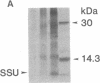
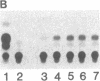
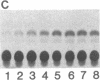
Abstract
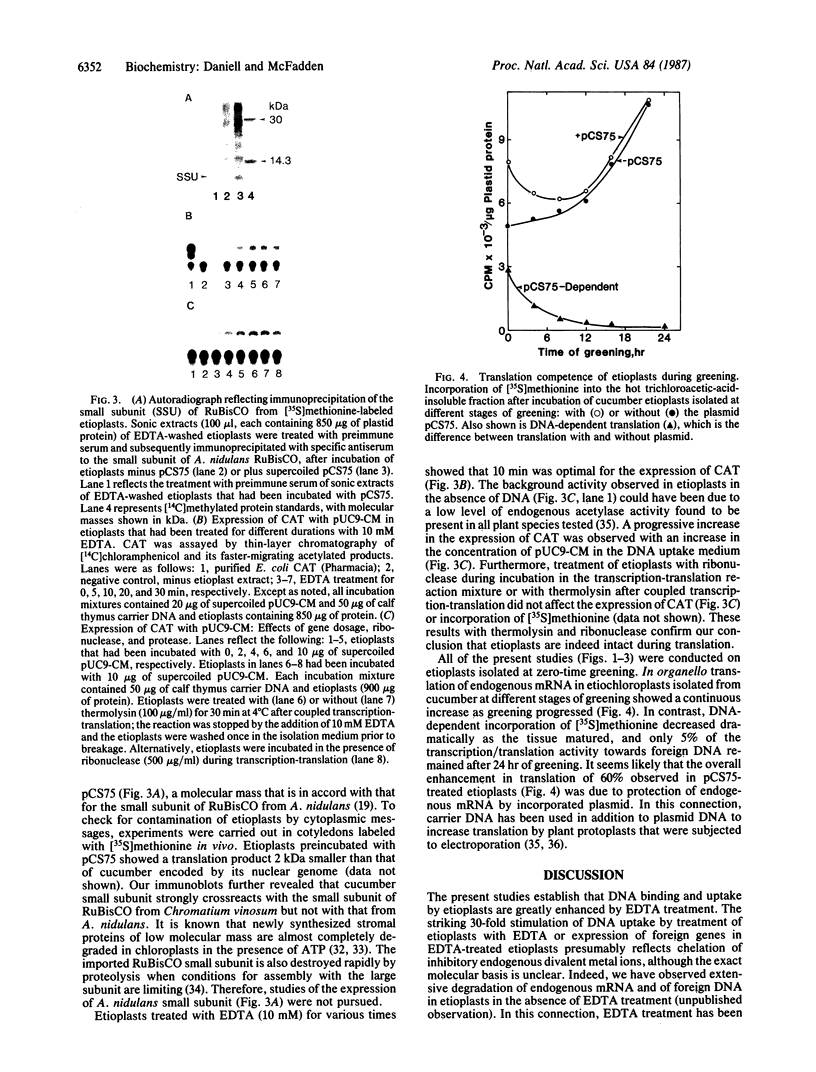
The uptake and expression by plastids isolated from dark-grown cucumber cotyledons (etioplasts) of two pUC derivatives, pCS75 and pUC9-CM, respectively carrying genes for the large small subunits of ribulose bisphosphate carboxylase/oxygenase of Anacystis nidulans or chloramphenicol acetyltransferase, is reported. Untreated etioplasts take up only 3% as much DNA as that taken up by EDTA-washed etioplasts after 2 hr of incubation with nick-translated [32P]-pCS75. The presence or absence of light does not affect DNA uptake, binding, or breakdown by etioplasts. Calcium or magnesium ions inhibit DNA uptake by 86% but enhance binding (23-200%) and breakdown (163-235%) of donor DNA by EDTA-treated etioplasts. Uncouplers that abolish membrane potential (delta psi), transmembrane proton gradient (delta pH), or both do not affect DNA uptake, binding, or breakdown by etioplast. However, both DNA uptake and binding are severely inhibited by ATP. Presumably this results from the hydrolysis of ATP, because the poorly hydrolyzable analog adenyl-5'-yl imidodiphosphate does not inhibit the uptake or binding of DNA by etioplasts. beta-Lactamase specified by the ampicillin resistance gene of pCS75 can be detected only in EDTA-treated etioplasts that have been incubated with the plasmid pCS75. After the incubation of EDTA-treated etioplasts with pCS75, immunoprecipitation using antiserum to the small subunit of ribulose bisphosphate carboxylase/oxygenase from A. nidulans reveals the synthesis of small subunits; these are smaller by 2 kDa than the cucumber small subunit encoded by the nuclear genome. Treatment of etioplasts with 10 mM EDTA shows a 10-min duration to be optimal for the expression of chloramphenicol acetyltransferase encoded by pUC9-CM. A progressive increase in the expression of this enzyme is observed with an increase in the concentration of pUC9-CM in the DNA uptake medium. The plasmid-dependent incorporation of [35S]methionine by EDTA-treated organelles declines markedly during cotyledon greening in vivo.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennett J., Radcliffe C. Plastid DNA replication and plastid division in the garden pea. FEBS Lett. 1975 Aug 15;56(2):222–225. doi: 10.1016/0014-5793(75)81096-9. [DOI] [PubMed] [Google Scholar]
- Buckley K. J., Hayashi M. Lytic activity localized to membrane-spanning region of phi X174 E protein. Mol Gen Genet. 1986 Jul;204(1):120–125. doi: 10.1007/BF00330198. [DOI] [PubMed] [Google Scholar]
