Abstract
Determining both the expression levels of mRNA and the regulation of its translation is important in understanding specialized cell functions. In this study, we describe both the expression profiles of cells within spatiotemporal domains of the Arabidopsis thaliana flower and the post-transcriptional regulation of these mRNAs, at nucleotide resolution. We express a tagged ribosomal protein under the promoters of three master regulators of flower development. By precipitating tagged polysomes, we isolated cell type-specific mRNAs that are probably translating, and quantified those mRNAs through deep sequencing. Cell type comparisons identified known cell-specific transcripts and uncovered many new ones, from which we inferred cell type-specific hormone responses, promoter motifs and coexpressed cognate binding factor candidates, and splicing isoforms. By comparing translating mRNAs with steady-state overall transcripts, we found evidence for widespread post-transcriptional regulation at both the intron splicing and translational stages. Sequence analyses identified structural features associated with each step. Finally, we identified a new class of noncoding RNAs associated with polysomes. Findings from our profiling lead to new hypotheses in the understanding of flower development.
Keywords: Arabidopsis, flower, intron, transcriptome, translation
Introduction
The development and function of plant tissues relies on constant interactions among distinct and nonequivalent cell types (Galbraith and Birnbaum, 2006; Nelson et al, 2008). To understand how cells work and how they interface with the environment, it is essential to acquire quantitative information on transcriptomes at cellular resolution. Recently, microarray analysis of transcriptomes has been extended to a cellular level of resolution by using laser microdissection (LM) or fluorescence-activated sorting (FAS; Galbraith and Birnbaum, 2006; Brady et al, 2007; Nelson et al, 2008; Jiao et al, 2009; Yadav et al, 2009). Although the number of transcriptome profiles at cellular resolution is still far from comprehensive, an early glimpse of the cellular transcriptional landscape seems to be information-rich for properties of both the genes from which the transcripts are derived, and of the cell types. Unfortunately, these methods have been limited by stresses associated with cellular separation and isolation procedures, and biases associated with mandatory RNA-amplification steps. To circumvent these problems, we combined a recently described translating ribosome affinity purification (TRAP) methodology (Zanetti et al, 2005; Heiman et al, 2008; Mustroph et al, 2009) with deep-sequencing (RNA-seq) to provide cell-level spatiotemporal maps for Arabidopsis early floral development at single-base resolution.
Flower development in Arabidopsis has been studied extensively and is one of the best-understood aspects of plant development. Although the organ formation process is as yet poorly understood, we have a relatively clear understanding of organ identity specification (for review, see Krizek and Fletcher, 2005). Genes controlling floral organ identity have been identified through genetic analysis of homeotic mutants and the ABC model was derived, in which three classes of regulatory genes, A, B and C, work in a combinatorial manner to confer organ identities of four whorls (Coen and Meyerowitz, 1991). Each class of regulatory gene is expressed in a specific and evolutionarily conserved domain, and the action of the class A, B and C genes is necessary for specification of organ identity (Figure 1A). However, we do not know exactly, beyond a few examples, which genes are expressed within each domain of the floral meristem or subsequently formed organ primordia at each morphologically defined stage during flower development.
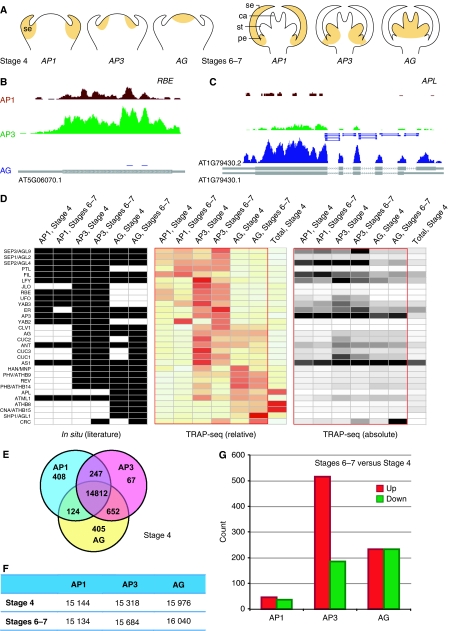
Figure 1.
Detection and quantification of cell-specific genes. (A) Diagrams showing the spatiotemporal domains being profiled. Sepal (se) primordia are visible at stage 4, and sepal, petal (pe), stamen (st) and carpel (ca) primordia are all visible at stages 6–7. (B) Translated mRNAs for AP1, AP3, and AG domains at flower stages 6–7 for a 1.0-kb region of chromosome-1 containing RBE, which is specific for AP1 and AP3 domains, and (C) a 4.2-kb region of chromosome-1 containing APL, which is highly expressed in the AG domain. TAIR annotated transcripts are shown as gray boxes at the bottom of (B) and (C) with ORFs highlighted as thick boxes. TAIR has two annotations for APL with the second form only detected in the AG domain. Reads covering exon–exon junctions are highlighted by short lines in (C). (D) Expression of previously characterized flower-specific genes. Genes were identified manually by searching PubMed abstracts followed by manual summarization of in situ hybridization data from each publication. Rows and columns were ordered manually. Relative expression levels were calculated by comparing each domain-specific expression with average expression across all domains. (E) Venn diagram of the cell domain-enriched genes that exhibited significant (⩾two-fold with P<0.001) up-regulation as compared with other domain(s) at stage 4. The numbers in middle areas indicate genes without domain-specific expression. (F) Count of genes with expression (RPKM ⩾1) for each spatiotemporal cell domain. (G) Differentially expressed (⩾two-fold with P<0.001) genes between stages 6–7 and stage 4 in each cell domains.
By epitope tagging a ribosomal protein in specific cellular domains, polysomes can be selectively and reproducibly purified from these A, B and C domains (Zanetti et al, 2005; Heiman et al, 2008; Mustroph et al, 2009). This strategy enables efficient purification of a cell-specific mRNA population, which is actively translating. This population is mapped and quantified by RNA-seq without RNA amplification (Wang et al, 2009; Marguerat and Bähler, 2010). RNA-seq is a newly developed approach to transcriptome profiling that exploits next-generation deep-sequencing technologies. Besides being far more precise and sensitive than microarrays, RNA-seq enables researchers to identify new features of transcriptomes and to refine transcript annotations. The combination of these technologies, which we term TRAP-seq, can detect and quantify gene expression with high sensitivity and reproducibility, can extend detection to single-base level gene structures, and can lead to the discovery of new genes and exons, all at cell type-specific resolution.
Besides cellular heterogeneity, the transcriptome is regulated at several steps through the life of mRNA molecules. Post-transcriptional regulation adds considerable richness and sophistication to gene regulation, and includes steps in intron splicing, nuclear export, storage, and degradation. Studies in plants, similar to those in yeast and animals, have found widespread regulation at least at two steps, intron splicing and translation state (for reviews, see Bailey-Serres et al, 2009; Simpson et al, 2010). These layers of regulation of the transcriptome are not directly available through traditional transcriptome profiling of total mRNA abundance, and make transcript abundance an imperfect proxy for corresponding protein abundance. Compared with the transcriptome, the translating transcriptome, which we call in this study the translatome, seems to be a better predictor of protein abundance in yeast and is expected to be so in other eukaryotes (Lu et al, 2007; Ingolia et al, 2009).
In addition to dissecting cell-specific transcriptomes, TRAP-seq can also be used to analyze post-transcriptional regulation. By comparing the translatome and transcriptome using a deep-sequencing approach, our data provide evidence for widespread intron retention (IR) and a dynamic range of translational controls as points of regulation during flower development. In addition, we observed several dozen polysome-associated noncoding RNAs (ncRNAs), which may imply new mechanisms of regulation of translation by RNA.
