Abstract
While directed cellular migration facilitates the coordinated movement of cells during development and tissue repair, the precise mechanisms regulating the interplay between the extracellular environment, the actin cytoskeleton and the overlying plasma membrane remain inadequately understood. The BAR domain family of lipid binding, actin cytoskeletal regulators are gaining greater appreciation for their role in these critical processes. BAR domain proteins are involved as both positive and negative regulators of endocytosis, membrane plasticity and directional cell migration. This review focuses on the functional relationship between different classes of BAR domain proteins and their role in guiding cell migration through regulation of the endocytic machinery. Competition for key signaling substrates by positive and negative BAR domain endocytic regulators appears to mediate control of directional cell migration, and may have wider applicability to other trafficking functions associated with development and carcinogenesis.
Key words: BAR domain, MIM, directed cell migration, competition, membrane dynamics, actin cytoskeleton, endocytosis
Introduction
Directed cellular migration facilitates the coordinated movement of cells throughout development and in wound repair in the adult. This critical developmental process is characterized by guidance cue signal reception, followed by changes in the cytoskeletal structure of the cell, the formation of new adhesion complexes, and removal of adhesions at the trailing end of the cell.1,2 Morphological changes in the cell depend on the cell's response to local migratory cues, which are used to guide the cells in a specific direction. The dynamic regulation of signal reception and in turn, specific changes in the cytoskeleton is the result of the precise coordination of extracellular signal reception and interplay between the plasma membrane and the actin cytoskeleton. Previous studies point to a central role for guidance receptor endocytosis in interpreting local migratory cues and relaying them to the underlying cytoskeleton. In cultured mammalian cells, localized receptor-mediated endocytosis and receptor recycling amplifies the guidance signal to focally activate key regulators of the cytoskeleton such as the GTPase Rac1.3 An area of intense interest is how spatially restricted guidance cues mediate localized receptor endocytosis and signaling to the cytoskeleton. Increasing attention has been directed towards the Bin/Amphiphysin/Rvs (BAR) superfamily of proteins and its role in regulating the precise coordination between endocytosis, vesicle trafficking, signal reception and changes in membrane morphology.4,5 Several excellent recent reviews give comprehensive overviews of the structure and function of the BAR domain superfamily.6,7 The purpose of this review is to examine recent data suggesting how BAR domain proteins function in regulating endocytosis, and in turn guided cellular migration.
The BAR domain superfamily of proteins was initially characterized as a conserved domain in the yeast Rvs161 and Rvs167 proteins and amphiphysin/BIN proteins.8–12 The three subfamilies are referred to as the BAR/N-BAR, the F-BAR (Fes/CIP4 homology) and the I-BAR (inverse or IMD homology).5,13 Structural analysis of the BAR domain has aided in our understanding of the involvement of these proteins in terms of endocytosis, intracellular trafficking and the development of internal and external tubular structures such as filopodia, podosomes and the T-tubules found in striated muscles.14–17 The banana-shaped BAR domain dimer has been shown to bind to membranes with a curvature of 11–15 nm, and preferentially binds to negatively charged lipids such as phosphatidylinositol (4,5) bis-phosphate (PI(4,5) P2).6,7 BAR domain proteins have been shown to bind to lipid membranes, and bend them to the curvature corresponding to the angle of their BAR domain dimer. Amphiphysins have been shown to mediate the formation of clathrin-coated pits during endocytosis, as well as create a lipid tubule between the membrane and the newly formed vesicle.18–22 Atomic model fitting of F-BAR family dimers to membranes reveals that residues along the concave surface of the BAR dimer actually bind to the membrane and force the membrane to conform to the crescent shape of the F-BAR dimer.23
Membrane remodeling into various shapes and structures appears underlies the functions of the wide variety of BAR domain proteins. Structural studies have revealed that BAR domain proteins have different inter-dimer angles that will conform membranes into characteristic shapes. For example, the classic BAR domain family members have convex inter-dimer angles and create inward protrusions of the membrane during endocytosis and the formation of T-tubules.19,24–29 By contrast, the I-BAR family and more recently, members of the F-BAR family have concave inter-dimer angles and have been shown to be involved in the creation of outward membrane projections such as filopodia, lamellipodia and podosomes.5,15,17,30–33 This suggests that the various BAR domain family members have contrasting effects on membrane shape and function that correlates with the shape of the dimer.
A key aspect of BAR domain function is the ability to functionally link plasma membrane events to the cytoskeleton. The actin cytoskeleton provides mechanical force, in addition to the BAR domain, necessary to drive cell migration, changes in the shape of the membrane, cell division and the intracellular trafficking of vesicles and endosomes. A large number of the BAR domain proteins contain a Src homology 3 (SH3) domain, which has been shown to bind to key actin cytoskeletal regulators such as WASP and dynamin (reviewed in ref. 34). These BAR domain proteins bind to and dimerize the WASP/WAVE proteins to activate them and subsequently promote Arp2/3-mediated actin polymerization.4,35 The BAR domain proteins arfaptin and IRSp53 have been shown to interact with the GTPase Rac and are required for the correct activation of the GTPase and the formation of filopodia respectively.12,15,36,37 Similarly, the Toca family has been shown to be involved in the recruitment of WASP and dynamin in the case of endocytosis, and for the activation of the GTPase Cdc42.17,35,38 These finding suggest that many of the BAR domain proteins are key regulators in fundamental regulatory networks that coordinate the interplay between extracellular signaling, BAR-mediated membrane deformation, downstream signaling and cell motility.
