Abstract
Bacillus thuringiensis subs israelensis (Bti) produces at least four different crystal proteins that are specifically toxic to different mosquito species and that belong to two non-related family of toxins, Cry and Cyt named Cry4Aa, Cry4Ba, Cry11Aa and Cyt1Aa. Cyt1Aa enhances the activity of Cry4Aa, Cry4Ba or Cry11Aa and overcomes resistance of Culex quinquefasciatus populations resistant to Cry11Aa, Cry4Aa or Cry4Ba. Cyt1Aa synergized Cry11Aa by their specific interaction since single point mutants on both Cyt1Aa and Cry11Aa that affected their binding interaction affected their synergistic insecticidal activity. In this work we show that Cyt1Aa loop β6-αE K198A, E204A and β7 K225A mutants affected binding and synergism with Cry4Ba. In addition, site directed mutagenesis showed that Cry4Ba domain II loop α-8 is involved in binding and in synergism with Cyt1Aa since Cry4Ba SI303-304AA double mutant showed decreased binding and synergism with Cyt1Aa. These data suggest that similarly to the synergism between Cry11Aa and Cyt1Aa toxins, the Cyt1Aa also functions as a receptor for Cry4Ba explaining the mechanism of synergism between these two Bti toxins.
1. Introduction
Bacillus thuringiensis subs israelensis (Bti) is a gram-positive bacterium that produces crystal toxins during its sporulation phase that are specifically toxic to different mosquito species as Anopheles sp, Aedes aegypti and Culex sp that are vectors of important human diseases as malaria, yellow and dengue fevers among others. Bti is used world wide in the control of these mosquito species, particularly in cases where resistance to chemical insecticides has developed. Despite the large use of Bti in mosquito control there are no reports of resistance to Bti in field conditions [3, 19].
The lack of insect resistance development to Bti is mainly due to the fact that Bti produces at least four different crystal proteins that belong to two non-related family of toxins, Cry and Cyt named Cry4Aa, Cry4Ba, Cry11Aa and Cyt1Aa. Interestingly the insecticidal activity of the isolated Bti toxins is magnitudes of order less toxic than the crystal inclusion containing all toxins [10, 17, 26]. This synergistic effect is mainly due to Cyt1Aa that enhances the activity of Cry4Aa, Cry4Ba or Cry11Aa [10, 17, 25]. Even more, it was shown that Cyt1Aa overcomes resistance of C. quinquefasciatus populations resistant to Cry11Aa, Cry4Aa or Cry4Ba [25].
Cry and Cyt toxins have a different three dimensional structures and mechanisms of action. Cry4Aa, Cry4Ba and Cry11Aa belong to the three-domain (3-D) family of Cry toxins that are composed of three discrete structural domains. Domain I is a seven α-helix bundle that is involved in membrane insertion, oligomerization and pore-formation while domain II and domain III are β-sheet domains involved in receptor interaction [4, 5]. In particular, the exposed loop regions in domain II are involved in receptor binding [4, 5]. In contrast, Cyt proteins have a single α–βdomain comprising of two outer layers of α-helix hairpins wrapped around a β-sheet [18]. Both proteins are synthesized as protoxins that are solubilized in the gut of susceptible dipteran insects and proteolytically activated by midgut proteases leading to activated toxins. For exerting their toxic effect, 3-D Cry toxins rely on the sequential binding with at least two different midgut protein receptor molecules that in lepidopteran insects have been identified as a transmembrane cadherin and glicosyl-phosphatidil-inositol (GPI)-anchored proteins as aminopeptidase-N (APN) and alkaline phosphatase (ALP) [5, 22]. Binding to cadherin facilitates further proteolytic cleavage of helix α-1 and formation of an oligomer structure that gains affinity to GPI-anchored receptors leading to membrane insertion and pore formation [5, 22]. Recently, cadherin, APN and ALP molecules have been identified as Cry11Aa or Cry4Ba binding molecules in different mosquito species suggesting a conserved mode of action of Cry toxins in dipteran insects [2, 8, 9, 12, 15, 27]. In contrast, activated Cyt1Aa do not rely on midgut proteins to bind to the midgut epithelium and directly interacts with membrane lipids resulting in membrane insertion and pore-formation [7].
Understanding the molecular mechanism of synergism of Cyt1Aa and Cry4 and Cry11Aa toxins could have important implications to develop strategies for resistance management of Cry toxins in agriculture and in mosquito control [6].
