Abstract
Cancer metastasis results from a multi-step cascading process that includes: 1) vascularization of the primary tumor; 2) detachment and invasion of cancer cells; 3) intravasation into lymphatic and blood vessels; 4) survival and arrest in the circulation; 5) extravasation into distant organs; and 6) colonization and growth of metastatic tumors. microRNAs (miRNAs) play critical roles in this multi-step process, both promoting and suppressing metastasis. This review updates the progress made in understanding the roles of miRNAs for invasion and metastasis during cancer progression. A specific miRNA signature of cancer metastasis is also reviewed.
Keywords: miRNAs, Cancer metastasis, Invasion, Cancer stem cells and angiogenesis
Introduction
For most solid malignancies, metastasis is the predominant cause of cancer death [1]. Elucidation of the molecular mechanisms that regulate the sequential steps of metastasis is critical for the reduction of mortality by cancer. Cancer metastasis is the process by which cancer cells spread from a primary tumor to other non-adjacent organs and tissues, forming viable secondary deposits of cancer. Cancer cells spread to other organs through a multi-step process that includes: 1) progressive vascularization and growth of the primary tumor; 2) detachment and invasion of cancer cells; 3) intravasation into lymphatic and blood vessels; 4) survival and arrest in the circulation; 5) extravasation into a new microenvironment; and 6) colonization and growth of metastatic tumors [1, 2].
Angiogenesis permits the initial growth of primary tumors and the survival and growth of metastatic tumor colonies [1, 2]. During detachment and invasion, cancer cells secrete matrix metalloproteinases (MMPs), which undergo an epithelial-mesenchymal transition (EMT) and acquire both motility and invasiveness [1, 3]. While EMT has been found to impact cancer cell intravasation, mesenchymal-epithelial transition (MET) may influence cancer cell extravasation. The properties of cancer stem cells (CSC) and organ-specific gene expression may dictate the successful rate of colonization of metastatic tumors.
The microRNAs (miRNAs) are small 19–25 nucleotides non-coding RNAs that can modulate gene expression by hybridizing to complementary target mRNAs, resulting in either translation inhibition or mRNA degradation [4–12]. Discovery of miRNAs has provided a novel mechanism for regulating human gene expression that impacts diverse biological and pathological processes, including development, cell proliferation, differentiation, apoptosis and tumorigenesis [6–11]. Recent data clearly demonstrates that miRNAs can function as both metastasic activators and suppressors by critically regulating various stages of migration and invasion [13, 14]. Several excellent reviews have covered this exciting area [15–18]. In the present review, recent progress made in understanding the roles of miRNAs in regulating the cancer invasion-metastasis cascade is highlighted. A specific miRNA signature of cancer metastasis is also reviewed.
miRNAs Modulate Cancer Angiogenesis
The formation of new blood vessels is essential for the initial growth of primary cancers that are more than 1–2 mm in diameter [1]. Once microcirculation is established, tumor-associated blood vessels provide an important route for cancer metastasis [1]. Recent studies suggest that miRNAs can regulate angiogenesis. Several miRNAs have been identified that exert proangiogenic or antiangiogenic effects and these are summarized in Table 1 and Fig. 1. miR-221 and miR-222 are highly expressed in endothelial cells [19]. Through directly regulating downstream targets, such as c-kit, p27Kip1, p57Kip2 and cyclin G1, miR-221 and miR-222 impact migration and proliferation of endothelial cells [20–22]. While c-kit has been well recognized for its importance in stem/progenitor cell proliferation, p27Kip1 has also shown to have oncogenic function to increase the numbers of stem/progenitor cells in the blood, retina, lung, and glial systems [23]. Thus, miR-221 and miR-222 may play direct roles in regulating the circulating endothelial progenitors. The miR-15a-16-1 cluster can promote apoptosis, as well as inhibit cell proliferation and VEGF expression by targeting Bcl-2, cyclin D1, wingless-type MMTV integration site family member 3A (WNT3A), AKT serine/threonine-protein-kinase (AKT3), ribosomal-protein-S6, MAP-kinases, and NF-kappaB activator MAP3KIP3 [24–27]. miR-122 targets a known promoter of metastasis, a disintegrin and metalloprotease 17 (ADAM17), and inhibits both tumor angiogenesis and cancer cell migration/invasion [28].
Table 1.
miRNAs involved in angiogenesis
| miRNAs | Targets | Molecular regulation | Deregulation in cancer | Refs |
|---|---|---|---|---|
| Anti-angiogenic miRNAs | ||||
| miR-221/222 | c-kit, cyclin G1, p27Kip1, p57Kip2 | Endothelial cell proliferation/migration | Decreased | [19–22] |
| miR-15a-16-1 | Bcl-2, CCND1, WNT3A, AKT3 S6, MAP3KIP3 | VEGF level, cell proliferation/survival | Decreased | [24–27] |
| miR-122 | ADAM17 | Unknown | Decreased | [28] |
| Pro-angiogenic miRNAs | ||||
| miR-17–92 cluster | TSP-1, CTGF, SIRT1, Rap-1, S1P1, MKK4, integrin | Endothelial cell proliferation/migration, proangiogenic molecules | Increased or Decreased | [39, 40] |
| miR-126 | SPRED1, PIK3R2 | VEGF level, endothelial cell proliferation | Decreased | [29, 30] |
| miR-296 | HGS | VEGFR2 and PDGFRβ | Increased | [34] |
| miR-378 | Sufu, Fus-1 | VEGF, Ang-1 and Ang-2 | Increased | [41] |
| miR-210 | Ephrin-A3 | Endothelial cell proliferation/migration | Increased | [45, 46] |
| miR-130a | HOXA5, GAX | Endothelial cells proliferation/migration, tube formation | Increased | [47] |
| miR-143-145 | ACE, Tpm4 | VSMCs, vessel wall | Increased | [48] |
ACE angiotensin-converting enzyme; ADAM17 a disintegrin and metalloprotease 17; AKT3 AKT serine/threonine-protein-kinase 3; Ang-1/2 angiopoietin-1/2; CCND1 cyclin D1; CTGF connective tissue growth factor; Fus-1 fusion 1 protein; GAX growth arrest homeobox; HGS the hepatocyte growth factor-regulated tyrosine kinase substrate; HOXA5 homeobox protein A5; MAP3KIP3 MAP-kinases, and NF-kappaB-activator; MKK4 the mitogen-activated kinase kinase 4; PDGFRβ platelet-derived growth factor (PDGF) receptor β; PIK3R2 phosphoinositol-3 kinase regulatory subunit 2; Rap-1 the small guanosine triphosphate-binding protein; S1P1 the sphingosine-1-phosphate receptor 1; S6 ribosomal-protein-S6; SIRT1 silent mating type information regulation 2 homolog 1; SPRED1 sprouty-related, EVH1 domain containing 1; Sufu suppressor of fused; Tpm4 tropomyosin 4; TSP1 thrombospondin-1; VEGF vascular endothelial growth factor; VEGFR2 VEGF receptor 2; VSMCs murine vascular smooth muscle cells; and WNT3A wingless-type MMTV integration site family member 3A
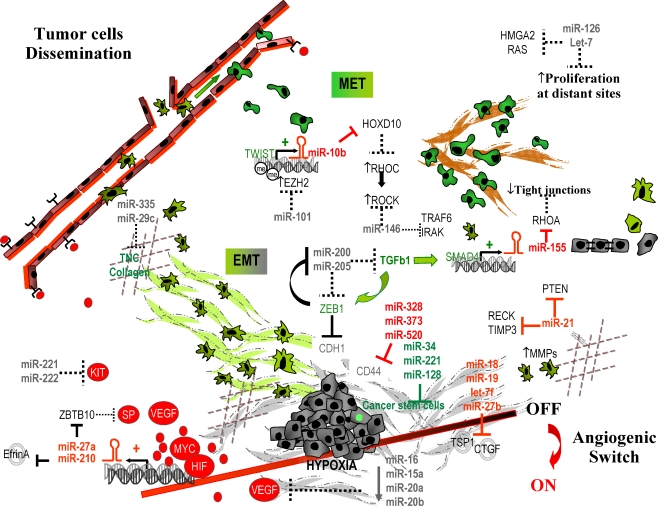
Fig. 1.
