1. Structure
Methylation of cytosine, to 5-methylcytosine, is an epigenetic gene regulatory mechanism with implications for ageing and disease in all tissues. More familiar aspects of gene regulation concern the binding of transcription factors to their cognate DNA sequences in gene promoters. However, epigenetic mechanisms also control gene expression without altering the genomic DNA sequence. This epigenome is literally “overlaid on the genome”. Fundamentally, epigenetic regulation controls the chromatin architecture to affect the access of DNA binding proteins to the DNA. Chromatin is comprised of genomic DNA associated with the nucleosome core histone proteins (H2A, H2B, H3, H4). The fundamental subunit of chromatin is the nucleosome, comprised of a histone octamer (two of each histone) around which the DNA is coiled for about 2 complete turns. DNA-methylation alters chromatin architecture through cross talk with the post-translational modification of histone proteins. We refer readers to a recent review of these and other epigenetic mechanisms relevant to eye development and disease (Cvekl and Mitton, 2010).
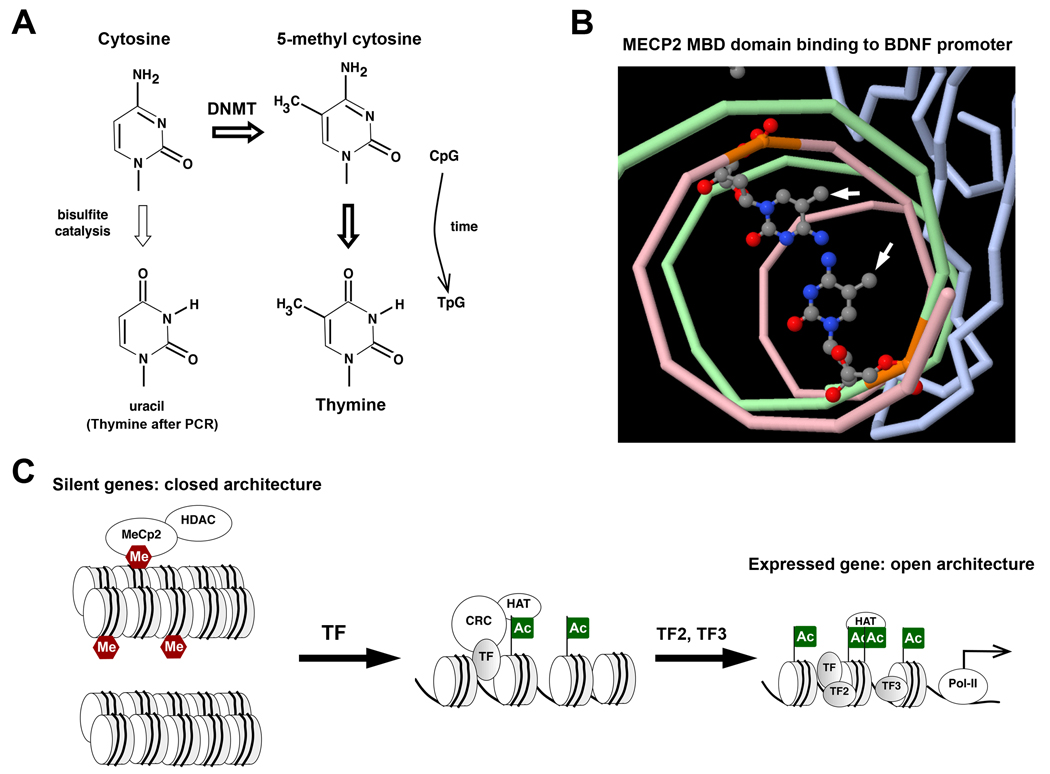
5-Methylcytosine and its conversions with cytosine, thymine, and uracil are illustrated in Figure 1A. In mammals, methylation of cytosine is most common at CpG dinucleotides (Methyl-CpG). Cytosines in both strands of the CpG motif are methylated. In the mammalian genome, the relative frequency of CpG dinucleotides is lower than expected, and CpG motifs are more prevalent in gene promoter regions. Methyl-CpG may deaminate to TpG over time, except where essential function maintains the motif. Methylation of non-CpG cytosines does occur, but the extent of this methylation is not yet known for most tissues.
Figure 1.
A. Cytosine, most often in CpG dinucleotides, can be methylated at position 5 of the ring by DNA methyl transferase (DNMT) to produce 5-methylcytosine. Deamination of 5-methylcytosine in non-essential CpG dinucleotides has resulted in their gradual conversion to TpG througout the the genome. Thus CpG motifs are more concentrated near transcription start sites. 5-methylcytosine is relatively more resistant to bisulfite catalyzed deamination than cytosine, forming a basis for biochemical determination of CpG methylation state. Non-methylated cytosine is converted to uracil, and is replaced by thymine during PCR based amplification.
