas GL, Kahn CE, Aguilar E, Dick EL, Schooler NR, et al. Superior temporal gyrus and the course of early schizophrenia: Progressive, static, or reversible? J Psychiatr Res. 1998;32:161–167. doi: 10.1016/s0022-3956(97)00038-1. [DOI] [PubMed] [Google Scholar]
- 45.Feinberg I. Schizophrenia: Caused by a fault in programmed synaptic elimination during adolescence? J Psychiatr Res. 1982;17:319–334. doi: 10.1016/0022-3956(82)90038-3. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki M, Nohara S, Hagino H, Kurokawa K, Yotsutsuji T, Kawasaki Y, et al. Regional changes in brain gray and white matter in patients with schizophrenia demonstrated with voxel-based analysis of MRI. Schizophr Res. 2002;55:41–54. doi: 10.1016/s0920-9964(01)00224-9. [DOI] [PubMed] [Google Scholar]
- 47.Arguello PA, Gogos JA. A signaling pathway AKTing up in schizophrenia. J Clin Invest. 2008;118:2018–2021. doi: 10.1172/JCI35931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bertolino A, Fazio L, Caforio G, Blasi G, Rampino A, Romano R, et al. Functional variants of the dopamine receptor D2 gene modulate prefronto-striatal phenotypes in schizophrenia. Brain. 2009;132:417–425. doi: 10.1093/brain/awn248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Catapano LA, Manji HK. G protein-coupled receptors in major psychiatric disorders. Biochim Biophys Acta. 2007;1768:976–993. doi: 10.1016/j.bbamem.2006.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalkman HO. The role of the phosphatidylinositide 3-kinase-protein kinase B pathway in schizophrenia. Pharmacol Ther. 2006;110:117–134. doi: 10.1016/j.pharmthera.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 51.Saiz PA, Garcia-Portilla MP, Arango C, Morales B, Alvarez V, Coto E, et al. Association study of serotonin 2A receptor (5-HT2A) and serotonin transporter (5-HTT) gene polymorphisms with schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2007;31:741–745. doi: 10.1016/j.pnpbp.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 52.Kulkarni G, Li H, Wadsworth WG. CLEC-38, a transmembrane protein with C-type lectin-like domains, negatively regulates UNC-40-mediated axon outgrowth and promotes presynaptic development in Caenorhabditis elegans. J Neurosci. 2008;28:4541–4550. doi: 10.1523/JNEUROSCI.5542-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quinn CC, Wadsworth WG. Axon guidance: Asymmetric signaling orients polarized outgrowth. Trends Cell Biol. 2008;18:597–603. doi: 10.1016/j.tcb.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brunet A, Datta SR, Greenberg ME. Transcription-dependent and -independent control of neuronal survival by the PI3K-Akt signaling pathway. Curr Opin Neurobiol. 2001;11:297–305. doi: 10.1016/s0959-4388(00)00211-7. [DOI] [PubMed] [Google Scholar]
- 55.Weiner OD. Regulation of cell polarity during eukaryotic chemotaxis: The chemotactic compass. Curr Opin Cell Biol. 2002;14:196–202. doi: 10.1016/s0955-0674(02)00310-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Komatsuzaki K, Dalvin S, Kinane TB. Modulation of G(ialpha(2)) signaling by the axonal guidance molecule UNC5H2. Biochem Biophys Res Commun. 2002;297:898–905. doi: 10.1016/s0006-291x(02)02277-5. [DOI] [PubMed] [Google Scholar]
- 57.Park D, Park I, Lee D, Choi YB, Lee H, Yun Y. The adaptor protein lad associates with the G protein beta subunit and mediates chemokine-dependent T-cell migration. Blood. 2007;109:5122–5128. doi: 10.1182/blood-2005-10-061838. [DOI] [PubMed] [Google Scholar]
