Abstract
Background
Invasive species pose a significant threat to global economies, agriculture and biodiversity. Despite progress towards understanding the ecological factors associated with plant invasions, limited genomic resources have made it difficult to elucidate the evolutionary and genetic factors responsible for invasiveness. This study presents the first expressed sequence tag (EST) collection for Senecio madagascariensis, a globally invasive plant species.
Methods
We used pyrosequencing of one normalized and two subtractive libraries, derived from one native and one invasive population, to generate an EST collection. ESTs were assembled into contigs, annotated by BLAST comparison with the NCBI non-redundant protein database and assigned gene ontology (GO) terms from the Plant GO Slim ontologies.
Key Results
Assembly of the 221 746 sequence reads resulted in 12 442 contigs. Over 50 % (6183) of 12 442 contigs showed significant homology to proteins in the NCBI database, representing approx. 4800 independent transcripts. The molecular transducer GO term was significantly over-represented in the native (South African) subtractive library compared with the invasive (Australian) library. Based on NCBI BLAST hits and literature searches, 40 % of the molecular transducer genes identified in the South African subtractive library are likely to be involved in response to biotic stimuli, such as fungal, bacterial and viral pathogens.
Conclusions
This EST collection is the first representation of the S. madagascariensis transcriptome and provides an important resource for the discovery of candidate genes associated with plant invasiveness. The over-representation of molecular transducer genes associated with defence responses in the native subtractive library provides preliminary support for aspects of the enemy release and evolution of increased competitive ability hypotheses in this successful invasive. This study highlights the contribution of next-generation sequencing to better understanding the molecular mechanisms underlying ecological hypotheses that are important in successful plant invasions.
Keywords: ESTs, genomics, invasive species, maternal effects, rapid adaptation, selection, Senecio madagascariensis
INTRODUCTION
Invasive plant species pose a significant threat to global economies, agriculture and biodiversity (Richardson and Pysek, 2006; Thuiller et al., 2006; Prentis et al., 2007), and they are recognized as a major component of human-induced global change (Wilson et al., 2009a). As international trade and travel continue to grow (Wilson et al., 2009a), it seems likely that alien plant invasions will continue to increase at a rapid rate. Understanding the process of invasion is critical to the management of invasive species and for identifying species that may pose a risk in the future. Despite progress towards understanding the ecology of plant invasions (Thuiller et al., 2006; Milton et al., 2007; Wilson et al., 2007, 2009b; Proches et al., 2008), relatively little headway has been made in identifying the evolutionary or genetic factors responsible (Sakai et al., 2001; Lee, 2002; Prentis et al., 2008).
Evolutionary changes in introduced populations were first recognized as an important process in biological invasions over 35 years ago (Baker, 1974). However, the majority of evolutionary research in invasion biology has been conducted within the past 10 years, and has largely concentrated on comparing genetic diversity between source and invasive populations (Bossdorf et al., 2005; Kang et al., 2007; Dlugosch and Parker, 2008; Prentis et al., 2009). Fewer studies have examined the role of adaptive evolution in biological invasion (Whitney et al., 2006; Dlugosch and Parker, 2008), and even fewer studies have attempted to identify the source of genetic and epigenetic variation underlying adaptation in an invasive species (Parisod et al., 2009).
Two hypotheses commonly invoked to explain the success of invasive species are the enemy release hypothesis (ERH) and the evolution of increased competitive ability (EICA). These hypotheses theorize that invasive species will escape the negative effects of their natural enemies in the introduced range (Blossey and Notzold, 1995; Keane and Crawley, 2002; Colautti et al., 2004; Orians and Ward, 2010), but they are supported in only approx. 50 % of studies (Bossdorf et al., 2005). Although these arguments are intuitive, the extent for evolution to occur following release from enemies in the introduced range depends on the extent to which natural enemies controlled the abundance of these species in the native range. For species that do escape the negative effects of their natural enemies in the introduced range, EICA predicts that natural selection will favour those genotypes with decreased allocation to defence and increased allocation to competitiveness. Although these hypotheses have been extensively tested ecologically, little research has investigated the underlying molecular mechanisms.
Research in this area has lagged largely because the kind of genomic or expressed sequence tag (EST) resources required to identify candidate genes or genomic changes associated with invasiveness are not yet developed for most invasive species. In fact, only few invasive species have had significant genomic resources developed for this purpose (Lai et al., 2006; Broz et al., 2007, 2009; Barker et al., 2008). Although genomic tools developed for crop or model plants such as rice or Arabidopsis may be applied to their weedy relatives, genomic resources must be developed specifically for the invasive species to best identify the genetic changes responsible for success in recipient environments and to manage invasive populations. For most non-model species, EST data have been traditionally obtained through generation of normalized complementary DNA (cDNA) libraries and Sanger sequencing (Bouck and Vision, 2007). However, next-generation sequencing approaches, such as massively parallel 454 pyrosequencing, can improve this process (Margulies et al., 2005; Cheung et al., 2006; Goldberg et al., 2006; Moore et al., 2006; Wicker et al., 2006; Huse et al., 2007) and have recently been used to rapidly and accurately characterize a transcriptome sequence in a non-model organism (Vera et al., 2008). Therefore, next-generation sequencing can be an effective method for developing large-scale genomic resources for non-model invasive organisms.
Senecio madagascariensis, a native of southern Africa, is a highly invasive plant that has been introduced to Australia, Japan, Argentina, Mexico and Hawaii (Radford et al., 2000; Le Roux et al., 2006; Lopez et al., 2008). Its polyploid form, Senecio inaequidens, is invasive throughout southern Europe (Lafuma et al., 2003; Cano et al., 2007; Lafuma and Maurice, 2007; Bossdorf et al., 2008; Monty et al., 2009; Monty and Mahy, 2009; Lachmuth et al., 2010). In Australia alone, annual losses of up to US$12 million have been directly linked to fireweed populations (Le Roux et al., 2006). Fireweed often colonizes disturbed habitats or overgrazed pastures throughout its introduced range where it competes strongly with native species and pasture plants for light, moisture and soil nutrients.