B. X-ray crystallograhic structure of the MECP2 methyl-DNA binding domain (MBD) bound to 5-methylcytosines within the BDNF promoter. Complimentary DNA strands are colored green and pink, the MBD domain is blue, and methylated cytosines are rendered as ball-and-stick structures. 5-Methyl groups (arrows) within the same CpG dinucleotide interact with the MDB on the same side of the DNA double helix. Image generated using structural coordinates from the PDB file 3C2I, http://www.rcsb.org/pdb/explore.do?structureId=3C2I. (PMID 18313390).
C. Silent genes have a closed chromatin architecture with tight nucleosome packing, characterized by non-acetylated histones. At some genes (top right), this is reinforced by DNA (CpG)-methylation (Me). Methyl-CpG-binding proteins (MECP2), recruit histone deacetylase (HDAC), preventing histone acetylation. Silent genes are activated by pioneer transcription factors (TF) that recruit chromatin-remodeling complexes (CRC) with histone acetyl-transferase (HAT), and DNA-demethylase activity if required. Histone acetylation (Ac) begins to unpack nucleosomes, providing access to additional transcription factors (TF2, TF3), and HATs. Some HATs bind acetylated side-chains and acetylate additional histone sites. The expressed gene (lower right) has an open chromatin architecture and provides access to RNA-polymerase-II (Pol-II) for transcription.
Several methods are commonly used to detect 5-methylcytosine. Methyl-cytosine specific antibodies can be used to capture methylated DNA fragments, which are subsequently quantified by qPCR or hybridization to DNA arrays. Other popular methods begin with the bisulfite-catalyzed deamination of cytosine to uracil under conditions where 5-methylcytosine remains unreactive (Fig. 1A). Methylation-specific PCR (MSP) requires two primer sets that amplify methylated or non-methylated targets after bisulfite conversion. Another method, bisulfite sequencing, requires PCR primers that do not contain CpG in their sequences (each C becomes T). The resulting PCR products can be directly sequenced to evaluate strand-specific methylation, or subcloned to sequence C/T conversions in a sampling of individual molecules. More recently, hydroxymethylcytosine has emerged as a chemical modification of 5-methylcytosine. Bisulfite conversion does not distinguish between the two. Commercial antibodies specific for hydroxymethylcytosine are now available.
2. Function
5-Methylcytosine impacts gene expression in two ways. First, the CpG motif may be part of a cognate binding site and methylation may block protein binding. One example of this is the chromatin insulator, CCCTC binding factor (CTCF). Secondly, methyl-DNA-binding proteins (MBPs) recruit repressor complexes containing histone deacetylases (HDACs) and histone methyltransferases (HMTs). MBPs are currently grouped into three families. The members of the MBD family (MECP2, MBD1, MBD2, MBD4), contain a methyl-CpG binding domain (MBD) and a transrepression domain (TRD). The members of the SRA family (UHRF1 and UHRF2) contain a methyl-CpG binding SRA domain, as well as a ubiquitin-like domain, a PHD zinc-finger and a ring-finger domain. Finally, the members of the Kaiso family (Kaiso, ZBTB4, ZBTB38) contain a triple Zn-finger domain (methyl-DNA binding) and a BTB domain. 5-Methylcytosines on both strands
Binding of the MDB domain, from MECP2, to a methylated CpG site in the BDNF promoter is illustrated in Figure 1B. Note that both 5-methyl groups of the same CpG site are oriented on the same side of the DNA double helix, and both interact with the MDB domain. MBPs, bound to methylated DNA, recruit complexes that include HDACs and HMTs. Thus DNA methylation promotes de-acetylation of histones, which favors the closed chromatin architecture of silent genes. (Fig. 1C) For example, lysine-9 (K9) of histone H3 is de-acetylated and trimethylated (H3K9Me3) in dense heterochromatin of the cell nucleus. De-acetylation may be reinforced by CpG methylation at some genes, which must be reversed to permit histone acetylation and an open achitecture for gene expression. (Fig. 1C)
DNA-methylation is essential for X-chromosome inactivation, genomic imprinting, and transcriptional silencing/activation during development. Mammals have three types of DNA-methyl transferases: DNMT1, DNMT2, and the related DNMT3a and 3b. DNMT1 contributes to the maintenance of DNA-methylation patterns during cell division by acting on hemi-methylated DNA. DNMT2 has weaker activity and targets tRNA. DNMT3a and DNMT3b can act on non-methylated DNA. How these proteins are targeted to DNA remains unclear. To reproduce template methylation patterns during DNA replication, DNMT1 can be recruited by UHRF1, which binds to hemi-methylated DNA. Similar to Dnmt1 knockout mice, Uhrf1 knockout mice die early in embryogenesis. During fertilization, the paternal and maternal genomes are demethylated, followed by new methylation patterns characteristic of emerging cell-types. Much of this developmental DNA methylation is dependent on DNMT3a and 3b. Combined knockout of Dnmt3a and Dnmt3b are embryonically lethal. In contrast to DNA-methylation, the mechanisms that de-methylate methyl-CpG are not fully resolved. Controversy exists regarding de-methylase activity by specific MDB proteins versus replacement of 5-methylcytosine nucleotides by components of DNA repair systems.
