Abstract
A promising therapeutic approach for Duchenne muscular dystrophy (DMD) is exon skipping using antisense oligonucleotides (AOs). In-frame deletions of the hinge 3 region of the dystrophin protein, which is encoded by exons 50 and 51, are predicted to cause a variety of phenotypes. Here, we performed functional analyses of muscle in the exon 52–deleted mdx (mdx52) mouse, to predict the function of in-frame dystrophin following exon 51-skipping, which leads to a protein lacking most of hinge 3. A series of AOs based on phosphorodiamidate morpholino oligomers was screened by intramuscular injection into mdx52 mice. The highest splicing efficiency was generated by a two-oligonucleotide cocktail targeting both the 5′ and 3′ splice sites of exon 51. After a dose-escalation study, we systemically delivered this cocktail into mdx52 mice seven times at weekly intervals. This induced 20–30% of wild-type (WT) dystrophin expression levels in all muscles, and was accompanied by amelioration of the dystrophic pathology and improvement of skeletal muscle function. Because the structure of the restored in-frame dystrophin resembles human dystrophin following exon 51-skipping, our results are encouraging for the ongoing clinical trials for DMD. Moreover, the therapeutic dose required can provide a suggestion of the theoretical equivalent dose for humans.
Introduction
Duchenne muscular dystrophy (DMD) is a severe muscle disorder characterized by mutations in the DMD gene that mainly disrupt the reading frame leading to the absence of functional protein.1 A related allelic disorder, Becker muscular dystrophy (BMD), which shows a much milder phenotype, typically results from shortened but in-frame transcripts of the DMD gene that allow expression of limited amounts of an internally truncated but partially functional protein.2 Antisense oligonucleotide (AO)-mediated exon-skipping therapy for DMD, which is a splice modification of out-of-frame dystrophin transcripts, has been demonstrated to exclude specific exons, thereby correcting the translational reading frame, resulting in the production of “Becker-like,” shortened but partially functional protein.3,4,5,6,7 As a result of exon skipping, DMD could be converted to the milder BMD.4
The principle underlying exon-skipping therapy for DMD has been demonstrated in cultured mouse or human cells in vitro.7,8,9,10,11,12,13 In addition, in vivo studies in murine or canine animal models have provided preclinical evidence for the therapeutic potential of AO-mediated exon-skipping strategies for DMD.11,14,15,16,17,18,19 However, the number of patients who have the same mutation as the mice or dogs is estimated to be quite low.20,21 On the other hand, a hot spot for deletion mutations between exons 45 and 55 accounts for >60% of DMD patients with deletion mutations.20,22 In particular, exon skipping that targets exon 51 is theoretically applicable to the highest percentage (13%) of DMD patients with an out-of-frame deletion mutation.20,23,24,25 Recently, efficient in-frame dystrophin expression following an exon 51-skipping approach has been successfully demonstrated in human subjects using local intramuscular AO injection.23,24
The functionality of the dystrophin protein produced by exon 51-skipping has been inferred by the identification of patients harboring the corresponding in-frame deletions (e.g., in BMD patients).6,25 In-frame deletions near hinge 3 (refs. 12,26,27), which is encoded by exons 50 and 51, are predicted to lead to BMD; however, the severity of this disease can vary considerably.25,28,29,30,31,32 Consequently, it is desirable to use an animal model to investigate the molecular functionality of in-frame dystrophin lacking hinge 3 following exon 51-skipping. In the exon 52–deficient mdx mouse (mdx52), exon 52 of the Dmd gene has been deleted by gene-targeting, resulting in the production of a premature termination codon in exon 53 (refs. 7,33). This mouse lacks dystrophin and shows dystrophic features as well as muscle hypertrophy.33 It would be meaningful in predicting whether exon 51-skipping led to an accumulation of BMD-like dystrophin that was able to correct the dystrophic histology and muscle function in the mdx52 mouse.11,23,27,34
Here, we showed the efficient restoration of dystrophin function: the reading frame was restored by exon 51-skipping, and we observed considerable amelioration of the skeletal muscle pathology and function in mdx52 mice. We describe the first successful effort at systemic rescue of in-frame dystrophin lacking hinge 3 and recovery of muscle function by AO-mediated exon 51-skipping in a mouse model.
Results
Two AOs targeting the 5′ and 3′ splice sites achieved efficient exon 51-skipping in mdx52 mice
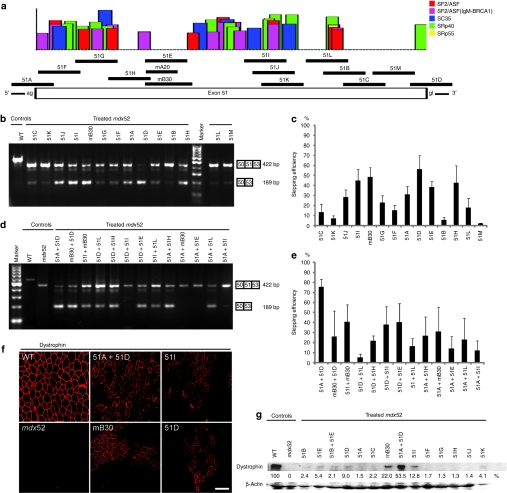
Skipping exon 51 of the murine Dmd gene in the mdx52 mouse corrects the open-reading frame, resulting in the production of truncated dystrophin that lacks two-thirds of the hinge 3 region and resembles human dystrophin following exon 51-skipping (Figure 1a).11,27,34 We first identified effective AO sequences by intramuscular injection into the tibialis anterior (TA) muscles of mdx52 mice.14 To optimize the screening dose in the TA muscle, Murine B30 (mB30) AO was injected into 8-week-old mdx52 mice at doses of 1–10 µg.15 The mB30 AO designed to skip murine exon 51 was based on human B30 (ref. 11), which targets human exon 51 (Figure 2a). Mice were euthanized 2 weeks after the injection; the TA muscles were isolated and analyzed by reverse transcription (RT)-PCR and immunohistochemistry. Using RT-PCR with primers flanking exons 50 and 53, the cDNA band equivalent to the mRNA missing exons 51 and 52 was detected. We found that mB30 restored dystrophin expression in a highly dose-dependent manner (Supplementary Figure S1). Then, we designed 13 AO sequences targeting either exonic sequences or exon/intron junctions of murine dystrophin exon 51 (refs. 9,11,35,36). The sequences and compositions of these AOs are described in Table 1 and Figure 2a. We then directly injected one or two of the 14 AOs into the TA muscle of mdx52 mice. Two weeks after the injection, we analyzed RNA fractions by RT-PCR and cryosections by immunohistochemistry and western blotting. Among the AOs examined, 51D, mB30, and 51I were shown by RT-PCR to be capable of inducing exon 51-skipping at a level approaching 50% (Figure 2b,c). 51D and 51I were designed to target the 5′ splice site and an exonic site of exon 51, respectively. It has been reported that a combination of two AOs directed at appropriate motifs in target exons induces more efficient exon skipping than that induced by a single injection.34,37 We therefore injected combinations of two AOs into the TA muscles of mdx52 mice, and found that a combination of two AOs, 51A plus 51D, showed ~75% skipping efficiency, the highest among the combinations that we examined by RT-PCR (Figure 2d,e). 51A is targeted to the 3′ splice site of exon 51. We then showed by immunohistochemistry that the combination of 51A plus 51D rendered 50–70% of the fibers dystrophin-positive in cross-sections (Figure 2f), and produced ~50% dystrophin expression on western blots compared with the normal control (Figure 2g). Taking these results together, we concluded that the co-injection of two AOs, 51A plus 51D, was the optimal combination to skip exon 51 of the murine Dmd gene.
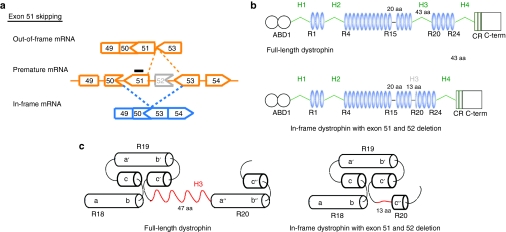
Figure 1.
