Abstract
Importance of the field
Cancer is a collection of diseases that arise from the progressive accumulation of genetic alterations in somatic cells. Genomic approaches have identified a great variety of genetic abnormalities associated with tumorigenesis, and molecular imaging and quantification assays have further elucidated the complex interactions within or between pathways. It is acknowledged that it is proteins, rather than genes, to fulfill most cellular functions; and signaling proteins largely operate through a large and complex network. To this end, cancer is mostly a pathway dysregulated disease – a small number of core pathways are dominate in aberrant cell growth leading to cancer. Thus, understanding the functional consequences of dysregulated and/or mutant signaling proteins in the context of native signaling networks is the frontier in cancer research.
Areas covered in this review
This article reviews why resonant waveguide grating (RWG) biosensor cellular assays are considered to be integrative in nature, and how RWG biosensor can be used for mining the surface markers of cancer cells, and discovering core pathway(s) of cancer receptor signaling.
What the reader will gain
The reader will gain an overview of cancer biology from pathway perspective, and have a glimpse of potential implications of integrative cellular assays, as promised by RWG biosensor, in cancer research and diagnosis.
Take home message
Successful approaches for developing next-generation anti-cancer therapies and diagnostic protocols should take into account that the dysregulation of oncogenic pathways is central to tumorigenesis. The biosensor cellular assays offer unprecedented advantage in characterizing cancer biology. However, significant challenges are also presented in deconvoluting and validating cellular mechanisms identified in cancer receptor signaling using these assays.
Keywords: Cancer signaling, cellular assays, dynamic mass redistribution, oncogene addiction, resonant waveguide grating biosensor
1. Introduction
Human cancer is considered to be a pathway dysregulated disease (1). The ability of tumor cells to outgrow their neighboring cells is often driven by constitutive activation of downstream proteins (2, 3). Genetic studies over several decades have discovered a wide range of tumor-associated genes and their mutations, many of which preferentially occur in signaling proteins involved in a small number of pathways (4–10). Genetic mutations are often enriched in positive regulatory loops (gain of function), and methylated genes in negative regulatory loops (loss of function), leading to the disruption of the normal cooperative behavior of cells and thus promoting tumor phenotypes (11, 12). A hallmark in the onset of cancer is how mutated proteins alter and govern signaling of cancer cells in the context of intracellular or intercellular signaling networks (13–15). Despite advances in the discovery of genetic and epigenetic mutations as well as in the molecular delineation of oncogenic pathways, little is known about the systems biology of cancers and how oncogenic signaling alters the propagation routes of a diverse array of receptor pathways in cancer. Therefore, means to investigate and characterize oncogenic pathways are crucial for further elucidating the molecular mechanisms of cancer, and developing next generation molecular target-based therapies.
One promising approach is resonant waveguide grating (RWG) biosensor cellular assay, which has recently emerged as a valuable technology for studying cell signaling in a variety of disease states including cancers (16–20). RWG biosensor measures non-invasively a diverse array of cellular responses, resulting in dynamic mass redistribution (DMR) signatures of cell biology, receptor signaling and drug pharmacology (21–22). Given its high sensitivity and integrative nature in measurements, RWG biosensor can be used as a platform for characterizing cancer signaling, including mining functional receptors, discovering core pathways downstream cancer receptor signaling, and characterizing cancer cells via drug responsiveness.
2. Resonant waveguide grating biosensors
RWG biosensor belongs to a family of label-free optical biosensors that is sensitive to alterations in local refractive index at or near the sensor surface (23). A RWG biosensor consists of three components: a biological component, a detector element, and a transducer associated with both components (Fig. 1).
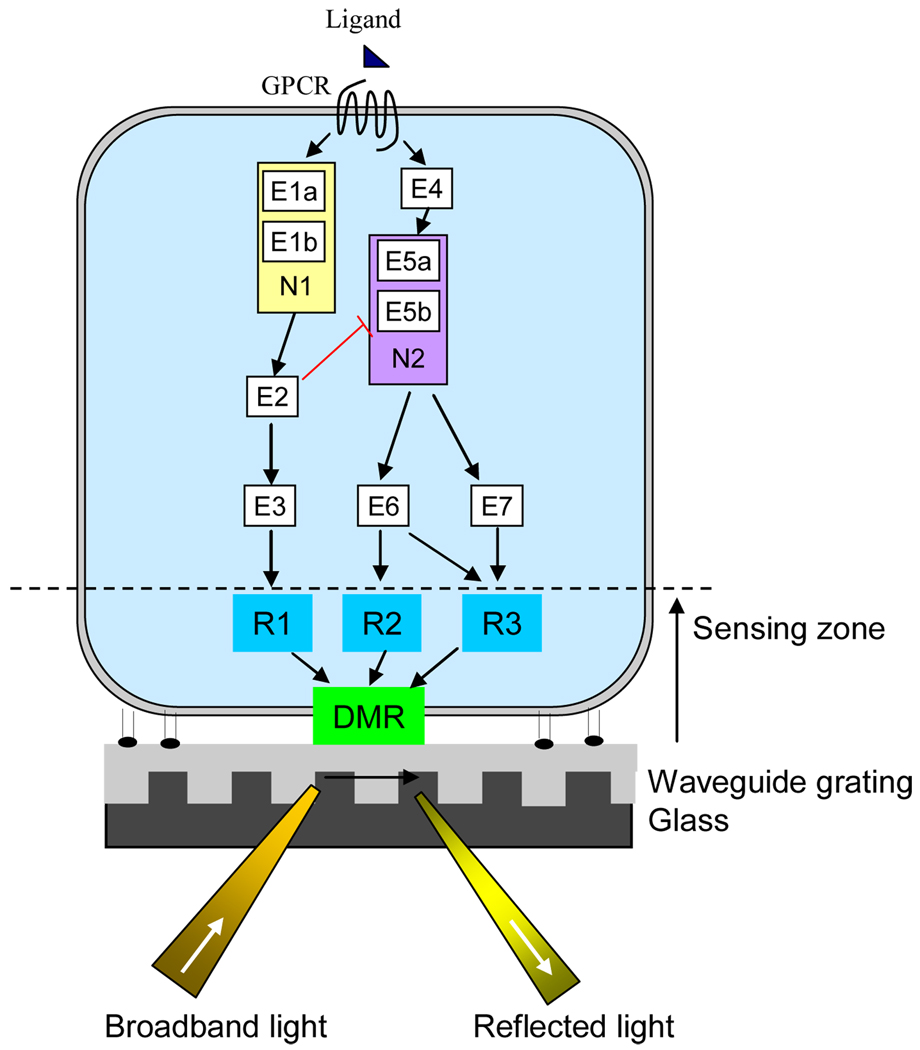
Figure 1.
