Abstract
Acrolein (AC) and 4-hydroxy-2-nonenal (HNE) are α,β-unsaturated aldehyde (enal) endogenous bis-electrophiles that arise from the oxidation of polyunsaturated fatty acids. AC is also found in high concentrations in cigarette smoke and automobile exhaust. These reactive enals covalently modify nucleic acids, to form exocyclic adducts, where the three-carbon hydroxypropano unit bridges the N1 and N2 -positions of deoxyguanosine (dG). The bifunctional nature of these enals enables them to undergo reaction with a second nucleophilic group and form DNA cross-links. These cross-linked enal adducts are likely to contribute to the genotoxic effects of both AC and HNE. We have developed a sensitive mass spectrometric method to detect cross-linked adducts of these enals in calf thymus DNA (CT DNA) treated with AC or HNE. The AC and HNE cross-linked adducts were measured by the stable isotope dilution method, employing a linear quadrupole ion trap mass spectrometer and consecutive reaction monitoring at the MS3 or MS4 scan stage. The lower limit of quantification of the cross-linked adducts is ~1 adduct per 108 DNA bases, when 50 µg of DNA is assayed. The cross-linked adducts occur at levels that are ~1–2% of the levels of the monomeric 1,N2-dG adducts in CT DNA treated with either enal.
Introduction
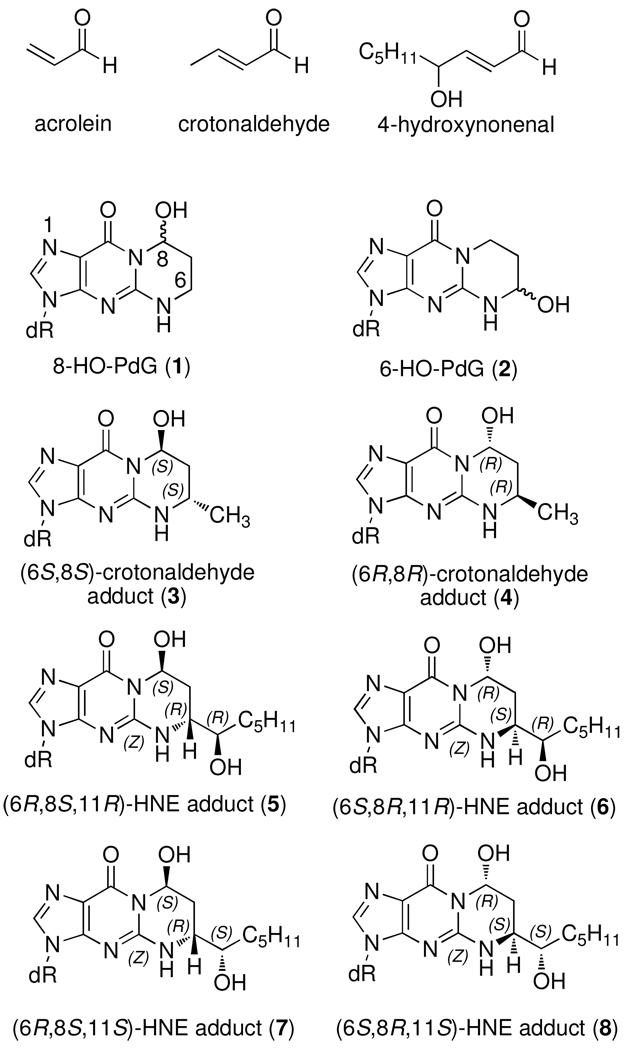
Acrolein (AC), crotonaldehyde, 4-hydroxy-2-nonenal (HNE) and other α,β-unsaturated aldehydes (enal) are endogenous bis-electrophiles that arise from the degradation of polyunsaturated fatty acids initiated by reactive oxygen species (1). Acrolein is also found in high concentrations in cigarette smoke and automobile exhaust (2). Enals are reactive electrophiles that covalently modify proteins, peptides, and nucleic acids. As a result, many enals have been shown to be cytotoxic and genotoxic (3–5).
The bifunctional nature of enals enables them to undergo reaction with two nucleophilic groups. In the case of deoxyguanosine (dG), this reactivity results in a so-called exocyclic adduct in which a three-carbon hydroxypropano unit bridges the N1 and N2 -positions (Figure 1). The reaction of AC with dG affords two regioisomeric products, the 8-hydroxy-1,N2 -propano-dG (1, 8-HO-PdG) and 6-hydroxy-1,N2-propano-dG (2, 6-HO-PdG) adducts in approximately a 2:1 ratio. Thus far, only the 8-hydroxy isomers have been identified from the higher enals. The reaction of crotonaldehyde with dG results in two stereoisomeric adducts (3 and 4), while HNE produces four stereoisomers (4–8). AC, crotonaldehyde, and HNE adducts of dG have been found in background levels in rodent and human DNA (6–11); high levels of the AC and HNE adducts have been detected in the brains of Alzheimer’s patients (12, 13) and both regioisomeric AC adducts were found in the lung tissue from smokers (14). Acrolein, crotonaldehyde, and HNE adducts have been demonstrated to be mutagenic when replicated in mammalian cells using site-specifically modified vector with G→T transversions being the predominate mutation observed. (5).
Figure 1.
Structures of AC, crotonaldehyde, 4-HNE, and their dG adducts
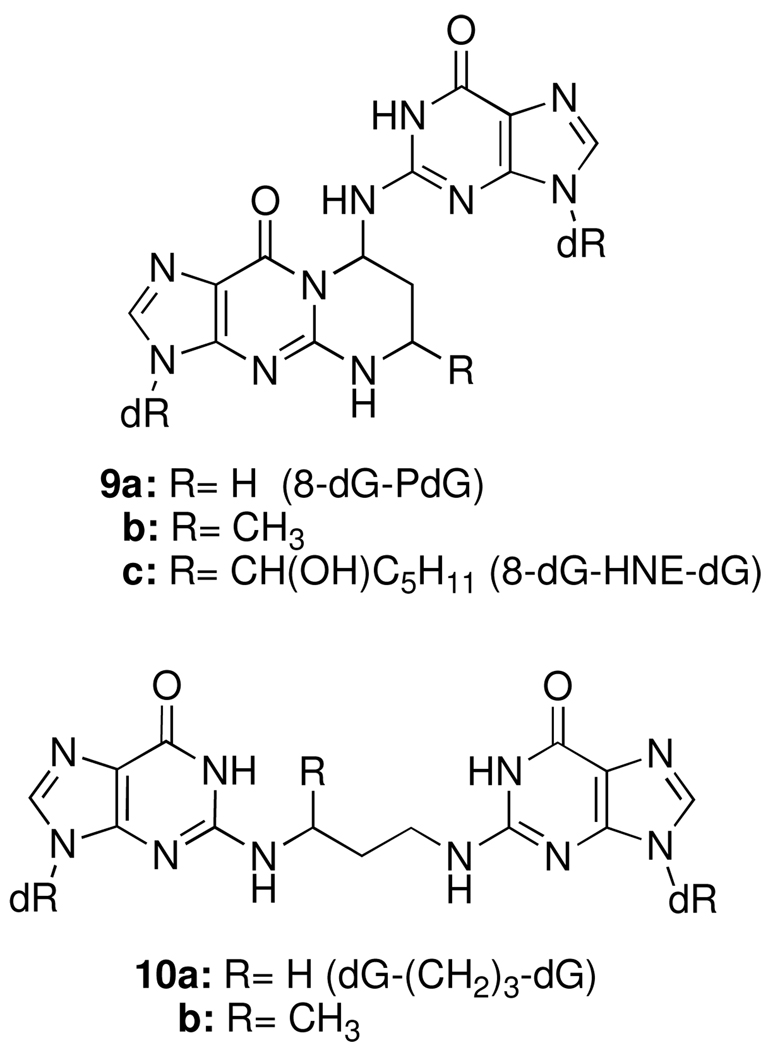
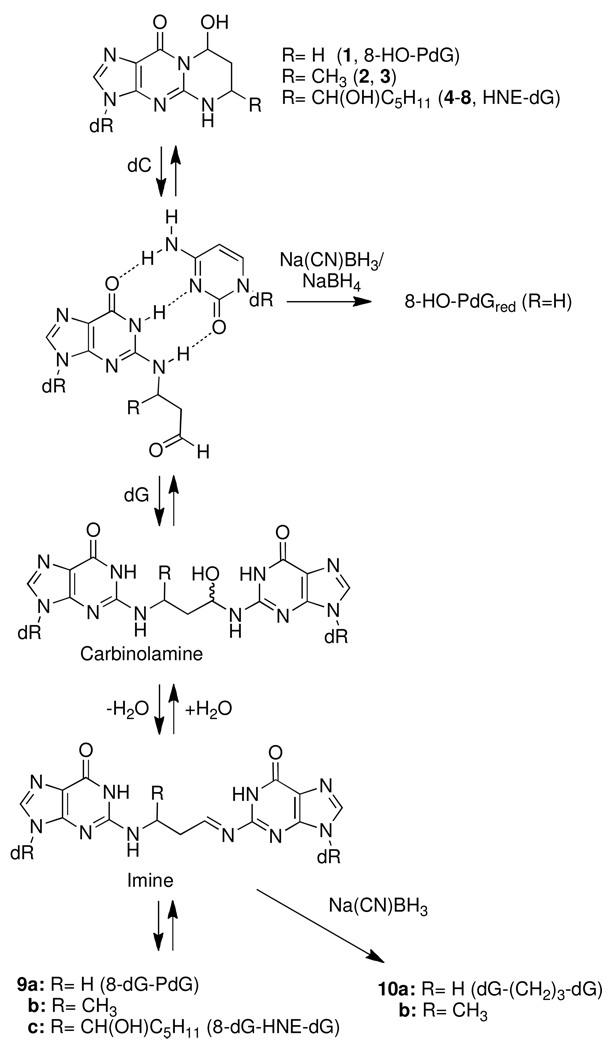
Kawanishi reported electrophoretic evidence that acrolein can cross-link DNA (15); however, the structure of the cross-link was not determined. Wang et al. identified a pyrimidopurinone crosslink from calf thymus DNA (CT DNA) treated with acetaldehyde (Figure 2, 9b) (16, 17). This product is formally a cross-link derived from crotonaldehyde and two dG’s; although crotonaldehyde is the aldol condensation product of acetaldehyde, it was not believed to be involved in the cross-linking reaction (18). Kozekov et al. demonstrated that oligonucleotides containing dG adducts of acrolein and crotonaldehyde formed interstrand DNA cross-links when site-specifically incorporated in a 5′-CpG-3′ sequence context (19, 20); the HNE adduct was subsequently shown to form interstrand cross-links in the same sequence (21). The cross-linking reaction was slow in each case, but most efficient for the AC adduct and highly dependent on the stereochemistry of the crotonaldehyde and HNE adducts (22). The corresponding pyrimidopurinones (9a–c) (Figure 2) were identified from the enzymatic digestion of the cross-links duplex. Treatment of AC and crotonaldehyde cross-linked duplexes with Na(CN)BH3 prior to digestion resulted in the reduced cross-links where the N2-positions of two dG’s is tethered by a three-carbon chain (10, dG-(CH2)3-dG) (19, 22).
Figure 2.
Structures of dG cross-linked adducts of AC, crotonaldehyde and HNE (9), and their chemically reduced analogs (10).
Many cellular processes involving DNA require transient strand separation. Interstrand cross-links are anticipated to interfere with transcription, replication, and DNA repair. The mechanism of action of many anti-cancer agents, such as nitrogen mustards, mitomycin C and cisplatin, are believed to involve DNA cross-links (23–26). Cell lines from patients with the rare genetic disorder Fanconi anemia are highly sensitive to DNA cross-linking agents, but not other DNA damaging agents; this suggests that cells have developed mechanisms to repair DNA crosslinks (26–29). We have hypothesized that interstrand DNA cross-links may contribute to the genotoxic effects of enals. We report here the development of a mass spectrometric method to detect dG enal cross-links in calf thymus DNA (CT DNA) treated with AC or HNE. The cross-linked adducts were quantitated by the stable isotope dilution method, using liquid chromatography-electrospray ionization/multi-stage tandem mass spectrometry (LC-ESI/MS/MSn) with a linear quadrupole ion trap mass spectrometer (LIT MS) and consecutive reaction monitoring at the MS3 or MS4 scan stage. We estimate that the cross-linked adducts are present at levels that are ~1% that of the monomeric 1,N2-dG adducts.