Strategy for exon 51-skipping in mdx52 mouse. (a) Exon 51-skipping by appropriate phosphorodiamidate morpholino oligomers, indicated by a black line, can restore the reading frame of dystrophin in the mdx52 mouse, which lacks exon 52 in the mRNA of the murine Dmd gene, leading to out-of-frame products. (b) The molecular structure of in-frame dystrophin lacking hinge 3 induced by exon 51-skipping is shown below the full-length dystrophin. The protein contains the actin-binding domain 1 (ABD1) at the N-terminus, the central rod domain containing 24 spectrin-like repeats (R1–24), four hinge domains (H1–4), a 20-amino acid insertion between spectrin-like repeats 15 and 16 (segment 5), the cysteine-rich domain (CR), and the C-terminal domain (C-term). The hinge 3 is encoded by exons 50 and 51; therefore, most of this region is lost after exon 51-skipping in the mdx52 mouse. (c) Predicted nested repeat model with one long helix, one short helix, and overlap between the “a” helix of the following repeat with the “b” and “c” helices of the preceding repeat, forming the triple helix. The predicted structure of full-length dystrophin (upper) and in-frame dystrophin with exon 51 and 52 deletion (lower).
Figure 2.
Local intramuscular injection into mdx52 mice. The restoration of dystrophin in the tibialis anterior (TA) muscles was examined 2 weeks after the injection of 10 µg of one or a combination of two antisense oligonucleotides (AOs). (a) Fourteen different AOs designed to skip exon 51 of the murine Dmd gene. Each AO targets either an exonic splicing enhancer (ESE) or the 5′ or 3′ splice site, indicated by black lines. The certainties of ESE sites according to ESEfinder 3.0 are indicated by colored boxes. Candidates for splicing enhancer-binding proteins are shown (red, SF2/ASF; purple, SF2/ASF (IgM-BRCA1); blue, SC35; green, SRp40; yellow, SRp55). A murine B30 AO (mB30) corresponding to human B30 was designed.13 (b) Effectiveness of the 14 different AOs for exon 51-skipping detected by RT-PCR. Representative data are shown. Skipped products (50–53) are compared with unskipped products (50–51–53). WT, wild-type mouse. (c) Quantitative analysis by RT-PCR of exon 51-skipping by 14 different AOs. The percentages of in-frame transcripts in each lane of b are shown. The data (n = 3) are presented as mean ± SEM. (d) Effectiveness of 13 different combinations of two AOs targeting exon 51 of the murine Dmd gene. Representative data are shown. Skipped products (50–53) are compared with unskipped products (50–51–53). mdx52, untreated mdx52 mouse. (e) Quantitative analysis of exon 51-skipping by 13 different combinations of two AOs. The percentages of in-frame transcripts in each lane of d are shown. The data (n = 3) are presented as mean ± SEM. (f) Immunohistochemical staining of dystrophin in TA muscle of WT, untreated and treated mdx52 mice. The results for AOs 51A plus 51D, 51I, mB30 and 51D are indicated. Dystrophin was detected with a rabbit polyclonal antibody P7. Bar = 100 µm. (g) Western blotting to detect expression of dystrophin in WT, untreated and treated mdx52 mice. Representative results for 10 single AOs and three combinations of two AOs. A quantitative analysis (see Materials and Methods) normalized to the expression of β-actin (upper panel), and western blotting to detect β-actin expression (lower panel) are shown. Dystrophin was detected with the Dys2 monoclonal antibody. Note that additional bands between the unskipped and skipped products are visible in some analyses. This is due to heteroduplex formation and has been described previously.13 bp, base pair.
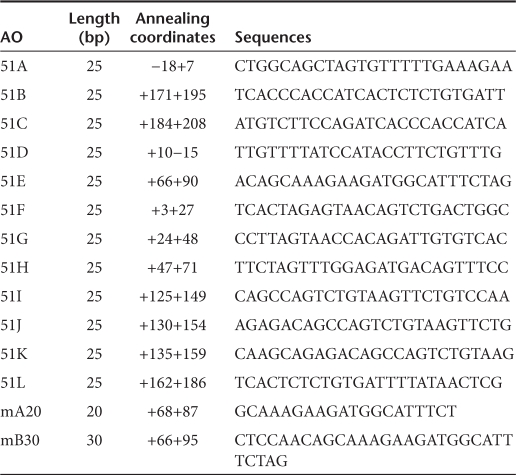
Table 1. Length, annealing coordinates, sequences of all AOs targeting mouse exon 51.
AOs restored body-wide dystrophin expression in a highly dose-dependent manner in mdx52 mice
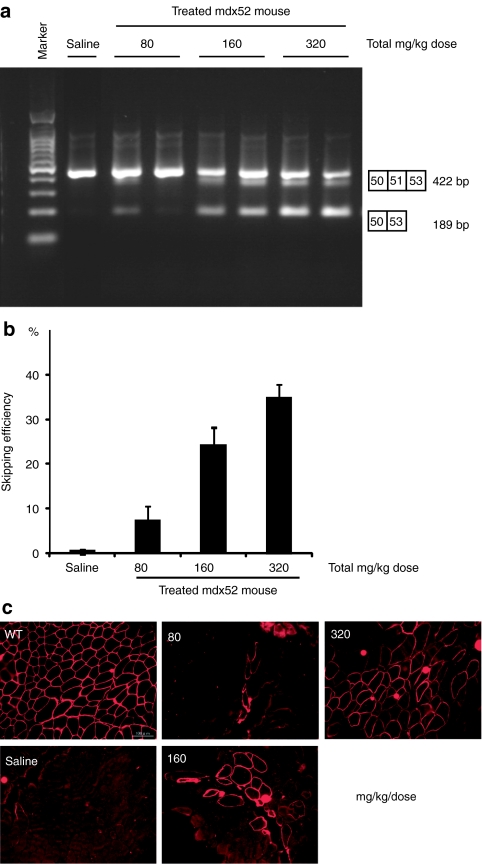
To examine the effect of systemic delivery, we intravenously injected a single dose of 51A plus 51D into 8-week-old mdx52 mice, at 80 (ref. 15), 160 or 320 mg/kg. Mice were euthanized 2 weeks after the injection; the muscles were isolated and analyzed by RT-PCR and the cryosections by immunohistochemistry. We found that the AOs restored body-wide dystrophin expression in a highly dose-dependent manner, with the 320 mg/kg dose showing ~45% skipping efficiency by RT-PCR (Figure 3a,b) and 45% dystrophin-positive fibers by immunohistochemistry (Figure 3c) in the gastrocnemius (GC) muscle.
Figure 3.
Dose-escalation study of systemic delivery of antisense oligonucleotides (AOs) to mdx52 mice. Restoration of dystrophin in the gastrocnemius muscle 2 weeks after single intravenous co-injections of 80, 160, or 320 mg/kg/dose of AO. Intravenous saline injection into Mdx52 mice was used as a control. (a) Detection of exon 51-skipped dystrophin mRNA by RT-PCR. Representative data are shown. Skipped products (50–53) are compared with unskipped products (50–51–53). The additional bands between the unskipped and skipped products is due to heteroduplex formation. (b) Quantitative analysis of exon 51-skipping by AO. The percentages of in-frame transcripts are shown. The data (n = 3) are presented as mean ± SEM. (c) Immunohistochemical staining of dystrophin in the quadriceps muscles of a treated mdx52 mouse. Dystrophin was detected with rabbit polyclonal antibody P7. Bar = 100 µm. bp, base pair.