A network of microRNAs and protein-coding genes involved in the multiple steps of cancer metastasis. For details see text
While some miRNAs can inhibit angiogenesis, other miRNAs can stimulate new vessel formation. miR-126 is an endothelial-specific miRNA, which modulates vascular endothelial growth factor (VEGF) levels and endothelial cell proliferation by directly repressing the sprouty-related protein SPRED1 and phosphoinositol-3 kinase regulatory subunit 2 (PIK3R2) [29, 30]. Knockout of miR-126 leads to loss of vascular integrity and neoangiogenesis [29, 30]. Despite its positive role in angiogenesis, miR-126 appears to inhibit cancer cell growth, proliferation, adhesion and invasion, and is downregulated in colon, lung and breast cancers (Table 2 and Fig. 1) [31–33]. miR-296 can modulate the expression of VEGF receptor 2 and platelet-derived growth factor (PDGF) receptor β by directly targeting the hepatocyte growth factor-regulated tyrosine kinase substrate (HGS), which mediates degradation of the growth factor receptors [34]. The miR-17-92 cluster, which contains miR-17, miR-18, miR-19a, miR-19b-1, 20a and miR-92-1, is the first oncogenic miRNAs identified in human [35]. Although the major known function of the miR-17-92 cluster is related to transcriptional factors c-Myc, E2F and their autoregulatory loop [36–38], this cluster also enhances tumor angiogenesis by targeting thrombospondin-1 (TSP1), connective tissue growth factor (CTGF) and a number of proangiogenic targets including the histone deacetylase SIRT1, the small guanosine triphosphate-binding protein Rap-1, the sphingosine-1-phosphate receptor 1 (S1P1), the mitogen-activated kinase kinase 4 (MKK4) and integrin subunits α5 and αv [39, 40]. miR-378 can induce Sonic hedgehog (Shh) signaling, expression of VEGF and angiopoietin-1 (Ang-1) and -2 (Ang-2) and promote large-diameter vessel formation by directly targeting two tumor suppressors, suppressor of fused (Sufu) and Fusion 1 protein (Fus-1) [41, 42]. miR-210, a hypoxia-induced miRNA identified by three independent reports [24, 43, 44], seems to play a crucial role in regulating tube formation and survival of endothelial cells under hypoxic conditions by depressing Ephrin-A3 [45, 46]. Two anti-angiogenic homeobox proteins, HOXA5 and growth arrest homeobox (GAX) in vascular endothelial cells, are regulated by miR-130a [47]. The miR-143-145 cluster promotes the contractile phenotype of murine vascular smooth muscle cells (VSMCs) and improves vascular wall support/integrity by regulating angiotensin-converting enzyme (ACE) and tropomyosin 4 (Tpm4) [48].
Table 2.
miRNAs involved in detachment and invasion
| miRNAs | Targets | Molecular regulation | Deregulation in cancer | Refs |
|---|---|---|---|---|
| miR-29c | collagen, laminin γ1, TDG, FUSIP1, SPARC | ↓ cancer cell invasion | Decreased | [84] |
| miR-31 | frizzled3, MMP16, integrin a5, radixin, RhoA | ↓ cancer cell adhesion/ migration/invasion | Decreased | [85] |
| miR-122 | ADAM17 | ↓ cancer cell migration/ invasion | Decreased | [28] |
| miR-126 | Crk, VCAM-1, PIK3R2,IRS-1 | ↓ cancer cell proliferation/ adhesion/ migration/ invasion | Decreased | [31–33] |
| miR-128 | Reelin, DCX, E2F3a | ↓ cancer cell migration/ proliferation | Decreased | [86, 87] |
| miR-146a | ROCK1 | ↓ cancer cell migration/ invasion | Decreased | [62] |
| miR-146b | MMP16 | ↓MMP, cancer cell invasion | Decreased | [61] |
| miR-200 family | ZEB1, ZEB2 | ↑E-cadherin, ↓TGFβ, ↓EMT, cancer cell migration/invasion | Decreased | [74–79] |
| miR-205 | ZEB2, PKCε | ↑E-cadherin, ↓vimentin, ↓EMT, cancer cell migration/invasion | Decreased | [76, 80] |
| miR-335 | SOX4, TNC | ↓ cancer cell migration/ invasion | Decreased | [82] |
| miR-10b | HOXD10 | ↑Rho C, ↑cancer cell adhesion/migration/invasion | Increased | [13] |
| miR-21 | Cdc25A, BTG2, LRRFIP1, TPM1, maspin, SPRY2, TAp63, HNRPK, PDCD4, MARCKS, TIMP3, PTEN, RECK | ↑MMPs, ↑cancer cell migration/invasion | Increased | [49–56, 58] |
| miR-29a | TTP | ↑EMT, cancer cell migration/invasion | Increased | [81] |
| miR-155 | RhoA | ↑TGF-EMT, ↑cancer cell adhesion/migration/invasion | Increased | [47] |
| miR-328 | CD44 | ↑Cancer cell adhesion/ migration/invasion | Increased | [64] |
| miR-373 | LATS2, CD44 | Oncogene, ↑cancer cell adhesion/migration/invasion | Increased | [65, 66] |
| miR-520c | CD44 | ↑Cancer cell adhesion/ migration/invasion | Increased | [65, 66] |
ADAM17 a disintegrin and metalloprotease 17; BTG2 BTG family, member 2; Crk v-crk sarcoma virus CT10 oncogene homolog (avian); DCX doublecortin; FUSIP1 FUS-interacting protein; Fzd3, frizzled3; HNRNPK heterogeneous nuclear ribonucleoprotein K; HOXD10 homeobox D10 gene; IRS1 insulin receptor substrate 1; ITGA5 integrin a5; LATS2, LATS large tumor suppressor, homolog 2 (Drosophila); LRRFIP1 leucine rich repeat (in FLII) interacting protein 1; MARCKS myristoylated alanine-rich protein kinase c substrate; MMP16 matrix metallopeptidase 16 (membrane-inserted); PDCD4 programmed cell death 4; PIK3R2 phosphoinositol-3 kinase regulatory subunit 2; PKCε protein kinase C epsilon; PTEN phosphatase and tensin homolog deleted on chromosome 10; RDX radixin; RECK reversion-inducing-cysteine-rich protein with kazal motifs; RhoA ras homolog gene family, member A; RhoC ras homolog gene family, member C; ROCK1 Rho-associated, coiled-coil containing protein kinase 1; SPARC secreted protein, acidic, cysteine-rich; SPRY2 sprouty2; Tap63 tumor protein p63; TDG thymine-DNA glycosylase; TGFβ transforming growth factor β; TIMP3 TIMP metallopeptidase inhibitor 3; TNC tenascin C; TPM1 tropomyosin 1; TTP tristetraprolin; VCAM-1 vascular cell adhesion molecule 1; ZEB1/2 zinc-finger E-box binding homeobox ½