Results
Profiling cell-type specific translatomes using TRAP-seq
To circumvent the low RNA yield and perturbation of the transcriptome caused by LM and FAS and to eliminate the need for specialized cell-isolation equipment, we extended the immunopurification of polysomes to the cell type-specific level and quantified cell-specific translating transcripts isolated by this means using RNA-seq. In brief, we used a fusion of the large subunit ribosomal protein L18 with N-terminal His and FLAG epitope tags (HF–RPL18) for efficient incorporation into polysomes and for immunopurification of all translating cellular mRNAs (Zanetti et al, 2005). HF–RPL18 was chosen because its global and cell-specific overexpression has no obvious effects on plant development even at the transcriptome level, because it accumulates uniformly in polysomes of different size, and because it maintains the integrity of immunopurified polysomes and associated mRNAs (Zanetti et al, 2005; Supplementary Figure S1). HF–RPL18 was expressed specifically in the A, B and C domains of early developing flowers, which were defined by the promoters of the APETALA1 (AP1), APETALA3 (AP3) and AGAMOUS (AG) genes (Figure 1A; Supplementary Figure S2). These domains cover the four whorls that will develop into sepals, petals, stamens, and carpels (Figure 1A). We focused on early flower development, during which floral patterning and the specification of floral organs is established. To achieve temporal specificity, we used a floral induction system to facilitate collection of early-stage flowers (Wellmer et al, 2006). The combination of domain-specific promoters and this floral induction system enabled fine spatiotemporal expression of HF–RPL18 (Supplementary Figure S2), which allowed isolation of translating mRNA in specific cellular domains, and at specific developmental stages. Flower stages 4 and 6–7 were selected for profiling because they coincide with prominent morphological changes in sepals and stamens, respectively, in addition to other fast-changing developmental processes (Smyth et al, 1990). It should be emphasized that co-precipitation with ribosomes is the only evidence for the translation of any RNAs in our experiments. In this study, these polysome-associated mRNAs are termed ‘translating mRNA’ for simplicity, but ribosome-associated nontranslated RNAs, although so far unknown, may exist.
Compared with LM and FAS, TRAP can not only isolate cell-specific translating mRNA but also easily achieve a high yield of RNA, which eliminates the need for bias-introducing and information-losing RNA amplification. We also confirmed that cell type-specific immunopurification is specific enough that essentially no RNA was isolated when using it with plants without the HF–RPL18. Owing to the high yield, it was possible for TRAP-purified mRNA to be subsequently mapped and quantified by deep sequencing. For each replicate, we made randomly primed cDNA from poly(A)+ polysome-associated RNA hydrolyzed to 200–300 nt in length to construct a sequencing library subject to Illumina sequencing. We obtained ∼10 million mapped 37- or 38-bp reads from each library, and assayed two independent libraries for each spatiotemporal sample (Supplementary Table S1). For the Arabidopsis genome, we observed this sequence depth is sufficient to reliably detect and measure rare, yet biologically relevant mRNA species. At this sequencing capacity, transcript detection was highly reproducible at and above 1.0 reads per kb of the transcript per million mapped reads of the transcriptome (RPKM) for a typical 2-kb transcript (Supplementary Figure S3A and B), a level of detection that we estimated corresponds to about 0.5 transcripts per cell based on literature values for the mRNA content of a plant cell (Goldberg et al, 1978) and a set of in vitro RNA spike-in standards (Supplementary Table S2), ranging from 500 to 10 000 nt in length (Supplementary Figure S3A). Across a range of abundance equivalent to ∼0.3–40 000 transcripts per cell, we typically obtained linear readouts from RNA-seq. Consistent with previous reports, we observed that technical replicate determinations of transcript abundance were highly reproducible (R>0.98; Supplementary Figure S3B for one example). In comparison, at this sequence depth microarrays with analog measurements had clear biases across this range, with limited sensitivity for genes of normal length (Mortazavi et al, 2008; Wang et al, 2009). The linear relationship of the same RNA spike-in standards was compromised in microarray readouts and high expression range was skewed partially by saturation (Supplementary Figure S3C).
Deep sequencing of the translatome provided single-base resolution measurement as illustrated in Figure 1. The gene RABBIT EARS (RBE) is a much-studied transcription factor that is specifically expressed at a modest level in AP1 and AP3 domains but not in the AG domain (Takeda et al, 2004). In contrast to only four reads found for the RBE transcript in the AG domain, evidence for RBE expression in the AP1 and AP3 domains consisted of 156 and 507 mapped sequence reads, respectively (Figure 1B). In a separate example, the low-expression gene ALTERED PHLOEM DEVELOPMENT (APL) was observed to be preferentially expressed in the AG domain as expected (Bonke et al, 2003). One of the two annotated splicing isoforms, AT1G79430.2, was consistent with all of the sequences obtained from flowers, a conclusion that is supported by 15 reads that cross splice junctions (Figure 1C). In addition, the flower-expressed APL transcript has a shorter 5′UTR than both splicing isoforms described in the Arabidopsis database TAIR.
Cellular-level properties uncovered through comparing cell-specific translatomes
Domain-specific translatomes for early flower development showed qualitative and quantitative differences consistent with functional specialization. We detected the expected patterns of expression for most genes that had been previously shown to be specifically expressed in the floral tissue. The in situ hybridization-based expression patterns of these flower-expressed genes from the literature (Supplementary Table S3) are summarized in Figure 1D, in parallel to absolute and relative expression values from our spatiotemporal translatome data set. The comparisons validate the translatome profiling. Clearly, our data set had more depth than in situ hybridization, and transcript levels of two genes can only be quantitatively and directly compared in our data set.
The translatome of each domain was distinct, consisting of transcripts from 15 134 to 16 040 genes (55.5–58.8% of the genome, Figure 1F), with the AG domain expressing more genes. We detected a significant portion of the genome differentially expressed among the tested spatial domains, which in total contribute to 11.4 and 5.6% of the translatome at stage 4 and stages 6–7, respectively (Figure 1E; Supplementary Figure S4; Supplementary Tables S4 and S5). Another significant portion of the genome was activated or repressed (4.7 and 3.0% of the translatome) during the developmental time course between stage 4 and stages 6–7 in at least one domain (Figure 1G; Supplementary Table S6). Notably, a large number of genes were activated in the AP3 domain, implying active organ determination and formation in the second and third whorl region of the developing flower.
When compared with mature flowers at stage 12 profiled as part of the AtGenExpress project (Schmid et al, 2005), we noticed a larger difference with ∼20% of stage 4 or 6–7 expressing genes not found at stage 12 in each domain and ∼15% of stage 12 expressing genes not found in earlier stages (Supplementary Figure S5). This finding implies that distinct transcriptional networks are used at early and late stages for each floral domain.
The comparison of our early developmental series of domain-specific translatomes provided a wealth of genes with candidate developmental roles. For example, we observed that the annotation of genes expressed preferentially in each domain corresponds in many cases to known physiological functions of the domain (Figure 2A). Notably, we observed ‘Petal Development’ and ‘Stamen Development’ were significantly enriched in the AP3 domain but not the development of the other two whorls. Moreover, many gene ontology (GO) categories were enriched, suggesting localized physiological functions that previously were uncharacterized (Supplementary Figure S6). For instance, chloroplast- and plastid-related GO terms were enriched in the AG domain, and this bias became greater as flower development proceeded.