The mechanism of synergism of Cyt1Aa and Cry11Aa was proposed to depend on the specific interaction between these toxins since single point mutations on Cyt1Aa affected its binding interaction with Cry11Aa and their synergism [20]. Also, it was shown that loop regions of domain II from Cry11Aa toxin that are involved in receptor binding were also involved in binding and synergism with Cyt1Aa [11, 20]. These data led to the hypothesis that Cyt1Aa functions as a membrane bound binding protein of Cry11Aa [20]. Furthermore, recent data showed that binding of Cry11Aa to Cyt1Aa facilitates formation of Cry11Aa oligomers that were efficient in pore-formation indicating that Cyt1Aa at least fulfils the role of the primary cadherin receptor that facilitates oligomer formation [21]. The mechanism of synergism of Cyt1Aa with Cry4Ba and Cry4Aa has not been analyzed. Although Cry4Ba and Cry11Aa share low amino acid sequence identity both bind to A. aegypti ALP receptor indicating that they share binding sites with at least this receptor [13]. In order to analyze if binding of Cry4Ba to Cyt1Aa was involved in their synergism, Cyt1Aa mutants that were previously shown to affect binding to Cry11Aa and in synergism were analyzed regarding to the binding of Cry4Ba and their effect on synergism against A. aegypti larae was also analyzed. Even more, Cry4Ba domain II loop regions were subject to mutagenesis and Cry4Ba mutants analyzed regarding their binding and synergism with Cyt1Aa.
Here we show that some Cyt1Aa mutants affected in binding and synergism with Cry11Aa are also affected in binding and synergism with Cry4Ba. Also, that Cry4Ba domain II loop α-8 is involved in its binding interaction and in synergism with Cyt1Aa. These data suggest that Cyt1Aa also functions as a specific binding protein for Cry4Ba explaining the mechanism of synergism between these two Bti toxins.
2. Methods
2.1 Growth of Bacillus thuringiensis, purification of Cry4Ba and Cyt1Aa crystal inclusions and toxin activation
For the production of Cry4Ba crystals and Cyt1A, acrystalliferous Bti strain Q2-81 containing plasmid pHT618 or pWF45 respectively were cultured for 3 days at 29°C and 200 rpm in nutrient broth sporulation medium supplemented with 10μg erythromycin per ml. Spores and inclusions produced by the Bt strains were harvested and washed three times with 0.3 M NaCl, 0.01 M EDTA, pH 8.0. For Cry4Ba activation, the spores and crystals were solubilized in carbonate buffer (sodium bicarbonate/carbonate 0.1M, pH 9.5) supplemented with 0.2% β-mercaptoethanol [pH 10.5], and activated with 1:20 w/w trypsin (Sigma-Aldrich Co.), for 16 h at 37°C. Cyt1Aa inclusions were solubilized in 50 mM Na2CO3, 10 mM DTT, pH 10.5 and activated with 1:30 proteinase K (Sigma-Aldrich Co.) w/w for 1 h at 30 C.
2.2 Site directed mutagenesis of Cry4Ba
Plasmid pHT618 was used as template for site directed loop α-8 mutagenesis using QuikChange®Multi following the manufacturer’s instructions (Stratagene). In this site directed mutagenesis system synthetic primers with the mutated sequence are annealed to previously thermal-denaturated plasmid template and amplified with PfuTurbo DNA polymerase to obtain single stranded mutated DNA. Non-mutated DNA template is digested with DpnI restriction enzyme that digest methylated DNA, prior to transformation of XL10-Gold E. coli cells. The following oligonucleotides were used for site directed mutagenesis of cry4Ba gene: α8M1 5’CAGCTTTAGTAGAATCTCCTGCTGCTGCATCTATAGCAGCACTGGAGGC3’; α8M2 5’GTAGAATCTCCTTCTAGTAAAGCTGCAGCAGCACTGGAGGCAGCACTTAC3';α8M35’CTAGTAAATCTATAGCAGCAGCGGCGGCAGCACTTACACGAGATGTTC3’. Mutants were sequenced and transformed into acrystalliferous Bt strain 407 as reported [11].