- 58.Vassilatis DK, Hohmann JG, Zeng H, Li F, Ranchalis JE, Mortrud MT, et al. The G protein–coupled receptor repertoires of human and mouse. Proc Natl Acad Sci U S A. 2003;100:4903–4908. doi: 10.1073/pnas.0230374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neville MJ, Johnstone EC, Walton RT. Identification and characterization of ANKK1: A novel kinase gene closely linked to DRD2 on chromosome band 11q23 and 1. Hum Mutat. 2004;23:540–545. doi: 10.1002/humu.20039. [DOI] [PubMed] [Google Scholar]
- 60.Reinoso BS, Undie AS, Levitt P. Dopamine receptors mediate differential morphological effects on cerebral cortical neurons in vitro. J Neurosci Res. 1996;43:439–453. doi: 10.1002/(SICI)1097-4547(19960215)43:4<439::AID-JNR5>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 61.Azdad K, Gall D, Woods AS, Ledent C, Ferre S, Schiffmann SN. Dopamine D2 and adenosine A2A receptors regulate NMDA-mediated excitation in accumbens neurons through A2A-D2 receptor heteromerization. Neuropsychopharmacology. 2009;34:972–986. doi: 10.1038/npp.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gosso FM, de Geus EJ, Polderman TJ, Boomsma DI, Posthuma D, Heutink P. Exploring the functional role of the CHRM2 gene in human cognition: Results from a dense genotyping and brain expression study. BMC Med Genet. 2007;8:66. doi: 10.1186/1471-2350-8-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arnold SE, Talbot K, Hahn CG. Neurodevelopment, neuroplasticity, and new genes for schizophrenia. Prog Brain Res. 2005;147:319–345. doi: 10.1016/S0079-6123(04)47023-X. [DOI] [PubMed] [Google Scholar]
- 64.Emamian ES, Hall D, Birnbaum MJ, Karayiorgou M, Gogos JA. Convergent evidence for impaired AKT1-GSK3beta signaling in schizophrenia. Nat Genet. 2004;36:131–137. doi: 10.1038/ng1296. [DOI] [PubMed] [Google Scholar]
- 65.Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 66.Kuruvilla R, Ye H, Ginty DD. Spatially and functionally distinct roles of the pi 3-K effector pathway during NGF signaling in sympathetic neurons. Neuron. 2000;27:499–512. doi: 10.1016/s0896-6273(00)00061-1. [DOI] [PubMed] [Google Scholar]
- 67.Liou YJ, Lai IC, Lin MW, Bai YM, Lin CC, Liao DL, et al. Haplotype analysis of endothelial nitric oxide synthase (NOS3) genetic variants and tardive dyskinesia in patients with schizophrenia. Pharmacogenet Genomics. 2006;16:151–157. doi: 10.1097/01.fpc.0000184958.05775.66. [DOI] [PubMed] [Google Scholar]
- 68.Heales SJ, Barker JE, Stewart VC, Brand MP, Hargreaves IP, Foppa P, et al. Nitric oxide, energy metabolism and neurological disease. Biochem Soc Trans. 1997;25:939–943. doi: 10.1042/bst0250939. [DOI] [PubMed] [Google Scholar]
- 69.Nuttall FQ, Gannon MC, Bai G, Lee EY. Primary structure of human liver glycogen synthase deduced by cDNA cloning. Arch Biochem Biophys. 1994;311:443–449. doi: 10.1006/abbi.1994.1260. [DOI] [PubMed] [Google Scholar]
- 70.Ruano G, Windemuth A, Kocherla M, Holford T, Fernandez ML, Forsythe CE, et al. Physiogenomic analysis of weight loss induced by dietary carbohydrate restriction. Nutr Metab. 2006;3:20. doi: 10.1186/1743-7075-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hallmayer J. Getting our AKT together in schizophrenia? Nat Genet. 2004;36:115–116. doi: 10.1038/ng0204-115. [DOI] [PubMed] [Google Scholar]