Repeated systemic delivery of AOs induced highly efficient in-frame dystrophin in skeletal muscles body-wide
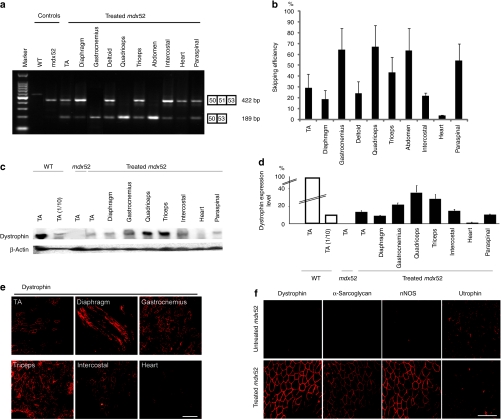
Next, we intravenously injected 320 mg/kg/dose of 51A plus 51D into 8-week-old mdx52 mice, seven times at weekly intervals. Two weeks after the final injection, whole-body skeletal muscles and the heart were examined. By RT-PCR, we identified cDNA bands corresponding to exon 51 having been skipped in nearly all skeletal muscles of treated mice (Figure 4a). The levels of skipping efficiency were variable: ~67% in the quadriceps (QC), 64% in the GC, 63% in the abdominal, 54% in the paraspinal, 43% in the triceps, 29% in the TA, 24% in the deltoid, 21% in the intercostal, 18% in the diaphragm, and 3% in the heart muscles (Figure 4b). Dystrophin expression was also evaluated by quantitative western blotting (Figure 4c). The expression levels in the QC, GC, and triceps muscles were the highest at 30–40% of normal levels. Those in the TA, intercostal, paraspinal, and diaphragm muscles showed modest expression at 10–20% of normal levels, whereas the dystrophin expression level in the heart was only 1% of normal levels (Figure 4d). We detected 60–80% dystrophin-positive fibers in all skeletal muscles by immunohistochemistry, most prominently in the QC, GC, and paraspinal muscles (Figure 4e). Furthermore, most of the nonpositive fibers in our study showed weak dystrophin signals.
Figure 4.
Repeated systemic delivery of antisense oligonucleotides (AOs) to mdx52 mice. The restoration of dystrophin in various muscles after seven weekly intravenous co-injections of 320 mg/kg/dose of AOs was examined. (a) Detection of exon 51-skipped dystrophin mRNA by RT-PCR. Representative data are shown. Skipped products (50–53) are compared with unskipped products (50–51–53). The additional bands between the unskipped and skipped products is due to heteroduplex formation. (b) Quantitative analysis of exon 51-skipping by AOs. The percentages of in-frame transcripts are shown. The data (n = 3) are presented as mean ± SEM. (c) Western blotting after AO injections to detect the expression of dystrophin (upper panel) and β-actin (lower panel) in the TA, diaphragm, gastrocnemius, quadriceps, triceps brachii, intercostal, heart, and paraspinal muscles of a treated mdx52 mouse. Representative results are shown. Dystrophin was detected with the Dys2 monoclonal antibody. (d) Quantitative analysis of dystrophin expression after AO injection. The data (n = 4) are presented as mean ± SEM. TA(1/10): 10% of WT samples. (e) Immunohistochemical staining of dystrophin in the TA, diaphragm, gastrocnemius, triceps brachii, intercostal, and heart muscles of a treated mdx52 mouse. Dystrophin was detected with rabbit polyclonal antibody P7. Bar = 100 µm. (f) Immunohistochemical staining of dystrophin, α-sarcoglycan, neuronal nitric oxide synthase (nNOS) and utrophin in the quadriceps muscle of an untreated mdx52 mouse (upper panel) and a treated mdx52 mouse (lower panel). Bar = 100 µm. mdx52, untreated mdx52 mouse; TA, tibialis anterior; WT, wild-type mouse.
We examined the expression of components included in the dystrophin–glycoprotein complex in the QC by immunohistochemistry. The expression of α-sarcoglycan correlated well with that of dystrophin (Figure 4f). We also observed the recovery of β-dystroglycan and α1-syntrophin expression at the sarcolemma (data not shown). On the other hand, utrophin expression was diminished in dystrophin-positive fibers (Figure 4f).
In-frame dystrophin largely lacking hinge 3 ameliorated skeletal muscle pathology
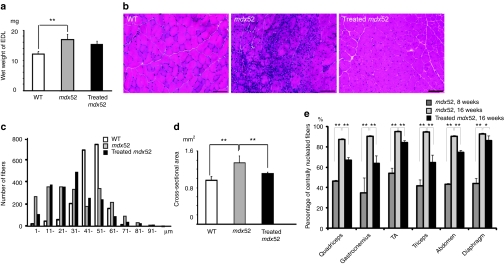
The mdx52 mice skeletal muscle shows hypertrophy and an increased ratio of centrally nucleated fibers.17 Two weeks after seven consecutive weekly i.v. injections of the combination of AOs, the wet weight of the extensor digitorum longus muscle tended to be slightly lower in treated mice than in untreated mice (Figure 5a). We observed less muscle degeneration and fewer cellular infiltrates in the treated TA muscle compared with the untreated TA muscle (Figure 5b). We then evaluated the detailed histological changes in the treated muscles and compared them with the changes in the untreated muscles. The fiber size variation in the treated TA muscle was less than that in the untreated TA muscle (Figure 5c). We found a significant decrease in the mean cross-sectional area of muscle fibers in treated mice compared with those in untreated mice (Figure 5d). The percentages of centrally nucleated fibers were lower in the triceps, GC, QC, and abdominal muscles than in the diaphragm and TA muscles (Figure 5e). These changes reflect the amelioration of muscle fiber hypertrophy and dystrophic changes in the treated mdx52 mice.
Figure 5.
Amelioration of pathology in body-wide muscles of mdx52 mice after seven weekly intravenous co-injections of 320 mg/kg/dose of antisense oligonucleotides. (a) Wet weight of the extensor digitorum longus (EDL) muscles of wild-type (WT), untreated and treated 16-week-old mdx52 mice. The data (n = 4) are presented as mean ± SEM. **P < 0.01. (b) Hematoxylin and eosin staining of cryosections in tibialis anterior (TA) muscle of WT, untreated and treated mdx52 mice. Bar = 100 µm. (c) Histogram of muscle fibers in the TA muscle of WT, untreated and treated 16-week-old mdx52 mice. (d) Cross-sectional area of EDL muscles of WT, untreated and treated 16-week-old mdx52 mice. The data (n = 4) are presented as mean ± SEM. **P < 0.01. (e) The ratio of centrally nucleated fibers in the quadriceps, gastrocnemius, TA, triceps brachii, abdominal, and diaphragm muscles of untreated 8-week-old (dark gray), 16-week-old mdx52 mice (light gray), and treated 16-week-old mdx52 mice (black). The data (n = 4) are presented as mean ± SEM. *P < 0.05; **P < 0.01. mdx52, untreated mdx52 mouse.
In-frame dystrophin largely lacking hinge 3 restored skeletal muscle function
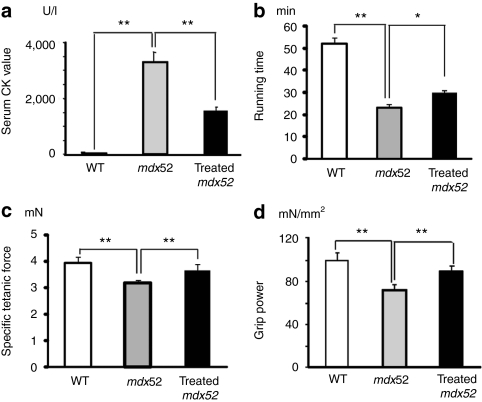
To examine the function of the AO-induced dystrophin, we evaluated skeletal muscle function with a battery of tests after seven weekly i.v. AO injections. The protection of muscle fibers against degeneration was supported by a significant reduction in serum creatine kinase levels in the treated mice (Figure 6a). Significant improvements in treadmill endurance (Figure 6b), maximum forelimb grip force (Figure 6c), and specific tetanic force of the extensor digitorum longus muscle (Figure 6d) were observed in treated mdx52 mice compared with nontreated mdx52 mice.
Figure 6.
Muscle function in mdx52 mice after seven weekly intravenous co-injections of 320 mg/kg/dose of antisense oligonucleotides. (a) Measurement of serum creatine kinase (CK) levels (IU/l). (b) Treadmill performance (min), (c) grip power test (mN/g), and (d) specific tetanic force of the extensor digitorum longus muscle (mN/mm2). Wild-type (WT), untreated (mdx52), and treated 16-week-old mdx52 mice were examined. The data (n = 4) are presented as mean ± SEM. *P < 0.05; **P < 0.01.