2.3 Enzyme Linked Immunosorbent Assay (ELISA)
ELISA plates, 96-wells, were incubated 12 h at 4 C with 250 ng/well of Cyt1Aa in 50 mM NaHCO3 pH 9.6, followed by five washes with PBS, 0.2% Tween 20. The plates were then incubated with PBS, 2% BSA (Sigma-Aldrich Co.) 0.2%/ Tween 20, for 1 h at 37 C and washed five times with buffer A (PBS, 0.1% Tween 20). The ELISA plates were incubated with different concentrations of Cry4Ba or Cry4Ba mutants for 2 h at 37 C and washed three times with buffer A. The Cry4Ba proteins bound to Cyt1Aa were detected with anti-Cry4Ba antibody (1:10000) 2 h at 37 C, followed by a secondary goat-anti-rabbit- antibody coupled to horseradish peroxidase (HRP) 1 h at 37 C (1:10000). The HRP enzymatic activity was revealed with a freshly prepared substrate 840 mg of o- phenylenediamine, 18 ml of H2O2 in 100 ml of 100 mM NaH2PO4, pH 5.09. The enzymatic reaction was stopped with 6 N HCl and the absorbance read at 490 nm with a Pharmacia LKB Ultraspec II.
2.4 Ligand blot assays
Three μg of Cry4Ba or Cry4Ba mutant proteins were separated in 10% SDS-PAGE and electrotransferred to Hybond-ECL membranes. After blocking with PBS-M (PBS, 5% skim milk), the membranes were incubated for 2 h with 5 nM of Cyt1Aa, and the bound protein was revealed with anti-Cyt1Aa antibody (1:70000) 2 h at 37 C, followed by a secondary goat-anti-rabbit-HRP antibody 1 h at 37 C (1:10000) and SuperSignal chemiluminescent substrate (Pierce). 2.5 Bioassays and synergism factors (SF). Twenty 4th-instar A. aegypti larvae in 100 ml of dechlorinated water were exposed to different concentrations of Cyt1Aa and Cry4Ba toxins tested at different protein ratios (1:1, 0.5:1 and 0.2:1). Positive (Bti) and negative controls [dechlorinated water] were included in the bioassay, and larvae examined 24 h after treatment. The mean lethal concentration (LC50) was estimated by Probit analysis using statistical parameters [14] after four independent assays (Polo-PC LeOra Software). The theoretical toxicity of each ratio mixture was evaluated according to Tabashnik’s equation [23], assuming a simple additive effect. The theoretical LC50 value is the mean of the intrinsic LC50 values of each component weighted by the ratio used in the mixture:
where rCyt1A and rCry4Ba are the Cyt1A and Cry4Ba protein proportions used in the final ratio of the mixture. LC50 (Cyt1A) and LC50 (Cry4Ba) are the LC50 values for each individual toxin. The SF was calculated by dividing the theoretical toxicity by the observed toxicity of the mixture in bioassays. SF values greater than 1 indicate synergism.
3. Results
3.1 Cyt1Aa single point mutations affect Cry4Ba binding and synergism
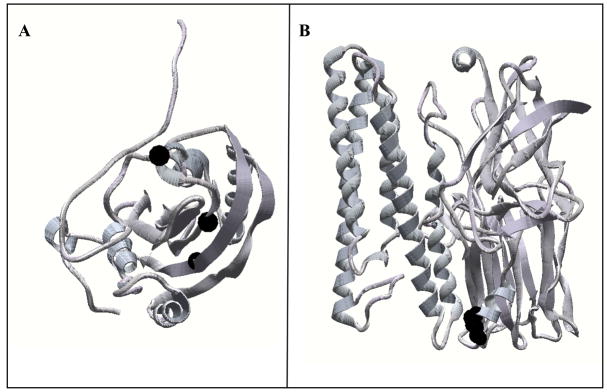
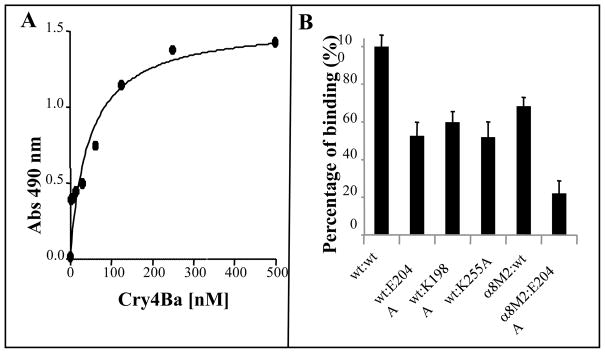
Previously, we isolated and characterized Cyt1Aa single point mutations in loop β6-αEor in β7 that are affected in their binding-interaction with Cry11Aa and their synergistic activity against A. aegypti insect larvae. In addition, Cyt1Aa loop β6-αE mutant K198A showed enhanced synergism with Cry11Aa [20]. Table 1 shows the mean lethal concentration (LC50) of Cyt1Aa and three Cyt1Aa mutants (K198A, E204A, K225A) that were in the range of the insecticidal activities previously reported [20]. To determine if the same Cyt1Aa amino acid regions involved in Cry11Aa binding and synergism were also involved on Cry4Ba synergism, we analyzed the synergism of Cyt1Aa mutants with Cry4Ba. Figure 1A shows a 3D model of the Cyt1Aa structure that was obtained by comparison to that of Cyt2Ab and the location of the Cyt1Aa residues K198A, E204E and K225A. The synergism factor (SF) of different ratio mixtures of wild type Cyt1Aa and Cry4Ba toxins was determined by bioassays against A. aegypti fourth instar larvae as described in Methods. For the two wild type proteins the highest SF value of 9 was found at the 0.2:1 Cyt1Aa:Cry4Ba ratio (Table 2). At 0.2:1 mixture ratio the Cyt1Aa mutants K198A, E204A and K225A showed a reduced SF value of 3.5, 2.6 and 1.8 respectively, indicating that these mutations affected the synergistic activity between Cyt1Aa and Cry4Ba.