- Carlson P. S. The use of protoplasts for genetic research. Proc Natl Acad Sci U S A. 1973 Feb;70(2):598–602. doi: 10.1073/pnas.70.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen D. M., Bedbrook J. R., Bogorad L., Rich A. Maize chloroplast DNA fragment encoding the large subunit of ribulosebisphosphate carboxylase. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5487–5491. doi: 10.1073/pnas.74.12.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi G., Broglie R., Edwards C., Chua N. H. Tissue-specific and light-regulated expression of a pea nuclear gene encoding the small subunit of ribulose-1,5-bisphosphate carboxylase. EMBO J. 1984 Aug;3(8):1671–1679. doi: 10.1002/j.1460-2075.1984.tb02031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H., Krishnan M., Uma Bai U., Gnanam A. An efficient and prolonged in vitro translational system from isolated cucumber etioplasts. Biochem Biophys Res Commun. 1986 Feb 26;135(1):248–255. doi: 10.1016/0006-291x(86)90969-1. [DOI] [PubMed] [Google Scholar]
- Daniell H., McFadden B. A. Characterization of DNA uptake by the cyanobacterium Anacystis nidulans. Mol Gen Genet. 1986 Aug;204(2):243–248. doi: 10.1007/BF00425505. [DOI] [PubMed] [Google Scholar]
- Daniell H., Ramanujam P., Krishnan M., Gnanam A., Rebeiz C. A. In vitro synthesis of photosynthetic membranes: I. Development of photosystem I activity and cyclic photophosphorylation. Biochem Biophys Res Commun. 1983 Mar 16;111(2):740–749. doi: 10.1016/0006-291x(83)90367-4. [DOI] [PubMed] [Google Scholar]
- Daniell H., Rebeiz C. A. Chloroplast culture. IX. Chlorophyll(ide) a biosynthesis in vitro at rates higher than in vivo. Biochem Biophys Res Commun. 1982 May 31;106(2):466–470. doi: 10.1016/0006-291x(82)91133-0. [DOI] [PubMed] [Google Scholar]
- Daniell H., Rebeiz C. A. Chloroplast culture. VIII. A new effect of kinetin in enhancing the synthesis and accumulation of protochlorophyllide in vitro. Biochem Biophys Res Commun. 1982 Jan 29;104(2):837–843. doi: 10.1016/0006-291x(82)90713-6. [DOI] [PubMed] [Google Scholar]
- Daniell H., Sarojini G., McFadden B. A. Transformation of the cyanobacterium Anacystis nidulans 6301 with the Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2546–2550. doi: 10.1073/pnas.83.8.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della-Cioppa G., Bauer S. C., Klein B. K., Shah D. M., Fraley R. T., Kishore G. M. Translocation of the precursor of 5-enolpyruvylshikimate-3-phosphate synthase into chloroplasts of higher plants in vitro. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6873–6877. doi: 10.1073/pnas.83.18.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm M., Taylor L. P., Walbot V. Expression of genes transferred into monocot and dicot plant cells by electroporation. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5824–5828. doi: 10.1073/pnas.82.17.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor J. J., Still C. C. Subcellular localization of rice leaf aryl acylamidase activity. Plant Physiol. 1983 May;72(1):80–85. doi: 10.1104/pp.72.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joenje H., Venema G. Different nuclease activities in competent and noncompetent Bacillus subtilis. J Bacteriol. 1975 Apr;122(1):25–33. doi: 10.1128/jb.122.1.25-33.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeker W. D., Kowalski D., Laskowski M., Sr Mung bean nuclease I. Terminally directed hydrolysis of native DNA. Biochemistry. 1976 Oct 5;15(20):4463–4467. doi: 10.1021/bi00665a020. [DOI] [PubMed] [Google Scholar]
