Summary
MicroRNAs (miRNAs) are endogenous ∼22-nucleotide RNAs that play important gene-regulatory roles by pairing to the mRNAs of protein-coding genes to direct their repression. Repression of these regulatory targets leads to decreased translational efficiency and/or decreased mRNA levels, but the relative contributions of these two outcomes have been largely unknown, particularly for endogenous targets expressed at low-to-moderate levels. Here, we use ribosome profiling to measure the overall effects on protein production and compare these to simultaneously measured effects on mRNA levels. For both ectopic and endogenous miRNA regulatory interactions, lowered mRNA levels account for most (≥84%) of the decreased protein production. These results show that changes in mRNA levels closely reflect the impact of miRNAs on gene expression and indicate that destabilization of target mRNAs is the predominant reason for reduced protein output.
Each highly conserved mammalian miRNA typically targets mRNAs of hundreds of distinct genes, such that as a class these small regulatory RNAs dampen the expression of most protein-coding genes to optimize their expression patterns1,2. When pairing to a target is extensive, a miRNA can direct destruction of the targeted mRNA through Argonaute-catalyzed mRNA cleavage3,4. This mode of repression dominates in plants5, but in animals all but a few targets lack the extensive pairing required for cleavage2.
The molecular consequences of the repression mode that dominates in animals are less clear. Initially miRNAs were thought to repress protein output with little or no influence on mRNA levels6,7. Then mRNA-array experiments revealed that miRNAs decrease the levels of many targeted mRNAs8-11. A revisit of the initially identified targets of Caenorhabditis elegans miRNAs showed that these transcripts also decrease in the presence of their cognate miRNAs12. The mRNA decreases are associated with poly(A)-tail shortening, leading to a model in which miRNAs cause mRNA deadenylation, which promotes de-capping and more rapid degradation through standard mRNA-turnover processes10,13-15. The magnitude of this destabilization, however, is usually quite modest, which has bolstered the lingering notion that with some exceptions (e.g., Drosophila miR-12 regulation of CG1001114) most repression occurs through translational repression, and that monitoring mRNA destabilization might miss many targets that are downregulated without detectable mRNA changes. Challenging this view are results of high-throughput analyses comparing protein and mRNA changes after introducing or deleting individual miRNAs16,17. An interpretation of these results is that the modest mRNA destabilization imparted by each miRNA:target interaction represents most of the miRNA-mediated repression16. We call this the “mRNA-destabilization” scenario and contrast it to the original “translational-repression” scenario, which posited decreased translation with relatively little mRNA change.
In the mRNA-destabilization scenario differences between protein and mRNA changes are mostly attributed to either measurement noise or complications arising from pre-steady-state comparisons of mRNA-array data, which measure differences at one moment in time, and proteomic data, which measure differences integrated over an extended period of protein synthesis. If either mRNA levels or miRNA activities change over the period of protein synthesis (or the period of metabolic labeling), correspondence between mRNA destabilization and protein decreases could become distorted. Another complication of proteomic datasets is that they preferentially examine more highly expressed proteins, whose repression might differ from more modestly expressed proteins. A recent study used mRNA arrays to monitor effects on both mRNA levels and mRNA ribosome density and occupancy, thereby providing a more sensitive analysis of changes in mRNA utilization and bypassing the need to compare protein and mRNA18. This array study supports the mRNA-destabilization scenario but examines the response to an ectopically introduced miRNA, leaving open the question of whether endogenous miRNA:target interactions might impart additional translational repression.
Ribosome profiling, a method that determines the positions of ribosomes on cellular mRNAs with subcodon resolution19, is based on deep sequencing of ribosome-protected mRNA fragments (RPFs) and thereby provides quantitative data on thousands of genes not detected by general proteomics methods. Moreover, ribosome profiling reports on the status of the cell at a particular time point, and thus generates results more directly comparable to mRNA-profiling results than does proteomics. We extended this method to human and mouse cells, thereby enabling a fresh look at the molecular consequences of miRNA repression.
Ribosome profiling in mammalian cells
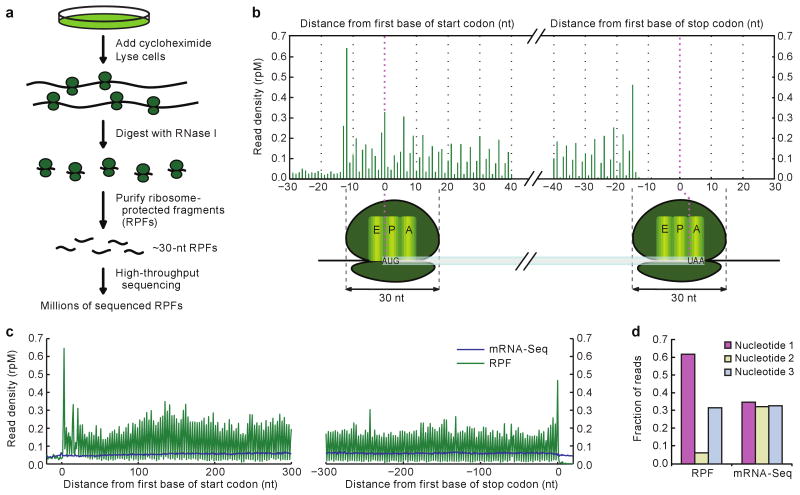
Ribosome profiling generates short sequence tags that each mark the mRNA coordinates of one bound ribosome19. The outline of our protocol for mammalian cells paralleled that used for yeast (Fig. 1a). Cells were treated with cycloheximide to arrest translating ribosomes. Extracts from these cells were then treated with RNase I to degrade regions of mRNAs not protected by ribosomes. The resulting 80S monosomes, many of which contained a ∼30-nucleotide RPF, were purified on sucrose gradients and then treated to release the RPFs, which were processed for Illumina high-throughput sequencing.