miRNAs Modulate Cancer Cell Detachment and Invasion
Cancer cell detachment, migration and invasion represent early steps in the metastatic cascade. Activation of MMPs, breakdown of extracellular matrix, EMT, an increase in cancer cell migration, and motility and invasiveness all occur during these early stages of tumor progression. Table 2 and Fig. 1 list the miRNAs that have been implicated in regulating these events. Several miRNAs have been found to promote cancer cell detachment and invasion. miR-21 is elevated in many tumors and promotes cancer cell proliferation, migration, detachment and invasion while suppressing apoptosis. Expression of miR-21 targets and decreases expression of multiple tumor suppressor genes including reversion-inducing-cysteine-rich protein with kazal motifs (RECK) [49], TIMP metallopeptidase inhibitor 3 (TIMP3) [49], phosphatase and tensin homolog deleted on chromosome 10 (PTEN) [50], tropomyosin 1 (TPM1) [51], programmed cell death 4 (PDCD4) [52, 53], heterogeneous nuclear ribonucleoprotein K (HNRPK) [54], tumor protein p63 (TAp63) [54], Sprouty2 (SPRY2) [55], maspin [56], myristoylated alanine-rich protein kinase C substrate (MARCKS) [57], leucine rich repeat (in FLII) interacting protein 1 (LRRFIP1) [58], BTG family, member 2 (BTG2) [59], and Cdc25A [60]. While miR-146b inhibits cancer cell migration and invasion by targeting MMP16 [61], miR-146a decreases cancer cell migration and invasion by targeting Rho-associated, coiled-coil containing protein kinase 1 (ROCK1) [62]. CD44 is a cell-cell and cell-extracellular matrix adhesion glycoprotein, which modulates adhesiveness, motility, matrix degradation, proliferation and cell survival [63]. Several miRNAs such as miR-328, miR-373 and miR-520c have been reported to regulate CD44 and promote cancer cell motility or invasion [64–66]. In addition, CD44 has recently been recognized as a cell surface marker of CSC and mesenchymal stem cells (MSC) [67–72]. Therefore, these CD44-regulating miRNAs may function in CSC and MSC. miR-373 has been shown to act as an oncogene to repress the tumor-suppressor LATS2 (large tumor suppressor, homolog 2, Drosophila) in human testicular germ cell tumors [73]. miR-10b, which can be induced by the pro-metastatic transcription factor Twist, can indirectly increase Ras homolog gene family, member C (Rho C), as well as motility and invasion by directly targeting homeobox D10 gene (HOXD10) [13].
Several miRNAs regulate EMT either negatively or positively (Table 2 and Fig. 1). Five members of the miR-200 family (miR-200a, miR-200b, miR-200c, miR-141 and miR-429) and miR-205 are able to increase E-cadherin expression, decrease vimentin expression, inhibit EMT, and prevent migration and invasion of cancer cells through post-transcriptional repression of zinc-finger E-box binding homeobox 1 (ZEB1) and ZEB2 (ZFHX1B) [74–78]. On the other hand, ZEB1 can directly suppress transcription of miR-200 family members and stabilize EMT and promote cancer cell invasion [79]. miR-205 has a function similar to members of the miR-200 family [76]. Additionally, miR-205 represses protein kinase C epsilon (PKCε) and prevent EMT [80]. miR-155 expression is increased in cancer cells and plays a positive role in transforming growth factor (TGF) β-induced EMT and cell migration and invasion by targeting RhoA [47]. miR-29a can promote EMT and cancer metastasis in cooperation with oncogenic Ras signaling by repressing the expression of tristetraprolin (TTP), a protein involved in the degradation of messenger RNAs with AU-rich 3′-untranslated regions [81].
A number of miRNAs negatively regulate the process of cancer cell detachment and invasion (Table 2). miR-126 appears to have multiple functions. As describe above, it supports neoangiogenesis. Yet, miR-126 is downregulated in cancers and inhibits cancer cell growth, adhesion, migration, and invasion through suppressing PIK3R2, insulin receptor substrate 1 (IRS-1), adhesion adaptor protein Crk, and vascular cell adhesion molecule 1 (VCAM-1) [31–33, 82, 83]. miR-335 can suppress cancer cell migration and invasion through targeting of the progenitor cell transcription factor SOX4 and extracellular matrix component tenascin C (TNC) [82]. miR-29c, which is inhibited in nasopharyngeal carcinomas, directly regulates a variety of genes that constitute extracellular matrix such as collagen 3A1, 4A1, 15A1, laminin γ1, thymine-DNA glycosylase (TDG), FUS-interacting protein (FUSIP1) and secreted protein, acidic, cysteine-rich (SPARC) [84]. miR-31 is able to repress a group of metastasis-promoting genes including frizzled3 (Fzd3), integrin a5 (ITGA5), MMP16, radixin (RDX), and RhoA [85]. miR-128 inhibits migration and proliferation of neuroblastoma or glioma cells by down-modulating the expression of Reelin, DCX [86] and E2F3a [87].
miRNAs Modulate Cancer Cell Intravasation
As a part of the process of metastasis, cancer cells intravasate by dissociating from other epithelial cancer cells, invading through the basement membrane and entering into blood vessels (not necessary for all types of tumors, e.g. lymphomas) [1, 3, 88]. The exact mechanisms that regulate this process, which requires collegenases [88], MMPs [89], urokinase plasminogen activator (uPA), uPA receptor (uPAR) [89] and EMT, are not clear. Therefore, some of the miRNAs that modulate detachment and invasion are also involved in regulating cancer cell intravasation. Oncogenic miR-21, for example, targets a number of tumor suppressor genes (Table 2) and is able to enhance cancer cell intravasation in addition to migration and cell proliferation [53].