BAR Proteins as Positive Regulators of Endocytosis
Directed cell migration uses local endocytosis and recycling of guidance receptors to amplify signaling to the cytoskeleton and provide for directionality. Endocytosis and the intracellular transport of endosomes and vesicles involved three major steps: (1) the internalization of cargo through the bending and deforming of the membrane, (2) pinching off of the newly formed vesicle from the overlying membrane, and (3) subcellular transport of the cargo to the correct intracellular compartment. These three well-defined steps characterize the major points of regulation by which large molecules from the extracellular environment are internalized and correctly directed to specific intracellular compartments. An emerging property within BAR domain proteins is the ability of some classes to promote endocytosis and others to antagonize it. Amphiphysins and endophilins, along with Tuba, have been shown to acts as scaffolds for actin cytoskeletal regulators to come together with endocytic proteins to promote the internalization of a newly formed vesicle.26,39–41
The amphiphysin and endophilin family of N-BAR proteins play a positive role in the endocytosis of activated receptor tyrosine kinases (RTK), as well as in the recycling of synaptic vesicles.19,22,24,42,43 Both of these proteins bind to key regulators of clathrin-mediated endocytosis, and have been shown to be crucial for the proper internalization of activated receptors (Fig. 1A–C). The N-BAR domain of these proteins has also been shown to tubulate membranes in vitro.12,29 The in vitro tubulation results suggest that the N-BAR proteins regulate the shape of the endosomal membrane, and by doing so, regulate the process of endocytosis itself. After the clathrin-coated pit has formed and invaginated, N-BAR proteins are likely to bind to the curved surface of the early endosome. This membrane binding contributes to further curvature of the membrane in order for the membrane to conform to the N-BAR dimer itself. Yeast and mammalian cell culture studies support the idea that dynamin and actin cytoskeletal regulators are recruited to the newly formed tubule in order to cause scission of the membrane and a pinching off of the early endosome from the membrane (Fig. 1D). The coating of the neck of the endosome allows N-BAR proteins to promote the recruitment of the cytoskeletal proteins (Fig. 1E). The timing of scission by dynamin, the creation and elongation of the neck and the movement provided by nucleation of actin filaments are all points at which this highly dynamic process may be regulated to give a specific molecular outcome.
Figure 1.
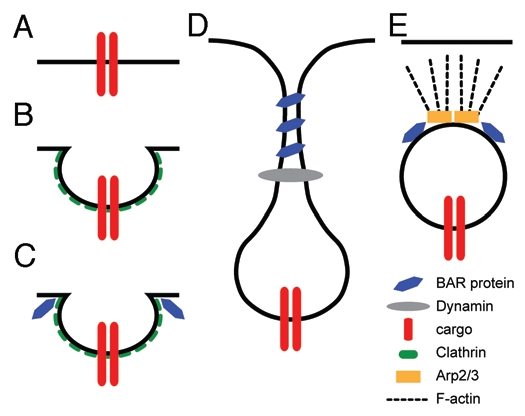
Model for the positive role of BAR domain proteins during endocytosis and vesicle trafficking. (A and B) extracellular cargo or receptors activated by ligand binding are targeted to clathrin-coated pits for internalization. (C) BAR domain proteins are recruited to the clathrin-coated pits to aid in assembly of the coat proteins and the initiation of endocytosis. The BAR proteins induce curvature of the membrane, reshaping the pit into a more spherical form. (D) BAR proteins further reshape the membrane by creating a long tubule between the plasma membrane and the newly formed vesicle. This newly formed tubule is the site for the action of Dynamin to facilitate scission of the vesicle from the overlying membrane. (E) BAR proteins also aid in the activation of the Arp2/3 complex, causing actin polymerization and vesicle trafficking further into the cytoplasm. Figure adapted from ref. 4.
Besides N-BAR proteins, some F-BAR family members also positively regulate endocytosis. Loss-of-function experiments in cell culture confirm the role for F-BAR proteins FBP–17, CIP4 or Toca-1 in promoting endocytosis of EGF, PDGF and transferrin.17,35,38,44–46 These proteins all regulate the activity of N-WASP, and are able to bind to dynamin, N-WASP and Cdc42 through their respective SH3 domains. It is thought that these proteins form coats, along with dynamin, that create a specific area for tubulation of the membrane to be linked with the scission machinery and actin polymerization. Another well-studied group of F-BAR proteins are the syndapins. These proteins contain SH3 domains, and regulate a number of endocytic events in various tissues.47–49 It has also been observed that syndapins can oligomerize through their N-terminal coiled-coil domain, and that this can create a concentrated area containing SH3 domains that would recruit dynamin and WASP. This in turn creates an area where the components necessary for endocytosis are all ready present or primed for endocytosis to occur.