Table 1.
Toxicity of Cy1Aa, Cry4Ba and toxin mutants
| Toxin | LC50a (ng/ml) |
|---|---|
| Cyt1Aa | 952.5 (602.5–1824.3) |
| K198A | 601.8 (466.3–792.7) |
| E204A | 103.9 (80.3–134.9) |
| K225A | 858.6 (548.5–1527.6) |
| Cry4Ba | 79.4 (61–102) |
| α8M1 | 117.2 (93.9–146.6) |
| α8M2 | 182.2 (77.9–358.8) |
Bioassays were performed with 10 early fourth-instar Ae. aegypti larvae in 100 ml H2O per dose. Three independent experiments of ten different doses each were done. Mean lethal 95% confidence limits are given in concentration (LC50) was estimated by Probit analysis. parentheses.
Figure 1.
Three dimensional structures of insecticidal toxins produced by Bacillus thuringiensis subs. israelensis. A. Top view of the three dimensional structure model of Cyt1Aa obtained by comparison to the three dimensional structure of Cyt2Ab. Residues K198, E204 and K225 are highlighted by space filling black figures. B. Three dimensional structure of Cry4Ba . Domain II loop a-8 303SI304 residues are highlighted by space filling black figures.
Table 2.
Synergism factor and binding affinities of mixtures of proteins at 0.2:1 ratio.
| Toxins Cyt1Aa:Cry4Ba | Predicted LC50 (ng/ml) | Experimental LC50 (ng/ml) | SFa |
|---|---|---|---|
| wt:wt | 75.2 | 8.5 (6.5–10.8)b | 8.8 |
| K198A:wt | 74.5 | 21.0 (10.1–30.1) | 3.5 |
| E204A:wt | 66.7 | 25.2 (20.1–32.3) | 2.6 |
| K225A:wt | 75.1 | 41.4 (31.1–52.3) | 1.8 |
| wt:α8M2 | 218.9 | 38.4 (32.7– 44.7) | 5.7 |
| E204:α8M2 | 158.3 | 77.2 (67.1–88.9) | 2 |
Synergism factor (Predicted LC50/Experimental LC50)
95% fiducial limits.
To determine if the effects of Cyt1Aa mutations on Cry4Ba synergism correlated with a decrease in their binding interaction, ELISA binding assays of Cry4Ba to Cyt1Aa mutants were performed. First, a saturation binding curve of Cyt1Aa and Cry4Ba was obtained, supporting the interaction between these two toxins and revealing an apparent binding affinity (Kd) of 47 nM (Fig. 2A). Figure 2B shows that Cyt1Aa mutants K198A, E204A and K225A showed a reduced binding with Cry4Ba in comparison with Cyt1Aa. These results show that synergism of Cry4Ba and Cyt1Aa correlate with their binding interaction.
Figure 2.
ELISA binding assays of Cry4Ba to Cyt1Aa. A. Saturation binding curve of Cry4Ba to Cyt1Aa. ELISA plates were coated with Cyt1Aa (250 ng/well) and revealed with different Cry4Ba concentrations followed by anti-Cry4Ba antibody and by a secondary goat-anti-rabbit- antibody coupled to horseradish peroxidase. B. ELISA plates were coated with Cyt1Aa or Cyt1Aa Cyt1Aa mutants K198A, E204A and K225A (250 ng/well) and revealed with 75 mM of Cry4Ba or loop α8M2 mutant as described bove. Data of three replicates with standard deviations.