Efficacy of repeated AO injection in mdx52 mice confirmed by gene expression array
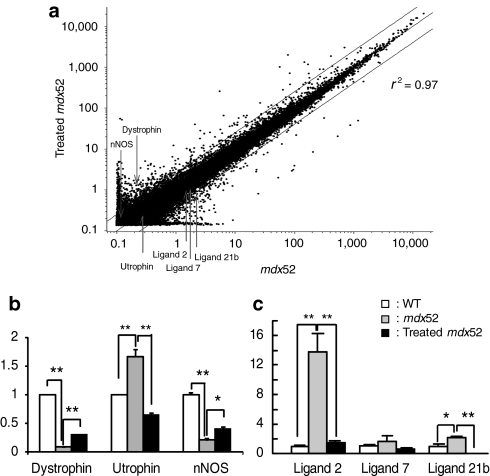
Gene expression array analysis has been widely used to profile gene expression for disease diagnosis and therapy due to its ability to interrogate every transcript in the genome simultaneously. Dystrophic TA muscle has been compared with normal TA muscle in human and mdx mice at various stages of the disease.23,24 To evaluate the gene expression profile of TA muscles following exon 51-skipping, we performed genome-wide gene expression analysis (Figure 7a). Gene expression array analysis showed that the gene expression profiles of TA muscles correlated well between the treated and untreated mdx52 mice (r2 = 0.97), and there was no unexpected downregulation of housekeeping genes or upregulation of stress-related proteins. We found that dystrophin-associated proteins such as dystrophin, neuronal nitric oxide synthase, and α1-syntrophin were upregulated, α-sarcoglycan and β-dystroglycan levels were unchanged, and utrophin was downregulated. We also found that inflammatory cytokines were downregulated in treated mdx52 mice. Quantitative RT-PCR following gene expression array analysis showed that dystrophin and neuronal nitric oxide synthase expression levels were 3.4 and 1.9 times higher than those in the untreated mdx52 mice, respectively; however, they were still only 33 and 40% of the normal levels, respectively (Figure 7b). Utrophin expression levels were upregulated in the untreated mdx52 mice, but downregulated in the treated mdx52 mice compared with wild-type (WT) mice (Figure 7b). In treated mdx52 mice, we observed reduced levels of several C-C class chemokine ligands (Ccls) such as Ccl7, Ccl21b, and Ccl2, which are small cytokines that induce the migration of monocytes and other cell types such as natural killer cells and dendritic cells (Figure 7c). This might reflect an improvement of the muscle inflammatory response in the treated mdx52 mice.
Figure 7.
Genome-wide expression analyses by gene expression array using tibialis anterior muscles. Gene expression array analysis after repeated intravenous co-injections of 320 mg/kg/dose of antisense oligonucleotides into mdx52 mice. (a) A scatter plot of global gene expression analyses by gene expression array in treated and untreated mdx52 mice. The upper line shows twofold changes and the lower line shows 0.5-fold changes in gene expression levels between treated and untreated mdx52 mice. The positions of dystrophin, utrophin, neuronal nitric oxide synthase (nNOS), and the C-C class chemokine ligands (Ccls) 2, 7, and 21b are indicated. (b,c) The gene expression levels of dystrophin, utrophin, nNOS (b) and CCLs 2, 7, and 21b (c) by quantitative PCR, in 16-week-old WT, untreated and treated mdx52 mice. The data (n = 3) are presented as mean ± SEM. *P < 0.05; **P < 0.01. mdx52, untreated mdx52 mouse; WT, wild-type mouse.
No detectable toxicity after repeated delivery of AOs into mdx52 mice
No signs of illness and no deaths were noted during the period of AO treatment. To further monitor any potential toxicities in the major organs induced by treatment with AOs, we compared a series of serum markers commonly used as indicators of liver and kidney dysfunction in WT, untreated and treated mdx52 mice. No significant differences were detected among the three groups in the levels of creatinine, blood urea nitrogen, aspartate amino transferase, alanine aminotransferase, total bilirubin, alkaline phosphatase, and γ-glutamyl transpeptidase (Supplementary Figure S2a). Histological examination of liver and kidney revealed no signs of tissue damage or increased monocyte infiltrations in treated mdx52 mice (Supplementary Figure S2b). These data confirm that this AO combination was nontoxic in this study.
In vitro exon 51-skipping in DMD 5017 cells with deletion of exons 45–50
We newly designed several AOs based on murine sequences: hAc (51Ac) targeting the 5′ splice site, and hDo1 (51D1) and hDo2 (51D2) targeting the 3′ splice site of human exon 51. The sequences and composition of the AO treatments are described in Supplementary Table S1. MyoD-converted fibroblasts (DMD 5017 cells) were examined after 48-hour incubation with a single or two AOs at a final concentration of 1, 5, or 10 µmol/l. Among the AOs examined, hAc plus hDo1, and B30 alone, were shown by RT-PCR to be capable of inducing exon 51-skipping at a level approaching 50 and 30%, respectively (Supplementary Figure S3). On the other hand, hAc, hDo1, or hDo2 alone were less effective at inducing exon 51-skipping (Supplementary Figure S3). These results suggest that the co-injection of two AOs could induce highly effective exon 51-skipping in DMD cells.
Discussion
This is the first report showing widespread induction of in-frame dystrophin lacking most of hinge 3 following exon 51-skipping, and clear recovery of muscle function to therapeutic levels in a DMD mouse model.
Exon skipping produces several forms of in-frame dystrophin that lack part of the molecular structure depending on the targeted exons.4,5,6 The molecular structure of dystrophin is composed of the actin-binding domain 1 at the N-terminus (ABD1), the central rod domain containing 24 spectrin-like repeats (R1–24), four hinge domains, a 20-amino acid insertion between spectrin-like repeats 15 and 16 (segment 5), the cysteine-rich domain, and the C-terminal domain (Figure 1b).27,38 Until now, the in-frame dystrophin formed following exon 23-skipping, which lacks half of the 6th spectrin-like repeat and part of the 7th, ameliorated the muscle pathology and function in mdx mice with a point mutation in exon 23 (refs. 15,16). These results were consistent with the fact that in-frame deletions of the central rod domain in humans typically lead to a mild BMD.6 However, the severity of BMD with in-frame deletions including hinge 3 can vary considerably.28,29,30,31,32,38 The hinges are proline-rich, nonrepeat segments that may confer flexibility to dystrophin.27 Among them, the hinge 3 region is encoded by exons 50 and 51, located between the 19th and 20th spectrin-like repeats, and is prone to deletion mutations.20,22 Recently, it was reported that hinge 3 is more important than hinge 2 in preventing muscle degeneration and promoting muscle maturation in the microdystrophinΔR4–R23/mdx transgenic mice.38 The in-frame dystrophin produced following exon-51-skipping is predicted to lack most of the hinge 3 region (Figure 1b); hinges 1, 2, 3, and 4 consist of 75, 50, 43, and 72 amino acid residues, respectively.27,38 The small segment of hinge 3 that remains following exon 51-skipping consists of 13 amino acid residues with two prolines and would not be predicted to function as a hinge (Figure 1c). This remaining fragment is very similar to segment 5, which consists of 20-amino acid residues with one proline and is located between the 15th and 16th spectrin-like repeats.27 Based on the molecular structure of dystrophin, the small segment of the hinge 3 might act as a “turn,” which is bound to the helix of the 20th repeat.27,38 Our results suggest that the hinge 3 region is more essential in the short microdystrophin (167 kDa) than in the almost full-length dystrophin lacking hinge 3 due to exon 51-skipping (420 kDa).38,39
In-frame dystrophin expression in skeletal muscle at 20% of normal levels produced moderate/mild BMD phenotypes.26 Moreover, restoration of 20–30% in-frame dystrophin expression resulted in protection from muscle degeneration and recovery of skeletal muscle function in the transgenic mdx mouse.40 As a first step, we screened optimal AO sequences including mB30 for skipping of exon 51 to induce in-frame dystrophin at up to 20% of normal dystrophin levels. To date, specific AO sequences have been assessed for their efficiency of exon skipping using cell-based experimental systems, with the optimal sequences then used for in vivo experiments.11,41,42 However, the in vivo efficacy of phosphorodiamidate morpholino oligomers (PMOs), which are rather difficult to deliver into mammalian cells in culture because of their neutral chemistry,11,19 could differ from that in vitro.14 Therefore, we used the mdx52 mouse to screen AO sequences for exon 51-skipping by intramuscular injection in vivo. We also showed that simultaneous delivery of two AO sequences directed against both the 3′ and the 5′ splice sites drove skipping of exon 51 more efficiently than any single or two AO sequences targeting exonic regions in the mdx52 mouse. This combination of AOs worked in a synergistic fashion, where the increase in activity was greater than the additive effect of each individual AO.14,34,37
We found that the skipping efficiency induced by systemically delivered PMO increased in proportion to the AO dose in the mdx52 mouse. A dose-dependent restoration of dystrophin expression in the muscles of mdx mice by systemically delivered PMO has also been reported.43 The therapeutic dose (320 mg/kg/dose) of exon 51-skipping AOs in the mdx52 mouse to induce 20–30% of normal dystrophin levels is approximately four times as much as the dose (80 mg/kg/dose) required for exon 23-skipping in the mdx mouse. We have to consider the possibility that the difference of the genetic background of mice between mdx52 (C57BL/6J) and mdx (C57BL/10) mice could influence the properties of exon skipping. Our results suggest that the therapeutic dose of AO required is different depending on which exon is being targeted. Because AO-mediated exon skipping is the first RNA-modulating therapeutic with this mechanism of action, this study using a DMD mouse model could provide a suggestion for human equivalent doses based on body surface area.44 Recently, press releases from both Prosensa for PRO051 and AVI Biopharma for AVI-4658 have revealed that DMD patients who received 2–6 and 2–20 mg/kg, respectively, induced specific exon 51-skipping and dystrophin expression in a dose-related manner (http://www.prosensa.eu/ press-room/press-releases/2009-09-14-PRO051-shows-favourable-results.php; http://investorrelations.avibio.com/ phoenix.zhtml?c=64231&p=irol-newsArticle&ID=1433350&highlight=).