Even within well-known families of vesicular trafficking proteins, BAR domains appear and aid in positively regulating endocytosis. The sorting nexins (SNX) are a group of proteins responsible for the correct regulation of intracellular traffic. To this date, there are 29 SNX proteins, 8 of which contain a BAR domain.50–53 These proteins are required to properly sort the early and late endosomes to several key subcellular compartments. SNX9 has been shown to localize to clathrin coated pits, bind to dynamin, WASP and synaptojanin, all proteins required for the correct internalization of endosomes.51,54–56
BAR Proteins as Negative
Regulators of Endocytosis Although many of the BAR domain proteins have been shown to act as positive regulators of endocytosis and vesicle trafficking, a growing number of BAR domain proteins act to inhibit these processes. One such example of a BAR domain protein that acts as a negative regulator is the F-BAR protein PICK1. PICK1 is a PDZ-containing protein that binds to a variety of membrane proteins, including the AMPA receptor subunits GluR2/3, an interaction that is required for the proper internalization of the AMPA receptor.57 PICK1 acts to inhibit Arp2/3-mediated actin assembly, which has been shown to be required for the proper organization of the actin cytoskeleton in order to establish appropriate morphology in neurons. PICK1 in the absence of the GluR2/3 cargo, maintains low level inhibition of actin polymerization, which prevents the formation of membrane protrusions during F-actin assembly. Upon NMDA-ligand binding to its receptor, PICK1-dependent inhibition is increased, resulting in a loss of the F-actin network in the area of the newly formed endosome and allowing the overlying membrane to invaginate.57
The PACSIN proteins, well-known cytoskeleton and membrane regulators, have been recently identified as containing an F-BAR domain.58 PACSINs have F-BAR domains as well as SH3 domains, and have been shown to bind to important endocytic regulators such as huntingtin, synapsin 1 and synaptojanin 1.59–62 PACSIN acts as an adaptor protein to bring together key regulators of the actin cytoskeleton to the endocytic machinery. PACSIN also binds to dynamin, and overexpression of PACSIN has been shown to inhibit endocytosis of the transferrin receptor by interfering with the normal function of dynamin.63
Another recent example is the BAR-dependent inhibition of clathrin-mediated endocytosis of the EGFR and Transferrin receptors by the I-BAR protein MIM. The I-BAR proteins MIM and IRSp53 have been shown to be involved in the formation of filopodia.15,33,36,64,65 Since these proteins have a BAR domain dimer conformation that is inverse to the other BAR domain family members, it is possible that they might act to inhibit or reduce endocytosis, cell migration and other processes.7,13,33,66 The negative regulation of endocytosis and the production of outward protrusions creates a situation in which BAR domain family members can act in opposition to one another with regards to the regulation of endocytosis, the actin cytoskeleton and the dynamics of the plasma membrane. Loss-of-function studies of MIM in both vertebrate cell culture and in Drosophila in vivo have shown that the loss of MIM protein leads to an increase in the internalization and receptor recycling of both the EGF and Transferrin receptors.67 Loss of the DMIM protein in Drosophila border cell cluster leads to an increase in the mobile fraction of the underlying actin cytoskeleton, suggesting that under wild type conditions, DMIM acts to stabilize the cortical actin cytoskeleton, preventing drastic changes to cell morphology (Fig. 2A and B).
Figure 2.
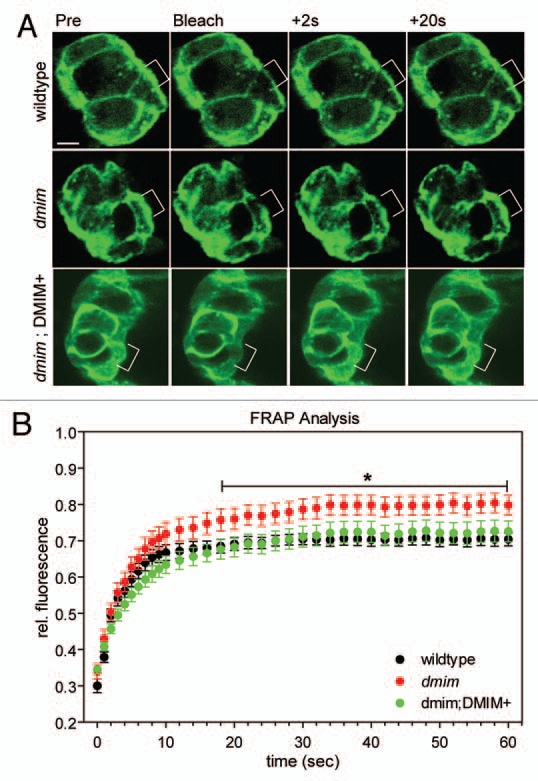
FRAP at the leading edge of migrating border cells. dmim is an I-BAR protein in Drosophila shown to inhibit endocytosis and regulate the migration of border cells and primordial germ cells.67 (A) Photobleaching of an early stage 9 Drosophila border cell clusters expressing UAS-moesin::GFP under 306-Gal4. A 5 µm diameter area was ablated at the leading edge (shown in white brackets) and images were captured at 1 second intervals after photobleaching. Scale bar = 5 µm. (B) Quantitation of FRAP time series shows that while there is only a slight difference in the rate of recovery, there is a significant difference in the mobile fraction of the mutant border cells when compared to wildtype and rescue. Data are represented as the mean ± SEM from 5 separate time series for each genotype. (*p < 0.01, t-test).
BAR Domain Competition
With the emerging theme of both positive and negative endocytosis regulators within the BAR family, how different BAR domain classes interact is needed to fully appreciate the functions these proteins. The structures of the individual BAR domains suggest that while the N- and F-BAR domain proteins create inward tubules and invaginations in the membrane, the I-BAR members create outward projections. These opposing structures and subsequent membrane formations lead to the intriguing possibility that proteins within the BAR domain superfamily regulate the precise timing and coordination of protein/membrane complexes through antagonism with one another. Since the BAR proteins appear to bind to similar cytoskeletal substrates in many cases, competition between BAR domain proteins over the same critical substrate could lead to the positive or negative regulation of a cellular process based on the status of the binding partner(s) to which the particular substrate binds.