Figure 1. Ribosome profiling in human cells captured features of translation.
a, Schematic diagram of ribosome profiling. Sequencing reproducibility and evidence for mapping to the correct mRNA isoforms are illustrated (Supplementary Fig. 1a, b). b, RPF density near the ends of ORFs, combining data from all quantified genes. Plotted are RPF 5′ termini, as reads per million reads mapping to genes (rpM). Illustrated below the graph are the inferred ribosome positions corresponding to peak RPF densities, at which the start codon was in the P site (left) and the stop codon was in the A site (right). The offset between the 5′ terminus of an RPF and the first nucleotide in the human ribosome A site was typically 15 nucleotides (nt). c, Density of RPFs and mRNA-Seq tags near the ends of ORFs in HeLa cells. RPF density is plotted as in panel b, except positions are shifted +15 nucleotides to reflect the position of the first nucleotide in the ribosome A site. Composite data are shown for ≥600-nucleotide ORFs that passed our threshold for quantification (≥100 RPFs and ≥100 mRNA-Seq tags). d, Fraction of RPFs and mRNA-Seq tags mapping to each of the three codon nucleotides in panel c.
We started with HeLa cells, performing ribosome profiling on miRNA- and mock-transfected cells. In parallel, poly(A)-selected mRNA from each sample was randomly fragmented, and the resulting mRNA fragments were processed for sequencing (mRNA-Seq) using the same protocol as that used for the RPFs. Sequencing generated 11–18 million raw reads per sample, of which 4–8 million were used for subsequent analyses because they uniquely matched a database of annotated pre-mRNAs and mRNA splice junctions (Supplementary Table 1).
Combining RPFs from HeLa-expressed mRNAs into one composite mRNA showed that ribosome profiling captured fundamental features of translation (Fig. 1b, c and Supplementary Fig. 1c). Although a few RPFs mapped to annotated 5′UTRs, which suggested the presence of ribosomes at upstream open reading frames (ORFs)19, the vast majority mapped to annotated ORFs. RPF density was highest at the start and stop codons, reflecting known pauses at these positions20. mRNA-Seq tags, in contrast, mapped uniformly across the length of the mRNA, as expected for randomly fragmented mRNA.
The most striking feature in the composite-mRNA analysis was the 3-nucleotide periodicity of the RPFs. In sharp contrast to the 5′ termini of the mRNA-Seq tags, which mapped to all three codon nucleotides equally, the RPF 5′ termini mostly mapped to the first nucleotide of the codon (Fig. 1d). This pattern, analogous to that observed in yeast19, is attributable to the RPFs capturing the movement of ribosomes along mRNAs—three nucleotides at a time. The protocol applied to mouse neutrophils generated ∼30-nucleotide RPFs with the same pattern (Supplementary Fig. 1d, e). Thus, ribosome profiling mapped, at subcodon resolution, the positions of translating ribosomes in human and mouse cells.
Similar repression regardless of target expression level
General features of translation and translational efficiency in mammalian cells will be presented elsewhere. Here, we focus on miRNA-dependent changes in protein production. Our HeLa-cell experiments examined the impact of introducing miR-1 or miR-155, both of which are not normally expressed in HeLa cells, and our mouse-neutrophil experiments examined the impact of knocking out mir-223, which encodes a miRNA highly and preferentially expressed in neutrophils21. These cell types and miRNAs were chosen because proteomics experiments using either the SILAC (stable isotope labeling with amino acids in cell culture) or pSILAC (a pulsed-labeled version of SILAC) methods had already reported the impact of each of these miRNAs on the output of thousands of proteins16,17.
Pairing to the miRNA seed (nucleotides 2–7) is important for target recognition, and several types of seed-matched sites, ranging in length from 6 to 8 nucleotides, mediate repression2. Ribosome-profiling and mRNA-Seq results showed the expected correlation between site length and site efficacy2 (Supplementary Fig. 2). Because the response of mRNAs with single 6-nucleotide sites was marginal and observed only in the miR-1 experiment, subsequent analyses focused on mRNAs with at least one canonical 7–8-nucleotide site.