Recently, miR-23b was reported to directly target uPA and c-Met and decrease migration and proliferation of human hepatocellular carcinoma (HCC) cells [90]. Raf kinase inhibitory protein (RKIP) as a metastasis suppressor inhibits breast tumor cell intravasation and bone metastasis by a signaling cascade that includes inhibition of MAP kinase (MAPK), decrease in transcription of LIN28 by c-Myc, expression of let-7, repression of high mobility group A2 (HMGA2, a chromatin remodeling protein), and activation of pro-invasive genes (Snail) [91]. It has been demonstrated that Tetraspanin CD151 (Tspan 24), an important regulator of laminin-binding integrins (alpha[6]beta[4], alpha[6]beta[1], and alpha[3]beta[1]), specifically regulates intravasation of the epidermoid carcinoma cells and fibrosarcoma cells in vivo without affecting cancer cell motility, proliferation and extravasation [92]. The Targetscan algorithm [93] has predicted that miR-124 and miR-506 regulate CD151, but this has yet to be confirmed. As describe above, miR-29c targets collagens [84] and miR-31 targets MMP16 [85]. It is likely that both miR-29c and miR-31 may also be involved in regulation of cancer cell intravasation.
miRNAs Modulate Cancer Cell Survival and Arrest in the Circulation
It is known that cancer cells can attach to capillaries in significantly greater numbers than normal fibroblasts [94]. Cells with high metastatic potential attach to the capillaries in greater numbers than cells with low metastatic potential [94]. The green fluorescent protein (GFP)-expressing cancer cells proliferate inside the blood vessels of the liver and the lung to express Ki-67 and MMP [95]. However, most of cancer cells that intravasate and enter into the circulation are still destroyed in the bloodstream by shear stress to the cells, as well as attack from the immune system [1–3]. It is possible that miRNAs that regulate cell proliferation and apoptosis [7] have some roles in this stage of cancer metastasis. miR-126 expression is often downregulated in cancers and is able to decrease leukocyte and possibly cancer cell adherence to endothelial cells by targeting VCAM-1 on endothelial cells [83]. Additionally, a number of miRNAs have been discovered to play critical roles in modulation of T and B lymphocytes activation, innate and adaptive immune responses [96, 97]. For example, miR-155 is required in conventional and regulatory T lymphocyte differentiation/activation, B lymphocyte development, macrophage response and toll-like receptors (TLR) response through targeting suppressor of cytokine signaling 1 (SOCS1), activation-induced cytidine deaminase (AID) and transcription factor c-Maf [96–98]. miR-223 is found to have crucial roles in regulating granulocyte proliferation and activation by regulating myeloid ELF-1-like factor (Mef2c) [99]. Cancer suppressing miR-146a can regulate TLR and cytokine signaling through a negative feedback regulation of TNF receptor-associated factor 6 (TRAF6) and IL-1 receptor-associated kinase 1 (IRAK1) genes [100]. miR-181, miR-17 ∼ 92 cluster and miR-150 are important regulators of the immune system and therefore could participate in cell survival and arrest in circulation [96, 97].
miRNAs Modulate Cancer Cell Extravasation
The ability of cancer cells to exit the capillaries and enter the parenchyma of organs is closely related to the ability of cancer cells to establish viable colonies in new sites as described in next section. Although a particular miRNA that specifically regulates cancer cell extravasation has not yet been identified, it is still believed that this step may also be regulated by miRNAs. Indeed, two miRNAs have been shown to be involved in this process. miR-31, which inhibits cancer cell detachment and invasion through repressing several metastasis-promoting genes (Fzd3, integrin a5, MMP16, radixin and RhoA) [85], can also impair extravasation of GFP-labeled breast cancer cells [85]. The precise mechanism by which miR-31 suppresses extravasation is not known, but may be associated with the repression of its targeted genes (Table 2). In addition to their effects in EMT, miR-200 family members are reported to promote a mesenchymal to epithelial cell transition by inhibiting ZEB2 expression, which enhances macroscopic metastases in mouse breast cancer cell lines [101]; however, the relationship between mesenchymal to epithelial cell transition and cancer cell extravasation is not clear yet.
miRNAs Modulate Metastatic Colonization
The last step of cancer metastasis is the establishment of macroscopic tumors at distant sites [1, 3]. According to the cancer “seed and soil” hypothesis [1], the properties of the “seed”, or cancer cells, and the “soil”, or cancer environment, determine the outcome of metastasis in a new organ. Consistent with this hypothesis, cancer stem cells could provide the “seed” [102]. Expression of metastasis progression genes at organ-specific locations could provide the “soil” [103]. Successful metastatic colonization may depend on 1) stem cell properties that ensure proliferation of cancer cells or dormancy in a new environment and/or 2) expression of site-specific genes that ensure cancer cells interact with and survive in a particular environment.
CSC, also called tumor-initiating cells represent a small subpopulation of cells identified in a variety of cancers that have the ability to self-renew and to develop into multi-lineage cells [104]. Dysregulation of stem cell self-renewal and differentiation is not only a likely requirement for the initiation of cancer, but also a likely requirement for cancer metastasis, especially in the stage of metastatic colonization [1, 3, 104]. Several signaling molecules and pathways have been shown to regulate the self-renewal, differentiation and survival of CSC, such as Notch, Hedgehog, Wnt/β-catenin, HMGA2, Bcl-2, Bmi-1, c-Myc and c-Met [102, 104, 105]. miRNA regulation is also essential in maintaining the stem cell population since depletion of Dicer-1 function results in early embryonic death and depletion of stem cells in mouse embryos [106]. Other evidence to support the roles of miRNAs in CSC is that a number of miRNAs have been found to critically regulate stem cell molecules and pathways as described above. For example, the miR-34 family (miR-34a, miR-34b and miR-34c) directly targets Notch, HMGA2, Bcl-2, CDK6, c-Met and c-Myc genes involved in the self-renewal and survival of cancer stem cells [66, 68, 107]. Expression of miR-34 family members inhibits cancer growth and metastasis [66, 68, 107]. Table 3 lists the important miRNAs that regulate CSC. Tumor suppressor let-7 can repress Ras and reduce self renewal of CSC [14, 108]. let-7 can also repress HMGA2 and enhance differentiation and transformation [14, 109]. The miR-15a-16-1 cluster can act as a negative regulator of cancer angiogenesis as described in the Angiogenesis section. Additionally, the miR-15a-16-1 cluster also suppresses the self-renewal and survival of cancer stem cells by targeting the oncogene Bcl-2 and WNT3A [24–27]. miR-199b-5p is downregulated in medulloblastoma and is regarded as tumor suppressor gene [110]. miR-199b-5p can reduce stem-cell-like (CD133+) subpopulation of medulloblastoma cells through its targeting of the transcription factor HES1 (hairy and enhancer of split 1) and the Notch pathway [110].