- Lurquin P. F., Kleinhofs A. Effects of chloramphenicol on plant cells: potential as a selectable marker for transformation studies. Biochem Biophys Res Commun. 1982 Jul 16;107(1):286–293. doi: 10.1016/0006-291x(82)91702-8. [DOI] [PubMed] [Google Scholar]
- Mulder J. A., Venema G. Isolation and partial characterization of Bacillus subtilis mutants impaired in DNA entry. J Bacteriol. 1982 Apr;150(1):260–268. doi: 10.1128/jb.150.1.260-268.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou-Lee T. M., Turgeon R., Wu R. Expression of a foreign gene linked to either a plant-virus or a Drosophila promoter, after electroporation of protoplasts of rice, wheat, and sorghum. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6815–6819. doi: 10.1073/pnas.83.18.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow D. W., DE Wet J. R., Helinski D. R., Howell S. H., Wood K. V., Deluca M. Transient and stable expression of the firefly luciferase gene in plant cells and transgenic plants. Science. 1986 Nov 14;234(4778):856–859. doi: 10.1126/science.234.4778.856. [DOI] [PubMed] [Google Scholar]
- Pardo A. D., Chereskin B. M., Castelfranco P. A., Franceschi V. R., Wezelman B. E. ATP requirement for mg chelatase in developing chloroplasts. Plant Physiol. 1980 May;65(5):956–960. doi: 10.1104/pp.65.5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt G. W., Mishkind M. L. Rapid degradation of unassembled ribulose 1,5-bisphosphate carboxylase small subunits in chloroplasts. Proc Natl Acad Sci U S A. 1983 May;80(9):2632–2636. doi: 10.1073/pnas.80.9.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H., Wiersma K., Venema G., Bron S. Transformation in Bacillus subtilis: a 75,000-dalton protein complex is involved in binding and entry of donor DNA. J Bacteriol. 1984 Mar;157(3):733–738. doi: 10.1128/jb.157.3.733-738.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabita F. R., Small C. L. Expression and assembly of active cyanobacterial ribulose-1,5-bisphosphate carboxylase/oxygenase in Escherichia coli containing stoichiometric amounts of large and small subunits. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6100–6103. doi: 10.1073/pnas.82.18.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nieuwenhoven M. H., Hellingwerf K. J., Venema G., Konings W. N. Role of proton motive force in genetic transformation of Bacillus subtilis. J Bacteriol. 1982 Aug;151(2):771–776. doi: 10.1128/jb.151.2.771-776.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]