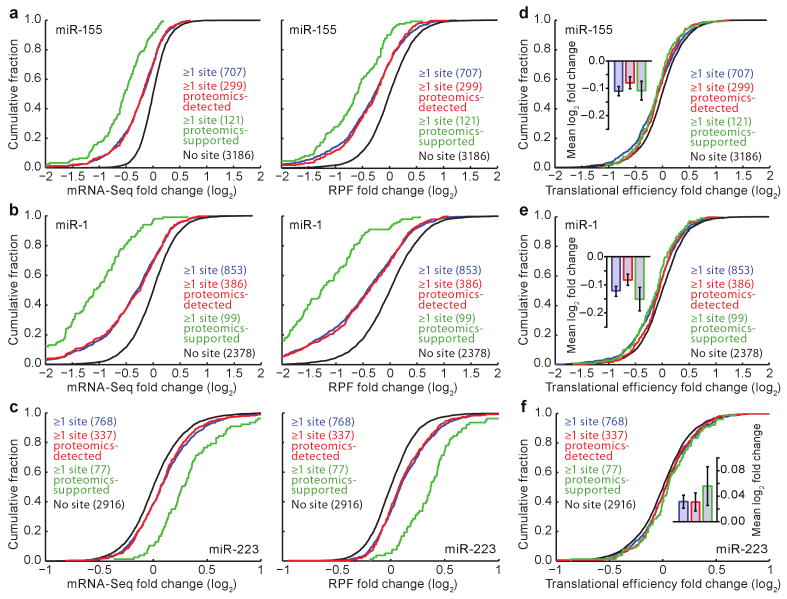
In the miR-155 experiment, mRNAs from 5103 distinct genes passed our read threshold for single-gene quantification (≥100 RPFs and ≥100 mRNA-Seq tags in the mock-transfection control). Genes with at least one 3′UTR site tended to be repressed following addition of miR-155, yielding fewer mRNA-Seq tags and fewer RPFs in the presence of the miRNA [Fig. 2a; P < 10−48 and 10−37, respectively, one-tailed Kolmogorov–Smirnov (K–S) test, comparing to genes with no site in the entire message]. Proteins from 2597 of the 5103 genes were quantified in the analogous pSILAC experiment17. The mRNA and RPF changes for the pSILAC-detected subset were no less pronounced than those of the larger set of analyzed genes (Fig. 2a; P = 0.70 and 0.62 for mRNA and RPF data, respectively, K–S test), which implied that the response of mRNAs of proteins detected by high-throughput quantitative proteomics accurately represented the response of all mRNAs. Analogous results were obtained in the miR-1 and miR-223 experiments (Fig. 2b, c; P < 10−10 for each comparison to genes with no site, and P > 0.56 for each comparison to the proteomics-detected subset). Furthermore, analyses of genes binned by expression level, which enabled inclusion of data from 11,000 distinct genes that ranged broadly in expression (more than 1000-fold difference between the first and last bins), confirmed that miRNAs do not repress their lowly expressed targets more potently than they do their more highly expressed targets (Supplementary Fig. 3).
Figure 2. MicroRNAs downregulated gene expression mostly through mRNA destabilization, with a small effect on translational efficiency.
a, Cumulative distributions of mRNA-Seq changes (left) and RPF changes (right) after introducing miR-155. Plotted are distributions for the genes with ≥1 miR-155 3′UTR site (blue), the subset of these genes detected in the pSILAC experiment (proteomics-detected, red), the subset of the proteomics-detected genes with proteins responding with log2-fold change ≤ −0.3 (proteomics-supported, green), and the control genes, which lacked miR-155 sites throughout their mRNAs (no site, black). The number of genes in each category is indicated in parentheses. b, Cumulative distributions of mRNA-Seq changes (left) and RPF changes (right) after introducing miR-1. Otherwise, as in panel a. c, Cumulative distributions of mRNA-Seq changes (left) and RPF changes (right) after deleting mir-223. Otherwise, as in panel a, with proteomics-supported genes referring to genes with proteins that responded with log2-fold change ≥ 0.3 in the SILAC experiment. d, Cumulative distributions of translational efficiency changes for the polyadenylated mRNA that remained after introducing miR-155. For each gene, the translational efficiency change was calculated by normalizing the RPF change by the mRNA-Seq change. For each distribution, the mean log2-fold change (± standard error) is shown (inset). e, Cumulative distributions of translational efficiency changes for the polyadenylated mRNA that remained after introducing miR-1. Otherwise, as in panel d. f, Cumulative distributions of translational efficiency changes for the polyadenylated mRNA that remained after deleting mir-223. Otherwise, as in panel d.
As these results indicated that restricting analyses to mRNAs with higher expression, by requiring either a minimal read count or a proteomics-detected protein, did not somehow distort the picture of miRNA targeting and repression, we focused on the mRNAs with at least one 3′UTR site and for which the proteomics detected a substantial change at the protein level. These sets of mRNAs were called “proteomics-supported targets” because they were expected to be highly enriched in direct targets of the miRNAs. Indeed, they responded more robustly to the introduction or ablation of cognate miRNAs (Fig. 2a–c; P < 10−5 for each comparison to proteomics-detected genes with sites). Because some 7–8-nucleotide seed-matched sites do not confer repression by the corresponding miRNA2,22, the proteomics-supported targets, which excluded most messages with nonfunctional sites, were the most informative for subsequent analyses.