3.2 Cry4Ba mutations in loop α-8 affect binding and synergism with Cyt1Aa
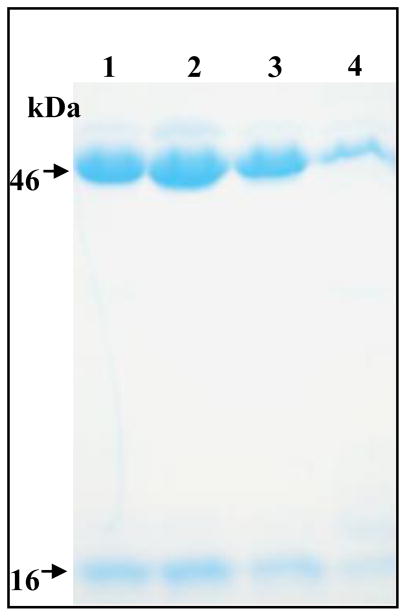
Domain II loops of Cry toxins have been recognized as important receptor binding regions in different Cry toxins [5, 11, 16, 24]. In the case of Cry11Aa, it was shown that Domain II loop α-8 was an important binding region with the ALP GPI-anchored receptor [11]. Also, this amino acid region in Cry11Aa was shown to be involved on Cyt1Aa binding and in synergism suggesting that Cyt1Aa functions as a Cry11Aa receptor [20]. To determine if Domain II loop regions of Cry4Ba are involved on Cyt1Aa binding and in synergism, we performed site directed mutagenesis of Cry4Ba Domain II loop α-8 since a synthetic peptide corresponding to this amino acid region competed the binding with ALP1 protein in contrast with synthetic peptides corresponding to the other Domain II loop regions (Reyes E. Z. and Soberón M. unpublished data). Figure 1B shows the 3D structure of Cry4Ba and the location of Domain II loop α-8. Three mutants of loop α-8 region (300SSKSIAALE308) were constructed, mutant α8M1 (SSK300-302AAA), mutant α8M2 (SI303-304AA) and mutant α8M3 (LE307-308AA). The three Cry4Ba mutants were transformed to the Bt 407 acrystaliferous strain and cultured until sporulation. Crystals from the three mutants were solubilized and activated with trypsin. From the solubization step it was clear that α8M3 mutant produced lower yields of the 130 kDa Cry4Ba protoxin (data not shown). Figure 3 shows that α8M1 and α8M2 produced two bands of 48 kDa and 18 kDa as wild type Cry4Ba toxin after trypsin treatment suggesting that these two proteins have no structural constrains. In contrast, α8M3 mutations affected protein stability since it produce very low levels of the expected bands after trypsin treatment and therefore α8M3 was not further analyzed. The toxicity of Cry4Ba and loop α8 mutants was analyzed in A. aegypti fourth instar larvae. Table 1 shows that α8M2 had a slight reduction in toxicity but with an overlap in the confidential limits with Cry4Ba toxicity while α8M1 showed a similar toxicity to A. aegypti larvae as Cry4Ba toxin.
Figure 3.
Proteolytic activation of Cry4Ba and Domain II loop α-8 mutants. Different Cry4Ba proteins (5 μg) were solubilized, treated with trypsin (1:20 W/W) and separated by 10% SDS-PAGE electrophoresis. Lane 1 Cry4Ba, lane 2 α8M1, lane 3 α8M2, lane 4 α8M3.
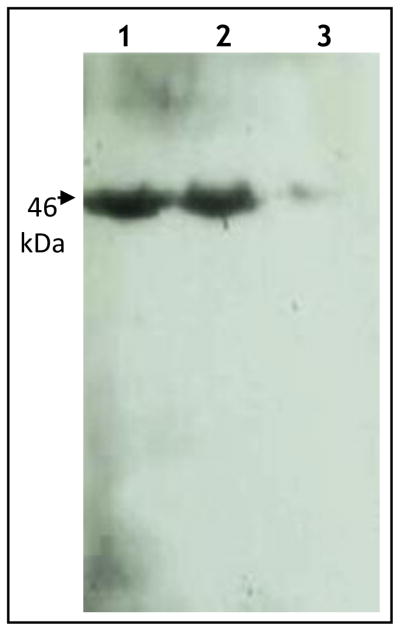
Binding of Cry4Ba and loop α8 mutants to Cyt1Aa was analyzed by ligand blot. Cry4Ba and the two loop α–8 mutants were subject to SDS-PAGE, blotted to a membrane and detected with Cyt1Aa followed by anti-Cyt antibody. Figure 4A shows a significant reduction in binding of α8M2 to Cyt1Aa in contrast with α8M1 that bound Cyt1Aa as wild type Cry4Ba. ELISA binding assays confirmed that α8M2 had a reduced binding to Cyt1Aa in comparison with α8M1 and Cry4Ba (Fig. 2B). Figure 2B also shows that Cry4Ba α8M2 showed very weak binding to the mutant Cyt1Aa E204A indicating that both Cyt1Aa and Cry4Ba mutations had an additive effect on the binding interaction of these proteins. Bioassays were performed to determine the SF factor of Cyt1Aa and α8M2 and also of the two mutants Cyt1Aa E204A and α8M2. Table 2 shows that Cyt1Aa-α8M2 showed a SF value of 5.7, while Cyt1AaE204A and α8M2 had a SF value of 2, indicating a direct correlation between Cry4Ba binding to Cyt1Aa and synergism.