Resonant waveguide grating biosensor for studying cancer signaling. The biosensor uses its intrinsic evanescent wave to characterize cancer signaling in live cells. Combining with chemical biology and molecular genetic approaches, it is possible to decode the core pathway(s) and critical nodes in receptor signaling in a cancer cell. An example is that binding of a GPCR agonist to its cognate receptor leads to two major pathways: the first one consists effectors E1a, E1b, E2, E3, leading to response R1, while another consists of E4, E5a, E5b, E6, and E7, leading to responses R2 and R3. The integration of the responses R1, R2 and R3 when occurred within the sensing zone of the biosensor leads to the DMR. The effectors E1a and E1b form the critical node N1, while E5a and E5b form another critical node N2. The intervention of these critical nodes by small molecules or genetic manipulations would have greater impact on the receptor DMR signal, than other signaling components.
The biological component is a live cell or a tissue cell for whole cell sensing. Anchorage dependent cell can be directly cultured onto the transducer surface to form an adherent layer of cells (16). Suspension cells (e.g., lymphoblastic leukemia cells) can be brought to closely in contact with the transducer surface via physical sedimentation or specific biochemical binding between the immobilized molecule and a cell surface molecule (17).
The RWG transducer is a nano-grating waveguide (24). For whole cell sensing, the biosensor is often considered as a three-layer waveguide configuration: a substrate with a diffractive grating, a high index of refraction waveguide coating, and a cell layer. Such a configuration supports both transverse magnetic (TM0) and transverse electric (TE0) modes. The TM0 mode has higher sensitivity and longer penetration depth (i.e., larger sensing volume) but relatively lower spatial resolution (~ tens of micrometers), comparing to the TE0 mode (21, 24). Thus, most RWG biosensors use the TM0 mode for whole cell sensing. The penetration depth is the distance from the sensor surface at which the electric field strength has decreased to 1/e of its initial value. The penetration depth of a biosensor can be variable, dependent on detection scheme (21, 24) and the biosensor configuration (25). The electromagnetic field, termed evanescent wave, is created by the diffraction grating coupled waveguide resonance (21). This indicates that the biosensor only samples the bottom portion of the cells contacting with the sensor surface.
The RWG detector exploits resonant coupling of light into a waveguide via the diffraction grating (23). When illuminated with broadband light at a fixed and nominally normal angle of incidence, these sensors reflect only a narrow band of wavelengths (resonant wavelength) that is a sensitive function of the local index of refraction of the biosensor (17). Since the local index of refraction is directly proportional to the density and distribution of biomass (e.g., proteins, molecular complexes) in live cells (26), the RWG can non-invasively detect stimulus-induced DMR in native cells. The DMR defines redistribution of cellular matters within the sensing volume. Such a redistribution is often not random; instead, it is tightly regulated and is often dynamic both spatially and temporally (27, 28). The biosensor simply acts a non-invasive monitor to record the DMR in real time. The DMR contains high information, and multiple parameters can be derived from a DMR signal and used for characterizing receptor signaling (24) and drug pharmacology (29). The DMR is common to almost all types of cells, and many (if not all) receptor signaling and cellular processes. This is because cell signaling often involves protein trafficking, microfilament remodeling, cell adhesion alterations and morphological changes of cells, all of which can lead to DMR. However, since cells vary in the relative stoichiometries of intracellular signaling components and the DMR assays detect such differences, the activation of a receptor may result in cellular background-dependent phenotypic responses. Therefore, it is not surprising to see in recent years that RWG biosensor cellular assays have found broad applications to a diverse array of cellular processes, including adhesion (22, 30), viral infection (31), proliferation (32) and apoptosis (33) of cells. These assays are also amenable to a wide range of receptors, including G protein-coupled receptors (GPCRs) (34, 35), ion channels (36), kinases (24, 37), enzymes (38), and structural proteins (39). Numerous studies have found that the DMR measurements are pathway-sensitive, and often reflect the complexity of receptor biology (40–45) and drug pharmacology (29, 46–48). In general, a DMR signal may contain contributions from protein trafficking, microfilament remodeling, and cell adhesion alterations (21), but different events may dominate different DMR signals. Thus it is possible to identify many critical nodes and core pathways in receptor signaling network (49) (Fig.1).
RWG biosensor systems including Epic® and BIND™ are commercially available nowadays (49). Both systems employ the wavelength interrogation configuration, in which a broadband light source is used for illumination, and the wavelength of the reflected light is recorded (17). Such a configuration is amenable to high throughput screening (HTS) since conventional HTS often uses microtiter plates having large footprint. Alternative angular interrogation configurations have also been explored, in which a single wavelength light at variable angles is used to illuminate the biosensor (21, 22, 24, 32). For example, optical waveguide lightmode spectroscopy (OWLS) monitors the incoupling angles by continuously changing the incident angle of the light and measuring the incoupled light intensity with a photodetector placed at the edge of the waveguide. The OWLS and other angular interrogation systems often have limited capability for parallel biosensing applications (17).
The Epic® system (Corning Inc) is the first optical biosensor that is amenable to microtiter plate-based HTS for both biomolecular interaction analysis and cell-based assays (17, 50). The system consists of a RWG detector, an external liquid handling accessory and a scheduler, such that it can process large numbers of microplates using end-point measurements for HTS, or using kinetic measurements for high information content screening. The detector utilizes a linear array of fiber optics to rapidly scan a whole microtiter plate, and to track changes in the central wavelength (resonant wavelength) of the biosensor resonant spectrum. However, since the system is a standard-alone reader system, full integration with external liquid handler is prerequisite for effective DMR assays. Furthermore, since the system is stationary without any CO2 control unit and maintained at 26°C for its internal temperature, it is obviously difficult to study cell signaling at variable temperatures, particularly under physiological conditions. Epic® biosensor microplates (typically 384well format) have appropriate surface coatings for different applications.
3. Probing cancer biology with small molecules
Advances in molecular genetics have identified numerous molecules that control living systems (51). Specific proteins can be suppressed or eliminated in living system via RNAi knockdown or gene knockout, respectively. Specific proteins can also be increased in concentration via gene expression, or altered via mutagenesis. Determining the functional consequences of these manipulations has greatly assisted our understanding of various diseases including cancer (52). Equally important is the use of small molecules for deciphering the molecular mechanisms underlying cancer development (53). Cellular interventions with small molecules are advantageous, because of the easiness in administration, the ability to temporally control the system, and the wide accessibility and coverage in biological target space of diverse small molecules. Furthermore, it is possible for small molecules to selectively perturb one of the multifaceted functions of a protein, resulting in a level of understanding of protein function that would not be possible through gene-based perturbation (54). Thus, small molecules can be used as probes to systematically map biological-activity space and to understand a cell system including cancer cells. Combining conventional molecular characterization assays (e.g., assays for monitoring alterations in quantity, location and phosphorylation state of signaling proteins) with small molecule probes will further enable mechanistic deconvolution and validation of cellular mechanisms of cancer signaling.