Table 3.
miRNAs involved in stem/progenitor cells phenotype
| miRNAs | Targets | Molecular regulation | Refs |
|---|---|---|---|
| Let-7 | Ras, HMGA2 | ↓CSC | [14, 108, 109] |
| miR-15a-16-1 | Bcl-2, WNT3A | ↓CSC | [24–27] |
| miR-34a/b/c | Notch, Bcl-2, HMGA2, CDK6, c-Met, c-Myc | ↓CSC | [66, 68, 107] |
| miR-128 | Bmi-1 | ↓CSC | [113] |
| miR-199b-5p | HES1/Notch | ↓CSC | [110] |
| miR-125b, miR-326, miR-324-5p | Smo Gli-1 Hedgehog | ↓CSC | [114] |
| miR-328 | CD44 | ↑CSC and MSC | [64, 67–72] |
| miR-373 | CD44 | ↑CSC and MSC | [65, 67–72] |
| miR-520c | CD44 | ↑CSC and MSC | [65, 67–72] |
Bmi1 BMI1 polycomb ring finger oncogene; CSC cancer stem cells; Gli1 GLI family zinc finger 1; HES1 hairy and enhancer of split 1, (Drosophila); HMGA2 high mobility group A2; MSC mesenchymal stem cells; SMO smoothened homolog (Drosophila); WNT3A wingless-type MMTV integration site family, member 3A
Bmi-1, a polycomb ring finger family member, is essential for self-renewal of CSC [111, 112]. Beside its roles in cell proliferation and migration as discussed above [86, 87], miR-128 can specifically block the self-renewal of glioma by targeting the Bmi-1 oncogene [113]. miR-125b and miR-326 are able to inhibit proliferation and to enhance differentiation of medulloblastoma cells through repressing Smoothened (Smo), an activator of the Hedgehog pathway [114]. miR-324-5p can also inhibit proliferation and enhance differentiation of medulloblastoma cells by repressing transcription factor Gli-1 (Gli family zinc finger 1) [114]. Conversely, a number of miRNAs can positively regulate CSC functions. The oncogenic miR-17–92 cluster is not only induced by transcriptional factors c-Myc, E2F [36–38], but also by Hedgehog signaling [115, 116], which suggests that this cluster may play a role in promoting self renewal of cancer stem cells. miR-328, miR-373, and miR-520c have been reported to regulate CSC surface marker CD44 and may promote CSC proliferation and motility [67–72].
Metastases from certain primary cancers display specific organ preference. For example, pancreatic and colorectal cancers often metastasize to the liver and lungs while prostate cancers frequently metastasize to bone [103]. Certain tumor types such as breast cancer remain dormant for long periods before developing metastasis, indicating that ‘speciation’ of cancer cells in microenvironments of a particular organ is needed [103]. This kind of organ-specific colonization of cancers may require expression of metastasis progression genes [103]. Indeed, pro-metastatic genes, such as cyclooxygenase 2 (COX2) and the epidermal growth factor receptor (EGFR) ligand HBEGF, are the mediators of cancer cell colonization in both brain and lung tissues [117]. The alpha2,6-sialyltransferase ST6GALNAC5 is specifically expressed in cancer cells that are able to pass through the blood-brain barrier [117]. miRNAs are well suited to play roles in expression of these organ-specific genes. Although no miRNA has been indicated in regulating organ-specific genes, it is likely that miRNAs’ roles in organ-specific cancer metastasis will be confirmed in the near future. Recently, miRNA profiling showed that downregulation of miR-30e-3p and miR-514 and upregulation of miRNAs, such as miR-199a*, miR-515-3p, miR-519d, miR-302c*, miR-517b, and miR-520f, miR-30a-5p, miR-518b, miR-523, miR-425-3p, and miR-519b, are important for metastatic colonization in melanoma cells [118].
miRNA Signature of Cancer Metastasis
The previously reviewed data clearly indicates that miRNAs critically regulate the process of cancer metastasis. Are there specific miRNA signatures for cancer metastasis? Based on available data, there indeed are a number of miRNAs strongly associated with cancer metastasis. In a prototypical mouse model of multistage tumorigenesis that involves the stepwise transformation of pancreatic β cells into pancreatic neuroendocrine carcinomas, down-regulation of the miR-200 family (miR-200a, miR-200b, miR-200c, miR-141 and miR-429) is found to be a metastasis-specific feature [119]. As described in a previous section, the miR-200 family negatively controls expression of ZEB1/2 (Table 2) and inhibits EMT [74–78]. As such, it makes sense that down-regulation of this family permits cancer cells to acquire aggressive EMT, invasion and metastasis. This finding is further supported by another report that illustrates a metastatic cancer miRNA signature in 43 metastatic lymph nodes of primary tumors (including colon, bladder, breast, and lung cancers) [120]. Down-regulation of miR-148a and miR-148b seems to be a common metastasis feature in HCC and pancreatic cancer [119, 121]. This observation is supported by the fact that miR-148 is specifically hypermethylated in metastatic cancers [107]. miR-148 is known to target DNMT3B and TGFB-induced factor homeobox 2 (TGIFB, a transcriptional repressor) [107, 122, 123]. The mechanism by which down-regulation of miR-148a and miR-148b promote cancer metastasis is not well known. Down-regulation of the miR-9 family (including miR-9-1, miR-9-2, and miR-9-3) also seems to be a common metastasis feature in a number of solid tumors [120, 121, 124]. This observation is also supported by the report that the miR-9 family is specifically hypermethylated in metastatic cancers [107]. Downregulation of the miR-9 family members is shown to activate NF-kappaB1, recruit DNA methyltransferase 1 and increase in DNA methylation [125–127]. Lastly, expression of the proangiogenic miR-210 (Table 1), a hypoxia-induced miRNA [24, 43, 44], has been shown to increase metastatic capability in both breast and pancreatic cancers [119, 128]. In addition, some miRNAs are reported to link to metastatic cancers at specific organs of origin [120]. For example, up-regulated miR-21 is reported to be involved in metastatic colon cancer; whereas, up-regulated miR-10b is reported to be involved in metastatic bladder cancer [120]. Nevertheless, further validation of these organ-specific miRNA signatures is needed. Taking together, three down-regulated and one up-regulated miRNAs have been confirmed to have a strong association with cancer metastasis (Table 4).