Figure 4.
Cry4Ba Domain II loop α-8 is involved on Cyt1Aa binding. Different Cry4Ba proteins (three μg) were activated by trypsin treatment separated by 10% SDS-PAGE electrophoresis and blotted into nylon membranes. Blots were revealed with 10 nM Cyt1Aa followed with anti-Cyt1Aa and by a secondary goat-anti-rabbit- antibody. Lane 1 Cry4Ba, lane 2 α8M1 and lane 3 α8M2.
4. Discussion
Previous work identified specific amino acids of Cyt1Aa located in loop β6-αEor in β7 that are affected in binding to Cry11Aa and synergism. In this work we analyzed the effect of three Cyt1Aa mutants in these structural regions regarding to its binding and synergism with Cry4Ba. The three Cyt1Aa mutants analyzed, K198A, E204A and K225A showed a reduction on synergism and a correlative reduction on Cry4Ba binding indicating that the same Cyt1Aa regions are involved on Cry11Aa and Cry4Ba binding interaction. However, the Cyt1Aa K198A mutant phenotype was different regarding its effect on Cry11Aa or Cry4Ba. In the case of Cry11Aa, Cyt1Aa K198A showed an enhancement on synergism and correlated with higher binding affinity to Cry11Aa [20], while in the case of Cry4Ba, this Cyt1Aa mutation reduced the binding interaction and synergism [Fig. 1]. This data shows that specific residues of Cyt1Aa binding epitope have a differential effect on its binding to different Cry toxins.
Regarding the Cry4Ba regions involved in Cyt1Aa interaction, we show that a Domain II loop α-8 mutation affected Cyt1Aa binding and synergism between these toxins. As mentioned earlier Domain II exposed loops have been shown to be important for receptor interaction in different Cry toxins [5, 11, 16, 24]. In the case of Cry4Ba it has been shown that Domain II exposed loops 1, 2 and 3 are involved in binding to A. aegypti BBMV and in toxicity [16, 24]. This is the first report of the mutagenesis of Cry4Ba loop α-8. The mutants isolated showed a non-significant marginal effect on toxicity to A. aegypti larvae. However, one double alanine mutant, α8M2 [SI303-304AA], showed reduced binding to Cyt1Aa and a correlative reduction in synergism. The fact that Domain II loops are involved in both receptor and Cyt1Aa binding is a clear indication that Cyt1Aa functions as a specific binding protein of Cry4Ba mediating its toxicity.
Based on the molecular mechanism proposed for the synergistic effect of Cyt1Aa on Cry11Aa, it was proposed that Bti is the first example of an insect pathogenic bacterium that carries a toxin and also its functional receptor, promoting toxin binding to target membranes and toxicity [20]. As shown in this manuscript, this mechanism also applies for Cry4Ba. This molecular mechanism of synergism could be important to develop tools for countering insect resistance [6]. It would be interesting to identify Cyt proteins active against lepidopteran or coleopteran insects and that could synergize other Cry toxins. Alternatively, an attractive strategy to select Cyt1Aa toxins that show synergism with other Cry toxins, such as lepidopteran or coleopteran specific Cry toxins, could be the engineering of the binding regions in Cyt1Aa, thus providing additional binding sites in Cyt1Aa that could promote synergism with other Cry toxins. This would be a promising development to counteract resistant insects or to control insects with low susceptibility to the known Cry toxins.