Recent molecular investigations have shown that Australian and Hawaiian fireweed populations are of South African origin (Scott et al., 1998; Le Roux et al., 2006), but were probably introduced to Australia from multiple sources in South Africa (Radford et al., 2000). Multiple introductions of genetically differentiated source populations provide opportunities for admixture in the introduced range, which has been proposed as a stimulus for adaptive evolution and invasiveness in many introduced plants (Ellstrand and Schierenbeck, 2000). Investigations of neutral genetic variation have found similar levels of genetic diversity in invasive Australian (E. E. Dormontt et al., University of Adelaide, Australia, unpubl. res.) and Hawaiian (Le Roux et al., 2010) populations compared with source populations from South Africa, indicating that this species has not gone through a strong bottleneck during population founding. High levels of neutral genetic variation in introduced populations (Le Roux et al., 2010) indicate that substantial standing genetic variation at loci underlying important traits may also have been introduced. Adaptive evolution in invasive species has been hypothesized to be dominated by alleles from standing genetic variation (Prentis et al., 2008), because favourable alleles are immediately available and usually occur at a greater frequency in populations than do neutral or deleterious alleles (Barrett and Schluter, 2008). However, it remains unclear whether admixture or adaptation from standing genetic variation was responsible for the successful invasion of S. madagascariensis.
As a first step towards understanding the molecular basis for adaptive evolution in invasive species, we have generated one normalized and two subtractive EST libraries representing one native South African and one invasive Australian population of S. madagascariensis. Here we describe the annotation of our S. madagascariensis ESTs by sequence comparison with the entire core protein database of NCBI. We also provide an initial list of candidate genes from our subtractive libraries that could be utilized in future experiments to test for correlations between patterns of gene expression and ecological or evolutionary factors that may be associated with successful invasions. In addition, we mined singlet contigs for microsatellites to be utilized in genome scan experiments to examine contemporary selection during the invasion of S. madagascariensis.
MATERIALS AND METHODS
Plant material
Senecio madagascariensis Poir. (fireweed) seeds were obtained from one native South African and one invasive Australian population. The South African population was collected from East London (33°02′S, 27°50′E), while seeds from the Australian population were collected from Springbrook National Park (28°08′21″S, 153°16′41″E). The Springbrook population was selected because it was an allopatric site identified by Prentis et al. (2007). This site contained no hybrid plants or seed and was isolated from any Senecio pinnatifolius population by more than 10 km. The seeds were placed in Petri dishes containing germination paper moistened with distilled water. Germination took place in a growth cabinet under standard culture conditions (12/12-h light–dark photoperiod at a constant temperature of 20 °C). Seedlings with fully expanded cotyledons were transferred and planted in 8-cm pots in California mix and returned to standard culture conditions in the growth cabinet.
Plants were exposed to a range of different treatments to maximize the number of different genes expressed for EST library construction (Table 1). Treatments included: (1) abiotic stresses – drought, abscisic acid (ABA), salt, heat, cold, frost, low/high nutrient, high/low/no light; (2) hormone treatments – methyl jasmonate (MJ), salicylic acid (SA), ethylene, auxin, cytokinins; and (3) control or no treatment. Plants were washed to remove soil particles, and from each treatment leaf and stem, flowers and roots were harvested separately at three growth stages: young plants, young flowering plants and old flowering plants. Six plants were harvested for each treatment at each growth stage. In addition, whole seedlings were harvested at germination (1 d), and at 7 and 14 d from the control plants. Individual plant tissues were wrapped in aluminium foil, snap-frozen in liquid nitrogen and stored at –80 °C until RNA extraction.
Table 1.
Treatment types, concentration of treatments, treatment applied and harvest times following treatments for Senecio madagascariensis plants stressed for mRNA extraction
| Treatment | Concentration of application | Application type | Harvest times |
|---|---|---|---|
| Salicylic acid | 1 mL of 10 mm | Spray | 5 h, 24 h |
| Abscisic acid | 1 mL of 2 mm | Spray | 5 h, 24 h |
| Auxin | 1 mL of 10 mm | Spray | 5 h, 24 h |
| Bric-a-brac protein | 1 mL of 2 mm | Spray | 5 h, 24 h |
| Methyl jasmonate | 400 µL of 400 mm | Contained area | 5 h, 24 h |
| Ethylene | 100 mL ethylene gas | Contained area | 5 h, 24 h |
| Salt stress NaCl | 25 mL 1 m NaCl | In soil | 24 h |
| Cold stress | 4 °C | N/A | 24 h |
| Heat stress | 40 °C in a glasshouse | N/A | 24 h |
| Dark Stress | 48 h in the dark | N/A | 48 h |
N/A, not applicable.
RNA extraction and cDNA synthesis
Total RNA was isolated from approx. 30 mg of each plant tissue using the Promega SV RNA isolation kit (Promega, Wisconsin, USA). Two to four replicates from each growth stage were combined to yield 5 µg or more of total RNA for each tissue at each growth stage under all stress treatments. Five micrograms of total RNA from each sample was combined to yield a total of 250 µg total RNA for both the Australian and the South African populations. Overall 5·5 µg of messenger RNA (mRNA) was purified from 250 µg total RNA using the Macherey-Nagel Nucleo Trap mRNA kit (Macherey-Nagel, Duren, Germany). The quality of this mRNA was visualized on a 1·5 % agarose gel.