Modest influence on translational efficiency
We next examined whether our results supported the translation-repression scenario, in which translation is repressed without a substantial mRNA decrease. In the characterized examples in which miRNAs direct translation inhibition, repression is reported to occur through either reduced translation initiation23-25 or increased ribosome drop-off26. Both of these mechanisms would lead to fewer ribosomes on target mRNAs and thus fewer RPFs from these mRNAs after accounting for changes in mRNA levels. To detect this effect, we accounted for changes in mRNA levels by incorporating the mRNA-Seq results. For example, for each quantified gene in the miR-155 experiment, we divided the change in RPFs by the change in mRNA-Seq tags (i.e., we subtracted the log2-fold changes). This calculation removed the component of the RPF change attributable to miRNA-dependent changes in poly(A) mRNA, leaving the residual change as the component attributable to a change in ribosome density, which we interpret as a change in “translational efficiency19”.
We observed a statistically significant decrease in translational efficiency for messages with miR-155 sites compared to those without, indicating that miRNA targeting leads to fewer ribosomes on target mRNAs that have not yet lost their poly(A)-tail and become destabilized (Fig. 2d, P = 0.003, K–S test). This decrease, however, was very modest. Even these proteomics-supported targets underwent only a 7% decrease in translational efficiency (−0.11 log2-fold change, Fig. 2d, inset), compared to a 33% decrease in polyadenylated mRNA (Fig. 2a). Analogous results were obtained for the miR-1 and miR-223 experiments (Fig. 2e, f; P = 0.001, P = 0.05, respectively). Thus, for both ectopic and endogenous regulatory interactions, only a small fraction of repression observed by ribosome profiling (11-16%) was attributable to reduced translational efficiency. At least 84% of the repression was attributable instead to decreased mRNA levels, a percentage somewhat greater than the ∼75% reported from array analyses of ectopic interactions18.
Analyses described thus far focused on messages with at least one 3′UTR site to the cognate miRNA, without considering whether or not the site was conserved in orthologous UTRs of other animals. When we focused on evolutionarily conserved sites1, the results were similar but noisier because the conserved sites, although more efficacious, were 3–13-fold less abundant (Supplementary Fig. 4). When changing the focus to messages with sites only in the ORFs, the results were also similar but again noisier because sites in the open reading frames are less efficacious16,17,22, which led to ∼70% fewer genes classified as proteomics-supported targets (Supplementary Fig. 5).
mRNA reduction consistently mirrored RPF reduction
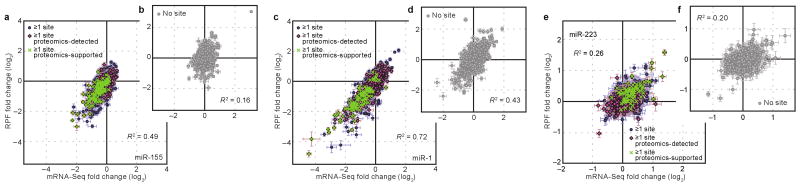
Analyses of fold-change distributions (Fig. 2) supported the mRNA-destabilization scenario for most targets, but still allowed for the possibility that the translational-repression scenario might apply to a small subset of targets. To search for evidence for a set of unusual targets undergoing translational repression without substantial mRNA destabilization, we compared the mRNA and ribosome-profiling changes for the 5103 quantifiable genes from the miR-155 experiment. Correlation between the two types of responses was strong for the messages with miR-155 sites, and particularly for those that were proteomics-supported targets (Fig. 3a, R2 = 0.49 and 0.63, respectively). A strong correlation was also observed for genes considered only after relaxing the expression cutoffs (Supplementary Fig. 6a). Any scatter that might have suggested that a few genes undergo translational repression without substantial mRNA destabilization strongly resembled the scatter observed in parallel analysis of genes without sites (Fig. 3b). The same was observed for the miR-1 experiment, but in this case the correlations were even stronger (R2 = 0.72 and 0.80, respectively), presumably because the increased response to the miRNA led to a correspondingly reduced contribution of experimental noise (Fig. 3c, d; Supplementary Fig. 6b). The same was also observed for the miR-223 experiment, with weaker correlations (R2 = 0.26 and 0.40, respectively) attributable to the reduced response to the miRNA and a correspondingly increased contribution of experimental noise (Fig. 3e, f). Supporting this interpretation, systematically increasing expression cutoffs, which retained data with progressively lower noise from stochastic counting fluctuations, progressively increased the correlation between RPF and mRNA-Seq changes (Supplementary Fig. 6c). We also examined messages with multiple sites to the cognate miRNA and found that they behaved no differently with regard to the relationship between mRNA-Seq and RPF changes (Supplementary Fig. 7). In summary, we found no evidence that countered the conclusion that miRNAs predominantly act to reduce mRNA levels of nearly all, if not all, targets.
Figure 3. Ribosome changes from miRNA targeting corresponded to mRNA changes.
a, Correspondence between ribosome (RPF) and mRNA (mRNA-Seq) changes after introducing miR-155, plotting data for the 707 quantified genes with at least one miR-155 3′UTR site (blue circles). Proteomics-detected targets and proteomics-supported targets are highlighted (pink diamonds and green crosses, respectively). Expected standard deviations (error bars) were calculated based on the number of reads obtained per gene and assuming random counting statistics. The R2 derived from Pearson's correlation of all data is indicated. b, Correspondence between ribosome and mRNA changes after introducing miR-155, plotting data for 707 genes randomly selected from the 3186 quantified genes lacking a miR-155 site anywhere in the mRNA. Otherwise, as in panel a. c and d, As in panels a and b, but plotting results for the miR-1 experiment. e and f, As in panels a and b, but plotting results for the miR-223 experiment.