A recent example of competition between two BAR domain proteins is seen in the interaction between the N-BAR protein endophilin and the I-BAR domain protein MIM for cortactin as a substrate.67–69 MIM binding to cortactin prevents binding to endophilin, which acts as a positive regulator and initiator of the internalization of activated EGFR. Competition with endophilin appears to underlie MIM function as a negative regulator of endocytosis, and allows MIM to attenuate the downstream activation of EGFR substrates. This competition has been implicated in the regulation of directional cell migration both in vertebrate cell culture and in vivo in the migration of the Drosophila border cell cluster.67
The competition between BAR domain proteins or between BAR domains and other lipid binding proteins may not only affect the morphology of the plasma membrane, but may affect the kinetics of downstream signaling events dependent on intracellular trafficking and correct subcellular localization. Several studies have shown that proper endocytic recycling and trafficking is crucial for the activation and activity of the small GTPase Rac, and that this process is required during cell migration for the spatial restriction of signaling at the leading edge of the migrating cell.3,4,70–72 The idea of competition between two BAR domain proteins creates a situation in which differential binding of a shared substrate could directly result in the correct spatial restriction of signaling events, changes in membrane morphology and cell motility in the correct direction. The migration of the Drosophila border cell cluster is a good example of a biological system dependent on the precise regulation of receptor activation and signaling through the endocytic machinery in order to mediate proper directed cell migration.73,74
From these studies, we propose two possible models by which competition between BAR domain proteins may regulate several key biological processes to affect directional cell migration. The first model is by which I-BAR proteins create membrane domains that dictate spatial limitations on receptor activation and internalization (Fig. 3A and B). BAR proteins may create specific areas along the plasma membrane that either allow for or inhibit the local activation of RTK signaling, and thus the downstream signaling of those pathways. It is the external stimuli that offsets the symmetry present between the sets of BAR domain proteins, and allows for polarization of the membrane, and the eventual formation of the leading and lagging edges of the cell. The creation of such a polarized cell membrane is crucial for the cells to properly migrate in a specific direction. Another mechanism by which BAR domain competition may affect directional migration is through the regulation of vesicle trafficking and the subcellular relocalization of various factors (Fig. 3C). This model, used by sorting nexins to regulate intracellular trafficking,12,52,53,75 postulates that the binding of different BAR domain proteins to the vesicle aid in the correct trafficking of that vesicle to various compartments within the cell. Two or more BAR proteins competing to bind to new vesicles could create a situation in which that vesicle can be directed to the lysosome for degradation and inactivation or could be redirected back to the cell surface during receptor recycling for increased signaling. Both models suggest a mechanism by which the competition between BAR domain proteins for binding to vesicles or substrates, allows for the precise regulation of directional cell migration.
Figure 3.
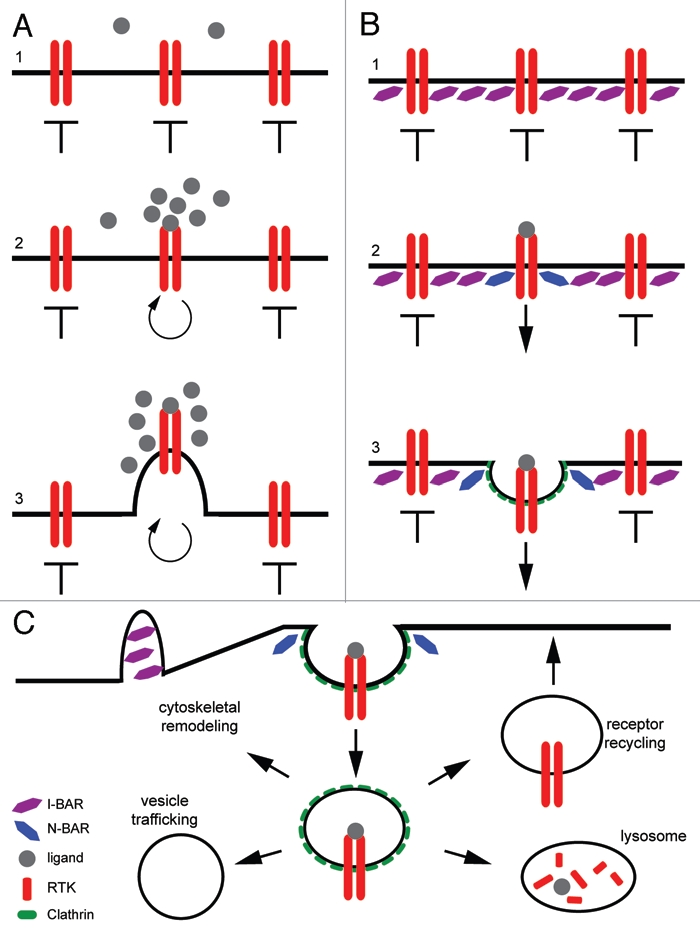
Proposed models of the mechanism by which BAR domain competition creates an edge detector for localized signaling events. (A) In this case, BAR domain proteins act to inhibit endocytosis and signaling across the plasma membrane. Upon ligand binding, activation of the receptor leads to the inactivation of the negative BAR protein and the relocalization or activation of the positive BAR proteins. This allows for the spatial localization of receptor activation and internalization at the point of highest ligand concentration, without inappropriate signaling events occurring laterally across the entirety of the membrane. This also leads to the creation of outward cellular projections and cytoskeletal remodeling in the direction of the guidance cue, leading to correct directional cell migration. (B) In a wildtype state, BAR protein a (purple) acts to inhibit endocytosis across the plasma membrane. Upon ligand binding and receptor activation, the inhibition provided by BAR protein A is overridden and internalization of the cargo proceeds. Internalization of the cargo is aided by BAR protein B (blue). An edge is setup between the two different BAR proteins such that internalization of the cargo only occurs where BAR protein B is active and BAR protein A has been inactivated. (C) After internalization of the cargo or activated receptor, BAR proteins act to specify the subcellular localization of the newly formed vesicle. The specific subcellular relocalization of that vesicle is dependent on the BAR protein(s) that bind to the vesicle membrane.