Two micrograms of mRNA was converted to double-stranded DNA using the Clontech SMART cDNA synthesis kit (Clontech, California, USA). The manufacturer's protocol was modified slightly by using the primers Trsa Mme1 and TS oligo in the first-strand synthesis step (Trsa Mme1 primer, CGCAGTCGGTACTCCAACTTTTTTTTTTTTTTTTTTTTVN; TS oligo primer, AAGCAGTGGTATCAACGCAGAGTACGCrGrGrG). PCR was performed for 17 cycles following this initial step for the synthesis and amplification of double-stranded cDNA. The protocol used 200 ng of template cDNA, Novagen's KOD polymerase (Novagen, Wisconsin, USA) and primers Trsa Mme1 PCR and TS_PCR (Trsa Mme1_PCR, CGCAGTCGGTACTCCAACTT; TS_PCR, AAGCAGTGGTATCAACGCAGAGT).
EST library construction
cDNA amplified from Australian S. madagascariensis was normalized according to the protocol supplied with the Evrogen Trimmer Direct cDNA Normalization Kit (Novagen, Wisconsin, USA). Poly A tails on the normalized cDNA were removed by the MmeI restriction enzyme. One half of the library was fragmented for pyrosequencing using dual restriction enzyme digests, while the other was sonicated. For the dual digest, 5 µg of cDNA was cleaved using enzyme RsaI while another 5 µg was cleaved using enzyme AluI. cDNA from these two reactions was purified using the Invitrogen PCR cleanup kit and then combined into a single library (Library 2). The other half of the library (10 µg) was fragmented by the Australian Genome Research Facility (AGRF) using a micro-sonicator (Library 1).
Two subtractive libraries were generated using a modified version of the Clontech PCR-Select™ cDNA Subtraction Kit (Clontech, California, USA). cDNA amplified from Australian and South African populations of S. madagascariensis was halved. On both samples two double digests were performed using MmeI (to remove Poly A tails) and either RsaI or AluI (for suppressive subtractive hybridization). Two subtractive libraries were generated (an RsaI library and an AluI library) of the Australian S. madagascariensis ecotype subtracted with the corresponding South African ecotype digests. The two resulting libraries were purified using the Invitrogen PCR cleanup kit (Invitrogen, California, USA) and then combined in equal concentrations to make one library (Library 3). Another library was generated in the same manner but using the reverse subtraction; the South African sample cDNA was subtracted with the Australian sample cDNA (Library 4).
Sequencing of EST libraries
Pyrosequencing libraries were sequenced using Roche 454 FLX technology at the AGRF. Sequence reads were scored for quality and those reads with acceptable thresholds (Q > 25) had adapter sequence removed. Reads were assembled into contigs using the GS De Novo Assembler Application provided by Roche Life Sciences. The software identified pairwise overlaps between reads and assembled reads into contigs from the overlap regions using default settings. Consensus basecalls of the contigs were generated by averaging the processed flow signals for each nucleotide included in the alignment.
Sequence analysis
Contigs from each library were used as BLASTx queries against the non-redundant (NR) database at NCBI, with an E-value cut-off of 1 × 10−5. To obtain an estimate of how many unique transcripts are likely to be represented in each of our libraries, we clustered contigs on the basis of their BLASTx hits. Contigs were clustered if they shared at least one ‘good hit’, defined as the best BLASTx hit and any subsequent hit with an E-value at least 99 % as good as that of the best hit. We refer to groups of clustered contigs as ‘clusters’ and to contigs that did not cluster with any other contig as ‘singlets’. Annotations were transferred from best BLASTx hits to each cluster or singlet.
Gene ontology (GO) terms from the Plant GO Slim (http://www.geneontology.org/GO.slims.shtml) ontologies were assigned to each contig using Blast2GO with default settings (Conesa et al., 2005). We then used WEGO (Ye et al., 2006) to determine the numbers of contigs annotated with each GO term for each library. We tested for significant enrichment of GO terms in different libraries using Fisher's exact tests, corrected for multiple tests using the false discovery rate as implemented in the R package fdrtool (Strimmer, 2008; R Team, 2009).
Microsatellite mining
We mined singlet contigs for microsatellites, using SSRIT (Temnykh et al., 2001; http://www.gramene.org/db/searches/ssrtool), which is a Perl script that identifies all microsatellite markers (simple sequence repeats, SSRs) within a set of sequences. We set the script to identify all possible di-, tri- and tetranucleotide repeats with a minimum of five subunits. Although some groups have used higher cut-offs (Kantety et al., 2002), our method of relaxing the threshold maximizes SSR discovery while still producing a high percentage of polymorphic loci (Pashley et al., 2006).
RESULTS
454 EST sequencing and contig assembly
From a single Roche 454 GS FLX run, a total of 221 746 EST sequences that exceeded our minimum quality thresholds were generated from four cDNA libraries constructed from various tissue types, growth stages and treatments (Table 1). The average size of these ESTs was 210 bp, generating 46·57 Mbp of sequence. The two subtractive libraries (Libraries 3 and 4) had a much greater quantity of ESTs that met our quality thresholds than either of the normalized libraries (Libraries 1 and 2; Table 2). All EST sequences were deposited in the short read archive of the NCBI database (short read archive ID for Library 1, SRX001877; Library 2, SRX001878; Library 3, SRX001880; Library 4, SRX001879) or can be obtained via email from the corresponding author. Assembly of high-quality ESTs formed 12 442 contig sequences of average length 264 bp, with an average depth of coverage of 5·68 sequences per nucleotide position. Some highly expressed contigs were sequenced in over 1000 reads. Overall our average contig length was longer (264 bp) and our average coverage substantially higher (5·67-fold) than most previous studies using 454 sequencing in non-model species: 197 bp at 2·3× coverage (Melitaea cinxia; Vera et al., 2008) and 245 bp (Eucalyptus grandis; Novaes et al., 2008).
Table 2.