Uniform changes along the length of the reading frame
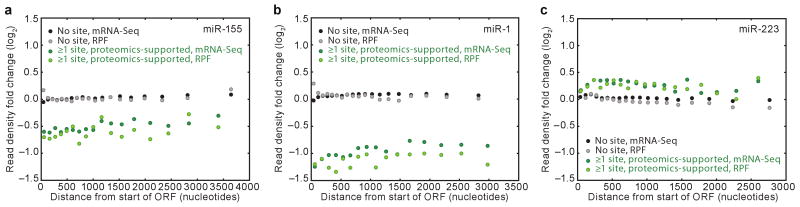
If miRNA targeting causes ribosomes to drop off the message after translating a substantial fraction of the ORF, then the RPF changes summed over the length of the ORF might underestimate the reduced production of full-length protein. Therefore, we re-examined the ribosome profiling data, which determines the location of ribosomes along the length of the mRNAs, thereby providing transcriptome-wide information that could detect ribosome drop-off. For highly expressed genes targeted in their 3′UTRs (e.g, TAGLN2 in the miR-1 experiment; Supplementary Fig. 8a), downregulation at the mRNA and ribosome levels was observed along the length of the ORF. In order to extend this analysis to genes with more moderate expression, we examined composite ORFs representing proteomics-supported targets and compared these to composite ORFs representing genes without sites. When miR-155 targets were compared to genes without sites, fewer mRNA-Seq tags were observed across the length of the composite ORF (Fig. 4a). RPFs tended to be further reduced (P = 0.007, one-tailed Mann–Whitney test), but without a systematic change in the magnitude of this additional reduction across the length of the ORF [P = 0.95, two-tailed Analysis of Covariance (ANCOVA) test]. Because ribosome drop-off would decrease the ribosome occupancy less at the beginning of the ORF than at the end, whereas inhibiting translation initiation would not, the observed uniform reduction supported mechanisms in which initiation was inhibited. Analogous results were observed in the miR-1 experiment (Fig. 4b; P = 0.002, for further reduction in RPFs; P = 0.85 for systematic change across the ORF). Evidence for drop-off was also not observed in the miR-223 experiment, although a change in translational efficiency was difficult to detect in this analysis, presumably because the miRNA-mediated changes were lower in magnitude (Fig. 4c). The same conclusions were drawn from analyses in which we first normalized for ORF length (Supplementary Fig. 9).
Figure 4. Ribosome and mRNA changes were uniform along the length of the ORFs.
a, Ribosome and mRNA changes along the length of ORFs after introducing miR-155. mRNA segments of quantified genes were binned based on their distance from the first nucleotide of the start codon, with the boundaries of the segments chosen such that each bin contained the same number of nucleotides (Supplementary Fig. 8b). Binning was done separately for mRNAs with no miR-155 site and proteomics-supported miR-155 targets. Fold changes in RPFs and mRNA-Seq tags mapping to each bin were then plotted with respect to the median distance of the central nucleotide of each segment from the first nucleotide of the start codon. Changes in RPFs and mRNA-Seq tags for mRNAs with no site (grey and black, respectively) and for proteomic-supported targets (light and dark green, respectively) are shown. Only bins with read contribution from ≥20 genes are shown (see Supplementary Fig. 8b). The ANCOVA test for systematic change across the ORF length was performed by first calculating the differences between RPF changes and mRNA-Seq changes for each group of genes, fitting lines through these changes in translational efficiency, then testing for a difference between the resulting slopes. b, As in panel a, but plotting results for the miR-1 experiment. c, As in panel a, but plotting results for the miR-223 experiment.
Implications for the mechanism of repression
For both ectopic and endogenous miRNA targeting interactions, the molecular consequences of miRNA regulation were most consistent with the mRNA-destabilization scenario. Although acquiring similar data on cell types beyond the two examined here will be important, we have no reason to doubt that our conclusion will apply broadly to the vast majority of miRNA targeting interactions. If indeed general, this conclusion will be welcome news to biologists wanting to measure the ultimate impact of miRNAs on their direct regulatory targets. Because the quantitative effects on translating ribosomes so closely mirrored the decreases in polyadenylated mRNA, the impact on protein production can be closely approximated using mRNA arrays or mRNA-Seq. Our results might also provide insight into the question of why some targets are more responsive to miRNAs than others; in the destabilization scenario, otherwise long-lived messages might undergo comparatively more destabilization than would constitutively short-lived ones.