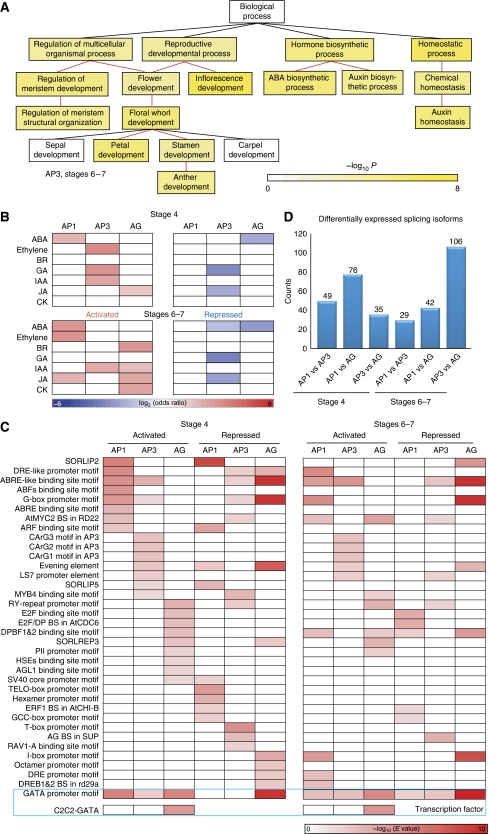
Figure 2.
The spatiotemporally regulated transcriptome during early flower development. (A) GO analysis identifies significantly overrepresented (P<0.001) gene categories under ‘Biological Process’ for the cell-specific transcripts in the AP3 domain at stages 6–7. Color bar: significance level for categories by hypergeometric test with FDR correction. (B) Domain-specific enrichment of hormone-responsive genes in each floral domain at stages 4 or 6–7. Red for upregulated genes and blue for downregulated genes. (C) Cell-specific enriched known cis-elements at flower stages 4 and 6–7. Candidate cis- and trans-transcriptional control cognate partners are shown in the blue box, in which GATA promoter motif and C2C2-GATA family transcription factors are enriched in floral domains. Only significantly overrepresented (P<0.001) classes by hypergeometric test with FDR correction are colored in (B) and (C). (D) Number of genes with splicing isoforms differentially expressed between any two floral domains at stages 4 and 6–7.
Translating transcripts for hormone-responsive genes were enriched in specific spatiotemporal domains during early flower development. We examined the sets of genes that respond to stimulation by seven phytohormones: abscisic acid, auxin, brassinosteroid, cytokinin, ethylene, gibberellins (GA), and jasmonic acid (JA). The sources and lists of phytohormone-responsive genes are provided in the Supplementary information and Supplementary Table S7. Genes in these classes showed cell type-specific patterns of enrichment, including known and new ‘activity centers’ for gene activation and repression within floral domains (Figure 2B). We observed that genes downregulated by GA were significantly enriched in the AP3 domain at stage 4 and stage 6–7, whereas genes upregulated by GA were enriched in the same domain at stage 4 only. Only genes downregulated by JA were found significantly enriched in the AP3 domain at both stages, although genes upregulated by JA were more abundant in the AG domain. The involvement of GA and JA in the AP3 domain for the development of stamens and petals is consistent with previous phenotypic studies of hormone-deficient mutants and with exogenous hormone-treatment experiments (Yu et al, 2004; Brioudes et al, 2009; Cheng et al, 2009), and our data suggested that these phytohormones act mainly through repressing their target genes in the AP3 domain.
Through genes coexpressed in a floral domain at a given stage, we attempted to identify promoter DNA motifs associated with cellular patterns of expression. We compared cis-element enrichment in the promoters of genes regulated in a domain-specific and stage-specific manner (Figure 2C). Enrichment of several known cis-elements, such as the AG-binding site motif, the AGL1-binding site motif, and three other CArG motifs (Huang et al, 1996; Riechmann et al, 1996; Tilly et al, 1998), was observed upstream of genes observed in domain-specific enriched or depleted gene sets, suggesting that gene activation and repression are equally important for flower development gene regulatory networks. The majority of these enriched cis-elements were observed in selected domains with a few exceptions used in multiple domains at both stages. For the most significantly abundant cis-element, the GATA box, we observed the highest enrichment in the AG domain at both flower development stages. Regulation of the GATA box is achieved by the binding of the conserved C2C2-GATA transcription factors with two GATA zinc-fingers (Reyes et al, 2004). In Arabidopsis, 29 C2C2-GATA family members have been identified, a few of which are involved in flower development (Zhao et al, 2004; Mara and Irish, 2008). We observed that this family of transcription factors was also highly expressed in early flower domains with a few members significantly enriched in the AG domain (Supplementary Figure S7), implying its function during flower development, especially in the inner whorls, as regulators of GATA box-containing genes. Such cell-specific coexpression analysis may prove to be a substantial aid in sorting true protein–DNA and protein–protein interaction partners from numerous candidates (Supplementary Figure S8).
Regulation of the translatome by splicing
Using single-base resolution translatome data, we successfully eliminated nontranslating transcripts, such as those with premature termination codons, which are potentially targeted for degradation by the nonsense-mediated mRNA decay surveillance machinery (Maquat, 2004), to enable genome-wide mapping of splicing. At cellular resolution, we were able to quantify the expression of each splicing isoform and observed the majority of genes had a dominant isoform expressed at each spatiotemporal point (Supplementary Table S8). In addition, we identified small numbers (29–106) of domain-specific alternative splicing (AS) events (Figure 2D). A high portion (40%) of this small set of cell-specific AS events did not alter the encoded protein, as they had alternative exons within the 5′ and/or 3′UTR regions. This finding reinforced the notation that AS is used by plants (Filichkin et al, 2010), although it may be less prevalent than in animals (Reddy, 2007).
In addition to annotated isoforms, we observed 29 new splicing isoforms during early flower development (Supplementary Table S9). As these splicing isoforms were not previously reported from the almost-saturated cDNA and EST collection efforts, they are likely truly specific to early flower development. Among them, 25 were observed only in selected domains or flower stages, which may explain the reason why they were previously missed.
To understand regulation by splicing, we further compared the translatome with the total transcriptome to find intron retention (IR) events. To this end, we profiled the translatome of stage 4 flowers from plants expressing HF-RPL18 under the ubiquitously expressed RPL18 promoter (Supplementary Figure S2D) using TRAP-seq, and compared this with the total transcriptome of the same plants using RNA-seq. In this study, IR is considered as the complete retention of an intron in a poly(A)+ transcript, which is likely associated with failure of the recognition of weak intron splice sites. From the translatome, we were able to distinguish IR from protein-coding AS isoforms regardless of existing annotations (for example, see Supplementary Figure S13G). Using the same detection criteria as we used for exons and transcripts, that is, ⩾1.0 RPKM, we detected 4130 introns in the total transcriptome or in the translatome (Supplementary Table S10). The majority (83.2%) of these retained introns were not observed in the translatome and were probably IR events instead of previously unknown events of AS (Figure 3A). These real IR events included ∼5% of all known introns in the genome.
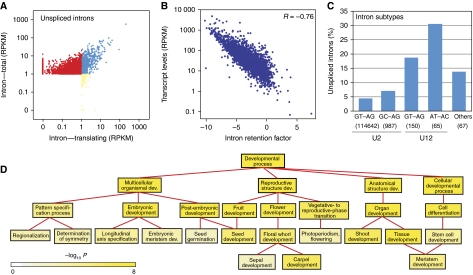
Figure 3.
Intron retention events detected during flower development. (A) Expression profiles of detected TAIR-annotated introns. Red dots represent introns detected in only total poly(A)+ mRNAs, whereas blue and yellow dots represent introns detected in both total and translating mRNAs, or only translating mRNAs. (B) Transcript levels are negatively correlated with IR levels (R=−0.76, P<10−23). IR levels are quantified by normalizing detected intron levels with neighboring exon levels. (C) Intron subtypes significantly differ in IR levels. Percentages of IR events for intron subtypes are shown. Number of introns for each subtype is included below subtype name. (D) GO analysis identifies significantly overrepresented (P<0.001) gene categories under ‘Developmental Process’ for the genes with IR events in floral organs at flower stage 4. Color bar: significance level for categories by hypergeometric test with FDR correction.