Judicious collections of small molecules can be made to modulating the functions of many different proteins, all in one experiment using HTS technologies. The assembly of ideal small molecule library is possible, partly because recent genomic landscape studies have identified many protein classes that are important to tumorigenesis (4–8), and partly because diverse small molecule modulators are available for many of these proteins. More important is that although cancer is highly heterogeneous, only a small number of core pathways are essential to cancer development (1, 5). This means that a relatively small library may be sufficient for characterizing cancer. Libraries can be assembled from available compounds with known effects on specific proteins and phenotypes, such as modulators for GPCRs, proteases, phosphatases, ion channels and kinases.
Phenotypic screens using chemical genetics have been used to characterize cancer cell biology (55). Phenotypic readouts, such as proliferation and migration, are often used as a metrics to characterize the activity and specificity of compounds, whereas the patterns of different compounds acting on a cancer cell are indicative for the type of cancers. Weinstein et al. examined the impacts of distinct chemicals on the proliferation of a panel of 60 tumor cell lines, and found that compounds with similar structures or similar mechanisms of action had similar phenotypic profiles (i.e., similar ability to inhibit the growth of a similar set of tumor cell lines) (56). However, these phenotypic screening assays are inherently vague, partly due to their readouts that are largely long term cellular responses (e.g., alterations in gene expression and/or growth rate), and partly due to the presence of many compensatory pathways that may lead to similar long-term responses. Since the DMR assays often measure early events in cell signaling and receptor activity is reported as a DMR signal indicative of pathways involved, combining multi-parametric biosensor cellular assays with small molecules should allow systematic mapping of biological-activity space in cancer, and thus greatly accelerate our understanding of cancer biology.
4. Mining the surface markers of cancer cells
Signal transduction originates at the cell surface membrane. Membrane bound proteins are highly abundant, comprising more than a third of all cellular proteins. These receptors are central to signal transduction, and many of them are often up-regulated or mutated in cancer cells (57). Mining the surface proteome of cancer cells is useful for identifying markers to classify tumors, to monitor disease progression, regression and recurrence, and to assess the responsiveness of cancer cells to therapy. Traditionally, cancer signatures are identified mostly via gene expression analysis which is inherently more detail-rich and specific than phenotypic assays. However, since DMR assays provide a functional and global readout of receptor signaling involving altered and/or mutated proteins in cancer cells, they offer a complementary view of cancer biology.
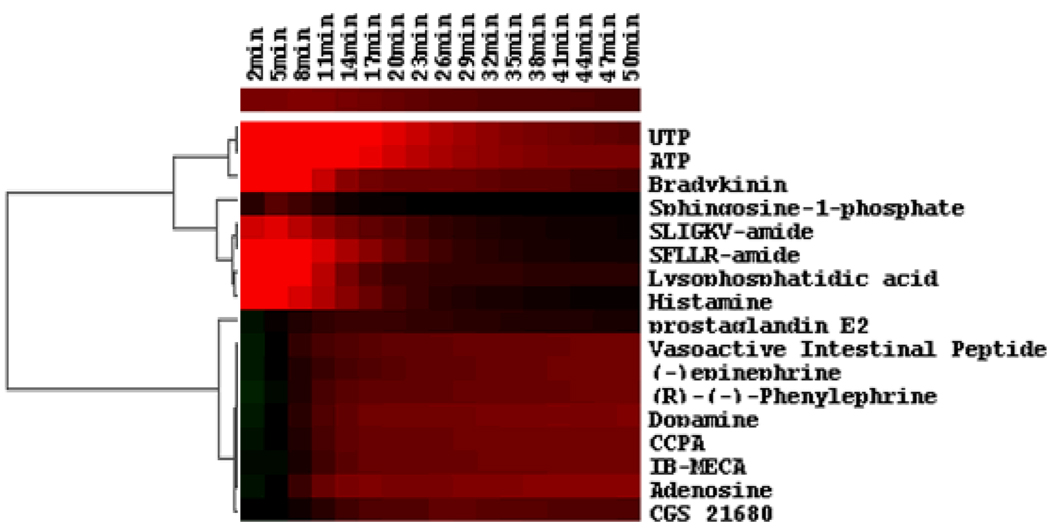
Large scale surveying functional receptors in cancer cells is feasible using RWG biosensor cellular assays (21, 34, 48). For example, to mine endogenous and functional GPCRs, a library of agonists for most of all known GPCRs (~220) can be assembled and used to stimulate a cancer cell. The DMR signal of each agonist can be recorded independently in real time. The patterns of DMR signals obtained can be clustered using similarity analysis – a technology that is widely used in gene expression analysis (49, 58). Results showed that the human skin carcinoma cell line A431 cells respond to a quite few of GPCR agonists (33, 40, 48, data unpublished). Figure 2 showed representative DMR signals, each with distinct characteristics (e.g., shape, kinetics, duration, and amplitude). For a subset of agonists tested, the heat-map generated with Hierarchical Euclidean clustering indicated that beside these agonists that did not trigger any obvious DMR signals, the other known GPCR agonist-induced DMR signals can be classified into two classes at the low resolution, each several sub-clusters. Based on the heat map shown in Fig. 3, a Gs-type DMR signal (29) was observed for the three known Gs-coupled β2-adrenergic receptor agonist epinephrine, dopamine and phenylephrine (29), and the four Gs-coupled adenosine A2B receptor agonist adenosine, IB-MECA, CCPA and CGS21680 (34), the Gs-coupled VIP1 agonist vasoactive intestinal peptide, and the Gs-coupled EP4 agonist prostaglandin E2 and prostaglandin D2. On the other hand, a distinct Gq-like DMR signal (34, 48) was observed for the Gq-coupled P2Y2 receptor agonist ATP and UTP (34), the Gq-coupled histamine receptor H1R agonist histamine (50), the Gq-dominant bradykinin B2 receptor agonist bradykinin (40), the Gq-dominant S1P receptors agonist sphinosine-1-phosphate, the Gq-dominant LPA receptors agonist LPA (34), and the Gq-dominant protease activated receptor PAR1 agonist SFLLR-amide and PAR2 agonist SLIGKV-amide (48). These functional DMR responses are correlated well with the expression of their corresponding receptor(s). These results suggest that DMR assays faithfully report functional receptors in the cells. However, it is known that many ligand molecules display polypharmacology. Thus, the assignments of ligand DMR signals to specific receptors need to be validated via several methods. First, a relatively large set of ligands for a specific receptor should be used, and their DMR signals can be compared to determine whether the receptor is expressed and functional in the cell. Second, pharmacology characterization using antagonists can also be used to determine the specificity of the agonist-receptor pair. Third, gene expression and gene manipulation (e.g., gene transfection or RNAi knockdown) can further validate the specificity of receptor-ligand pair.