Table 4.
miRNA signature of cancer metastasis
| miRNAs | Molecular regulation | Deregulation in cancer | Refs |
|---|---|---|---|
| miR-9 family | ↓NF-kappaB1 ↓DNA methylation, | Down-regulated | [120, 121, 124] |
| miR-148a, miR-148b | ↓DNMT3B, ↓TGIFB | Down-regulated | [107, 119, 121] |
| miR-200 family | ↑E-cadherin, ↓EMT, ↓cancer cell migration/ invasion | Down-regulated | [119, 120] |
| miR-210 | hypoxia-induced, ↑angiogenesis | Up-regulated | [119, 128] |
DNMT3B DNA (cytosine-5-)-methyltransferase 3 beta; EMT epithelial-mesenchymal transition; TGIFB TGFB-induced factor homeobox 2
Future Perspectives
Each of the steps in cancer metastasis can be regulated by different miRNAs. Five key issues remain to be addressed in the future studies. The first issue is to validate which miRNAs are critically involved in cancer metastasis (Tables 1∼3 and Fig. 1). Identification of miRNA targets is just the beginning of this exploration. For example, more attention needs to be paid to exploring the molecular mechanisms by which miRNAs regulate their targets and associated signaling pathways. The second is to explore which miRNAs play important roles in the regulation of cancer stem cells. Cancer stem cells have been found to be critically involved in intravasation, extravasation and organ-specific metastasis. The third is to further identify the cancer type-specific miRNA signatures of metastasis. The forth is to translate laboratory observations regarding miRNA-regulated cancer metastasis into the development of markers for prognosis, as well as new approaches for treatments. And the last is to develop new techniques and technology for miRNA detection, whereby, it will be useful to develop a reliable in situ hybridization method for detecting miRNAs on formalin-fixed paraffin embedded tissues. In addition, the efficiency of in vivo delivery of miRNA remains to be improved, but the future looks promising.
Acknowledgements
The authors thank Dr. Milena Nicoloso for drawing the Fig. 1 and Sherri De Jesus for her technical editing expertise. George A. Calin, a Fellow, is supported by The University of Texas M. D. Anderson Research Trust, The University of Texas System Regents Research Scholar Program, and by the Ladjevardian Regents Research Scholar Fund. This study was supported in part by a grant from the Anne and Henry Zarrow Foundation and by NIH grant 1R01CA135444. The authors acknowledge that all related articles are not cited in this article due to the space limit.
Contributor Information
Xiao-Feng Le, Phone: +1-713-7454353, Email: xfle@mdanderson.org.
George A. Calin, Phone: +1-713-7925461, Email: gcalin@mdanderson.org
References
- 1.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 2.Bockhorn M, Jain RK, Munn LL. Active versus passive mechanisms in metastasis: do cancer cells crawl into vessels, or are they pushed? Lancet Oncol. 2007;8:444–448. doi: 10.1016/S1470-2045(07)70140-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinberg RA. The biology of cancer. 1. New York: Garland Science, Taylor & Francis Group; 2007. [Google Scholar]
- 4.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 5.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 8.Zorio E, Medina P, Rueda J, et al. Insights into the role of microRNAs in cardiac diseases: from biological signalling to therapeutic targets. Cardiovasc Hematol Agents Med Chem. 2009;7:82–90. doi: 10.2174/187152509787047676. [DOI] [PubMed] [Google Scholar]
- 9.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 10.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 11.Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. Blood. 2006;108:3646–3653. doi: 10.1182/blood-2006-01-030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spizzo R, Nicoloso MS, Croce CM, Calin GA. SnapShot: MicroRNAs in cancer. Cell. 2009;137:586–586. doi: 10.1016/j.cell.2009.04.040. [DOI] [PubMed] [Google Scholar]
- 13.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 14.Yu F, Yao H, Zhu P, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 15.Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009;69:7495–7498. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bracken CP, Gregory PA, Khew-Goodall Y, Goodall GJ. The role of microRNAs in metastasis and epithelial-mesenchymal transition. Cell Mol Life Sci. 2009;66:1682–1699. doi: 10.1007/s00018-009-8750-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma L, Weinberg RA. Micromanagers of malignancy: role of microRNAs in regulating metastasis. Trends Genet. 2008;24:448–456. doi: 10.1016/j.tig.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs-the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 19.Poliseno L, Tuccoli A, Mariani L, et al. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 20.Sage C, Nagel R, Egan DA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felli N, Fontana L, Pelosi E, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci USA. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gramantieri L, Fornari F, Callegari E, et al. MicroRNA involvement in hepatocellular carcinoma. J Cell Mol Med. 2008;12:2189–2204. doi: 10.1111/j.1582-4934.2008.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Besson A, Hwang HC, Cicero S, et al. Discovery of an oncogenic activity in p27Kip1 that causes stem cell expansion and a multiple tumor phenotype. Genes Dev. 2007;21:1731–1746. doi: 10.1101/gad.1556607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hua Z, Lv Q, Ye W, et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS ONE. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonci D, Coppola V, Musumeci M, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 26.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roccaro AM, Sacco A, Thompson B, et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood. 2009;113:6669–6680. doi: 10.1182/blood-2009-01-198408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai WC, Hsu PW, Lai TC, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 29.Fish JE, Santoro MM, Morton SU, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S, Aurora AB, Johnson BA, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crawford M, Brawner E, Batte K, et al. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem Biophys Res Commun. 2008;373:607–612. doi: 10.1016/j.bbrc.2008.06.090. [DOI] [PubMed] [Google Scholar]
- 32.Guo C, Sah JF, Beard L, Willson JK, Markowitz SD, Guda K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer. 2008;47:939–946. doi: 10.1002/gcc.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Du YY, Lin YF, et al. The cell growth suppressor, mir-126, targets IRS-1. Biochem Biophys Res Commun. 2008;377:136–140. doi: 10.1016/j.bbrc.2008.09.089. [DOI] [PubMed] [Google Scholar]
- 34.Wurdinger T, Tannous BA, Saydam O, et al. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell. 2008;14:382–393. doi: 10.1016/j.ccr.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 36.Rinaldi A, Poretti G, Kwee I, et al. Concomitant MYC and microRNA cluster miR-17-92 (C13orf25) amplification in human mantle cell lymphoma. Leuk Lymphoma. 2007;48:410–412. doi: 10.1080/10428190601059738. [DOI] [PubMed] [Google Scholar]