Only a limited positive correlation (R=0.22) was observed when plotting absolute intron levels with the levels of corresponding transcripts (Supplementary Figure S9). Thus, we compared the normalized intron levels, which were based on neighboring exon levels and reflected the percentages of unspliced introns, and the levels of corresponding transcripts (Figure 3B). A highly significant negative correlation (R=−0.76) was revealed, which suggested that IR is more often observed in weakly expressed transcripts, perhaps as a means to control protein levels.
Similar to other eukaryotes, plant pre-mRNA splicing is a nuclear process that involves the recognition of exon–intron junctions by the spliceosome (Brown and Simpson, 1998; Maquat, 2004; Reddy, 2007; Schuler, 2008). The spliceosome is assembled from small nuclear RNAs (snRNAs) and associated proteins that make up the small nuclear ribonucleoprotein particles (snRNPs) that recognize and splice introns. There are at least two classes of pre-mRNA introns, based on the splicing machinery that catalyzes the reaction: U2 snRNP-dependent introns and U12 snRNP-dependent introns. Each class can be further divided into subtypes according to their terminal dinucleotides at both ends of the introns. Following the classification of Sheth et al, 2006, we observed the U2 GU-AG subtype makes up the majority (98.5%) of all introns, whereas U12 introns comprise a small fraction, only of 0.06% of all introns. These subtypes of introns showed dramatic difference in IR rate (Figure 3C). U12 introns were observed to be ∼5 times less efficiently spliced than the U2 class. Within each class, GU-AG dinucleotides seemed to be cleaved more efficiently than other dinucleotide pairs.
One distinguishing feature of plant introns, compared with introns from vertebrates and yeast, is their UA richness. Although the exact role of UA richness remains to be fully understood, it is required for efficient splicing of both U2 and U12 introns (Goodall and Filipowicz, 1989; Lewandowska et al, 2004). At the whole genome scale, we confirmed the UA richness of both intron classes. Furthermore, we observed that UA richness is slightly lower in those introns with detectable IR (Supplementary Figure S10).
Other sequence features may also influence IR. As an example, longer transcripts were clearly spliced more efficiently than shorter transcripts, which were enriched with retained introns (Supplementary Figure S11A). Similarly, transcripts with only one or a few introns showed more IR events than transcripts with greater numbers of introns (Supplementary Figure S11B).
To reveal possible general relationships between functions of genes and IR events, we searched for common biological themes among transcripts with IR using GO term analysis. The distribution of IR event involvement was observed to be significantly biased (P<0.001) in about half of the categories of molecular function, cellular component, and biological process (Supplementary Figure S12), such as chloroplast, plasma membrane, and response to abiotic or biotic stimulus. Genes involved in developmental processes were also observed to be enriched among RNAs with IR events. A closer inspection of developmental processes genes revealed that IR events were involved in most major processes to different extents (Figure 3D). For example, several mRNAs involved in meristem and flower development were observed to be characterized by IR, such as AGL24, PHB, PHV, and SEP1/2/3/4 (Supplementary Figure S13). The IR events were observed in both cognate and distinct introns of members of conserved SEP1/2/3/4 and PHB/PHV families.
Regulation of translation at the transcriptome scale
We next extended our analysis to understand post-transcriptional regulation at the translational level by quantifying mRNA association with ribosomes. In the same stage 4 flowers, we detected the presence of 99.7% of the total transcriptome as ribosome bound. In addition, 10 transcripts were detected only in the translatome. Although the abundance of translating mRNAs was generally correlated with the level of total mRNA, over- and underrepresentation of translating transcripts was evident for many genes in stage 4 flowers (Figure 4A). We used as cutoff criterion a translating/total mRNA ratio >2 for enriched and a translating/total mRNA ratio <0.5 for depleted (using a P<0.001 in both cases). A total of 2043 transcripts (10.7% of all expressed genes) was found as highly translated, and a total of 2800 genes (14.7% of all expressed genes) was found as weakly translated (Figure 4A). Using P<0.001 as the only criterion, a total of 6755 (35.4% of all expressed genes) were observed to be differentially expressed between translatome and transcriptome.
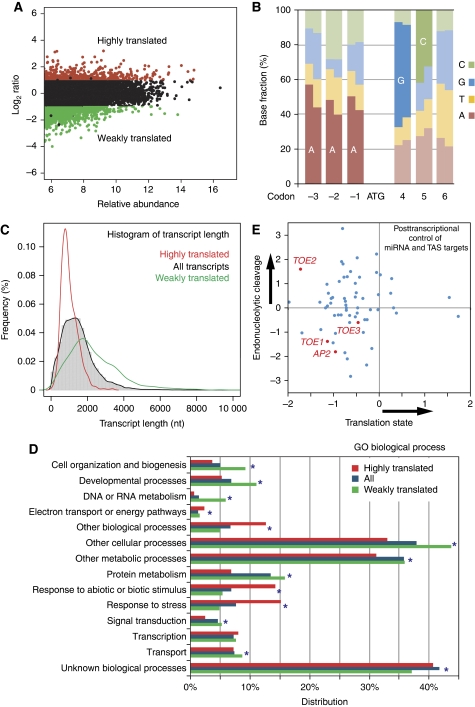
Figure 4.
Translational regulation during flower development. (A) M-A plot in which log2-transformed ratios of translating versus total mRNA signal for stage 4 flowers are plotted against log-intensity averages. Transcripts with statistically different levels in the translating and total RNA populations (P<0.001) and a ratio above two are considered as highly translated and are shown in red. Transcripts with statistically different levels and ratio below 0.5 were considered as weakly translated and are shown in green. Plants expressing HF–RPL18 under the RPL18 promoter were used for translatome profiling. (B) Biases around the start codon shown as the fractions of bases. At each base (rectangle), the left half represents the fraction of each base in highly translated genes and the right half represents the fraction of each base in weakly translated genes. Enriched bases are labeled. (C) Enhanced translation for short transcripts. A transcript length distribution of all expressed genes is shown by the histogram and the black line, while red and green lines show distributions of highly and weakly translated transcripts, respectively. (D) Relationship between gene function and translation levels. All genes detected as expressed for ‘Biological Process’ categories (Total) are compared with highly translated (red) and weakly translated (green) transcripts. ‘Distribution’ refers to the percentages of genes annotated to descriptive terms in a particular GO category divided by all genes. Categories with FDR corrected hypergeometric test P<0.001 are marked with stars. (E) Estimated regulation modes of miRNA target genes. Scatterplot of uncapping levels (endonucleolytic cleavage) of miRNA and ta-siRNA (TSA) target mRNAs versus translation efficiency (translation inhibition) of the same mRNAs in stage 4 flower tissues. Targets of miR172, including AP2 and AP2-like mRNAs are highlighted in red.
To investigate whether noncoding and coding sequences can influence translation, we explored possible structural properties of mRNAs with respect to the degree to which they are observed to be bound to ribosomes. To find possible influences on translation by sequences flanking the AUG start codon, we surveyed sequence biases at each position of a gene using all annotated genes. We observed that the sequences flanking the AUG are all biased, with bases from −3 to −1 and the two codons after AUG being most significant. Such bias in Arabidopsis is similar to the Kozak sequence in mammals (Kozak, 2005), although the sequence seems to be distinct. Such a bias is more evident (P<0.01) for all positions near the AUG codon in the group of highly ribosome-bound genes than in the group of weakly ribosome-bound genes (Figure 4B), suggesting the significance of this plant Kozak sequence in enhancing association of RNAs with ribosomes. This sequence is very similar to a previously identified preferred initial codon context for highly translated Arabidopsis rosette leave transcripts during dehydration stress (Kawaguchi and Bailey-Serres, 2005). However, we did not observe a significant overlap between transcripts under translational regulation between these two data sets (Supplementary information), implying translational state is tissue- and environmental-specifically regulated. Nevertheless, the plant Kozak sequence probably has basal functions for translation initiation.