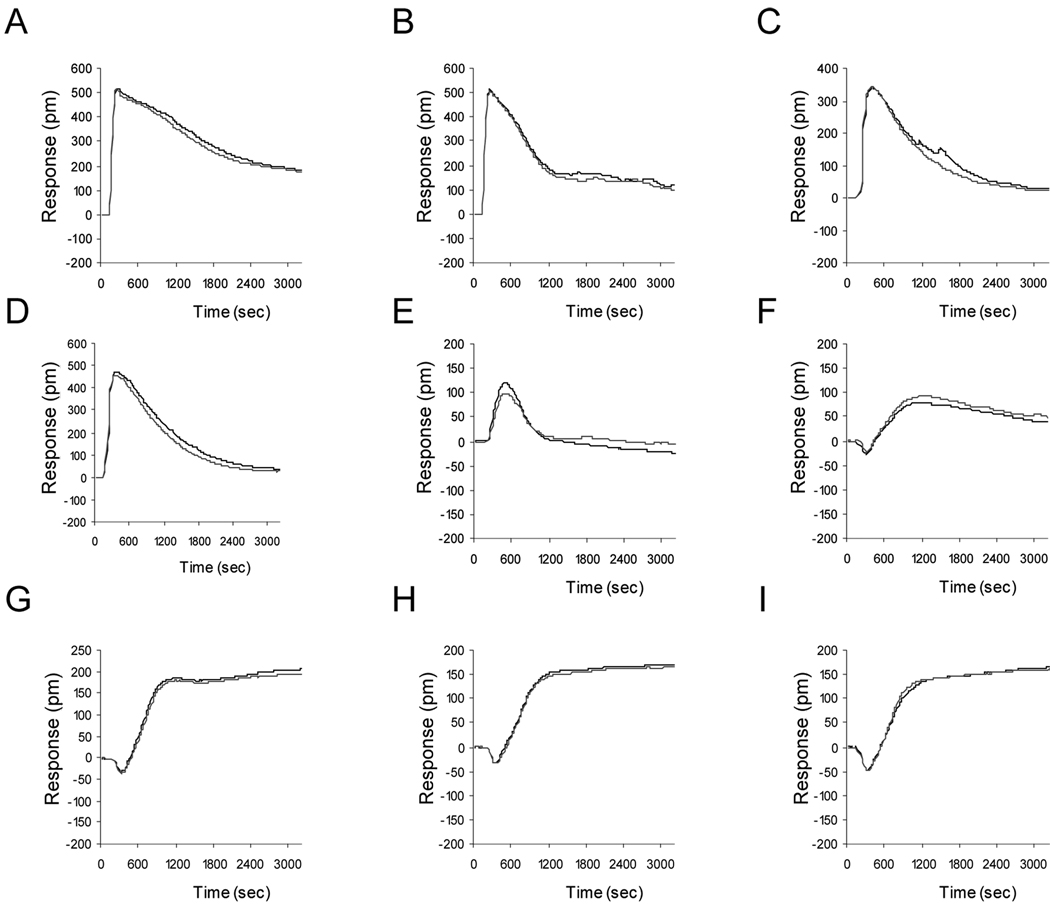
Figure 2.
Representative DMR signals of A431 cells upon stimulation with distinct GPCR agonists. The GPCR agonists include (A) ATP for the Gq-coupled P2Y2 receptor; (B) bradykinin for the Gq-dominant bradykinin B2 receptor; (C) SLIGKV-amide for the Gq-dominant protease activated receptor PAR2; (D) SFLLR-amide for the Gq-dominant protease activated receptor PAR1; (E) sphinosine-1-phosphate for the Gq-dominant S1P2 and S1P5 receptors; (F) PGE2 for the Gs-coupled prostaglandin EP4 receptor; (G) and (H) adenosine and CCPA, respectively, both for the Gs-coupled adenosine A2B, and (I) phenylephrine for the Gs-coupled β2-adrenergic receptor. Each agonist was assayed at 10 µM in duplicate. The A431 cells were cultured using serum medium overnight, followed by 20hr starvation with serum-free medium. After washing with assay buffer, the confluent A431 cells were directly assayed with Epic® system.
Figure 3.
The heat map classification of the DMR signals of quiescent A431 cells induced by diverse GPCR agonists. The heat map was generated using the Euclidean hierarchical cluster analysis (ref. 48–57). The real responses of all DMR signals at discrete time points, as indicated, were used as the basis for similarity analysis. The amplitude and direction is indicated by color – the red indicates a positive value, the green a negative value, and the black a value close to zero.
RWG biosensor is also amenable to label-free cell attachment assays for identifying the presence of specific surface markers of cancer (30). Here, probe molecules that recognize specific antigens presented at the cancer cell surface are pre-immobilized onto the sensor surface, and are used as fishing baits to detect, and enrich the antigen presenting cancer cells. For example, a RWG biosensor has been used to identify specific antigen positive cells, including carcinoembryonic antigen (CEA) expressing cells (59). CEA is a glycosyl phosphatidyl inositol-cell surface anchored glycoprotein involved in cell adhesion, and is one of the most widely used tumor markers worldwide. CEA testing is mainly used as a tumor marker to identify recurrences after surgical resection, or localize cancer spread, particularly for gastrointestinal and colorectal malignancy (60).
5. Probing core pathway(s) in cancer receptor signaling via chemical tools and RWG biosensor readouts
Common to cancers are the hallmarks related to survival and proliferation in foreign environments (13), evading immune surveillance (14), and stress phenotypes (15). Tumorigenesis starts with a single cell, and arises through a multistage, mutagenic process that involves genetic alterations resulting in the gain-of-function mutation, amplification, and/or over-expression of key oncogenes, and the loss-of-function mutation, deletion, and/ or epigenetic silencing of key tumor suppressors (11). Gain-of-function mutations in positive regulatory loops directly lead to hyperactivity and tumor aberrant growth, whereas loss-of-function mutations in negative regulatory loops result in the removal of the restraints necessary to prevent aberrant growth and survival or genomic instability, thus leading to tumorigenesis (61–64). Cancer cells also induce angiogenesis and metastasis which are important to clinical manifestation of cancer (65).
The hallmark of cancer is its complexity in genetic and epigenetic abnormalities (1). In many cancers, there are a few of frequently mutated oncogenes and tumor suppressors such as PI3K, Ras, p53, and PTEN. For example, genetic alterations in the PIK3CA gene encoding p110a PI3K and in related pathway genes are presented in >30% of colon and breast cancers (4). Deletion or epigenetic silencing of tumor suppressor genes is also evident in many cancers. An example is the deletion of the tumor suppressor and lipid phosphatase PTEN (66). PTEN normally acts to constrain PI3K signaling, and thus cancer bearing PTEN deletion is likely to be sensitive to PI3K inhibitors. Malignant carcinomas also harbor a complex combination of infrequent mutations, many of which are thought to drive the cancer phenotype (4–7). Stratton and colleagues estimate that individual mutations in as many as 20% of all kinases can play an active role in tumorigenesis (67).