Experimental Procedures
Materials
AC was purchased from Aldrich. HNE was prepared according to a literature procedure (30). [15N5]-dG (>98% isotopic purity) was purchased from Cambridge Isotope Laboratory, Inc. [2H6]-1,3-diaminopropane hydrochloride (>98.8% isotopic purity) was purchased from CDN Isotopes. [2H11]-1-bromopentane (>98% isotopic purity was purchased from Isotec (Sigma-Aldrich). Oligonucleotides containing the 8-HO-PdG and HNE-dG adducts were prepared as previously described (21,31). Calf thymus (CT) DNA, dG, adenosine deaminase (Type X from calf spleen), DNase I (Type IV, bovine pancreas), alkaline phosphatase (from E. coli), and nuclease P1 (from Penicillium citrinum) were purchased from Sigma (St. Louis, MO). Phosphodiesterase I (from Crotalus adamanteus venom) was from GE Healthcare (Piscataway, NJ). HyperSep™ filter SpinTips C18 (20 mg) were from Thermo Scientific (Palm Beach, FL). The isotopic purity of the 15N and 2H-labeled standards was > 99.5%.
HPLC Separations
The purification of nucleosides and oligonucleotides and the analysis of reaction mixtures and nucleosides obtained from enzymatic digestions were performed on a Beckman HPLC system (32 Karat software version 7.0, pump module 125) with a diode array UV detector (module 168) monitoring at 260 nm. An analytical Waters YMC ODS-AQ column (250 mm × 4.6 mm i.d., 1.5 mL/min) was used for monitoring the reactions and a semi-preparative column (250 mm × 10 mm i.d., 5 mL/min) was used for purification of the desired products. The mobile phases were H2O and CH3CN for nucleosides and 0.1 M aqueous ammonium formate and CH3CN for oligonucleotides and some nucleosides with the following gradients: Gradient A: initially 1% acetonitrile, 1–10% acetonitrile over 15 min, 10–20% acetonitrile over 5 min, hold for 5 min, 20–100% acetonitrile over 3 min, hold for 2 min, and then back to 1% acetonitrile over 3 min. Gradient B: initially 1% acetonitrile, 1–5% acetonitrile over 5 min, 5–6% acetonitrile over 15 min, 6–100% acetonitrile over 3 min, hold for 2 min, and then back to 1% acetonitrile over 3 min. Gradient C: initially 18% acetonitrile, 18–26% acetonitrile over 38 min, and then back to 18% acetonitrile over 2 min.
6-HO-PdG/[15N5]-6-HO-PdG and 8-HO-PdG/[15N5]-8-HO-PdG
Acrolein 31.3 µL (0.468 mmol, 3 eqv.) was added in three portion over one hour to a solution of dG•H2O (44.5 mg, 0.156 mmol) and L-arginine (81.6 mg, 0.486 mmol) in degassed phosphate buffer (5 mL, 0.1 M, pH8) at 37° C, and reaction was incubated overnight. The dG was completely consumed in the reaction and three products were observed in nearly equal amounts. The reaction mixture was extracted with ethyl acetate (3 × 5 mL), and then purified by HPLC using Gradient B. The first two peaks were collected together and were the two diastereoisomers of 6-HO-PdG (18 mg, 35.5%). The third peak was collected separately and was 8-HO-PdG (11 mg, 21.8%). The 8- and 6-HO-PdG were identified based on comparison of the retention time with known standards (7, 32). 8-HO-PdG: UV: λmax 260 nm (ε=11650, H2O). 6-HO-PdG, UV: λmax 260 nm (ε=10,880, H2O).
The reaction was perform with (15N5)-dG(5mg) and the isotopic purity of [15N5]-6-HO-PdG and [15N5]-8-HO-PdG was > 99.5% by LC-MS/MS.
N2-(3-Hydroxypropyl)-PdG (8-HO-PdGred)
8-HO-PdG (2.0 mg) in 1:1 (v/v) H2O/methanol (200 µL) was treated with sodium borohydride (3.0 mg in 39.6 µL 1N NaOH) (32). The mixture was stirred at room temperature for 8 h and then neutralized with 5% acetic acid. HPLC purification (Gradient B) afforded 8-HO-PdGred (1.5 mg, 75%). UV: λmax 260 nm (ε= 13,551, H2O).
The reaction was performed with (15N5)-8-HO-PdG (0.5 mg) and the isotopic purity of (15N5)-8-HO-PdGred was > 99.5% by LC-MS/MS.
N1-(3-Hydroxypropyl)-dG (6-HO-PdGred)
NaBH4 (6 mg, 50 equiv) was added in three portions to a solution of 6-HO-PdG (1.0 mg) in phosphate buffer (200 µL. 50 mM, pH 7.0) over 1 h. The reaction was followed by HPLC and the starting material was completely consumed after 24 h. The reaction was neutralized with 5% acetic acid, and then was purified by HPLC (Gradient A) to afford 6-HO-PdGred (0.5 mg, 50%). UV: λmax 260 nm (ε= 11,180, H2O).
1,N2-Propano-dG (PdG)
A solution of 6-HO-PdG (1.0 mg) and Na(CN)BH3 (3 mg, 15 equiv) in phosphate buffer (200 µL, 50 mM, pH 7.0) was stirred for 24 h at 45°C, after which time HPLC analysis showed the reaction to be complete. Purification by HPLC (Gradient A) afforded PdG (0.6 mg, 60%). UV: λmax 260 nm (ε= 13,190, H2O).
[15N5]-1,N2-Propano-dG ([15N5]-PdG)
[15 N5]-dG (1 mg, 3.7 µmol) and anhydrous K2CO3 (5 mg) were placed in glass tube with dry DMSO (300 µL). After 30 min at 45° C 1,3-dibromopropane (0.82 mg, 0.41 µL, 4 µmol) was added and the reaction was kept at 45° C for 24 h. HPLC showed that the dG completely consuming the starting material and the product was purified by HPLC (Gradient A) to afford [15N5]-PdG (0.7 mg, 61%).
[2H6]-1,3-Bis(2'-deoxyguanosin-N2-yl)propane (dG-(CH2)3-dG)
The [2H6]-1,3-diaminopropane hydrochloride (5 mg, 32.8 µmol) was added to excess of O6-(trimethylsilyl)ethyl)-2-fluoro-2’-deoxyinosine (25 mg, 67.5 µmol), in DMSO (150 µL) and diisopropylethylamine (200 µL). The mixtures were stirred at 55°C for 24 h. The reactions were stopped and the solvents were evaporated under vacuum. The residues were dissolved in 5% acetic acid (1 mL) and stirred at room temperature for 1 h, neutralized, followed by HPLC purification (Gradient A) afforded [2H6]-dG-(CH2)3-dG (10.5 mg, 55%). UV λmax 260 nm (ε= 23,700, H2O).
[2H11]-4-HNE
Mg(0) turnings (90 mg, 3.6 mmol) and dry ethyl ether (2 mL) were placed in a three-necked flask. A few drops of a solution of [2H11]-1-bromopentane (420 mg, 2.8 mmol) in dry ether (4 mL) were added, and the flask was gently heated until the reaction started. After the initiation of Grignard reagent formation, the remaining bromide solution was added dropwise. The reaction mixture was heated at reflux for 30 min, and then cooled to 0°C. A solution of fumaraldehyde monodimethyl acetal (300 mg, 2.3 mmol) in dry ether (2 mL) was added dropwise. The monoacetal was obtained by the partial acid hydrolysis of the fumaraldehyde dimethyl acetal (500 mg, 2.8 mmol), using Amberlyst-15 catalyst (0.12 g) in acetone (11.5 mL) and H2O (0.1 mL). The reaction was stirred for 6 min, and then filtered through a bed of anhydrous sodium carbonate and solvent removed. After all of the monoacetal had been added, the stirring was continued for an additional 2 h at 0°C. The reaction was quenched with saturated aqueous NH4Cl and extracted with diethyl ether. The extracts were dried with MgSO4, filtered and evaporated. Purification on a Biotage medium pressure chromatography apparatus using an M25 column and eluting with 0–5% methanol in dichloromethane afforded the dimethyl acetal of [2H11]-HNE. The dimethyl acetal was hydrolyzed with 2% H2SO4 (4.2 mL) for 1h at room temperature. The solution was neutralized with 1M NaOH and extracted with diethyl ether. The extracts was dried with MgSO4, filtered and evaporated. Purification on a Biotage medium pressure chromatography apparatus using an M25 column and eluting with 0–3% methanol in dichloromethane afforded [2H11]-4-HNE (280 mg, 78%).
[2H11]-HNE-dG
[2H11]-4-HNE (17.6 mg, 0.105 mmol) was added to a solution of dG•H2O (30 mg, 0.105 mmol) and L-arginine (55 mg, 0.315 mmol) in degassed phosphate buffer (4 mL, 0.1 M, pH 8.0) at 37 °C. Two additional portions of [2H11]-4-HNE were added after 15 and 60 min. The reaction vessel was sealed and heated at 37° C for 24h. After cooling, the unreacted HNE and polymerized products were extracted with ethyl acetate (3 × 5 mL). Purification of the aqueous phase by HPLC (Gradient C) afforded [2H11]-HNE-dG (16 mg, 35%) as a mixture of four stereoisomers. UV: λmax 260 nm (ε= 12,000, H2O).
[2H11]-8-dG-HNE-dG
[2H11]-HNE-dG (3 mg) and dG(6 mg) were dissolved in DMSO (200 µL). The reaction mixture was heated at 100 °C for 5 days, and then the solvent was evaporated under vacuum (21). The residue was dissolved in sodium phosphate buffer (500 µL, pH 7.0, 0.05 M) and purified by HPLC (Gradient C) to afford [2H11]-8-dG-HNE-dG (0.4 mg, 8%). UV λmax 260 nm (ε= 24,200, H2O).
Cross-linking of the 5′-GCTAGC(8-HO-PdG)AGTCC-3′ • 5′-GGACTCGCTAGC-3′ Duplex
The 5′-GCTAGC(8-HO-PdG)AGTCC-3′ oligonucleotide (1.0 OD, 8.1 nmol, MW = 3,700.6) was hybridized with complementary strand (1.1 OD, 8.91 nmol, MW = 3,646.4) and incubated in phosphate buffer (200 µL, 50 mM, pH 7.0 containing 1M KCl) at 45 °C. The cross-linking reaction was monitored by CGE; after 10 day the level of interstrand cross-link was ~50% (19, 20). The reaction was treated with Na(CN)BH3 (0.1 M final concentration). Additional Na(CN)BH3 (0.2 M final concentration) was added after 30 min, then again after 1h (0.3 M final concentration). The sample was incubated at room temperature for 24 h, and then quenched with 5% CH3CO2H. The reaction was filtered through a spin column (Bio-Spin 6 Tris Columns from BioRad) and lyophilized.
Cross-linking of the 5′-GCTAGC(8-HNE-dG)AGTCC-3′ • 5′-GGACTCGCTAGC-3′ Duplex
The 5′-GCTAGC(8-HNE-dG)AGTCC-3′ oligonucleotide (1.5 OD, 12.15 nmol, MW = 3,802.5) was hybridized with complementary strand (1.6 OD, 12.96 nmol, MW = 3,646.4) and incubated in phosphate buffer (200 µL, 50 mM, pH=7.0 containing 1M KCl) at 45° C. The cross-linking reaction was monitored by CGE; the level of cross-link was estimated to be ~50 %, based upon UV response, after 28 days (21).
Capillary Gel Electrophoresis (CGE)
Electrophoretic analyses were carried out using a Beckman P/ACE MDQ instrument system (using 32 Karat software, version 5.0) monitored at 260 nm on a 31.2 cm × 100 µm eCAP capillary with samples applied at 10 kV and run at 9 kV. The capillary was packed with their 100-R gel (for ss-DNA) using the Tris-borate buffer system containing 7 M urea.