- 37.Sylvestre Y, Guire V, Querido E, et al. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 38.Aguda BD, Kim Y, Piper-Hunter MG, Friedman A, Marsh CB. MicroRNA regulation of a cancer network: consequences of the feedback loops involving miR-17-92, E2F, and Myc. Proc Natl Acad Sci USA. 2008;105:19678–19683. doi: 10.1073/pnas.0811166106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dews M, Homayouni A, Yu D, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonauer A, Carmona G, Iwasaki M, et al (2009) MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science 2009 324:1710–1713 [DOI] [PubMed]
- 41.Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA. 2007;104:20350–20355. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pola R, Ling LE, Silver M, et al. The morphogen Sonic hedgehog is an indirect angiogenic agent upregulating two families of angiogenic growth factors. Nat Med. 2001;7:706–711. doi: 10.1038/89083. [DOI] [PubMed] [Google Scholar]
- 43.Kulshreshtha R, Ferracin M, Wojcik SE, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hebert C, Norris K, Scheper MA, Nikitakis N, Sauk JJ. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol Cancer. 2007;6:5. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pulkkinen K, Malm T, Turunen M, Koistinaho J, Yla-Herttuala S. Hypoxia induces microRNA miR-210 in vitro and in vivo ephrin-A3 and neuronal pentraxin 1 are potentially regulated by miR-210. FEBS Lett. 2008;582:2397–2401. doi: 10.1016/j.febslet.2008.05.048. [DOI] [PubMed] [Google Scholar]
- 46.Fasanaro P, D’Alessandra Y, Stefano V, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong W, Yang H, He L, et al. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28:6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parmacek MS. MicroRNA-modulated targeting of vascular smooth muscle cells. J Clin Invest. 2009;119:2526–2528. doi: 10.1172/JCI40503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gabriely G, Wurdinger T, Kesari S, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 52.Lu Z, Liu M, Stribinskis V, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PubMed] [Google Scholar]
- 53.Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 54.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 55.Sayed D, Rane S, Lypowy J, et al. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol Biol Cell. 2008;19:3272–3282. doi: 10.1091/mbc.E08-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang D, Fan GC, Zhou X, et al. Over-expression of CXCR4 on mesenchymal stem cells augments myoangiogenesis in the infarcted myocardium. J Mol Cell Cardiol. 2008;44:281–292. doi: 10.1016/j.yjmcc.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li T, Li D, Sha J, Sun P, Huang Y. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem Biophys Res Commun. 2009;383:280–285. doi: 10.1016/j.bbrc.2009.03.077. [DOI] [PubMed] [Google Scholar]
- 58.Li Y, Li W, Yang Y, et al. MicroRNA-21 targets LRRFIP1 and contributes to VM-26 resistance in glioblastoma multiforme. Brain Res. 2009;1286:13–18. doi: 10.1016/j.brainres.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 59.Tang YF, Zhang Y, Li XY, Li C, Tian W, Liu L. Expression of miR-31, miR-125b-5p, and miR-326 in the adipogenic differentiation process of adipose-derived stem cells. OMICS. 2009;13:331–336. doi: 10.1089/omi.2009.0017. [DOI] [PubMed] [Google Scholar]
- 60.Wang P, Zou F, Zhang X, et al. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res. 2009;69:8157–8165. doi: 10.1158/0008-5472.CAN-09-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao B, Guo J, Miao Y, et al. Detection of miR-106a in gastric carcinoma and its clinical significance. Clin Chim Acta. 2009;400:97–102. doi: 10.1016/j.cca.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 62.Lin SL, Chiang A, Chang D, Ying SY. Loss of mir-146a function in hormone-refractory prostate cancer. RNA. 2008;14:417–424. doi: 10.1261/rna.874808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marhaba R, Zoller M. CD44 in cancer progression: adhesion, migration and growth regulation. J Mol Histol. 2004;35:211–231. doi: 10.1023/b:hijo.0000032354.94213.69. [DOI] [PubMed] [Google Scholar]
- 64.Wang CH, Lee DY, Deng Z, et al. MicroRNA miR-328 regulates zonation morphogenesis by targeting CD44 expression. PLoS ONE. 2008;3:e2420. doi: 10.1371/journal.pone.0002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang Q, Gumireddy K, Schrier M, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 66.Li Y, Guessous F, Zhang Y, et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69:7569–7576. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Horst D, Kriegl L, Engel J, Kirchner T, Jung A. Prognostic significance of the cancer stem cell markers CD133, CD44, and CD166 in colorectal cancer. Cancer Invest. 2009;27:844–850. doi: 10.1080/07357900902744502. [DOI] [PubMed] [Google Scholar]
- 68.Ji Q, Hao X, Zhang M, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS ONE. 2009;4:e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin EH, Jiang Y, Deng Y, Lapsiwala R, Lin T, Blau CA. Cancer stem cells, endothelial progenitors, and mesenchymal stem cells: “seed and soil” theory revisited. Gastrointest Cancer Res. 2008;2:169–174. [PMC free article] [PubMed] [Google Scholar]
- 70.Palapattu GS, Wu C, Silvers CR, et al. Selective expression of CD44, a putative prostate cancer stem cell marker, in neuroendocrine tumor cells of human prostate cancer. Prostate. 2009;69:787–798. doi: 10.1002/pros.20928. [DOI] [PubMed] [Google Scholar]
- 71.Takaishi S, Okumura T, Tu S, et al. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells. 2009;27:1006–1020. doi: 10.1002/stem.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu Z, Hao X, Yan M, et al (2009) Cancer stem/progenitor cells are highly enriched in CD133(+)CD44(+) population in hepatocellular carcinoma. Int J Cancer 2009. doi:10.1002/ijc.24868 [DOI] [PubMed]
- 73.Voorhoeve PM, Sage C, Schrier M, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 74.Hurteau GJ, Carlson JA, Spivack SD, Brock GJ. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007;67:7972–7976. doi: 10.1158/0008-5472.CAN-07-1058. [DOI] [PubMed] [Google Scholar]
- 75.Christoffersen NR, Silahtaroglu A, Orom UA, Kauppinen S, Lund AH. miR-200b mediates post-transcriptional repression of ZFHX1B. RNA. 2007;13:1172–1178. doi: 10.1261/rna.586807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 77.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burk U, Schubert J, Wellner U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gandellini P, Folini M, Longoni N, et al. miR-205 Exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cepsilon. Cancer Res. 2009;69:2287–2295. doi: 10.1158/0008-5472.CAN-08-2894. [DOI] [PubMed] [Google Scholar]