- 72.Bayle FJ, Leroy S, Gourion D, Millet B, Olie JP, Poirier MF, Krebs MO. 5HTTLPR polymorphism in schizophrenic patients: Further support for association with violent suicide attempts. Am J Med Genet B Neuropsychiatr Genet. 2003;119B:13–17. doi: 10.1002/ajmg.b.10037. [DOI] [PubMed] [Google Scholar]
- 73.Lesch KP, Zeng Y, Reif A, Gutknecht L. Anxiety-related traits in mice with modified genes of the serotonergic pathway. Eur J Pharmacol. 2003;480:185–204. doi: 10.1016/j.ejphar.2003.08.106. [DOI] [PubMed] [Google Scholar]
- 74.Wojcikowski J, Daniel WA. Identification of factors mediating the effect of the brain dopaminergic system on the expression of cyto-chrome P450 in the liver. Pharmacol Rep. 2008;60:966–971. [PubMed] [Google Scholar]
- 75.Graham KA, Cho H, Brownley KA, Harp JB. Early treatment-related changes in diabetes and cardiovascular disease risk markers in first episode psychosis subjects. Schizophr Res. 2008;101:287–294. doi: 10.1016/j.schres.2007.12.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ferno J, Raeder MB, Vik-Mo AO, Skrede S, Glambek M, Tronstad KJ, et al. Antipsychotic drugs activate SREBP-regulated expression of lipid biosynthetic genes in cultured human glioma cells: A novel mechanism of action? Pharmacogenomics J. 2005;5:298–304. doi: 10.1038/sj.tpj.6500323. [DOI] [PubMed] [Google Scholar]
- 77.Mimmack ML, Ryan M, Baba H, Navarro-Ruiz J, Iritani S, Faull RL, et al. Gene expression analysis in schizophrenia: Reproducible up-regulation of several members of the apolipoprotein L family located in a high-susceptibility locus for schizophrenia on chromosome. 22. Proc Natl Acad Sci U S A. 2002;99:4680–4685. doi: 10.1073/pnas.032069099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wen F, Tan J. Effects of phenothiazine drugs on serum levels of apolipoproteins and lipoproteins in schizophrenic subjects. Acta Pharmacol Sin. 2003;24:1001–1005. [PubMed] [Google Scholar]
- 79.McCudden CR, Hains MD, Kimple RJ, Siderovski DP, Willard FS. G-protein signaling: Back to the future. Cell Mol Life Sci. 2005;62:551–577. doi: 10.1007/s00018-004-4462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kunugi H, Kato T, Fukuda R, Tatsumi M, Sakai T, Nanko S. Association study of C825T polymorphism of the G-protein b3 subunit gene with schizophrenia and mood disorders. J Neural Transm. 2002;109:213–218. doi: 10.1007/s007020200018. [DOI] [PubMed] [Google Scholar]
- 81.Luedecking-Zimmer E, DeKosky ST, Chen Q, Barmada MM, Kamboh MI. Investigation of oxidized LDL-receptor 1 (OLR1) as the candidate gene for Alzheimer’s disease on chromosome 12. Hum Genet. 2002;111:443–451. doi: 10.1007/s00439-002-0802-7. [DOI] [PubMed] [Google Scholar]
- 82.Meda SA, Jagannathan K, Gelernter J, Calhoun VD, Liu J, Stevens MC, Pearlson GD. A pilot multivariate parallel ICA study to investigate differential linkage between neural networks and genetic profiles in schizophrenia [published online ahead of print November 26] Neuroimage. 2009 doi: 10.1016/j.neuroimage.2009.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.St Clair D. Copy number variation and schizophrenia. Schizophr Bull. 2009;35:9–12. doi: 10.1093/schbul/sbn147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Allen AJ, Griss ME, Folley BS, Hawkins KA, Pearlson GD. Endo-phenotypes in schizophrenia: A selective review. Schizophr Res. 2009;109:24–37. doi: 10.1016/j.schres.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]