Chemical Modification of DNA with AC
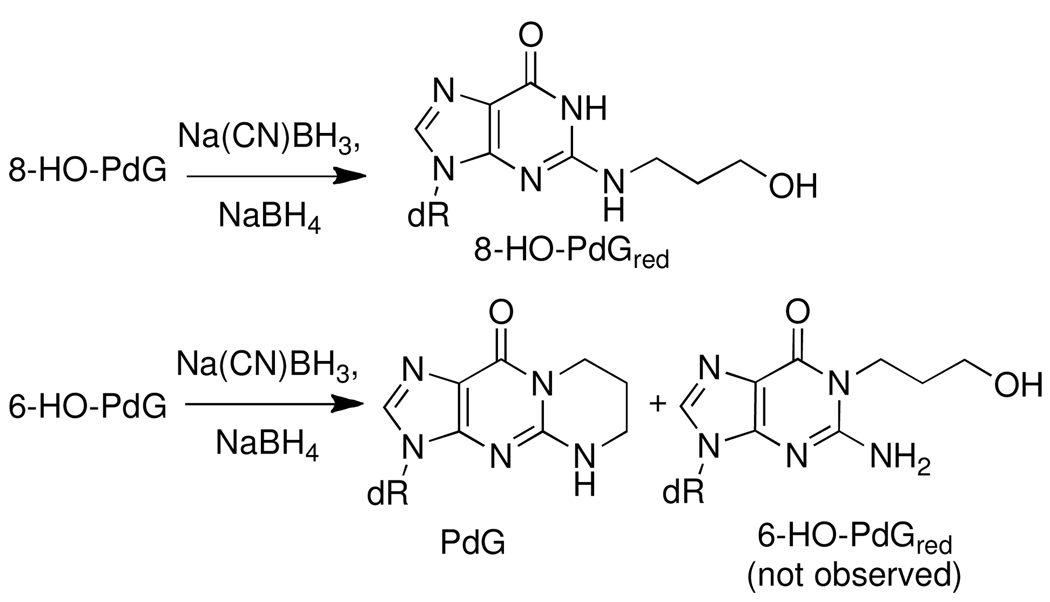
CT DNA (0.33 mg/0.33 mL) in 50 mM sodium phosphate buffer (pH 7.0) was treated with 0.1, 0.5, 1 or 5 mol of AC per mol of DNA base for 0, 1, 2, 3, 6 or 10 days, at 37 °C. The unreacted AC and its by-products were removed from the DNA by 3 solvent extractions with an equal volume of ethyl acetate. The AC-cross-linked adduct 8-dG-AC-dG (9a) was chemically reduced by treatment of the CT DNA with Na(CN)BH3 (10 µL of 3.3 M stock solution). The mixture was incubated at room temperature for 15 min. Thereafter, another 10 µL of the Na(CN)BH3 solution was added and the reduction continued for 15 min. Finally, another 10 µL of the Na(CN)BH3 solution was added after 1 h, and the reduction reaction stirred overnight at room temperature. The DNA solution was then quenched with 5% CH3CO2H (~50 µL) to attain a pH value between 7.0 and 7.5. The treatment of AC-modified DNA with Na(CN)BH3 completely reduced the 8-HO-PdG cross-link 8-dG-AC-dG to its ring-opened, reduced form dG-(CH2)3-dG (10) (Figure 2) However, 6-HO-PdG and 8-HO-PdG only underwent partial reduction. Therefore, a second chemical reduction with NaBH4 was done, following enzymatic digestion of the DNA, as described below, to reduce both HO-PdG adducts (Scheme 2). The AC-DNA was precipitated by addition of 0.1 volumes of 5 M NaCl, followed by 1.5 mL C2H5OH. The DNA filament was washed with C2H5OH:H2O (7:3) and air-dried.
Scheme 2.
Chemical reduction of AC and HNE-DNA adducts by Na(CN)BH3/NaBH4
Chemical Modification of DNA with HNE
CT DNA (1 mg/mL in 50 mM sodium phosphate buffer pH 7.0) was reacted with 2 mol of HNE per mol DNA base. The time course of reaction proceeded for 0, 1, 7, 14, or 21 days at 37 °C. Unreacted HNE and its by-products were removed from the DNA by extraction with an equal volume of ethyl acetate (3×), and the DNA was isolated as described above, except that the DNA was not treated with reducing agents.
Enzymatic Digestion of AC and HNE-DNA Adducts and Interstrand Cross-linked Adducts
The enzyme digestion conditions used for the hydrolysis of AC-modified DNA (10 – 50 µg, in 5 mM BisTris-HCl buffer (pH 7.1, 50 µL) employed DNAse I (13 U) for 1.5 h at 37 °C, followed by incubation with nuclease P1 (200 mU, 0.1 U/µL in 1 mM ZnCl2) for 3 h, and then by treatment with alkaline phosphatase (100 mU) and phosphodiesterase (4 mU) for 18 h (33). Thereafter, the DNA digest was treated with adenine deaminase (15 mU), for 90 min at room temperature. A second chemical reduction was then done, by treatment of the digest with NaBH4 (3 × 6 µL of 2 M solution prepared in 1N NaOH), over a time period of 20 min. The treatment with NaBH4 completely reduced 8-HO-PdG to its ring-opened form N2-(3-hydroxypropyl)-2′-deoxyguanosine (8-HO-PdGred). Reduction of the 6-HO-PdG resulted in 1,N2-(1,3-propano)-dG (PdG), presumably through the corresponding N5-C6 imine intermediate (14); N1-(3-hydroxypropyl)-2′-deoxyguanosine (6-HO-PdGred) from the reduction of the ring-opened aldehyde was not observed (Scheme 1). The excess NaBH4 was then quenched with 5% CH3CO2H (~50 µL), to attain a pH value of 7.0 – 7.5.
Scheme 1.
Synthesis of the isotopically labeled internal standards
The 8-dG-HNE-dG cross-link (9c) could not be reduced with either Na(CN)BH3 or NaBH4. Moreover, the cross-link underwent partial hydrolysis to form HNE-dG, during the digestion procedure employed for AC-modified CT DNA (unpublished observations). Therefore, the DNA digestion conditions were modified to conserve the 8-dG-HNE-dG cross-link. The amounts of several of the enzymes used for the digestion of DNA were increased, and the time of digestion was decreased to 2.5 h. CT DNA (50 µg in 5 mM BisTris-HCl buffer (pH 7.1, 50 µL)) was simultaneously digested with DNAse I (10 U), nuclease P1 (1.8 U, 0.1 U/µL in 1 mM ZnCl2), alkaline phosphatase (120 mU), and phosphodiesterase I (200 mU), for 2.5 h at 37 °C.
Solid Phase Extraction (SPE) of AC- and HNE-DNA Adducts
Thereafter, the DNA digest was diluted with high-purity water (200 µL; Burdick and Jackson), and the adducts were purified by SPE, using HyperSep™ filter SpinTips. The AC-modified DNA digests were applied to a SpinTip, which was placed on a vacuum manifold and prewashed with CH3OH containing 0.1% HCO2H (2 × 0.25 mL), followed by H2O (2 × 0.25 mL). The SpinTips were then washed with H2O (2 × 0.25 mL), to remove non-modified 2′-deoxynucleosides. The desired adducts were then eluted with CH3OH containing 0.1% HCO2H (0.3 mL) into silylated glass insert capillary LC vials (Microliter Analytical Supplies, Suwanee, GA). Samples were evaporated to dryness by vacuum centrifugation and reconstituted in H2O (20 µL). The same procedure was employed for HNE-modified DNA, except that the SpinTips were washed with 0.1% HCO2H containing 10% CH3OH (2 × 0.25 mL), instead of H2O (2 × 0.25 mL). The HNE-DNA samples were reconstituted in 1:1 H2O:DMSO (20 µL).
Recovery Experiments on 8-HO-PdG and HNE-dG Cross-links in Duplex DNA
The 5′- GCTAGCXAGTCC-3′ • 5′-GGACTCGCTAGC-3′ duplex (X= 8-HO-PdG), which was incubated for 10 days and contained about 50% of the 8-dG-AC-dG cross-link, was used to assess the stability and recovery of the 8-dG-AC-dG cross-link during the enzyme digestion procedure for CT DNA. The duplex was treated with Na(CN)BH3 to form the reduced dG-(CH2)3-dG cross-linked oligomer duplex (vide supra). The recovery of dG-(CH2)3-dG cross-link during the enzyme digestion of CT DNA was then determined by LC-ESI/MS/MSn. The 5′-GCTAGCXAGTCC-3′ • 5′-GGACTCGCTAGC-3′ duplex (2.9 pmol, X= 8-HO-PdG), pre-reduced with Na(CN)BH3 (vide supra), 29 pmol of [2H6]-dG-(CH2)3-dG, 29 pmol [15N5]-8-HO-PdG, and 29 pmol of [15N5]-8-HO-PdGred were added to CT DNA (25 µg, 75 nmol DNA bases, to achieve a final level of 3.9 dG-AC-dG cross-linked adducts per 105 bases, assuming 100% of adduct is cross-linked in the duplex DNA. The samples were digested, treated with NaBH4, and processed by SPE as described above for the AC-modified CT DNA.
Similarly, the same duplex 5′-GCTAGCXAGTCC-3′ • 5′-GGACTCGCTAGC-3′ duplex containing the (6S,8R,11S)-HNE-dG was incubated for 28 days and level of interstrand crosslink was estimated to be 50%, by CGE with UV detection. The cross-linked duplex (15.1 or 151 fmol) and [2H11]-8-dG-HNE-dG standard (15.1 fmol) were added to CT DNA (50 µg DNA, 151 nmol DNA bases, to achieve a level of 1 or 10 8-dG-HNE-dG cross-linked adducts per 107 bases, assuming 100% of adduct is cross-linked in the duplex DNA. The samples were digested and processed by SPE as previously described for the HNE-modified DNA. The estimates of both the AC and HNE cross-linked adducts determined by LC-ESI/MS/MSn ranged between 40–60% of the amount of duplex added to CT DNA (Table 2), which is consistent with the estimate of crosslink formation (50%) observed by gel electrophoresis. Thus, the stable isotope dilution method adequately compensates for partial reversion of the cross-linked adducts back to the monomers, during the chemical reduction and/or enzyme digestion procedures.
Table 2.
Estimates of Levels of interstrand 8-dG-AC-dG (9a) and 8-dG-HNE-dG (10c) Cross-links Recovered from DNA following Enzyme Digestion.
|
aLevel of spiking 8-dG-AC- dG cross-link in CT DNA |
dG-(CH2)3-dG cross-link measured |
8-HO-PdGred level measured |
|---|---|---|
| 3.9 adducts per 105 bases | 2.1 ± 0.04 per 105 bases | 2.2 ± 0.06 per 105 bases |
|
aLevel of spiking 8-dG-HNE-dG cross-link in CT DNA |
8-dG-HNE-dG cross-link level measured |
|---|---|
| 1.0 adduct per 107 bases | 6.2 ± 2.6 per 108 bases |
| 1.0 adduct per 106 bases | 3.6 ± 1.0 per 107 bases |
We assume that only the crosslinked adducts is present in the oligonucleotide duplex N = 4 independent measurements
S1 nuclease treated CT DNA
CT DNA (25 mg, 2 mg/mL, A260/A280 1.85) in sodium acetate buffer (50 mM, pH 4.5) with NaCl (0.28 M) and ZnSO4 (4.5 mM) was treated with S1 Nuclease (10 000 U, Promega) for 30 min at 37°C. The reaction was quenched with EDTA (500µL, 100 mM) and precipitated with ice-cold ethanol (30 mL). The solution was centrifuged at 7000g for 10 min. The pellet was wash with 70% and 100% ethanol, and dried in a centrifugal evaporator for 5 min. The CT DNA was dissolved in sodium phosphate buffer (5 mL, 50 mM, pH 7.0) and added ProCipitate™ solution (2.5 mL, Biotech Support Group, LLC) to remove the protein. The suspension was gently mixed by inversion for 5 min at room temperature and then the sample was centrifuged at 3000g for 15 min. The supernatant was removed and NaCl solution (300 µL, 5M) added. The CT DNA precipitated with ice-cold ethanol (10 mL), and centrifuged at 7000g for 10 min. The CT DNA was dissolved in sodium phosphate buffer (5 mL, 50 mM, pH 7.0) with NaCl (300 mM), precipitated with ice-cold ethanol (10 mL), and centrifuged at 7000g for 10 min. The pellet was wash with 70% and 100% ethanol, and dried in a centrifugal evaporator for 5 min. The CT DNA was dissolved in water, passed through a 30 cm Sephadex G25 column eluting with water, and lyophilized to dryness (15 mg by A260). A sample was dissolved in sodium phosphate buffer (50 mM, pH 7.0) and the A260/A280 value was 1.90.