Translation repression and mRNA destabilization are sometimes coupled27, which raises the possibility that the miRNA-mediated mRNA destabilization might be a consequence of translational repression. If so, a greater fraction of the repression might be attributable to decreased translational efficiency if the effects were analyzed sooner after introducing a miRNA. However, the fraction attributable to decreased translational efficiency remained small when repeating the analysis using samples from 12 hours (rather than 32 hours) after introducing miR-155 or miR-1 (Supplementary Fig. 10, Supplementary Table 2). Although results at earlier time points cannot rule out rapid destabilization as a consequence of translational repression, our results revealing such small decreases in translational efficiency for target mRNAs strongly imply that even if destabilization were secondary to translational repression, it would be this destabilization (i.e., the reduced availability of mRNA for subsequent rounds of translation) that would exert the greatest impact on protein production. Moreover, miRNA-mediated mRNA de-adenylation, which is the best-characterized mechanism of miRNA-mediated mRNA destabilization, can occur with or without translation of an ORF(refs 10,13,15,28), which suggests that the miRNA-mediated destabilization does not result from translational repression and indicates that translational repression could occur after the initial de-adenylation signal. Perhaps the miRNA-induced poly(A)-tail interactions that eventually trigger de-adenylation also cause the closed circular form of the mRNA to open up, thereby inhibiting translation initiation. This inhibition would occur before de-adenylation is complete, as polyadenylated mRNAs seem to be translationally repressed (Fig. 2d–f).
Another consideration is that, as done previously16-18, we equated mRNA destabilization to the loss of polyadenylated mRNA. Thus, transcripts that have lost their poly(A) tails might still be present but underrepresented in our mRNA-Seq of poly(A)-selected mRNA. In certain cell types, most notably oocytes, such transcripts can be stable and eventually tailed by a cytoplasmic polyadenylation complex to become translationally competent29. In the typical somatic cell, however, deadenylated transcripts are not translated and are instead rapidly decapped and/or degraded. Thus, our consideration of deadenylated transcripts as operational and functional equivalents of degraded transcripts seems appropriate. One possibility, though, is that mRNAs that were deadenylated while being translated will yield some RPFs from ribosomes that initiated when the poly(A) tails were intact but will not yield mRNA-Seq tags. However, a narrowing of the differences between changes in RPFs and mRNA-Seq tags through this process is expected to have been very small, since the vast majority of RPFs should derive from mRNAs with poly(A) tails.
A way that our results might still be reconciled with the translation-repression scenario would be if ribosome profiling missed the bulk of translation repression because translation was repressed without reducing the density of ribosomes on the targeted messages, i.e., if reduced initiation was coupled with correspondingly slower elongation. However, direct evidence for slower elongation has not been reported in any miRNA studies, and it seems unlikely that decreases in initiation and elongation rates would so frequently be so closely matched so as to yield such minor differences in apparent translational efficiency for so many messages. Moreover, translational repression without changes in ribosome density would cause the changes measured by proteomics to exceed those measured by ribosome profiling. The same would hold for cotranslational degradation of nascent polypeptides, another proposed mechanism for miRNA-mediated repression7,30. Arguing strongly against both of these possibilities, we found that changes measured by proteomics were not greater than those measured by ribosome profiling (Supplementary Fig. 11).
Although the changes we observed in translational efficiency were consistent with slightly reduced translation of the targeted messages, such changes could also occur without any miRNA-mediated translational repression. If some fraction of the polyadenylated mRNA was in a cellular compartment sequestered away from the compartment containing both miRNAs and ribosomes, then preferential destabilization of the mRNA in the miRNA/ribosome compartment would lead to an observed decrease in translational efficiency without a need to invoke translational repression. For example, to the extent that mature mRNAs awaiting transport to the cytoplasm reside in the nucleus where they presumably would not be subject to either miRNA-mediated destabilization or translation, the reduction of mRNA-Seq tags would not match the reduction of RPFs, and the more pronounced RPF reduction would indicate decreased ribosome density even in the absence of translational repression. Heterologous reporter mRNAs, some of which have lent support to the translational-repression scenario, might be particularly prone to nuclear accumulation. With this consideration in mind, the observed miRNA-dependent reductions in translational efficiency might be considered upper limits on the magnitude of translational repression.
Although we cannot determine the precise amount of miRNA-mediated translational repression, we can reliably say that the pervasive and dominant miRNA-mediated translational repression with persistence of repressed mRNAs, which had been widely anticipated, has not materialized. Instead, the outcome of regulation is predominantly mRNA destabilization, as first suggested by analyses of proteomic data16. We cannot rule out a few interactions for which there is substantial translational repression with little or no mRNA destabilization, but if these exist, they would be rare outliers. For such outliers, miRNAs might be working in concert with other mRNA-binding factors such that the action of the other factors depends on miRNA binding. Such outliers with readily detectable translational repression would be the most attractive subjects of mechanistic studies. The mechanism of translational repression might differ for different messages, depending on the identity of the cooperating factors, perhaps helping to explain the diversity of reported mechanisms by which miRNAs translationally repress their targets31. Understanding these potential elaborations of miRNA-mediated repression would be important, as is a more thorough mechanistic understanding of the predominant reason for reduced protein output, which is mRNA destabilization.