Future Perspectives
Competition between pairs of BAR domain proteins competing for common substrates may underlie other key cellular processes. An assumption of the model is that BAR proteins bound to substrate can exerts either a positive or negative regulation of that process. This competition also sets up the possibility that functions of either the positive or negative regulator can be removed through an external stimulus, such as a guidance cue, to promote endocytosis. Moreover, BAR domain competition also suggest that possibility of multiple redundant pairs of regulators controlled by different stimuli. Removal or overexpression of either the positive or negative regulator would imbalance endocytosis, resulting in abnormal directional migration. However, removal of both regulators leads to compensation by another pair of proteins and restoration of that balance and a wildtype phenotype, such as that seen where endophilin and MIM compete for cortactin as a substrate in cell migration. When both MIM and cortactin are removed, wild type migration occurs. Future experiments testing the functions and competition amongst other BAR domain proteins will determine the generality of the competition model.
Acknowledgements
Work in the author's laboratory was supported by a NSF graduate research fellowship to G.A.Q. and a NIH/NIAMS grant to A.E.O. (R01 AR052785).
Abbreviations
- BAR
bin/amphiphysin/rvs domain
- I-BAR
inverse BAR domain
- F-BAR
fes/CIP4 homology BAR domain
- MIM
missing-in-metastasis
- WASP
wiskott-aldrich syndrome protein
- WAVE
wiskott-aldrich verprolin protein
- SH3
src homology 3 domain
- SNX
sorting nexin
- RTK
receptor tyrosine kinase
- IRSp53
insulin receptor substrate of 53 KDa
- srGAP2
slit-robo GTPase activating protein
- EGFR
epidermal growth factor receptor
- CD2AP
CD2 associated proteins
- EHD2
eps14 homology domain
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/12123
References
- 1.Blaser H, Eisenbeiss S, Neumann M, Reichman-Fried M, Thisse B, Thisse C, et al. Transition from non-motile behaviour to directed migration during early PGC development in zebrafish. J Cell Sci. 2005;118:4027–4038. doi: 10.1242/jcs.02522. [DOI] [PubMed] [Google Scholar]
- 2.Friedl P, Hegerfeldt Y, Tusch M. Collective cell migration in morphogenesis and cancer. The International journal of developmental biology. 2004;48:441–449. doi: 10.1387/ijdb.041821pf. [DOI] [PubMed] [Google Scholar]
- 3.Palamidessi A, Frittoli E, Garre M, Faretta M, Mione M, Testa I, et al. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell. 2008;134:135–147. doi: 10.1016/j.cell.2008.05.034. [DOI] [PubMed] [Google Scholar]
- 4.Dawson JC, Legg JA, Machesky LM. Bar domain proteins: a role in tubulation, scission and actin assembly in clathrin-mediated endocytosis. Trends Cell Biol. 2006;16:493–498. doi: 10.1016/j.tcb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Frost A, De Camilli P, Unger VM. F-BAR proteins join the BAR family fold. Structure. 2007;15:751–753. doi: 10.1016/j.str.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Saarikangas J, Zhao H, Lappalainen P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol Rev. 2010;90:259–289. doi: 10.1152/physrev.00036.2009. [DOI] [PubMed] [Google Scholar]
- 7.Saarikangas J, Zhao H, Pykalainen A, Laurinmaki P, Mattila PK, Kinnunen PK, et al. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr Biol. 2009;19:95–107. doi: 10.1016/j.cub.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 8.Balguerie A, Sivadon P, Bonneu M, Aigle M. Rvs167p, the budding yeast homolog of amphiphysin, colocalizes with actin patches. J Cell Sci. 1999;112:2529–2537. doi: 10.1242/jcs.112.15.2529. [DOI] [PubMed] [Google Scholar]
- 9.Colwill K, Field D, Moore L, Friesen J, Andrews B. In vivo analysis of the domains of yeast Rvs167p suggests Rvs167p function is mediated through multiple protein interactions. Genetics. 1999;152:881–893. doi: 10.1093/genetics/152.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott K, Sakamuro D, Basu A, Du W, Wunner W, Staller P, et al. Bin1 functionally interacts with Myc and inhibits cell proliferation via multiple mechanisms. Oncogene. 1999;18:3564–3573. doi: 10.1038/sj.onc.1202670. [DOI] [PubMed] [Google Scholar]
- 11.Ge K, Prendergast GC. Bin2, a functionally nonredundant member of the BAR adaptor gene family. Genomics. 2000;67:210–220. doi: 10.1006/geno.2000.6216. [DOI] [PubMed] [Google Scholar]