Recent work in Escherichia coli revealed a correlation between secondary structure stability of local mRNA sequences near the start codon and mRNA translation efficiency (Kudla et al, 2009). To find out whether Arabidopsis coding sequences have similar effects on binding to ribosomes, we computed the predicted minimum free energy associated with the secondary structure of specific regions of each mRNA molecule. A moving window analysis identified that sequences right after the start codon affected binding. Folding energy of the first 60 nucleotides (including the start codons) showed one of the highest correlations with ribosome binding efficiency (R=0.15, P<1E−4, Supplementary Figure S14; Supplementary Table S11). Although predicted folding energy could only explain a small portion of the variation in binding efficiency, it was a significant factor. Consistent with our finding, a number of structure features of transcripts that affect folding, including UTR length, GC contents, and secondary structure, were previously observed to be affecting ribosome loading by Kawaguchi and Bailey-Serres (2005).
We identified the transcript length, which is highly correlated with ORF length, as a more significant factor affecting the likelihood that an RNA will bind ribosomes. By comparing the length distribution of all genes, highly translated genes, and weakly translated genes, it was evident that the class of highly translated genes was enriched in short genes and depleted in long genes (Figure 4C). Selective redistribution of the affinity tag within the mRNA pool during polysome isolation should have been prevented because polysomes were stabilized with cycloheximide in the extraction buffers and during the affinity purification steps. This relationship between transcript length and translation was not likely due to nonrandom association between transcript length and gene physiological functions either. The same inverse relationship was observed when functional groups of genes with significant length bias were excluded. Such an unexpected observation that ribosome density decreases with increasing transcript length was previously reported in yeast, flies, mammalian cell culture, and plants using velocity sedimentation-isolated polysomes (Arava et al, 2003; Kawaguchi and Bailey-Serres, 2005; Lackner et al, 2007; Qin et al, 2007; Hendrickson et al, 2009). The recent observation that ribosomes are approximately three times enriched on the first 30–40 nt of a yeast ORF may explain this phenomenon (Ingolia et al, 2009).
To find possible general relationships between functions of genes and translational control, we searched for common biological themes in highly and weakly translated genes using GO term analysis. The translation state was observed to be significantly biased (P<0.001) in the majority of the categories of molecular function, cellular component, and biological process (Figure 4D; Supplementary Figure S15). For example, many biological processes, including developmental ones, were observed to be enriched in weakly translated genes, whereas genes responsive to external stimuli and stress were observed to be enriched in highly translated genes. Similarly, transcription factors were observed to be enriched in highly translated genes, whereas most other molecular function groups, such as kinases were enriched in weakly translated genes. Closer inspection of genes responsive to abiotic stimulus and genes encoding chloroplast thylakoid-located proteins revealed that different levels of strong translation were observed in most subgroups (Supplementary Figure S15).
MicroRNAs (miRNA) post-transcriptionally regulate gene expression by interfering with a target transcript's stability, translation, or both. Although the importance of miRNAs has been widely appreciated, the relative contributions of endonucleolytic cleavage or translation state of the target mRNAs to the overall effects on protein synthesis remain to be elucidated (Brodersen et al, 2008). At the genome scale, the translation-profiling approach measured the translation of all mRNAs including miRNA targets. In addition, our previous work on uncapped mRNA profiling quantified the degradation of all mRNAs in early developing flowers (Jiao et al, 2008). By combining translation with relative uncapped mRNA levels in the same stage 4 flowers, we sought to dissect the respective contributions of translational inhibition and mRNA decay to miRNA regulation (Figure 4E). By plotting these two measurements, it is evident that most miRNA target transcripts had below-average levels of translation, which might be due to miRNA-mediated translation inhibition. Among others, AP2 and AP2-like mRNAs as targets of miR172 were all found weakly translated (Figure 4E), which is consistent with previous reports (Aukerman and Sakai, 2003; Chen, 2004). On the other hand, a large portion of these miRNA targets was observed to be enriched in the uncapped form, which could be the product of miRNA-mediated endonucleolytic cleavage. The effects of these two regulation modes of miRNA on their cognate genes did not show competition but rather synergistic effects in many cases.
Identification of polysome-associated ncRNAs
In addition to protein-coding mRNAs and small RNAs, there are additional classes of mRNA-like molecules, which have gained attention recently. Such mRNA-like classes include nonprotein-coding RNAs (ncRNAs), transposable element (TE)-related transcripts, and pseudogene transcripts (Jiao and Deng, 2007; Ben Amor et al, 2009; Kurihara et al, 2009). TE-related genes and pseudogenes do not have obvious biological functions, although they are widespread in the Arabidopsis genome. ncRNAs are mRNA-like RNAs that do not encode proteins.
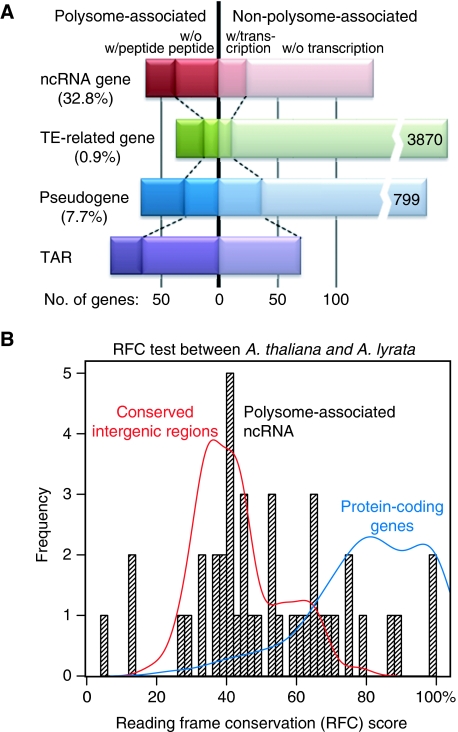
We sought to explore the transcription and even possible translation, or at least ribosome association, of these mRNA-like RNA classes. To simplify our analysis, we excluded all natural antisense transcript RNAs that arise from the strands opposite the coding strands of mRNAs. In stage 4 flowers, we detected close to half of the ncRNAs, ∼1% of TE-related genes, and ∼10% of pseudogenes. In addition, we found ∼150 transcriptionally active regions (TARs), which are at least 100 nt in length (Figure 5A). Among these expressed mRNA-like transcripts, a major portion was observed to be associated with polysomes for each class. Taken together, 32.8% of all ncRNAs were polysome associated in this flower sample. In contrast, only 0.9% of TE-related RNAs and 7.7% of pseudogene RNAs were co-purified with polysomes. Strikingly, 34 out of those 57 polysome-associated ncRNAs were observed to be differentially expressed among the AP1, AP3, and AG domains or between stages (Supplementary Table S12), which implies developmental regulation of these ncRNAs, and therefore possible regulatory roles during flower development.
Figure 5.