Cancer is largely a pathway dysregulated disease (1, 5). Some cancers apparently depend on the continued activity of certain oncogenes for maintenance of the malignant phenotype (2). Evidence for such oncogene addiction is amounting. First, in a transgenic mouse model, switching on the c-myc oncogene in the hematopoietic cells led to the development of T-cell and myeloid leukemias. However, the phenotype of leukemia cells was reversed upon myc silencing (68). Similar was found in mouse models for c-myc-driven skin papillomas, and osteosarcomas (69, 70), and for BCR-ABL-induced leukemia (71). In human colorectal cancer cells bearing a K-Ras mutation, somatic knockout of this oncogene led to reversion of the transformed phenotype and abrogated the ability of these cells to form tumors in nude mice (72). Second, global surveying several tumors including breast and colorectal cancers at genomic level has found that many genetic abnormalities are mostly associated with a small set of biological processes and biochemical pathways, and different tumors share some common dysregulated pathways (4–7). Cancer signaling network mapping studies also revealed two oncogene-signaling blocks that are enriched in gene mutations and tend to collaborate in most tumor types – p53 (composed of p53, p14, Rb, BRAC1 and BRAC2, etc.) and Ras (Ras, PI3K and EGFR, etc.) blocks (73). In 592 tumors analyzed, at least 2 signaling gene mutations, one from the p53 block and the other from the Ras block, are necessary for tumorigenesis and further support the notion that both the prevention of cell death (p53 block) and the promotion of cell proliferation (Ras or other blocks) are necessary to generate most tumors. The third, and the most convincing, evidence is from some experimental research and clinical settings in which molecule targeted therapy has demonstrated therapeutic efficacy – the inactivation of a single or a few of oncogenes appears largely impair the growth and survival of cancer (15). Successful examples include the receptor tyrosine kinase HER-2 antagonizing antibody drug Herceptin for breast cancer (74), and several oncogenic protein kinase inhibitor drugs such as imatinib/Gleevec (75), gefitinib (76) and erlotinib (77). Imatinib targets the bcr-abl oncogene in chronic myeloid leukemia and also targets the c-kit oncogene in gastrointestinal stromal tumors. Gefitinib and erlotinib target the EGFR in non–small cell lung carcinoma, pancreatic cancer, and glioblastoma. Taken together, these studies suggest that a limited number of central molecular pathways are crucial to cancer development. Because of that as well as the clinical efficacy of molecule targeted therapy is tied to accurate identification of the state of oncogene addiction in specific cancers, RWG biosensor cellular assays provide alternative means to identify and better define these core pathways.
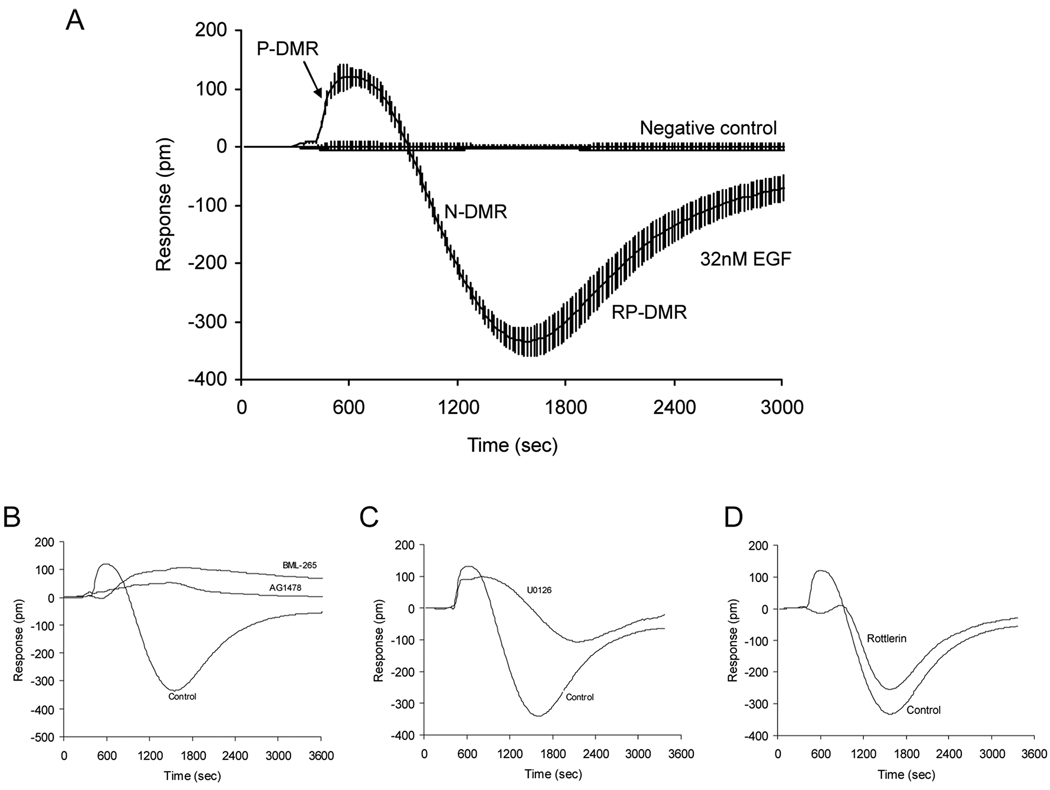
Epidermal growth factor receptors (EGFR) belong to a family of receptor tyrosine kinases, and are one of the most frequently mutated proto-oncogenes in many cancers (57). RWG biosensor cellular assays were used to map the signaling and its network interactions of EGFR in A431 cells (24). A431 is well-known for its over-expression of EGFR. A previous study using the EGF DMR signal in A431 cells as a readout showed that EGFR signaling is cellular status dependent – quiescent cells respond more robustly to EGF than proliferating cells. Chemical biology studies, based on the modulation profiles of an array of known modulators, also linked several targets and cellular processes to the EGFR signaling. The EGFR signaling was found to require its intrinsic tyrosine kinase activity and to be mostly originated from the internalized receptors. The EGFR signaling also led to actin remodeling, dynamin and clathrin dependent receptor internalization, and MEK pathway-mediated cell detachment (possibly via FAK). To further determine the core pathways of EGFR signaling, a judicious selection of kinase inhibitors was made to examine their impacts on the EGF DMR signal in quiescent A431 cells (data unpublished). Figure 4 shows the different sensitivity of the EGF DMR signal in quiescent A431 cells to distinct kinase modulators. As expected, the EGFR tyrosine kinase inhibitors, A1478 and BML-265, almost completely blocked the EGF signal. However, the MEK1/2 inhibitor U0126 selectively attenuated the late DMR event, and the protein kinase C (PKC) inhibitor rottlerin selectively blocked the early DMR event. These results suggest that distinct pathways preferentially occur at different time domain during signaling – the PKC pathway plays important role in the early response, but the MAPK pathways dominate in the late cellular response upon the activation of EGFR.
Figure 4.