In this study, the dystrophin expression level in treated mdx52 mice was restored to roughly 20–30% of normal levels, and creatine kinase levels and skeletal muscle function significantly recovered in the treated mice. These findings show that the in-frame dystrophin lacking most of hinge 3 ameliorates dystrophic histology and functional phenotypes in mdx52 mice as well as the dystrophin produced following exon 23-skipping or microdystrophin in mdx mice.15,16 On the other hand, the treadmill endurance of the treated mdx52 mice was still considerably inferior to that of WT mice compared with the clear recovery of forelimb grip force and specific tetanic force of the extensor digitorum longus muscle. These findings are similar to the data produced using microdystrophin in the mdx mouse.39 Two possibilities remain to explain the incomplete recovery: insufficient dystrophin expression or defective dystrophin molecular structure due to exon 51-skipping. To examine these possibilities, we are now trying to increase the level of dystrophin expression using a high dose of PMO and using peptide-conjugated PMO in mdx52 mice.
The amelioration of the histopathology demonstrated by the reduction of centrally nucleated fibers in treated mdx52 mice was marked in the muscles with high levels of dystrophin expression, and the effect was modest in the muscles with low levels of dystrophin, in accordance with a previous report.15,16 It is noteworthy that high levels of dystrophin expression were seen in severely degenerated muscles, and that low levels of expression were found in less affected muscles. We suggest the possibility that the antigravity muscles, such as the QC, GC, triceps brachii, abdominal, and paraspinal muscles45 efficiently incorporated PMO into the muscle fibers. We showed that the antigravity muscles were mainly affected in mdx52 mice during the period that we examined; therefore, those severely affected muscles had taken up the most AO. Our data support the fragile membrane hypothesis that has been proposed as the background for PMO incorporation into dystrophic muscle.46,47 This hypothesis also explains the inefficient incorporation of PMO into the diaphragm muscle where dystrophic changes chronically persisted, as previously reported.15,16
To screen for changes in gene expression levels influenced by the in-frame dystrophin, we performed gene expression array analysis. The inflammatory chemokines Ccl2, Ccl7, and Ccl21b, which were downregulated after treatment, play important roles in the migration of macrophages, CD4+ and CD8+ T cells to muscle in mdx mice.48 It is also known that depletion of these cells from mdx mice decreased the sarcolemmal damage.49 These data showed that the inflammatory process, which could aggravate the pathology, was prevented in mdx52 mice. Measurement of the chemokines might be a beneficial index of the therapeutic effects on mdx52 mice.
In conclusion, this report describes the first successful effort at systemic rescue of in-frame dystrophin lacking most of hinge 3 and muscle function by PMO-mediated exon 51-skipping in a mouse model. Because the structure of the in-frame dystrophin lacking most of hinge 3 in mice resembles human dystrophin following exon 51-skipping, our results are extremely encouraging as regards the ongoing systemic clinical trials for DMD. In addition, the therapeutic dose in DMD model mice provides a suggestion of the theoretical equivalent dose in humans.
Materials and Methods
Animals. Exon 52–deficient X chromosome–linked muscular dystrophy (mdx52) mice were produced by a gene-targeting strategy and maintained at our facility.33 The mice have been backcrossed to the C57BL/6J WT strain for more than eight generations. Eight-week-old male mdx52 and WT mice were used in this study. All experimental protocols in this study were approved by The Experimental Animal Care and Use Committee of the National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP), Tokyo, Japan.
Antisense sequences and delivery methods. Thirteen AOs for targeted skipping of exon 51 during dystrophin pre-mRNA splicing in mice were comprehensively designed to anneal to the 5′ splice site (51A), the 3′ splice site (51D), and other intraexonic regions (51B, 51C, 51E, 51F, 51G, 51H, 51I, 51J, 51K, 51L, 51M). The sequences and positions of the AOs are described in Table 1. mB30, which corresponds to human B30, was also specifically designed for this study.11 To design these sequences, we referred to previously published sequences and considered GC content and secondary structure to avoid self- and heterodimerization.9,11 All sequences were synthesized using a morpholino backbone (Gene Tools, Philomath, OR). Primers for RT-PCR and sequencing analysis were synthesized by Operon Biotechnologies (Tokyo, Japan) and are listed in Supplementary Table S1.
Ten micrograms of PMO were injected into each TA muscle of mdx52 mice. Muscles were obtained 2 weeks after the intramuscular injection. To examine the optimal therapeutic dose, a total of 80 (ref. 15), 160 or 320 mg/kg/dose of AO was injected into the tail vein of mdx52 mice singly. Muscles were isolated 2 weeks after the systemic injection and analyzed by RT-PCR and the cryosections by immunohistochemistry. Following the dose-escalation study, a 320 mg/kg dose of PMO in 200 µl of saline,50 or 200 µl saline, was injected into the tail vein of mdx52 mice or WT mice, seven times at weekly intervals. The mice were examined 2 weeks after the final injection. Muscles were dissected immediately, snap-frozen in liquid nitrogen–cooled isopentane and stored at −80 °C for RT-PCR, immunohistochemistry, western blotting, and gene expression array analysis. Liver and kidney were also frozen in liquid nitrogen and stored at −80 °C for pathological analysis.