- 12.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, et al. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 13.Suetsugu S, Toyooka K, Senju Y. Subcellular membrane curvature mediated by the BAR domain super-family proteins. Semin Cell Dev Biol. 2009 doi: 10.1016/j.semcdb.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Razzaq A, Robinson IM, McMahon HT, Skepper JN, Su Y, Zelhof AC, et al. Amphiphysin is necessary for organization of the excitation-contraction coupling machinery of muscles, but not for synaptic vesicle endocytosis in Drosophila. Genes Dev. 2001;15:2967–2979. doi: 10.1101/gad.207801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmed S, Goh WI, Bu W. I-BAR domains, IRSp53 and filopodium formation. Semin Cell Dev Biol. 2009 doi: 10.1016/j.semcdb.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 16.Bharti S, Inoue H, Bharti K, Hirsch DS, Nie Z, Yoon HY, et al. Src-dependent phosphorylation of ASAP1 regulates podosomes. Mol Cell Biol. 2007;27:8271–8283. doi: 10.1128/MCB.01781-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bu W, Chou AM, Lim KB, Sudhaharan T, Ahmed S. The Toca-1-N-WASP complex links filopodial formation to endocytosis. J Biol Chem. 2009;284:11622–11636. doi: 10.1074/jbc.M805940200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takei K, Slepnev VI, Haucke V, De Camilli P. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat Cell Biol. 1999;1:33–39. doi: 10.1038/9004. [DOI] [PubMed] [Google Scholar]
- 19.Arkhipov A, Yin Y, Schulten K. Membrane-bending mechanism of amphiphysin N-BAR domains. Biophys J. 2009;97:2727–2735. doi: 10.1016/j.bpj.2009.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallop JL, McMahon HT. BAR domains and membrane curvature: bringing your curves to the BAR. Biochem Soc Symp. 2005:223–231. doi: 10.1042/bss0720223. [DOI] [PubMed] [Google Scholar]
- 21.Meunier B, Quaranta M, Daviet L, Hatzoglou A, Leprince C. The membrane-tubulating potential of amphiphysin 2/BIN1 is dependent on the microtubule-binding cytoplasmic linker protein 170 (CLIP-170) Eur J Cell Biol. 2009;88:91–102. doi: 10.1016/j.ejcb.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 22.Pant S, Sharma M, Patel K, Caplan S, Carr CM, Grant BD. AMPH-1/Amphiphysin/Bin1 functions with RME-1/Ehd1 in endocytic recycling. Nat Cell Biol. 2009;11:1399–1410. doi: 10.1038/ncb1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimada A, Niwa H, Tsujita K, Suetsugu S, Nitta K, Hanawa-Suetsugu K, et al. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 24.Blood PD, Swenson RD, Voth GA. Factors influencing local membrane curvature induction by N-BAR domains as revealed by molecular dynamics simulations. Biophys J. 2008;95:1866–1876. doi: 10.1529/biophysj.107.121160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blood PD, Voth GA. Direct observation of Bin/amphiphysin/Rvs (BAR) domain-induced membrane curvature by means of molecular dynamics simulations. Proc Natl Acad Sci USA. 2006;103:15068–15072. doi: 10.1073/pnas.0603917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui H, Ayton GS, Voth GA. Membrane binding by the endophilin N-BAR domain. Biophys J. 2009;97:2746–2753. doi: 10.1016/j.bpj.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferguson S, Raimondi A, Paradise S, Shen H, Mesaki K, Ferguson A, et al. Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev Cell. 2009;17:811–822. doi: 10.1016/j.devcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frost A, Unger VM, De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallop JL, Jao CC, Kent HM, Butler PJ, Evans PR, Langen R, et al. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006;25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aspenstrom P. Roles of F-BAR/PCH proteins in the regulation of membrane dynamics and actin reorganization. Int Rev Cell Mol Biol. 2009;272:1–31. doi: 10.1016/S1937-6448(08)01601-8. [DOI] [PubMed] [Google Scholar]
- 31.Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, et al. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–1004. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Machesky LM, Johnston SA. MIM: a multifunctional scaffold protein. J Mol Med. 2007;85:569–576. doi: 10.1007/s00109-007-0207-0. [DOI] [PubMed] [Google Scholar]
- 33.Mattila PK, Pykalainen A, Saarikangas J, Paavilainen VO, Vihinen H, Jokitalo E, et al. Missing-inmetastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J Cell Biol. 2007;176:953–964. doi: 10.1083/jcb.200609176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masuda M, Mochizuki N. Structural characteristics of BAR domain superfamily to sculpt the membrane. Semin Cell Dev Biol. doi: 10.1016/j.semcdb.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 35.Fricke R, Gohl C, Dharmalingam E, Grevelhorster A, Zahedi B, Harden N, et al. Drosophila Cip4/Toca-1 integrates membrane trafficking and actin dynamics through WASP and SCAR/WAVE. Curr Biol. 2009;19:1429–1437. doi: 10.1016/j.cub.2009.07.058. [DOI] [PubMed] [Google Scholar]