Acknowledgments
We thank Lizbeth Cabrera, for technical assistance. Research was funded in part through grants from the National Institutes of Health, 1R01 AI066014, DGAPA/UNAM IN218608 and IN210208-N, CONACyT U48631-Q. IRdE received a José Castillejo postdoctoral grant (Spanish Ministry of Education and Science), and a grant for mobility for Teaching and Research Staff of Public University of Navarre, Spain (UPNA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Pablo Emiliano Cantón, Instituto de Biotecnología, Universidad Nacional Autónoma de México. Apdo. postal 510-3, Cuernavaca 62250, Morelos, Mexico.
Esmeralda Zanicthe Reyes, Instituto de Biotecnología, Universidad Nacional Autónoma de México. Apdo. postal 510-3, Cuernavaca 62250, Morelos, Mexico.
Iñigo Ruiz de Escudero, Departamento de Producción Agraria, Universidad Pública de Navarra, Campus Arrosadía. 31006. Pamplona, España.
Alejandra Bravo, Instituto de Biotecnología, Universidad Nacional Autónoma de México. Apdo. postal 510-3, Cuernavaca 62250, Morelos, Mexico.
Mario Soberón, Instituto de Biotecnología, Universidad Nacional Autónoma de México. Apdo. postal 510-3, Cuernavaca 62250, Morelos, Mexico.
References
- 1.Abdullah MA, Alzate O, Mohammad M, McNall RJ, Adang MJ, Dean DH. Introduction of Culex toxicity into Bacillus thuringiensis Cry4Ba by protein engineering. Appl Environm Microbiol. 2003;69:5343–5353. doi: 10.1128/AEM.69.9.5343-5353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdullah MA, Valaitis AP, Dean DH. Identification of a Bacillus thuringiensis Cry11Ba toxin-binding aminopeptidase from the mosquito, Anopheles quadrimaculatus. BMC Biochem. 2006;7:16. doi: 10.1186/1471-2091-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker N. Bacterial control of vector-mosquitoes and black flies. In: Charles JF, Delécluse A, Nielsen-LeRoux C, editors. Entomopathogenic bacteria: from laboratory to field application. Kluwer Academic Publishers; 2000. p. 383. [Google Scholar]
- 4.Boonserm P, Davis P, Ellar DJ, Li J. Crystal Structure of the Mosquito-larvicidal Toxin Cry4Ba and Its Biological Implications. J Mol Biol. 2005;348:363–382. doi: 10.1016/j.jmb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Bravo A, Gill SS, Soberón M. Mode of action of Bacillus thuringiensis toxins and their potential for insect control. Toxicon. 2007;49:423–435. doi: 10.1016/j.toxicon.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bravo A, Soberón M. How to cope with resistance to Bt toxins? Trends in Bioetch. 2008;26:573–579. doi: 10.1016/j.tibtech.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Butko P. Cytolytic toxin Cyt1A and its mechanism of membrane damage: Data and hypotheses. Appl Environ Microbiol. 2003;69:2415–2422. doi: 10.1128/AEM.69.5.2415-2422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Aimanova KG, Fernández LE, Bravo A, Soberón M, Gill SS. Aedes aegypti cadherin serves as a putative receptor of the Cry11Aa toxin from Bacillus thuringiensis subsp. Israelensis Biochem J. 2009;424:191–200. doi: 10.1042/BJ20090730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Aimanova KG, Pan S, Gill SS. Identification of the Aedes aegypti aminopeptidase N as a putative receptor of Bacillus thuringiensis Cry11A toxin. Insect Biochem Mol Biol. 2009;39:688–696. doi: 10.1016/j.ibmb.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crickmore N, Bone EJ, Wiliams JA, Ellar D. Contribution of individual components of the d-endotoxins crystal to the mosquitocidal activity of Bacillus thuringiensis subsp. israeliensis. FEMS Microbiol Lett. 1995;131:249–254. [Google Scholar]
- 11.Fernández LE, Pérez C, Segovia L, Rodríguez MH, Gill SS, Bravo A, Soberón M. Cry11Aa toxin from Bacillus thuringiensis binds its receptor in Aedes aegypti mosquito larvae trough loop α-8 of domain II. FEBS Lett. 2005;579:3508–3514. doi: 10.1016/j.febslet.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 12.Fernández LE, Aimanova KG, Gill SS, Bravo A, Soberón M. A GPI-anchored alkaline phosphatase is a functional midgut receptor of Cry11Aa toxin in Aedes aegypti larvae. Biochem J. 2006;394:77–84. doi: 10.1042/BJ20051517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernández LE, Martinez-Anaya C, Lira E, Chen J, Evans J, Hernández-Martínez S, Lanz-Mendoza H, Bravo A, Gill SS, Soberón M. Cloning and epitope mapping of Cry11Aa-binding sites in the Cry11Aa-receptor alkaline phosphatase from Aedes aegypti. Biochemistry. 2009;48:8899–8907. doi: 10.1021/bi900979b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finney D. Probit analysis. United Kingdom: Cambridge University Press; 1971. [Google Scholar]