RT-PCR and sequencing of cDNA. Total RNA was extracted from cells or frozen tissue sections using TRIzol (Invitrogen, Carlsbad, CA) from treated mdx52 mice, and from WT and untreated mdx52 mice, which were used as controls, respectively. Two hundred nanograms of total RNA template was used for RT-PCR with a QuantiTect Reverse Transcription kit (Qiagen, Crawley, UK) according to the manufacturer's instructions. The cDNA product (1 µl) was then used as the template for PCR in a 25 µl reaction with 0.125 units of TaqDNA polymerase (Qiagen). The reaction mixture comprised 10× PCR buffer (Roche, Basel, Switzerland), 10 mmol/l of each dNTP (Qiagen), and 10 µmol/l of each primer. The primer sequences were Ex50F 5′-TTTACTTCGGGAGCTGAGGA-3′ and Ex53R 5′-ACCTGTTCGGCTTCTTCCTT-3′ for amplification of cDNA from exons 50–53. The cycling conditions were 95 °C for 4 minutes, then 35 cycles of 94 °C for 1 minute, 60 °C for 1 minute, 72 °C for 1 minute, and finally 72 °C for 7 minutes. The intensity of PCR bands was analyzed by using ImageJ software (http://rsbweb.nih.gov/ij/), and skipping efficiency was calculated by using the following formula [(the intensity of skipped band) / (the intensity of skipped band + the intensity of unskipped band)].51 After the resulting PCR bands were extracted using a gel extraction kit (Qiagen), direct sequencing of PCR products was performed by the Biomatrix Laboratory (Chiba, Japan).
Immunohistochemistry, and hematoxylin and eosin staining. At least ten 8 µm cryosections were cut from flash-frozen muscles at 100 µm intervals. The serial sections were stained with polyclonal rabbit antibody P7 against the dystrophin rod domain (a gift from Qi-Long Lu, Carolinas Medical Center, Charlotte, NC), anti-α-sarcoglycan monoclonal rabbit antibody (Novocastra Laboratories, Newcastle, UK), anti-β-dystroglycan monoclonal mouse antibody (Novocastra Laboratories), anti-α1-syntrophin monoclonal mouse antibody (Novocastra Laboratories), antineuronal nitric oxide synthase polyclonal rabbit antibody (Zymed, San Francisco, CA), and antiutrophin polyclonal rabbit antibody (UT-2). Alexa-488 or 568 (Molecular Probes, Cambridge, UK) was used as a secondary antibody. 4′,6-diamidino-2-phenylindole containing a mounting agent (Vectashield; Vector Laboratories, Burlingame, CA) was used for nuclear counterstaining. The maximum number of dystrophin-positive fibers in one section was counted, and the TA muscle fiber sizes were evaluated using a BZ-9000 fluorescence microscope (Keyence, Osaka, Japan). Hematoxylin and eosin (H&E) staining was performed using Harris H&E.
Western blotting. Muscle protein from cryosections was extracted with lysis buffer as described previously.14 Two to twenty micrograms of protein were loaded onto a 5–15% XV Pantera Gel (DRC, Tokyo, Japan). The samples were transferred onto an Immobilon polyvinylidene fluoride membrane (Millipore, Billerica, MA) by semidry blotting at 5 mA/mm2 for 1.5 hours. The membrane was incubated with the C-terminal monoclonal antibody Dys2 (Novocastra) at room temperature for 1 hour. The bound primary antibody was detected by horseradish peroxidase–conjugated goat anti-mouse IgG (Cedarlane, Burlington, ON) and SuperSignal chemiluminescent substrate (Pierce, Rockford, IL). Anti-β-actin antibody was used as a loading control. Signal intensity of detected bands of the blots were quantified using ImageJ software and normalized to the loading control.
Serum creatine kinase levels and toxicity tests. Four treated mdx52 mice were examined for toxic effects of PMO before injection, 1 week after the third injection, and 2 weeks after the last injection. Blood was taken from the tail artery and centrifuged at 3,000g for 10 minutes. The biochemical markers creatine kinase, electrolytes (sodium, potassium, and chloride ions), blood urea nitrogen, total bilirubin, alkaline phosphatase, aspartate transaminase, and alanine transaminase were assayed as described previously.14 The histology of the liver, lung, and kidney was examined microscopically on cryosections.
Functional testing. The mice were placed on a flat MK-680S treadmill (Muromachi Kikai, Tokyo, Japan) and forced to run at 5 m/minute for 5 minutes. After 5 minutes, the speed was increased by 1 m/minute every minute. The test was stopped when the mouse was exhausted and did not attempt to remount the treadmill, and the time to exhaustion was determined.
The grip strength of the mice was assessed by a grip strength meter (MK-380M; Muromachi Kikai). The mice were held 2 cm from the base of the tail, allowed to grip a woven metal wire with their forelimbs, and pulled gently until they released their grip. Five sequential tests for the exerted force were carried out for each mouse, with 5-second intervals, and the data were averaged.
The extensor digitorum longus muscles were kept in Krebs–Henseleit solution at 25 °C and stimulated with a pair of platinum electrodes using an electronic stimulator (SEN-3301; Nihon Kohden, Tokyo, Japan). A Thermal Arraycorder (WR300; Graphtec, Yokohama, Japan) was used to control the stimulation and to record the force of the muscle contraction. Measurement of the specific tetanic force was performed as previously described.39
Gene expression array analysis. TA muscles from treated mdx52, age-matched WT, and untreated mdx52 mice (n = 3 each) were used for these experiments. Total RNA was purified using an RNeasy mini kit (Qiagen) according to the manufacturer's protocol. Gene expression array analysis was performed by the branch of the Agilent Technologies (Santa Clara, CA) in Japan. Three whole mouse genome oligo microarrays 44K (Agilent Technologies) were used in this study. Global normalization was performed to compare genes from chip to chip using GeneSpring 9.0 (Tomy Digital Biology, Denver, CO). All data quality controls were performed and met the Affymetrix quality assessment guidelines. Data analysis was performed using GeneSpring 9.0 (Tomy Digital Biology). Differentially expressed genes were selected if they passed Welch's t-test, a parametric test in which the variance is not assumed to be equal. P < 0.01 (with correction for multiple testing by the Benjamini and Hochberg method for the false discovery rate) and a 5% cutoff were used; a change of at least twofold between any two of the groups used in this study was considered significant.
Quantitative real-time PCR. For genes selected for the gene expression array, we used the same RNA that was isolated for the gene expression array and prepared cDNA using SuperScript III Reverse Transcriptase (Invitrogen). Real-time PCR was performed using a SYBR Premix Ex Taq II kit (Takara, Tokyo, Japan). Expression values were normalized to 18S rRNA expression and shown as a fold increase in the treated mdx52, untreated mdx52, and age-matched WT samples.
Transfection of cultured cells with AO. DMD 5017 cells were obtained from Coriell Cell Repositories (Camden, NJ). Fibroblasts were cultured in 20% growth medium, containing Dulbecco's modified Eagle's medium and F-12 in a 1:1 mixture (Invitrogen), 20% fetal bovine serum (SAFC Biosciences, Lenexa, KS) and 1% penicillin/streptomycin (Sigma-Aldrich, St Louis, MO). Then, fluorescence-activated cell sorting sorted MyoD-enhanced green fluorescent protein-positive fibroblasts (MyoD-converted fibroblasts) were cultured in differentiation medium, containing Dulbecco's modified Eagle's medium/F-12 in a 1:1 mixture (Invitrogen), 2% horse serum (Invitrogen), and 1% penicillin/streptomycin (Sigma-Aldrich). hDo1, hDo2, and hAc were designed (Table 1) and synthesized by Gene Tools. MyoD-converted fibroblasts were transfected with a single or two AOs at a final concentration of 10 µmol/l. EndoPorter (Gene Tools) was added to give a final concentration of 6 µmol/l. After 48-hour incubation with the AOs, total RNA was extracted from MyoD-converted fibroblasts using Trizol (Invitrogen).
Statistical analysis. Statistical differences were assessed by one-way analysis of variance with differences among the groups assessed by a Tukey comparison. All data are reported as mean values ± SEM. The level of significance was set at P < 0.05.
SUPPLEMENTARY MATERIAL Figure S1. Dose-escalation study of exon 51-skipping by local intramuscular injection into mdx52 mice. Figure S2. Examination of adverse effects after systemic delivery of antisense oligonucleotides (AOs). Figure S3. In vitro exon 51-skipping in DMD 5017 cells with deletion of exons 45–50. Table S1. Length, annealing coordinates, sequences of all AOs targeting human exon 51.