LC/MS Parameters
Chromatography was performed with an Agilent 1100 Series capillary LC system (Agilent Technologies, Palo Alto, CA) equipped with an Aquasil C18 column (0.32 × 250 mm) from Thermo Fisher (Bellafonte, PA). Samples (2 or 6 µL) were injected, and adducts were separated with the following gradients. For AC-DNA adducts, the solvent conditions were held at 100% A (solvent composition: 0.01% HCO2H) for 1 min, followed by a linear gradient to 30% B (solvent composition: 95% CH3CN containing 0.01% HCO2H) over 40 min at a flow rate of 6 µL/min. For HNE-modified CT DNA adducts, the solvent conditions commenced at 90% A (solvent composition: 0.01% HCO2H) and 10% B (95% CH3CN containing 0.01% HCO2H) and a linear gradient was conducted and arrived at 100% B at 30 min. The mass spectrometer (MS) was a linear quadrupole ion trap LTQ MS (ThermoElectron, San Jose, CA), and Xcalibur version 2.07 software was used for data manipulations. Analyses were conducted in the positive ionization mode and employed an Advance nanospray source from Michrom Bioresource Inc. (Auburn, CA).
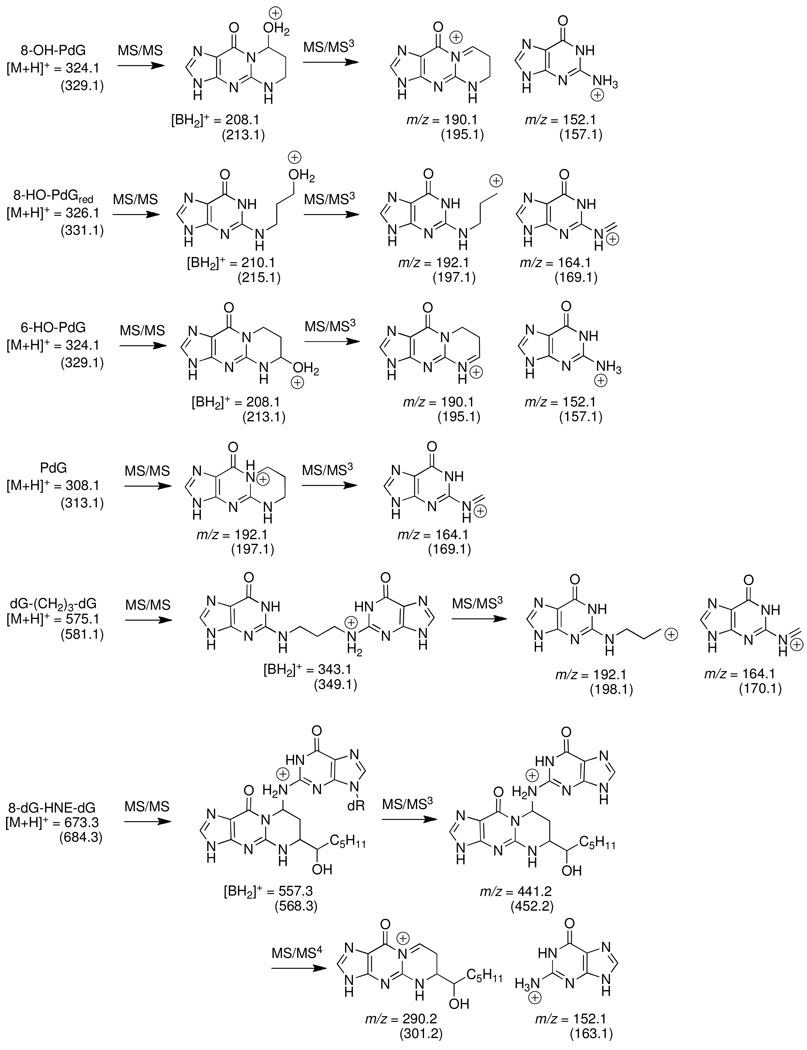
MS fragmentations are shown in Figure 3. The product ions obtained at the MS2 scan stage for all of the monomeric adducts [M+H-116]+ were the aglycone adducts [BH2]+, which arise through the loss of deoxyribose from the protonated molecules. The top two most abundant ions produced in the MS3 scan stage, obtained by fragmentation of the [BH2]+ adducts, were used for quantitative measurements of the monomeric adducts. For the reduced dG-(CH2)3-dG cross-link, the product ions monitored at the MS3 scan stage were obtained by collision-induced dissociation of the aglycone, following the loss of the two deoxyribose moieties at the MS2 scan stage [M+H]+ → [M+H–232]+. The 8-dG-HNE-dG cross-link was measured at the the MS4 scan stage, by monitoring the transitions due to the successive loss of the two deoxyribose moieties at the MS2 and MS3 scan stages: [M+H]+ → [M+H-116]+ → [M+H–232]+. The proposed structures of the product ions of the adducts, produced at the MS/MSn scan stages, are presented in Figure 3, and the LC-ESI/MS/MSn parameter settings employed for adduct measurements are summarized in Table 1.
Figure 3.
Proposed structures of principal product ions arising at the MS/MSn scan stages. Mass is parentheses are the 15N or 2H labeled internal standards. The sites of isotopic incorporation are noted on Scheme 1.
Table 1.
LTQ/MS/MSn Parameter Settings for Analysis of Enal Adducts
| MSn | Adduct | m/z | Isolation width (m/z) |
CE | Q | Activation time (ms) |
Ions Monitored at final MS/MSn scan stage |
|---|---|---|---|---|---|---|---|
| MS/MS | 6-HO-PdG 8-HO-PdG |
324.1 | 5 | 22 | 0.35 | 10 | |
| MS/MS3 | 208.1 | 2 | 34 | 0.35 | 10 | 152.1, 190.1 | |
| MS/MS | [15N5]-6-HO-PdG [15N5]-8-HO-PdG |
329.1 | 5 | 22 | 0.35 | 10 | |
| MS/MS3 | 213.1 | 2 | 34 | 0.35 | 10 | 157.1, 195.1 | |
| MS/MS | 6-HO-PdGred 8-HO-PdGred |
326.1 | 5 | 25 | 0.35 | 10 | |
| MS/MS3 | 210.1 | 2 | 35 | 0.35 | 10 | 164.2, 192.2 | |
| MS/MS | [15N5]-6-HO-PdGred [15N5]-8-HO-PdGred |
331.1 | 5 | 25 | 0.35 | 10 | |
| MS/MS3 | 215.1 | 2 | 35 | 0.35 | 10 | 169.1, 197.2 | |
| MS/MS | PdG | 308.1 | 5 | 25 | 0.35 | 10 | |
| MS/MS3 | 192.1 | 2 | 35 | 0.35 | 10 | 164.2, 192.2 | |
| MS/MS | [15N5]PdG | 315.1 | 5 | 25 | 0.35 | 10 | |
| MS/MS3 | 197.1 | 2 | 35 | 0.35 | 10 | 169.1, 197.1 | |
| MS/MS | 8-PdG-(CH2)3-dG | 575.1 | 5 | 30 | 0.35 | 10 | |
| MS/MS3 | 343.2 | 2 | 32 | 0.35 | 10 | 164.2, 192.1 | |
| MS/MS | [2H6]-8-PdG-(CH2)3-dG | 581.1 | 5 | 30 | 0.35 | 10 | |
| MS/MS3 | 349.2 | 2 | 32 | 0.35 | 10 | 166.1, 198.1 | |
| MS/MS | HNE-dG | 424.2 | 4 | 18 | 0.35 | 30 | |
| MS/MS3 | 308.1 | 2 | 25 | 0.35 | 30 | 152.1, 290.1 | |
| MS/MS | [2H11]-HNE-dG | 425.2 | 4 | 18 | 0.35 | 30 | |
| MS/MS3 | 319.2 | 2 | 25 | 0.35 | 30 | 152.1, 301.1 | |
| MS/MS | dG-HNE-dG | 673.1 | 5 | 20 | 0.25 | 50 | |
| MS/MS3 | 557.1 | 4 | 20 | 0.25 | 30 | ||
| MS/MS4 | 441.1 | 3 | 20 | 0.25 | 30 | 152.1, 290.1 | |
| MS/MS | [2H11]-dG-HNE-dG | 684.2 | 5 | 20 | 0.25 | 50 | |
| MS/MS3 | 568.1 | 4 | 20 | 0.25 | 30 | ||
| MS/MS4 | 452.3 | 3 | 20 | 0.25 | 30 | 152.1, 301.1 | |
Representative optimized instrument tuning parameters were as follows: capillary temperature 185 °C; source spray voltage 1.5 kV and source current 100 µA (HO-PdG’s and dG-(CH2)3-dG adducts); source spray voltage 4.0 kV; source current 180 µA (HNE-dG and 8-dG-HNE-dG adducts); no sheath gas, sweep gas or auxiliary gas was employed; capillary voltage 39 V; tube lens voltage 95 V; and in-source fragmentation 10 V. Helium was used as the collision damping gas in the ion trap and was set at a pressure of 1 mTorr. One µscan was used for data acquisition. The automatic gain control settings were full MS target 30,000 and MSn target 10,000, and the maximum injection time was 10 ms. With these MS parameters, about 12 – 15 scans were acquired for each adduct and its corresponding internal standard.
Calibration Curves
Calibration curves were constructed in triplicate for each calibrant level, using CT DNA (50 µg, 151 nmol DNA bases). For the AC-DNA adduct calibration curve, the internal standards of the monomeric adducts (non-reduced and reduced) were added at a level of 1.1 adducts per 105 bases (for CT DNA treated with AC at 0.1 and 0.5 mol AC per mol DNA bases), or 2.2 adducts per 105 bases (for CT DNA treated with AC at 1 and 5 mol AC per mol DNA bases). The unlabeled monomeric adducts were added at the following levels: 0, 0.3, 1.1, 3.3, 6.6, 9.9, 33 or 77 adducts per 105 bases. The [2H6]-dG-(CH2)3-dG was added to the DNA at a level of 1.2 adducts per 106 bases, and the unlabeled dG-(CH2)3-dG was added at the following levels: 0, 0.2, 0.6, 0.9, 3.8, 5.7, 19, or 44 adducts per 106 bases. The DNA adducts and internal standards were added to the DNA prior to enzyme digestion and SPE. For estimates of AC adducts in untreated CT-DNA, the HO-PdG internal standards were added at a level of 2.0 adducts per 106 bases, the reduced internal standards were added at a level of 1.0 adducts per 106 bases, and the amount [2H6]-dG-(CH2)3-dG remained at a level of 1.2 adducts per 106 bases.
For the HNE-dG calibration curves, CT DNA (50 µg, 151 nmol DNA bases) was spiked with [2H11]-HNE-dG (15.1 pmol, or 1 adduct per 107 bases, mixture of four stereoisomers), and unlabeled HNE-dG was added at levels of 0, 0.5, 3, 10, 30, 100 or 300 adducts per 107 bases. The [2H11]-8-dG-HNE-dG was spiked into the same DNA sample at a level of 1 adduct per 107 bases and the unlabeled 8-dG-HNE-dG was added at levels of 0, 0.05, 0.1, 0.3, 0.6, 1 or 3 adducts per 107 bases. Because of the instability of 8-dG-HNE-dG, the DNA adducts and internal standards were added to the DNA, following enzyme digestion and were processed by SPE; this procedure allowed us to account for potential ion suppression matrix effects.
The calibration curves were fitted to a straight line (area of response of the adduct/area of response of the internal standard versus the amount of adduct/amount of internal standard), using ordinary least-squares with equal weightings and forcing the Y intercept through the origin. The calibration curve of HNE-dG was non-linear, and the response of the signal of the adduct approached saturation at the highest levels. Therefore, a quadratic equation was used to fit this calibration curve. The coefficient of determination (r2) values of the slopes of calibration curves exceeded 0.98. Representative calibration curves are shown in Supporting Information, Figure S-1.