The characteristics of the EGF DMR signal in quiescent A431 cells. (A) Real time EGF DMR signal in comparison with the negative control (i.e., the response of cells stimulated with the assay vehicle only), each error bar representing the standard deviation of 32 replicates. The EGF DMR proceeds in three phases: an initial positive-DMR (P-DMR), followed by a negative-DMR (N-DMR) and a recovery positive DMR (RP-DMR). (B) to (D) The sensitivity of the EGF DMR signal to different modulators, AG1478 and BML-265 (B), U0126 (C) and rottlerin (D). The control is the EGF response of cells pretreated with the assay vehicle only. In all experiments, EGF was at 32nM, whereas the rest compounds used to pretreat the cells were at 10µM.
In another recent RWG biosensor study, Du and her colleagues found that distinct cancer cell lines responded differently to EGF – squamous cell carcinoma of the head and neck (SCCHN) cell line UPCI-37B exhibited a rapid rise in DMR signal, while lung adenocarcinoma cell line A549 showed a biphasic DMR profile (37). Pathway deconvolution using chemical biology approach revealed that the EGF-induced DMR signal in the SCCHN cell is insensitive to inhibitors targeting the Ras/Raf/MAPK pathway, but is completely blocked by the two PI3K inhibitors LY 294002 and wortmannin, indicating that PI3K is a critical mediator of the EGFR signal in this cancer cell. Moreover, they found that the EGF-induced DMR signal in SCCHN cancer cells can also be completely suppressed by the two EGFR inhibitor drugs, gefitinib and erlotinib. Both gefitinib and erlotinib have been approved by the FDA for the treatment of NSCLC and are under phase II clinical evaluation for use in SCCHN (78). These results suggest that the biosensor cellular assays are applicable to many oncogenic pathways for the discovery of novel therapeutic agents targeting various cancers. It is worthy noting that small kinase inhibitors are well-known for their polypharmacological properties. Thus, several inhibitors for a single kinase should be used, together with conventional cell biology studies, to ascertain the roles and specificity of the kinase in receptor signaling (24).
6. Conclusions
Optical biosensors including RWG biosensors have evolved from a research tool for biomolecular interaction analysis to a high throughput and high content screening platform for whole cell sensing. Together with chemical biology and chemical genetics, these biosensors enable systematically mining endogenous receptors in cancer cells, and to elucidate critical nodes and core pathways of cancer signaling networks. The ability to assay endogenous receptors without any manipulations makes it possible to characterize cancer cells including primary cells, and to identify responsive therapeutics for specific cancers. Next generation RWG biosensors will have higher spatial resolution (25, 79–80), enabling non-invasively measure cancer signaling in single cells including rare cancer cells, and in mixed populations of cancer cells such as tissue cells, reprogrammed cells, and unpurified primary cells.
7. Expert opinion
Cancer is the collection of complex genetic diseases. The molecular target-based therapies for cancer today is primarily for preexisting diseases, typically late in their progression. Next generation anti-cancer medicine will move toward predictive and preventive modes. This requires understanding of cancer development at the levels of genome, systems biology and single cell. Advances in gene sequencing technologies will allow individuals to have their genomes sequenced, which, in turn, allow mapping the genomic landscape of potential genetic alterations. Multi-parametric molecular diagnostics via blood analysis will make early diagnosis of cancer possible and will become a routine procedure in clinical laboratories. Advances in systems biology and systems pharmacology approaches will enable the extensive correlations of genetic alterations with cancer development, determine the unlined mechanisms separating normal from pathological processes, and identify the personalized therapies. Improvements in single cell assay technologies will allow characterization of cancerous cells at early stages in the context of dysregulated pathways and drug responsiveness.
Understanding the systems biology and pharmacology is the new frontier in cancer research. Conventional genomic approaches have been very fruitful for the discovery of genetic and epigenetic mutations (1–6). Since it is proteins, but not genes, that fulfill most biological functions of cells, the functional consequences of these genetic abnormalities are still largely uncharted by these approaches. The wide adoption of recombinant DNA technologies has made molecular characterization assays possible to delineate many oncogenic pathways. However, it is the large and complex network, rather than linear pathways, in which signaling proteins mostly operate. The integrative cellular assays, as promised by RWG biosensor, open new possibility to bridge mutated/dysregulated targets, pathways and cellular phenotypic responses associated with cancer. The biosensor cellular assays are well-suited for pathway profiling, since the DMR signal is common to many cellular processes, and signaling downstream many receptors. The target/pathway specificity can be achieved by chemical biology approaches – together with increasing numbers of small molecules for expanded biological space as well as incorporation of molecular characterization assays, the wide uses of small molecules will improve the resolution and quality of DMR assays. The DMR signal also represents a novel phenotypic response of cells, and is associated with many cellular processes related to cancer biology including signaling, trafficking and metastasis. It becomes clear that these assays not only allow systematic determination of signaling proteins and core pathways pathologically relevant to cancer, but also enable detailed characterization of cancerous cells and drug responsiveness.
However, RWG biosensor cellular assays are still in infancy. A challenge in adopting the biosensor cellular assays is that RWG biosensor, label-free biosensor in general, is largely non-specific. Many cellular processes and pathways can result in detectable DMR signals. However, due to the widely existed polypharmacology of many drug molecules, and the limited numbers of possible DMR signals which are obviously much less than the rich biological-activity spaces, the exact cellular mechanisms that lead to detectable DMR signal often need to be deconvoluted. Thus, novel assay designs and methodologies are required to further advance biosensor cellular assays for both basic research and industrial applications.
Article highlights
Functional genomics and gene sequencing has led to discovery of a wide array of genetic abnormalities. Functional studies have helped to sort out pathogenically relevant alterations. As our understanding of cancer development advances, it has become clear that cancer is a pathway dysregulated disease.
The ability to study mutated and/or dysregulated signaling proteins in the native signaling networks is crucial to advance cancer research, to develop next generation therapies and diagnostic protocols.
Resonant waveguide grating biosensor cellular assay emerged recently as a promising platform for cancer research – it allows systematically surveying functional receptors and pathways, and determining core pathways downstream cancer receptor signaling, in combination with chemical and biological tools.
Assay methodologies in development, together with next-generation biosensor technologies, will allow the characterization of primary cancer cells and cancer stem cells at high resolution and with high information content.
This box summarizes key points contained in the article.
Acknowledgments
Declaration of interest:
Y Fang is an employee and shareholder of Corning Inc and was partially supported by a National Institutes of Health Grant 5U54MH084691.