Assembly statistics of the Senecio madagascariensis ESTs, including number of ESTs, number of large contigs, mean contig size, largest contig and total contig for the four libraries separately and all libraries combined
| Assembly statistics | Library 1 | Library 2 | Library 3 | Library 4 | All libraries |
|---|---|---|---|---|---|
| Number of ESTs | 36 856 | 26 896 | 76 661 | 81 333 | 221 746 |
| No. of large contigs (≥500 bp) | 156 | 71 | 81 | 277 | 804 |
| Mean contig size (bp) | 236 | 227 | 254 | 262 | 264 |
| Largest contig | 1210 | 822 | 1028 | 1129 | 1575 |
| Total contigs | 3786 | 2859 | 2388 | 5769 | 12 442 |
Library 1, sonicated normalized library; Library 2, restriction enzyme digested normalized library; Library 3, South African subtractive library; Library 4, Australian subtractive library; All Libraries, assembly statistics for all libraries combined.
Functional annotation of contigs
Among the 12 442 contigs, 6183 (49·7 %) had hits exceeding our significance threshold (E-value <1 × 10−5) in the NR database (Table 3). Of the 6183 contigs that had a significant hit in the NR database, 3946 were singlets and 2237 formed 872 clusters of contigs (contigs sharing best or near-best BLAST hits). The number of contigs that formed a cluster ranged from two to 22, with an average of 2·4 ESTs per cluster.
Table 3.
BLASTx analysis statistics of the Senecio madagascariensis EST libraries against the NCBI NR database
| Library | Total contigs | Singlet contigs | Clustered contigs | No. of clusters | No-hit contigs |
|---|---|---|---|---|---|
| 1 | 3786 | 1389 | 265 | 124 | 2132 |
| 2 | 2859 | 912 | 158 | 75 | 1789 |
| 3 | 2388 | 754 | 430 | 168 | 1204 |
| 4 | 5769 | 2169 | 1134 | 437 | 2466 |
| Total | 12442 | 3946 | 2237 | 872 | 6259 |
Library 1, sonicated normalized library; Library 2, restriction enzyme digested normalized library; Library 3, South African subtractive library; Library 4, Australian subtractive library.
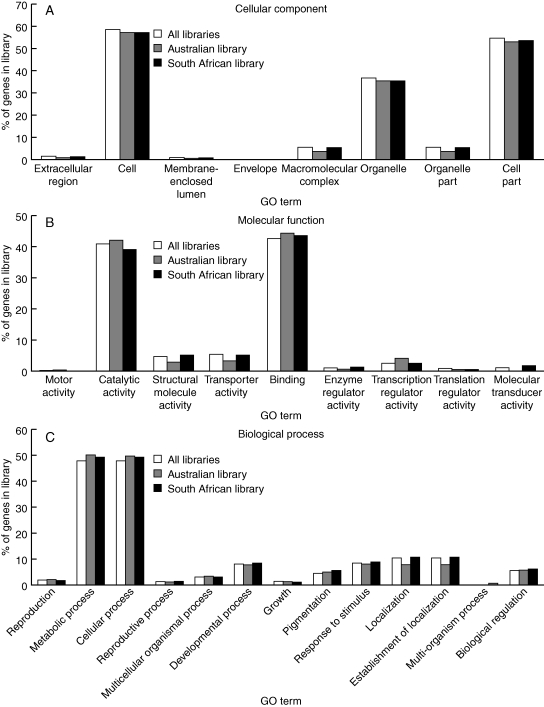
Overall, the contigs were quite diverse in the types of transcripts present, based on the number of GO terms assigned to contigs (Fig. 1). Given the diversity in GO terms assigned, it appears we were successful in capturing a number of functional gene categories in our libraries.
Fig. 1.
Proportional representation of GO terms in the Australian, South African and total EST libraries for (A) cellular component, (B) molecular function and (C) biological process domains.
To investigate which categories of genes are over-represented in the subtractive libraries, we tested for significant enrichment of GO terms in Libraries 3 and 4 relative to contigs assembled from all libraries, and also between Libraries 3 and 4. No GO terms were over-represented in subtractive libraries relative to the normalized library after correction for multiple tests. The molecular transducer activity GO term was significantly (P < 0·001) enriched in Library 4 (South African population) compared with Library 3 (Australian population) after correction for multiple tests (Fig. 1B; see Table 4 for molecular transducer gene list). We undertook a literature search of the genes and gene families identified with the molecular transducer activity GO term from the South African library. Over 40 % of these genes respond to biotic stimuli and have been implicated in defence response to viral, fungal and bacterial pathogens.
Table 4.
Molecular transducer GO class genes that were over-represented in the native range EST library compared with the invasive range EST library
| Gene | Contig ID | Length (bp) |
|---|---|---|
| Phototropic-responsive NPH3 family protein | contig00533; contig01103; contig02636; contig02080 | 294; 241; 243; 156 |
| Histidine phosphotransfer protein | contig00734 | 211 |
| Pyruvate dehydrogenase kinase | contig00887 | 234 |
| Ethylene insensitive 4 | contig01144 | 241 |
| Root phototropism protein | contig03698 | 240 |
| Guanine nucleotide-binding protein beta | contig03914 | 241 |
| Phototropin B1 blue light photoreceptor | contig03963; contig05165 | 240; 164 |
| PAC motif-containing protein | contig04546; contig04557 | 370; 474 |
| Vacuolar sorting receptor protein | contig00082 | 200 |
| Somatic embryogenesis receptor kinase 1 | contig00379; contig00577; contig05630; contig05684 | 356; 439; 586; 172 |
| Histidine kinase 3 | contig00384; contig01470; contig01600 | 548; 242; 485 |
| Eukaryotic translation initiation factor 3 | contig00424; contig01524 | 704; 178 |
| ER lumen retaining receptor | contig01095; contig01221; contig02806 | 241; 237; 404 |
| LrgB-like family protein | contig01889 | 241 |
| Somatic embryogenesis receptor-like kinase2 | contig02285 | 241 |
| Phytosulfokine receptor precursor | contig03383 | 241 |
| Interleukin-1 receptor-associated kinase | contig03394 | 186 |
| Transmembrane BAX inhibitor motif-containing protein | contig03442 | 241 |
| Leaf senescence related protein | contig04299 | 206 |
| Signal recognition particle receptor protein | contig05496 | 228 |
‘Gene’ is the best BLAST hit in the NCBI database with an E-value cut-off of 1 × 10−5. ‘Contig ID’ is the number assigned to each contig in our database and ‘length’ is the length of individual contigs.