- 81.Gebeshuber CA, Zatloukal K, Martinez J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep. 2009;10:400–405. doi: 10.1038/embor.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tavazoie SF, Alarcon C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sengupta S, Boon JA, Chen IH, et al. MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc Natl Acad Sci USA. 2008;105:5874–5878. doi: 10.1073/pnas.0801130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Valastyan S, Reinhardt F, Benaich N, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 86.Evangelisti C, Florian MC, Massimi I, et al (2009) MiR-128 up-regulation inhibits Reelin and DCX expression and reduces neuroblastoma cell motility and invasiveness. FASEB J 2009 23:4276–4287 [DOI] [PubMed]
- 87.Huang ZM, Yang J, Shen XY, et al. MicroRNA expression profile in non-cancerous colonic tissue associated with lymph node metastasis of colon cancer. J Dig Dis. 2009;10:188–194. doi: 10.1111/j.1751-2980.2009.00384.x. [DOI] [PubMed] [Google Scholar]
- 88.Weiss L, Orr FW, Honn KV. Interactions of cancer cells with the microvasculature during metastasis. FASEB J. 1988;2:12–21. doi: 10.1096/fasebj.2.1.3275560. [DOI] [PubMed] [Google Scholar]
- 89.Kim J, Yu W, Kovalski K, Ossowski L. Requirement for specific proteases in cancer cell intravasation as revealed by a novel semiquantitative PCR-based assay. Cell. 1998;94:353–362. doi: 10.1016/s0092-8674(00)81478-6. [DOI] [PubMed] [Google Scholar]
- 90.Salvi A, Sabelli C, Moncini S, et al. MicroRNA-23b mediates urokinase and c-met downmodulation and a decreased migration of human hepatocellular carcinoma cells. FEBS J. 2009;276:2966–2982. doi: 10.1111/j.1742-4658.2009.07014.x. [DOI] [PubMed] [Google Scholar]
- 91.Dangi-Garimella S, Yun J, Eves EM, et al. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009;28:347–358. doi: 10.1038/emboj.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zijlstra A, Lewis J, Degryse B, Stuhlmann H, Quigley JP. The inhibition of tumor cell intravasation and subsequent metastasis via regulation of in vivo tumor cell motility by the tetraspanin CD151. Cancer Cell. 2008;13:221–234. doi: 10.1016/j.ccr.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 94.Repesh LA, Fitzgerald TJ. Interactions of tumor cells with intact capillaries: a model for intravasation. Clin Exp Metastasis. 1984;2:139–150. doi: 10.1007/BF00052414. [DOI] [PubMed] [Google Scholar]
- 95.Li C, Feng Y, Coukos G, Zhang L (2009) Therapeutic microRNA strategies in human cancer. AAPS J 2009 11:747–757 [DOI] [PMC free article] [PubMed]
- 96.Lu LF, Liston A. MicroRNA in the immune system, microRNA as an immune system. Immunology. 2009;127:291–298. doi: 10.1111/j.1365-2567.2009.03092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rhee KJ, Wu S, Wu X, et al. Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infect Immun. 2009;77:1708–1718. doi: 10.1128/IAI.00814-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tili E, Croce CM, Michaille JJ. miR-155: on the crosstalk between inflammation and cancer. Int Rev Immunol. 2009;28:264–284. doi: 10.1080/08830180903093796. [DOI] [PubMed] [Google Scholar]
- 99.Johnnidis JB, Harris MH, Wheeler RT, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 100.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dykxhoorn DM, Wu Y, Xie H, et al. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS ONE. 2009;4:e7181. doi: 10.1371/journal.pone.0007181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea—a paradigm shift. Cancer Res. 2006;66:1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. [DOI] [PubMed] [Google Scholar]
- 103.Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 104.Al-Hajj M, Clarke MF. Self-renewal and solid tumor stem cells. Oncogene. 2004;23:7274–7282. doi: 10.1038/sj.onc.1207947. [DOI] [PubMed] [Google Scholar]
- 105.Welm AL, Kim S, Welm BE, Bishop JM. MET and MYC cooperate in mammary tumorigenesis. Proc Natl Acad Sci USA. 2005;102:4324–4329. doi: 10.1073/pnas.0500470102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 107.Lujambio A, Calin GA, Villanueva A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 109.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Garzia L, Andolfo I, Cusanelli E, et al. MicroRNA-199b-5p impairs cancer stem cells through negative regulation of HES1 in medulloblastoma. PLoS ONE. 2009;4:e4998. doi: 10.1371/journal.pone.0004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Park IK, Qian D, Kiel M, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 112.Liu S, Dontu G, Mantle ID, et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66:6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Godlewski J, Nowicki MO, Bronisz A, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 114.Ferretti E, Smaele E, Miele E, et al. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 2008;27:2616–2627. doi: 10.1038/emboj.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Northcott PA, Fernandez LA, Hagan JP, et al. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009;69:3249–3255. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Uziel T, Karginov FV, Xie S, et al. The miR-17∼92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc Natl Acad Sci USA. 2009;106:2812–2817. doi: 10.1073/pnas.0809579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bos PD, Zhang XH, Nadal C, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mueller DW, Rehli M, Bosserhoff AK. miRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. J Invest Dermatol. 2009;129:1740–1751. doi: 10.1038/jid.2008.452. [DOI] [PubMed] [Google Scholar]
- 119.Olson P, Lu J, Zhang H, et al. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009;23:2152–2165. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Baffa R, Fassan M, Volinia S, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219:214–221. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 121.Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 122.Duursma AM, Kedde M, Schrier M, Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Melhuish TA, Gallo CM, Wotton D. TGIF2 interacts with histone deacetylase 1 and represses transcription. J Biol Chem. 2001;276:32109–32114. doi: 10.1074/jbc.M103377200. [DOI] [PubMed] [Google Scholar]
- 124.Laios A, O’Toole S, Flavin R, et al. Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol Cancer. 2008;7:35. doi: 10.1186/1476-4598-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Guo LM, Pu Y, Han Z, et al. MicroRNA-9 inhibits ovarian cancer cell growth through regulation of NF-kappaB1. FEBS J. 2009;276:5537–5546. doi: 10.1111/j.1742-4658.2009.07237.x. [DOI] [PubMed] [Google Scholar]
- 126.Hsu PY, Deatherage DE, Rodriguez BA, et al. Xenoestrogen-induced epigenetic repression of microRNA-9-3 in breast epithelial cells. Cancer Res. 2009;69:5936–5945. doi: 10.1158/0008-5472.CAN-08-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bazzoni F, Rossato M, Fabbri M, et al. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci USA. 2009;106:5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Foekens JA, Sieuwerts AM, Smid M, et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci USA. 2008;105:13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]