- 36.Lim KB, Bu W, Goh WI, Koh E, Ong SH, Pawson T, et al. The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J Biol Chem. 2008;283:20454–20472. doi: 10.1074/jbc.M710185200. [DOI] [PubMed] [Google Scholar]
- 37.Suetsugu S, Murayama K, Sakamoto A, Hanawa-Suetsugu K, Seto A, Oikawa T, et al. The RAC binding domain/IRSp53-MIM homology domain of IRSp53 induces RAC-dependent membrane deformation. J Biol Chem. 2006;281:35347–35358. doi: 10.1074/jbc.M606814200. [DOI] [PubMed] [Google Scholar]
- 38.Kakimoto T, Katoh H, Negishi M. Regulation of neuronal morphology by Toca-1, an F-BAR/EFC protein that induces plasma membrane invagination. J Biol Chem. 2006;281:29042–29053. doi: 10.1074/jbc.M604025200. [DOI] [PubMed] [Google Scholar]
- 39.Cestra G, Kwiatkowski A, Salazar M, Gertler F, De Camilli P. Tuba, a GEF for CDC42, links dynamin to actin regulatory proteins. Methods Enzymol. 2005;404:537–545. doi: 10.1016/S0076-6879(05)04047-4. [DOI] [PubMed] [Google Scholar]
- 40.Salazar MA, Kwiatkowski AV, Pellegrini L, Cestra G, Butler MH, Rossman KL, et al. Tuba, a novel protein containing bin/amphiphysin/Rvs and Dbl homology domains, links dynamin to regulation of the actin cytoskeleton. J Biol Chem. 2003;278:49031–49043. doi: 10.1074/jbc.M308104200. [DOI] [PubMed] [Google Scholar]
- 41.Masuda M, Takeda S, Sone M, Ohki T, Mori H, Kamioka Y, et al. Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms. EMBO J. 2006;25:2889–2897. doi: 10.1038/sj.emboj.7601176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhatia VK, Madsen KL, Bolinger PY, Kunding A, Hedegard P, Gether U, et al. Amphipathic motifs in BAR domains are essential for membrane curvature sensing. EMBO J. 2009;28:3303–3314. doi: 10.1038/emboj.2009.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin Y, Arkhipov A, Schulten K. Simulations of membrane tubulation by lattices of amphiphysin N-BAR domains. Structure. 2009;17:882–892. doi: 10.1016/j.str.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 45.Takano K, Toyooka K, Suetsugu S. EFC/F-BAR proteins and the N-WASP-WIP complex induce membrane curvature-dependent actin polymerization. EMBO J. 2008;27:2817–2828. doi: 10.1038/emboj.2008.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toguchi M, Richnau N, Ruusala A, Aspenstrom P. Members of the CIP4 family of proteins participate in the regulation of platelet-derived growth factor receptor-beta-dependent actin reorganization and migration. Biol Cell. 2009 doi: 10.1042/BC20090033. [DOI] [PubMed] [Google Scholar]
- 47.Dharmalingam E, Haeckel A, Pinyol R, Schwintzer L, Koch D, Kessels MM, et al. F-BAR proteins of the syndapin family shape the plasma membrane and are crucial for neuromorphogenesis. J Neurosci. 2009;29:13315–11327. doi: 10.1523/JNEUROSCI.3973-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halbach A, Morgelin M, Baumgarten M, Milbrandt M, Paulsson M, Plomann M. PACSIN 1 forms tetramers via its N-terminal F-BAR domain. FEBS J. 2007;274:773–782. doi: 10.1111/j.1742-4658.2006.05622.x. [DOI] [PubMed] [Google Scholar]
- 49.Wang Q, Navarro MV, Peng G, Molinelli E, Lin Goh S, Judson BL, et al. Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin. Proc Natl Acad Sci USA. 2009;106:12700–12705. doi: 10.1073/pnas.0902974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carlton J, Bujny M, Peter BJ, Oorschot VM, Rutherford A, Mellor H, et al. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high-curvature membranes and 3-phosphoinositides. Curr Biol. 2004;14:1791–1800. doi: 10.1016/j.cub.2004.09.077. [DOI] [PubMed] [Google Scholar]
- 51.Shin N, Ahn N, Chang-Ileto B, Park J, Takei K, Ahn SG, et al. SNX9 regulates tubular invagination of the plasma membrane through interaction with actin cytoskeleton and dynamin 2. J Cell Sci. 2008;121:1252–1263. doi: 10.1242/jcs.016709. [DOI] [PubMed] [Google Scholar]
- 52.van Weering JR, Verkade P, Cullen PJ. SNX-BAR proteins in phosphoinositide-mediated, tubular-based endosomal sorting. Semin Cell Dev Biol. 2009 doi: 10.1016/j.semcdb.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yarar D, Surka MC, Leonard MC, Schmid SL. SNX9 activities are regulated by multiple phosphoinositides through both PX and BAR domains. Traffic. 2008;9:133–146. doi: 10.1111/j.1600-0854.2007.00675.x. [DOI] [PubMed] [Google Scholar]
- 54.Haberg K, Lundmark R, Carlsson SR. SNX18 is an SNX9 paralog that acts as a membrane tubulator in AP-1-positive endosomal trafficking. J Cell Sci. 2008;121:1495–1505. doi: 10.1242/jcs.028530. [DOI] [PubMed] [Google Scholar]