Microsatellite markers for mapping and population studies
A total of 190 SSRs from 185 different contigs were detected in the 3946 singlet contigs. This produced an overall SSR discovery rate of 4·8 %, or one microsatellite in every 20 singlet contigs. The most abundant repeat motifs were di-nucleotide repeats (101), closely followed by tri-nucleotide repeats (86) but very few tetra-nucleotide repeats (three).
DISCUSSION
Here we have described an EST collection generated by 454 pyrosequencing and de novo assembly to characterize the transcriptome of a non-model invasive species, S. madagascariensis. The assembly of long contigs with an average length of 264 bp allowed us to achieve high annotation success for species with a lack of genomic resources. Sequencing of two subtractive libraries and a single normalized library also allowed initial characterization of gene expression differences between the native and introduced ranges of S. madagascariensis. The molecular transducer GO term was significantly over-represented in the native (South African) subtractive library compared with the invasive library. Significantly, many of the genes from this GO term are involved in response to biotic stimulus or are defence response proteins.
Gene annotation and candidate markers for invasiveness
Gene annotation against the NR database in NCBI allowed us to annotate approx. 4800 independent transcripts. From these annotated transcripts we identified a number of genes that may underlie traits of evolutionary and ecological interest. Most notable are genes from the molecular transducer activity GO category that are significantly under-represented in the Australian EST library. Although this result is highly significant, other techniques, such as real-time PCR and large-scale sequence analysis, may be used to provide additional support for this finding. Over 40 % of genes and gene families annotated with the molecular transducer activity term in the South African library respond to biotic stimulus and have been implicated in defence responses to fungal, bacterial and viral pathogens (Thomma et al., 1999; Trusov et al., 2006; Hopkins et al., 2008; Watanabe and Lam, 2008; Zhu et al., 2009).
The under-representation of molecular transducer genes in the invasive range library indicates that they may be associated with important evolutionary responses or maternal effects to natural enemies or other biotic stimuli in invasive populations of S. madagascariensis. An under-representation of defence response proteins in the introduced range provides preliminary support for aspects of the ERH and EICA hypotheses in this successful invasive. It has been hypothesized that differences in the natural enemy community between the native and introduced ranges of invasive plants can promote a range of evolutionary responses in the allocation of resources to plant defence and growth (Blossey and Notzold, 1995; Joshi and Vrieling, 2005; Orians and Ward, 2010). For example, an examination of phytochemicals in the invasive range of wild parsnip has provided evidence for increased production of defence compounds that coincide with the introduction of its specialist herbivore (Zangerl and Berenbaum, 2005). Other recent studies have shown that defence and stress genes are downregulated in invasive populations of sunflower and starthistle (Lai et al., 2008; Stewart et al., 2009). Selection for decreased expression of defence genes in the introduced range of S. madagascariensis may explain why genes from the molecular transducer GO category are underrepresented in the Australian library. Alternatively, environmental maternal effects associated with escaping natural enemies in the native range may also explain the under-representation of molecular transducer genes in the Australian library. Both explanations require that introduced populations of S. madagascariensis have escaped their specialist natural enemies.
One assumption of the ERH has been supported in the Australian range of S. madagascariensis (White et al., 2008a). This study demonstrated that a native herbivore prefers native congeners over the introduced S. madagascariensis and that foliage damage was significantly greater on the native species, S. pinntifolius, than on S. madagascariensis (White et al., 2008a). Although this study fulfils some aspects of the ERH it does not demonstrate that natural enemy damage is reduced in the introduced range compared with the native range. Therefore, other studies should investigate whether the negative impacts of natural enemies are decreased in the introduced range of S. madagascariensis. Furthermore, to confirm whether selection or maternal effects are responsible for the observed pattern of under-representation of molecular transducer genes in the Australian library, candidate genes need to be tested for evidence of recent selection and gene expression differences associated with maternal environments.
EST-derived microsatellite markers
We found 190 EST-associated microsatellites suitable for use as genetic markers in future population-level and mapping studies. The rate of microsatellite discovery in our project (4·8 %) is similar to those of most previous studies in plants, which typically range between 2 and 5 % when using similar search parameters (Kantety et al., 2002). Many studies have found in the order of 80–90 % of EST-derived microsatellite markers to be polymorphic (Bandopadhyay et al., 2004; Ellis et al., 2006; Pashley et al., 2006; Gasic et al., 2009). These microsatellites are likely to be highly transferable across closely related species, as has been found in previous studies (Gutierrez et al., 2005; Pashley et al., 2006; Ellis and Burke, 2007). Therefore, these markers may provide a resource for another highly invasive species, S. inaequidens (Lafuma and Maurice, 2007; Bossdorf et al., 2008; Cano et al., 2009; Monty et al., 2009; Monty and Mahy, 2009), and for hybridization studies with the Australian native, S. pinnatifolius (Prentis et al., 2007).
Conclusions
Senecio madagascariensis and its closely related congener S. inaequidens are highly invasive species that are becoming increasingly used as model species with which to investigate hypotheses in invasion biology (Radford and Cousens, 2000; Lafuma et al., 2003; Cano et al., 2007; Lafuma and Maurice, 2007; Prentis et al., 2007; White et al., 2008a, b, c; Bossdorf et al., 2008; Monty et al., 2009; Monty and Mahy, 2009, 2010). The use of genome scans and gene expression approaches to determine if invasion success may have a molecular genetic basis has been constrained by a lack of genomic resources to identify candidate genes or genomic changes associated with invasiveness. Here, we report an EST collection for S. madagascariensis that provides a first step toward understanding the genetic bases of invasion success. Our data provide large-scale EST resources that will further enhance S. madagascariensis as a model species for examining hypotheses about invasion success. This resource will aid microarray design and sequencing projects and provides a large number of initial candidate gene markers that can help to answer a number of leading hypotheses in invasion biology.