Results
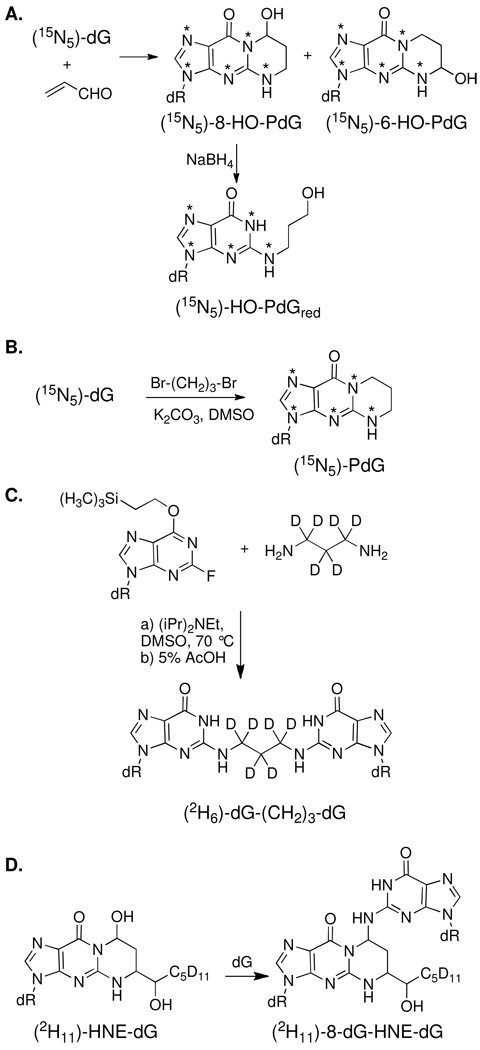
Synthesis of the Isotopically-Labeled Internal Standards
Isotopically labeled standards were synthesized according to Scheme 1. [15N5]-dG was reacted with AC to afford a mixture of the [15N5]-8-HO-PdG and [15N5]-6-HO-PdG regioisomers, which were separated by HPLC (Scheme 1A) (32). Reduction of [15N5]-8-HO-PdG with NaBH4 provided the [15N5]-N2-(3-hydroxypropyl)-dG (HO-PdGred). [15N5]-PdG was synthesized according to a literature procedure from [15N5]-dG and 1,3-dibromopropane (Scheme 1B) (34). The reduced AC cross-linked adduct, [2H6]-dG-(CH2)3-dG (10a) was synthesized by the reaction of [2H6]-1,3-diaminopropane with excess equivalents of O6-(trimethylsilyl)ethyl)-2-fluoro-2′-deoxyinosine (Scheme 1C). [2H11]-HNE was synthesized from [2H11]-1-bromopentane and fumaraldehyde bis(dimethylacetal) as previously described (30, 35, 36). [2H11]-HNE was then reacted with dG using arginine to promote the addition reaction to afford [2H11]-HNE-dG as a mixture of four stereoisomers (37–39). The [2H11]-HNE-dG adduct mixture was reacted with dG to afford the [2H11]-HNE-dG pyrimidopurinone cross-link standard (9c, 8-dG-HNE-dG) as a mixture of stereoisomers (Scheme 1D) (21). The isotopic purity of the labeled standards was > 99.5%.
LC-ESI/MS/MSn Characterization of AC and HNE DNA Adducts with a Linear Quadrupole Ion Trap MS (LIT MS)
DNA adducts of AC and HNE were characterized and quantified by LC-ESI/MS/MSn with a LIT MS. The 8-HO-PdG (1), 6-HO-PdG (2), HNE-dG (5–8), the unreduced 8-dG-AC-dG (9a) and reduced dG-(CH2)3-dG (10a) cross-links were analyzed at the MS3 scan stage, whereas the unreduced 8-dG-HNE-dG cross-link (9c) (Figure 1 and Figure 2) was assayed at the MS4 scan stage. The structures of the proposed principal product ions of these enal adducts, produced at the different MS/MSn scan stages, are depicted in Figure 3, and the MS parameters employed for the analyses of the adducts are summarized in Table 1.
As was previously noted (14), the use of adenine deaminase was critical for the successful analysis of 6-HO-PdG and 8-HO-PdG. The response of the signals for both HO-PdG adducts was very poor, probably due to co-elution with dA or other components, when this enzyme was omitted from the digestion reaction. The response of the signal for the 8-dG-AC-dG (9a) cross-link was weak under the LC-ESI/MS conditions employed, and the adduct underwent extensive deribosylation in the MS source. Because of the poor response 8-dG-AC-dG cross-link combined with its reversion back to 8-HO-PdG during the enzyme digestion of CT DNA, we reduced the cross-link prior to enzyme digestion. The chemically reduced dG-(CH2)3-dG (10a) cross-link was stable towards the enzyme digestion procedure (unpublished observations). Moreover, the response of the signal of the reduced dG-(CH2)3-dG (10a) cross-link, by LC-ESI/MS, was at least 5-fold greater than the response of the signal of 8-dG-AC-dG (9a).
The 8-dG-HNE-dG cross-link (9c) was resistant to chemical reduction. The isolation widths at all MSn scan stages were increased to maximize the level of sensitivity for the unreduced 8-dG-HNE-dG cross-link (9c), cross-link; however, the wide isolation widths employed resulted in the spillover of some isobaric interference at the MS3 scan stage. Therefore, a MS4 scan stage was employed to filter out this interference. The high dissociation efficiency of the LTQ MS permitted scanning of the 8-dG-HNE-dG cross-link at the MS4 scan stage with little loss of total ion counts across the MSn scan stages (unpublished observations).
Estimates of Interstrand dG-(CH2)3-dG (10a) and 8-dG-HNE-dG (9c) Cross-links in Oligonucleotide Duplexes Containing 5′-CpG-3′ Sequences
The 8-HO-PdG and (6S,8R,11S)-HNE-dG adducts were site-specifically incorporated into 12-mer oligonucleotides as previously described (19,21). The modified oligonucleotides, in which the adduct was situated in a 5′-CpG-3′ local sequence context, were hybridized to its complementary strand and incubated to allow interstrand cross-link formation. The 8-dG-AC-dG cross-link reached ~50% after 10 days, whereas the 8-dG-HNE-dG cross-link reached ~50% after 28 days as judged by CGE analysis with UV detection. These cross-linked oligonucleotide duplexes were utilized to optimize enzymatic digestion conditions for recovery of the intact cross-links. The oligonucleotide duplexes were diluted with unmodified CT DNA (3.8 adducts per 105 bases for 8-dG-AC-dG cross-link and 1 adduct per 106 or 1 adduct per 107 bases for the 8-dG-HNE-dG cross-link), and then digested with phosphodiesterase I, phosphodiesterase II, nuclease P1, and adenosine deaminase (for AC-modified DNA) (vide supra). The AC-dG containing duplex was treated with Na(CN)BH3 prior to enzyme digestion to reduce the dG-AC-dG cross-link (9a) to the reduced dG-(CH2)3-dG cross-link (10a). The Na(CN)BH3 treatment also partially reduced the 8-HO-PdG to the N2-(3-hydroxypropyl)-dG (8-HO-PdGred). After DNA digestion, NaBH4 was added to convert any remaining 8-HO-PdG to 8-HO-PdGred to simplify the analysis. Reduction of the 8-dG-HNE-dG cross-link with NaB(CN)H3 or NaBH4 was slow and incomplete; therefore analysis of the 8-dG-HNE-dG was performed on the unreduced pyrimidopurinone (9c). The reduced dG-(CH2)3-dG and unreduced 8-dG-HNE-dG cross-links in the spiked CT DNA were found to be ~50% of the total adduct level (Table 2) as determined by the isotope dilution method and LC-ESI/MS/MSn with the LIT MS. Thus, the estimates of AC and HNE cross-linked adducts obtained by quantitative LC/MS methods are similar to the levels of cross-linked adducts characterized by HPLC and CGE with UV detection.
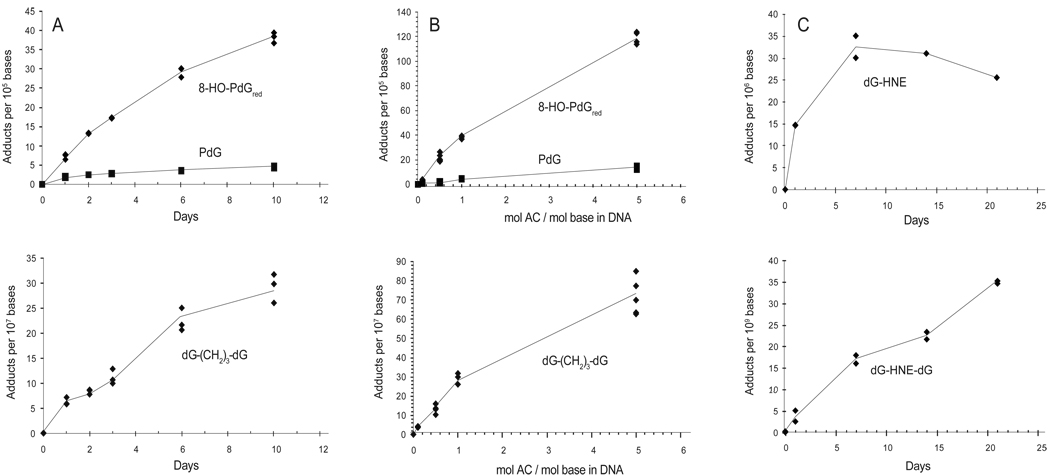
Time Course for the Formation of 6-HO-PdG, 8-HO-PdG, the Reduced dG-(CH2)3-dG (10a) Cross-link, HNE-dG and the Unreduced 8-dG-HNE-dG (9c) Cross-link in CT DNA Treated with AC or HNE
CT DNA was treated with 0, 0.1, 0.5, 1, and 5 mol AC per mol base in CT DNA, and the formation of the dG adducts, including the cross-links, was monitored over 10 days. The formation of the HNE-dG adduct and its cross-link was examined by treating CT DNA with 2 mol HNE per mol DNA base for a period of time up to 21 days. The data are summarized in Figure 4. In untreated CT DNA, the amount of 6-HO-PdG and 8-HO-PdG was estimated at a level of 3.0 ± 0.5 and 4.9 ± 1.0 adducts per 107 DNA bases (mean ± SD, N = 3), respectively. After the Na(CN)BH3 reduction, digestion, and the NaBH4 reduction protocol, which also reduces the 6-HO-PdG adduct to PdG, the amounts of PdG and 8-HO-PdGred due to background levels of 6-HO-PdG and 8-HO-PdG adducts in CT DNA were estimated to be 3.1 ± 0.8 and 5.3 ± 0.5 adducts per 107 DNA bases (mean ± SD, N = 3), respectively. The background level of the HNE-dG adduct in untreated CT DNA was below the limit of quantification (LOQ) (~3 adducts per 109 bases).
Figure 4.
Time course of 6-HO-PdG, 8-HO-PdG, 8-dG-AC-dG (measured as the reduced, 8-dG-(CH2)3-dG cross-link), HNE-dG and 8-dG-HNE-dG cross-link formation in CT DNA. Panel A: 1 mol AC reacted per mol base in CT DNA over a time period of 0, 1, 2, 3, 6 and 10 days; Panel B: Levels of 6-HO-PdG, 8-HO-PdG and 8-dG-AC-dG (measured as the reduced, 8-dG-(CH2)3-dG cross-link) formed in CT DNA treated with 0.1, 0.5, 1 or 5 mol AC per mol base in CT DNA for 10 days; and Panel C: Time course of HNE-dG and 8-dG-HNE-dG cross-link formation in CT DNA treated with 2 mol HNE per mol base in CT DNA over a time period up to 21 days.The upper panels depict the monomeric DNA adducts and lower panels depict the cross-linked DNA adducts.
The level of 8-HO-PdG formed was about 5 to 10-fold greater than the amount of 6-HO-PdG formed in AC-treated CT DNA at all doses (Figure 4A and 4B). As previously noted (7), the reactivity of AC with CT DNA was considerably greater than HNE (Figure 4C). On a per mol basis (mol enal per mol base in CT DNA), the extent of 8-HO-PdG adduct formation was ~20-fold or greater than the extent of HNE-dG adduct formation. The amounts of the 8-dG-AC-dG (9a) adduct, measured as the reduced dG-(CH2)3-dG (10a) cross-link, and the unreduced 8-dG-HNE-dG (9c) cross-link were low and accounted for ~1% of the total adducts of each enal. The 8-dG-AC-dG (9a) cross-link formation reached its maximum level between days 7 and 10, whereas the level of the 8-dG-HNE-dG (9c) cross-link continued to increase during the 3-week incubation period (Figure 4A and 4C). The formation of both the 8-dG-AC-dG (9a) and the 8-dG-HNE-dG (9c) cross-links continued to increase during the incubation period (Figure 4A and 4C). The percentage of the reduced 8-dG-AC-dG (9a) cross-link to total AC-DNA adducts (8-HO-PdGred and PdG) formed was relatively constant over all dose treatments of AC, at the 10-day time point (Figure 4B).