Methods Summary
HeLa cells were transfected with 100 nM miRNA duplex as described17 and harvested 12 and 32 h later. Haematopoietic progenitors were isolated from wild-type (WT) and mir-223 knockout (KO) male mice and cultured in media containing granulocyte colony-stimulating factor (G-CSF) and stem cell factor (SCF) as described16 for six days before harvesting. Just before harvesting, translation was arrested using cycloheximide for 8 min at 37 °C. Harvested cells were partitioned into two portions for ribosome profiling and mRNA profiling. Ribosome profiling was performed as outlined in Fig. 1a. For mRNA profiling, poly(A)+ mRNA was randomly fragmented by partial alkaline hydrolysis and size-selected RNA fragments were used to construct libraries for high-throughput sequencing. Illumina sequencing reads were mapped using the Bowtie short-read mapping program32. An iterative mapping strategy was adopted to obtain unique genome-matching and splice junction-spanning reads. A set of non-redundant transcripts served as our reference transcript database, which was used to map splice junction-spanning reads, quantify gene expression, and quantify RPF and mRNA-Seq changes.
Methods
Transfections and neutrophil culture
HeLa cells were transfected with 100 nM miRNA duplex as described17. At 12 and 32 h post-transfection, cycloheximide (100 ug/ml) was added to arrest translation, and after incubating 8 min at 37 °C, cells were harvested in ice-cold PBS supplemented with cycloheximide. For each transfection, cells from six 6-cm dishes were combined and then split into two portions for mRNA profiling and ribosome profiling. Haematopoietic progenitors were isolated from two 3-month-old WT male mice and two 3-month-old mir-223 KO male mice and cultured in IMDM media containing granulocyte colony-stimulating factor (G-CSF) and stem cell factor (SCF) as described16. On day 6, cycloheximide (100 ug/ml) was added to arrest translation. After incubating 8 min at 37 °C, cells were harvested in ice-cold PBS supplemented with cycloheximide and split into two portions for mRNA profiling and ribosome profiling.
Ribosome footprinting and RPF purification
Cells were pelleted and resuspended in ice-cold lysis buffer (10 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 100 mM KCl, 1% Triton X-100, 2 mM DTT, 100 ug/ml cycloheximide, 500 U/ml RNasin, 1 × complete protease inhibitor). The lysis mixture was homogenized six times with a 26-gauge needle at 4 °C and centrifuged at 5000 rpm for 8 min. The supernatant was snap-frozen for later use or processed immediately. RNase I (Ambion, final concentration, 0.5–1.0 U/ul) was added to the cell extract, and the reaction was incubated for 30 min on a shaker at room temperature (∼25 °C). Digested extracts were layered onto 11-ml 10–50% linear sucrose gradients that were prepared by horizontal diffusion and centrifuged in an SW-41Ti rotor at 36,000 rpm for 2 h. Gradients were fractionated by upward displacement with 60% sucrose on a gradient fractionator (Brandel). Monosome fractions were pooled, concentrated using Ultra-4 centrifugal filters with Ultracel-100 membranes (Amicon) by centrifuging at 1900 × g for 30 min at 4 °C. Ice-cold release buffer (20 mM HEPES-KOH, 100 mM KCl, 1 mM EDTA, 2 mM DTT, 20 U/ml SUPERase·In, Ambion) was then added to the retentate, and the mixture was incubated on ice for 10 min to release mRNA fragments from ribosomal subunits, after which the mixture was again centrifuged at 1900 × g for 15 min at 4 °C. The filtrate was then supplemented with SDS to 1% and treated with proteinase K (200 ug/ml) for 30 min at 42 °C. RNA was extracted with acid phenol:chloroform (pH 4.5, Ambion), ethanol-precipitated and resuspended in water. Pilot experiments, using nuclease-protection assays like those performed for yeast samples19, showed that the lengths of mammalian RPFs centered at ∼30 nucleotides. Therefore, RPFs were gel-purified on a denaturing 10% polyacrylamide-urea gel, excising the region corresponding to 27–33 nucleotides, with the intent of avoiding abundant ribosomal RNA degradation fragments that were 26 and 35 nucleotides in length.
mRNA fragmentation and microarrays
Total RNA was isolated using TRI Reagent (Ambion) and poly(A)+ mRNA was isolated using oligo(dT) DynaBeads (Invitrogen) according to manufacturers' instructions. Alkaline fragmentation buffer (2 mM EDTA, 10 mM Na2CO3, 90 mM NaHCO3, pH ≈ 9.3) was added to an equal volume of the purified mRNA and the reaction incubated for 20 min at 95 °C. Ice-cold stop solution (final 0.3 M NaOAc, pH 5.2, with GlycoBlue co-precipitant, Ambion) was then added, and RNA was ethanol precipitated. RNA fragments from ∼25–45 nucleotides were gel-purified on a denaturing 10% polyacrylamide-urea gel. Each sample of total RNA was also analyzed by microarray profiling, using the Affymetrix platform: Human Genome U133 Plus 2.0 Array, or Mouse Genome 430 2.0 Array.