Genome-wide surveys of DNA-methylation patterns during organogenesis are not available for most organs; however, there are examples involving individual gene promoters in the vertebrate eye. Expression of the Rpb3 gene in maturing photoreceptors correlates to de-methylation of CpG dinucleotides in the gene’s promoter, while the promoter remains hyper-methylated in non-photoreceptor neurons. During lens embryogenesis, the rat gamma-D-crystallin gene and the avian delta-crystallin gene also display promoter DNA-methylation changes consistent with the model in Figure 1C.
Targeted disruptions of the DNMT family members in zebrafish have demonstrated a requirement for all three DNMT enzymes in normal ocular development. DNMT1 knockdown affects the terminal differentiation of several tissues in zebrafish embryos including the neural retina. Similarly DNMT2 and DMNT3 are required for normal neurogenesis and retinal development (Rai et al., 2009).
3. Disease involvement
Mutations to DNMT3b cause the autosomal recessive disorder Immunodeficiency, Centromere instability and Facial anomalies (ICF) syndrome. DNA-methylation is also essential for genomic imprinting, where the paternal or maternal allele is silenced. For example, insulin-like growth factor-2 is expressed from the paternal allele. Approximately 7% of expressed genes show random monoallelic expression. Defects in genomic imprinting are involved in Prader-Willi syndrome, Angleman syndrome, Beckmith-Widermann syndrome and some cancers.
Age and environmental contributions to the etiology of several diseases including cancers, rheumatoid arthritis, fibrosis, heart and neurodegenerative diseases, may be mediated by changes in 5-methylcytosine patterns. Studies of monozygotic (MZ) twins demonstrate that several age-related diseases, including AMD, have significant non-genetic contributions. MZ-twins can present discordant phenotypes, which correlate to differences in DNA-methylation and gene expression patterns. The extent of these epigenetic differences increases with age. Environmental factors, such as diet, also change DNA-methylation patterns during ageing. It is intriguing that age-related changes to DNA-methylation at some gene promoters are similar to those found in the younger diseased tissue.
While genome-wide studies of epigenetic regulation are not available for ocular tissues, there are interesting examples of epigenetic dysregulation of DNA methylation in particular. Methylation-silencing of the p16 tumor suppressor gene (CDKN2A) was reported in some uveal melanomas (Merbs and Sidransky, 1999). Hyper-methylation of the CDKN2A promoter and elevated expression of DNMT3b were also correlated with diminished expression of CDKN2A in pterygium tissue removed from conjuntiva (Chen et al., 2006). This is similar to the hyper-methylation silencing of several tumor suppressor genes reported in some non-ocular cancers.
4. Future studies
Genome-wide investigations of DNA-methylation are required to determine if age-related changes in gene expression correlate to changes in promoter DNA-methylation. Many genes decrease expression in the ageing human retina, including the estrogen receptor (ER). Hyper-methylation of the ER gene promoter correlates to its reduced expression in non-ocular tissues, suggesting a similar epigenetic mechanism may be present in ageing retina. Tissue-specific studies of DNA-methylation during development will be required to reveal which genes are hyper-methylated prior to their expression (like Rbp3) and which are not. Towards this goal, we are currently pursing genome-wide ChIP analysis of 5-methylcytosine around transcription start sites for correlation with our currently available maps of RNA-Polymerase-II binding during photoreceptor maturation (http://www.molvis.org/molvis/v16/a32/). Obtaining this information will reveal DNA-methylation regulated genes that could be more susceptible to environmental effects or pharmacological manipulation of DNA-methylation. Inhibitors of DNMT activity can ameliorate tissue damage in animal models of inflammatory joint disease, and some are under evaluation in cancer clinical trials. Other drugs currently on the market (i.e. valproic acid) are under evaluation for previously unrealized effects on gene expression, through changes to histone-acetylation and DNA-methylation.
Acknowledgments
Support EY014626 (Mitton), OU Center for Biomedical Research Fellowship, OU-Beaumont Multidisciplinary Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chen PL, Cheng YW, Chiang CC, Tseng SH, Chau PS, Tsai YY. Hypermethylation of the p16 gene promoter in pterygia and its association with the expression of DNA methyltransferase 3b. Mol. Vis. 2006;12:1411–1416. [PubMed] [Google Scholar]
- Cvekl A, Mitton KP. Epigenetic regulatory mechanisms in vertebrate eye development and disease. Heredity. 2010;105:135–151. doi: 10.1038/hdy.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merbs SL, Sidransky D. Analysis of p16 (CDKN2/MTS-1/INK4A) alterations in primary sporadic uveal melanoma. Invest. Ophthalmol. Vis. Sci. 1999;40:779–783. [PubMed] [Google Scholar]
- Rai K, Jafri IF, Chidester S, James SR, Karpf AR, Cairns BR, Jones DA. Dnmt3 and G9a cooperate for tissue-specific development in zebrafish. J. Biol. Chem. 2009;285:4110–4121. doi: 10.1074/jbc.M109.073676. [DOI] [PMC free article] [PubMed] [Google Scholar]