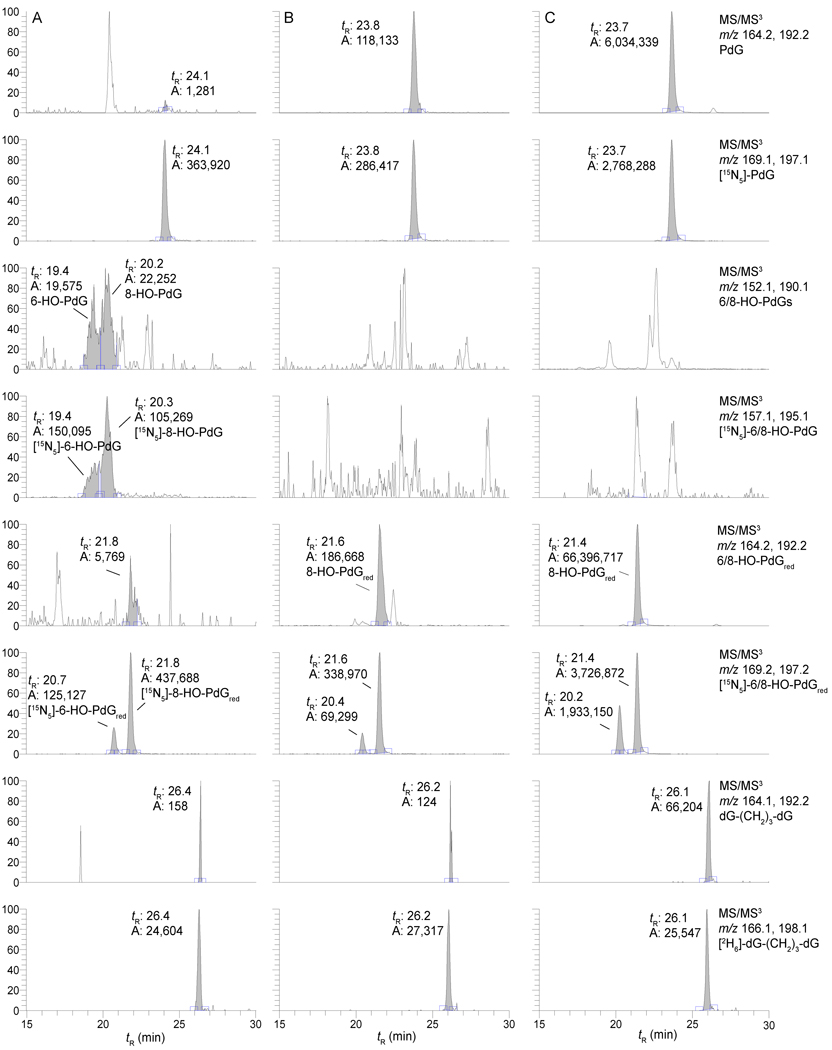
Representative reconstructed ion chromatograms of the LC-ESI/MS/MS3 traces for the adducts assayed from untreated CT DNA, with and without reduction with Na(CN)BH3 and NaBH4, and AC-treated CT DNA (1 mol AC per mol base in CT DNA), followed by reduction, are presented in Figure 5. The reconstructed ion chromatograms of the LC-ESI/MS/MS3 trace for HNE-dG adducts and the reconstructed ion chromatograms for the LC-ESI/MS/MS4 trace for the 8-dG-HNE-dG cross-link are shown in Figure 6. The cross-linked adducts are readily observed in the AC- and HNE-treated CT DNA samples.
Figure 5.
A reconstructed ion chromatogram of the LC-ESI/MS/MS3 traces of CT-DNA treated with AC. Panel A depicts untreated CT DNA; Panel B depicts untreated CT DNA chemically reduced with Na(CN)BH3 and NaBH4; Panel C depicts CT DNA treated with 1 mol AC per mol base in CT DNA for 10 days, followed by chemical reduction. For untreated DNA, the level of spiking with non-reduced monomer internal standards was 2.0 adducts per 106 bases, reduced monomer internal standards were added at a level of 1.0 adducts per 106 bases, and the spiking level with [2H6]-dG-(CH2)3-dG was 1.2 adducts per 106 bases. The level of spiking with monomeric internal standards was 2.2 adducts per 105 bases and the spiking level with [2H6]-dG-(CH2)3-dG was 1.2 adducts per 106 bases for the AC-treated CT DNA.
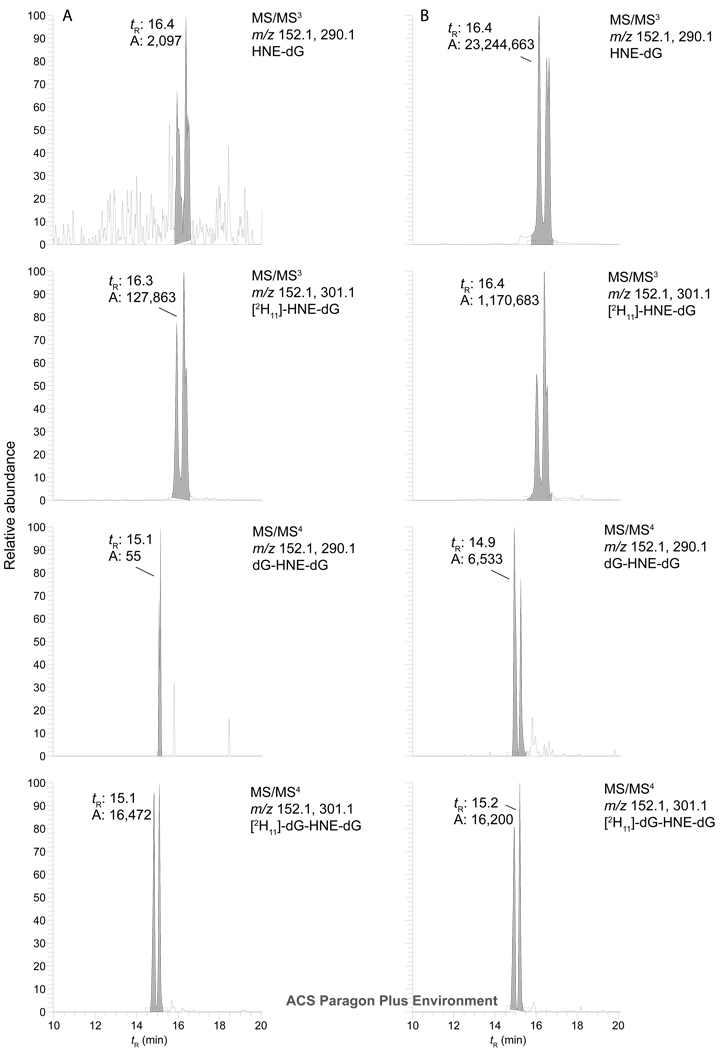
Figure 6.
Reconstructed ion chromatograms of the LC-ESI/MS/MSn traces of CT-DNA treated with HNE. A reconstructed ion chromatogram of the LC-ESI/MS/MS3 trace for HNE-dG and the reconstructed ion chromatogram of the LC-ESI/MS/MS4 trace for the 8-dG-HNE-dG cross-link. Both [2H11]-HNE-dG and [2H11]-8-dG-HNE-dG cross-link were added to CT DNA at a level of 1 adduct per 107 bases. Panel A depicts untreated CT DNA and Panel B depicts CT DNA treated with 2 mol of HNE per mol base in CT DNA.
There are conflicting reports regarding the usage of the ratio of the UV absorbance at 260:280 nm (A260/A280) to assess CT DNA purity. The A260/A280 of the CT DNA used in this study was 1.87, which according to one estimate indicates ~50% protein content (40). However, other reports state that an A260/A280 ratio > 1.80 is regarded as highly pure DNA, i.e. protein-free (41–43). Enals react readily with nucleophilic sidechains of lysine, cysteine, and histidine residues and the formation of protein adducts and therefore could influence the adduct level observed in our study. Moreover, the 8-HO-PdG and HNE-dG adducts readily form cross-links with peptides (44). It is possible that the formation of DNA-protein cross-links is more favorable than DNA cross-links, resulting in lower than expected levels for the 8-dG-AC-dG and 8-dG-HNE-dG cross-links. Of course, the formation of protein adducts and DNA-protein cross-links of AC and HNE are expected to occur in cells. We were also concerned that single-stranded regions of CT DNA could influence the results since they might be expected to be more reactive toward AC and HNE, and adducts in single-stranded regions would be unable to form interstrand crosslinks.
To address these concerns, the CT DNA was treated with S1 nuclease to remove single stranded regions then precipitated. The S1 nuclease and any other residual proteins were removed by an ion-pairing interaction to an insoluble, cross-linked poly-carboxylic acid polymer (ProCipitate™), followed by centrifugation. The CT DNA was precipitated from the supernatant solution, re-dissolved and precipitated again before passing through a Sephadex G-25 ion-exchange column. The A260/A280 ratio of this purified CT DNA was 1.90. The modification of the purified CT DNA with AC was repeated. After 2 and 10 days, the CT DNA was treated with Na(CN)BH3/NaBH4 and digested as described earlier (Table 3). A 2-fold increase in the level of the reduced dG-(CH2)3-dG cross-link (2% versus 1%) relative to the 8-HO-PdGred adduct formed was observed from the CT DNA that was purified by S1 nuclease and ProCipitate™ treatment versus untreated CT DNA. There was a modest decrease (~10%) in the amount of 8-HO-PdGred formed from the purified CT DNA. However, the ratio of the 8-HO-PdGred and PdG increased by ~5-fold from the purified CT DNA and was attributed to a 5-fold reduction in the amount of 6-HO-PdG formed.
Table 3.
AC-DNA Adduct Formation in Untreated CT DNA versus CT DNA that was “Purified” by Treatment with S1 Nuclease, followed by ProCipitate™.
| Sample | Days | 8-HO-PdGred× 10−5 | dG-(CH2)3-dG × 10−7 | PdG × 10−6 |
|---|---|---|---|---|
| CT DNA | 2 | 3.9 ± 0.2* | 3.6 ± 0.6** | 14.8 ± 0.9** |
| Purified- CT DNAa |
2 | 3.5 ± 0.1 | 7.6 ± 0.9 | 2.6 ±0.2 |
| CT DNA | 10 | 11.3 ± 1.3* | 11.4 ± 1.3** | 33.7 ± 4.5** |
| Purified-CT DNAa |
10 | 9.7 ± 1.6 | 21.9 ± 3.9 | 7.3 ± 0.9 |
N = 6 independent DNA binding experiments with AC
Unpaired t test (2-tailed)
P < 0.014: AC adduct formation from CT DNA versus Purified CT DNA
P < 0.001: AC adduct formation from CT DNA versus Purified DNA
Discussion
A variety of bifunctional chemicals have been demonstrated to cause interstrand DNA crosslinks; the interstrand cross-links make up a small fraction of the total DNA adducts formed (typically 1–5%), with the majority of the lesions being the corresponding uncross-linked, monomeric adducts, followed by intrastrand cross-links (23–25). Additionally, these lesions can also form DNA-protein cross-links (45–47). The relatively small contribution of interstrand cross-linked adducts to the total adduct level is likely due to spatial reasons, as many adducts cannot conform to the geometrical requirements for interstrand cross-link formation. Nonetheless, the cellular response to such bifunctional agents is usually ascribed to the minor interstrand DNA cross-link. It has been estimated that as few as 20–40 interstrand cross-links can be lethal to mammalian cells, while a single interstrand cross-link may be sufficient to kill bacteria and yeast cells (23, 24, 26). Thus, biologically relevant levels of interstrand cross-links may be below the limit of detection by current analytical methods.
NMR studies of the 8-HO-PdG adducts, the major DNA adduct of AC, demonstrated that this adduct can undergo a ring opening reaction in duplex DNA when paired opposite dC, which is presumably driven by the potential for Watson-Crick pairing (48). The ring-opened N2-(3-oxopropyl)-dG adduct possesses a reactive aldehyde group in the DNA minor groove, which can react with other nucleophilic groups to form DNA cross-links (Scheme 3). We demonstrated that oligonucleotides containing dG adducts of AC, crotonaldehyde, and HNE form interstrand DNA cross-links when site-specifically incorporated in a 5′-CpG-3′ sequence context (5, 22). The cross-linking chemistry of the AC, crotonaldehyde, and HNE was characterize by NMR spectroscopy and revealed that the chemical nature of the cross-link is a carbinolamine (22). However, the AC and crotonaldehyde cross-links can be reduced with Na(CN)BH3 (19, 20), suggesting that the carbinolamines are in equilibrium with low concentrations of the imines, which are below the detection limit of NMR spectroscopy. We observed that interstrand crosslink formation was highly dependent on the stereochemistry of the crotonaldehyde-dG and HNE-dG adducts; the (6R,8S)-crotonaldehyde-dG and (6S,8R,11S)-HNE-dG adducts formed interstrand cross-links when incorporated in a 5′-CpG-3′ sequence context, while other stereoisomers formed cross-links at significantly lower levels or not at all (19, 21). NMR analysis suggested that the adduct stereochemistry was critical for orienting the reactive aldehyde toward the N2-amino group of the dG that participated in the cross-linking reaction (49–52).