Small-RNA library preparation
Libraries for Illumina sequencing were prepared as described33 but with the following modifications. Because RPFs and alkaline fragmentation products terminate with a 5′-hydroxyl and a 3′-phosphate, they were 3′-dephosphorylated with polynucleotide kinase (PNK, New England Biolabs) for 6 h at 37 °C in dephosphorylation buffer (100 mM MES-NaOH, pH 5.5, 10 mM MgCl2, 10 mM β-mercaptoethanol, 300 mM NaCl, 0.5 U/ul enzyme) and desalted (Microspin G-25 column, Amersham) before ligation to the 3′ adaptor. Gel-purified 3′-ligation products were then 5′-phosphorylated with PNK, according to manufacturer's instructions, before the 5′-ligation step. Despite steps taken to minimize ribosomal RNA contamination, our ribosome-profiling libraries were initially contaminated by high levels of rRNA (ranging from 60–93%). To enrich for RPFs, DNA from each library was amplified for an additional six cycles and then gel-purified on a 90% formamide, 8% acrylamide denaturing gel. With this additional step, ribosomal RNA contamination was reduced to 40–54%.
Sequence analyses
Illumina sequencing reads were mapped to the reference genome (hg18 for human, mm9 for mouse) with the Bowtie short-read mapping program32 using the first 25 nucleotides as the ‘seed’ region. Reads with multiple equivalent hits to the genome were discarded, as were reads that mapped to ribosomal RNA and other annotated noncoding RNAs. To allow for a miscalled residue within the seed region, reads that had failed to map when allowed no seed mismatches were fed into Bowtie again, this time allowing for one seed mismatch. To capture reads uniquely spanning splice junctions, reads that failed to map to the genome were mapped to a set of reference transcripts, using the same two-stage iterative mapping and again discarding those with multiple equivalent hits. These uniquely transcript-matching reads were combined with the genome-matching reads for subsequent analyses. To compile the set of reference transcripts we started from only curated coding transcripts (entries with NM accession numbers) in the RefSeq database (refFlat files, generated on August 9, 2009, were downloaded from the UCSC Genome Browser, http://genome.ucsc.edu). Of these, transcripts with incomplete coding sequences or those that could be potential substrates of nonsense-mediated decay were filtered out. If a gene had multiple isoforms remaining after this filtering, the longest isoform was picked to represent it. This non-redundant set of mRNAs from unique genes then served as our reference transcript database. Reads of ambiguous origin, such as a read that could derive from either of two different overlapping genes, were discarded. Of the remaining reads, those that could be unambiguously assigned to an exon or intron from a gene represented in our reference transcript database were attributed to that gene. The reference transcript databases for both human and mouse will be available for anonymous download at http://web.wi.mit.edu/bartel/pub/publication.html.
Quantification of gene expression
A modified version of reads per kilobase exon model per million mapped reads (rpkM) was used to quantify gene expression. The original rpkM, developed for RNA-Seq34, was calculated as such: R = (109·C/N·L), where C is the number of mapped reads in a gene's exons, N is the total number of reads mapped (library size), and L is the length of the sum of the exons in nucleotides. To prevent ribosomal RNA contamination in the RPF libraries from skewing our measurements of gene expression, the library size was taken to be the total number of reads mapping to all the exons and introns of our reference transcript database (N′). Because we were interested in comparing mRNA-level and translation-level expression, the length of the open reading frame was taken to be the feature length of each gene (L′) and we only included reads mapping to coding exons (C′) in our quantification. Hence, rpkM in this study refers to R′ = (109·C′/N′·L′). Fold changes were calculated by dividing the normalized gene expression value in the experimental condition by the same measure in the control condition. For the cumulative-distribution plots, the median of the distribution of genes without seed matches (No site) was subtracted from all the fold changes (including those from messages with sites). This normalization caused our reported fold-change distributions of the genes without sites to center on zero. Thresholds for gene quantification, when applied, were applied to the mock transfection data set or the mir-223 KO data set.
Supplementary Material
Acknowledgments
We thank F. Camargo, C. Jan, J. Kim and C. Petersen for advice and discussions, R. Green and O. Rissland for comments on the manuscript, and the Whitehead Institute's Genome Technology Core for sequencing and microarray profiling. This work was supported by grants from the NIH (D.P.B. and J.S.W.). H.G. was supported by the Agency for Science, Technology and Research, Singapore. N.T.I. was supported by a Ruth L. Kirschstein National Research Service Award (GM080853). D.P.B and J.S.W. are investigators of the Howard Hughes Medical Institute.
Footnotes
Author Contributions H.G. performed the experiments and analyzed the data, with input from the other authors. H.G., J.S.W., and D.P.B. contributed to the design of the study, and all authors contributed to preparation of the manuscript.
Author Information Small-RNA sequencing data and array data were deposited in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under accession number GSE22004.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 5.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 6.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 7.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 8.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 9.Krutzfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 10.Giraldez AJ, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 11.Rehwinkel J, et al. Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster. Mol Cell Biol. 2006;26:2965–2975. doi: 10.1128/MCB.26.8.2965-2975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bagga S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 13.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Behm-Ansmant I, et al. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eulalio A, et al. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 18.Hendrickson DG, et al. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sachs MS, et al. Toeprint analysis of the positioning of translation apparatus components at initiation and termination codons of fungal mRNAs. Methods. 2002;26:105–114. doi: 10.1016/S1046-2023(02)00013-0. [DOI] [PubMed] [Google Scholar]
- 21.Johnnidis JB, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 22.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pillai RS, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 24.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci U S A. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chendrimada TP, et al. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 26.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 27.Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 28.Eulalio A, et al. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat Rev Mol Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- 30.Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- 31.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of posttranscriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 32.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grimson A, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.