ACKNOWLEDGEMENTS
We thank Ed Gilding, Ian Godwin and Peter Mather for comments on earlier versions of the manuscript, and Zeinab Galal for assistance with GenBank submissions. This manuscript was improved by comments from Pat Heslop-Harrison and two anonymous reviewers. Funding for this work was provided by the Australian Research Council (grant number DP0664967) awarded to A.J.L. and P.M.S.
LITERATURE CITED
- Baker HG. The evolution of weeds. Annual Review of Ecology, Evolution, and Systematics. 1974;5:1–24. [Google Scholar]
- Bandopadhyay R, Sharma S, Rustgi S, et al. DNA polymorphism among 18 species of Triticum–Aegilops complex using wheat EST-SSRs. Plant Science. 2004;166:349–356. [Google Scholar]
- Barker MS, Kane NC, Matvienko M, et al. Multiple paleopolyploidizations during the evolution of the Compositae reveal parallel patterns of duplicate gene retention after millions of years. Molecular Biology and Evolution. 2008;25:2445–2455. doi: 10.1093/molbev/msn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RDH, Schluter D. Adaptation from standing genetic variation. Trends in Ecology & Evolution. 2008;23:38–44. doi: 10.1016/j.tree.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Blossey B, Notzold R. Evolution of increased competitive ability in invasive nonindigenous plants – a hypothesis. Journal of Ecology. 1995;83:887–889. [Google Scholar]
- Bossdorf O, Auge H, Lafuma L, Rogers WE, Siemann E, Prati D. Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia. 2005;144:1–11. doi: 10.1007/s00442-005-0070-z. [DOI] [PubMed] [Google Scholar]
- Bossdorf O, Lipowsky A, Prati D. Selection of preadapted populations allowed Senecio inaequidens to invade Central Europe. Diversity and Distributions. 2008;14:676–685. [Google Scholar]
- Bouck A, Vision T. The molecular ecologist's guide to expressed sequence tags. Molecular Ecology. 2007;16:907–924. doi: 10.1111/j.1365-294X.2006.03195.x. [DOI] [PubMed] [Google Scholar]
- Broz AK, Broeckling CD, He JB, Dai X, Zhao PX, Vivanco JM. A first step in understanding an invasive weed through its genes: an EST analysis of invasive Centaurea maculosa. BMC Plant Biology. 2007;7(25) doi: 10.1186/1471-2229-7-25. doi:10.1186/1471-2229-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz AK, Manter DK, Bowman G, Muller-Scharer H, Vivanco JM. Plant origin and ploidy influence gene expression and life cycle characteristics in an invasive weed. BMC Plant Biology. 2009;9(33) doi: 10.1186/1471-2229-9-33. doi:10.1186/1471-2229-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colautti RI, Ricciardi A, Grigorovich IA, MacIsaac HJ. Is invasion success explained by the enemy release hypothesis? Ecology Letters. 2004;7:721–733. [Google Scholar]
- Cano L, Escarre J, Sans FX. Factors affecting the invasion success of Senecio inaequidens and S. pterophorus in Mediterranean plant communities. Journal of Vegetation Science. 2007;18:281–288. [Google Scholar]
- Cano L, Escarre J, Vrieling K, Sans FX. Palatability to a generalist herbivore, defence and growth of invasive and native Senecio species: testing the evolution of increased competitive ability hypothesis. Oecologia. 2009;159:95–106. doi: 10.1007/s00442-008-1182-z. [DOI] [PubMed] [Google Scholar]
- Cheung F, Haas BJ, Goldberg SMD, May GD, Xiao YL, Town CD. Sequencing Medicago truncatula expressed sequenced tags using 454 Life Sciences technology. BMC Genomics. 2006;7(272) doi: 10.1186/1471-2164-7-272. doi:10.1186/1471-2164-7-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Dlugosch KM, Parker IM. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Molecular Ecology. 2008;17:431–449. doi: 10.1111/j.1365-294X.2007.03538.x. [DOI] [PubMed] [Google Scholar]
- Ellis JR, Burke JM. EST-SSRs as a resource for population genetic analyses. Heredity. 2007;99:125–132. doi: 10.1038/sj.hdy.6801001. [DOI] [PubMed] [Google Scholar]
- Ellis JR, Pashley CH, Burke JM, McCauley DE. High genetic diversity in a rare and endangered sunflower as compared to a common congener. Molecular Ecology. 2006;15:2345–2355. doi: 10.1111/j.1365-294X.2006.02937.x. [DOI] [PubMed] [Google Scholar]
- Ellstrand NC, Schierenbeck KA. Hybridization as a stimulus for the evolution of invasiveness in plants? Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7043–7050. doi: 10.1073/pnas.97.13.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasic K, Han YP, Kertbundit S, et al. Characteristics and transferability of new apple EST-derived SSRs to other Rosaceae species. Molecular Breeding. 2009;23:397–411. [Google Scholar]
- Goldberg SMD, Johnson J, Busam D, et al. A Sanger/pyrosequencing hybrid approach tor the generation of high-quality draft assemblies of marine microbial genomes. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11240–11245. doi: 10.1073/pnas.0604351103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MV, Patto MCV, Huguet T, Cubero JI, Moreno MT, Torres AM. Cross-species amplification of Medicago truncatula microsatellites across three major pulse crops. Theoretical and Applied Genetics. 2005;110:1210–1217. doi: 10.1007/s00122-005-1951-6. [DOI] [PubMed] [Google Scholar]
- Hopkins MT, Lampi Y, Wang TW, Liu ZD, Thompson JE. Eukaryotic translation initiation factor 5A is involved in pathogen-induced cell death and development of disease symptoms in Arabidopsis. Plant Physiology. 2008;148:479–489. doi: 10.1104/pp.108.118869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biology. 2007;8(R143) doi: 10.1186/gb-2007-8-7-r143. doi:10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi J, Vrieling K. The enemy release and EICA hypothesis revisited: incorporating the fundamental difference between specialist and generalist herbivores. Ecology Letters. 2005;8:704–714. [Google Scholar]