Scheme 3.
Cross-linking chemistry of enal adducts of dG
Our findings show that the amounts of 8-dG-AC-dG and 8-dG-HNE-dG cross-links formed in CT DNA occur at levels on the order of 1–2% that of the monomeric adducts. While analyses of AC or HNE treated CT DNA demonstrated that these bis-electrophiles induce DNA cross-links, the structural nature of the cross-link – interstrand versus intrastrand – is lost once the DNA has undergone enzymatic digestion. Our previous studies showed that the (6S,8R,11S)-HNE-dG (8) stereoisomers forms interstrand DNA cross-links in a 5′-CpG-3′ sequence contexts, while no evidence of cross-links was observed for the other three stereoisomers (5–7). Only one 8-dG-HNE-dG cross-link was observed when the enzyme digested oligonucleotide duplex containing the (6S,8R,11S)-HNE-dG adduct was analyzed by LC-ESI/MS/MSn (Supplementary Information, Figure S-2). Interestingly, at least two isomers of the 8-dG-HNE-dG cross-link were detected in HNE-treated CT DNA (Figure 6). This observation suggests that both interstrand and intrastrand cross-linked adducts are formed in CT DNA, and both types of crosslinks also likely to occur in AC-treated CT DNA as well (53).
Further purification of the CT DNA prior to AC resulted in a modest increase in cross-links formation (2% versus 1%). The purification involved treatment of the CT DNA with S1nuclease to remove single-stranded regions, removal of any residual proteins with ProCipitate™, and ion-exchange chromatography. Interestingly, the ratio of 8-HO-PdG and 6-HO-PdG adducts (observed as 8-HO-PdGred and PdG, respectively) increased by ~5-fold for the modification of the purified CT DNA. The formation of 8-HO-PdG and 6-HO-PdG involves initial Michael adduct of either the N2- or N1-position of dG to the β-carbon of AC. Therefore, the reactivity of AC with double-stranded DNA is expected to favor the 8-HO-PdG adduct since N2-position is accessible from the minor groove. These observations are consistent with the S1 nuclease treatment removing single-stranded regions of the CT DNA. The removal of any residual proteins from CT DNA with ProCipitate™ appeared to have marginal effects on the levels of AC adduct formation. This study was not repeated with HNE since the increase in AC cross-link formation was relatively small (2-fold) and HNE does not form the 6-HO regioisomer.
A goal of this study was to establish a lower limit of detection of the AC and HNE cross-links relative to the level of uncross-linked 8-HO-PdG and HNE-dG adducts, which are formed in mammalian cells. For example, high levels of both the 6-HO-PdG and 8-HO-PdG have been detected in DNA from brain tissues of Alzheimer’s disease patients (54, 55). The levels of 6-HO-PdG and 8-HO-PdG ranged from 2,800 to 5,100 adducts per 109 bases. Both regioisomeric AC adducts were also found in the lung tissue from smokers at levels ranging from 10 to 160 HO-PdG adducts per 109 bases (14). The average level of the 6-HO-PdG isomers was slightly higher than the 8-HO-PdG adduct in the 30 human lung samples examined. This is an interesting observation since formation of the 8-HO-PdG adduct is predicted to be in duplex DNA and may reflect differential repair of the between the two AC adducts. The 6- and 8-HO-PdG adducts were also found in human placental and leukocyte DNA at levels of 108 and 78 adducts per 108 bases (56); the regioisomers were integrated together in these analyses. The HNE-dG adduct was also identified in DNA from brain tissue of Alzheimer’s disease patients at levels ranging from 190 to 1260 adducts per 109 bases (57). The levels of these enal adducts in brain tissue are high in comparison to the adduct levels measured in lung tissue. If appropriate measures are not employed during the isolation of DNA from tissues, artifactual enal DNA adduct formation can occur and result in artificially high levels of DNA adducts (58). However, both AC and HNE have been reported to be elevated in brain regions of subjects with mild cognitive impairment and late-stage Alzheimer disease, suggesting that enal-DNA adduct formation in the brain may well be greater than in other organs (12). If the proportion of AC and HNE cross-linked dG-adducts formed relative to the monomeric adducts in enal-modified CT DNA are similar to the proportion that occurs in DNA in cells, and depending on the rate that the cross-linked adducts are removed by repair pathway (26–28), we could expect to detect the 8-dG-AC-dG cross-link adduct at levels ranging from 2–200 adducts per 109 bases in brain tissue of Alzheimer’s patients noted above. The levels of the 8-dG-HNE-dG cross-link may be present at levels of several adducts per 109 bases, which is close to the limit of current detection of MS. Indeed, background levels of a cross-linked adduct derived from the reaction of endogenously produced formaldehyde with the N6 atoms of two dA molecules, have been measured at levels of several adducts per 109 DNA bases in rat liver (59). Thus, it may be possible to detect these enal cross-links from brain tissues of Alzheimer patients or tissue from laboratory animals that are deficient in essential cross-links repair factors (60).
In conclusion, we have developed a mass spectrometric method for the detection and quantitation of dG cross-links of AC and HNE in CT DNA. Our current objective is to determine if either one of these DNA cross-links is formed in vivo.
Supplementary Material
Acknowledgements
NIH supported this work through research grant P01 ES05355 (C.J.R. and R. J. T.) and center grant P30 ES00267 (C.J.R.).
Abbreviations
- AC
acrolein
- 4-HNE
trans-4-hydroxy-2-nonenal
- CT DNA
calf thymus DNA
- dG
2'-deoxyguanosine
- dA
2'-deoxyadenosine
- dN
2'-deoxynucleoside
- 6-HO-PdG
3-(2'-deoxy-β-D-erythro-pentofuranosyl)-5,6,7,8-tetrahydro-6-hydroxypyrimido[1,2-a]purin-10(3H)-one
- 8-HO-PdG
3-(2'-deoxy-β-D-erythro-pentofuranosyl)-5,6,7,8-tetrahydro-8-hydroxypyrimido[1,2-a]purin-10(3H)-one
- dG-(CH2)3-dG
N',N''-1,3-propanediylbis-(2′-deoxyguanosine)
- 8-dG-AC-dG
N-[3-(2-deoxy-β-D-erythro-pentofuranosyl)-3,5,6,7,8,10-hexahydro-10-oxopyrimido[1,2-a]purin-8-yl]-2′-deoxyguanosine
- 6-HO-PdGred
N1-(3-hydroxypropyl)-2′-deoxyguanosine
- PdG
3-(2-deoxy-β-D-erythro-pentofuranosyl)-5,6,7,8-tetrahydro-pyrimido1,2-a]purin-10(3H)-one
- 8-HO-PdGred
N 2-(3-hydroxypropyl)-2′-deoxyguanosine
- HNE-dG
3-(2-deoxy-β-D-erythro-pentofuranosyl)-5,6,7,8-tetrahydro-8-hydroxy-6-(1-hydroxyhexyl)-pyrimido[1,2-a]purin-10(3H)-one
- 8-dG-HNE-dG
N[3-(2-deoxy-β-D-erythro-pentofuranosyl)-3,5,6,7,8,10-hexahydro-6-(1-hydroxyhexyl)-10-oxopyrimido[1,2-a]purin-8-yl]-2′-deoxyguanosine
- CGE
capillary gel electrophoresis
- LOQ
limit of quantification
- LC-ESI/MS/MSn
liquid chromatography-electrospray ionization/multi-stagetandem mass spectrometry
- LIT MS
linear quadrupole ion trap mass spectrometry;
- SPE
solid phase extraction
Footnotes
Supporting Information.
Calibration curves and reconstructed ion profiles. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 2.Stevens JF, Maier CS. Acrolein: Sources, metabolism, and biomolecular interactions relevant to human health and disease. Mol. Nutr. Food Res. 2008;52:7–25. doi: 10.1002/mnfr.200700412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esterbauer H. Cytotoxicity and genotoxicity of lipid-oxidation products. Am. J. Clin. Nutr. 1993;57 doi: 10.1093/ajcn/57.5.779S. 779S–785S. [DOI] [PubMed] [Google Scholar]
- 4.Niki E. Lipid peroxidation: Physiological levels and dual biological effects. Free Radic. Biol. Med. 2009;47:469–484. doi: 10.1016/j.freeradbiomed.2009.05.032. [DOI] [PubMed] [Google Scholar]
- 5.Minko I, Kozekov I, Harris T, Rizzo C, Lloyd R, Stone M. Chemistry and biology of DNA containing 1,N2-deoxyguanosine adducts of the α,β-unsaturated aldehydes acrolein, crotonaldehyde, and 4-hydroxynonenal. Chem. Res. Toxicol. 2009;22:759–778. doi: 10.1021/tx9000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung FL, Nath RG, Nagao M, Nishikawa A, Zhou GD, Randerath K. Endogenous formation and significance of 1,N2-propanodeoxyguanosine adducts. Mutat. Res. 1999;424:71–81. doi: 10.1016/s0027-5107(99)00009-3. [DOI] [PubMed] [Google Scholar]
- 7.Chung FL, Chen HJ, Nath RG. Lipid peroxidation as a potential endogenous source for the formation of exocyclic DNA adducts. Carcinogenesis. 1996;17:2105–2111. doi: 10.1093/carcin/17.10.2105. [DOI] [PubMed] [Google Scholar]
- 8.Chung FL, Zhang L, Ocando JE, Nath RG. Role of 1,N2-propanodeoxyguanosine adducts as endogenous DNA lesions in rodents and humans. IARC Sci. Publ. 1999:45–54. [PubMed] [Google Scholar]
- 9.Nath RG, Chung FL. Detection of exocyclic 1,N2-propanodeoxyguanosine adducts as common DNA lesions in rodents and humans. Proc. Nati. Acad. Sci. U.S.A. 1994;91:7491–7495. doi: 10.1073/pnas.91.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nath RG, Ocando JE, Chung FL. Detection of 1,N2-propanodeoxyguanosine adducts as potential endogenous DNA lesions in rodent and human tissues. Cancer Res. 1996;56:452–456. [PubMed] [Google Scholar]
- 11.Nath RG, Ocando JE, Guttenplan JB, Chung FL. 1,N2-propanodeoxyguanosine adducts: Potential new biomarkers of smoking-induced DNA damage in human oral tissue. Cancer Res. 1998;58:581–584. [PubMed] [Google Scholar]
- 12.Bradley MA, Markesbery WR, Lovell MA. Increased levels of 4-hydroxynonenal and acrolein in the brain in preclinical Alzheimer disease. Free Radic. Biol. Med. 2010;48:1570–1576. doi: 10.1016/j.freeradbiomed.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lovell MA, Markesbery WR. Oxidative DNA damage in mild cognitive impairment and late-stage Alzheimer's disease. Nucleic Acids Res. 2007;35:7497–7504. doi: 10.1093/nar/gkm821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang S, Villalta PW, Wang M, Hecht SS. Detection and quantitation of acrolein-derived 1,N2-propanodeoxyguanosine adducts in human lung by liquid chromatography-electrospray ionization-tandem mass spectrometry. Chem. Res. Toxicol. 2007;20:565–571. doi: 10.1021/tx700023z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawanishi M, Matsuda T, Nakayama A, Takebe H, Matsui S, Yagi T. Molecular analysis of mutations induced by acrolein in human fibroblast cells using supF shuttle vector plasmids. Mutat. Res. 1998;417:65–73. doi: 10.1016/s1383-5718(98)00093-x. [DOI] [PubMed] [Google Scholar]
- 16.Wang M, McIntee EJ, Cheng G, Shi Y, Villalta PW, Hecht SS. Identification of DNA adducts of acetaldehyde. Chem. Res. Toxicol. 2000;13:1149–1157. doi: 10.1021/tx000118t. [DOI] [PubMed] [Google Scholar]
- 17.Lao Y, Hecht SS. Synthesis and properties of an acetaldehyde-derived oligonucleotide interstrand cross-link. Chem. Res. Toxicol. 2005;18:711–721. doi: 10.1021/tx0497292. [DOI] [PubMed] [Google Scholar]