Polysome-associated ncRNAs. (A) Summary of numbers and percentages of detection for ncRNAs, TE-related RNAs, pseudogene RNAs, and RNAs from TARs in stage 4 flower tissues. The vertical line separates genes observed associated with ribosomes, or not, in our analysis. Additional genes with detection in total mRNA population are labeled as ‘w/transcription’. Polysome-associated transcripts supported by proteomics data (Baerenfaller et al, 2008; Castellana et al, 2008) are labeled as ‘w/peptide’. Percentages of polysome-associated transcripts for each category are listed in parentheses. Number of nonpolysome-associated TE-related genes or pseudogenes is listed at the right end of each bar. (B) Most polysome-associated ncRNAs lack coding capacity. A RFC score distribution of all these ncRNAs is shown by the histogram, while red and blue lines show distributions of conserved intergenic regions and protein-coding genes (with UTRs) within the similar length range of the ncRNAs, respectively. RFC scores were calculated as described previously (Clamp et al, 2007) based on alignments between the Arabidopsis thaliana TAIR9 build and Arabidopsis lyrata v1.0 release build using the VISTA pipeline (Frazer et al, 2004).
Using an independent polysome extraction approach, we separated polysomes of different sizes by differential centrifugation through sucrose density gradients. Using similar stage 4 floral tissues without the HF–RPL18 transgene, we obtained two polysome fractions, nonpolysomal (NP) for 40S and 60S ribosomal subunits, and 80S monosomes, and multiple ribosome (PS) with two or more ribosomes (Supplementary Figure S16A). Semi-quantitative RT–PCR analysis confirmed the association of ncRNA with ribosomes (34 out of 35) and determined the relative proportion of ncRNA in the NP and PS fractions. Most ncRNA were observed in both, although clear differential distribution between NP and PS fractions was evident for about two-thirds of them (Supplementary Figure S16B). There were 16 ncRNAs more abundantly represented in the PS portion, with three almost exclusively in the PS fraction. Another six ncRNAs were observed to be more associated with the NS portion.
The ncRNAs could either be new regulatory RNAs, or they could be simply misannotated protein-coding mRNAs with noncanonical ORFs. To further investigate the coding capacity of these ncRNAs, we used the draft sequence of Arabidopsis lyrata, a close A. thaliana relative, to identify possible ORFs and to distinguish protein-coding mRNAs and ncRNAs. Reading frame conservation (RFC) score, which reflects the percentage of nucleotides reading frame of which is conserved between two species (Clamp et al, 2007), was calculated for each ncRNA. The RFC scores for confirmed protein-coding mRNAs and conserved intergenic regions within a similar size range were also calculated as positive and negative controls (Figure 5B). This coding capacity analysis independently confirmed that most of these annotated ncRNAs lack protein-coding capacity with only a few potential exceptions (Supplementary Table S12). The molecular functions of the large number of ribosome-associated ncRNAs remain unknown.
Discussion
Widely used high-throughput transcriptome-profiling approaches have been successful in dissecting gene regulatory networks. Still, transcriptome profiling is often compromised by limited resolution at least at two levels. First, the cell-specific transcriptome in multicellular organisms is usually difficult to study, although a cell is considered as the basic structural and functional unit of all living organisms. Second, mRNA has a complicated life cycle from transcription to decay, which is indistinguishable by traditional RNA profiling. In this study, we sought to dissect the transcriptional networks controlling early Arabidopsis flower development without these limitations.
Using cell type-specific expression of an epitope-tagged ribosomal protein in selected cell types, we were able to purify translating mRNAs from those cells with little contamination from other cell types. This approach is noninvasive; the interaction between ribosome and mRNA can be strongly stabilized during purification (Obrig et al, 1971); this approach is high in yield; and it does not require specialized equipment such as fluorescence-activated cell sorters or laser-capture microdissection apparati. Because of the high yield of cell-specific translating mRNAs, deep sequencing can be used without bias-introducing RNA amplification. When combined with an inducible floral system, this TRAP-seq approach was used to dissect early flower development by profiling spatiotemporal domains within the floral meristem. Multiple lines of evidence confirmed the specificity of this approach, including detecting the expression in expected domains but not in other domains for well-studied flower marker genes (Figure 1B–D), confirming known physiological functions (Figure 2A), locating related phytohormone centers in specific domains (Figure 2B), and rediscovering cis-elements in floral domain-specific transcripts (Figure 2C).
We provide numerous examples from flower development in which a spatiotemporal map of rigorously comparable cell-specific translatomes makes possible new views of the properties of cell domains not evident in data obtained from whole organs or tissues, including patterns of transcription and cis-regulation, new physiological differences among cell domains and between flower stages, putative hormone-active centers, and AS events specific for flower domains (Figure 2, Supplementary Figures S6–S8). Such findings may provide new targets for reverse genetics studies and may aid in the formulation and validation of interaction and pathway networks.
Consistent with an earlier transcriptome profiling during early Arabidopsis flower development (Wellmer et al, 2006), distinct sets of early expressed genes were observed in later flower stages (Supplementary Figure S5). This can be contributing to distinct developmental programs activated at early and late flower stages. For example, the vast majority of those organ-specific transcripts expressed in the reproductive floral organs in late stages are probably primarily involved in sporogenesis. Homeotic genes and a small number of other genes, however, are probable regulators that orchestrate distinct downstream cascades at different stages.
There is a growing appreciation for the integral role that post-transcriptional regulation has in the control of plant development, and in a plant's response to the environment. It has long been reported that for a number of plant genes, a lack of correlation has been observed between mRNA levels and protein synthesis. Such observations suggest that post-transcriptional regulation may be involved (Gallie, 1993). Using informatics and genomics tools, post-transcriptional regulation has recently been studied in plants at two key control points, intron splicing (Ner-Gaon et al, 2004; Simpson et al, 2008; Zhang et al, 2008; Filichkin et al, 2010), or translation state (Kawaguchi et al, 2004; Kawaguchi and Bailey-Serres, 2005; Branco-Price et al, 2008; Piques et al, 2009). However, integration of these two related control points is desired. For example, RNAs with splicing defects that are not translated into protein can be misidentified as new splicing variants.
In this study, we integratively profiled these two key post-transcriptional control points. From our translatome-wide profiling, we (i) confirmed that both post-transcriptional regulation mechanisms were used by a large portion of the transcriptome; (ii) identified a number of cis-acting features within the coding or noncoding sequences that affected splicing or translation state; and (iii) revealed correlation between each regulation mechanism and gene function. Our transcriptome-wide surveys, in combination with our previous genome-scale study of uncapped mRNAs subject to decay (Jiao et al, 2008), have extended our current understanding of post-transcriptional regulation in flower development and have highlighted target genes transcripts of which are probably under extensive post-transcriptional regulation.
As one example of post-transcriptional regulators, miRNA, can affect both the translation and stability of mRNAs in plants and animals. Although a growing number of miRNA and target mRNAs are reported using experimental and computational approaches, little is known about the mechanisms used for each miRNA-target RNA pair. Traditionally, target cleavage was considered the main mode of regulation. However, a recent report pointed out the possibility of widespread translational inhibition by plant miRNA (Brodersen et al, 2008). The combination of our translation state profiling and previous uncapping level profiling could serve as a quantitative indicator of different balance points between transcript cleavage and translation inhibition (Figure 5).
Compared with the transcriptome, the translatome should be a better predictor of protein abundance, which is supported by recent study in yeast (Ingolia et al, 2009). Using TRAP-seq, we were able to discriminate actively translated mRNAs from those being stored or under degradation. In addition, we could identify real splicing variants instead of splicing errors that could not be translated into peptides. Nonetheless, post-translational regulation, such as protein degradation, may substantially affect protein abundance. Understanding post-translational regulation would require genome-wide profiling of protein abundance and modification, which is beyond the scope of this study.