Acknowledgments
We thank Eric Hoffman and Terence Partridge for insightful discussions about this study. We thank Kouichi Tanaka and Hiroko Hamazaki for their support, and Mikiharu Yoshida, Michihiro Imamura, Masanori Kobayashi, Jing Hong Shin, Yuko Shimizu, Norio Motohashi, Erika Yada, Katsutoshi Yuasa, and Morizono Hiroki for useful discussions and technical assistance. We thank Qi-Long Lu for supplying the antibody to dystrophin (P7). This work was supported by Grants-in-Aid for Research on Nervous and Mental Disorders (19A-7), Health and Labor Sciences Research Grants for Translation Research (H19-Translational Research-003 and H21-Clinical Research-015), and Health Sciences Research Grants for Research on Psychiatry and Neurological Disease and Mental Health (H18-kokoro-019) from the Ministry of Health, Labour and Welfare of Japan. This work was performed in Kodaira, Tokyo, Japan.
Supplementary Material
Dose-escalation study of exon 51-skipping by local intramuscular injection into mdx52 mice.
Examination of adverse effects after systemic delivery of antisense oligonucleotides (AOs).
In vitro exon 51-skipping in DMD 5017 cells with deletion of exons 45–50.
Length, annealing coordinates, sequences of all AOs targeting human exon 51.
REFERENCES
- Hoffman EP, Brown RH., Jr, and, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H., and, Kunkel LM. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- Wilton SD, Fall AM, Harding PL, McClorey G, Coleman C., and, Fletcher S. Antisense oligonucleotide-induced exon skipping across the human dystrophin gene transcript. Mol Ther. 2007;15:1288–1296. doi: 10.1038/sj.mt.6300095. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Bremmer-Bout M, Janson AA, den Dunnen JT, van Ommen GJ., and, van Deutekom JC. Targeted exon skipping as a potential gene correction therapy for Duchenne muscular dystrophy. Neuromuscul Disord. 2002;12 Suppl 1:S71–S77. doi: 10.1016/s0960-8966(02)00086-x. [DOI] [PubMed] [Google Scholar]
- Matsuo M, Masumura T, Nishio H, Nakajima T, Kitoh Y, Takumi T, et al. Exon skipping during splicing of dystrophin mRNA precursor due to an intraexon deletion in the dystrophin gene of Duchenne muscular dystrophy kobe. J Clin Invest. 1991;87:2127–2131. doi: 10.1172/JCI115244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntoni F, Torelli S., and, Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–740. doi: 10.1016/s1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- Dunckley MG, Manoharan M, Villiet P, Eperon IC., and, Dickson G. Modification of splicing in the dystrophin gene in cultured Mdx muscle cells by antisense oligoribonucleotides. Hum Mol Genet. 1998;7:1083–1090. doi: 10.1093/hmg/7.7.1083. [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Bremmer-Bout M, Janson AA, Ginjaar IB, Baas F, den Dunnen JT, et al. Antisense-induced exon skipping restores dystrophin expression in DMD patient derived muscle cells. Hum Mol Genet. 2001;10:1547–1554. doi: 10.1093/hmg/10.15.1547. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, De Winter CL, Janson AA, Kaman WE, Van Ommen GJ, Den Dunnen JT, et al. Functional analysis of 114 exon-internal AONs for targeted DMD exon skipping: indication for steric hindrance of SR protein binding sites. Oligonucleotides. 2005;15:284–297. doi: 10.1089/oli.2005.15.284. [DOI] [PubMed] [Google Scholar]
- Wilton SD, Lloyd F, Carville K, Fletcher S, Honeyman K, Agrawal S, et al. Specific removal of the nonsense mutation from the mdx dystrophin mRNA using antisense oligonucleotides. Neuromuscul Disord. 1999;9:330–338. doi: 10.1016/s0960-8966(99)00010-3. [DOI] [PubMed] [Google Scholar]
- Arechavala-Gomeza V, Graham IR, Popplewell LJ, Adams AM, Aartsma-Rus A, Kinali M, et al. Comparative analysis of antisense oligonucleotide sequences for targeted skipping of exon 51 during dystrophin pre-mRNA splicing in human muscle. Hum Gene Ther. 2007;18:798–810. doi: 10.1089/hum.2006.061. [DOI] [PubMed] [Google Scholar]
- Beggs AH, Hoffman EP, Snyder JR, Arahata K, Specht L, Shapiro F, et al. Exploring the molecular basis for variability among patients with Becker muscular dystrophy: dystrophin gene and protein studies. Am J Hum Genet. 1991;49:54–67. [PMC free article] [PubMed] [Google Scholar]
- Aartsma-Rus A, Janson AA, Kaman WE, Bremmer-Bout M, den Dunnen JT, Baas F, et al. Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum Mol Genet. 2003;12:907–914. doi: 10.1093/hmg/ddg100. [DOI] [PubMed] [Google Scholar]
- Yokota T, Lu QL, Partridge T, Kobayashi M, Nakamura A, Takeda S, et al. Efficacy of systemic morpholino exon-skipping in Duchenne dystrophy dogs. Ann Neurol. 2009;65:667–676. doi: 10.1002/ana.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter J, Lou F, Rabinowitz A, Yin H, Rosenfeld J, Wilton SD, et al. Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nat Med. 2006;12:175–177. doi: 10.1038/nm1345. [DOI] [PubMed] [Google Scholar]
- Lu QL, Mann CJ, Lou F, Bou-Gharios G, Morris GE, Xue SA, et al. Functional amounts of dystrophin produced by skipping the mutated exon in the mdx dystrophic mouse. Nat Med. 2003;9:1009–1014. doi: 10.1038/nm897. [DOI] [PubMed] [Google Scholar]
- Bremmer-Bout M, Aartsma-Rus A, de Meijer EJ, Kaman WE, Janson AA, Vossen RH, et al. Targeted exon skipping in transgenic hDMD mice: a model for direct preclinical screening of human-specific antisense oligonucleotides. Mol Ther. 2004;10:232–240. doi: 10.1016/j.ymthe.2004.05.031. [DOI] [PubMed] [Google Scholar]
- Fletcher S, Honeyman K, Fall AM, Harding PL, Johnsen RD., and, Wilton SD. Dystrophin expression in the mdx mouse after localised and systemic administration of a morpholino antisense oligonucleotide. J Gene Med. 2006;8:207–216. doi: 10.1002/jgm.838. [DOI] [PubMed] [Google Scholar]
- Gebski BL, Mann CJ, Fletcher S., and, Wilton SD. Morpholino antisense oligonucleotide induced dystrophin exon 23 skipping in mdx mouse muscle. Hum Mol Genet. 2003;12:1801–1811. doi: 10.1093/hmg/ddg196. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Fokkema I, Verschuuren J, Ginjaar I, van Deutekom J, van Ommen GJ, et al. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum Mutat. 2009;30:293–299. doi: 10.1002/humu.20918. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Van Deutekom JC, Fokkema IF, Van Ommen GJ., and, Den Dunnen JT. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34:135–144. doi: 10.1002/mus.20586. [DOI] [PubMed] [Google Scholar]
- Prior TW, Bartolo C, Pearl DK, Papp AC, Snyder PJ, Sedra MS, et al. Spectrum of small mutations in the dystrophin coding region. Am J Hum Genet. 1995;57:22–33. [PMC free article] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- Kinali M, Arechavala-Gomeza V, Feng L, Cirak S, Hunt D, Adkin C, et al. Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 2009;8:918–928. doi: 10.1016/S1474-4422(09)70211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helderman-van den Enden AT, Straathof CS, Aartsma-Rus A, den Dunnen JT, Verbist BM, Bakker E, et al. Becker muscular dystrophy patients with deletions around exon 51; a promising outlook for exon skipping therapy in Duchenne patients. Neuromuscul Disord. 2010;20:251–254. doi: 10.1016/j.nmd.2010.01.013. [DOI] [PubMed] [Google Scholar]