- 18.Nechev LV, Kozekov I, Harris CM, Harris TM. Stereospecific synthesis of oligonucleotides containing crotonaldehyde adducts of deoxyguanosine. Chem. Res. Toxicol. 2001;14:1506–1512. doi: 10.1021/tx0100690. [DOI] [PubMed] [Google Scholar]
- 19.Kozekov ID, Nechev LV, Moseley MS, Harris CM, Rizzo CJ, Stone MP, Harris TM. DNA interchain cross-links formed by acrolein and crotonaldehyde. J. Am. Chem. Soc. 2003;125:50–61. doi: 10.1021/ja020778f. [DOI] [PubMed] [Google Scholar]
- 20.Kozekov ID, Nechev LV, Sanchez A, Harris CM, Lloyd RS, Harris TM. Interchain cross-linking of DNA mediated by the principal adduct of acrolein. Chem. Res. Toxicol. 2001;14:1482–1485. doi: 10.1021/tx010127h. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Kozekov ID, Harris TM, Rizzo CJ. Site-specific synthesis and reactivity of oligonucleotides containing stereochemically defined 1,N2-deoxyguanosine adducts of the lipid peroxidation product trans-4-hydroxynonenal. J. Am. Chem. Soc. 2003;125:5687–5700. doi: 10.1021/ja0288800. [DOI] [PubMed] [Google Scholar]
- 22.Stone MP, Cho Y-J, Huang H, Kim H-Y, Kozekov ID, Kozekova A, Wang H, Minko IG, Lloyd RS, Harris TM, Rizzo CJ. Interstrand DNA cross-links induced by α,β-unsaturated aldehydes derived from lipid peroxidation and environmental sources. Acc. Chem. Res. 2008;41:793–804. doi: 10.1021/ar700246x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schärer OD. DNA interstrand crosslinks: Natural and drug-induced DNA adducts that induce unique cellular responses. Chembiochem. 2005;6:27–32. doi: 10.1002/cbic.200400287. [DOI] [PubMed] [Google Scholar]
- 24.Grillari J, Katinger H, Voglauer R. Contributions of DNA interstrand crosslinks to aging of cells and organisms. Nucleic Acids Res. 2007;35:7566–7575. doi: 10.1093/nar/gkm1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Efimov VA, Fedyunin SV, Chakhmakhcheva OG. Cross-linked nucleic acids: Formation, structure, and biological function. Russ. J. Bioorg. Chem. 2010;36:49–72. doi: 10.1134/s1068162010010061. [DOI] [PubMed] [Google Scholar]
- 26.Noll D, Mason T, Miller P. Formation and repair of interstrand cross-links in DNA. Chem. Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: Molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2:483–490. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- 28.Niedernhofer LJ, Lalai AS, Hoeijmakers JHJ. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123:1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 29.Minko IG, Harbut MB, Kozekov ID, Kozekova A, Jakobs PM, Olson SB, Moses RE, Harris TM, Rizzo CJ, Lloyd RS. Role for DNA polymerase κ in the processing of N2 -N2 -guanine interstrand cross-links. J. Biol. Chem. 2008;283:17075–17082. doi: 10.1074/jbc.M801238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grée R, Tourbah H, Carrié R. Fumaraldehyde monodimethyl acetal: An easily accessible and versatile intermediate. Tetrahedron Lett. 1986;27:4983–4986. [Google Scholar]
- 31.Nechev LV, Harris CM, Harris TM. Synthesis of nucleosides and oligonucleotides containing adducts of acrolein and vinyl chloride. Chem. Res. Toxicol. 2000;13:421–429. doi: 10.1021/tx990167+. [DOI] [PubMed] [Google Scholar]
- 32.Chung FL, Young R, Hecht SS. Formation of cyclic 1,N2-propanodeoxyguanosine adducts in DNA upon reaction with acrolein or crotonaldehyde. Cancer Res. 1984;44:990–995. [PubMed] [Google Scholar]
- 33.Goodenough AK, Schut HAJ, Turesky RJ. Novel LC-ESI/MS/MSn method for the characterization and quantification of 2'-deoxyguanosine adducts of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by 2-D linear quadrupole ion trap mass spectrometry. Chem. Res. Toxicol. 2007;20:263–276. doi: 10.1021/tx0601713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marinelli ER, Johnson F, Iden CR, Yu PL. Synthesis of 1,N2-(1,3-propano)-2'-deoxyguanosine and incorporation into oligodeoxynucleotides: A model for exocyclic acrolein-DNA adducts. Chem. Res. Toxicol. 1990;3:49–58. doi: 10.1021/tx00013a009. [DOI] [PubMed] [Google Scholar]
- 35.Rees MS, van Kuijk FJGMAN, Siakotos AN, Mundy BP. Improved synthesis of various isotope labeled 4-hydroxyalkenals and peroxidation intermediates. Synth. Commun. 1995;25:3225–3236. [Google Scholar]
- 36.Chandra A, Srivastava S. A synthesis of 4-hydroxy-2-trans-nonenal and 4-(3H) 4-hydroxy-2-trans-nonenal. Lipids. 1997;32:779–782. doi: 10.1007/s11745-997-0100-6. [DOI] [PubMed] [Google Scholar]
- 37.Sako M, Yaekura I. A convenient preparative method for the 1,N2-cyclic adducts of guanine nucleosides and nucleotides with crotonaldehyde. Tetrahedron. 2002;58:8413–8416. [Google Scholar]
- 38.Winter CK, Segall HJ, Haddon WF. Formation of cyclic adducts of deoxyguanosine with the aldehydes trans-4-hydroxy-2-hexenal and trans-4-hydroxy-2-nonenal in vitro. Cancer Res. 1986;46:5682–5686. [PubMed] [Google Scholar]
- 39.Wang H, Rizzo CJ. Stereocontrolled syntheses of all four stereoisomeric 1,N2-deoxyguanosine adducts of the lipid peroxidation product trans-4-hydroxynonenal. Org. Lett. 2001;3:3603–3605. doi: 10.1021/ol016810c. [DOI] [PubMed] [Google Scholar]
- 40.Glasel JA. Validity of nucleic acid purities monitored by 260nm/280nm absorbance ratios. BioTechniques. 1995;18:62–63. [PubMed] [Google Scholar]
- 41.Maniatis JA, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 42.Manchester KL. Value of A260/A280 ratios for measurement of purity of nucleic acids. BioTechniques. 1995;19:208–209. [PubMed] [Google Scholar]
- 43.Manchester KL. Use of UV methods for measurement of protein and nucleic acid concentrations. BioTechniques. 1996;20 doi: 10.2144/96206bm05. 968-967. [DOI] [PubMed] [Google Scholar]
- 44.Kurtz AJ, Lloyd RS. 1,N2-deoxyguanosine adducts of acrolein, crotonaldehyde, and trans-4-hydroxynonenal cross-link to peptides via Schiff base linkage. J. Biol. Chem. 2003;278:5970–5976. doi: 10.1074/jbc.M212012200. [DOI] [PubMed] [Google Scholar]
- 45.Barker S, Weinfeld M, Murray D. DNA-protein crosslinks: Their induction, repair, and biological consequences. Mutat. Res. 2005;589:111–135. doi: 10.1016/j.mrrev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 46.Permana PA, Snapka RM. Aldehyde-induced protein-DNA crosslinks disrupt specific stages of SV40 DNA replication. Carcinogenesis. 1994;15:1031–1036. doi: 10.1093/carcin/15.5.1031. [DOI] [PubMed] [Google Scholar]
- 47.Kuykendall JR, Bogdanffy MS. Efficiency of DNA-histone crosslinking induced by saturated and unsaturated aldehydes in vitro. Mutat. Res. 1992;283:131–136. doi: 10.1016/0165-7992(92)90145-8. [DOI] [PubMed] [Google Scholar]
- 48.de los Santos C, Zaliznyak T, Johnson F. NMR characterization of a DNA duplex containing the major acrolein-derived deoxyguanosine adduct γ-OH-1,-N2-propano-2'-deoxyguanosine. J. Biol. Chem. 2001;276:9077–9082. doi: 10.1074/jbc.M009028200. [DOI] [PubMed] [Google Scholar]
- 49.Cho Y, Wang H, Kozekov I, Kurtz A, Jacob J, Voehler M, Smith J, Harris T, Lloyd R, Rizzo C, Stone M. Stereospecific formation of interstrand carbinolamine DNA cross-links by crotonaldehyde- and acetaldehyde-derived α-CH3-γ-OH-1,N2-propano-2'-deoxyguanosine adducts in the 5'-CpG-3' sequence. Chem. Res. Toxicol. 2006;19:195–208. doi: 10.1021/tx050239z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho Y-J, Kozekov ID, Harris TM, Rizzo CJ, Stone MP. Stereochemistry modulates the stability of reduced interstrand cross-links arising from R-and S-α-CH3-γ-OH-1,N2-propano-2'-deoxyguanosine in the 5'-CpG-3' DNA sequence. Biochemistry. 2007;46:2608–2621. doi: 10.1021/bi061381h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho Y-J, Wang H, Kozekov ID, Kozekova A, Kurtz AJ, Jacob J, Voehler M, Smith J, Harris TM, Rizzo CJ, Lloyd RS, Stone MP. Orientation of the crotonaldehyde-derived N2-[3-oxo-1(S)-methyl-propyl]-dG DNA adduct hinders interstrand cross-link formation in the 5'-CpG-3' sequence. Chem. Res. Toxicol. 2006;19:1019–1029. doi: 10.1021/tx0600604. [DOI] [PubMed] [Google Scholar]
- 52.Huang H, Wang H, Qi N, Lloyd RS, Rizzo CJ, Stone MP. The stereochemistry of trans-4-hydroxynonenal-derived exocyclic 1,N2-2'-deoxyguanosine adducts modulates formation of interstrand cross-links in the 5'-CpG-3' sequence. Biochemistry. 2008;47:11457–11472. doi: 10.1021/bi8011143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanchez AM, Kozekov ID, Harris TM, Lloyd RS. Formation of inter-and intrastrand imine type DNA-DNA cross-links through secondary reactions of aldehydic adducts. Chem. Res. Toxicol. 2005;18:1683–1690. doi: 10.1021/tx0500528. [DOI] [PubMed] [Google Scholar]
- 54.Liu X, Lovell MA, Lynn BC. Development of a method for quantification of acrolein-deoxyguanosine adducts in DNA using isotope dilution-capillary LC/MS/MS and its application to human brain tissue. Anal. Chem. 2005;77:5982–5989. doi: 10.1021/ac050624t. [DOI] [PubMed] [Google Scholar]
- 55.Nam DT, Arseneault M, Murthy V, Ramassamy C. Potential role of acrolein in neurodegeneration and in Alzheimer's disease. Curr. Mol. Pharmacol. 2010;3:66–78. [PubMed] [Google Scholar]
- 56.Chen HJ, Lin WP. Simultaneous quantification of 1,N2-propano-2'-deoxyguanosine adducts derived from acrolein and crotonaldehyde in human placenta and leukocytes by isotope dilution nanoflow LC nanospray ionization tandem mass spectrometry. Anal. Chem. 2009;81:9812–9818. doi: 10.1021/ac9019472. [DOI] [PubMed] [Google Scholar]
- 57.Liu X, Lovell MA, Lynn BC. Detection and quantification of endogenous cyclic DNA adducts derived from trans-4-hydroxy-2-nonenal in human brain tissue by isotope dilution capillary liquid chromatography nanoelectrospray tandem mass spectrometry. Chem. Res. Toxicol. 2006;19:710–718. doi: 10.1021/tx0502903. [DOI] [PubMed] [Google Scholar]
- 58.Emami A, Dyba M, Cheema AK, Pan J, Nath RG, Chung FL. Detection of the acrolein-derived cyclic DNA adduct by a quantitative 32P-postlabeling/solid-phase extraction/HPLC method: Blocking its artifact formation with glutathione. Anal. Biochem. 2008;374:163–172. doi: 10.1016/j.ab.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang M, Cheng G, Villalta PW, Hecht SS. Development of liquid chromatography electrospray ionization tandem mass spectrometry methods for analysis of DNA adducts of formaldehyde and their application to rats treated with N-nitrosodimethylamine or 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Chem. Res. Toxicol. 2007;20:1141–1148. doi: 10.1021/tx700189c. [DOI] [PubMed] [Google Scholar]
- 60.Niedernhofer L, Odijk H, Budzowska M, van Drunen E, Maas A, Theil A, de Wit J, Jaspers N, Beverloo H, Hoeijmakers J, Kanaar R. The structure-specific endonuclease Ercc1-XPF is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell. Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.