Finally, we reported the finding of a large number of polysome-associated ncRNAs. About one-third of all annotated ncRNA in the Arabidopsis genome were observed to be co-purified with polysomes. Coding capacity analysis confirmed that most of them are real ncRNA without conserved ORFs. Recently, ncRNAs have been observed to be involved in small RNA formation and are involved in the stress response (Ben Amor et al, 2009). Many ncRNAs were also observed as antisense transcripts to mRNAs and are under nonsense-mediated mRNA decay pathway control (Kurihara et al, 2009). The group of polysome-associated ncRNA reported in this study is a potential new addition to the expanding riboregulator catalog; they could have roles in translational regulation during early flower development.
Materials and methods
Generation of plant lines
The Arabidopsis thaliana ecotype Landsberg erecta was used in this study. Details of the cloning steps and screening procedures of transformants by in situ hybridization, GUS staining, and western blotting are provided in Supplementary Materials and methods.
Purification of mRNA from HF–RPL18 plants
Plants were grown on a soil:vermiculite:perlite mixture under constant illumination with a light intensity range of 80–100 μmol/m2/s at 20°C. Immediately after the onset of bolting, inflorescences of plants with 35S:AP1-GR ap1 cal were treated with a solution containing 1 μM dexamethasone (Sigma-Aldrich, St Louis, MO), 0.01% (v/v) ethanol, and 0.015% (v/v) Silwet L-77. Inflorescence tissue was collected under a dissecting microscope 3 or 5 days after dexamethasone treatment using jewelers forceps (Wellmer et al, 2006). Two independent sets of biological samples were pooled for each replicate.
For transcriptome profiling, total RNA was extracted using RNeasy kit (Qiagen, Valencia, CA) as previously described (Jiao et al, 2008). For translatome profiling, tissue was frozen in liquid nitrogen, powdered using a chilled mortar and pestle, and homogenized in ice-cold polysome extraction buffer (200 mM Tris–HCl (pH 9.0), 200 mM KCl, 36 mM MgCl2, 25 mM ethylene glycol tetraacetic acid, 1 mM dithiothreitol, 50 μg/ml cycloheximide, 50 μg/ml chloramphenicol, 1% Igepal CA-630, 1% Brig-35, 1% Triton X-100, 1% Tween-20, 2% polyoxyethylene (10) tridecyl ether, 1% sodium deoxycholate, 0.5 mg/ml heparin, and recombinant RNase inhibitors). After incubation on ice for 10 min, homogenates were centrifuged for 10 min at 16 000 g and 4°C to pellet insoluble cell debris. Anti-FLAG M2 affinity gel (Sigma-Aldrich, St Louis, MO) was added to the supernatant, and the mixture was incubated at 4°C for at least 30 min. Gels were subsequently collected by centrifugation, and washed three times with polysome wash buffer (200 mM Tris–HCl (pH 9.0), 200 mM KCl, 36 mM MgCl2, 25 mM ethylene glycol tetraacetic acid, 5 mM dithiothreitol, 50 μg/ml cycloheximide, and 50 μg/ml chloramphenicol). Polysomes were eluted by resuspension of the washed gel in polysome wash buffer containing 3 × FLAG peptide. A final elution was immediately used to extract the bound rRNA and mRNA using an RNeasy kit (Qiagen, Valencia, CA), which was subjected to two rounds of hybridization to oligo(dT)-coated Dynal magnetic beads (Invitrogen, Carlsbad, CA).
cDNA preparation and sequencing
Sequencing libraries were prepared from poly(A)+ RNA as described previously (Mortazavi et al, 2008) with reduced RNA input (1–10 μg total RNA). Libraries were sequenced as 37- or 38-mers using Genome Analyzer II (Illumina, San Diego, CA) with standard settings.
Read mapping and expression quantification
The Arabidopsis genome built and annotated gene set were downloaded from The Arabidopsis Information Resource (ftp://ftp.arabidopsis.org/home/tair/Sequences/). Additional ncRNA annotation from Ben Amor et al (2009) was added. After removing reads containing sequencing adapters, we mapped reads to the Arabidopsis TAIR9 genome build with BOWTIE (Langmead et al, 2009) allowing up to two mismatches. Reads that fail to be mapped to aligned by de novo mapped using BLAT (Kent, 2002) against the same genome build to find splice junctions. Remaining reads were further mapped to the TAIR9 genome annotation cDNA sequences with BOWTIE allowing up to two mismatches to find more annotated splice junctions.
The gene locus expression levels were measured in the RPKM unit, and were calculated based on mapping outputs using ERANGE (Mortazavi et al, 2008). Expression levels of splicing isoforms were further estimated using rSeq (Jiang and Wong, 2009). The cutoff value of determining significant expression or translation was 1.0 RPKM based on spike-in controls and reproducibility (Supplementary Figure S3). TARs were identified by making contiguous those positions in which reads were aligned above a coverage depth threshold, which was equivalent to 1.0 RPKM in expression levels, and were required to be adjacent with gaps less than 50 nt.
Differential expression analysis was performed using edgeR (Robinson et al, 2010). Data were modeled as negative binomial distributed because our data set contains biological replicates instead of technique replicates. Differential expression among splicing isoforms was identified using rSeq (Jiang and Wong, 2009). The multiple testing errors were addressed using the false discovery rate (FDR). Differential expression cutoff was set as above two-fold changes in expression and adjusted P<0.001. To alleviate the bias in the detection of differential expression influenced by the transcript length (Oshlack and Wakefield, 2009; Young et al, 2010), we added the expression ratio cutoff (two-fold).
Accession numbers
Supplementary Material
Supplementary Materials and Methods, Supplementary Figures.