- Hoffman EP, Kunkel LM, Angelini C, Clarke A, Johnson M., and, Harris JB. Improved diagnosis of Becker muscular dystrophy by dystrophin testing. Neurology. 1989;39:1011–1017. doi: 10.1212/wnl.39.8.1011. [DOI] [PubMed] [Google Scholar]
- Koenig M., and, Kunkel LM. Detailed analysis of the repeat domain of dystrophin reveals four potential hinge segments that may confer flexibility. J Biol Chem. 1990;265:4560–4566. [PubMed] [Google Scholar]
- Carsana A, Frisso G, Tremolaterra MR, Lanzillo R, Vitale DF, Santoro L, et al. Analysis of dystrophin gene deletions indicates that the hinge III region of the protein correlates with disease severity. Ann Hum Genet. 2005;69 Pt 3:253–259. doi: 10.1046/j.1529-8817.2005.00160.x. [DOI] [PubMed] [Google Scholar]
- Baumbach LL, Chamberlain JS, Ward PA, Farwell NJ., and, Caskey CT. Molecular and clinical correlations of deletions leading to Duchenne and Becker muscular dystrophies. Neurology. 1989;39:465–474. doi: 10.1212/wnl.39.4.465. [DOI] [PubMed] [Google Scholar]
- Gillard EF, Chamberlain JS, Murphy EG, Duff CL, Smith B, Burghes AH, et al. Molecular and phenotypic analysis of patients with deletions within the deletion-rich region of the Duchenne muscular dystrophy (DMD) gene. Am J Hum Genet. 1989;45:507–520. [PMC free article] [PubMed] [Google Scholar]
- Koenig M, Beggs AH, Moyer M, Scherpf S, Heindrich K, Bettecken T, et al. The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am J Hum Genet. 1989;45:498–506. [PMC free article] [PubMed] [Google Scholar]
- Coral-Vázquez R, Arenas D, Cisneros B, Peñaloza L, Kofman S, Salamanca F, et al. Analysis of dystrophin gene deletions in patients from the Mexican population with Duchenne/Becker muscular dystrophy. Arch Med Res. 1993;24:1–6. [PubMed] [Google Scholar]
- Araki E, Nakamura K, Nakao K, Kameya S, Kobayashi O, Nonaka I, et al. Targeted disruption of exon 52 in the mouse dystrophin gene induced muscle degeneration similar to that observed in Duchenne muscular dystrophy. Biochem Biophys Res Commun. 1997;238:492–497. doi: 10.1006/bbrc.1997.7328. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Kaman WE, Weij R, den Dunnen JT, van Ommen GJ., and, van Deutekom JC. Exploring the frontiers of therapeutic exon skipping for Duchenne muscular dystrophy by double targeting within one or multiple exons. Mol Ther. 2006;14:401–407. doi: 10.1016/j.ymthe.2006.02.022. [DOI] [PubMed] [Google Scholar]
- De Angelis FG, Sthandier O, Berarducci B, Toso S, Galluzzi G, Ricci E, et al. Chimeric snRNA molecules carrying antisense sequences against the splice junctions of exon 51 of the dystrophin pre-mRNA induce exon skipping and restoration of a dystrophin synthesis in Delta 48-50 DMD cells. Proc Natl Acad Sci USA. 2002;99:9456–9461. doi: 10.1073/pnas.142302299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aartsma-Rus A, van Vliet L, Hirschi M, Janson AA, Heemskerk H, de Winter CL, et al. Guidelines for antisense oligonucleotide design and insight into splice-modulating mechanisms. Mol Ther. 2009;17:548–553. doi: 10.1038/mt.2008.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams AM, Harding PL, Iversen PL, Coleman C, Fletcher S., and, Wilton SD. Antisense oligonucleotide induced exon skipping and the dystrophin gene transcript: cocktails and chemistries. BMC Mol Biol. 2007;8:57. doi: 10.1186/1471-2199-8-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks GB, Judge LM, Allen JM., and, Chamberlain JS. The polyproline site in hinge 2 influences the functional capacity of truncated dystrophins. PLoS Genet. 2010;6:e1000958. doi: 10.1371/journal.pgen.1000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura M, Sakamoto M, Ikemoto M, Mochizuki Y, Yuasa K, Miyagoe-Suzuki Y, et al. AAV vector-mediated microdystrophin expression in a relatively small percentage of mdx myofibers improved the mdx phenotype. Mol Ther. 2004;10:821–828. doi: 10.1016/j.ymthe.2004.07.025. [DOI] [PubMed] [Google Scholar]
- Wells DJ, Wells KE, Asante EA, Turner G, Sunada Y, Campbell KP, et al. Expression of human full-length and minidystrophin in transgenic mdx mice: implications for gene therapy of Duchenne muscular dystrophy. Hum Mol Genet. 1995;4:1245–1250. doi: 10.1093/hmg/4.8.1245. [DOI] [PubMed] [Google Scholar]
- Mitrpant C, Adams AM, Meloni PL, Muntoni F, Fletcher S., and, Wilton SD. Rational design of antisense oligomers to induce dystrophin exon skipping. Mol Ther. 2009;17:1418–1426. doi: 10.1038/mt.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Yin H, Camelliti P, Betts C, Moulton H, Lee H, et al. In vitro evaluation of novel antisense oligonucleotides is predictive of in vivo exon skipping activity for Duchenne muscular dystrophy. J Gene Med. 2010;12:354–364. doi: 10.1002/jgm.1446. [DOI] [PubMed] [Google Scholar]
- Wu B, Lu P, Benrashid E, Malik S, Ashar J, Doran TJ, et al. Dose-dependent restoration of dystrophin expression in cardiac muscle of dystrophic mice by systemically delivered morpholino. Gene Ther. 2010;17:132–140. doi: 10.1038/gt.2009.120. [DOI] [PubMed] [Google Scholar]
- Reagan-Shaw S, Nihal M., and, Ahmad N. Dose translation from animal to human studies revisited. FASEB J. 2008;22:659–661. doi: 10.1096/fj.07-9574LSF. [DOI] [PubMed] [Google Scholar]
- de Lateur BJ., and, Giaconi RM. Effect on maximal strength of submaximal exercise in Duchenne muscular dystrophy. Am J Phys Med. 1979;58:26–36. [PubMed] [Google Scholar]
- Shivers RR., and, Atkinson BG. The dystrophic murine skeletal muscle cell plasma membrane is structurally intact but “leaky” to creatine phosphokinase. A freeze-fracture analysis. Am J Pathol. 1984;116:482–496. [PMC free article] [PubMed] [Google Scholar]
- Heemskerk H, de Winter C, van Kuik P, Heuvelmans N, Sabatelli P, Rimessi P, et al. Preclinical PK and PD studies on 2'-O-methyl-phosphorothioate RNA antisense oligonucleotides in the mdx mouse model. Mol Ther. 2010;18:1210–1217. doi: 10.1038/mt.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JD, Guo W, Merriam AP, Khanna S, Cheng G, Zhou X, et al. Persistent over-expression of specific CC class chemokines correlates with macrophage and T-cell recruitment in mdx skeletal muscle. Neuromuscul Disord. 2003;13:223–235. doi: 10.1016/s0960-8966(02)00242-0. [DOI] [PubMed] [Google Scholar]
- Demoule A, Divangahi M, Danialou G, Gvozdic D, Larkin G, Bao W, et al. Expression and regulation of CC class chemokines in the dystrophic (mdx) diaphragm. Am J Respir Cell Mol Biol. 2005;33:178–185. doi: 10.1165/rcmb.2004-0347OC. [DOI] [PubMed] [Google Scholar]
- Lu QL, Rabinowitz A, Chen YC, Yokota T, Yin H, Alter J, et al. Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles. Proc Natl Acad Sci USA. 2005;102:198–203. doi: 10.1073/pnas.0406700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitali P, Heemskerk H, Vossen RH, Ferlini A, den Dunnen JT, ‘t Hoen PA, et al. 2010Accurate quantification of dystrophin mRNA and exon skipping levels in Duchenne Muscular Dystrophy Lab Investepub ahead of print). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dose-escalation study of exon 51-skipping by local intramuscular injection into mdx52 mice.
Examination of adverse effects after systemic delivery of antisense oligonucleotides (AOs).
In vitro exon 51-skipping in DMD 5017 cells with deletion of exons 45–50.
Length, annealing coordinates, sequences of all AOs targeting human exon 51.