- Kang M, Buckley YM, Lowe AJ. Testing the role of genetic factors across multiple independent invasions of the shrub Scotch broom (Cytisus scoparius) Molecular Ecology. 2007;16:4662–4673. doi: 10.1111/j.1365-294X.2007.03536.x. [DOI] [PubMed] [Google Scholar]
- Kantety RV, La Rota M, Matthews DE, Sorrells ME. Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Molecular Biology. 2002;48:501–510. doi: 10.1023/a:1014875206165. [DOI] [PubMed] [Google Scholar]
- Keane RM, Crawley MJ. Exotic plant invasions and the enemy release hypothesis. Trends in Ecology & Evolution. 2002;17:164–170. [Google Scholar]
- Lachmuth S, Durka W, Schurr FM. The making of a rapid plant invader: genetic diversity and differentiation in the native and invaded range of Senecio inaequidens. Molecular Ecology. 2010;19:3952–3967. doi: 10.1111/j.1365-294X.2010.04797.x. doi:10.1111/j.1365-294X.2010.04797.x. [DOI] [PubMed] [Google Scholar]
- Lafuma L, Maurice S. Increase in mate availability without loss of self-incompatibility in the invasive species Senecio inaequidens (Asteraceae) Oikos. 2007;116:201–208. [Google Scholar]
- Lafuma L, Balkwill K, Imbert E, Verlaque R, Maurice S. Ploidy level and origin of the European invasive weed Senecio inaequidens (Asteraceae) Plant Systematics and Evolution. 2003;243:59–72. [Google Scholar]
- Lai Z, Gross BL, Zou Y, Andrews J, Rieseberg LH. Microarray analysis reveals differential gene expression in hybrid sunflower species. Molecular Ecology. 2006;15:1213–1227. doi: 10.1111/j.1365-294X.2006.02775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Kane NC, Zou Y, Rieseberg LH. Natural variation in gene expression between wild and weedy populations of Helianthus annuus. Genetics. 2008;179:1881–1890. doi: 10.1534/genetics.108.091041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux JJ, Wieczorek AM, Ramadan MM, Tran CT. Resolving the native provenance of invasive fireweed (Senecio madagascariensis Poir.) in the Hawaiian Islands as inferred from phylogenetic analysis. Diversity and Distributions. 2006;12:694–702. [Google Scholar]
- Le Roux J, Wieczorek A, Tran C, Vorsino A. Disentangling the dynamics of invasive fireweed (Senecio madagascariensis Poir. species complex) in the Hawaiian Islands. Biological Invasions. 2010;12:2251–2264. [Google Scholar]
- Lee CE. Evolutionary genetics of invasive species. Trends in Ecology & Evolution. 2002;17:386–391. [Google Scholar]
- Lopez MG, Wulff AF, Poggio L, Xifreda CC. South African fireweed Senecio madagascariensis (Asteraceae) in Argentina: relevance of chromosome studies to its systematics. Botanical Journal of the Linnean Society. 2008;158:613–620. [Google Scholar]
- Margulies M, Egholm M, Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milton SJ, Wilson JRU, Richardson DM, et al. Invasive alien plants infiltrate bird-mediated shrub nucleation processes in arid savanna. Journal of Ecology. 2007;95:648–661. [Google Scholar]
- Monty A, Mahy G. Clinal differentiation during invasion: Senecio inaequidens (Asteraceae) along altitudinal gradients in Europe. Oecologia. 2009;159:305–315. doi: 10.1007/s00442-008-1228-2. [DOI] [PubMed] [Google Scholar]
- Monty A, Mahy G. Evolution of dispersal traits along an invasion route in the wind-dispersed Senecio inaequidens (Asteraceae) Oikos. 2010;119:1563–1570. [Google Scholar]
- Monty A, Lebeau J, Meerts P, Mahy G. An explicit test for the contribution of environmental maternal effects to rapid clinal differentiation in an invasive plant. Journal of Evolutionary Biology. 2009;22:917–926. doi: 10.1111/j.1420-9101.2009.01728.x. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Dhingra A, Soltis PS, et al. Rapid and accurate pyrosequencing of angiosperm plastid genomes. BMC Plant Biology. 2006;6(17) doi: 10.1186/1471-2229-6-17. doi:10.1186/1471-2229-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novaes E, Drost DR, Farmerie WG, et al. High-throughput gene and SNP discovery in Eucalyptus grandis, an uncharacterized genome. BMC Genomics. 2008;9(312) doi: 10.1186/1471-2164-9-312. doi:10.1186/1471-2164-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orians CM, Ward D. Evolution of plant defenses in nonindigenous environments. Annual Review of Entomology. 2010;55:439–459. doi: 10.1146/annurev-ento-112408-085333. [DOI] [PubMed] [Google Scholar]
- Parisod C, Salmon A, Zerjal T, Tenaillon M, Grandbastien M, Ainouche M. Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in Spartina. New Phytologist. 2009;184:1003–1015. doi: 10.1111/j.1469-8137.2009.03029.x. [DOI] [PubMed] [Google Scholar]
- Pashley CH, Ellis JR, McCauley DE, Burke JM. EST databases as a source for molecular markers: lessons from Helianthus. Journal of Heredity. 2006;97:381–388. doi: 10.1093/jhered/esl013. [DOI] [PubMed] [Google Scholar]
- Prentis PJ, White EM, Radford IJ, Lowe AJ, Clarke AR. Can hybridization cause local extinction: a case for demographic swamping of the Australian native Senecio pinnatifolius by the invasive Senecio madagascariensis? New Phytologist. 2007;176:902–912. doi: 10.1111/j.1469-8137.2007.02217.x. [DOI] [PubMed] [Google Scholar]
- Prentis PJ, Wilson JRU, Dormontt EE, Richardson DM, Lowe AJ. Adaptive evolution in invasive species. Trends in Plant Science. 2008;13:288–294. doi: 10.1016/j.tplants.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Prentis PJ, Sigg D, Raghu S, Dhileephan K, Pavasovic A, Lowe AJ. Understanding invasion history: genetic structure and diversity of two globally invasive plants and implications for their management. Diversity and Distributions. 2009;15:822–830. [Google Scholar]