Supplementary Tables
Acknowledgments
We thank J Bailey-Serres, T Laux, I Moore, and F Wellmer for seeds and constructs; X Zhang and S-O Shan for invaluable help during polysome isolation; A Mortazavi, B Williams, B Wold, and H Jiang for advice on deep sequencing and data processing; H Amrhein, L Schaeffer, and D Trout for professional technical assistance; and W Li, A Roeder, K Sugimoto, and other members of the Meyerowitz lab for comments on the paper. This study was supported by the Edelman Discovery Fund of the California Institute of Technology (CIT) to EMM, by NSF 2010 Project grant MCB-0929349 to EMM and YJ, and by the Millard and Muriel Jacobs Genetics and Genomics Laboratory at CIT.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D (2003) Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 100: 3889–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941 [DOI] [PubMed] [Google Scholar]
- Ben Amor B, Wirth S, Merchan F, Laporte P, d'Aubenton-Carafa Y, Hirsch J, Maizel A, Mallory A, Lucas A, Deragon JM, Vaucheret H, Thermes C, Crespi M (2009) Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res 19: 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonke M, Thitamadee S, Mahonen AP, Hauser MT, Helariutta Y (2003) APL regulates vascular tissue identity in Arabidopsis. Nature 426: 181–186 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Sorenson R, Juntawong P (2009) Getting the message across: cytoplasmic ribonucleoprotein complexes. Trends Plant Sci 14: 443–453 [DOI] [PubMed] [Google Scholar]
- Branco-Price C, Kaiser KA, Jang CJ, Larive CK, Bailey-Serres J (2008) Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J 56: 743–755 [DOI] [PubMed] [Google Scholar]
- Brioudes F, Joly C, Szecsi J, Varaud E, Leroux J, Bellvert F, Bertrand C, Bendahmane M (2009) Jasmonate controls late development stages of petal growth in Arabidopsis thaliana. Plant J 60: 1070–1080 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Brown JW, Simpson CG (1998) Splice site selection in plant pre-mRNA splicing. Annu Rev Plant Physiol Plant Mol Biol 49: 77–95 [DOI] [PubMed] [Google Scholar]
- Castellana NE, Payne SH, Shen Z, Stanke M, Bafna V, Briggs SP (2008) Discovery and revision of Arabidopsis genes by proteogenomics. Proc Natl Acad Sci USA 105: 21034–21038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Song S, Xiao L, Soo HM, Cheng Z, Xie D, Peng J (2009) Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet 5: e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Fry B, Kamal M, Xie X, Cuff J, Lin MF, Kellis M, Lindblad-Toh K, Lander ES (2007) Distinguishing protein-coding and noncoding genes in the human genome. Proc Natl Acad Sci USA 104: 19428–19433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC (2010) Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 20: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res 32: W273–W279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith DW, Birnbaum K (2006) Global studies of cell type-specific gene expression in plants. Annu Rev Plant Biol 57: 451–475 [DOI] [PubMed] [Google Scholar]
- Gallie DR (1993) Posttranscriptional regulation of gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 44: 77–105 [Google Scholar]
- Goldberg RB, Hoschek G, Kamalay JC, Timberlake WE (1978) Sequence complexity of nuclear and polysomal RNA in leaves of the tobacco plant. Cell 14: 123–131 [DOI] [PubMed] [Google Scholar]
- Goodall GJ, Filipowicz W (1989) The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell 58: 473–483 [DOI] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, Greengard P, Heintz N (2008) A translational profiling approach for the molecular characterization of CNS cell types. Cell 135: 738–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO (2009) Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol 7: e1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Tudor M, Su T, Zhang Y, Hu Y, Ma H (1996) DNA binding properties of two Arabidopsis MADS domain proteins: binding consensus and dimer formation. Plant Cell 8: 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wong WH (2009) Statistical inferences for isoform expression in RNA-Seq. Bioinformatics 25: 1026–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Deng XW (2007) A genome-wide transcriptional activity survey of rice transposable element-related genes. Genome Biol 8: R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Riechmann JL, Meyerowitz EM (2008) Transcriptome-wide analysis of uncapped mRNAs in Arabidopsis reveals regulation of mRNA degradation. Plant Cell 20: 2571–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Tausta SL, Gandotra N, Sun N, Liu T, Clay NK, Ceserani T, Chen M, Ma L, Holford M, Zhang HY, Zhao H, Deng XW, Nelson T (2009) A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nat Genet 41: 258–263 [DOI] [PubMed] [Google Scholar]
- Kawaguchi R, Bailey-Serres J (2005) mRNA sequence features that contribute to translational regulation in Arabidopsis. Nucleic Acids Res 33: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, Girke T, Bray EA, Bailey-Serres J (2004) Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J 38: 823–839 [DOI] [PubMed] [Google Scholar]
- Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (2005) Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361: 13–37 [DOI] [PubMed] [Google Scholar]
- Krizek BA, Fletcher JC (2005) Molecular mechanisms of flower development: an armchair guide. Nat Rev Genet 6: 688–698 [DOI] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB (2009) Coding-sequence determinants of gene expression in Escherichia coli. Science 324: 255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Matsui A, Hanada K, Kawashima M, Ishida J, Morosawa T, Tanaka M, Kaminuma E, Mochizuki Y, Matsushima A, Toyoda T, Shinozaki K, Seki M (2009) Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc Natl Acad Sci USA 106: 2453–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner DH, Beilharz TH, Marguerat S, Mata J, Watt S, Schubert F, Preiss T, Bähler J (2007) A network of multiple regulatory layers shapes gene expression in fission yeast. Mol Cell 26: 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowska D, Simpson CG, Clark GP, Jennings NS, Barciszewska-Pacak M, Lin CF, Makalowski W, Brown JW, Jarmolowski A (2004) Determinants of plant U12-dependent intron splicing efficiency. Plant Cell 16: 1340–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Vogel C, Wang R, Yao X, Marcotte EM (2007) Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol 25: 117–124 [DOI] [PubMed] [Google Scholar]
- Maquat LE (2004) Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5: 89–99 [DOI] [PubMed] [Google Scholar]
- Mara CD, Irish VF (2008) Two GATA transcription factors are downstream effectors of floral homeotic gene action in Arabidopsis. Plant Physiol 147: 707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguerat S, Bähler J (2010) RNA-seq: from technology to biology. Cell Mol Life Sci 67: 569–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R (2004) Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J 39: 877–885 [DOI] [PubMed] [Google Scholar]
- Nelson T, Gandotra N, Tausta SL (2008) Plant cell types: reporting and sampling with new technologies. Curr Opin Plant Biol 11: 567–573 [DOI] [PubMed] [Google Scholar]
- Obrig TG, Culp WJ, McKeehan WL, Hardesty B (1971) The mechanism by which cycloheximide and related glutarimide antibiotics inhibit peptide synthesis on reticulocyte ribosomes. J Biol Chem 246: 174–181 [PubMed] [Google Scholar]
- Oshlack A, Wakefield MJ (2009) Transcript length bias in RNA-seq data confounds systems biology. Biol Direct 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piques M, Schulze WX, Hohne M, Usadel B, Gibon Y, Rohwer J, Stitt M (2009) Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol Syst Biol 5: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Ahn S, Speed TP, Rubin GM (2007) Global analyses of mRNA translational control during early Drosophila embryogenesis. Genome Biol 8: R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS (2007) Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol 58: 267–294 [DOI] [PubMed] [Google Scholar]
- Reyes JC, Muro-Pastor MI, Florencio FJ (2004) The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol 134: 1718–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Krizek BA, Meyerowitz EM (1996) Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc Natl Acad Sci USA 93: 4793–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schuler MA (2008) Splice site requirements and switches in plants. Curr Top Microbiol Immunol 326: 39–59 [DOI] [PubMed] [Google Scholar]
- Sheth N, Roca X, Hastings ML, Roeder T, Krainer AR, Sachidanandam R (2006) Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res 34: 3955–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson CG, Fuller J, Maronova M, Kalyna M, Davidson D, McNicol J, Barta A, Brown JW (2008) Monitoring changes in alternative precursor messenger RNA splicing in multiple gene transcripts. Plant J 53: 1035–1048 [DOI] [PubMed] [Google Scholar]
- Simpson CG, Manthri S, Raczynska KD, Kalyna M, Lewandowska D, Kusenda B, Maronova M, Szweykowska-Kulinska Z, Jarmolowski A, Barta A, Brown JW (2010) Regulation of plant gene expression by alternative splicing. Biochem Soc Trans 38: 667–671 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Matsumoto N, Okada K (2004) RABBIT EARS, encoding a SUPERMAN-like zinc finger protein, regulates petal development in Arabidopsis thaliana. Development 131: 425–434 [DOI] [PubMed] [Google Scholar]
- Tilly JJ, Allen DW, Jack T (1998) The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development 125: 1647–1657 [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer F, Alves-Ferreira M, Dubois A, Riechmann JL, Meyerowitz EM (2006) Genome-wide analysis of gene expression during early Arabidopsis flower development. PLoS Genet 2: e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Girke T, Pasala S, Xie M, Reddy GV (2009) Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc Natl Acad Sci USA 106: 4941–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Ito T, Zhao Y, Peng J, Kumar P, Meyerowitz EM (2004) Floral homeotic genes are targets of gibberellin signaling in flower development. Proc Natl Acad Sci USA 101: 7827–7832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti ME, Chang IF, Gong F, Galbraith DW, Bailey-Serres J (2005) Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol 138: 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Byrnes JK, Gal TS, Li W-H, Borevitz JO (2008) Whole genome transcriptome polymorphisms in Arabidopsis thaliana. Genome Biol 9: R165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Medrano L, Ohashi K, Fletcher JC, Yu H, Sakai H, Meyerowitz EM (2004) HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell 16: 2586–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods, Supplementary Figures.
Supplementary Tables