- 55.Pylypenko O, Ignatev A, Lundmark R, Rasmuson E, Carlsson SR, Rak A. A combinatorial approach to crystallization of PX-BAR unit of the human Sorting Nexin 9. J Struct Biol. 2008;162:356–360. doi: 10.1016/j.jsb.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 56.Yarar D, Waterman-Storer CM, Schmid SL. SNX9 couples actin assembly to phosphoinositide signals and is required for membrane remodeling during endocytosis. Dev Cell. 2007;13:43–56. doi: 10.1016/j.devcel.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 57.Rocca DL, Martin S, Jenkins EL, Hanley JG. Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nat Cell Biol. 2008;10:259–271. doi: 10.1038/ncb1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kessels MM, Qualmann B. The syndapin protein family: linking membrane trafficking with the cytoskeleton. J Cell Sci. 2004;117:3077–3086. doi: 10.1242/jcs.01290. [DOI] [PubMed] [Google Scholar]
- 59.Modregger J, DiProspero NA, Charles V, Tagle DA, Plomann M. PACSIN 1 interacts with huntingtin and is absent from synaptic varicosities in presymptomatic Huntington's disease brains. Hum Mol Genet. 2002;11:2547–2558. doi: 10.1093/hmg/11.21.2547. [DOI] [PubMed] [Google Scholar]
- 60.Qualmann B, Roos J, DiGregorio PJ, Kelly RB, Syndapin I. a synaptic dynamin-binding protein that associates with the neural Wiskott-Aldrich syndrome protein. Mol Biol Cell. 1999;10:501–513. doi: 10.1091/mbc.10.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qualmann B, Kessels MM, Kelly RB. Molecular links between endocytosis and the actin cytoskeleton. J Cell Biol. 2000;150:111–116. doi: 10.1083/jcb.150.5.f111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qualmann B, Kelly RB. Syndapin isoforms participate in receptor-mediated endocytosis and actin organization. J Cell Biol. 2000;148:1047–1062. doi: 10.1083/jcb.148.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Modregger J, Ritter B, Witter B, Paulsson M, Plomann M. All three PACSIN isoforms bind to endocytic proteins and inhibit endocytosis. J Cell Sci. 2000;113:4511–4521. doi: 10.1242/jcs.113.24.4511. [DOI] [PubMed] [Google Scholar]
- 64.Bompard G, Sharp SJ, Freiss G, Machesky LM. Involvement of Rac in actin cytoskeleton rearrangements induced by MIM-B. J Cell Sci. 2005;118:5393–5403. doi: 10.1242/jcs.02640. [DOI] [PubMed] [Google Scholar]
- 65.Sawallisch C, Berhorster K, Disanza A, Mantoani S, Kintscher M, Stoenica L, et al. The insulin receptor substrate of 53 kDa (IRSp53) limits hippocampal synaptic plasticity. J Biol Chem. 2009;284:9225–9236. doi: 10.1074/jbc.M808425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang C, Hoelzle M, Disanza A, Scita G, Svitkina T. Coordination of membrane and actin cytoskeleton dynamics during filopodia protrusion. PLoS One. 2009;4:5678–5678. doi: 10.1371/journal.pone.0005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Quinones G, Jin J, Oro A. I-BAR Protein Antagonism of Endocytosis Mediates Directional Sensing During Guided Cell Migration. J Cell Biol. 2010:189. doi: 10.1083/jcb.200910136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dikic I. CIN85/CMS family of adaptor molecules. FEBS Lett. 2002;529:110–115. doi: 10.1016/s0014-5793(02)03188-5. [DOI] [PubMed] [Google Scholar]
- 69.Lynch DK, Winata SC, Lyons RJ, Hughes WE, Lehrbach GM, Wasinger V, et al. A Cortactin-CD2-associated protein (CD2AP) complex provides a novel link between epidermal growth factor receptor endocytosis and the actin cytoskeleton. J Biol Chem. 2003;278:21805–21813. doi: 10.1074/jbc.M211407200. [DOI] [PubMed] [Google Scholar]
- 70.Cotton M, Claing A. G protein-coupled receptors stimulation and the control of cell migration. Cell Signal. 2009;21:1045–1053. doi: 10.1016/j.cellsig.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 71.Falguieres T, Luyet PP, Gruenberg J. Molecular assemblies and membrane domains in multivesicular endosome dynamics. Exp Cell Res. 2009;315:1567–1573. doi: 10.1016/j.yexcr.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 72.Jones MC, Caswell PT, Norman JC. Endocytic recycling pathways: emerging regulators of cell migration. Curr Opin Cell Biol. 2006;18:549–557. doi: 10.1016/j.ceb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 73.Bianco A, Poukkula M, Cliffe A, Mathieu J, Luque CM, Fulga TA, et al. Two distinct modes of guidance signalling during collective migration of border cells. Nature. 2007;448:362–365. doi: 10.1038/nature05965. [DOI] [PubMed] [Google Scholar]
- 74.Jekely G, Sung HH, Luque CM, Rorth P. Regulators of endocytosis maintain localized receptor tyrosine kinase signaling in guided migration. Dev Cell. 2005;9:197–207. doi: 10.1016/j.devcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 75.Liu H, Liu ZQ, Chen CX, Magill S, Jiang Y, Liu YJ. Inhibitory regulation of EGF receptor degradation by sorting nexin 5. Biochem Biophys Res Commun. 2006;342:537–546. doi: 10.1016/j.bbrc.2006.01.179. [DOI] [PubMed] [Google Scholar]