- Proches S, Wilson JRU, Richardson DM, Rejmanek M. Searching for phylogenetic pattern in biological invasions. Global Ecology and Biogeography. 2008;17:5–10. [Google Scholar]
- R Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2009. [Google Scholar]
- Radford IJ, Cousens RD. Invasiveness and comparative life-history traits of exotic and indigenous Senecio species in Australia. Oecologia. 2000;125:531–542. doi: 10.1007/s004420000474. [DOI] [PubMed] [Google Scholar]
- Radford IJ, Muller P, Fiffer S, Michael PW. Genetic relationships between Australian fireweed and south African and Madagascan populations of Senecio madagascariensis Poir. and closely related Senecio species. Australian Systematic Botany. 2000;13:409–423. [Google Scholar]
- Richardson DM, Pysek P. Plant invasions: merging the concepts of species invasiveness and community invasibility. Progress in Physical Geography. 2006;30:409–431. [Google Scholar]
- Sakai AK, Allendorf FW, Holt JS, et al. The population biology of invasive species. Annual Review of Ecology and Systematics. 2001;32:305–332. [Google Scholar]
- Scott LJ, Congdon BC, Playford J. Molecular evidence that fireweed (Senecio madagascariensis, Asteraceae) is of South African origin. Plant Systematics and Evolution. 1998;213:251–257. [Google Scholar]
- Stewart CN, Tranel PJ, Horvath DP, et al. Evolution of weediness and invasiveness: charting the course for weed genomics. Weed Science. 2009;57:451–462. [Google Scholar]
- Strimmer K. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24:1461–1462. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
- Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Research. 2001;11:1441–1452. doi: 10.1101/gr.184001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Tierens KFMJ, Broekaert WF. Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiology. 1999;121:1093–1101. doi: 10.1104/pp.121.4.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuiller W, Richardson DM, Rouget M, Proches S, Wilson JRU. Interactions between environment, species traits, and human uses describe patterns of plant invasions. Ecology. 2006;87:1755–1769. doi: 10.1890/0012-9658(2006)87[1755:ibesta]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Trusov Y, Rookes JE, Chakravorty D, Armour D, Schenk PM, Botella JR. Heterotrimeric G proteins facilitate Arabidopsis resistance to necrotrophic pathogens and are involved in jasmonate signaling. Plant Physiology. 2006;140:210–220. doi: 10.1104/pp.105.069625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera JC, Wheat CW, Fescemyer HW, et al. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Molecular Ecology. 2008;17:1636–1647. doi: 10.1111/j.1365-294X.2008.03666.x. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Lam E. BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis. Journal of Biological Chemistry. 2008;283:3200–3210. doi: 10.1074/jbc.M706659200. [DOI] [PubMed] [Google Scholar]
- White EM, Sims NM, Clarke AR. Test of the enemy release hypothesis: the native magpie moth prefers a native fireweed (Senecio pinnatifolius) to its introduced congener (S. madagascariensis) Austral Ecology. 2008a;33:110–116. [Google Scholar]
- White EM, Wilson JC, Clarke AR. Diversity and abundance of arthropod floral visitor and herbivore assemblages on exotic and native Senecio species. Plant Protection Quarterly. 2008b;23:90–98. [Google Scholar]
- White EM, Wilson JC, Clarke AR. Plant–pollinator interactions in sympatric exotic and native Senecio species: is facilitation or competition for pollinators occurring? Plant Protection Quarterly. 2008c;23:120–126. [Google Scholar]
- Whitney KD, Randell RA, Rieseberg LH. Adaptive introgression of herbivore resistance traits in the weedy sunflower Helianthus annuus. American Naturalist. 2006;167:794–807. doi: 10.1086/504606. [DOI] [PubMed] [Google Scholar]
- Wicker T, Schlagenhauf E, Graner A, Close TJ, Keller B, Stein N. 454 sequencing put to the test using the complex genome of barley. BMC Genomics. 2006;7(275) doi: 10.1186/1471-2164-7-275. doi:10.1186/1471-2164-7-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JRU, Richardson DM, Rouget M, et al. Residence time and potential range: crucial considerations in modelling plant invasions. Diversity and Distributions. 2007;13:11–22. [Google Scholar]
- Wilson JRU, Dormontt EE, Prentis PJ, Lowe AJ, Richardson DM. Something in the way you move: dispersal pathways affect invasion success. Trends in Ecology & Evolution. 2009a;24:136–144. doi: 10.1016/j.tree.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Wilson JRU, Dormontt EE, Prentis PJ, Lowe AJ, Richardson DM. Biogeographic concepts define invasion biology. Trends in Ecology & Evolution. 2009b;24:586–586. doi: 10.1016/j.tree.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Ye J, Fang L, Zheng HK, et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Research. 2006;34:W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangerl AR, Berenbaum MR. Increase in toxicity of an invasive weed after reassociation with its coevolved herbivore. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15529–15532. doi: 10.1073/pnas.0507805102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu HF, Li GJ, Ding L, et al. Arabidopsis extra large G-protein 2 (XLG2) interacts with the G beta subunit of heterotrimeric G protein and functions in disease resistance. Molecular Plant. 2009;2:513–525. doi: 10.1093/mp/ssp001. [DOI] [PMC free article] [PubMed] [Google Scholar]