- 15.Hua G, Zhang R, Abdullah MA, Adang MJ. Anopheles gambiae cadherin AgCad1 binds the Cry4Ba toxin of Bacillus thuringiensis israelensis and a fragment of AgCad1 synergizes toxicity. Biochem. 2008;47:5101–5110. doi: 10.1021/bi7023578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khaokhiew T, Angsuthanasombat Ch, Promptmas Ch. Correlative effect on the toxicity of three surface-exposed loops in the receptor-binding domain of the Bacillus thuringiensis Cry4Ba toxin. FEMS Microbiol Lett. 2009;300:139–145. doi: 10.1111/j.1574-6968.2009.01774.x. [DOI] [PubMed] [Google Scholar]
- 17.Khasdan V, Ben-Dov E, Manasherob R, Boussiba S, Zaritsky A. Toxicity and synergism in transgenic Escheichia coli expressing four genes from Bacillus thuringiensis subsp. israeliensis. Environ Microbiol. 2001;3:798–806. doi: 10.1046/j.1462-2920.2001.00253.x. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Pandelakis AK, Ellar D. Structure of the mosquitocidal d-endotoxin CytB from Bacillus thuringiensis sp. kyushuensis and implications for membrane pore formation. J Mol Biol. 1996;257:129–152. doi: 10.1006/jmbi.1996.0152. [DOI] [PubMed] [Google Scholar]
- 19.Margalith Y, Ben-Dov E. Biological control by Bacillus thuringiensis subsp. israeliensis. In: Rechcigl JE, Rechcigl NA, editors. Insect pest management: techniques for environmental protection. CRC Press; 2000. p. 243. [Google Scholar]
- 20.Pérez C, Fernandez LE, Sun J, Folch JL, Gill SS, Soberón M, Bravo A. Bacillus thuringiensis subsp. israeliensis Cyt1Aa synergizes Cry11Aa toxin by functioning as a membrane-bound receptor. Proc Natl Acad Sci. 2005;102:18303–18308. doi: 10.1073/pnas.0505494102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pérez C, Muñoz-Garay C, Portugal LC, Sánchez J, Gill SS, Soberón M, Bravo A. Bacillus thuringiensis subsp. israelensis Cyt1Aa enhances activity of Cry11Aa toxin by facilitating the formation of a pre-pore oligomeric structure. Cell Microbiol. 2007;9:2931–2937. doi: 10.1111/j.1462-5822.2007.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soberón M, Gill SS, Bravo A. Signaling versus punching hole: How do Bacillus thuringiensis toxins kill insect midgut cells? Cell Mol Life Sci. 2009;66:1337–1349. doi: 10.1007/s00018-008-8330-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabasnik BE. Evaluation of sinergismo hmong Bacillus thuringiensis toxins. Appl Environ Microbiol. 1992;58:3343–3346. doi: 10.1128/aem.58.10.3343-3346.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuntitippawan T, Boonserm P, Katzenmeier G, Angsuthanasombat C. Targeted mutagenesis of loop residues in the receptor-binding domain of the Bacillus thuringiensis Cry4Ba toxin affects larvicidal activity. FEMS Microbiol Lett. 2005;242:325–332. doi: 10.1016/j.femsle.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 25.Wirth MC, Georghiou GP, Federeci BA. CytA enables CryIV endotoxins of Bacillus thuringiensis to overcome high levels of CryIV resistance in the mosquito, Culex quinquefasciatus. Proc Natl Acad Sci. 1997;94:10536–10540. doi: 10.1073/pnas.94.20.10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu D, Johnson JJ, Federeci BA. Synergism of mosquitocidal toxicity between CytA and CryIVD proteins using inclusions produced from cloned genes of Bacillus thuringiensis. Mol Microbiol. 1994;13:965–972. doi: 10.1111/j.1365-2958.1994.tb00488.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhang R, Hua G, Andacht TM, Adang MJ. A106-kDa aminopeptidase is a putative receptor for Bacillus thuringiensis Cry11Ba toxin in the mosquito Anopheles gambiae. Biochem. 2008;47:11263–11272. doi: 10.1021/bi801181g. [DOI] [PMC free article] [PubMed] [Google Scholar]