Abbreviations
- GPCR
G protein coupled receptor
- EGFR
epidermal growth factor receptor
- DMR
dynamic mass redistribution
- RWG
resonant waveguide grating
- HTS
high throughput screening
Bibliography
- 1.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 2.Weinstein IBCancer. Addiction to oncogenes–the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 3.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- 4.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 5.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Nature. 2008;455:1061–1068. [Google Scholar]
- 7.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlabach MR, Luo J, Solimini NL, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin J, Gan CM, Zhang X, et al. A multidimensional analysis of genes mutated in breast and colorectal cancers. Genome Res. 2009;17:1304–1318. doi: 10.1101/gr.6431107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Velculescu VE. Defining the blueprint of the cancer genome. Carcinogenesis. 2008;29:1087–1091. doi: 10.1093/carcin/bgn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahn WC, Weinberg RA. Modelling the molecular circuitry of cancer. Nat Rev Cancer. 2002;2:331–341. doi: 10.1038/nrc795. [DOI] [PubMed] [Google Scholar]
- 12.Gymnopoulos M, Elsliger MA, Vogt PK. Rare cancer-specific mutations in PIK3CA show gain of function. Proc Natl Acad Sci USA. 2007;104:5569–5574. doi: 10.1073/pnas.0701005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 14.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang Y. Label-free cell-based assays with optical biosensors in drug discovery. Assays Drug Dev Technol. 2006;4:583–595. doi: 10.1089/adt.2006.4.583. [DOI] [PubMed] [Google Scholar]
- 17.Fang Y. Non-invasive optical biosensor for probing cell signaling. Sensors. 2007;7:2316–2329. doi: 10.3390/s7102316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenakin T. Interrogating 7TM receptors: does texture in the question yield greater texture in the answer? J Receptors Signal Transduction. 2009;29:132–139. doi: 10.1080/10799890903050829. [DOI] [PubMed] [Google Scholar]
- 19.Kenakin T. Cellular assays as portals to seven-transmembrane receptor-based drug discovery. Nat Rev Drug Discov. 2009;8:617–626. doi: 10.1038/nrd2838. [DOI] [PubMed] [Google Scholar]
- 20.Fang Y, Frutos AG, Verkleeren R. Label-free cell assays for GPCR screening. Comb Chem HTS. 2008;11:357–369. doi: 10.2174/138620708784534789. [DOI] [PubMed] [Google Scholar]
- 21.Fang Y, Ferrie AM, Fontaine NH, et al. Resonant waveguide grating biosensor for living cell sensing. Biophys J. 2006;91:1925–1940. doi: 10.1529/biophysj.105.077818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramsden JJ, Horvath R. Optical biosensors for cell adhesion. J Receptors Signal Transduction. 2009;29:211–223. doi: 10.1080/10799890903064119. [DOI] [PubMed] [Google Scholar]
- 23.Tiefenthaler K, Lukosz W. Sensitivity of grating couplers as integrated-optical chemical sensors. J Opt Soc Am B. 1989;6:209–220. [Google Scholar]
- 24.Fang Y, Ferrie AM, Fontaine NH, Yuen PK. Characteristics of dynamic mass redistribution of EGF receptor signaling in living cells measured with label free optical biosensors. Anal Chem. 2005;77:5720–5725. doi: 10.1021/ac050887n. [DOI] [PubMed] [Google Scholar]
- 25.Horvath R, Cottier K, Pedersen HC, Ramsden JJ. Mutlidepth screening of living cells using optical waveguide. Biosensors Bioelectronics. 2008;24:799–804. doi: 10.1016/j.bios.2008.06.059. [DOI] [PubMed] [Google Scholar]
- 26.Barer R, Joseph S. Refractometry of living cells. Part I. basic principles. Quart J Microsc Science. 1954;95:399–423. [Google Scholar]
- 27.Kholodenko BN. Four-dimensional organization of protein kinase signaling cascades: the roles of diffusion, endocytosis and molecular motors. J Exper Biol. 2003;206:2073–2082. doi: 10.1242/jeb.00298. [DOI] [PubMed] [Google Scholar]
- 28.Shumay E, Gavi S, Wang HY, Malbon CC. Trafficking of β2-adrenergic receptors: insulin and β2-agonists regulate internalization by distinct cytoskeletal pathways. J Cell Sci. 2004;117:593–600. doi: 10.1242/jcs.00890. [DOI] [PubMed] [Google Scholar]
- 29.Fang Y, Ferrie AM. Label-free optical biosensor for ligand-directed functional selectivity of acting on β2 adrenoceptor in living cells. FEBS Lett. 2008;582:558–564. doi: 10.1016/j.febslet.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 30.Fang Y. Label-free biosensor cellular assays for cell adhesion. J Adhesion Sci Technol. 2010;24:1011–1021. [Google Scholar]
- 31.Owens RM, Wang Q, You JA, et al. Real-time quantitation of viral replication and inhibitor potency using a label-free optical biosensor. J Receptors Signal Transduction. 2009;29:195–201. doi: 10.1080/10799890903079919. [DOI] [PubMed] [Google Scholar]
- 32.Ramsden JJ, Li SY, Prenosil JE, Heinzle E. Optical method for measurement of number and shape of attached cells in real time. Cytometry. 1995;19:97–102. doi: 10.1002/cyto.990190202. [DOI] [PubMed] [Google Scholar]
- 33.Voros J, Graf R, Kenausis GL, et al. Feasibility study of an online toxicological sensor based on the optical waveguide technique. Biosensors Bioelectronics. 2000;15:423–429. doi: 10.1016/s0956-5663(00)00102-0. [DOI] [PubMed] [Google Scholar]
- 34.Fang Y, Li G, Ferrie AM. Non-invasive optical biosensor for assaying endogenous G protein-coupled receptors in adherent cells. J Pharmacol Toxicol Methods. 2007;55:314–322. doi: 10.1016/j.vascn.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Henstridge CM, Balenga NAB, Schroder R, et al. GPR55 ligands promote receptor coupling to multiple signalling pathways. Br J Pharmacol. 2010;160:604–614. doi: 10.1111/j.1476-5381.2009.00625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flemin MR, Kaczmarek LK. Use of optical biosensors to detect modulation of Slack potassium channels by G protein-coupled receptors. J Receptors Signal Transduction. 2009;29:173–181. doi: 10.1080/10799890903056883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du Y, Li Z, Li L, et al. Distinct growth factor-induced dynamic mass redistribution (DMR) profiles for monitoring oncogenic signaling pathways in various cancer cells. J Receptors Signal Transduction. 2009;29:182–194. doi: 10.1080/10799890902976933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tran E, Fang Y. Label-free optical biosensor for probing integrative role of adenylyl cyclase in G protein-coupled receptor signaling. J Receptors Signal Transduction. 2009;29:154–162. doi: 10.1080/10799890903052544. [DOI] [PubMed] [Google Scholar]
- 39.Fang Y, Ferrie AM, Li G. Probing cytoskeleton modulation by optical biosensors. FEBS Lett. 2005;579:4175–4180. doi: 10.1016/j.febslet.2005.06.050. [DOI] [PubMed] [Google Scholar]
- 40.Fang Y, Li G, Peng J. Optical biosensor provides insights for bradykinin B2 receptor signaling in A431 cells. FEBS Lett. 2005;579:6365–6374. doi: 10.1016/j.febslet.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 41.Peters MF, Vaillancourt F, Heroux M, et al. Comparing label-free biosensors for pharmacological screening with cell-based functional assays. Assay Drug Dev Technol. 2010;8:219–227. doi: 10.1089/adt.2009.0232. [DOI] [PubMed] [Google Scholar]
- 42.Schroeder R, Merten N, Mathiesen JM, et al. The C-terminal tail of CRTH2 is a key molecular determinant that constrains Gαi- and downstream-signaling cascade activation. J Biol Chem. 2009;284:1324–1336. doi: 10.1074/jbc.M806867200. [DOI] [PubMed] [Google Scholar]
- 43.Lee PH, Gao A, van Staden C, et al. Evaluation of dynamic mass redistribution technology for pharmacological studies of recombinant and endogenously expressed G protein-coupled receptors. Assay Drug Dev Technol. 2008;6:83–93. doi: 10.1089/adt.2007.126. [DOI] [PubMed] [Google Scholar]
- 44.Cooper MA. Signal transduction profiling using label-free biosensors. J Receptors Signal Transduction. 2009;29:224–233. doi: 10.1080/10799890903047825. [DOI] [PubMed] [Google Scholar]
- 45.Rocheville M, Jerman JC. 7TM pharmacology measured by label-free: a holistic approach to cell signaling. Curr Opin Pharmacol. 2009;9:643–649. doi: 10.1016/j.coph.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 46.Jiang J, Ganesh T, Du Y, et al. Neuroprotection by selective allosteric potentiators of the EP2 prostaglandin receptor. Proc Natl Acad Sci USA. 2010;107:2307–2312. doi: 10.1073/pnas.0909310107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kebig A, Kostenis E, Mohr K, Mohr-Andra M. An optical dynamic mass redistribution assay reveals biased signaling of dualsteric GPCR activators. J Receptors Signal Transduction. 2009;29:140–145. doi: 10.1080/10799890903047437. [DOI] [PubMed] [Google Scholar]
- 48.Fang Y, Ferrie AM. Optical biosensor differentiates signaling of endogenous PAR1 and PAR2 in A431 cells. BMC Cell Biol. 2007;8:e24. doi: 10.1186/1471-2121-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fang Y. Label-free receptor assays. Drug Discov Today Technol. 2010 doi: 10.1016/j.ddtec.2010.05.001. DOI: 10.1016/j.ddtec.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tran E, Fang Y. Duplexed label-free G protein-coupled receptor assays for high throughput screening. J Biomol Screen. 2008;13:975–985. doi: 10.1177/1087057108326141. [DOI] [PubMed] [Google Scholar]
- 51.Stockwell BR. Chemical genetics: ligand-based discovery of gene function. Nat Rev Genet. 2000;1:116–125. doi: 10.1038/35038557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 53.Stockwell BR. Exploring biology with small organic molecules. Nature. 2004;432:846–854. doi: 10.1038/nature03196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuruvilla FG, Shamji AF, Sternson SM, et al. Dissecting glucose signalling with diversity-oriented synthesis and small-molecule microarrays. Nature. 2002;416:653–657. doi: 10.1038/416653a. [DOI] [PubMed] [Google Scholar]
- 55.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 56.Weinstein JN, Myers TG, O'Connor PM, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- 57.Sharma SV, Bell DW, Settleman J, Haber DA. 2Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 7:169–181. doi: 10.1038/nrc2088. 007. [DOI] [PubMed] [Google Scholar]
- 58.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zourob M, Elwary S, Fan X, et al. Label-Free Detection with the Resonant Mirror Biosensor. Methods in Molecular Biology: Biosensors and Biodetection. 2008;503:89–138. doi: 10.1007/978-1-60327-567-5_6. [DOI] [PubMed] [Google Scholar]
- 60.Duffy MJ. Carcinoembryonic antigen as a marker for colorectal cancer: is it clinically useful? Clin. Chem. 2001;47:624–630. [PubMed] [Google Scholar]
- 61.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 62.Ventura A, Kirsch DG, McLaughlin ME, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 63.Xue W, Zender L, Miething C, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaiser JCancer. First pass at cancer genome reveals complex landscape. Science. 2006;313:1370. doi: 10.1126/science.313.5792.1370. [DOI] [PubMed] [Google Scholar]
- 65.Kim KJ, Li B, Winer J, et al. Inhibition of vascular endothelial growth factor-induced angiogenesis suppresses tumour growth in vivo. Nature. 1993;362:841–844. doi: 10.1038/362841a0. [DOI] [PubMed] [Google Scholar]
- 66.Obata K, Morland SJ, Watson RH, et al. Frequent PTEN/MMAC mutations in endometrioid but not serous or mucinous epithelial ovarian tumors. Cancer Res. 1998;58:2095–2097. [PubMed] [Google Scholar]
- 67.Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 69.Jain M, Arvanitis C, Chu K, et al. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297:102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- 70.Pelengaris S, Littlewood T, Khan M, et al. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell. 1999;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- 71.Huettner CS, Zhang P, van Etten RA, Tenen DG. Reversibility of acute B-cell leukaemia induced by BCR-ABL1. Nat Genet. 2000;24:57–60. doi: 10.1038/71691. [DOI] [PubMed] [Google Scholar]
- 72.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260:85–88. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- 73.Cui Q, Ma Y, Jaramillo M, et al. A map of human cancer signaling. Mol Systems Biol. 2007;3:152. doi: 10.1038/msb4100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hudis CA. Trastuzumab--mechanism of action and use in clinical practice. New Engl J Med. 2007;357:39–51. doi: 10.1056/NEJMra043186. [DOI] [PubMed] [Google Scholar]
- 75.Druker BJ, Lydon NB. Lessons learned from the development of an Abl tyrosine kinase inhibitor for chronic myelogenous leukemia. J Clin Invest. 2000;105:3–7. doi: 10.1172/JCI9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cragg MS, Kuroda J, Puthalakath H, et al. Gefitinib-induced killing of NSCLC cell lines expressing mutant EGFR requires BIM and can be enhanced by BH3 mimetics. PLoS Med. 2007;4:e316. doi: 10.1371/journal.pmed.0040316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Petty TL. Determinants of tumor response and survival with erlotinib in patients with non-small-cell lung cancer. J Clinical Oncology. 2003;1:3–4. doi: 10.1200/JCO.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 78.Taguchi T, Tsukuda M, Imagawa-Ishiguro Y, et al. Involvement of EGFR in the response of squamous cell carcinoma of the head and neck cell lines to gefitinib. Oncol Rep. 2008;19:65–71. [PubMed] [Google Scholar]
- 79.Peterson AW, Halter M, Tona A, et al. Surface plasmon resonance imaging of cells and surface-associated fibronectin. BMC Cell Biol. 2009;10:e16. doi: 10.1186/1471-2121-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ziblat R, Lirtsman V, Davidov D, Aroeti B. Infrared surface plasmon resonance: a novel tool for real time sensing of variations in living cells. Biophys J. 2006;90:2592–2599. doi: 10.1529/biophysj.105.072090. [DOI] [PMC free article] [PubMed] [Google Scholar]